Abstract
Scientists are rapidly developing synthetic gene drive elements intended for release into natural populations. These are intended to control or eradicate disease vectors and pests, or to spread useful traits through wild populations for disease control or conservation purposes. However, a crucial problem for gene drives is the evolution of resistance against them, preventing their spread. Understanding the mechanisms by which populations might evolve resistance is essential for engineering effective gene drive systems. This review summarizes our current knowledge of drive resistance in both natural and synthetic gene drives. We explore how insights from naturally occurring and synthetic drive systems can be integrated to improve the design of gene drives, better predict the outcome of releases, and understand genomic conflict in general.
Keywords: CRISPR-Cas9, fitness costs, meiotic drive, population suppression, selfish genetic elements, sex ratio distorter, transposable element, Wolbachia
1. Introduction
Organisms require networks of cooperating genes. Generally, alleles spread through populations by increasing the reproductive success of the organism as a whole. However, some alleles, defined here as drivers, selfishly bias reproduction to increase their own representation in the next generation, at a cost to the rest of the genome (Burt and Trivers 2006). For example, ‘segregation distorters’ are a type of driver that subvert the usual rules of Mendelian inheritance in such a way that they are inherited by over 50% of the descendants of heterozygous individuals, and occur naturally in many species includingplants, fungi, nematodes, insects and mice (Lindholm et al. 2016). Another example is drive by mitochondria, the key endosymbiont of eukaryotes, which damage male function in many hermaphroditic plants (Burt and Trivers 2006). This loss of male function diverts resources to seed production, enhancing transmission of the mitochondrial genome, which is typically uniparentally transmitted through ovules but not pollen. Selfish genetic elements likely occur in all species, and can have major impacts on the evolution and ecology of their hosts (Burt and Trivers 2006).
Crucially, the super-Mendelian rate at which gene drivers are transmitted over generations can allow them to spread through populations despite costs. This has inspired researchers to propose using gene drives to solve major biological challenges related to public health, the environment and agriculture (Burt 2014; Champer et al. 2016; Piaggio et al. 2017; Raban et al. 2020). Two broad types of gene drives have been proposed: population suppression gene drives and population replacement gene drives. Population suppression gene drives can be employed when reduction or elimination of a population (e.g., of disease vectors, agricultural pests, or invasive species) is desired. Replacement gene drives offer the potential to alter existing populations for human benefit, for example by spreading alleles or endosymbionts that reduce the ability of mosquitoes to transmit malaria. Strains of the intracellular bacterium Wolbachia reduce the ability of mosquitoes to transmit dengue and other viruses. Wolbachia strains have already been successfully deployed in Australia and elsewhere, spreading through populations by creating mating incompatibilities that disproportionately reduce the fitness of females that do not carry Wolbachia, and reducing the threat of dengue (Nazni et al. 2019; Ryan et al. 2020). New synthetic population suppression and replacement drive systems are being created with increasing regularity, highlighting the enormous promise of CRISPR-Cas9 and other new molecular tools for editing genomes (Champer et al. 2016).
However, gene drives impose costs, certainly on outcompeted alleles, and often on the individual as a whole. Costs at the individual level can arise directly via the mechanism of transmission, for example the costly death of gametes that carry rival alleles, or because the driver carries costs such as associated low fitness alleles or metabolic costs in driving endosymbionts (Burt and Trivers 2006). The resulting selection can lead to the rapid evolution of resistance traits that prevent the driver from spreading. As a result, many natural drivers have been completely suppressed, only showing drive when crossed into distant relatives that do not carry suppressor alleles (McDermott and Noor 2010; Courret et al. 2019). This research suggests that we should expect synthetic gene drives, especially those with large fitness effects, to select for resistance, which will potentially undermine their ability to spread, modify, or suppress populations (Unckless et al. 2017; Barrett et al. 2019; Holman 2019).
For synthetic gene drives to be effectively deployed, we urgently need to understand how quickly resistance will arise. Does resistance usually arise through selection on pre-existing genetic variation, or does it more often involve novel mutations that appear once drive has reached a high frequency? What fraction of natural gene drives reach fixation, go extinct, reach a stable polymorphism, or are fully suppressed, and how can we address this question given the difficulties of detection once a gene drive has fixed or been lost? Does resistance to drive typically involve the same fundamental mechanism (e.g. loss of the driver’s target, or “defusing” of the driver by interfering RNAs) across species and types of driver, or is the resistance mechanism highly idiosyncratic?
In this review we synthesize what is known about how resistance evolves against both natural and synthetic drives, and point out gaps in our knowledge. We begin by reviewing how resistance has evolved in well-studied natural systems, examining resistance that interferes directly with the molecular mechanisms of drive, and then resistance through behavior and life history. We then turn to the current evidence regarding resistance to synthetic drives. Finally, we discuss the implications for the design of “evolution proof” synthetic gene drives.
2. Resistance to gene drives in natural systems
In any drive system, selection for resistance will act on the target locus itself, genes linked to the target, and in some cases on the entire genome. Generally speaking, selection for resistance at the target and linked loci becomes stronger with more biased transmission, while the strength of selection for resistance on the rest of the genome increases with higher fitness loss for the organism (Figure 1). These two are often positively related leading to strong selection for resistance at both the target locus and genome wide. We classify resistance as adaptations that reduce the spread of drive elements either by 1) interfering with the molecular mechanism of drive (which we term as “suppression” in this Review) or by 2) altering some aspect of behavior or life history of carriers which in turn reduces the ability of a driver to spread. We use these categories to structure our review of known drive resistance factors, incorporating natural and synthetic drive systems.
Figure 1.
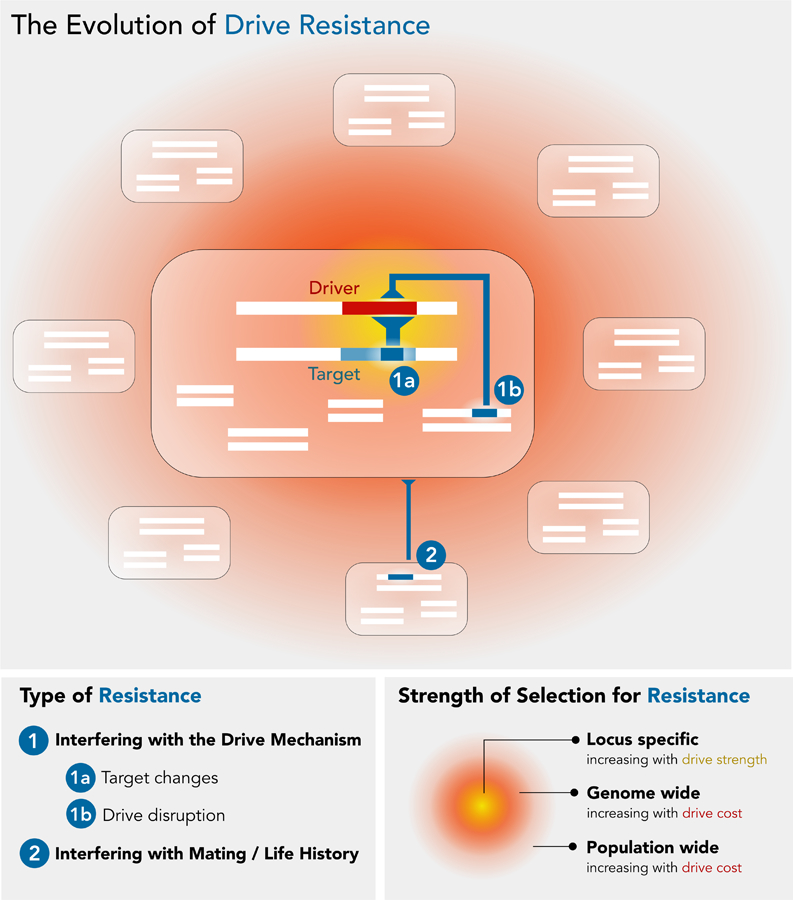
The evolutionary impact of a gene drive, as measured by the magnitude and location of costs imposed (yellow/red gradients). Boxes represent individuals, white rectangles are chromosomes within the organism. Drive creates selection pressure for the three drive resistance mechanisms discussed in this review (blue). The selection pressure for drive resistance is highest at the target locus itself (1a), where rivalling homologous genes suffer both from reduced transmission due to drive (yellow) and (potential) fitness costs to the organism (red). Selection pressure on unlinked loci throughout the genome to disrupt drive will be a function of organismal drive costs (1b). Finally, gene drive may create selection for mechanisms that suppress the drive at the population level (2).
2.1. Mutations at the target site & suppression of drive machinery
One way to evolve resistance is to modify the target of drive so that it is no longer susceptible. For example, a gene drive that spreads itself by targeting a specific sequence of nucleotides or peptides might impose selection that favors genotypes carrying an altered sequence. Below, we review the evidence for this mode of resistance in nature. For a brief overview of the biological differences between the natural gene drives, see Table 1.
Table 1.
A highly simplified view of mechanisms and associated costs for the gene drive systems discussed in this paper. Please note, all systems are considerably more diverse than described here.
| Gene drive system | Mechanism | Key effects |
|---|---|---|
| Gamete killer | Drives by killing or damaging gametes that do not carry the driving chromosome. | Reduces sperm number. If on a sex chromosome, can bias population sex ratios. |
| Female meiotic drive | Drive chromosome manipulates meiosis so rival chromosomes are disproportionately discarded in the polar bodies. | Costs relatively unknown, but some well-studied systems associated with low fitness. |
| Transposable elements | Drive sequences copy themselves into other locations in the genome. | Largely deleterious due to gene disruption and DNA breakage. |
| Genetic incompatibility systems | Factors inherited via cytoplasm such as organelles and endosymbionts increase the fitness of females at a cost to males. Mechanisms are extremely diverse. | Effects can include loss of male function, death of offspring, feminization. Can be highly costly. |
| Homing based systems | Induce targeted double strand DNA breaks that copy and insert the drive construct during DNA repair. | Effects depend on design. Can include sterility, offspring sex ratio bias, disease resistance. |
| Medea-like systems | Chromosomes bearing a set of lethal loci in which each suppresses the other, killing offspring that do not inherit drive. | Reduced viability if not all loci are inherited. Reduced offspring production. |
2.1.1. Sex chromosome linked gamete killers
Naturally-occurring ‘gamete killer’ meiotic drivers have often been found on sex chromosomes, where they cause distortion in the transmission of the heterogametic sex (Hurst and Pomiankowski 1991). The evolution of sex chromosome drivers is facilitated by the differentiation between X and Y chromosomes (and Z/W). Driver alleles arising on a well-differentiated sex chromosome therefore have potential targets at many sites that are never linked to the driver: for example, an X-linked driver could promote its own transmission by destroying gametes containing a particular Y-linked locus (Jaenike 2001). Sex-linked drivers generate especially strong selection for resistance because they alter the population sex ratio. A bias in the population sex ratio creates strong selection favoring individuals/genotypes that produce relatively more of the under-represented sex (Fisher 1930). This ‘Fisherian sex ratio selection’ confers an additional fitness benefit to alleles that confer resistance to drive, in populations showing a biased sex ratio due to the presence of a sex-linked driver. We therefore expect to see rapid evolution of resistance against sex-linked drivers (Hurst and Pomiankowski 1991).
We illustrate this using sex chromosome drive systems in Drosophila simulans. In the Paris Sex Ratio (SR) system, two X-linked drivers together prevent the disjunction of the Y sister chromatids in the second meiotic division. One of these encodes HP1D2, a protein that binds Y chromosome heterochromatin in premeiotic cells, suggesting it targets repeated DNA sequences (Helleu et al. 2016). The Y chromosome of D. simulans exhibits substantial variation in resistance to Paris SR drive, with a wide continuum of phenotypes from high susceptibility (95% female progeny) to complete resistance (50% female progeny) (Montchamp-Moreau et al. 2001). These more or less resistant Y chromosomes show extensive structural rearrangements affecting satellite sequences, which strongly suggests that resistance occurs through changes in the target repeat sequences (Helleu et al. 2019). In addition, Paris SR is suppressed by yet unidentified autosomal loci (Courret et al. 2018).
The Winters SR is another sex ratio distorting system in D. simulans, with a different drive phenotype to Paris SR, killing sperm after meiosis (Tao et al. 2007a). An X-linked gene, Dox, and likely its progenitor Mdox, are involved in drive (Tao et al. 2007a). Winters SR is typically entirely suppressed by high frequencies of the autosomal suppressor locus Nmy. Nmy arose from a retrotransposed inverted repeat of Dox (Tao et al. 2007b) and produces an antisense RNA that represses Dox and Mdox through the RNA interference pathway (Lin et al. 2018).
Geographic variation in frequencies of drivers and suppressors has been found in both Paris and Winters systems, including populations that appear completely suppressed (Atlan et al. 1997; Kingan et al. 2010; Bastide et al. 2013). Under complete suppression, drivers have no transmission advantage but might still impose organismal fitness costs leading to a resultant decline in driver frequency. The Nmy suppressor of Winters SR is dominant and almost at fixation, rendering the driver effectively cryptic, leading to the accumulation of loss-of-function driver mutants (Kingan et al. 2010). Both systems show genetic evidence of positive selection on the drivers, in the Winters systems about 3,000 years ago (Kingan et al. 2010) and in the Paris system within the last century (Derome et al. 2008; Bastide et al. 2013). Paris SR, as well as its genetic suppressors, have been recently increasing in frequency in the Middle East (Bastide et al. 2013), and rapid declines in SR frequency have been observed in completely suppressed populations (Atlan et al. 1997; Bastide et al. 2011).
These two SR systems illustrate empirically the dynamic nature of the spread of drivers, followed by the rise of suppressors, and then loss of drivers that evolve in a continuous cycle of “red queen” dynamics. However, while many meiotic drive systems we observe in nature have arrived at such a dynamic equilibrium, others have not. There is some evidence that drive can cause extinction, at least in local populations (Pinzone and Dyer 2013). Other drive systems seem to occur at stable frequencies in different populations, sometimes in geographical clines, for reasons that are not well understood, and there is some evidence that this stability can last for hundreds of generations (Price et al. 2014; Price et al. 2019).
2.1.2. Autosomal gamete killers
Autosomal gamete-killing meiotic drivers function by killing gametes that carry alternative alleles (Bravo Núñez et al. 2018b). Some of the best studied systems are the spore-killers in various fungal species. First, in Neurospora, an RNA interference-based genome defense mechanism has been shown to be a suppressor of spore-killing alleles (Svedberg et al. 2020). Secondly, there are multiple copies of drivers in the filamentous fungus Podospora anserina, one of which is a known suppressor (Grognet et al. 2014). Likewise, the wtf gene family in Schizosaccharomyces pombe encodes a poison-antidote drive system and has up to 42 copies throughout fission yeast genomes (López Hernández and Zanders 2018; Eickbush et al. 2019). Of these copies, some are intact meiotic drivers (alleles that encode both a poison and antidote), some are apparent pseudo-genes, and some are alleles that encode only the antidote (Bravo Núñez et al. 2020). These “antidote-only” wtfs act as suppressors of their selfish wtf homologs and are likely to be maintained by selection for resistance to the latter (Bravo Núñez et al. 2018a). The amplification of the different multi-copy spore-killer systems in fungal genomes is representative of the cycles of amplification of drivers and suppressors often seen in genetic conflict. Patterns of duplication of drivers and suppressors have also been observed on sex chromosomes in Mus musculus house mice (Soh et al. 2014).
Another well studied system is Segregation Distorter (SD) in Drosophila melanogaster, which contains a driver, enhancers of drive, and a target site, found in a region of low recombination (Larracuente and Presgraves 2012). Males heterozygous for SD and a sensitive wild type chromosome suffer chromatin condensation defects and dysfunction in wildtype sperm. The target site consists of a large block of tandem repeats. The number of copies of the tandem repeat correlates with sensitivity to drive, and alleles with fewer than ~300 repeats are insensitive to drive (Wu et al. 1988). There is substantial variation in target copy number in D. melanogaster populations across the globe. Frequencies of SD are low in natural populations, suggesting a balanced polymorphism, but evidence for genetic sweeps of SD instead suggest rapid turnover of SD chromosomes, either because of competition between SD variants or arms races with suppressors (Brand et al. 2015). Unlinked genetic suppressors are known (Hiraizumi and Thomas 1984), but they have not been studied at the molecular genetic level.
2.1.3. Female meiotic drive
Female meiotic drive exploits asymmetry in female meiosis to influence which homolog of the chromosome pair is distributed to the egg nucleus as opposed to the excluded polar bodies. Thus, the fitness of the non-driving homolog is reduced, but costs to the organism are small in terms of gamete production. If costs are negligible, then female drivers might readily spread and fix, since only a small region of the genome close to the drive locus would be under selection to evolve resistance. However, in Mimulus monkeyflowers, female drivers impose fitness costs when homozygous (Fishman and Kelly 2015). In maize (Zea mays), the Kindr (Ab10) driving knobs system has heterozygous and homozygous fitness costs in seed set and weight (Higgins et al. 2018). Resistant alleles block expression of the Kindr complex, and are characterized by small interfering RNAs and DNA methylation (Dawe et al. 2018).
2.1.4. Transposable elements
Transposable elements (“TEs”) are DNA sequences that can change their location within a genome, often copying themselves in the process (Feschotte and Pritham 2007). They have been found in prokaryotes, eukaryotes and even giant viruses (Sun et al. 2015). Transposition is generally deleterious to the individual, resulting in DNA breakage and potentially ectopic recombination, as well as potentially disrupting genes (Feschotte and Pritham 2007). Mechanisms for suppressing TEs are diverse, and many have ancient origins, such as genome methylation which silences TE expression.
Typically TE invasions follow a cycle, with a novel TE invading a species, or a TE already in the genome escaping suppression (Bousios and Gaut 2016). This TE rapidly replicates in the genome of the species, imposing costs, which select for suppression. This invasion and suppression can occur extremely quickly. In Drosophila melanogaster, a DNA-based TE invaded in the early 1950s and had spread worldwide by the 1980s (Anxolabéhère et al. 1988). In around the year 2000 this TE jumped to the closely related D. simulans, and spread even faster worldwide through that species (Hill et al. 2016). RNAi suppression of the TE evolved extremely rapidly in both species, resulting in the TE being largely suppressed in D. simulans populations within two decades of invasion. This fast evolution of suppression is facilitated by piRNA clusters in animals that appear to perform a defensive function against TEs (Czech et al. 2018), similar to the CRISPR libraries that provide adaptive immune defense against viruses and plasmid gene drivers in bacteria (Barrangou and Marraffini 2014). When a TE attacks the organism, sequences from the invading TE are recruited to the piRNA clusters, providing a DNA template that guides RNAi silencing of that TE, preventing it from further replication (Brennecke et al. 2007). The maintenance of these genomic regions as defenses against TEs suggests it is possible that other genomic regions may also be maintained over evolutionary time because they defend against TEs or other selfish genetic elements.
2.1.5. Genetic incompatibility systems
Cytoplasmic incompatibility can occur between nuclear and mitochondrial DNA (mtDNA), as mtDNA is transmitted almost exclusively from mother to offspring. The most widely-recorded example of cytoplasmic incompatibility is cytoplasmic male sterility, in which hermaphroditic plants are rendered male-sterile and are functionally female. Cytoplasmic male sterility is very widely distributed amongst angiosperm plant species, with populations consisting of both hermaphroditic and female plants (Touzet and Budar 2004). Nuclear suppressors that restore male fertility (called Rf genes) are commonly found within cytoplasmic male sterility systems. Many Rfs are members of the pentatricopeptide repeat protein family, involved in processing and editing RNA (Gaborieau et al. 2016). They typically act by binding directly to the mitochondrial transcripts, interfering with the production of male sterility proteins (Chen and Liu 2014). Rfs show evidence of rapid evolution and diversification (Fujii et al. 2011) suggesting ongoing cycles of conflict with cytoplasmic male sterility genes.
Male-killing caused by some Wolbachia bacteria, also inherited via cytoplasm, provide a demonstration of how quickly suppression can spread. Pacific island populations of the butterfly Hypolimnas bolina are infected with a Wolbachia strain that causes the death of the sons of infected females (Dyson et al. 2002). This benefits infected daughters due to decreased larval competition with siblings, allowing Wolbachia to reach extremely high frequencies, resulting in populations with fewer than one male per hundred females (Dyson and Hurst 2004). A nuclear gene which rescues the male embryos recently appeared and has spread rapidly; in the Samoan Hypolimnas population, an equal population sex ratio was restored over the course of 8–10 generations (a single year) after resistance reached the island (Charlat et al. 2007; Hornett et al. 2014).
In another example, feminizing Wolbachia in the woodlouse Armadillidium vulgare often reach very high frequencies within populations, such that the only males present come from eggs that by chance do not inherit sufficient Wolbachia to convert them into females (Leclercq et al. 2016). In these highly female-biased populations, the normal ZW sex determination system is defunct, with Wolbachia dose effectively controlling the sex of individuals. This can lead to the loss of the female-determining W chromosome; all individuals are ZZ genetic males, but this state is overwritten by the feminization imposed by Wolbachia, suggesting that the Z and autosomes have been unable to evolve resistance. In some populations a novel W sex chromosome has appeared; remarkably, this neo-W chromosome is a former autosome that now carries a near-complete copy of the Wolbachia genome. This neo-W is thought to have spread through outcompeting Wolbachia feminized ZZ individuals (Cordaux and Gilbert 2017). Given the likely cost of incorporating a bacterial genome, this illustrates that suppression of gene drives can involve high costs and major genomic rearrangements. Despite these examples of suppression in Wolbachia, suppressors against driving organelles and endosymbionts in animals seem to be rare. It is not clear why this is the case, given that cytoplasmic male sterility systems in plants are often suppressed.
2.1.6. Systems where suppression has not been found
Although mutations have allowed resistance to evolve in many systems, there are examples of both sex-linked and autosomal drivers for which little or no suppression has been found. For example, in the well-studied t haplotype of house mice, distorter loci are bound together in inversions and cause dysregulation of development in sperm carrying the wildtype target allele (Herrmann and Bauer 2012; Lindholm et al. 2019). Suppression of the t haplotype has not been found in wild populations (Ardlie and Silver 1996), although transmission differences have been reported in crosses between laboratory strains (Bennett et al. 1983; Gummere et al. 1986). In one closely-monitored study population the t haplotype declined and went extinct within eight years, which is thought to be due to negative density-dependent effects on fitness (Manser et al. 2011) and positive density-dependent effects on dispersal (Runge and Lindholm 2018), rather than suppressors of t. The combination of strong distortion and lack of evidence of suppression has led to plans to develop a synthetic sex chromosome driver from the t haplotype by adding a male sex-determining gene (Sry) to the t, for the purpose of controlling invasive house mouse populations on islands (Backus and Gross 2016; Campbell et al. 2019).
Similarly, the sex ratio-distorting X chromosome drive system in Drosophila pseudoobscura has been studied for almost a century, yet no evidence has been found of target site variation leading to suppression, or indeed any factors that reduce drive strength (Price et al. 2019). This is puzzling given that SR reaches 30% frequency in populations in the Southwestern USA, imposes significant costs on the males that carry it, and has apparently existed for hundreds of thousands of years (Kovacevic and Schaeffer 2000), providing ample time for the evolution of resistance. In the related species D. subobscura, only an extremely weak suppressor of drive has been found, again despite a high frequency of drive in natural populations and substantial costs of drive (Verspoor et al. 2018). The same lack of suppressors occurs in Teleopsis dalmanni stalk-eyed flies which again have a high frequency SR drive system which imposes significant viability costs in males and females (Finnegan et al. 2019) and is estimated to be a million years old (Reinhardt et al. 2014). The hybridizing species D. testacea and D. neotestacea each bear driving X chromosomes, but the former shows strong autosomal suppression (Keais et al. 2020), while the latter shows no evidence of suppression at all (Pinzone and Dyer 2013). Surprisingly, in the known Drosophila species with SR gamete killing drive systems only about half have evidence of genetic suppression of drive (Courret et al. 2019; Price et al. 2019), even though many of these systems are thought to have existed for tens if not hundreds of thousands of generations (Price et al. 2019).
The existence of multiple well-studied, ancient drive systems showing no evidence of suppression of drive is a mystery. Does this indicate that genetic suppression is unlikely to evolve? One possibility is that the locus that confers susceptibility to drive is small, providing a small mutational target. However, many drivers impose broad costs across the genome (Hamilton 1967; Dyer and Hall 2019; Finnegan et al. 2019; Larner et al. 2019; Zanders and Unckless 2019), so loci throughout the genome are predicted to evolve to resist costly gene drives. Here, the lack of resistance mechanisms cannot be due to the small size of the mutational target, suggesting the involvement of other evolutionary constraints. Perhaps effective resistance to the gene drive requires multiple mutations that are not individually beneficial, making resistance evolution less likely. Drive could also target essential sites in the genome that are constrained from evolving, or repetitive DNA that is continually re-created by mutation or transposition, as is thought to be the case for the satellite locus Rsp that is targeted by the SD gene drive in D. melanogaster (Courret et al. 2019). Another possibility is that some gene drives are involved in ongoing coevolutionary arms races with resistance loci, such that the supposedly unresistable gene drives that we observe are those that have temporarily outpaced their suppressors for a short span of evolutionary time. The Hypolimnas example appears to provide an example of this: the costs of Wolbachia sex ratio distortion were high and Wolbachia was very common, yet for at least a century there was no sign of resistance to the drive. When a resistance allele appeared, it rapidly spread across the species’ range within a few decades (Hornett et al. 2014).
2.2. Behavioural and life history resistance against drive
One explanation for lack of direct suppression of the mechanism of drive is the evolution of indirect resistance involving behavioral or life history changes. For example, self-medication in which a Wolbachia infected individual might reduce their titre by exposing themselves to heat that impairs Wolbachia, or feeding on an antibiotic rich diet (Snook et al. 2000; Abbott 2014; Shikano 2017) is a possible but untested idea. There may be many unexplored life history or behavioural ways to resist drive.
One of the best known ideas is that non-carriers may avoid drive carriers as mates, preventing offspring from inheriting harmful drivers, and improving offspring fitness. Theoretical models support this idea (Lande and Wilkinson 1999; Reinhold et al. 1999; Randerson et al. 2000; Manser et al. 2017). However, this requires a trait that reliably reveals the presence or absence of drive (Lande and Wilkinson 1999; Manser et al. 2017). However, evidence of mate avoidance of drive carriers is weak or absent from the majority of systems studied. For example, in species where Wolbachia induces cytoplasmic incompatibility, uninfected females lose any eggs fertilized by Wolbachia-infected males. Despite these costs, there is very little evidence for females avoiding mating with Wolbachia infected males (Champion de Crespigny and Wedell 2007). Likewise, the only test of populations infected with male-killing Wolbachia in D. innubila also found no evidence that males prefer to mate with uninfected females (Sullivan and Jaenike 2006). There is also little evidence for female preference against male Drosophila carrying SR drive despite decades of research in several species (Price and Wedell 2008). In house mice, wildtype females do not avoid mating with t haplotype-bearing males (Lenington and Coopersmith 1992; Manser et al. 2015; Sutter and Lindholm 2016), while t-bearing females have been found to avoid t-bearing males in some (Lenington and Coopersmith 1992) but not all studies (Manser et al. 2015; Sutter and Lindholm 2016).
In these gene drive systems, it is not obvious that any phenotypic characters reliably signal Wolbachia or drive carrier status, which may explain the lack of mate preference. In stalk-eyed flies, female preference is for males with large eyespan, and males carrying SR have a smaller average eyespan (Wilkinson et al. 1998; Johns et al. 2005; Cotton et al. 2014), providing a ready-made trait that can distinguish drive from non-drive carrying males. However, other species of stalk-eyed fly that lack meiotic drive also show female mate preference for exaggerated male eyespan (e.g. Diasemopsis meigenii, Cotton et al. 2006), and it has yet to be demonstrated whether mate preference has been strengthened for avoidance of drive carriers. Disentangling general condition-dependent mate preferences from evolved resistance to drive through avoidance of mating with drive carriers can be highly challenging.
In the Winters SR system of D. simulans, the strength of drive declines from 93% to 60% daughters when males are reared at high temperatures, and older males also show a decline in drive (Tao et al. 2007b). This could promote females evolving a preference for males unlikely to have strong drive due to these non-genetic causes (i.e. high temperature reared or older males), but to date this has not been examined, although age based mate choice is common in Drosophila and other organisms (Verspoor et al. 2015). Furthermore, D. simulans females disproportionately discard sperm of Paris SR males after mating (Angelard et al. 2008). It is not known whether this post-copulatory selection has evolved due to the benefits of reducing the likelihood of drive bearing offspring, or is a general mechanism selecting against mates that transfer small ejaculates. Perhaps the most convincing evidence for mate choice against drivers comes from feminizing Wolbachia in Armadillidium vulgare. In populations with high Wolbachia frequency, males are rare, and males will benefit from mating with uninfected females who produce more sons. In this case, males have been found to preferentially mate with ZW uninfected females, rather than genetically male ZZ individuals who have been feminized (Moreau et al. 2001). Whether this has suppressed Wolbachia frequency in populations has not been established. In general, the lack of choice against drive carriers may be due to evolutionary pressure to reduce detectability, with the least detectable gene drive alleles outcompeting rival variants, but this remains to be investigated.
Another route for drive-susceptible females to avoid siring offspring with drive carriers is by increasing the intensity of sperm competition. In several systems of gamete-killing male meiotic drive, drive-carrying males are inferior sperm competitors, because of a reduction in sperm number and quality (Price and Wedell 2008). For example, in controlled experimental matings, t-carrying males gain only 12% of paternity when a female mates with both a t-carrying and wildtype male (Sutter and Lindholm 2015). Females could therefore mate with several males indiscriminately and rely on sperm competition to suppress fertilization by drive sperm (Haig and Bergstrom 1995). An increase in the propensity to mate with multiple males could evolve as a form of resistance to the presence of a driver within the population. Multiple mating potentially evolves more easily than pre-copulatory mate choice, as no discrimination between driver-carrier and driver-free individuals is required (Haig and Bergstrom 1995). The evolution of higher remating rates in response to the presence of a sex ratio distorter was seen within 10 generations in a laboratory experiment using D. pseudoobscura (Price et al. 2008). So it is possible that in polyandrous species, sperm competition reduces the success of gamete killers enough that selection for direct genetic suppression is reduced. As yet, there is no concrete evidence for this in nature.
3. Resistance in synthetic systems
Synthetic gene drive systems provide some of the most informative studies of the evolution of resistance through sequence changes at target sites. They allow observation of the process of resistance evolution. Sequencing can reveal standing variation and mutations that confer resistance, identify the original resistant individuals and provide empirical evidence of the speed with which resistance arises in the laboratory and, possibly in the future, under field conditions.
3.1. Mutations at the target site and suppression of drive machinery
3.1.1. Homing based drive systems
Many newly engineered systems are based on homing drives that mimic the mode of propagation of homing endonuclease genes (HEGs), a class of naturally-occurring selfish genetic elements found in bacteria, fungi, and other organisms (Burt and Trivers 2006). These encode DNA-cleaving enzymes that generate double strand DNA breaks at target sites with a specific nucleotide sequence as a result of which the sequence is converted into a HEG+ allele. Homing potentially converts all target sites in all members of a population. The challenge in generating synthetic drive systems based on HEG proteins (Windbichler et al. 2011) has been alleviated by the production of homing systems based on CRISPR-Cas9 constructs.
Unlike natural homing endonucleases, which tolerate some variation in the fidelity of individual nucleotides in the target sequence, CRISPR-Cas9 cleavage requires a near-perfect match between the ~20 base pair guide RNA sequence and the genomic target site. Hence single nucleotide differences at target sites can confer resistance. These arise at a high rate, simply through the action of the gene drive. The double strand breaks induced by Cas9 are typically resolved by homology directed repair, inserting a copy of the homing agent sequence into the target site. Alternatively, double strand breaks undergo non-homologous end joining. This process often results in imprecise repair, increasing the rate of mutation at the target site without insertion of the gene drive. These novel alleles will confer resistance as they have a different sequence, and may preserve gene function. In laboratory experiments with flies and mosquitoes, resistance to CRISPR-Cas9 homing drives emerges rapidly, in particular when the driver targets single sites (Gantz et al. 2015; Champer et al. 2017; Hammond et al. 2017; Hammond et al. 2018; KaramiNejadRanjbar et al. 2018). Functional target gene mutants can be generated at considerable frequency within one generation by in-frame indels (KaramiNejadRanjbar et al. 2018). One approach to delay the evolution of resistance at the target site is to design targets at highly conserved regions in which sequence variation, including in-frame indels, cannot be tolerated because any change is associated with high fitness costs (Kyrou et al. 2018). Alternatively, a suite of sites can be targeted by the drive construct.
When the aim is gene replacement rather than population suppression, gene drives are designed to have low fitness costs and avoid disruption of normal host gene function. This should constrain selection for resistance alleles. But the “cargo” of replacement genes is unlikely to be cost-free. Examples of cargoes include genes that encode resistance or susceptibility to disease or toxins, and genes that alter sexual phenotype. All of these will carry costs and in the long term they are expected to be lost due to the spread of loss-of-function mutations. When loss-of-function is caused by deletion, this may even enhance gene drive spread (i.e. of a null allele); replacement gene drives are only useful as long as the cargo remains intact. The assumption is that the replacement gene will spread and persist sufficiently long to provide its public health benefit (Beaghton et al. 2017). Other types of cargo may be more resilient to loss, for example where the cargo is beneficial to the organism, such as thermal tolerance genes or symbionts (Piaggio et al. 2017). Finally, expression of the endonuclease is unlikely to be without fitness cost and thus subject to mutational decay. But this will mostly come to play at the point when the drive construct has already successfully propagated itself in a population. These constraints have hardly been investigated, but seem likely to place limits on the spread and effectiveness of homing gene drives.
3.1.2. Synthetic sex ratio distorters
The X chromosome is the target in engineered systems that aim at distorting the sex ratio towards males. One approach, inspired by the mode of action of natural sex-distorters in the mosquitoes Aedes aegypti and Culex pipiens (Wood and Newton 1991), operates by targeting the X-linked rDNA cluster with an endonuclease operating in spermatogenesis (Galizi et al. 2016). The lack of target site resistance, at least when observed at the limited scale of population cage experiments, reflects the use of extremely conserved rDNA target sequences which are present in hundreds of copies on the X chromosome, although even this cannot completely remove the possibility of resistance evolving. Gene drive systems targeting the heterogametic sex chromosome have only been investigated theoretically (Holman 2019; Prowse et al. 2019) and in preliminary experiments in a house mice system (Prowse et al. 2019).
3.1.3. Wolbachia
The cytoplasmic incompatibility wMelPop strain of Wolbachia was originally isolated from a laboratory screen of D. melanogaster, where it shortens lifespan (Min and Benzer 1997). It was proposed as a tool for biocontrol of dengue in Aedes aegypti mosquitoes as viruses are only transmitted by older mosquitoes. In addition, Wolbachia directly inhibits replication of the dengue virus (Walker et al. 2011). wMelPop and other Wolbachia strains have been successfully transinfected into Aedes aegypti and their spread is driven by cytoplasmic incompatibility. The spread of wMelPop into natural populations has proven challenging as this strain inflicts a suite of costly effects on its host (Nguyen et al. 2015) but strains with fewer deleterious effects have proven more successful (Hoffmann et al. 2011; Nazni et al. 2019).
There are several avenues to the evolution of resistance to Wolbachia infection. Mosquitoes could evolve resistance to cytoplasmic incompatibility or Wolbachia itself, Wolbachia infections could attenuate over time (there are many examples of weak effects in natural Wolbachia infections (Caragata et al. 2019)), or arboviruses could evolve to bypass the inhibitory effect of Wolbachia (Bull and Turelli 2013). Alternatively, as high temperature can eliminate Wolbachia infections, it might be possible for mosquitoes to suppress infections by altering their temperature preferences (Ross et al. 2019a). However, a trial introduction of Wolbachia has seen maintenance of strong cytoplasmic incompatibility and relatively stable frequencies in Australian field populations for seven years since their release, suggesting this may be unlikely, or at least slow to evolve (Ross et al. 2019b; Ryan et al. 2020). After nearly a decade of use, there is as yet no evidence of any type of resistance evolving and the ability to block dengue virus has not been lost (Ross et al. 2019b; Ryan et al. 2020). A further question is whether Wolbachia and dengue will enter a co-evolutionary arms race against one another in these populations.
3.1.4. Medea and underdominance-like systems
Medea-like systems encode a maternal toxin and zygotic antidote, killing offspring that do not inherit the Medea gene drive (Beeman et al. 1992). Synthetic underdominance systems are conceptually similar, consisting of a set of lethal loci, each associated with a suppressor of the other (Davis et al. 2001). Individuals inheriting only one of the loci carry a lethal locus, but not its suppressor, resulting in reduced viability or fertility. Resistance to these systems is likely to occur via changes to the toxin’s target. For example, an underdominant maternal-effect lethal introduced into the soft-fruit pest D. suzukii depends on a miRNA toxin and a zygotic antidote to function and will be impaired by variation at the miRNA binding site. Indeed, a recent survey shows natural variation in the miRNA toxin target sites (Buchman et al. 2018a). Population cage experiments found that the Medea drive was unable to persist in populations likely due to a combination of significant fitness costs of the driver as well as standing variation in resistance present in the cage populations (Buchman et al. 2018a).
In addition to target-site mutation, Medea and similar toxin-antidote systems could also encounter resistance through driver inactivation either through direct suppression or the spread of antidote-only alleles due to mutational inactivation of toxin production. The single study investigating the stability of a D. melanogaster underdominance system found no evidence of resistance evolution over >200 generations (Reed et al. 2018). Finally, there has been recent theoretical proof-of-principle of other Medea-like systems that rely on either CRISPR-Cas9 transcriptional overactivation of an endogenous target gene as the “toxin” and an insensitive copy of that target as the “antidote” or CRISPR-Cas9 cleavage as the “toxin” and resistant target gene as the “antidote” (Champer et al. 2019). These too will face similar types of resistance (e.g., target-site mutation, driver inactivation). They are not in principle different from other synthetic gene drive systems that utilize CRISPR-Cas9, although their development is still at an early stage and not advanced enough for empirical investigation of resistance evolution.
4. The strength of selection for resistance across gene drive systems
The strength of selection against a driver can vary dramatically between drive mechanisms and targets. At one extreme, a synthetic driver aimed at killing carriers or preventing reproduction, or distorting sex ratios will create extremely strong genome wide selection for resistance against drive. In contrast, a biased gene converter that carries no cost to the organism will select for resistance at the target locus and linked sites, but have no effect on the rest of the genome. Drivers may themselves have a range of harmful pleiotropic effects, or be in linkage with deleterious alleles (Burt and Trivers 2006). Fitness loss is often observed in both males and females, especially when drivers are homozygous (Hamilton 1967; Dyer and Hall 2019; Finnegan et al. 2019; Larner et al. 2019; Zanders and Unckless 2019).
To understand the strength of selection against novel drivers, we need to know their fitness consequences in the field. There is currently a lack of such information for virtually all considered synthetic gene drives. One of the few systems where such information is readily available is for Wolbachia-carrying Aedes mosquitoes. The fitness costs associated with Wolbachia infection have been shown to be exacerbated under field conditions. As an example, the wMelPop Wolbachia strain, which invaded mosquito populations in semi-field cage trials, failed in several field trials because infected females had unexpectedly reduced egg viability in the field (Nguyen et al. 2015). This emphasizes the need for field studies of the fitness of drive carriers for the use of gene drive in natural populations.
The spatial structure of target populations is likely to be an important factor in deciding the fate of a gene drive system, as well as the way resistance may arise or spread. For example, Noble et al. (2018) showed that moderate amounts of gene flow between neighboring populations is sufficient for a HEG-based replacement gene drive to spread between populations, even when resistance systematically arises in each individual population. More generally, we expect not only population genetic structure but also landscape and ecological characteristics to significantly impact the fate of a gene drive. Abiotic barriers (highways, open fields) have been shown to impede the spread of Wolbachia infections due to the limited dispersal ability of Aedes mosquitoes (Schmidt et al. 2017).
We can also imagine the evolution of tolerance to drive – meaning that the rest of the genome mitigates the deleterious effects of drive without directly interfering with the drive mechanism. For example, in stalk-eyed flies males with drive invest more in testes to compensate for the loss of half of their sperm caused by the driver (Meade et al. 2020). Such changes do not interfere with drive and may actually enhance its spread. They lessen the deleterious costs of drive to the rest of the genome even though they do not improve fitness for the target chromosome. This reduction in the costs of the gene drive potentially reduces the strength of selection to suppress the driver.
There has been surprisingly little consideration of how all these processes interact when a new driver evolves or enters a population. Does the evolution of an effective defense mechanism against a driver preclude the evolution of other defences? There may be some parallels with the evolution of multiple defences against predators and parasites, which suggests multiple defenses commonly evolve (Broom et al. 2010).
5. Strategies for designing synthetic drive systems to reduce resistance
Although resistance to synthetic gene drive elements cannot be entirely prevented, it can be anticipated and reduced by careful gene drive design. In particular, the study of both natural and synthetic gene drive systems has taught us some important lessons about how resistance may arise in the face of various drive systems, and what we can do to minimize different types of resistance.
Many gene drives require a target, and evolved changes to this target are an obvious route to resistance. Recent work on CRISPR-Cas9 based homing endonuclease genes (HEGs) and synthetic Medea elements has highlighted the substantial impact that pre-existing and drive-generated target site changes can have on preventing the spread of synthetic drivers (Gantz et al. 2015; Champer et al. 2017). There are several design strategies that may help prevent target site resistance. Firstly, targets in essential and/or highly conserved sequences/genes may be less tolerant of sequence variability/polymorphism, and thus less likely to harbor pre-existing resistance alleles, or to tolerate novel mutational variation (Buchman et al. 2018b). For homing-based systems, gene drives could home into genes that are essential, so that incorrect homing events (e.g., non-homologous end joining) result in lethal products (Esvelt et al. 2014; Bull and Malik 2017; Kyrou et al. 2018). A second strategy would be to target multiple sites. The same principle applies to Medea or other systems with “toxins” that act on specific sequence regions (Champer et al. 2017; Marshall et al. 2017; Noble et al. 2017; Champer et al. 2018). Combining multiple mechanism e.g. a suppressive gene drive that also distorts the sex ratio could be another way to delay the emergence of resistance (Simoni et al. 2020).
It is also critical to make the driver as stable as possible. For example, reducing the size of a CRISPR-Cas9 HEG transgene increases the likelihood that it will copy itself correctly, and integrating such a drive into endogenous genes may help achieve this goal (Nash et al. 2019; Hoermann et al. 2020). Clearly this may trade off with the benefits of more complicated drivers that reduce resistance evolution by attacking multiple loci. Additionally, repetitive DNA sequence (such as from multiple sgRNA or miRNA backbones) can reduce stability (Bzymek and Lovett 2001; Simoni et al. 2014; Marshall et al. 2017), and reduction of such repetitiveness can protect against recombination and possible loss of a part of the drive element. It is also important to take into consideration the inherent evolutionary stability of integral gene driver components and mechanisms. For example, using a smaller protein than Cas9 in the drive mechanism could reduce the chance of mutations that inactivate the driver. Additionally, the endogenous homology directed repair process required for CRISPR-Cas9 HEG function may be error-prone and lead to driver loss of function (Hammond et al. 2016; Oberhofer et al. 2018). Conversely, miRNA or chromosomal rearrangement-based systems may be more evolutionarily stable because they do not rely on large exogenous proteins and error-prone repair pathways to function.
Minimizing any fitness costs of the driver is also likely to reduce selection for resistance. Genomic insertion sites are associated with different costs, so transgenes inserting at a low cost site may create less selection for resistance. It is also advisable to reduce pleiotropic impacts of gene drive, as this can create resistance alleles in some systems. For example, work on CRISPR-Cas9 HEGs suggests that expression of the nuclease in somatic cells can lead to off target site mutation which reduces the spread of the driver (Gantz et al. 2015; Champer et al. 2017; Hammond et al. 2017; Beaghton et al. 2019).
It is also important to remember the ecology of the target species, as this may offer novel ideas for making a gene drive system durable, or reveal weaknesses only present in the field. For example, extremely high temperatures in Australia in 2019 may have impaired the transmission of the temperature sensitive wMel Wolbachia strain used to combat dengue in Queensland mosquitoes. Synthetic drives designed in benign laboratory conditions may struggle in the field during extreme environmental conditions. If a gene drive is unable to penetrate some areas of an environment, due to conditions that prevent drive function or increase its costs, this could provide ideal circumstances for resistance to evolve.
Finally, it is essential to choose the right gene drive for the job. Certain types of drive (e.g., translocations) are much less likely to face resistance, but may spread more slowly than drives that bias segregation (Champer et al. 2016; Buchman et al. 2018b). Additionally, population suppression drives will face considerably stronger evolutionary pressures in terms of resistance than replacement drives (Eckhoff et al. 2017; Prowse et al. 2017; KaramiNejadRanjbar et al. 2018). However, resistance will not always be an impediment to gene drive deployment. For example, if the goal is short term transformation of a population, then long term evolution of resistance against the gene drive may not matter (Unckless et al. 2017). Resistance in non-target populations may make gene drives less likely to spread accidentally (Esvelt et al. 2014). If the target population carries only susceptible alleles, but surrounding populations have a mix of susceptible and resistant alleles, the driver may also be unable to successfully spread to non-target populations (Sudweeks et al. 2019).
6. Conclusions
The evolution of resistance is a key problem in the design and use of gene drives. It is a major challenge faced by natural gene drive systems but remains poorly understood. Resistance based on interference can arise very rapidly, within a single generation, but in some natural systems does not appear to have evolved despite long timeframes.
As illustrated by this review, mechanisms of resistance are very diverse. Although we understand some of the mechanisms that can resist drivers, we rarely have a clear understanding of the forces underlying individual resistance pathways, nor the biological and ecological factors that determine which resistance type or mechanism is more likely to be selected in a given situation. In the context of applied control programs using specific gene drive approaches, the current lack of knowledge means that robust predictions cannot be made about the types of resistance that are most likely to arise that might impede success.
Nevertheless, combining work from natural and synthetic drive provides an opportunity for a new depth of understanding. The study of synthetic drive systems is limited to short evolutionary timescales, and typically small population sizes. In contrast, natural drive systems have evolved over thousands of generations, and several are ancient. Thus, synthetic drives provide a unique opportunity to understand what happens at short time scales when a novel driver arises, with unprecedented understanding of the mechanisms of drive and resistance. Techniques borrowed from synthetic technologies could allow researchers working on natural drive to create constructs based on their driver of interest, and directly test hypotheses related to evolutionary history and fate of natural drive. Conversely, a better understanding of the mechanisms of natural drivers, particularly those that appear hard to suppress, may lead to new generations of synthetic drivers that mimic the successful traits of the natural drivers. Lessons from both synthetic and natural drivers can be combined to produce safer, more effective drive systems.
Acknowledgements
Support from grant IZSEZ0_180460 from the Swiss National Science Foundation and a Progress Meetings in Evolutionary Biology grant from the European Society for Evolutionary Biology allowed us to meet in Arolla, Switzerland, to initiate this paper. Participation of AS, AM and JR was supported by the Swiss National Science Foundation (P300PA_177906, P300PA_177830 and 31003A_160328 respectively), of AML by the National Institutes of Health (NIH R35GM119515) and a Steven Biggar and Elisabeth Asaro Fellowship in Data Science, TARP by UKRI (NERC NE/P002692/1) and AP by UKRI (NERC NE/R010579/1, EPSRC EP/F500351/1 and EP/I017909/1). The authors thank Wolf Blanckenhorn and two anonymous reviewers for their comments on the paper.
Footnotes
Conflicts of Interest Statement
The authors state they have no conflicts of interest to declare.
References
- Abbott J (2014). Self-medication in insects: current evidence and future perspectives. Ecological Entomology, 39(3), 273–280. doi: 10.1111/een.12110 [DOI] [Google Scholar]
- Angelard C, Montchamp-Moreau C, & Joly D (2008). Female-driven mechanisms, ejaculate size and quality contribute to the lower fertility of sex-ratio distorter males in Drosophila simulans. BMC Evolutionary Biology, 8(1), 326. doi: 10.1186/1471-2148-8-326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anxolabéhère D, Kidwell MG, & Periquet G (1988). Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Molecular Biology and Evolution, 5(3), 252–269. doi: 10.1093/oxfordjournals.molbev.a040491 [DOI] [PubMed] [Google Scholar]
- Ardlie K, & Silver LM (1996). Recent evolution of mouse t haplotypes at polymorphic microsatellites associated with the t complex responder (Tcr) locus. Genetic Research, 67, 1–10. doi: 10.1017/S0016672300033425 [DOI] [PubMed] [Google Scholar]
- Atlan A, Merçot H, Landre C, & Montchamp-Moreau C (1997). The Sex-ratio trait in Drosophila simulans: geographical distribution of distortion and resistance. Evolution, 51(6), 1886–1895. doi: 10.1111/j.1558-5646.1997.tb05111.x [DOI] [PubMed] [Google Scholar]
- Backus G, & Gross K (2016). Genetic engineering to eradicate invasive mice on islands: Modeling the efficiency and ecological impacts. Ecosphere, 7, e01589. doi: 10.1002/ecs2.1589 [DOI] [Google Scholar]
- Barrangou R, & Marraffini Luciano A. (2014). CRISPR-Cas Systems: prokaryotes upgrade to adaptive immunity. Molecular Cell, 54(2), 234–244. doi: 10.1016/j.molcel.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LG, Legros M, Kumaran N, Glassop D, Raghu S, & Gardiner DM (2019). Gene drives in plants: opportunities and challenges for weed control and engineered resilience. Proceedings of the Royal Society B: Biological Sciences, 286(1911), 20191515. doi: 10.1098/rspb.2019.1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide H, Cazemajor M, Ogereau D, Derome N, Hospital F, & Montchamp-Moreau C (2011). Rapid rise and fall of selfish Sex-Ratio X chromosomes in Drosophila simulans: Spatiotemporal analysis of phenotypic and molecular data. Molecular Biology and Evolution, 28(9), 2461–2470. doi: 10.1093/molbev/msr074 [DOI] [PubMed] [Google Scholar]
- Bastide H, Gérard PR, Ogereau D, Cazemajor M, & Montchamp-Moreau C (2013). Local dynamics of a fast-evolving sex-ratio system in Drosophila simulans. Molecular Ecology, 22(21), 5352–5367. doi: 10.1111/mec.12492 [DOI] [PubMed] [Google Scholar]
- Beaghton A, Hammond A, Nolan T, Crisanti A, Godfray HCJ, & Burt A (2017). Requirements for driving antipathogen effector genes into populations of disease vectors by homing. Genetics, 205(4), 1587–1596. doi: 10.1534/genetics.116.197632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaghton AK, Hammond A, Nolan T, Crisanti A, & Burt A (2019). Gene drive for population genetic control: non-functional resistance and parental effects. Proceedings of the Royal Society B: Biological Sciences, 286(1914), 20191586. doi: 10.1098/rspb.2019.1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeman R, Friesen K, & Denell R (1992). Maternal-effect selfish genes in flour beetles. Science, 256(5053), 89–92. doi: 10.1126/science.1566060 [DOI] [PubMed] [Google Scholar]
- Bennett D, Alton A, & Artzt K (1983). Genetic analysis of transmission ratio distortion by t-haplotypes in the mouse. Genetical research, 41(1), 29–45. [DOI] [PubMed] [Google Scholar]
- Bousios A, & Gaut BS (2016). Mechanistic and evolutionary questions about epigenetic conflicts between transposable elements and their plant hosts. Current Opinion in Plant Biology, 30, 123–133. doi: 10.1016/j.pbi.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Brand CL, Larracuente AM, & Presgraves DC (2015). Origin, evolution, and population genetics of the selfish Segregation Distorter gene duplication in European and African populations of Drosophila melanogaster. Evolution, 69(5), 1271–1283. doi: 10.1111/evo.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Núñez MA, Lange JJ, & Zanders SE (2018a). A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver’s antidote. PLOS Genetics, 14(11), e1007836. doi: 10.1371/journal.pgen.1007836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Núñez MA, Nuckolls NL, & Zanders SE (2018b). Genetic villains: killer meiotic drivers. Trends in Genetics, 34(6), 424–433. doi: 10.1016/j.tig.2018.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo Núñez MA, Sabbarini IM, Eickbush MT, Liang Y, Lange JJ, Kent AM, & Zanders SE (2020). Dramatically diverse Schizosaccharomyces pombe wtf meiotic drivers all display high gamete-killing efficiency. PLoS Genetics, 16(2), e1008350. doi: 10.1371/journal.pgen.1008350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, & Hannon GJ (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell, 128(6), 1089–1103. doi: 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Broom M, Higginson AD, & Ruxton GD (2010). Optimal investment across different aspects of anti-predator defences. Journal of Theoretical Biology, 263(4), 579–586. doi: 10.1016/j.jtbi.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Buchman A, Marshall JM, Ostrovski D, Yang T, & Akbari OS (2018a). Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. Proceedings of the National Academy of Sciences of the United States of America, 115(18), 4725–4730. doi: 10.1073/pnas.1713139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman AB, Ivy T, Marshall JM, Akbari OS, & Hay BA (2018b). Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in Drosophila. ACS Synthetic Biology, 7(5), 1359–1370. doi: 10.1021/acssynbio.7b00451 [DOI] [PubMed] [Google Scholar]
- Bull JJ, & Malik HS (2017). The gene drive bubble: New realities. PLOS Genetics, 13(7), e1006850. doi: 10.1371/journal.pgen.1006850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, & Turelli M (2013). Wolbachia versus dengue: Evolutionary forecasts. Evolution, Medicine, and Public Health, 2013(1), 197–207. doi: 10.1093/emph/eot018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A (2014). Heritable strategies for controlling insect vectors of disease. Philosophical Transactions of the Royal Society B: Biological Sciences, 369(1645), 20130432. doi: doi: 10.1098/rstb.2013.0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, & Trivers R (2006). Genes in Conflict: the Biology of Selfish Genetic Elements. Cambridge: Harvard University Press. [Google Scholar]
- Bzymek M, & Lovett ST (2001). Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proceedings of the National Academy of Sciences, 98(15), 8319–8325. doi: 10.1073/pnas.111008398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KJ, Saah JR, Brown PR, Godwin J, Gould F, Howald GR, … Shiels A (2019). A potential new tool for the toolbox: assessing gene drives for eradicating invasive rodent populations In Veitch CR, Clout MN, Martin AR, Russell JC & West CJ (Eds.), Island invasives: scaling up to meet the challenge (pp. 6–14). Gland, Switzerland: IUCN. [Google Scholar]
- Caragata EP, Tikhe CV, & Dimopoulos G (2019). Curious entanglements: interactions between mosquitoes, their microbiota, and arboviruses. Current Opinion in Virology, 37, 26–36. doi: 10.1016/j.coviro.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J, Buchman A, & Akbari OS (2016). Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nature Reviews Genetics, 17(3), 146–159. doi: 10.1038/nrg.2015.34 [DOI] [PubMed] [Google Scholar]
- Champer J, Kim I, Champer SE, Clark AG, & Messer PW (2019). Performance analysis of novel toxin-antidote CRISPR gene drive systems. bioRxiv, 628362. doi: 10.1101/628362 [DOI] [PMC free article] [PubMed]
- Champer J, Liu J, Oh SY, Reeves R, Luthra A, Oakes N, … Messer PW (2018). Reducing resistance allele formation in CRISPR gene drive. Proceedings of the National Academy of Sciences, 115(21), 5522–5527. doi: 10.1073/pnas.1720354115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer J, Reeves R, Oh SY, Liu C, Liu J, Clark AG, & Messer PW (2017). Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLOS Genetics, 13(7), e1006796. doi: 10.1371/journal.pgen.1006796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion de Crespigny FE, & Wedell N (2007). Mate preferences in Drosophila infected with Wolbachia? Behavioral Ecology and Sociobiology, 61(8), 1229. doi: 10.1007/s00265-007-0353-y [DOI] [Google Scholar]
- Charlat S, Hornett EA, Fullard JH, Davies N, Roderick GK, Wedell N, & Hurst GDD (2007). Extraordinary flux in sex ratio. Science, 317(5835), 214–214. doi: 10.1126/science.1143369 [DOI] [PubMed] [Google Scholar]
- Chen L, & Liu Y-G (2014). Male sterility and fertility restoration in crops. Annual Review of Plant Biology, 65(1), 579–606. doi: 10.1146/annurev-arplant-050213-040119 [DOI] [PubMed] [Google Scholar]
- Cordaux R, & Gilbert C (2017). Evolutionary significance of Wolbachia-to-animal horizontal gene transfer: female sex determination and the f element in the isopod Armadillidium vulgare. Genes, 8(7), 186. doi: 10.3390/genes8070186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AJ, Földvári M, Cotton S, & Pomiankowski A (2014). Male eyespan size is associated with meiotic drive in wild stalk-eyed flies (Teleopsis dalmanni). Heredity, 112(4), 363–369. doi: 10.1038/hdy.2013.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton S, Rogers DW, Small J, Pomiankowski A, & Fowler K (2006). Variation in preference for a male ornament is positively associated with female eyespan in the stalk-eyed fly Diasemopsis meigenii. Proceedings of the Royal Society B: Biological Sciences, 273(1591), 1287–1292. doi: 10.1098/rspb.2005.3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courret C, Chang C-H, Wei KH-C, Montchamp-Moreau C, & Larracuente AM (2019). Meiotic drive mechanisms: lessons from Drosophila. Proceedings of the Royal Society B: Biological Sciences, 286(1913), 20191430. doi: 10.1098/rspb.2019.1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courret C, Gérard PR, Ogereau D, Falque M, Moreau L, & Montchamp-Moreau C (2018). X-chromosome meiotic drive in Drosophila simulans: a QTL approach reveals the complex polygenic determinism of Paris drive suppression. Heredity. doi: 10.1038/s41437-018-0163-1 [DOI] [PMC free article] [PubMed]
- Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, & Hannon GJ (2018). piRNA-guided genome defense: from biogenesis to silencing. Annual Review of Genetics, 52(1), 131–157. doi: 10.1146/annurev-genet-120417-031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Bax N, & Grewe P (2001). Engineered underdominance allows efficient and economical introgression of traits into pest populations. Journal of Theoretical Biology, 212(1), 83–98. doi: 10.1006/jtbi.2001.2357 [DOI] [PubMed] [Google Scholar]
- Dawe RK, Lowry EG, Gent JI, Stitzer MC, Swentowsky KW, Higgins DM, … Hiatt EN (2018). A kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell, 173(4), 839–850.e818. doi: 10.1016/j.cell.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Derome N, Baudry E, Ogereau D, Veuille M, & Montchamp-Moreau C (2008). Selective sweeps in a 2-locus model for sex-ratio meiotic drive in Drosophila simulans. Molecular Biology and Evolution, 25(2), 409–416. doi: 10.1093/molbev/msm269 [DOI] [PubMed] [Google Scholar]
- Dyer K, & Hall D (2019). Fitness consequences of a non-recombining sex-ratio drive chromosome can explain its prevalence in the wild. Proceedings of the Royal Society B: Biological Sciences. doi: 10.1098/rspb.2019.2529 [DOI] [PMC free article] [PubMed]
- Dyson EA, & Hurst GDD (2004). Persistence of an extreme sex-ratio bias in a natural population. Proceedings of the National Academy of Sciences of the United States of America, 101(17), 6520–6523. doi: 10.1073/pnas.0304068101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson EA, Kamath MK, & Hurst GDD (2002). Wolbachia infection associated with all-female broods in Hypolimnas bolina (Lepidoptera: Nymphalidae): evidence for horizontal transmission of a butterfly male killer. Heredity, 88(3), 166–171. doi: 10.1038/sj.hdy.6800021 [DOI] [PubMed] [Google Scholar]
- Eckhoff PA, Wenger EA, Godfray HCJ, & Burt A (2017). Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proceedings of the National Academy of Sciences, 114(2), E255–E264. doi: 10.1073/pnas.1611064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush MT, Young JM, & Zanders SE (2019). Killer meiotic drive and dynamic evolution of the wtf gene family. Molecular Biology and Evolution, 36(6), 1201–1214. doi: 10.1093/molbev/msz052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Smidler AL, Catteruccia F, & Church GM (2014). Concerning RNA-guided gene drives for the alteration of wild populations. eLife, 3, e03401. doi: 10.7554/eLife.03401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, & Pritham EJ (2007). DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics, 41(1), 331–368. doi: 10.1146/annurev.genet.40.110405.090448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan SR, White NJ, Koh D, Camus MF, Fowler K, & Pomiankowski A (2019). Meiotic drive reduces egg-to-adult viability in stalk-eyed flies. Proceedings of the Royal Society B: Biological Sciences, 286(1910), 20191414. doi: 10.1098/rspb.2019.1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1930). The Genetical Theory of Natural Selection. Oxford: Oxford University Press. [Google Scholar]
- Fishman L, & Kelly JK (2015). Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution, 69(5), 1208–1218. doi: 10.1111/evo.12661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Bond CS, & Small ID (2011). Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proceedings of the National Academy of Sciences, 108(4), 1723–1728. doi: 10.1073/pnas.1007667108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborieau L, Brown GG, & Mireau H (2016). The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. [Review]. Frontiers in Plant Science, 7(1816). doi: 10.3389/fpls.2016.01816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizi R, Hammond A, Kyrou K, Taxiarchi C, Bernardini F, O’Loughlin SM, … Crisanti A (2016). A CRISPR-Cas9 sex-ratio distortion system for genetic control. [Article]. Scientific Reports, 6, 31139. doi: 10.1038/srep31139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, & James AA (2015). Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proceedings of the National Academy of Sciences of the United States of America, 112(49), E6736–E6743. doi: 10.1073/pnas.1521077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grognet P, Bidard F, Kuchly C, Tong LCH, Coppin E, Benkhali JA, … Silar P (2014). Maintaining two mating types: Structure of the mating type locus and its role in heterokaryosis in Podospora anserina. Genetics, 197(1), 421–432. doi: 10.1534/genetics.113.159988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummere GR, McCormick PJ, & Bennett D (1986). The influence of genetic background and the homologous chromosome 17 on t-haplotype transmission ratio distortion in mice. Genetics, 114, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D, & Bergstrom C (1995). Multiple mating, sperm competition and meiotic drive. Journal of Evolutionary Biology, 8. doi: 10.1046/j.1420-9101.1995.8030265.x [DOI] [Google Scholar]
- Hamilton WD (1967). Extraordinary sex ratios. Science, 156, 477–488. [DOI] [PubMed] [Google Scholar]
- Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, … Nolan T (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology, 34(1), 78–83. doi: 10.1038/nbt.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, Kyrou K, Gribble M, Karlsson X, Morianou I, Galizi R, … Nolan T (2018). Improved CRISPR-based suppression gene drives mitigate resistance and impose a large reproductive load on laboratory-contained mosquito populations. bioRxiv, 360339. doi: 10.1101/360339 [DOI]
- Hammond AM, Kyrou K, Bruttini M, North A, Galizi R, Karlsson X, … Nolan T (2017). The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLOS Genetics, 13(10), e1007039. doi: 10.1371/journal.pgen.1007039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleu Q, Courret C, Ogereau D, Burnham KL, Chaminade N, Chakir M, … Montchamp-Moreau C (2019). Sex-Ratio meiotic drive shapes the evolution of the Y chromosome in Drosophila simulans. Molecular Biology and Evolution, 36(12), 2668–2681. doi: 10.1093/molbev/msz160 [DOI] [PubMed] [Google Scholar]
- Helleu Q, Gérard PR, Dubruille R, Ogereau D, Prud’homme B, Loppin B, & Montchamp-Moreau C (2016). Rapid evolution of a Y-chromosome heterochromatin protein underlies sex chromosome meiotic drive. Proceedings of the National Academy of Sciences, 113(15), 4110–4115. doi: 10.1073/pnas.1519332113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG, & Bauer H (2012). The mouse t-haplotype: a selfish chromosome – genetics, molecular mechanism, and evolution In Macholán M, Baird SJE, Munclinger P & Piálek J (Eds.), Evolution of the house mouse. Cambridge: Cambridge University Press. [Google Scholar]
- Higgins DM, Lowry EG, Kanizay LB, Becraft PW, Hall DW, & Dawe RK (2018). Fitness costs and variation in transmission distortion associated with the abnormal chromosome 10 meiotic drive system in maize. Genetics, 208(1), 297–305. doi: 10.1534/genetics.117.300060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T, Schlötterer C, & Betancourt AJ (2016). Hybrid dysgenesis in Drosophila simulans associated with a rapid invasion of the P-element. PLOS Genetics, 12(3), e1005920. doi: 10.1371/journal.pgen.1005920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraizumi Y, & Thomas AM (1984). Suppressor systems of Segregation Distorter (SD) chromosomes in natural populations of Drosophila melanogaster. Genetics, 106(2), 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoermann A, Tapanelli S, Capriotti P, Masters EKG, Habtewold T, Christophides GK, & Windbichler N (2020). Converting endogenous genes of the malaria mosquito into simple non-autonomous gene drives for population replacement. bioRxiv, 2020.2005.2009.086157. doi: 10.1101/2020.05.09.086157 [DOI] [PMC free article] [PubMed]
- Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, … O’Neill SL (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature, 476(7361), 454–457. doi: 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- Holman L (2019). Evolutionary simulations of Z-linked suppression gene drives. Proceedings of the Royal Society B: Biological Sciences, 286(1912), 20191070. doi: 10.1098/rspb.2019.1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornett EA, Moran B, Reynolds LA, Charlat S, Tazzyman S, Wedell N, … Hurst GDD (2014). The evolution of sex ratio distorter suppression affects a 25 cM genomic region in the butterfly Hypolimnas bolina. PLOS Genetics, 10(12), e1004822. doi: 10.1371/journal.pgen.1004822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, & Pomiankowski A (1991). Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics, 128(4), 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J (2001). Sex chromosome meiotic drive. Annual Reviews of Ecology and Systematics, 32, 25–49. [Google Scholar]
- Johns PM, Wolfenbarger LL, & Wilkinson GS (2005). Genetic linkage between a sexually selected trait and X chromosome meiotic drive. Proceedings of the Royal Society B, 272(1576), 2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KaramiNejadRanjbar M, Eckermann KN, Ahmed HMM, Sánchez C, H. M., Dippel S, Marshall JM, & Wimmer EA (2018). Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proceedings of the National Academy of Sciences, 115(24), 6189–6194. doi: 10.1073/pnas.1713825115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keais GL, Lu S, & Perlman SJ (2020). Autosomal suppression and fitness costs of an old driving X chromosome in Drosophila testacea. Journal of Evolutionary Biology, 00, 1–10. doi: 10.1111/jeb.13596 [DOI] [PubMed] [Google Scholar]
- Kingan SB, Garrigan D, & Hartl DL (2010). Recurrent selection on the Winters sex-ratio genes in Drosophila simulans. Genetics, 184(1), 253–265. doi: 10.1534/genetics.109.109587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic M, & Schaeffer SW (2000). Molecular population genetics of X-linked genes in Drosophila pseudoobscura. Genetics, 156(1), 155–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, … Crisanti A (2018). A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nature Biotechnology, 36, 1062. doi: 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, & Wilkinson GS (1999). Models of sex-ratio meiotic drive and sexual selection in stalk-eyed flies. Genetical Research, 74(3), 245–253. doi: Doi: 10.1017/s0016672399004218 [DOI] [Google Scholar]
- Larner W, Price T, Holman L, & Wedell N (2019). An X-linked meiotic drive allele has strong, recessive fitness costs in female Drosophila pseudoobscura. Proceedings of the Royal Society B: Biological Sciences. doi: 10.1098/rspb.2019.1534 [DOI] [PMC free article] [PubMed]
- Larracuente AM, & Presgraves DC (2012). The selfish Segregation Distorter gene complex of Drosophila melanogaster. Genetics, 192(1), 33–53. doi: 10.1534/genetics.112.141390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Thézé J, Chebbi MA, Giraud I, Moumen B, Ernenwein L, … Cordaux R (2016). Birth of a W sex chromosome by horizontal transfer of Wolbachia bacterial symbiont genome. Proceedings of the National Academy of Sciences, 113(52), 15036–15041. doi: 10.1073/pnas.1608979113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenington S, & Coopersmith C (1992). Genetic basis of mating preferences in wild house mice. American Zoologist, 32, 40–47. [Google Scholar]
- Lin C-J, Hu F, Dubruille R, Vedanayagam J, Wen J, Smibert P, … Lai EC (2018). The hpRNA/RNAi pathway ss essential to resolve intragenomic conflict in the Drosophila male germline. Developmental Cell, 46(3), 316–326.e315. doi: 10.1016/j.devcel.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm A, Sutter A, Künzel S, Tautz D, & Rehrauer H (2019). Effects of a male meiotic driver on male and female transcriptomes in the house mouse. Proceedings of the Royal Society B: Biological Sciences, 286, 20191927. doi: 10.1098/rspb.2019.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm AK, Dyer KA, Firman RC, Fishman L, Forstmeier W, Holman L, … Price TAR (2016). The ecology and evolutionary dynamics of meiotic drive. Trends in Ecology & Evolution, 31(4), 315–326. doi: 10.1016/j.tree.2016.02.001 [DOI] [PubMed] [Google Scholar]
- López Hernández JF, & Zanders SE (2018). Veni, vidi, vici: the success of wtf meiotic drivers in fission yeast. Yeast, 35(7), 447–453. doi: 10.1002/yea.3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser A, König B, & Lindholm AK (2015). Female house mice avoid fertilization by t haplotype incompatible males in a mate choice experiment. Journal of Evolutionary Biology, 28(1), 54–64. doi: 10.1111/jeb.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser A, Lindholm AK, König B, & Bagheri HC (2011). Polyandry and the decrease of a selfish genetic element in a wild house mouse population. Evolution, 65(9), 2436–2447. doi: 10.1111/j.1558-5646.2011.01336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser A, Lindholm AK, & Weissing FJ (2017). The evolution of costly mate choice against segregation distorters. Evolution, 2817–2828. doi: 10.1111/evo.13376 [DOI] [PubMed]
- Marshall JM, Buchman A, Sánchez CHM, & Akbari OS (2017). Overcoming evolved resistance to population-suppressing homing-based gene drives. Scientific Reports, 7(1), 3776. doi: 10.1038/s41598-017-02744-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott S, & Noor M (2010). The role of meiotic drive in hybrid male sterility. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 365(1544), 1265–1272. doi: 10.1098/rstb.2009.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade L, Finnegan SR, Kad R, Fowler K, & Pomiankowski A (2020). Maintenance of fertility in the face of meiotic drive. The American Naturalist, 195(4), 743–751. doi: 10.1086/707372 [DOI] [PubMed] [Google Scholar]
- Min K-T, & Benzer S (1997). Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proceedings of the National Academy of Sciences, 94(20), 10792–10796. doi: 10.1073/pnas.94.20.10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montchamp-Moreau C, Ginhoux V, & Atlan A (2001). The Y chromosomes of Drosophila simulans are highly polymorphic for their ability to suppress sex-ratio drive. Evolution, 55(4), 728–737. [DOI] [PubMed] [Google Scholar]
- Moreau J, Bertin A, Caubet Y, & Rigaud T (2001). Sexual selection in an isopod with Wolbachia-induced sex reversal: males prefer real females. Journal of Evolutionary Biology, 14(3), 388–394. doi: 10.1046/j.1420-9101.2001.00292.x [DOI] [Google Scholar]
- Nash A, Urdaneta GM, Beaghton AK, Hoermann A, Papathanos PA, Christophides GK, & Windbichler N (2019). Integral gene drives for population replacement. Biology Open, 8(1), bio037762. doi: 10.1242/bio.037762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazni WA, Hoffmann AA, NoorAfizah A, Cheong YL, Mancini MV, Golding N, … Sinkins SP (2019). Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. Current Biology, 29(24), 4241–4248.e4245. doi: 10.1016/j.cub.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Nguyen HL, Nguyen TY, Vu SN, Tran ND, Le TN, … Hoffmann AA (2015). Field evaluation of the establishment potential of wmelpop Wolbachia in Australia and Vietnam for dengue control. Parasites & Vectors, 8(1), 563. doi: 10.1186/s13071-015-1174-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C, Olejarz J, Esvelt KM, Church GM, & Nowak MA (2017). Evolutionary dynamics of CRISPR gene drives. Science Advances, 3(4). doi: 10.1126/sciadv.1601964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhofer G, Ivy T, & Hay BA (2018). Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proceedings of the National Academy of Sciences of the United States of America, 115(40), E9343–E9352. doi: 10.1073/pnas.1805278115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggio AJ, Segelbacher G, Seddon PJ, Alphey L, Bennett EL, Carlson RH, … Wheeler K (2017). Is it time for synthetic biodiversity conservation? Trends in Ecology & Evolution, 32(2), 97–107. doi: 10.1016/j.tree.2016.10.016 [DOI] [PubMed] [Google Scholar]
- Pinzone CA, & Dyer KA (2013). Association of polyandry and sex-ratio drive prevalence in natural populations of Drosophila neotestacea. Proceedings of the Royal Society B: Biological Sciences, 280(1769), 20131397. doi: 10.1098/rspb.2013.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T, Verspoor R, & Wedell N (2019). Ancient gene drives: an evolutionary paradox. Proceedings of the Royal Society B: Biological Sciences. doi: 10.1098/rspb.2019.2267 [DOI] [PMC free article] [PubMed]
- Price TA, & Wedell N (2008). Selfish genetic elements and sexual selection: their impact on male fertility. Genetica, 132. doi: 10.1007/s10709-007-9173-2 [DOI] [PubMed] [Google Scholar]
- Price TAR, Bretman A, Gradilla AC, Reger J, Taylor ML, Giraldo-Perez P, … Wedell N (2014). Does polyandry control population sex ratio via regulation of a selfish gene? Proceedings of the Royal Society B: Biological Sciences, 281(1783), 20133259. doi: 10.1098/rspb.2013.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TAR, Hodgson DJ, Lewis Z, Hurst GDD, & Wedell N (2008). Selfish genetic elements promote polyandry in a fly. Science, 322(5905), 1241–1243. doi: 10.1126/science.1163766 [DOI] [PubMed] [Google Scholar]
- Prowse TAA, Adikusuma F, Cassey P, Thomas P, & Ross JV (2019). A Y-chromosome shredding gene drive for controlling pest vertebrate populations. eLife, 8, e41873. doi: 10.7554/eLife.41873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowse TAA, Cassey P, Ross JV, Pfitzner C, Wittmann TA, & Thomas P (2017). Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proceedings of the Royal Society B: Biological Sciences, 284(1860), 20170799. doi: 10.1098/rspb.2017.0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raban RR, Marshall JM, & Akbari OS (2020). Progress towards engineering gene drives for population control. The Journal of Experimental Biology, 223(Suppl 1), jeb208181. doi: 10.1242/jeb.208181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerson JP, Jiggins FM, & Hurst LD (2000). Male killing can select for male mate choice: a novel solution to the paradox of the lek. Proceedings of the Royal Society of London. Series B: Biological Sciences, 267(1446), 867–874. doi: 10.1098/rspb.2000.1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed F, A., Aquino-Michaels T, G. , Costantini MS, Láruson ÁJ, & Sutto JT (2018). RPM-Drive: A robust, safe, and reversible gene drive system that remains functional after 200+ generations. arXiv, 1806.05304.
- Reinhardt JA, Brand CL, Paczolt KA, Johns PM, Baker RH, & Wilkinson GS (2014). Meiotic drive impacts expression and evolution of X-linked genes in stalk-eyed flies. PLOS Genetics, 10(5), e1004362. doi: 10.1371/journal.pgen.1004362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold K, Engqvist L, Misof B, & Kurtz J (1999). Meiotic drive and evolution of female choice. Proceedings of the Royal Society of London. Series B: Biological Sciences, 266(1426), 1341–1345. doi: 10.1098/rspb.1999.0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PA, Ritchie SA, Axford JK, & Hoffmann AA (2019a). Loss of cytoplasmic incompatibility in Wolbachia-infected Aedes aegypti under field conditions. PLoS Neglected Tropical Diseases, 13(4), e0007357. doi: 10.1371/journal.pntd.0007357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PA, Turelli M, & Hoffmann AA (2019b). Evolutionary ecology of Wolbachia releases for disease control. Annual Review of Genetics, 53(1). doi: 10.1146/annurev-genet-112618-043609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge J-N, & Lindholm AK (2018). Carrying a selfish genetic element predicts increased migration propensity in free-living wild house mice. Proceedings of the Royal Society B: Biological Sciences, 285(1888), 20181333. doi: 10.1098/rspb.2018.1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P, Turley A, Wilson G, Ryan P, Turley A, Wilson G, … O’Neill S (2020). Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in Cairns and surrounding locations in northern Queensland, Australia [version 2; peer review: 2 approved]. Gates Open Research, 3, 1547. doi: 10.12688/gatesopenres.13061.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TL, Barton NH, Rašić G, Turley AP, Montgomery BL, Iturbe-Ormaetxe I, … Turelli M (2017). Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biology, 15(5), e2001894. doi: 10.1371/journal.pbio.2001894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano I (2017). Evolutionary ecology of multitrophic interactions between plants, insect herbivores and entomopathogens. Journal of Chemical Ecology, 43(6), 586–598. doi: 10.1007/s10886-017-0850-z [DOI] [PubMed] [Google Scholar]
- Simoni A, Hammond AM, Beaghton AK, Galizi R, Taxiarchi C, Kyrou K, … Crisanti A (2020). A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nature Biotechnology. doi: 10.1038/s41587-020-0508-1 [DOI] [PMC free article] [PubMed]
- Simoni A, Siniscalchi C, Chan Y-S, Huen DS, Russell S, Windbichler N, & Crisanti A (2014). Development of synthetic selfish elements based on modular nucleases in Drosophila melanogaster. Nucleic Acids Research, 42(11), 7461–7472. doi: 10.1093/nar/gku387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook RR, Cleland SY, Wolfner MF, & Karr TL (2000). Offsetting effects of Wolbachia infection and heat shock on sperm production in Drosophila simulans: analyses of fecundity, fertility and accessory gland proteins. Genetics, 155(1), 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YQS, Alföldi J, Pyntikova T, Brown LG, Graves T, Minx PJ, … Page DC (2014). Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell, 159(4), 800–813. doi: 10.1016/j.cell.2014.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudweeks J, Hollingsworth B, Blondel DV, Campbell KJ, Dhole S, Eisemann JD, … Lloyd AL (2019). Locally fixed alleles: A method to localize gene drive to island populations. Scientific Reports, 9(1), 15821. doi: 10.1038/s41598-019-51994-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J, & Jaenike J (2006). Male-killing Wolbachia and male mate choice: a test with Drosophila innubila. Evolutionary Ecology Research, 8, 91–102. [Google Scholar]
- Sun C, Feschotte C, Wu Z, & Mueller RL (2015). DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biology, 13(1), 38. doi: 10.1186/s12915-015-0145-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter A, & Lindholm AK (2015). Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proceedings of the Royal Society B: Biological Sciences, 282(1811), 20150974. doi: 10.1098/rspb.2015.0974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter A, & Lindholm AK (2016). No evidence for female discrimination against male house mice carrying a selfish genetic element. Current Zoology, 62(6), 675–586. doi: 10.1093/cz/zow063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svedberg J, Vogan AA, Rhoades NA, Sarmarajeewa D, Jacobson DJ, Lascoux M, … Johannesson H (2020). An introgressed gene causes meiotic drive in Neurospora sitophila. bioRxiv, 2020.2001.2029.923946. doi: 10.1101/2020.01.29.923946 [DOI] [PMC free article] [PubMed]
- Tao Y, Araripe L, Kingan SB, Ke Y, Xiao H, & Hartl DL (2007a). A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biology, 5(11), e293. doi: 10.1371/journal.pbio.0050293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Masly JP, Araripe L, Ke Y, & Hartl DL (2007b). A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biology, 5(11), e292. doi: 10.1371/journal.pbio.0050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzet P, & Budar F (2004). Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends in Plant Science, 9(12), 568–570. doi: 10.1016/j.tplants.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Unckless RL, Clark AG, & Messer PW (2017). Evolution of resistance against CRISPR/Cas9 gene drive. Genetics, 205(2), 827–841. doi: 10.1534/genetics.116.197285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspoor RL, Cuss M, & Price TAR (2015). Age-based mate choice in the monandrous fruit fly Drosophila subobscura. Animal Behaviour, 102, 199–207. doi: 10.1016/j.anbehav.2015.01.024 [DOI] [Google Scholar]
- Verspoor RL, Smith JML, Mannion NLM, Hurst GDD, & Price TAR (2018). Strong hybrid male incompatibilities impede the spread of a selfish chromosome between populations of a fly. Evolution Letters, 2(3), 169–179. doi: 10.1002/evl3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, … Hoffmann AA (2011). The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. [10.1038/nature10355]. Nature, 476(7361), 450–453. doi: 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
- Wilkinson GS, Presgraves DC, & Crymes L (1998). Male eye span in stalk-eyed flies indicates genetic quality by meiotic drive suppression. Nature, 391(6664), 276–279. doi: 10.1038/34640 [DOI] [Google Scholar]
- Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, … Crisanti A (2011). A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature, 473. doi: 10.1038/nature09937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RJ, & Newton ME (1991). Sex-ratio distortion caused by meiotic drive in mosquitoes. The American Naturalist, 137(3), 379–391. [Google Scholar]
- Wu C-I, Lyttle TW, Wu M-L, & Lin G-F (1988). Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster. Cell, 54(2), 179–189. [DOI] [PubMed] [Google Scholar]
- Zanders SE, & Unckless RL (2019). Fertility costs of meiotic drivers. Current Biology, 29(11), R512–R520. doi: 10.1016/j.cub.2019.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]


