Abstract
Background
Genetic variation at a multigene cluster at chromosome 3p21.31 and the ABO blood group have been associated with the risk of developing severe COVID-19, but the mechanism remains unclear. Complement activation has been associated with COVID-19 severity.
Objective
The aim of this study was to examine whether chromosome 3p21.31 and the ABO variants are linked to the activation of the complement cascade in COVID-19 patients.
Methods
We considered 72 unrelated European hospitalized patients with genetic data and evaluation of circulating C5a and soluble terminal complement complex C5b-9 (SC5b-9). Twenty-six (36.1%) patients carried the rs11385942 G>GA variant and 44 (66.1%) non-O blood group associated with increased risk of severe COVID-19.
Results
C5a and SC5-b9 plasma levels were higher in rs11385949 GA carriers than in non-carriers (P = 0.041 and P = 0.012, respectively), while C5a levels were higher in non-O group than in O group patients (P = 0.019). The association between rs11385949 and SC5b-9 remained significant after adjustment for ABO and disease severity (P = 0.004) and further correction for C5a (P = 0.018). There was a direct relationship between upper airways viral load and SC5b-9 in carriers of the rs11385949 risk allele (P = 0.032), which was not observed in non-carriers.
Conclusions
The rs11385949 G>GA variant, tagging the chromosome 3 gene cluster variation and predisposing to severe COVID-19, is associated with enhanced complement activation, both with C5a and terminal complement complex, while non-O blood group with C5a levels. These findings provide a link between genetic susceptibility to more severe COVID-19 and complement activation.
Keywords: COVID-19, Complement, SC5b-9, C5a, rs11385942 G>GA variant, ABO group
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection displays a wide range of clinical manifestations spanning from an asymptomatic condition, observed in almost 50% of individuals with evidence of upper respiratory viral replication [1], to coronavirus disease 2019 (COVID-19) [2,3]. The disease can be associated with minor upper airway manifestations or interstitial pneumonia that can evolve into severe and life-threatening acute respiratory distress syndrome (ARDS) [[2], [3], [4], [5]]. An exaggerated “hyper-immune” response to lower respiratory airways and systemic infection is believed to be involved in ARDS pathogenesis [6]. Furthermore, COVID-19 is associated with a range of non-respiratory conditions affecting multiple organs, including the heart, circulatory system, kidney and the liver, whose pathogenesis involves endothelial activation and the immune response against SARS-CoV-2 [7,8].
Older age, male sex, and dysmetabolism predispose to severe COVID-19 [9,10]. We showed that inherited genetic factors contribute to determine the predisposition to COVID-19 related respiratory failure [11]. The main risk factor for severe respiratory disease was identified at a chromosome 3 gene cluster locus, encompassing variation at several candidate genes potentially implicated in regulating bronchial physiology, viral entry, and immune response. These include SCL6A20 (solute carrier family 6, member 20), LZTFL1 (leucine zipper transcription factor like 1), FYCO1 (FYVE [ Fab 1, YOTB, Vac 1, EEA1] and coiled-coil domain autophagy adaptor 1) and the chemokine receptors CCR9 (C-C motif chemokine receptor 9), CXCR6 (C-X-C motif chemokine receptor 6), XCR1 (X-C motif chemokine receptor 1), with a possible impact extended on expression of the neighboring CCR1, CCR2 and CCR3 [[11], [12], [13]]. However, due to the high linkage disequilibrium of this region inherited from Neanderthals [14], it was not yet possible to pinpoint the causal variant(s), and the mechanism underlying the association between chromosome 3 cluster variation and severe COVID-19 remains to be defined. In the FOGS study, a recently characterized cohort of a referral center with genetic characterization, we showed the chromosome 3 variation (the top hit rs11385942 G>GA variant) was associated with the susceptibility to develop COVID-19 with respiratory failure [11,15]. Although chromosome 3 gene cluster variation has repeatedly been confirmed to represent the main genetic risk factor for COVID-19 severity [11,12,16], in the FOGS cohort rs11385942 did not have a major impact on biomarkers of systemic inflammation, including C reactive protein, ferritin, IL-6 and circulating neutrophils and lymphocytes [17]. Furthermore, ABO blood group was involved in the susceptibility to COVID-19, with carriage of blood O conferring modest protection. Again a sound mechanistic explanation for such association was not yet reported [11,12,[18], [19], [20]].
In patients with severe disease, activation of the complement cascade, which may be facilitated by genetic predisposition, has been linked with inflammation grade and activation of coagulation, a typical feature of COVID-19 leading to thrombotic complications [[21], [22], [23]]. This process may be directly triggered by the SARS-CoV-2 Spike protein [24] as well as by the antibodies against the virus themselves. We recently showed that in hospitalized patients complement activation was associated with disease severity and perturbation of the endothelium, a main target of SARS-CoV-2 infection [7,25]. Since it is not known whether the susceptibility to the activation of the complement pathway following SARS-CoV-2 infection contributes the inter-individual variability in COVID-19 severity, the aim of this study was to examine the possible association between complement activation and chromosome 3 gene cluster variation and/or ABO blood group in a well characterized cohort of hospitalized COVID-19 patients.
2. Patients and methods
2.1. Patients
We considered 72 patients, who were enrolled in both the FOGS [11,17] and the COHERENT studies [25] at the Fondazione IRCCS Ca’ Granda Milan, between March 16th and April 29th, 2020 (at the beginning of the 1st wave in Italy), for whom both genetic data and characterization of complement activation were available. To avoid confounders, we restricted the analysis to unrelated individuals of European descent [11,17]. These patients were selected by the larger case-control FOGS cohort of 508 cases and 890 controls up to June 2020, whose clinical features were previously reported [17]. The severity of COVID-19 was graded on the basis of clinical, radiological and laboratory data as follows: mild when the patients were kept under observation with low intensity care, moderate when they required intermediate care with non-invasive ventilation, or severe when they were admitted to an intensive care unit for mechanical ventilation [23,25].
Nasopharingeal swabs were tested for the presence of SARS-CoV-2 using a One-Step Reverse Transcription Real-Time PCR kit (GeneFinder COVID-19 Plus RealAmp kit, ELITe, platform InGenius ELITech, Torino, Italy) according to manufacturer instructions, as previously described [25]. The InGenius platform displays a sensibility of 1000 cp/ml and a specificity of 100%. A gene was considered detected when the cycle threshold (Ct) value resulted ≤ 45 while the assay was considered negative for a Ct value > 45.
The study was approved by the Ethics committee of the Fondazione IRCCS Ca’ Granda and was carried out in conformity with the 2013 revision of the Declaration of Helsinki and the code of Good Clinical Practice. All participants gave their written informed consent.
2.2. Genetic studies
The rs11385942 G>GA variant and ABO blood group were determined as previously described [11]; blood typing was confirmed by direct immunophenotyping in >95% of cases (100% concordance), while rs11385942 was confirmed by direct typing by Taqman assays in 16 cases (100% concordance) [17].
2.3. Complement activation products
Complement activation status was determined on the day of admission, as previously described [25]. Plasma levels of soluble C5b-9 (SC5b-9) were measured using a solid-phase assay (MicroVue Complement SC5b-9 Plus EIA kit, Quidel Corporation, San Diego, CA, USA); intra- and inter-assay coefficients of variation (CVs) were respectively 6.8% and 13.1%; the lower detection limit was 3.7 ng/mL. Plasma C5a levels were measured using an immunoenzymatic method (MicroVue Complement C5a EIA, Quidel Corporation); intra- and inter-assay CVs of <12%; the lower detection limit was 0.01 ng/ml.
2.4. Statistical analysis
Comparisons were performed by fitting outcomes with univariate and multivariate general linear models. Models were adjusted for confounding factors and genetic factors, as specified. Not normally distributed variables were log transformed before entering the models. Statistical analyses were carried out using the JMP professional 15.0 (SAS Institute, Cary, NC, USA) and R statistical analysis software version 4.0.2 (http://www.R-project.org/). P values < 0.05 were considered statistically significant.
3. Results
3.1. Study cohort
The clinical features of patients, stratified by carriage of the rs11385942 variant, are shown in Table 1 . In the present cohort, which includes all the disease stages, the rs11385942 GA risk allele tended to be associated with the group of severe COVID-19. Baseline biochemical features, including inflammatory and coagulation indices and upper airways viral load, were also comparable between carriers and non-carriers. However, risk variant carriers tended to be younger and had a significantly lower burden of cardiometabolic comorbidities (p = 0.0045).
Table 1.
Clinical features of the selected cohort of 72 European unrelated patients hospitalized for severe COVID-19 with concomitant evaluation of genetic risk factors and of complement status activation. Patients were stratified by the presence of rs11385942 insertion–deletion GA or G variant at locus 3p21.31; all carriers were heterozygous for the GA variant.
| rs11385942 |
P value | ||
|---|---|---|---|
| G/G | G/GA | ||
| N (%) | 46 (63.9) | 26 (36.1) | |
| Age, years mean ± SD | 69.6 ± 10.1 | 64.8 ± 14.2 | 0.15 |
| Sex, male/female (female %) | 28/18 (39.1) | 17/9 (34.6) | 0.80 |
| Blood group O/A/B/AB (O/A/B/AB %) | 17/21/4/4 (40.0/45.6/9.7/9.7) | 11/15/0/0 (42.3/57.7/0/0) | 0.051 |
| Severity* | 0.47 | ||
| Mild (%) | 18 (39.1) | 9 (34.6) | |
| Moderate (%) | 15 (32.6) | 7 (26.9) | |
| Severe (%) | 13 (28.3) | 10 (38.5) | |
| Comorbidities | |||
| Smoking, yes (%) | 7 (15.2) | 3 (11.5) | 0.74 |
| Previous heart disease, yes (%) | 12 (26.1) | 2 (7.7) | 0.069 |
| Renal failure, yes (%) | 6 (13.0) | 1 (3.8) | 0.41 |
| Diabetes, yes (%) | 10 (21.7) | 3 (11.5) | 0.35 |
| Hypertension, yes (%) | 23 (50.0) | 9 (34.6) | 0.23 |
| Obesity (%) | 8 (18.2) | 2 (8.3) | 0.47 |
| Cardiometabolic comorbidities, N mean ± SD | 0.7 ± 0.7 | 1.3 ± 1.0 | 0.0045 |
| Treatments, N^ mean ± SD | 2.11 ± 1.12 | 2.36 ± 0.81 | 0.30 |
| Outcomes | |||
| Mortality (%) | 8 (17.4) | 5 (19.2) | 1.0 |
| Stroke (%) | 3 (7.3) | 0 | 0.28 |
| Venous thrombosis (%) | 2 (4.6) | 1 (4.2) | 1.0 |
| Baseline biochemistry | |||
| Viral load, upper respiratory tract Ct mean ± SD | 28.3 ± 6.4 | 28.7 ± 5.8 | 0.76 |
| Hb, g/l mean ± SD | 12.7 ± 1.9 | 12.8 ± 2.2 | 0.55 |
| Platelets, 103/mm3 mean ± SD | 242 ± 80 | 275 ± 129 | 0.26 |
| Neutrophils/lymphocytes ratio mean ± SD | 7.23 ± 6.55 | 6.97 ± 6.70 | 0.87 |
| GGT, IU/l median (IQR) | 31 (15–63) | 51 (30–117) | 0.056 |
| AST, IU/l median (IQR) | 33 (19–61) | 35 (23–65) | 0.59 |
| ALT, IU/l median (IQR) | 56 (31–85) | 50 (36–84) | 0.95 |
| LDH, IU/l mean ± SD | 304 ± 128 | 349 ± 125 | 0.32 |
| CRP, mg/dl mean ± SD | 10.5 ± 6.8 | 9.0 ± 7.7 | 0.46 |
| Procalcitonin, mg/dl mean ± SD | 0.65 ± 1.31 | 0.45 ± 0.73 | 0.54 |
| Fibrinogen, μg/l mean ± SD | 548 ± 160 | 632 ± 184 | 0.11 |
| IL-6, pg/ml median (IQR) | 51 (36–64) | 80 (33–202) | 0.27 |
| Ferritin, ng/ml median (IQR) | 868 (515–1527) | 1291 (445–2104) | 0.37 |
| PT, % | 1.15 ± 0.20 | 1.18 ± 0.14 | 0.42 |
| aPTT, % | 0.97 ± 0.13 | 1.04 ± 0.25 | 0.17 |
Data are shown as absolute number, mean ± SD, median (IQR), prevalence (%), as required. *Severity: Severe: requirement of mechanical ventilation or ECMO; Moderate: CPAP or high flux oxygen ventilation; Mild: low flux oxygen support. ^among low-molecular weight heparin, corticosteroids, hydroxycloroquine, remdesivir, anakinra. Ct: cycle threshold of reverse transcriptase-polymerase chain reaction. ALT: aspartate aminotransferase. AST: alanine aminotransferase. GGT: gamma-glutamyl transpeptidase. LDH: Lactate dehydrogenase. CRP: C reactive protein.
3.2. Impact of rs11385942 on complement activation
Circulating C5a levels were not significantly associated with available circulating biomarkers of inflammation and tissue damage. On the other hand, terminal complement complex SC5b-9 levels were directly associated with markers of tissue damage, including AST (P = 0.038), GGT (P = 0.047), and nearly associated with LDH (P = 0.055), and CPK (P = 0.078), but not associated with ALT (P > 0.5). Among markers of inflammation, SC5b-9 levels were nearly associated with ferritin (P = 0.057), but not with CRP, IL-6 and neutrophils/lymphocytes ratio (p > 0.5).
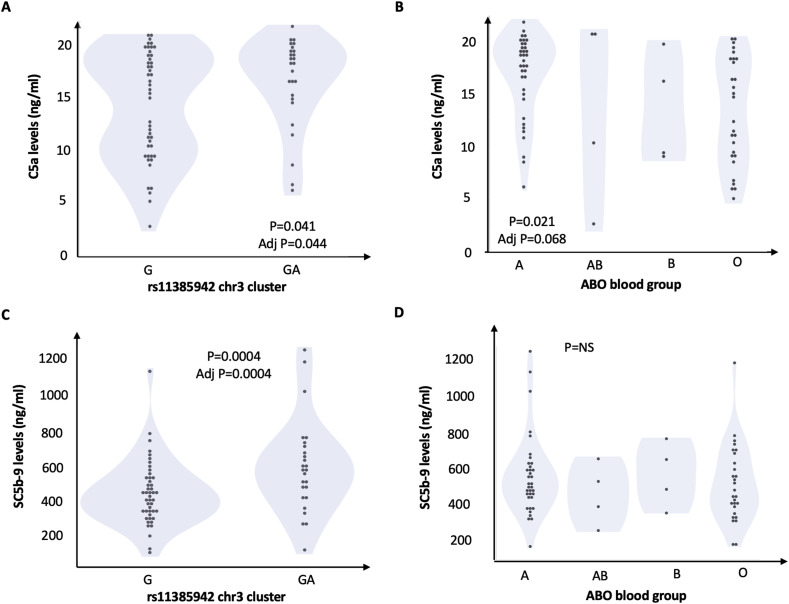
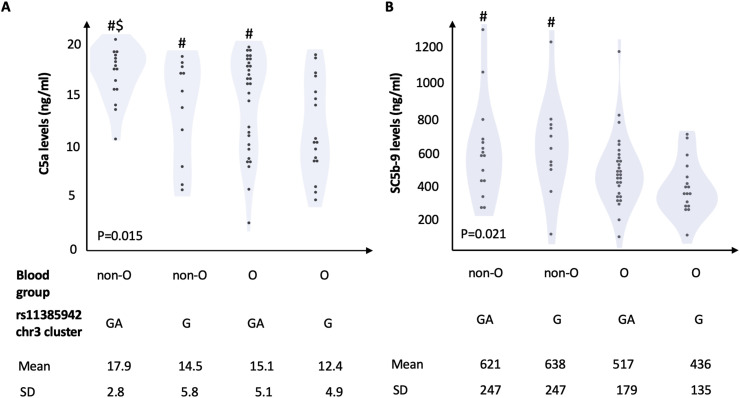
The impact of genetic predisposition on complement activation at baseline (hospital admission) is shown in Fig. 1 and in Table 2 . Plasma levels of both C5a and SC5-b9 were significantly higher in carriers of rs11385949 GA than in non-carriers (P = 0.041 and P = 0.012, respectively) and were higher in patients of non-O group than in patients carrying the O group, but only C5a reached statistical significance (P = 0.019), as shown in Table 2. The association between rs11385949 GA and SC5b-9 remained significant after adjustment for ABO and disease severity (P = 0.004) and after further correction for C5a levels (estimate 66 ± 65, P = 0.018 for rs11385949 GA allele; estimate 11 ± 5, P = 0.023 for C5a levels; other predictors were not significant). The combined effect of rs11385949 GA allele and O blood group on complement activation products (C5a and SC5b-9) is presented in Fig. 2 . Complement activation was higher in non-O blood group patients irrespective of rs11385949 (p < 0.05), while C5a levels were higher in rs11385949 GA carriers both in blood group O and non-O patients (p < 0.05).
Fig. 1.
Impact of genetic predisposition to COVID-19 on complement activation. Impact of rs11385942 G>GA and ABO on circulating C5a (panel A and B, respectively) and SC5b-9 (panel C and D, respectively). At generalized linear models unadjusted (P values), and multivariate models considering rs11385942, ABO blood groups, and disease severity (adj P).
Table 2.
Impact of the genetic background on complement activation at baseline and follow-up (30 days after admission) in 72 hospitalized patients with COVID-19. Follow-up data are available in a subset of 26 patients.
| C5a, ng/ml |
||||||
|---|---|---|---|---|---|---|
| rs11385942 |
P value | Non-O blood group |
P value | |||
| G | GA | Yes | No | |||
| Baseline | 14.1 ± 5.1 (n = 46) | 16.5 ± 5.3 (n = 26) | 0.041 | 16.1 ± 4.6 (n = 44) | 13.2 ± 5.1 (n = 28) | 0.019 |
| Follow-up | 13.4 ± 3.3 (n = 16) | 10.2 ± 4.2 (n = 10) | 0.054 | 11.3 ± 4.0 (n = 13) | 13.1 ± 3.7 (n = 13) | 0.25 |
| Differencea | −1.7 ± 14.3 | −5.8 ± 3.1 | 0.025 | −4.2 ± 15.2 | −2.3 ± 12.5 | 0.043 |
| SC5b-9, ng/ml |
||||||
|---|---|---|---|---|---|---|
| rs11385942 |
P value | Non-O blood group |
P value | |||
| G | GA | Yes | No | |||
| Baseline | 487 ± 167 (n = 46) | 628 ± 243 (n = 26) | 0.012 | 552 ± 208 (n = 44) | 516 ± 209 (n = 28) | 0.47 |
| Follow-up | 371 ± 158 (n = 16) | 367 ± 103 (n = 10) | 0.93 | 366 ± 165 (n = 13) | 372 ± 109 (n = 13) | 0.91 |
| Differencea | −121 ± 432 | −220 ± 477 | 0.17 | −142 ± 443 | −176 ± 454 | 0.63 |
For matched pairs (only patients with available follow-up were included); p = 0.001 and p < 0.0001 in the whole series for a decrease at follow-up of C5a and SCC5b-9, respectively.
Fig. 2.
Combined impact of rs11385942 G>GA and non-O blood group on complement activation. P values are reported for ANOVA test; #P < 0.05 vs. group O, rs11385942 G allele; $ P < 0.05 vs. group O, rs11385942 GA allele.
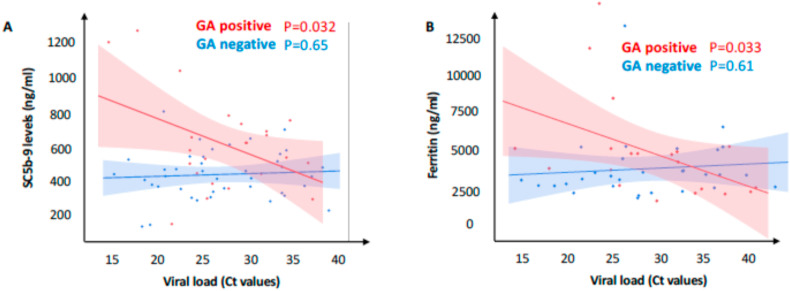
The relationship between upper airways viral load and terminal complement complex in patients stratified by rs11385949 GA is shown in Fig. 3 A. Remarkably, there was a significant direct relationship between upper airways viral load and SC5b-9 levels in carriers of the GA risk allele (estimate 18.8 ± 8.4 ng/ml of SC59b-9 per cycle at which the viral nucleic acids were detected by real-time PCR (cycle threshold); P = 0.032). The negative correlation of SC5b-9 levels with cycle threshold indicates a positive correlation with the log2 viral load, which was not observed in non-carriers despite the larger statistical power (estimate −1.4 ± 3.2 ng/ml of SC59b-9 per cycle; P = 0.65). Notably, we also observed a specific correlation between viral load and ferritin (shown in Fig. 3B) only in carriers of rs11385949 GA (estimate −118 ± 52 ng/ml of ferritin per cycle threshold; P = 0.033), but not in non-carriers (estimate 15 ± 30 ng/ml of ferritin per cycle threshold; P = 0.61).
Fig. 3.
Correlation between upper airways viral load, ferritin and soluble complement terminal complex (SC5b-9) in COVID-19 patients stratified by carriage of the rs11385942 GA risk variant. Relationship between upper respiratory viral load (Ct threshold) and SC5b-9 (A), and ferritin (B). Regression lines and 95% c.i. are shown; associations were tested at linear regression models.
In the subset of patients where the information was available the impact of rs11385949 and ABO blood group on complement activation disappeared at follow-up (at 30 days after admission during remission of symptoms in surviving patients) with a larger decrease in at risk genotypes for C5a (Table 2).
Overall, these data show that complement activation, which parallels COVID-19 severity, is associated with rs11385949 GA that is a recognized risk factor for the development of severe respiratory involvement.
4. Discussion
In this study, we examined whether the genetic predisposition to severe COVID-19 with respiratory failure, in particular the rs11385949 G>GA variant at the chromosome 3 gene cluster [[11], [12], [13]], influences complement activation in hospitalized patients. We were prompted by evidence that: a) the mechanism linking rs11385949 with respiratory failure in individuals infected by SARS-CoV-2 is still unclear, but may encompass modulation of the recruitment/activation of inflammatory cells [11], b) the complement cascade is activated during COVID-19 paralleling disease severity [21,22,25,26].
The main finding was that rs11385949 was associated with increased circulating C5a and SC5b-9. In line with the hypothesis that the mechanism linking chromosome 3 variation with the susceptibility to develop more severe COVID-19 encompasses complement activation in response to SARS-CoV-2 infection, SC5b-9 levels correlated with upper respiratory airways viral load specifically in carriers of the rs11385949 GA risk allele. The loss of association during remission in a patients’ subset suggests that the rs11385949 GA risk allele is associated with complement activation only when the SARS-CoV-2 infection is present. Remarkably, SC5b-9 levels correlated with biomarkers of tissue damage, e.g. biomarkers of necrosis such as LDH, but not with classic markers of systemic inflammation, suggesting that activation of the complement cascade may contribute in an independent manner in determining lung damage. This hypothesis is consistent with experimental evidence that SARS-CoV-2 Spike protein is able to trigger complement activation [24], whose inhibition prevents severe lung injury in mouse models of SARS coronavirus infection 1 [27,28].
In this study, terminal complement complex was also associated with higher GGT, possibly reflecting biliary damage, as biliary cells have been highlighted as a possible target for SARS-CoV-2 infection due to high expression of ACE2 [29]. However, the mechanism whereby chromosome 3 cluster variation facilitates complement activation during SARS-CoV-2 infection remains to be determined. Although we did not observe an impact of rs11385949 GA on upper respiratory airways viral load, it cannot be ruled out that the variant may facilitate viral replication in lower respiratory airways or other organs. However, we found that in carriers of the rs11385949 GA risk variant, viral load parallels not only complement activation but also ferritin, reflecting dysregulation of the immune response with increased monocyte/macrophage activation [30]. As rs11385949 tags genetic variation of a large gene cluster encoding several chemokine receptors directly influencing their expression (e.g. by upregulating CCR2), it could be speculated that it affects the recruitment of inflammatory cells in the lower airways or in vascular tissues, but additional studies are required to clarify the molecular pathways.
Interestingly, in the present cohort we found that rs11385949 GA risk allele carriers developed COVID-19 in the presence of a less severe burden of cardiometabolic comorbidities, a major risk factor for this condition [9,10]. Although complement activation, endothelialithis, systemic inflammation and thrombotic complications increase together with the severity of COVID-19 and are all induced in patients with the most severe disease manifestations [8,25], this observation highlights a potential source of heterogeneity among patients in the initial trigger of this process culminating with respiratory failure. Indeed, in those at increased risk due to carriage of chromosome 3 variation, although inflammatory pathways and the coagulation cascade are also activated, the complement cascade may play a role in sustaining aggressive disease even in the absence of cardiometabolic risk factors mediating endothelial damage and thrombosis through alternative pathways.
In parallel, we examined the impact of ABO blood group, as group O was associated with reduced risk of COVID-19, whereas the protection towards more severe outcomes in hospitalized patients is still disputed and may be modulated by genetic modifiers [11,12,[18], [19], [20]]. Interestingly, we found that group O is associated with reduced C5a and SC5b-9 levels, but only C5a reached statistical significance. It could be therefore speculated that patients with non-O group are more prone to an exaggerated activation of complement, possibly leading to a progression of tissue damage towards more severe lung disease.
Although we were able to correlate genetic background with a biomarkers panel in a well characterized cohort, limitations of this study encompass the retrospective monocentric design, limited sample size, lack of evaluation of individuals of non-European ancestry, and lack of assessment of the impact on an extended panel of cytokines and effector molecules. Independent replication in larger cohorts should therefore be obtained before ruling out a consensual minor impact of genetic predisposition on inflammatory markers and being able to draw reliable conclusions.
5. Conclusions
In summary, we found that the rs11385949 G>GA variant, tagging the chromosome 3 gene cluster variation responsible for severe COVID-19 predisposition, is associated with complement pathway activation. Indeed, rs11385949 impacted on both C5a and SC5b-9, while non-O blood group only on C5a levels. Furthermore, we found that complement activation and circulating ferritin parallel viral replication specifically in carriers of the rs11385949 risk variant. These findings provide a possible pathway (i.e. complement activation) linking genetic susceptibility to severe COVID-19 with a stronger inflammatory response that may be critical for disease pathogenesis. At the same time, they are consistent with a causal role of complement activation in determining lung damage. Inhibition of complement activation should be further investigated as therapeutic strategy for severe COVID-19, particularly in carriers of chromosome 3 gene cluster variant.
Author contributions
Luca Valenti: Conceptualization, Investigation, Data Curation, Writing - Original Draf; Samantha Griffini: Investigation, Writing - Review & Editing; Giuseppe Lamorte: Investigation, Writing - Review & Editing; Elena Grovetti: Investigation, Writing - Review & Editing; Sara Colonia Uceda Renteria: Investigation, Writing - Review & Editing; Francesco Malvestiti: Investigation, Writing - Review & Editing; Luigia Scudeller: Investigation, Writing - Review & Editing; Alessandra Bandera: Investigation, Writing - Review & Editing; Flora Peyvandi: Investigation, Writing - Review & Editing; Daniele Prati: Investigation, Writing - Review & Editing; Pier Luigi Meroni: Investigation, Writing - Review & Editing; Massimo Cugno: Investigation, Data Curation, Writing - Review & Editing, Supervision.
Funding
Ricerca Finalizzata Ministero della Salute RF-2016-02364358; The European Union (EU) Programme Horizon 2020 (under grant agreement No. 777377) to LV. Bando Ricerca COVID-19- Ministero della salute: COVID-2020-12371808 to PLM.
Declaration of competing interest
The authors declare that they have no relevant conflicts of interest.
References
- 1.Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the lombardy region. Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 8.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. J. Am. Med. Assoc. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwok S., Adam S., Ho J.H., Iqbal Z., Turkington P., Razvi S., et al. Obesity: a critical risk factor in the COVID-19 pandemic. Clin Obes. 2020;10 doi: 10.1111/cob.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severe Covid G.G., Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., et al. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shelton J.F., Shastri A.J., Ye C., Weldon C.H., Filshtein-Somnez T., Coker D., et al. Trans-ethnic analysis reveals genetic and non-genetic associations with COVID-19 susceptibility and severity. medRxiv. 2020 doi: 10.1101/2020.09.04.20188318. [DOI] [PubMed] [Google Scholar]
- 13.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020:2020. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 14.Zeberg H., Paabo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 15.Bandera A., Aliberti S., Gualtierotti R., Baldini M., Blasi F., Cesari M., et al. COVID-19 Network: the response of an Italian Reference Institute to research challenges about a new pandemia. Clin. Microbiol. Infect. 2020;26:1576–1578. doi: 10.1016/j.cmi.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in Covid-19. Nature. 2020 doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 17.Bianco C., Baselli G., Malvestiti F., Santoro L., Pelusi S., Manunta M., et al. Genetic insight into COVID-19-related liver injury. Liver Int. 2020 doi: 10.1111/liv.14708. [DOI] [PubMed] [Google Scholar]
- 18.Valenti L., Villa S., Baselli G., Temporiti R., Bandera A., Scudeller L., et al. Association of ABO blood group and secretor phenotype with severe COVID-19. Transfusion. 2020;60:3067–3070. doi: 10.1111/trf.16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnkob M.B., Pottegard A., Stovring H., Haunstrup T.M., Homburg K., Larsen R., et al. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020;4:4990–4993. doi: 10.1182/bloodadvances.2020002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoiland R.L., Fergusson N.A., Mitra A.R., Griesdale D.E.G., Devine D.V., Stukas S., et al. The association of ABO blood group with indices of disease severity and multiorgan dysfunction in COVID-19. Blood Adv. 2020;4:4981–4989. doi: 10.1182/bloodadvances.2020002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvelli J., Demaria O., Vely F., Batista L., Chouaki Benmansour N., Fares J., et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cugno M., Meroni P.L., Gualtierotti R., Griffini S., Grovetti E., Torri A., et al. Complement activation in patients with COVID-19: a novel therapeutic target. J. Allergy Clin. Immunol. 2020;146:215–217. doi: 10.1016/j.jaci.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J., Yuan X., Chen H., Chaturvedi S., Braunstein E.M., Brodsky R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D inhibition. Blood. 2020;136:2080–2089. doi: 10.1182/blood.2020008248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cugno M., Meroni P.L., Gualtierotti R., Griffini S., Grovetti E., Torri A., et al. Complement activation and endothelial perturbation parallel COVID-19 severity and activity. J. Autoimmun. 2020:102560. doi: 10.1016/j.jaut.2020.102560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regis Peffault de L., Anne B., Etienne L., Thibault D., Armance M., Lionel G., et al. Complement C5 inhibition in patients with COVID-19 - a promising target? Haematologica. 2020;105:2847–2850. doi: 10.3324/haematol.2020.260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gralinski L.E., Sheahan T.P., Morrison T.E., Menachery V.D., Jensen K., Leist S.R., et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.01753-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y., Zhao G., Song N., Li P., Chen Y., Guo Y., et al. Blockade of the C5a-C5aR axis alleviates lung damage in hDPP4-transgenic mice infected with MERS-CoV. Emerg. Microb. Infect. 2018;7:77. doi: 10.1038/s41426-018-0063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J., Aghemo A., Forner A., Valenti L. COVID-19 and liver disease. Liver Int. 2020;40:1278–1281. doi: 10.1111/liv.14470. [DOI] [PubMed] [Google Scholar]
- 30.Bermejo-Martin J.F., Gonzalez-Rivera M., Almansa R., Micheloud D., Tedim A.P., Dominguez-Gil M., et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit. Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]