Abstract
Biomimetic bone regeneration methods which demonstrate both clinical and manufacturing feasibility, as alternatives to autogenic or allogenic bone grafting, remain a challenge to the field of tissue engineering. Here, we report the pro-osteogenic capacity of exosomes derived from human dental pulp stem cells (hDPSCs) to facilitate bone marrow stromal cell (BMSC) differentiation and mineralization. To support their delivery, we engineered a biodegradable polymer delivery platform to improve the encapsulation and the controlled release of exosomes on a tunable time scale from poly(lactic-co-glycolic acid) (PLGA) and poly(ethylene glycol) (PEG) triblock copolymer microspheres. Our delivery platform integrates within three-dimensional tissue engineering scaffolds to enable a straightforward surgical insertion into a mouse calvarial defect. We demonstrate the osteogenic potential of these functional constructs in vitro and in vivo. Controlled release of osteogenic hDPSC-derived exosomes facilitates osteogenic differentiation of BMSCs, leading to mineralization to a degree which is comparable to exogenous administration of the same exosomes in human and mouse BMSCs. By recruiting endogenous cells to the defects and facilitating their differentiation, the controlled release of osteogenic exosomes from a tissue engineering scaffold demonstrates accelerated bone healing in vivo at 8 weeks. Exosomes recapitulate the advantageous properties of mesenchymal stem/progenitor cells, without manufacturing or immunogenic concerns associated with transplantation of exogenous cells. This biomaterial platform enables exosome-mediated bone regeneration in an efficacious and clinically relevant way.
Keywords: tissue engineering, exosome, controlled release, drug delivery, extracellular vesicle
Graphical Abstract
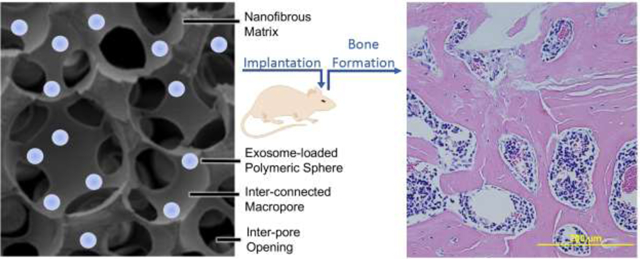
1. Introduction
Annually, over $21 billion is spent on the treatment of bone fractures in the United States, impacting about one million patients annually, where over half experience delayed healing or nonunion [1]. Physical impairments resulting from bone injury can lead to significant morbidity and socio-economic cost. Critical sized bone defects which do not heal spontaneously require surgical intervention to prevent potentially debilitating implications. In these critical-sized defects endogenous cells require exogenous guidance to bridge a defect and replace substantial bone loss, in the forms of biologic cues and an artificial matrix [2]. The goal of bone tissue engineering is to design a trajectory by which these large defects heal to restore the tissue’s function [3, 4]. Bone grafting is considered the current clinical standard for treating large defects, with limitations [5]. Autografts struggle to provide adequate amounts of bone which can be harvested and lead to donor site discomfort and morbidity. Allografts have pathogenic concerns, reduced structural integrity from commercial processing, and ethical concerns with cadaver bone use [6]. Significant progress in the field of bone tissue engineering has been made in recent years to design bone-grafting alternatives. Transplantation of mesenchymal stem/progenitor cells has been demonstrated to enhance bone formation [7]. However, the rate of clinical translation of these therapies is low due to manufacturing and regulatory concerns, scalability, immunogenicity of grafts and constructs, efficacy, safety, and cost [8]. We aim to design a three-dimensional tissue engineering construct which recapitulates the advantageous properties of transplanted stem/progenitor cells while circumventing translational hurdles associated with cell-based therapies, through the local, controlled release of stem/progenitor cell-derived exosomes (EXOs) and extracellular vesicles (EVs) to guide bone regeneration.
Cell-derived EXO/EVs, as well as synthetic liposomes, have received considerable consideration for therapeutic applications across a number of diseases and clinical indications. Exosomes are lipid bilayer-bound vesicles with diameters in the range of 50–150 nm and expressing a set of specific surface markers [9–11], which play important roles in autocrine and paracrine signaling [12, 13]. Since the goal of this work was to develop delivery systems for EXO/EVs instead of studying properties specific to pure exosomes, for description simplicity we use the term “exosomes (EXOs)” loosely to include EXOs and similar sized EVs throughout this manuscript. EXOs carry proteins and RNA, including microRNAs (miRNAs), which are characteristic of their donor cell identity and culture microenvironment [14, 15]. Similar to stem cells, EXOs have shown important therapeutic potentials in cardiac disease, neurogenesis, osteochondral defect and tooth tissue repair [11, 16–18]. Advantageously, EXOs have a better safety profile than stem cells in terms of tumorigenicity and immunogenicity [17, 19]. Clinically, exosome administration is limited to intravenous infusion or direct injection. Exosomes circulate the body rapidly and accumulate in the liver and spleen [20], requiring a high dose to reach therapeutic efficacy. Additionally, there is risk of off-target effects and short duration of any benefits. In order to realize the advantageous properties of stem cell-derived exosomes, a delivery platform is required for their efficient therapeutic delivery in a clinically and biologically relevant manner
Drug delivery systems allow for controlled release kinetics, single administration instead of routine dosing, reduced circulating drug concentration, and improved patient compliance in the clinical setting [21]. Poly(lactic-co-glycolic acid) (PLGA), a synthetic biodegradable copolymer, is widely used in controlled release systems because of its tunable degradation rate and safe degradation biproducts [22]. The double emulsion technique is the most well-known method to encapsulate proteins and hydrophilic small molecules in polymeric microspheres [23]. An important consideration in developing controlled release platforms for complex biologics is maintaining their integrity and bioactivity throughout encapsulation and release. Recent progress in the field of drug delivery has led to the development of controlled release systems for small molecules, proteins, and plasmids [24–26]. To fill space in a large bony defect, a tissue engineering scaffold serves as an artificial matrix to retain and organize cells. Previously our lab has developed nanofibrous, microporous tissue engineering scaffolds from poly(l-lactic acid) (PLLA) [27, 28]. Nanofibers which mimic the native extracellular matrix are fabricated by a thermally induced phase separation process; interconnected macropores are made using a sugar sphere porogen. The high surface area and porosity of these three-dimensional constructs are ideal for functionalizing their surface with a delivery modality [29], yielding a three-dimensional implantable construct.
The ideal exosome-delivery solution which demonstrates a high administration efficacy would allow for local and controlled release of exosomes, protecting their bioactivity. We hypothesize that a scaffold with an integrated delivery system for osteogenesis-inducing exosomes is sufficient to attract resident cells and cause their differentiation towards endogenous bone repair, without the need for transplanted stem cells. In this study, a biodegradable platform technology for the controlled release of stem cell-derived exosomes is developed and validated. Stem/progenitor cells from dental and craniofacial tissue are conveniently obtainable and have been shown to regenerate mineralized dental tissues [30–33]. We hypothesize that exosomes derived from dental pulp stem cells are also osteogenic and can be utilized to facilitate bone regeneration. Human dental pulp stem cell-derived exosomes are delivered using triblock copolymer microspheres immobilized on a nanofibrous PLLA scaffold to a critical size mouse calvarial bone defect in vivo and result in bone regeneration, without the need for transplantation of exogenous stem cells.
2. Materials and Methods
2.1. Materials
Polyethylene glycol (Mw = 1,000 g/mol), methyl-polyethylene glycol (MW = 1,000 g/mol), α-bromoisobutyryl bromide (BIBB), polyvinyl alcohol (PVA) (Aldrich); L-lactide (Altasorb); glycolide, stannous octanoate (Sn(Oct)2), 2-hydroxyethyl methacrylate (HEMA), azobisisobutyronitrile (AIBN) triethylamine (TEA), copper (I) bromide (CuBr), N,N,N′,N″,N″-pentamethyldiethylenetriamine (PMDETA), Span80, ethyl acetate, alkaline phosphatase staining kit (Sigma); nile blue acrylamide (Polysciences); tetrahydrofuran (THF), diethyl ether, 9-anthracecenylmethyl methacrylate, hexane, methanol, methylene chloride, isopropanol, Dow Sylgard 184 polydimethylsiloxane, TRIzol, micro BCA protein assay kit, Alexa-Fluor 555 phalloidin, ProLong Gold Anti-Fade, Alizarin Red S staining solution, conjugated secondary antibody (Fisher Scientific); D-Trehalose, D-fructose (Oakwood Chemical); mineral oil (Alfa Aesar); deuterated NMR solvents (CDCl3, D2O) (Acros Organics); chloroform (VWR Chemical); ethanol (Decon Labs); dialysis tubing (3,500 MW cutoff) (Spectrum Labs); AZ1518 photoresist, SU-8 photoresist (MicroChem), 3,3,3-trifluoropropyltrichlorosilane (Sigma Aldrich); Electron-microscopy grade paraformaldehyde, electron-microscopy grade glutaraldehyde (EMS Diasum); Methyl cellulose (TCI America); Dulbecco’s Modified Eagle Medium, fetal bovine serum (FBS), exosome-depleted FBS, penicillin, streptomycin, phosphate buffered saline (PBS), β-glycerophosphate, ascorbic acid, dexamethasone (Gibco); All-In-One RT MasterMix (Applied Biological Materials Inc.); 7500 Real Time PCR System, PowerUp Sybr Green Master Mix (Applied Biosystems); EpiQuik Whole Cell Extraction Kit (Epigentek); 4–12% Bis-Tris gels (NuPAGE); PVDF membrane (BioRad); BrightStar ECL detection kit (Alkali Scientific); Formvar-carbon coated electron microscopy grid (Ted Pella); miRNeasy Mini Kit (Qiagen).
2.2. Poly(ethylene glycol) and poly(l-lactic-co-glycolic acid) Block Copolymer Synthesis
Triblock PLGA-PEG-PLGA and diblock PEG-PLGA were synthesized from HO-PEG-OH and H3C-PEG-OH initiators, respectively. Stoichiometric amounts of the monomers: l-lactide and glycolide were added to a round-bottom flask with commercial PEG (Mw=1,000 g/mol), and Sn(Oct)2 (monomer/SnOct2 = 100). The system was purged with positive nitrogen pressure for 30 minutes, then heated to 120°C, where the reaction proceeded by stannous-catalyzed ring opening polymerization (ROP) for two hours [34].
Polymers are abbreviated as PLXGYA-PEG-PLXGYA where X and Y represent percentage of lactide and glycolide, respectively. In the case of PL85G15-PEG-PL85G15A: 1.25 g HO-PEG-OH, 4.25 g L-lactide, 0.75 g glycolide and 112 μL Sn(Oct)2 were added to a round bottom flask with magnetic stirring and positive nitrogen inflow, then heated to 120°C to melt. The reaction proceeded for two hours then was quenched by opening to air and cooling. The crude product was dissolved in a minimum amount of chloroform, precipitated into 5-times excess volume of cold methanol, and collected by suction filtration, twice. 1H NMR (400 MHz, CDCl3): δ 1.5 ppm, s, sp3 CH3; 3.6 ppm, m, sp3 CH2; 4.8 ppm, m, glycolide sp3 CH2; 5.2 ppm, m, lactide sp3 CH.
2.3. Nile Blue-PLLA Synthesis
Acrylic-end functionalized PLLA was synthesized from hydroxyethyl methacrylate (HEMA) initiator (0.8 mmol, 86 μL) and L-lactide (40 mmol, 5.760 g), with Sn(Oct)2 (112 μL) in a ring opening polymerization reaction (ROP) at 120°C, in an inert nitrogen environment [35]. After two hours, the resulting HEMA-PLLA polymer was exposed to air, cooled, then dissolved in 20 mL chloroform and precipitated in 100 mL cold methanol and the product was collected, repeated twice. 1H NMR (400 MHz, CDCl3): δ 1.6 ppm, s, sp3 CH3; 5.1 ppm, q, sp3 CH; 5.8 ppm, dt, sp2 CH; 6.1 ppm, dd, sp2 CH; 6.4 ppm, dd, sp2 CH.
HEMA-PLLA (1.40 g), nile blue acrylamide (0.012 mmol, 0.005 g), and freshly recrystallized AIBN (0.06 mmol, 0.0098 g, recrystallized from methanol) were dissolved in 10 mL dioxane at 70°C and allowed to react overnight. Solvent was removed by rotary evaporation, the resulting residue was dissolved in a minimum amount of chloroform and precipitated into cold methanol, then collected by suction filtration. This purification step was repeated until the methanol is no longer visibly colored by unreacted dye (approximately four times). 1H NMR (400 MHz, CDCl3): δ 1.5 ppm, s, sp3 CH3; 5.1 ppm, q, sp3 CH; 7.5–8.5 ppm, m, sp2 CH.
2.4. 9-Anthracenylmethyl methacrylate-PEG-PLLA Synthesis
Br-PEG-OH was synthesized from HO-PEG-OH (10 g Mw = 1,000 g/mol) dissolved with TEA (2.2 mL) in anhydrous THF (50 mL), purged under nitrogen for 30 minutes to remove residual oxygen. The reaction was submerged in an ice bath and α-bromoisobutyryl bromide (1.20 mL) was slowly added dropwise with a syringe. The solution was stirred for 24 hours and warmed to room temperature gradually as the ice melted. After 24 hours, solvent was removed by rotary evaporation. The resulting residue was dissolved in a minimum amount of dichloromethane (DCM), precipitated into cold diethyl ether (5x excess volume), and collected by suction filtration. The resulting Br-PEG-OH was lyophilized to a white powder. 1H NMR (400 MHz, D2O): δ 3.07 ppm, q, -CH2-Br; 3.5–3.6 ppm, m, -CH2CH2O-.
To make Br-PEG-PLLA, Br-PEG-OH (1.24 g) was combined with L-lactide monomer (5 g) and Sn(Oct)2 (112 μL) in a round bottom flask, purged with nitrogen for 30 minutes, with stirring. After 30 minutes, the mixture was heated to 120°C for two hours, to carry out ring opening polymerization of PLLA from the terminal hydroxyl group of the PEG macroinitiator. The solid residue was dissolved in a minimum amount of chloroform, precipitated into cold methanol (5x volume excess), and collected by suction filtration to be dried in a vacuum chamber. 1H NMR (400 MHz, CDCl3): δ 1.56 ppm, sp3 CH3; 3.1 ppm, q, -CH2-Br; 3.6–3.65 ppm, m, -CH2CH2O-; 4.7–4.8 ppm, m, glycolide sp3 CH2; 5.1–5.2 ppm, m, lactide, sp3 CH.
9-anthracecenylmethyl methacrylate was polymerized from the terminal bromide via atom transfer radical polymerization (ATRP). Br-PEG-PLLA (2.0 g), 9-anthracenylmethyl methacrylate (0.1 g), and CuBr (0.04 g) was added to a round bottom flak and purged with nitrogen for one hour. Separately N,N,N′,N′′,N′′-pentamethyldiethylenetriamine (PMDETA) initiator (200 μL) was dissolved in deionized water (40 mL) and purged with nitrogen to remove residual oxygen. The PMDETA-water solution was added via a syringe, through a rubber septum, to the round bottom flask containing the polymer and stirred for 24 hours at room temperature. The resulting solution was dialyzed against deionized water using 3,500 Mw cutoff dialysis tubing, changing the water daily for three days, then the solution was frozen to −80°C and lyophilized. 1H NMR (400 MHz, CDCl3): δ 1.56 ppm, sp3 CH3; 3.6–3.65 ppm, m, -CH2CH2O-; 4.7–4.8 ppm, m, glycolide sp3 CH2; 5.1–5.2 ppm, m, lactide, sp3 CH, 5.6 ppm, d, sp3 CH2 ; 6.1–6.2 ppm, m, sp2 CH; 6.5, d, sp2 CH2; 7.5–7.7 ppm, dq, sp2 CH; 8.2 ppm, d, sp2 CH.
2.5. Nuclear Magnetic Spectroscopy:
1H spectra were recorded with an Inova 400 (Varian), operating at 400 MHz at room temperature, using CDCl3 or D2O as solvent.
2.6. Gel Permeation Chromatography:
Polymer samples were dissolved in tetrahydrofuran (THF, 5 mg/mL) and filtered using a 200-um syringe filter. GPC analysis was performed on a Shimadzu GPC system containing three columns in series with refractive index and diode array detectors. The GPC was calibrated with narrow polydispersity polystyrene standards and the molecular weights are reported as polystyrene equivalents.
2.7. Polydimethylsiloxane Microfluidic Device Design and Fabrication:
The microfluidic device was drawn to scale using computer aided design software (AutoCAD 2017, Autodesk). These geometries consist of two inline flow focusing junctions fed by inlets for one oil and two aqueous phases. Following each junction is a straight channel to allow for droplet stabilization. Droplets were collected through a single outlet into an external reservoir.
Designs were converted to single polygons using K-Layout (0.25.3). Masks were fabricated on a soda lime glass (90 mm thick) with low reflective chrome, using AZ1518 photoresist on a Heidelberg uPG 501 Mask Maker by direct writing.
Master molds were fabricated by traditional photolithography. SU-8 2050 photoresist was spin-coated onto a 4” silicon wafer on an interlocked spinner (Solitec) for 60 seconds, to a thickness of 26 μm. Following a soft bake, the photoresist was exposed through the mask described above, using a MJB45 aligner. The SU-8 was baked post-exposure, then developed using SU-8 developer, and finally rinsed with isopropyl alcohol and dried with N2. The height and profile of the resulting photoresist was validated using a profilometer (Dektak) and differential interference contrast microscope (Olympus BX51). Finally, the SU-8 was hard-baked for 15 minutes at 150°C to further crosslink the polymer.
The surface of the master mold was coated with a releasing agent (0.5 mL, 3,3,3-trifluoropropyltrichlorosilane), then held under vacuum. PDMS (Sylgard 184 kit) was prepared at a crosslinking density of 17.5% (40.0 g elastomer, 7.0 g curing agent) and mixed well. Prior to casting, the PDMS was degassed in a benchtop desiccator for 15–30 minutes, until bubbles were gone. The PDMS mixture was poured onto the coated master mold to desired thickness and degassed again after casting. PDMS was cured overnight on a hot plate at 100°C.
The cured PDMS was carefully removed from the master mold and cut to individual microfluidic chips using a razor blade. 1/32” inlet and outlet holes were made using a biopsy punch. PDMS devices were covalently bound to glass microscope slides by plasma bonding. The PDMS surface was cleaned with Scotch tape and isopropyl alcohol; glass slides were rinsed with isopropyl alcohol and dried with N2. A Glen Plasma Cleaner was used to activate PDMS and glass surfaces using a prescribed recipe (Glen recipe 20). After the process was completed, PDMS devices were immediately aligned and pressed into the glass microscope slides. Completed devices were cured at 50°C on a hotplate overnight before use.
2.8. Computational Modeling of Microfluidic Device
Simulations of fluid flow in our microfluidic devices were carried out with COMSOL Multiphysics (5.3a, University of Michigan CAEN license) using a Two-Phase Flow Phase Field Model. AutoCAD designs of microfluidic devices were imported as dxf files. The single flow functioning junction model contains three inlets for two solvent phases (discreet aqueous and oil) and one outlet; the dual flow focusing model of the complete device contained five inlets for three solvent inlets (discrete water, oil, continuous aqueous). Immiscible fluids were injected into the channels through their respective inlets, and their flows at the inlet were assumed as fully developed laminar flow. Solvent parameters were defined based on PubChem physical characteristics: viscosity, density, and surface tension; liquid-liquid surface tension coefficients were from experimental evidence in the literature [36]. Droplet formation was a function of the interface surface tension coefficient and flow rate ratio.
2.9. Primary Cells and Culture Conditions
Primary human dental pulp stem cells (hDPSCs) were isolated from healthy human third molars, extracted from patients 16–20 years of age at the University of Michigan School of Dentistry following literature and our own prior publications [30–33, 37, 38]. The protocol for isolating hDPSCs was performed according to a protocol approved by the Institutional Review Board at the University of Michigan, and protocol reported previously[30, 37]. After the first passage, cells were cultured in maintenance media (Dulbecco’s Modified Eagle Medium (α-MEM) containing 10% fetal bovine serum (FBS) and antibiotics (100U/mL penicillin G; 0.1mg/mL streptomycin, GIBCO)). Mouse bone marrow stromal cells (mBMSCs) were obtained from wild-type male C57BL/6 mice, 6–8 weeks old, by aspiration of the femoral and tibia bone marrow using a 26-gauge syringe needle [39]. mBMSCs were cultured with maintenance media (α-MEM containing 20% FBS and antibiotics (100U/ml penicillin G; 0.1mg/ml streptomycin)). All cells were cultured at 37° under 5% CO2 in a humidified environment. Media was changed every two days and cells were used at passages 2–5. For three-dimensional cell culture experiments, the poly(l-lactic acid) nanofibrous scaffolds were sterilized and wet by 70% ethanol for 10 minutes under weak vacuum, and subsequently for 30 mins under ambient pressure. Then the scaffolds were washed with phosphate buffered saline (PBS) three times and twice with maintenance media.
2.10. Osteoinductive Culture and Exosome Isolation by Ultracentrifugation
Human DPSCs were cultured in osteoinductive media (OM) for 14 days. OM consists of α-MEM media with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and osteoinductive supplements (100 nM dexamethasone, 10 mM β-glycerophosphate, and 50 μg/mL ascorbic acid). Their mineralization status was confirmed by Alizarin Red staining. One million DPSCs were initially seeded in a 100mm Petri dish with 10 ml media and grown to about 3–4 million cells when reaching confluency. The culture media was changed every other day. More exosomes were produced at the mineralizing stage. From the 15th day, the media was changed to exosome-free osteogenic media (EXO-free OM, OM with exosome-depleted FBS (Gibco)). The conditioned media (CM) was collected every two days until the 28th day.
hDPSC-derived osteoinductive exosomes (OS-EXOs) were isolated from CM by differential centrifugation as reported in the literature [40]. Briefly, the CM was centrifuged at 300 g for 10 minutes, 2,000 g for 10 minutes, then 10,000 g for 30 minutes to remove cells and large debris. Then the CM was filtered through a 0.22-μm filter (GVS North America) to remove smaller residual debris. The supernatant was concentrated at 4,000 g in a 15 mL Amicon Ultra-15 Centrifugal Filter Unit with a molecular weight cut off (MWCO) of 100kD (Millipore Sigma) for 10 minutes. Subsequently the CM was ultra-centrifuged at 100,000 g for 70 minutes. Finally, the pellet was resuspended and centrifuged at 100,000 g for another 70 minutes. The pellet was resuspended with 200 μL PBS and the concentration of exosomes was determined by microBCA protein assay kit (Thermo Fisher Scientific) before storage at −80°C until use. About 300 μg exosomes were obtained at the end from the initial one million hDPSCs, where the osteoinductive supplements were depleted during the repeated centrifuging and washing with PBS.
2.11. Exosome Physical Characterization by Nanoparticle Tracking Analysis
Average hydrodynamic diameter and concentration of exosomes in solution was characterized by nanoparticle tracking analysis (NTA) using a NanoSight NS3000 (Malvern). Samples were agitated briefly in a gentle sonic bath to break up aggregates, were diluted in particle-free PBS and immediately analyzed. NTA was carried out using a 488 nm laser in scatter mode, using a syringe pump (Harvard Apparatus) to control fluid flow through the manufacturer’s flow plate. Five, 60-second videos were recorded at a concentration sufficient to obtain a minimum of 200 tracks per video and averaged for each sample analyzed. Analysis was carried out using Malvern NanoSight NTA software v3.2. Four independent samples from each time point were measured.
2.12. Fluorescent Labelling and Visualization of Cellular Uptake of Exosomes
Exosomes were labelled with DiO Green (Thermo Fisher Scientific) using a modified membrane-labelling method reported in the literature [41]. Briefly, 10 μg/mL OS-EXOs were incubated with DiO reagent for 20 minutes, condensed using Amicon 100kD filter, washed with PBS and ultracentrifuged three times. 50,000 mBMSCs were seeded on a glass cover slide and cultured in a 24-well-plate. An aliquot of 8 μg labeled OS-EXOs was added to 800 μl culture media and incubated for 30 minutes. After washing with PBS, Alexa-Fluor 555 phalloidin was used to stain the cytoskeleton (F-actin, 1:35; 1% BSA), per the manufacturer’s guidelines. Mounting media containing DAPI was used to stain nuclei (ProLong Gold Anti-Fade). Constructs were observed using confocal microscopy (Nikon Eclipse C1).
2.13. Alkaline Phosphatase Assay
The alkaline phosphatase assay was conducted with a staining kit (Sigma-Aldrich) according to manufacturer’s instructions. Briefly, the cells were washed with PBS three times before staining. Cells were fixed by alkaline phosphatase fixative (66% acetone, 25.6% citrate buffer, 8.4% formaldehyde) for 30 seconds, and the fixative was decanted. Cells were then washed twice by deionized water and incubated with staining solution for 15 minutes in the dark. Finally, cells were washed by deionized water twice and imaged using an Olympus light microscope with digital camera.
2.14. Alizarin Red Assay
The cells were washed by PBS 3 times before staining, fixed with 10% formalin for ten minutes, and washed by deionized water three times. Cells were stained with Alizarin Red S staining solution (20 g/L, pH 4.2) for 15 minutes. After staining the cells were washed by deionized water three times and imaged using an Olympus light microscope with digital camera.
2.15. Gene Expression Analysis by Polymerase Chain Reaction
Total RNA was isolated using Trizol Reagent (Thermo Fisher Scientific), following a standard protocol [42]. The RNA was reverse transcribed using 5X All-In-One RT MasterMix (Applied Biological Materials Inc.) and subsequently amplified using 7500 Real Time PCR System (Applied Biosystems) with PowerUp Sybr Green Master Mix (Applied Biosystems), and primers as shown in Table 1. Relative mRNA expression levels were normalized with respect to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, and expression is calculated according to ΔΔCt method [43].
2.16. Western Blot Analysis
Western blot analysis was performed according to standard protocol [44]. Briefly, exosomes and cells were lysed, and proteins were extracted by EpiQuik Whole Cell Extraction Kit (Epigentek) following manufacturer’s instruction, and the protein concentration was determined by micro BCA protein assay kit (Thermo Fisher Scientific). 30 μg total protein was denatured and then separated on 4–12% Bis-Tris gels (NuPAGE). Then the proteins were transferred from gel to a polyvinylidene difluoride (PVDF) membrane and blocked by 5% BSA solution. After that membrane was incubated with primary antibody in 5% BSA solution at 4°C overnight and washed with Tris-buffered saline with 0.1% Tween 20 (TBST) three times, 5 minutes each. Then the membrane was incubated with a conjugated secondary antibody in 5% BSA solution at room temperature for one hour, and washed by TBST for three times, five minutes per wash. The signal development was done with BrightStar ECL detection kit (Alkali Scientific) and images were acquired after developing in a darkroom.
2.17. EXO-MS Fabrication by a Microfluidic Device
Fluids were injected from reservoirs using three Fluigent Flow EZ pressure-driven flow controllers (2,000 mbar). Reservoirs were connected to the microfluidic chip using 1/32” FEP (fluorinated ethylene-propylene copolymer) solvent-resistant tubing (Cole Parmer). The PDMS device was mounted on a binocular microscope (AmScope) to visualize droplet formation. Videos and photos were recorded using a 5-megapixel USB camera (AmScope).
Exosomes isolated from DPSC conditioned media were isolated as described and concentrated to at least 2,000 μg/mL in PBS. The amount of OS-EXO solution was calculated assuming 85% encapsulation efficiency based on experience, and dose to match 1 μg/mL treatment regimen when matched for cell number. D-Trehalose, a hydrophilic sugar which acts as a stabilizer, was added to make a 2 mM solution and was the discrete aqueous phase. PLGA-b-PEG-b-PLGA copolymer was dissolved in ethyl acetate (2% wt/v) as the discrete oil phase. 0.01% (wt/wt) polyvinyl alcohol (PVA) in MilliQ water was the continuous aqueous phase. Solutions were flowed into the PDMS microfluidic device as described above, with precise control of their pressures (Fluigent FlowEZ). FEP tubing from the outlet flew into a 25 mL round bottom flask which contains 0.01% (w/w) PVA; resulting droplets were collected and stirred with a magnetic stir bar. After 10 mg particles were collected (determined by amount of discrete polymer phase consumed), particles were transferred to a beaker in a fume hood, with stirring, where organic solvent was allowed to slowly evaporate. Particles were collected by centrifugation (15,000 rpm, 12 minutes) and washed with MilliQ water five times before being frozen at −80°C and lyophilized.
2.18. Scanning Electron Microscopy Characterization
Particles and scaffolds were fixed to specimen mounts by carbon tape. In preparation for observation by scanning electron microscopy (SEM), samples were sputter-coated with gold (120s, DeskII, Denton Vacuum) and observed at 5 kV and a working distance of 10 mm.
2.19. Polymer Sphere Size Characterization
EXO-MS particles fabricated via our microfluidic device were measured by laser diffraction using a particle analyzer (Malvern Mastersizer 2000 with Hydro 2000S wet dispersion unit). Particles were suspended in MilliQ water (2–5 mg/mL) and added to the sample reservoir, with stirring and sonication. Standard refractive indices of 1.590 and 1.330 were used for PLGA (particles) and water, respectively. Samples were measured in triplicate.
2.20. Fabrication of Nanofibrous Tissue Engineering Scaffold
Nanofibrous, microporous three-dimensional tissue engineering scaffolds made of poly(L-lactic acid) (PLLA) were fabricated as previously described [27]. Briefly, a sugar sphere template was prepared by emulsifying molten D-fructose in hot mineral oil, with Span80 surfactant. After emulsifying, the mixture was quenched and mineral oil was exchanged for hexane, sugar spheres were collected. Spheres were separated by size using sieves, packed into Teflon molds and heat treated at 37°C for 10 minutes to anneal the spheres. Hexane was removed under vacuum, and polymer solution (PLLA, 10% wt/v in THF, 60°C) was cast into the mold. The cast polymer scaffolds were immediately frozen at −80°C for a minimum of 48 hours to induce phase separation, responsible for nanofiber formation. After 48 hours, THF was exchanged for hexane at room temperature overnight, then scaffolds were removed from molds and sugar was leached in deionized water. Scaffolds were cut to size (8 mm diameter × 1.5 mm height for subcutaneous implantation, 2.7 mm diameter × 1 mm height for calvarial defect), lyophilized, and sterilized with ethylene oxide gas prior to in vitro and in vivo experiments.
2.21. Solvent-wetting Method of Attaching EXO-MS to Nanofibrous Scaffolds
Exosome-releasing spheres (EXO-MS) were physically attached to the surface of nanofibrous, microporous tissue engineering scaffolds by a post-seeding method [45]. Scaffolds and particles were fabricated separately by the protocols described above. 1.2 mg particles were suspended in 500 μL hexane with 0.1% Span80, kept in suspension on a rocker for 30 minutes, then dried in a vacuum chamber. Particles were resuspended in 150 μL a 97:3 hexane:THF mixture which facilitates physical attachment between the PLGA particles and PLLA scaffold, both of which are soluble in THF, but not hexane. A low amount of THF was used to prevent loss of the nanofibrous surface structure of the scaffold or disrupt the particle morphology. 15 μL of suspended particles were seeded to the scaffold, rocked for 30 minutes at a low speed, at room temperature. This seeding procedure was repeated until a total of 1.0 mg particles were suspended evenly on both sides of the scaffold. Scaffolds were kept in a vacuum chamber to dry until a consistent mass was achieved.
2.22. Sterilization of Biomaterial Constructs
Prior to further in vitro or in vivo testing, all biomaterial constructs were sterilized using ethylene oxide (Andersen).
2.23. Evaluation of Controlled Release of Exosomes
The controlled release of exosomes from biomaterial constructs was evaluated in phosphate buffered saline (PBS, 0.1 M, pH 7.4). 1.0 mg of EXO-MS, or a scaffold seeded with 1.0 mg of EXO-MS, were incubated in 1.0 mL PBS and shaken at 10 rpm in a 37°C incubator. At designated time points (2 h, 4 h, 8 h, 12 h, 24 h, 48 h, 72 h, 96 h, 7 d, 10 d, 14 d, 3 w, 4 w, 5 w, 6 w, 7 w, 8 w, 9 w, 10 w, 11 w, 12 w) the PBS was collected, frozen at −80°C until subsequent analysis by NTA, and replaced with fresh PBS. EXO release was plotted as cumulative release as a function of time (n=4 per time point).
2.24. Transmission Electron Microscopy
Whole-mount exosome morphology and membrane integrity was evaluated by transmission electron microscopy. Exosome samples were prepared according to a published protocol [46, 47]. Briefly, EXO-MS eluent containing OS-EXOs were mixed with an equal volume of 4% electron microscopy-grade paraformaldehyde and 8 μL was incubated on a Formvar-carbon coated grid for 20 minutes. Grids were washed with PBS and transferred to a clean surface, then fixed with 50 μL of 1% EM-grade glutaraldehyde. Grids were washed eight times in fresh deionized water and stained with 1% uranyl acetate solution (pH 3.5) for 3 minutes and stained with 2% wt/v methyl cellulose for 5 minutes. Excess solution was removed with a piece of filter paper. Grids were imaged within 24 hours of staining. Samples were observed on a JEOL JEM 1400 TEM at 80.0 kV and 20,000x magnification.
2.25. RNA Isolation from Released Exosomes
EXO-MS eluent from release profile experiments was concentrated at 12,000 g for 90 minutes and 4°C. RNA was extracted from released exosomes using a phenol-based method [48]. TRIzol was used to extract total RNA, in accordance with the manufacturers protocol. RNA concentration was measured by absorbance at 260 nm using a Beckman DU640 spectrophotometer.
2.26. Infrared Spectroscopy of 3D Constructs
Three dimensional nanofibrous PLLA scaffolds were functionalized with EXO-MS, cultured with mBMSCs for up to three weeks, and fixed with 4% PFA. Fourier-transformed mid-infrared (FTIR) spectra were acquired using a Perkin-Elmer Spectrum GX, with microscope accessory, using a KBr beam-splitter and liquid nitrogen-cooled MCT-B detector, which covers the range from 4,000 cm−1 to 700 cm−1. The following parameters were used for each acquisition--resolution: 4 cm−1, J-step: 4 cm−1, interval: 1.00 cm−1, 50 scans per site. Spectra were collected from ten representative 625 μm2 areas of the construct surface and averaged for each sample.
2.27. Colorimetric Mineralization Assay
The colorimetric mineralization assay was conducted by the modified o-cresolphthalein complexone method. Briefly, the cell/scaffold constructs were washed by PBS three times for 5 minutes each, then homogenized in 250 μL 0.5 N Hydrochloric acid. The calcium extraction was performed by shaking overnight at 4 °C. The total calcium content was determined by calcium assay kit (Pointe Scientific Inc) following the manufactures’ instructions.
2.28. Energy-dispersive X-ray Spectroscopy (EDX) of 3D Constructs
Three dimensional constructs cultured for up to three weeks were fixed with 4% PFA and sputter-coated with gold (120s, DeskII, Denton Vacuum). Samples were observed using a JEOL JSM-7800FLV scanning electron microscope equipped with an Oxford XMaxN 80 mm2 silicon-drift energy-dispersive X-ray spectrometer. Ten spectra were acquired from representative locations in each sample using Oxford Aztec v3.3 EDS acquisition and processing software and averaged. The same samples were used for FTIR and EDX analysis.
2.29. Subcutaneous Implantation in Mice
200,000 primary mBMSCs were seeded on nanofibrous scaffolds functionalized with OS-EXO loaded EXO-MS, blank EXO-MS, or left virgin. Constructs were cultured in maintenance media for 24 hours before subcutaneous implantation in 8- to 10-week-old C57BL/6 mice (Charles River) following a protocol approved by the University of Michigan Institutional Animal Care and Use Committee (IACUC). Briefly, the mice were randomly divided into three groups.
Mice were anaesthetized by inhalation of isoflurane (1–5%). One midsagittal incision was made on the dorsa of each mouse. Two subcutaneous pockets were made by blunt dissection, one on each side. In total four cell-scaffold constructs were implanted into one mouse with two cell-scaffold constructs placed cephalad and caudad in each pocket. The incisions were closed with surgical staples and animals were given carprofen to manage pain. The mice were euthanized by CO2 asphyxiation followed by bilateral pneumothorax 8-weeks post operation. The implants were dissected and prepared for analysis. For histology, samples were fixed in 4% PFA prior to paraffin embedding and serial sectioning. For gene expression analysis, samples were subjected to total RNA extraction in TRIzol.
2.30. Calvarial Defect in Mice
All procedures were approved by University of Michigan IACUC. Briefly, 8- to 10-week-old C57BL/6 mice (Charles River) were allocated into 3 groups randomly. The mice were anaesthetized by inhalation of isoflurane (1–5%), and a midsagittal incision was made on the scalp, followed by blunt dissection to expose the parietal calvarial bone. A 2.7 mm craniotomy was performed using a trephine burr centered on the parietal calvarial bone and suture. A sterilized three-dimensional construct, without cells, was placed to fill the defect, and the incision was then closed with 4–0 sutures (Ethilon) and animals were given carprofen to manage pain. The mice were euthanized by CO2 asphyxiation followed by bilateral pneumothorax 8-weeks post operation; calvaria were harvested for further micro-CT and histological studies.
2.31. Microcomputed Tomography Analysis of Bone Formation
For the analysis of calvarial bone, a fixed global threshold of 180 was applied to the microcomputed tomography (μCT) scan. 3D reconstruction of the skull and quantification of the newly formed bone was performed. A 2.7 mm round region centered on the defect was determined and the bone volume (BV) and bone mineral density (BMD) in the area were measured using Scanco software (Scanco μCT 100).
2.32. Histological Analyses
Samples were fixed in 4% PFA and demineralized (calvaria only). After dehydration step, samples were embedded in paraffin and serial sections of 5 μm thickness were cut. Sections were prepared according to standard protocols for: hematoxylin and eosin (H&E), Masson’s trichrome, and von Kossa staining. All sections were evaluated using a light microscope and camera.
2.33. Statistical Methods
All data are reported as mean ± standard deviation. To determine statistical significance of observed differences between experimental groups, the Student’s t-test was used. p < 0.05 is used to determine significance. All analysis was carried out in IBM SPSS v23.0 and GraphPad Prism v8.
3. Results
3.1. Isolation of Exosomes from Human Dental Pulp Stem Cells
Human dental pulp stem cells (hDPSCs) are multipotent and can be differentiated towards adipogenic, osteogenic, and chondrogenic fates [30, 33, 38, 49]. Primary hDPSCs were cultured under osteogenic conditions for two weeks, then their osteogenic exosomes (OS-EXOs) were isolated by ultracentrifugation following literature [11, 40]. The physical and molecular identity of these exosomes was assessed to determine the purity of the exosome isolate following literature and our previous publication [11, 46, 50]. Based on nanoparticle tracking analysis (NTA), particles in the exosome fraction had an average hydrodynamic diameter of about 135 nm (Fig 1A, B) with a few minor shoulders reflecting extracellular vesicles outside the size range of the exosomes. Western blot analysis demonstrated that the OS-EXO fraction was highly enriched for characteristic CD9, Alix, and Tsg101 proteins (Fig 1C), which were much less enriched in the total cell lysate. Such EXOs were also shown to strongly express CD63 and CD81 [11]. β-actin was highly enriched in the cell lysate, but not the exosome lysate (Fig 1C). Freshly isolated OS-EXOs were labelled using a lipophilic dye (DiO) for 30 minutes and shown to be rapidly up taken by recipient primary mouse bone marrow stem cells (mBMSCs) using confocal microscopy (Fig 1D). OS-EXOs were up taken and localized with the recipient cell cytoplasm.
Fig. 1.
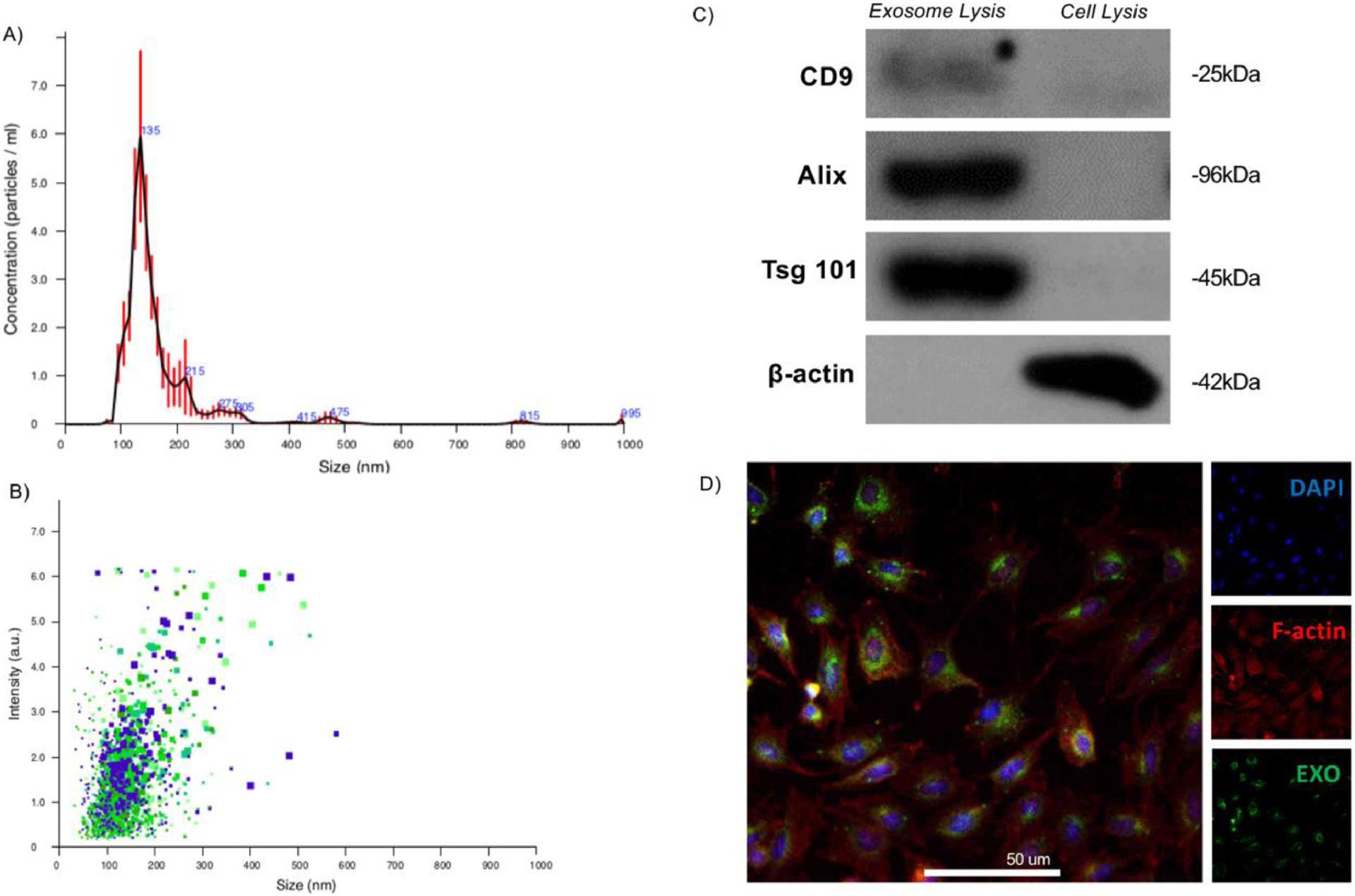
Isolation and characterization of exosomes derived from dental pulp stem cells (DPSCs). Exosomes were physically characterized by their size using nanotracking analysis, indicating that freshly isolated exosomes are of the characteristic hydrodynamic diameter (davg = 135 nm, A), and uniformly sized (B). Molecular profiling by Western blot indicates that the isolated exosomes are highly enriched for characteristic exosome surface markers not abundant in the total cell isolate (C). Freshly isolated exosomes were fluorescently labelled, as described, and their uptake by recipient primary mouse bone marrow stromal cells (mBMSCs) after 30 minutes incubation and visualized by confocal microscopy (D; blue: DAPI, red: F-actin, green: membrane-fluorescent exosomes; scale: 50 μm).
3.2. Characterization of the Osteogenic Potential of hDPSC-derived OS-EXOs
We hypothesized that exosomes derived from primary hDPSCs under osteogenic, or mineralization conditions would be a potent tool to modulate the mineralization and osteogenic differentiation of recipient primary mBMSCs. Primary mBMSCs were treated with 1 μg/mL and 5 μg/mL OS-EXOs, compared to osteogenic and growth media alone for up to two weeks. As early as three days in culture, treatment with OS-EXOs increased ALP activity compared to osteogenic and growth media (Fig 2A, 2B); ALP activity was consistently higher in OS-EXO treated cells during the seven-day culture. At 14 and 21 days in culture, exogenous administration of OS-EXOs increased mBMSC mineralization; an increased dose of OS-EXOs resulted in higher mineralization, particularly at 14 days (Fig 2C, 2D). Our data suggests that OS-EXOs may accelerate in vivo bone healing in a dose-dependent manner. The similar therapeutic benefit of OS-EXOs was shown in primary human BMSCs in vitro at 14 and 21 days (Sup Fig 1).
Fig. 2.
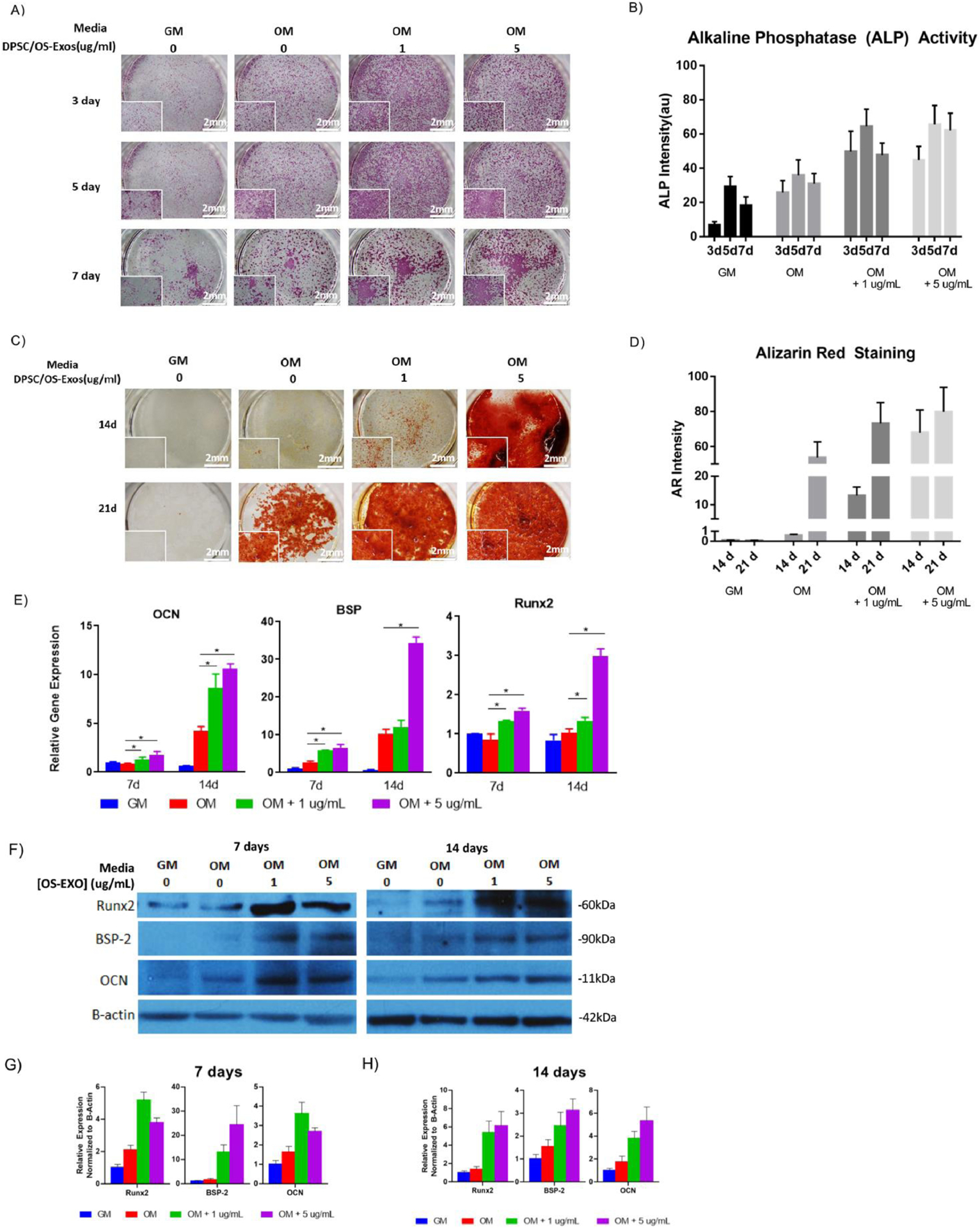
In vitro characterization of the osteogenic potential of DPSC-derived exosomes (OS-EXO) in primary mBMSCs. In as few as three days, OS-EXO increased alkaline phosphatase (ALP) activity (A, B), and increased calcium deposition at 14 and 21 days, indicated by Alizarin red staining (C, D), compared to osteogenic or growth media alone. Exogenous OS-EXO administration in vitro caused significant osteogenesis at the transcriptional (E) and translational levels (F, G, H). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 (n=4 for Western blotting and all other analyses, representative images presented).
Increases in mineralization were accompanied by upregulation in the expression of osteogenic markers including osteocalcin (OCN), bone sialoprotein (BSP) and RUNX Family Transcription Factor 2 (RUNX2) (Fig 2E), and increased protein secretion of the same gene products (Fig 2F, 2G, 2H).
Treatment with exogenous OS-EXOs increased phosphorylation of Erk in a dose-dependent manner and upregulated Runx2 after 30 minutes (Fig 3A&B). mBMSCs were treated with U1026, an inhibitor of Erk phosphorylation to probe the involvement of Erk phosphorylation in OS-EXOs mediated Runx2 upregulation [51]. In control experiments, without treatment by OS-EXOs, U1026 treatment suppressed p-Erk and Runx2 compared with untreated control. Treatment by 5 μg/mL OS-EXOs upregulated p-Erk and Runx2 protein levels; when treated with U1026 expression of p-Erk and Runx2 decreased simultaneously (Fig 3A&C). In contrast, OS-EXO treatment did not alter Smad1 phosphorylation (Sup Fig 2). Based on observed changes in Runx2 and p-Erk translation upon treatment with OS-EXOs alone or in combination with U1026, our results implicate that OS-EXOs induce osteogenic protein expression through an Erk-dependent way.
Fig. 3.
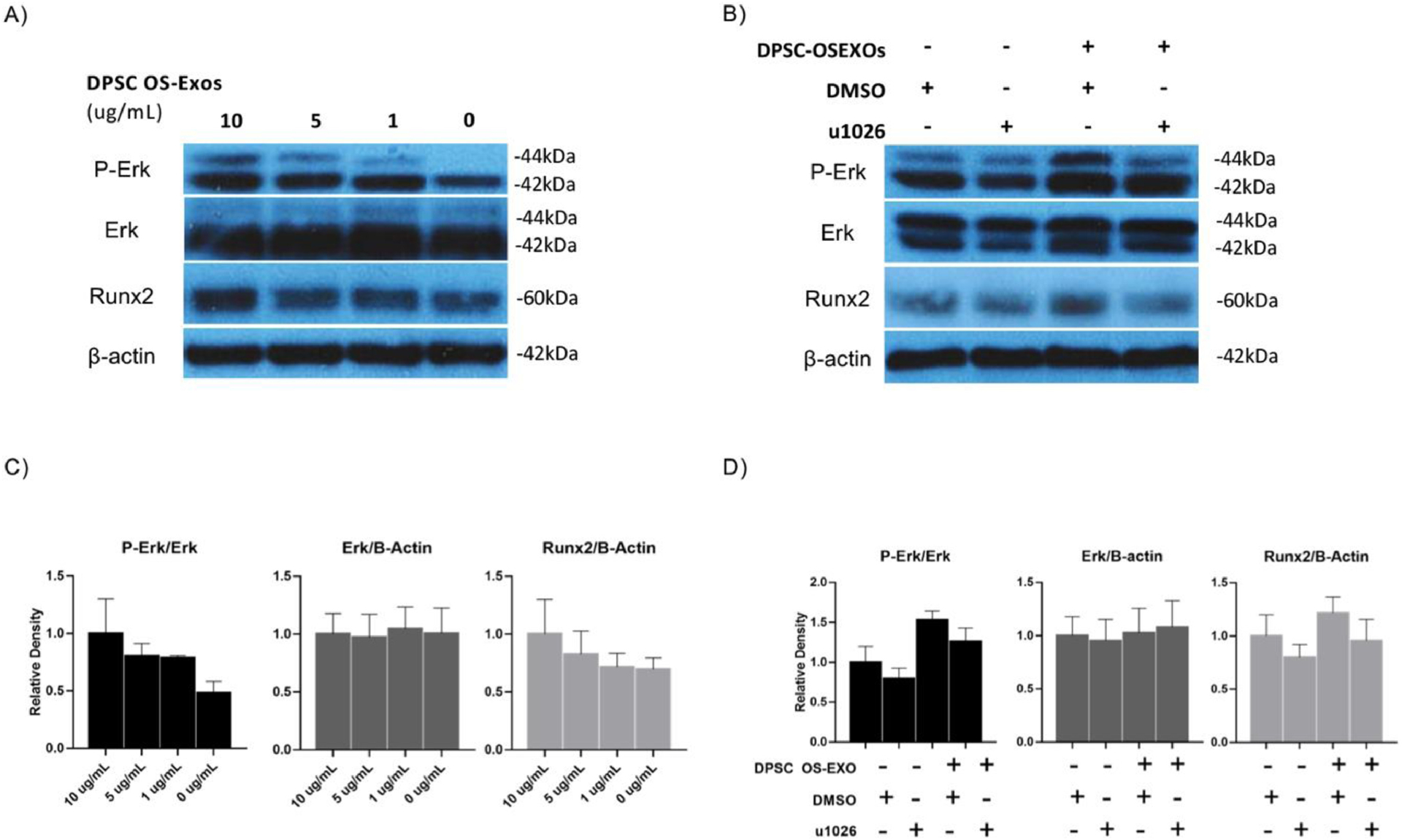
Osteogenic exosomes (OS-EXO) derived from DPSCs upregulated mBMSCs Runx2 level through ERK signaling. (A,C): OS-EXO upregulated mBMSCs Runx2 and ERK phosphorylation at 1、5、10 ug/ml concentrations; (B,D): With presence of ERK inhibitor u1026, the ERK phosphorylation and Runx2 level were both downregulated. N=4 for Western blotting analysis and representative images presented.
3.3. Droplet-based Microfluidics Facilitating EXO-MS Formation by Self-Assembly
Previously our group and others have used double emulsion methods to encapsulate small molecules, proteins and miRNA in poly(aliphatic ester) including poly(l-lactic-co-glycolic acid) (PLGA) microspheres, which degrade by hydrolysis [23, 26, 39, 52]. There were significant challenges to encapsulate large biologic species such as exosomes using such platform, where block copolymers were needed [53]. Here, droplet-based microfluidics was used to further facilitate EXO-MS fabrication as shown in the scheme (Fig 4A) and the microfluidic devices were fabricated from polydimethylsiloxane (PDMS) using soft-lithography methods. The microfluidic device was designed with three inlets, for each of the discrete cargo-containing aqueous phase, discrete polymer-containing organic phase, and continuous aqueous phase (Fig 4B, Sup. Fig. 3). The immiscible water and organic phases contacted at each of two flow-focusing junctions (Fig 4C).
Fig. 4.
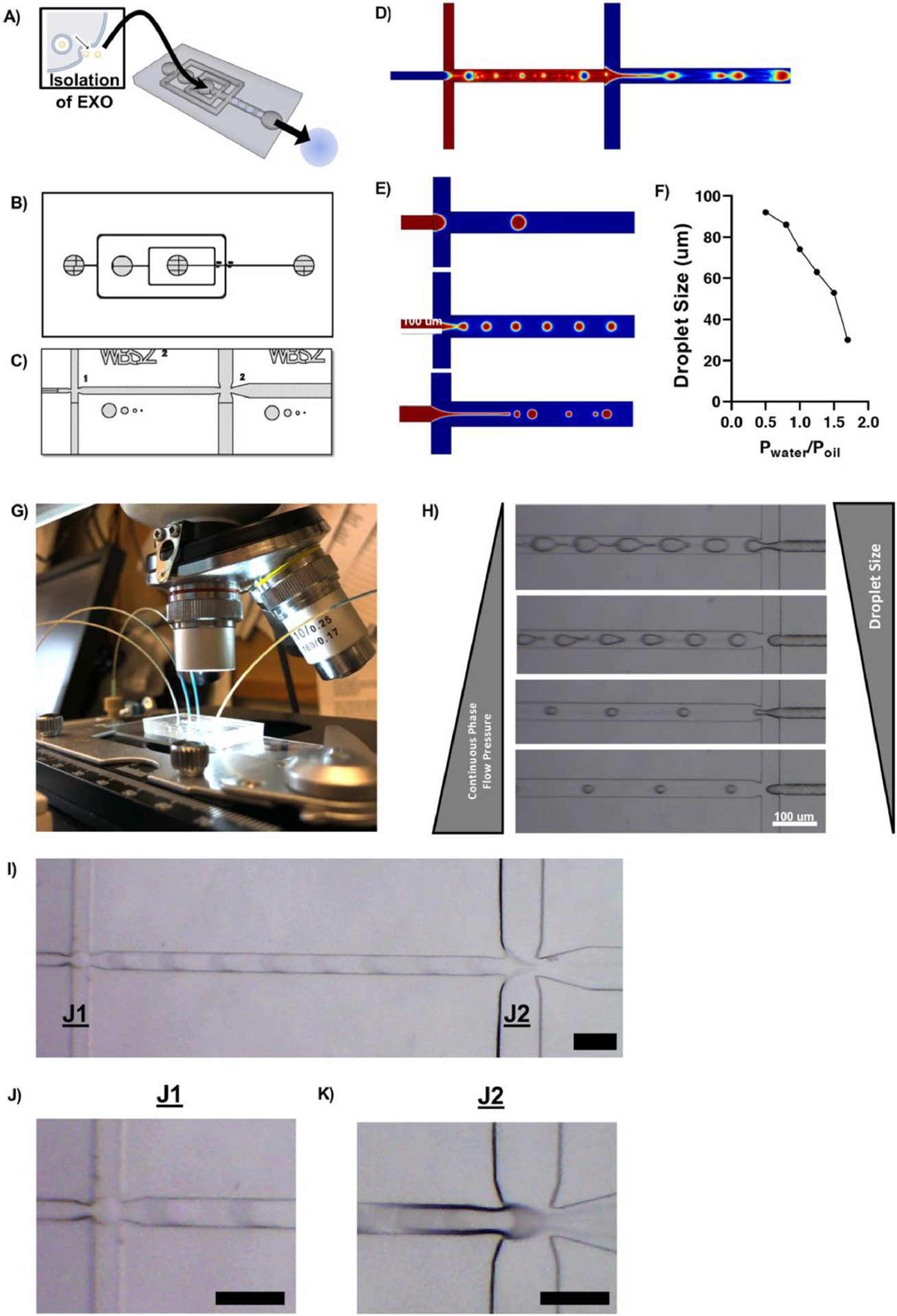
Development of a dual flow-focusing junction microfluidic device to facilitate exosome encapsulation. The device was designed to facilitate the formation of a water/oil/water double emulsion (A) via two flow focusing junctions (B) where immiscible solvents contacted and form droplets which later become particles (C). CAD drawings of the device were used to simulate fluid flow using COMSOL, demonstrating the formation of droplets at both junctions (D, E). Droplet size was controlled by the relative flow rates of two immiscible solvents when they contacted at the flow focusing junction (F). PDMS devices were interfaced with pressure-driven pumps and droplet formation was visualized under a light microscope (G, H, I, J, K). Representative images shown. Scale = 100 μm.
COMSOL fluid dynamics simulations using a phase field method demonstrated droplet formation at each junction, controlled by the interfacial surface tension between immiscible aqueous and organic phases at each of the two flow focusing junctions (Fig 4D). The resulting droplets were the product of a water/organic/water (w/o/w) emulsion and were discrete compartments where the aqueous phases were physically separated by an organic, polymer-containing layer. Small diameter channels restricted the Reynolds number of fluids to cause laminar flow, and the size of droplets at each junction was controlled by the relative flow rates of solutions at the flow focusing junction (Fig 4E, 4F).
Experimentally, fluid flow was controlled by three independent pressure-driven pumps which interfaced with the PDMS device through small-diameter solvent-resistant tubing (Fig 4G)[54]. In agreement with computational predictions, droplet size was controlled by the relative pressures of the interacting phases. At a constant discrete phase pressure, as the continuous phase flow pressure was increased droplet size was decreased (Fig 4H). Droplet formation in the device was visualized by light microscope, demonstrating the rapid formation of discrete droplets at each flow-focusing junction (Fig 4I, Sup Movie 1). At Junction 1 (J1), OS-EXOs suspended in PBS were discretized in a matrix of PLGA-PEG-PLGA, an amphiphilic triblock copolymer with distinct hydrophilic and hydrophobic blocks, in ethyl acetate (2% wt/v) to form a series of water/organic (w/o) droplets (Fig 4J). At Junction 2 (J2), the w/o droplet becomes discretized in dilute polyvinyl alcohol (PVA, 0.01% wt/v), a non-ionic surfactant which stabilized the organic-aqueous interface during droplet formation in the jet of newly formed w/o/w droplets (Fig 4K).
Droplets were collected from the device in a round-bottom flask and ethyl acetate was evaporated to yield solid polymeric spheres which were uniform in size, with smooth, non-porous surface morphology (Fig 5A). EXO-MS were fabricated with fluorescently-labelled copolymer blocks, as described in the Methods section, and visualized by confocal microscopy (Fig 5B). The particles consisted largely of a disperse PLGA matrix (red), the main component of the triblock PLGA-PEG-PLGA copolymer. An inner layer of PEG formed a thin spherical layer (blue), wherein OS-EXOs were localized in the particle (green). EXO-MS fabricated by droplet microfluidics could yield smaller and more uniformly sized particles than those fabricated by a standard mechanical emulsion technique (Fig 5C&D).
Fig 5.
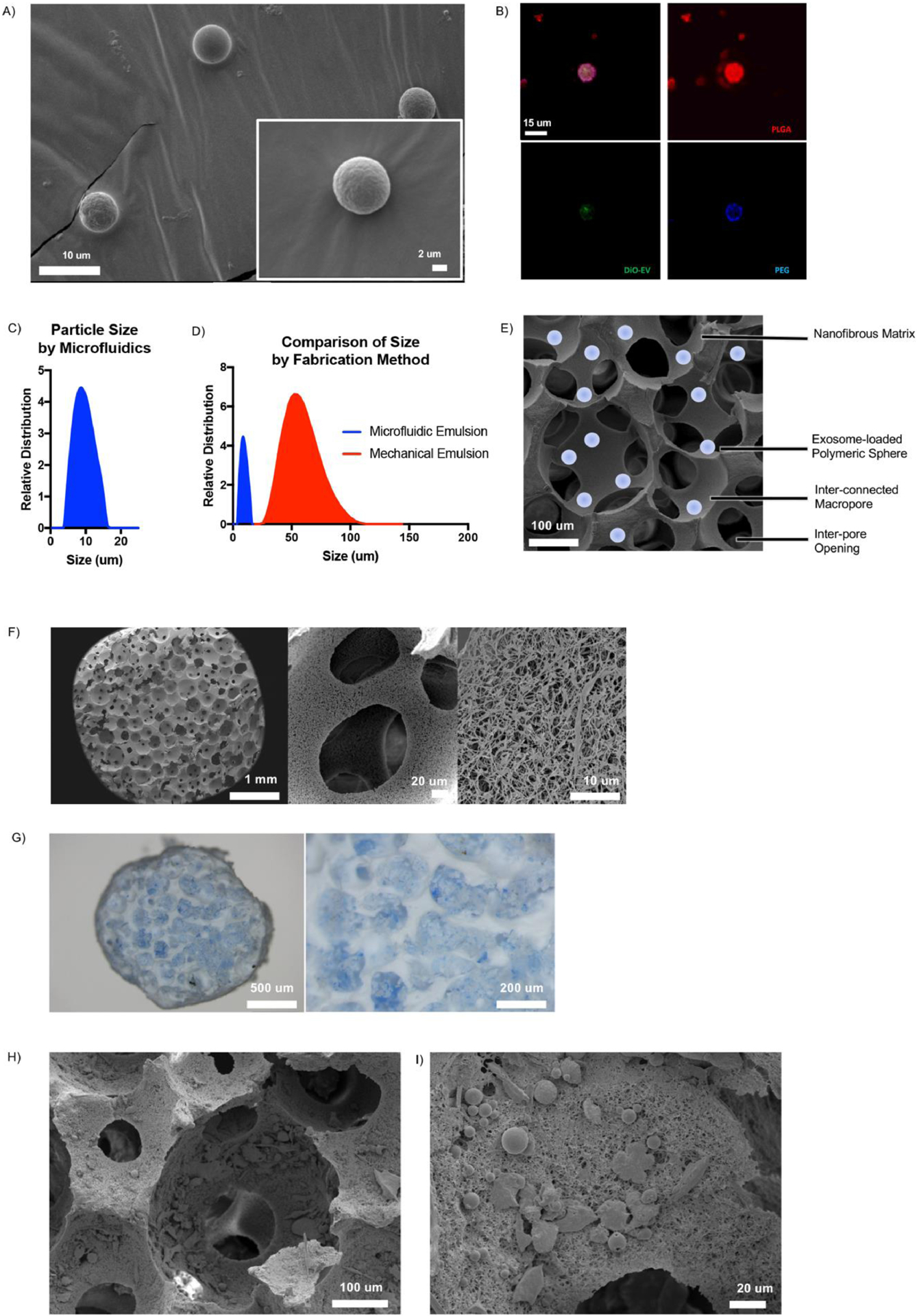
Characterization of EXO-MS. Scanning electron microscopy images of EXO-MS prepared by microfluidic double emulsion method showed smooth, uniformly sized spheres (A). EXO-MS made from PLGA-PEG-PLGA block copolymer were mixed with fluorescently labelled polymer blocks and visualized by confocal microscopy; PEG (blue) segments self-assembled on the inside of the particle to form a sphere wherein OS-EXO (green) localize, among a disperse PLGA (red) microspheres (B; scale: 15 um). The size of EXO-MS was measured by a laser diffraction method (C, davg = 7.70 um). Compared to self-assembling particles from the same polymer made under mechanical emulsion conditions, particles made by a microfluidic platform were significantly smaller and less disperse (D). EXO-MS were attached to poly(l-lactic acid) (PLLA) tissue engineering scaffolds, as shown in the schematic (E; scale: 100 um). Scaffolds were a matrix of interconnected macropores with a nanofibrous surface texture (F). EXO-MS were seeded onto the pore surface of the scaffold using a solvent-wetting method, visualized by light microscopy (G, dyed blue for visual contrast) and scanning electron microscopy (H, I).
3.4. Three-Dimensional Exosome-Releasing Tissue Engineering Constructs
EXO-MS are sufficiently small such that they can be physically attached to the surface of a tissue engineering scaffold, as shown schematically in Fig 5E. Three-dimensional scaffolds were fabricated from poly(l-lactic acid) (PLLA) (Fig 5F), which have a nanofibrous surface texture to mimic the native collagen extracellular matrix with an average fiber diameter of 50–500 nm, the result of thermally induced phase separation of PLLA [28]. In this work, the scaffolds were fabricated with 250–425 μm sized spherical macropores using a sugar sphere templating technique [27].
The high surface area of interconnected macropores is ideal to be functionalized with inductive cues which guide tissue formation in an efficient way. EXO-MS were attached to the surface of scaffold pores using a post-fabrication seeding method (Fig 5G) [24]. Observed by light microscopy, particles (colored blue for visualization) were distributed evenly throughout the pores of the matrix. EXO-MS were sufficiently small such that they did not occlude interconnections between pores which would otherwise limit mass transport and cell migration throughout the construct (Fig 5H&I).
3.5. Characterizing the Release of OS-EXOs from EXO-MS
OS-EXO encapsulating polymeric microspheres (EXO-MS) were fabricated with four different formulations of triblock copolymer. PLGA-b-PEG-b-PLGA was synthesized at lactide/glycolide percentages of 100/0, 85/15, 70/30, and 50/50, all with the same molecular weight (5 kDa). EXO-MS were incubated in PBS and aliquots analyzed for OS-EXOs. Cumulative release from EXO-MS in PBS showed an initial burst release followed by a nearly linear release profile, delivering a consistent dose of exosomes for up to 10 weeks (Fig 6A). Within the first day 18% was released from PL85G15A EXO-MS; following this initial burst release, the release rate became nearly constant, which is correlated to the hydrophilicity of the PLGA copolymer segments. The burst release was more prominent in more hydrophilic compositions compared to more hydrophobic compositions suggesting that they were more easily hydrolyzed.
Fig. 6.
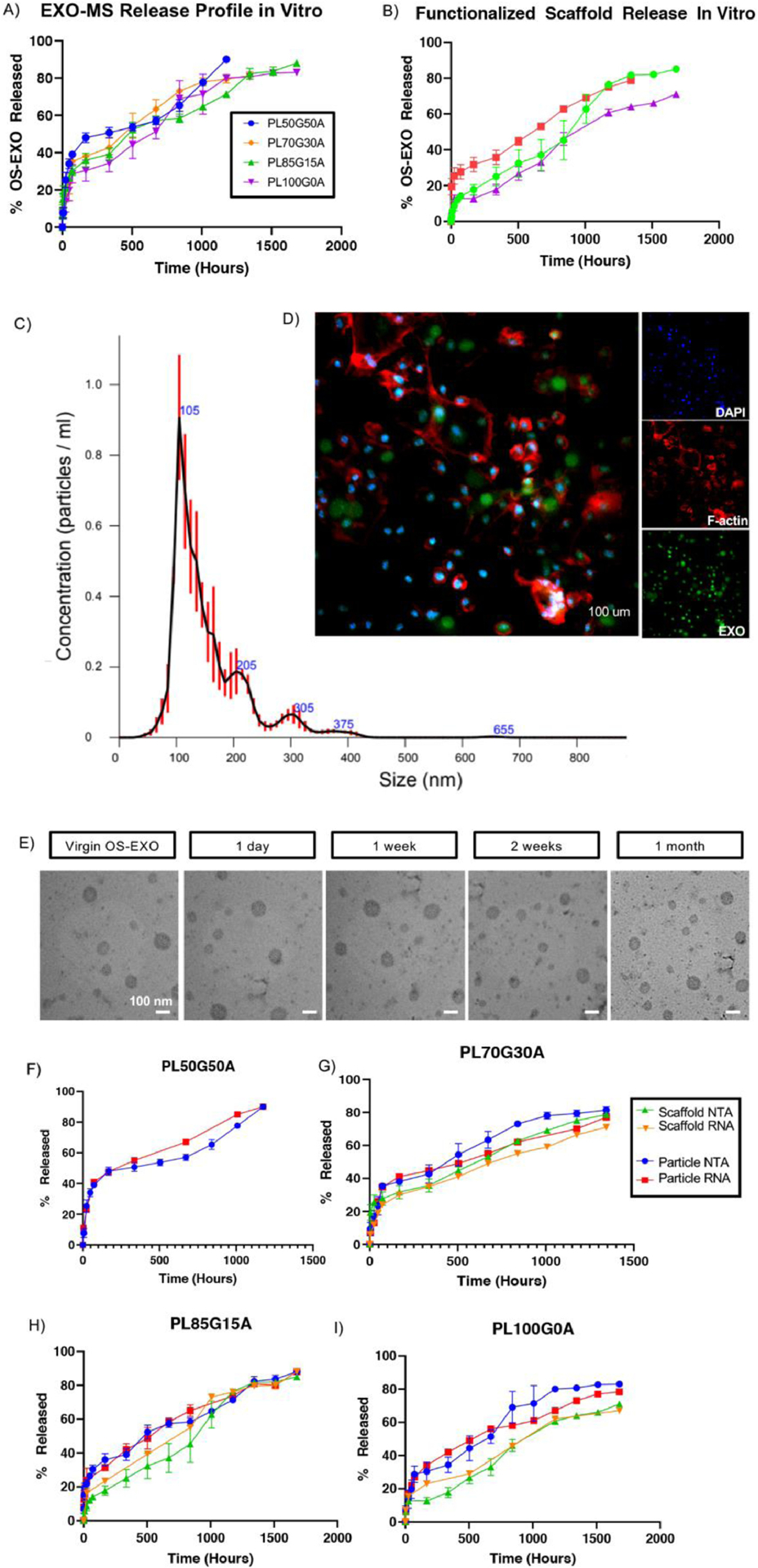
Evaluation of the controlled release of OS-EXO from EXO-MS. The release profile of OS-EXO was modulated by the hydrophilicity of the copolymer (A). After attachment of EXO-MS to PLLA scaffolds, the release of OS-EXO became more linear (B). OS-EXO released from EXO-MS at 2 weeks maintained their characteristic size (davg = 105 nm), evaluated by nanoparticle tracking analysis (C). Primary mBMSCs were treated with EXO-MS eluent (collected at 2 weeks) which contained fluorescently labeled OS-EXO for 30 minutes, and the uptake of OS-EXO by recipient cells was visualized by confocal microscopy (D; green: fluorescent-membrane of OS-EXO, red: F-actin, blue: DAPI; scale: 100 um). Aliquots of in vitro EXO-MS eluent were collected and subjected to transmission electron microscopy (TEM) analysis at prescribed time points (E; scalebars: 100 nm). Aliquots of in vitro EXO-MS eluent were subjected to RNA extraction at each release time point. Quantity of RNA correlates well to particle number (using NTA) for cases of both release from EXO-MS, and release from EXO-MS attached to PLLA scaffolds (F, G, H, I).
EXO-MS attached to PLLA scaffolds had a dampened release profile compared to EXO-MS suspended freely in solution (Fig 6B). The release profiles showed a significantly decreased initial burst release (7% release in 24 hours) and more linear, consistent controlled release throughout the duration, again correlated with relative hydrophilicity of the PLGA copolymer segments. Compared to free EXO-MS, we observed that EXO-MS attached to scaffolds released for a longer period of time despite the same initial cargo loading and polymer composition. PL85G15A composition was chosen for subsequent in vitro and in vivo study because it maximized the nearly linear delivery.
3.6. OS-EXO Integrity following Encapsulation and Release
At designated time points, the conditioned OS-EXO-containing PBS was withdrawn and frozen at −80°C for storage, and replaced with fresh PBS before analysis. Released OS-EXO concentration was assayed using NTA (nanotracking analysis). OS-EXOs maintained their characteristic hydrodynamic diameter throughout release (Fig 6C, Sup Fig 4). Fluorescently-labelled OS-EXOs, eluted from EXO-MS, were readily up taken by recipient mBMSCs within 30 minutes (Fig 6D). To visualize membrane integrity, eluted exosomes were fixed and stained for transmission electron microscopy. Compared to virgin OS-EXOs, released OS-EXOs maintained their spherical shape, size, and membrane integrity, attributed to the addition of D-trehalose to the discrete exosome-containing aqueous phase. Transmission electron microscopy analysis confirmed that exosomes maintained their morphology throughout release (Fig 6E). A stabilized lipid membrane was important to maintain the integrity of the biologic cargo.
miRNA and nucleotide cargo are critical to the physiologic function of exosomes in vitro and in vivo. Extracted miRNA from various time points was assayed for concentration. The amount of RNA extracted from eluted OS-EXOs at each release time point matched well to OS-EXO particle number, from NTA, for free EXO-MS and EXO-MS attached to nanofibrous scaffolds (Fig 6F–I).
3.7. In vitro Bioactivity of Functional EXO-MS Eluent
In vitro bioactivity was investigated over the course of three weeks. mBMSCs were cultured on nanofibrous scaffolds functionalized with EXO-MS but no exogenous OS-EXO treatment, blank nanofibrous scaffolds plus exogenous OS-EXO treatments of 1 μg/mL or 5 μg/mL, and blank nanofibrous scaffolds without any exosome treatment, in osteogenic and growth media. Fourier-transformed infrared spectroscopy (FTIR) confirmed the presence of calcium hydroxyapatite mineral (Ca10(PO4)6(OH)2) based on characteristic peaks in the spectra [55]. Constructs exposed to EXO-MS or exogenous OS-EXOs showed strong reflectance of PO43− (1,185–900 cm−1, 1,035–1,025 cm−1), compared to the positive and negative controls (Fig 7A, 7F). Carbonate can substitute for OH moieties in apatite (877 cm−1) or phosphate (871 cm−1). Its bands appeared in the treated constructs, much more intensely than in the negative control. The combination of these signals was indicative of hydroxyapatite mineralization, which was confirmed by colorimetric calcium assay at 14 and 21 days of culture (Fig 7B, 7G). At 14 days, the EXO-MS-functionalized construct showed significantly greater mineralization than exogenous administration, which together were greater than the positive and negative control (Fig 7B). By 21 days, the EXO-MS-functionalized constructs were mineralized similarly to the constructs receiving exogenous treatment; the OS-EXO treated groups were mineralized significantly more than the positive and negative controls (Fig 7G).
Fig. 7.
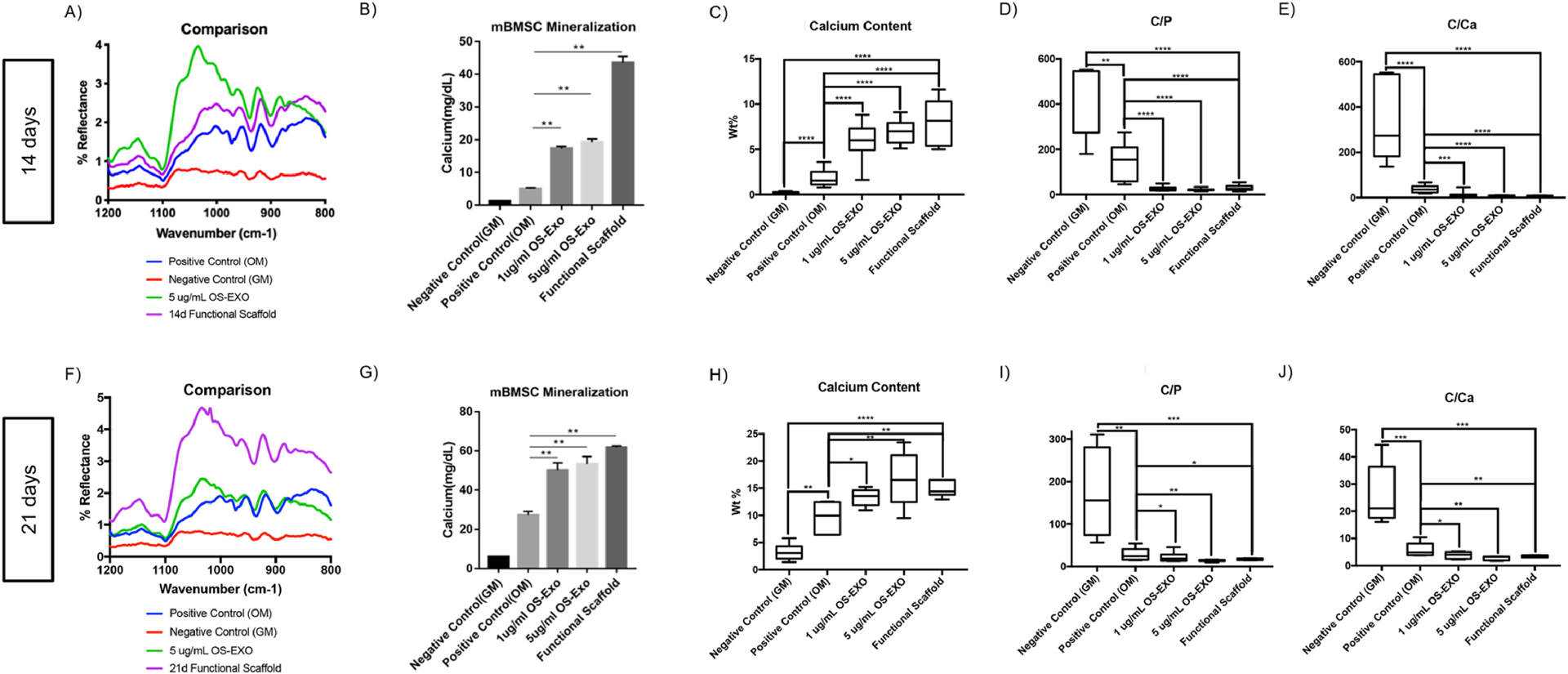
Mineral characterization of cell-laden 3D constructs cultured in vitro. At 14 and 21 days of primary mBMSC cell culture, Fourier-transformed infrared spectroscopy (FTIR) spectra indicated that EXO-MS laden PLLA scaffolds and exogenous administration of OS-EXO yielded calcium hydroxyapatite mineralization (A, F), confirmed my colorimetric calcium assay (B, G). Elemental analysis by energy-dispersive x-ray spectroscopy shows increased calcium content (C, H), decreased carbon-to-phosphorus ratio (D, I), and decreased carbon-to-calcium ratio (E, J) for EXO-MS functionalized scaffolds compared to unfunctionalized nanofibrous scaffolds alone. Mineralization of functionalized scaffold constructs is comparable to those treated by exogenous administration of OS-EXO at 1 μg/mL and 5 ug/mL over the course of three weeks. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Energy dispersive X-ray spectroscopy (EDX) was used to quantitatively determine the spatial distribution of elements in the constructs in tissue culture [56]. Compared to the untreated controls, EXO-MS-functionalized constructs and constructs receiving exogenous OS-EXO treatment showed increased calcium content (Fig 7C, 7H), and significantly decreased C/P (Fig 7D, 7I) and C/Ca (Fig 7E, 7J) ratios at 14 and 21 days, indicating that the calcium phosphate (CaP) content increased, and the proportion of organic components decreased as the mineral was deposited.
3.8. Subcutaneous Implantation of EXO-MS-functionalized Scaffolds
Constructs were seeded with primary mBMSCs and implanted subcutaneously in mice for 8 weeks. Histological analysis by Hematoxylin and Eosin staining indicated that EXO-MS functionalized scaffolds supported cells similarly to blank nanofibrous scaffolds (Fig 8, Sup. Fig. 5). Masson’s Trichrome staining showed increased ECM deposition in OS-EXO-loaded EXO-MS (OS-EXO-MS) constructs compared to blank EXO-MS constructs or blank scaffolds, attributing this difference to the controlled release of OS-EXO cargo. Here, blank EXO-MS were polymer microspheres loaded with PBS/trehalose without exosomes to serve as a control for EXO-MS, which were polymer microspheres loaded with exosomes in PBS/trehalose. Von Kossa staining was more intense in OS-EXO-MS constructs, indicating early mineralization caused by OS-EXO, not present in blank EXO-MS constructs nor blank scaffolds. The controlled release of OS-EXO also caused upregulation of bone sialoprotein as assessed by immunohistochemistry.
Fig. 8.
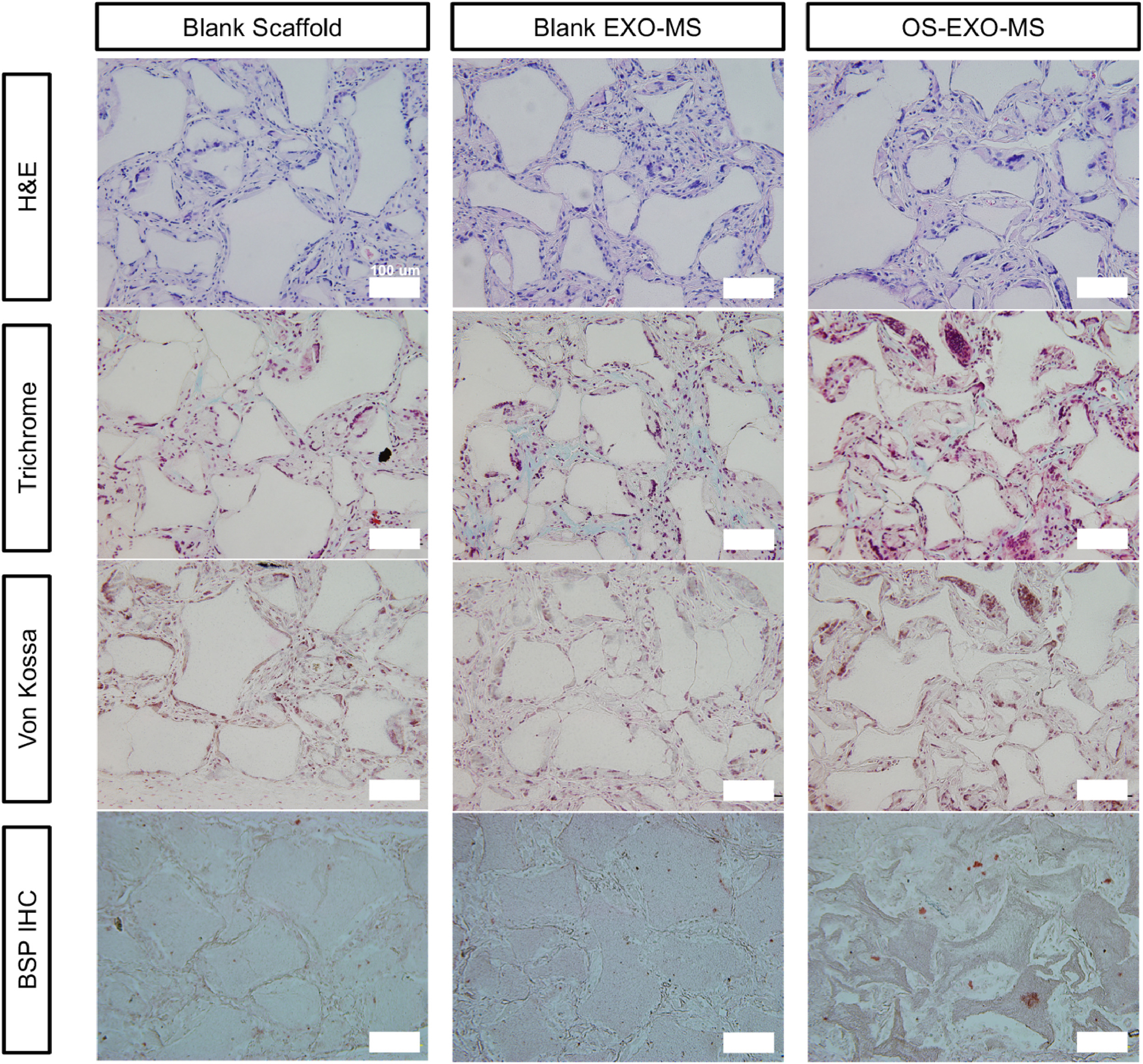
Histology of subcutaneous implants. Functionalized scaffolds implanted subcutaneously for eight weeks showed that cells infiltrated the construct well and occupied the interconnected spherical macropores (Hematoxylin and Eosin) equally well among nanofibrous scaffolds, scaffolds loaded with blank EXO-MS, and scaffolds functionalized with OS-EXO-MS. Masson’s trichrome staining showed increased extracellular matrix deposition in OS-EXO-MS functionalized scaffolds; von Kossa staining shows increased calcium deposition by calcium nodule formation in the OS-EXO-MS functionalized scaffolds. Immunohistochemical analysis for bone sialoprotein (BSP) showed increased BSP protein level as a result of OS-EXO-MS loaded scaffolds. Scale: 100 μm.
3.9. Mouse Calvarial Bone Defect Repair by EXO-MS Loaded Scaffolds
OS-EXO-MS-functionalized scaffolds were assessed as a method of cell-free bone regeneration in vivo in a calvarial defect model. A 2.7-mm round defect was created in the mouse skull. OS-EXO-MS-functionalized scaffold was used to fill the defect; blank EXO-MS scaffold, and blank scaffold were used as controls. H&E and trichrome staining showed that the OS-EXO-MS functionalized scaffold contained cell-laden collagen-rich matrix, filled the defect with significant marrow-containing bone tissue, and was integrated with the host tissue at 8 weeks (Fig 9A). Untreated controls did not show this integration; while detectable cells migrated into the construct, they did not form robust osseous tissue. Additionally, no signs of inflammation were noted, indicating that the constructs, made of biodegradable and biocompatible polymers which degrade over time, were safe and did not have harmful side effects. Three-dimensional micro-computed tomography (μCT) reconstructions of the skulls showed that localized OS-EXO delivery by EXO-MS resulted in the best regenerative outcome among treatment groups (Fig 9B). OS-EXO-MS-functionalized scaffold group showed significant mineralized bone volume in the defect site (Fig 9C), and bone mineral density in the mineralized areas was similar between the groups (Fig 9D).
Fig. 9.

EXO-MS functionalized scaffold leading to bone regeneration in vivo 8 weeks after implantation without the transplantation of exogenous cells. Hematoxylin & Eosin staining and Masson’s trichrome staining indicated marrow-containing bone tissue formation as a result of OS-EXO treatment, in contrast to minimal bone formation in untreated controls (A). Microcomputed tomography (μCT) of calvaria at 8 weeks (B; scale: 5 μm) indicated significant bone filling in OS-EXO-MS functionalized scaffold, compared to control scaffold and blank MS loaded scaffold (C). There was no significant difference in bone mineral density in the mineralized areas among the groups (D). * p < 0.05, ** p < 0.01
4. Discussion
Traditional bone tissue engineering strategies have historically relied on the exogenous transplantation of stem/progenitor cells with scaffold into a defect site to catalyze tissue neogenesis. Despite the significant progress achieved, there is limited evidence to support the long-term survival and differentiation of transplanted stem/progenitor cells in tissue engineering constructs [57]. It is believed that transplanted stem cells have a strong paracrine effect even if they don’t survive transplantation. Some cell-secreted components are likely involved in signaling cascades which direct regeneration. Additionally, there are well-known immunogenic concerns over donor cells, particularly in geriatric and pediatric patients, associated with stem cell therapies [58, 59]. Therefore, cell-free methods of bone regeneration which recapitulate the biologic properties of stem/progenitor cells to initiate healing and tissue regeneration are advantageous [24, 39, 45, 52, 60]. Here, we investigated the potential of exosomes derived from mineralizing cells as a new way to induce bone regeneration in vivo, without requiring the transplantation of exogenous cells. There have been efforts to utilize EXOs derived from various cell sources either alone or in combination with scaffolds to enhance bone and other tissue regeneration. For example, EXOs from iPSC-derived MSCs have been combined with tricalcium phosphate to improve bone regeneration [40]. EXOs derived from gingival or adipose-derived MSCs have been incorporated in polymer or polymer/calcium silicates composite scaffolds to enhance bone regeneration [61, 62]. EXOs from different sources have also been incorporated into hydrogels or hydrogel sponges for bone regeneration [63, 64]. While positive effect of EXO/EVs have been demonstrated, controlling the release duration and kinetics of EXO remains a challenge [65]. Given that the kinetics of exosome delivery impacts tissue regeneration [66], we engineered an implantable controlled release platform for their sustained delivery to maximize their therapeutic benefit in this work.
In order to realize the biologic capabilities of cell-derived exosomes to facilitate tissue regeneration in the clinical setting, a platform which allows for their controlled delivery and maintains their biologic activity is needed. We report that: i) human DPSCs release exosomes which are readily isolated, regulating phenotypic changes in recipient BMSCs by upregulating Erk phosphorylation and enhancing mineralization in both mouse and human BMSCs; ii) exosomes can be encapsulated in a biodegradable polymer vehicle, EXO-MS, by utilizing block copolymer self-assembly, in a way that maintains their cargo and biologic potency; iii) exosomes are released from the polymeric microspheres (EXO-MS) in a controlled fashion, and the kinetics of their release is tunable by varying the polymer composition; and iv) the controlled delivery of OS-EXO via EXO-MS in vivo facilitates calvarial bone regeneration by recruiting endogenous cells near the defect site and direct their differentiation and mineralization to form new bone tissue. We believe that this biomaterial technology, which utilizes the exosomes rather the source stem cells, may have significant translational and clinical advantages over traditional cell-based regenerative therapies.
A three-dimensional matrix that can support cell infiltration and vascularization is critical to support tissue neogenesis [39, 67]. Our scaffolds have good mechanical properties, nanofibrous surface texture, and well interconnected macropores which are shown to be osteogenic in vitro and in vivo, supporting cellular proliferation and protein adsorption [68–70]. The highly porous surfaces throughout these constructs make them an ideal candidate to be functionalized with delivery moieties, such as EXO-MS, which provide inductive cues for regeneration. OS-EXOs are released from the three-dimensional tissue engineering constructs with a well-controlled rate and duration; the amount of OS-EXOs delivered is modulated by the amount of EXO-MS incorporated into the scaffold. Additionally, mesenchymal stem/progenitor cell-derived exosomes are known to be pro-migratory [71]. The early release of OS-EXOs likely attracts endogenous cells from nearby host tissue to the defect site to secret an early extracellular matrix within the construct which helps orchestrate the formation of new bone tissue.
Microfluidic droplet generation and polymeric self-assembly are critical to the controlled properties of EXO-MS, both in their size and facilitating controlled release of OS-EXOs. The small and uniform size of EXO-MS is advantageous to subtly attach the delivery component to the scaffold pore walls. Microfluidic droplet formation driven by periodic accretion of pressure forms a free droplet from the dispersed phase within a continuous phase. Under restricted fluid flow surface tension is dominant over viscous forces, and small droplets are formed in the device channels [72]. We have also demonstrated that concentrated surfactants used in mechanical emulsions are not necessary in microfluidic emulsions.
A core challenge to the delivery of complex biomolecules is maintaining their bioactivity throughout encapsulation and release [73]. The hydrophilic and hydrophobic blocks of the triblock copolymer used to fabricate EXO-MS can associate with the aqueous and organic phases, respectively, to form a core-shell microsphere which creates an exosome compartment on its interior. In this way they act like a surfactant. Harsh mechanical techniques such as probe sonication, commonly used in double emulsion methods, are destructive to exosomes [74]. The PEG-block of the copolymer associates with the OS-EXO containing aqueous phase to facilitate its compartmentalization and discretization from the bulk phase. Ethyl acetate is chosen because of its low interfacial surface tension with water (6.8 × 10−3 N/m). The polymer also likely protects exosomes from oxidizing agents and enzymes in vivo. Combined with dilute D-trehalose stabilizer (2 mM), the physical and molecular integrity of OS-EXOs is preserved. Finally, our results indicate that degradation products of PLGA do not significantly impact OS-EXOs stability. In this way, OS-EXOs are sufficiently preserved and able to modulate downstream activity of cells in the same fashion as exogenous administration in vitro, through a single platform which is easily implanted in vivo and allows for local, controlled delivery.
Microfluidics also offers a high degree of batch-to-batch consistency, compared to other mechanical emulsion techniques. Emulsions typically result in a wide distribution of droplet sizes [75]. Particle size and more specifically surface area to volume ratio affect the release profile [76]. A small, immediate burst provides an immediate therapeutic dose to the defect site, likely influencing migration and differentiation of early migrant cells into the construct. The following prolonged, nearly zero-order release provides constant exposure which promotes gradual healing and bone tissue neogenesis, in a similar way to exogenous administration. Cells are shown to be saturable by exosomes [77]. Therefore, a large burst release is wasteful, and this notion of saturation provides rationale to maximize the sustained release. Since PLGA is rather hydrophobic, large exosomes cannot diffuse through the matrix as easily as small molecules or even macromolecules. Therefore OS-EXOs are released as the polymer sphere is hydrolyzed and becomes porous on a larger scale, maximizing the linear zero-order release for the majority of the EXO-MS’s functional lifespan.
We have demonstrated that exosomes from mineralizing hDPSCs (OS-EXOs) could enhance MSC osteogenic differentiation in vitro and bone repair in vivo. OS-EXOs induce the osteogenic activity of BMSCs as early as 3 days (increased ALP activity) and into late stages (increased mineralization output) in both mouse and human cells. Consistently, our in vivo models demonstrated the OS-EXOs released from scaffold share the same bioactivity by accelerating the secretion of extracellular matrix in subcutaneously implanted constructs, and craniofacial bone repair in a calvarial defect.
In recent years, exosomes derived from mesenchymal stem cells have shown great promise in accelerating musculoskeletal tissue healing. Most of these reports focus on exosomes derived from naïve MSCs which facilitate tissue repair by enhancing cell migration, survival or proliferation in a dose-dependent manner, but require relatively high concentrations in vitro (10–100 μg/mL) [17, 78]. Enhanced regeneration is likely to be achieved by greater cell infiltration. The application of naïve exosomes may be limited by high cost and unknown side-effects associated with high doses required in vivo. Here we report a low therapeutic dose (1–5 μg/mL) of OS-EXOs which specifically enhances new bone regeneration by inducing osteogenic differentiation. Additionally, because hDPSCs are isolated from extracted teeth, they are an easily accessible mesenchymal stromal cell source compared to bone marrow or adipose tissue which require invasive biopsies.
Compared with naïve exosomes, the cargo in exosomes derived from mature cells changes as a function of differentiation [79, 80]. The osteogenic-specific gene expression and protein synthesis in EXO-MS treated cells is superior compared to treatment by osteogenic media alone. Runx2 is a key osteogenic differentiation regulator previously shown to be implicated in exosome-cell interactions, and enhance the transcriptional activity and stabilization of Runx2 in osteogenic differentiation [81–83]. We demonstrated that with OS-EXO treatment, Runx2 was upregulated with a synchronous increase in Erk phosphorylation. The Erk pathway is known to be involved in cell-exosome interactions as well as the activation and stabilization of Runx2 [83, 84]. Interestingly, the BMP related Smad1 pathway is not involved. Since Runx2 coordinates multiple pathways which may incur complex crosstalk during osteogenic differentiation, a more detailed mechanism of the specific signaling pathways involved in OS-EXO-mediated osteogenesis merits further investigation to identify OS-EXO-specific miRNA cargo. We have additionally demonstrated that exosomes derived from human cells are able to modulate the phenotype of recipient cells similarly in both humans and mice, which agrees with recent literature indicating cross-species efficacy of exosomes [85].
Our ability to encapsulate and deliver stem cell-derived exosomes demonstrates clear translational potential towards exciting clinical developments in regenerative medicine. Donor cell phenotype is carefully selected to direct recipient cells on a designated trajectory, through their exosomal miRNA cargo. Following this rationale, we are able to hack into endogenous repair mechanisms and potentially circumvent costly stem cell transplantation therapies. In support of this notion, exosome-mediated cell-free therapy has a number of manufacturing and regulatory advantages compared to stem cell therapies [19, 86, 87]. Of importance, EXO-MS and scaffold constructs can be conveniently sterilized by ethylene oxide gas.
5. Conclusions
Exosomes derived from hDPSCs are encapsulated in triblock PLGA-PEG-PLGA microspheres utilizing a microfluidic device, which are subsequently incorporated into a nanofibrous PLLA scaffold. The controlled delivery of the exosomes from the scaffold in a critical-size bone defect recruits endogenous cells and stimulates bone tissue neogenesis in vivo, without the need for exogenous stem cell transplantation. This platform facilitates the local activity of exosomes, which bear many of the favorable properties of stem cells, without off-target effects or immunogenic and tumorigenic concerns. Exosomes could facilitate a new wave of potential therapies in regenerative medicine; their clinical success will be highly dependent on methods developed for their efficient delivery. Our results provide strong evidence to suggest that exosomes derived from mineralizing hDPSCs provide pro-mineralization cues to drive local stem/progenitor cells towards osteogenic differentiation in vitro and in vivo.
Supplementary Material
Acknowledgments
The work was carried out in the Polymeric Biomaterials and Tissue Engineering Laboratory at the University of Michigan. The authors thank Tianming Du and Youlong Xie for their assistance and discussion in TEM analysis and cell culture studies. The authors would like to acknowledge the financial support from the NIH R01-DE022327 (PXM), R01-DE27662 (PXM), T32 DE-007057 (WBS). ZZ was partially supported by China Scholarship Council (CSC201706240064) for living expenses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors have no competing financial or other interests to disclose related to the present work.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Buza JA 3rd, Einhorn T, Bone healing in 2016, Clin Cases Miner Bone Metab 13(2) (2016) 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nauth A, McKee MD, Einhorn TA, Watson JT, Li R, Schemitsch EH, Managing bone defects J Orthop Trauma 25(8) (2011) 462–6. [DOI] [PubMed] [Google Scholar]
- [3].Langer R, Vacanti JP, Tissue engineering, Science 260(5110) (1993) 920–6. [DOI] [PubMed] [Google Scholar]
- [4].Ma PX, Biomimetic materials for tissue engineering, Adv Drug Deliv Rev 60(2) (2008) 184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Laurencin CT, Ambrosio AM, Borden MD, Cooper JA Jr., Tissue engineering: orthopedic applications, Annu Rev Biomed Eng 1 (1999) 19–46. [DOI] [PubMed] [Google Scholar]
- [6].Baroli B, From natural bone grafts to tissue engineering therapeutics: Brainstorming on pharmaceutical formulative requirements and challenges, J Pharm Sci 98(4) (2009) 1317–75. [DOI] [PubMed] [Google Scholar]
- [7].De Kok IJ, Peter SJ, Archambault M, van den Bos C, Kadiyala S, Aukhil I, Cooper LF, Investigation of allogeneic mesenchymal stem cell-based alveolar bone formation: preliminary findings, Clin Oral Implants Res 14(4) (2003) 481–9. [DOI] [PubMed] [Google Scholar]
- [8].Heathman TR, Nienow AW, McCall MJ, Coopman K, Kara B, Hewitt CJ, The translation of cell-based therapies: clinical landscape and manufacturing challenges, Regen Med 10(1) (2015) 49–64. [DOI] [PubMed] [Google Scholar]
- [9].Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr., Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr., Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, J Extracell Vesicles 7(1) (2018) 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Witwer KW, Van Balkom BWM, Bruno S, Choo A, Dominici M, Gimona M, Hill AF, De Kleijn D, Koh M, Lai RC, Mitsialis SA, Ortiz LA, Rohde E, Asada T, Toh WS, Weiss DJ, Zheng L, Giebel B, Lim SK, Defining mesenchymal stromal cell (MSC)-derived small extracellular vesicles for therapeutic applications, J Extracell Vesicles 8(1) (2019) 1609206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Swanson WB, Gong T, Zhang Z, Eberle M, Niemann D, Dong R, Rambhia KJ, Ma PX, Controlled release of odontogenic exosomes from a biodegradable vehicle mediates dentinogenesis as a novel biomimetic pulp capping therapy, J Control Release 324 (2020) 679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C, Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes), J Biol Chem 262(19) (1987) 9412–20. [PubMed] [Google Scholar]
- [13].Tkach M, Thery C, Communication by Extracellular Vesicles: Where We Are and Where We Need to Go, Cell 164(6) (2016) 1226–1232. [DOI] [PubMed] [Google Scholar]
- [14].Fevrier B, Raposo G, Exosomes: endosomal-derived vesicles shipping extracellular messages, Curr Opin Cell Biol 16(4) (2004) 415–21. [DOI] [PubMed] [Google Scholar]
- [15].Quesenberry PJ, Aliotta JM, Cellular phenotype switching and microvesicles, Adv Drug Deliv Rev 62(12) (2010) 1141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kishore R, Khan M, More Than Tiny Sacks: Stem Cell Exosomes as Cell-Free Modality for Cardiac Repair, Circ Res 118(2) (2016) 330–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang S, Chuah SJ, Lai RC, Hui JHP, Lim SK, Toh WS, MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity, Biomaterials 156 (2018) 16–27. [DOI] [PubMed] [Google Scholar]
- [18].Sharma P, Mesci P, Carromeu C, McClatchy DR, Schiapparelli L, Yates JR, Muotri AR, Cline HT, Exosomes regulate neurogenesis and circuit assembly, Proceedings of the National Academy of Sciences 116(32) (2019) 16086–16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rani S, Ryan AE, Griffin MD, Ritter T, Mesenchymal Stem Cell-derived Extracellular Vesicles: Toward Cell-free Therapeutic Applications, Mol Ther 23(5) (2015) 812–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire CA, Chen JW, Tannous BA, Breakefield XO, Dynamic Biodistribution of Extracellular Vesicles in Vivo Using a Multimodal Imaging Reporter, ACS Nano 8(1) (2014) 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Langer R, Drug delivery and targeting, Nature 392(6679 Suppl) (1998) 5–10. [PubMed] [Google Scholar]
- [22].Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V, PLGA-based nanoparticles: An overview of biomedical applications, Journal of Controlled Release 161(2) (2012) 505–522. [DOI] [PubMed] [Google Scholar]
- [23].Wei G, Pettway GJ, McCauley LK, Ma PX, The release profiles and bioactivity of parathyroid hormone from poly(lactic-co-glycolic acid) microspheres, Biomaterials 25(2) (2004) 345–52. [DOI] [PubMed] [Google Scholar]
- [24].Wei G, Jin Q, Giannobile WV, Ma PX, The enhancement of osteogenesis by nanofibrous scaffolds incorporating rhBMP-7 nanospheres, Biomaterials 28(12) (2007) 2087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Feng G, Zhang Z, Dang M, Zhang X, Doleyres Y, Song Y, Chen D, Ma PX, Injectable nanofibrous spongy microspheres for NR4A1 plasmid DNA transfection to reverse fibrotic degeneration and support disc regeneration, Biomaterials 131 (2017) 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Feng K, Sun H, Bradley MA, Dupler EJ, Giannobile WV, Ma PX, Novel antibacterial nanofibrous PLLA scaffolds, J Control Release 146(3) (2010) 363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wei G, Ma PX, Macroporous and nanofibrous polymer scaffolds and polymer/bone-like apatite composite scaffolds generated by sugar spheres, J Biomed Mater Res A 78(2) (2006) 306–15. [DOI] [PubMed] [Google Scholar]
- [28].Ma PX, Zhang R, Synthetic nano-scale fibrous extracellular matrix, J Biomed Mater Res 46(1) (1999) 60–72. [DOI] [PubMed] [Google Scholar]
- [29].Jin Q, Ma PX, Giannobile WV, Platelet-Derived Growth Factor Delivery via Nanofibrous Scaffolds for Soft-Tissue Repair, Adv Skin Wound Care 1 (2010) 375–381. [PMC free article] [PubMed] [Google Scholar]
- [30].Kuang R, Zhang Z, Jin X, Hu J, Gupte MJ, Ni L, Ma PX, Nanofibrous spongy microspheres enhance odontogenic differentiation of human dental pulp stem cells, Adv Healthc Mater 4(13) (2015) 1993–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kuang R, Zhang Z, Jin X, Hu J, Shi S, Ni L, Ma PX, Nanofibrous spongy microspheres for the delivery of hypoxia-primed human dental pulp stem cells to regenerate vascularized dental pulp, Acta Biomater 33 (2016) 225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang J, Liu X, Jin X, Ma H, Hu J, Ni L, Ma PX, The odontogenic differentiation of human dental pulp stem cells on nanofibrous poly(L-lactic acid) scaffolds in vitro and in vivo, Acta Biomaterialia 6 (2010) 3856–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang W, Dang M, Zhang Z, Hu J, Eyster TW, Ni L, Ma PX, Dentin regeneration by stem cells of apical papilla on injectable nanofibrous microspheres and stimulated by controlled BMP-2 release, Acta Biomater 36 (2016) 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ryner M, Stridsberg K, Albertsson A-C, von Schenck H, Svensson M, Mechanism of Ring-Opening Polymerization of 1,5-Dioxepan-2-one andl-Lactide with Stannous 2-Ethylhexanoate. A Theoretical Study, Macromolecules 34(12) (2001) 3877–3881. [Google Scholar]
- [35].Liu X, Ma PX, The nanofibrous architecture of poly(L-lactic acid)-based functional copolymers, Biomaterials 31(2) (2010) 259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Demond AH, Lindner AS, Estimation of Interfacial-Tension between Organic Liquids and Water, Environ Sci Technol 27(12) (1993) 2318–2331. [Google Scholar]
- [37].Gronthos S, Mankani M, Brahim J, Robey P, Shi S, Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo, Proceedings of the National Academy of Sciences of the United States of America 97(25) (2000) 13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang J, Ma H, Jin X, Hu J, Liu X, Ni L, Ma PX, The effect of scaffold architecture on odontogenic differentiation of human dental pulp stem cells, Biomaterials 32(31) (2011) 7822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang X, Li Y, Chen YE, Chen J, Ma PX, Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects, Nat Commun 7 (2016) 10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang J, Liu X, Li H, Chen C, Hu B, Niu X, Li Q, Zhao B, Xie Z, Wang Y, Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signaling pathway, Stem Cell Res Ther 7(1) (2016) 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Jung KO, Jo H, Yu JH, Gambhir SS, Pratx G, Development and MPI tracking of novel hypoxia-targeted theranostic exosomes, Biomaterials 177 (2018) 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rio DC, Ares M Jr., Hannon GJ, Nilsen TW, Purification of RNA using TRIzol (TRI reagent), Cold Spring Harb Protoc 2010(6) (2010) pdb prot5439. [DOI] [PubMed] [Google Scholar]
- [43].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods 25(4) (2001) 402–8. [DOI] [PubMed] [Google Scholar]
- [44].Mahmood T, Yang PC, Western blot: technique, theory, and trouble shooting, N Am J Med Sci 4(9) (2012) 429–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wei G, Jin Q, Giannobile WV, Ma PX, Nano-fibrous scaffold for controlled delivery of recombinant human PDGF-BB, J Control Release 112(1) (2006) 103–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Thery C, Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids, Current Protocols in Cell Biology, John Wiley & Sons, Inc; 2006, pp. 3.22.1–3.22.29. [DOI] [PubMed] [Google Scholar]
- [47].Asadi J, Ferguson S, Raja H, Hacker C, Marius P, Ward R, Pliotas C, Naismith J, Lucocq J, Enhanced imaging of lipid rich nanoparticles embedded in methylcellulose films for transmission electron microscopy using mixtures of heavy metals, Micron 99 (2017) 40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tang YT, Huang YY, Zheng L, Qin SH, Xu XP, An TX, Xu Y, Wu YS, Hu XM, Ping BH, Wang Q, Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum, Int J Mol Med 40(3) (2017) 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nuti N, Corallo C, Chan BM, Ferrari M, Gerami-Naini B, Multipotent Differentiation of Human Dental Pulp Stem Cells: a Literature Review, Stem Cell Rev 12(5) (2016) 511–523. [DOI] [PubMed] [Google Scholar]
- [50].van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R, Optical and non-optical methods for detection and characterization of microparticles and exosomes, J Thromb Haemost 8(12) (2010) 2596–607. [DOI] [PubMed] [Google Scholar]
- [51].Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM, Identification of a novel inhibitor of mitogen-activated protein kinase kinase, J Biol Chem 273(29) (1998) 18623–32. [DOI] [PubMed] [Google Scholar]
- [52].Dang M, Koh AJ, Jin X, McCauley LK, Ma PX, Local pulsatile PTH delivery regenerates bone defects via enhanced bone remodeling in a cell-free scaffold, Biomaterials 114 (2017) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Swanson W, Gong T, Zhang Z, Xiu K, Eberle M, Ma PX, Controlled Release of Odontogenic Exosomes from a Biodegradable Vehicle Mediates Dentinogenesis as a Novel Biomimetic Pulp Capping Therapy, Under Review (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Boudot T, Nicolas Petit N, Ritty S, Kieffer C, Flow-rate control in a microfluidic device, 2012. [Google Scholar]
- [55].Sonou T, Ohya M, Yashiro M, Masumoto A, Nakashima Y, Ito T, Mima T, Negi S, Kimura-Suda H, Shigematsu T, Mineral Composition of Phosphate-Induced Calcification in a Rat Aortic Tissue Culture Model, J Atheroscler Thromb 22(11) (2015) 1197–206. [DOI] [PubMed] [Google Scholar]
- [56].Henmi A, Okata H, Anada T, Yoshinari M, Mikami Y, Suzuki O, Sasano Y, Bone matrix calcification during embryonic and postembryonic rat calvarial development assessed by SEM-EDX spectroscopy, XRD, and FTIR spectroscopy, J Bone Miner Metab 34(1) (2016) 41–50. [DOI] [PubMed] [Google Scholar]
- [57].Kitami M, Kaku M, Rocabado JM, Ida T, Akiba N, Uoshima K, Prolonged Survival of Transplanted Osteoblastic Cells Does Not Directly Accelerate the Healing of Calvarial Bone Defects, J Cell Physiol 231(9) (2016) 1974–82. [DOI] [PubMed] [Google Scholar]
- [58].Poulos J, The limited application of stem cells in medicine: a review, Stem Cell Res Ther 9(1) (2018) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Diederichs S, Shine KM, Tuan RS, The promise and challenges of stem cell-based therapies for skeletal diseases: stem cell applications in skeletal medicine: potential, cell sources and characteristics, and challenges of clinical translation, Bioessays 35(3) (2013) 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dang M, Saunders L, Niu X, Fan Y, Ma PX, Biomimetic delivery of signals for bone tissue engineering, Bone Res 6 (2018) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Diomede F, Gugliandolo A, Cardelli P, Merciaro I, Ettorre V, Traini T, Bedini R, Scionti D, Bramanti A, Nanci A, Caputi S, Fontana A, Mazzon E, Trubiani O, Three-dimensional printed PLA scaffold and human gingival stem cell-derived extracellular vesicles: a new tool for bone defect repair, Stem Cell Res Ther 9(1) (2018) 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gandolfi MG, Gardin C, Zamparini F, Ferroni L, Esposti MD, Parchi G, Ercan B, Manzoli L, Fava F, Fabbri P, Prati C, Zavan B, Mineral-Doped Poly(L-lactide) Acid Scaffolds Enriched with Exosomes Improve Osteogenic Commitment of Human Adipose-Derived Mesenchymal Stem Cells, Nanomaterials (Basel) 10(3) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shi Q, Qian Z, Liu D, Sun J, Wang X, Liu H, Xu J, Guo X, GMSC-Derived Exosomes Combined with a Chitosan/Silk Hydrogel Sponge Accelerates Wound Healing in a Diabetic Rat Skin Defect Model, Front Physiol 8 (2017) 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhang K, Zhao X, Chen X, Wei Y, Du W, Wang Y, Liu L, Zhao W, Han Z, Kong D, Zhao Q, Guo Z, Han Z, Liu N, Ma F, Li Z, Enhanced Therapeutic Effects of Mesenchymal Stem Cell-Derived Exosomes with an Injectable Hydrogel for Hindlimb Ischemia Treatment, ACS Appl Mater Interfaces 10(36) (2018) 30081–30091. [DOI] [PubMed] [Google Scholar]
- [65].Akbari A, Jabbari N, Sharifi R, Ahmadi M, Vahhabi A, Seyedzadeh SJ, Nawaz M, Szafert S, Mahmoodi M, Jabbari E, Asghari R, Rezaie J, Free and hydrogel encapsulated exosome-based therapies in regenerative medicine, Life Sci 249 (2020) 117447. [DOI] [PubMed] [Google Scholar]
- [66].Henriques-Antunes H, Cardoso RMS, Zonari A, Correia J, Leal EC, Jimenez-Balsa A, Lino MM, Barradas A, Kostic I, Gomes C, Karp JM, Carvalho E, Ferreira L, The Kinetics of Small Extracellular Vesicle Delivery Impacts Skin Tissue Regeneration, ACS Nano 13(8) (2019) 8694–8707. [DOI] [PubMed] [Google Scholar]
- [67].Gupte MJ, Swanson WB, Hu J, Jin X, Ma H, Zhang Z, Liu Z, Feng K, Feng G, Xiao G, Hatch N, Mishina Y, Ma PX, Pore size directs bone marrow stromal cell fate and tissue regeneration in nanofibrous macroporous scaffolds by mediating vascularization, Acta Biomater 82 (2018) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Woo KM, Chen VJ, Ma PX, Nano-fibrous scaffolding architecture selectively enhances protein adsorption contributing to cell attachment, J Biomed Mater Res A 67(2) (2003) 531–7. [DOI] [PubMed] [Google Scholar]
- [69].Smith LA, Liu X, Hu J, Ma PX, The enhancement of human embryonic stem cell osteogenic differentiation with nano-fibrous scaffolding, Biomaterials 31(21) (2010) 5526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hu J, Smith LA, Feng K, Liu X, Sun H, Ma PX, Response of human embryonic stem cell-derived mesenchymal stem cells to osteogenic factors and architectures of materials during in vitro osteogenesis, Tissue Eng Part A 16(11) (2010) 3507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhu Y, Jia Y, Wang Y, Xu J, Chai Y, Impaired Bone Regenerative Effect of Exosomes Derived from Bone Marrow Mesenchymal Stem Cells in Type 1 Diabetes, Stem Cells Transl Med 8(6) (2019) 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Seemann R, Brinkmann M, Pfohl T, Herminghaus S, Droplet based microfluidics, Rep Prog Phys 75(1) (2012) 016601. [DOI] [PubMed] [Google Scholar]
- [73].Wang L, Liu Y, Zhang W, Chen X, Yang T, Ma G, Microspheres and microcapsules for protein delivery: strategies of drug activity retention, Curr Pharm Des 19(35) (2013) 6340–52. [DOI] [PubMed] [Google Scholar]
- [74].Sukreet S, Silva BVRE, Adamec J, Cui J, Zempleni J, Sonication and Short-term Incubation Alter the Content of Bovine Milk Exosome Cargos and Exosome Bioavailability (OR26-08-19), Current Developments in Nutrition 3(Supplement_1) (2019). [Google Scholar]
- [75].Prileszky TA, Ogunnaike BA, Furst EM, Statistics of droplet sizes generated by a microfluidic device, AIChE Journal 62(8) (2016) 2923–2928. [Google Scholar]
- [76].Busatto C, Pesoa J, Helbling I, Luna J, Estenoz D, Effect of particle size, polydispersity and polymer degradation on progesterone release from PLGA microparticles: Experimental and mathematical modeling, International Journal of Pharmaceutics 536(1) (2018) 360–369. [DOI] [PubMed] [Google Scholar]
- [77].Huang C-C, Narayanan R, Alapati S, Ravindran S, Exosomes as biomimetic tools for stem cell differentiation: Applications in dental pulp tissue regeneration, Biomaterials 111 (2016) 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang W, Bai X, Zhao B, Li Y, Zhang Y, Li Z, Wang X, Luo L, Han F, Zhang J, Han S, Cai W, Su L, Tao K, Shi J, Hu D, Cell-free therapy based on adipose tissue stem cell-derived exosomes promotes wound healing via the PI3K/Akt signaling pathway, Exp Cell Res 370(2) (2018) 333–342. [DOI] [PubMed] [Google Scholar]
- [79].Xu JF, Yang GH, Pan XH, Zhang SJ, Zhao C, Qiu BS, Gu HF, Hong JF, Cao L, Chen Y, Xia B, Bi Q, Wang YP, Altered microRNA expression profile in exosomes during osteogenic differentiation of human bone marrow-derived mesenchymal stem cells, PLoS One 9(12) (2014) e114627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang X, Omar O, Vazirisani F, Thomsen P, Ekstrom K, Mesenchymal stem cell-derived exosomes have altered microRNA profiles and induce osteogenic differentiation depending on the stage of differentiation, PLoS One 13(2) (2018) e0193059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Franceschi RT, Xiao G, Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways, J Cell Biochem 88(3) (2003) 446–54. [DOI] [PubMed] [Google Scholar]
- [82].Jun JH, Yoon WJ, Seo SB, Woo KM, Kim GS, Ryoo HM, Baek JH, BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity, J Biol Chem 285(47) (2010) 36410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, Franceschi RT, MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1, J Biol Chem 275(6) (2000) 4453–9. [DOI] [PubMed] [Google Scholar]
- [84].Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, Franceschi RT, Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor, J Biol Chem 284(47) (2009) 32533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO, Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells, Nat Cell Biol 9(6) (2007) 654–9. [DOI] [PubMed] [Google Scholar]
- [86].Gimona M, Pachler K, Laner-Plamberger S, Schallmoser K, Rohde E, Manufacturing of Human Extracellular Vesicle-Based Therapeutics for Clinical Use, Int J Mol Sci 18(6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lener T, Gimona M, Aigner L, Borger V, Buzas E, Camussi G, Chaput N, Chatterjee D, Court FA, Del Portillo HA, O’Driscoll L, Fais S, Falcon-Perez JM, Felderhoff-Mueser U, Fraile L, Gho YS, Gorgens A, Gupta RC, Hendrix A, Hermann DM, Hill AF, Hochberg F, Horn PA, de Kleijn D, Kordelas L, Kramer BW, Kramer-Albers EM, Laner-Plamberger S, Laitinen S, Leonardi T, Lorenowicz MJ, Lim SK, Lotvall J, Maguire CA, Marcilla A, Nazarenko I, Ochiya T, Patel T, Pedersen S, Pocsfalvi G, Pluchino S, Quesenberry P, Reischl IG, Rivera FJ, Sanzenbacher R, Schallmoser K, Slaper-Cortenbach I, Strunk D, Tonn T, Vader P, van Balkom BW, Wauben M, Andaloussi SE, Thery C, Rohde E, Giebel B, Applying extracellular vesicles based therapeutics in clinical trials - an ISEV position paper, J Extracell Vesicles 4 (2015) 30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


