Abstract
Numerous cancer drivers have been identified, but they are specific to a given cancer type and condition; universal cancer drivers and universal cancer mechanisms still remain largely unclear. Here, we identified the deadliest universal drivers for all cancers via developing algorithms to analyze massive RNAseqs and clinical data from The Cancer Genome Atlas (TCGA). In general, noncoding RNAs primarily serve as the most important inducers and suppressors for all types of cancers. In particular, pseudogenes are primary inducers, and specifically the antisense RNA RP11−335K5.2 serves as the most universal cancerous driver, independently of the cancer type and condition. Therefore, noncoding RNAs, instead of proteins as conventionally thought, primarily drive cancer, which establishes a novel field for future cancer research and therapy.
Keywords: Noncoding RNA, pseudogene, antisense RNA, cancer, driver, review
All cancers arise from genome abnormalities that share a certain degree of commonality (1-4), which is based on universal regulators for all cancers. Understanding these cancer regulators will contribute to the understanding of the fundamental common mechanisms of tumorigenesis and the development of therapeutic strategies against all cancers.
Numerous studies have aimed to identify universal cancerous drivers (2-6). Both genome sequences, functional genomics and biochemistry have been employed. It has been hypothesized that DNA mutations play a major role in driving tumorigenesis and mutations conserved across all cancers act as universal cancer drivers. Projects based on this hypothesis have produced a large number of patient DNA samples from various cancer types and identified thousands of mutations in both protein-coding regions and noncoding RNAs (2,6,7); however, they have not found consensus mutations across all cancer types. Most DNA mutations are specific to individual patients. For example, KRAS has been shown to be one of the most mutated genes in lung cancer, however, a KRAS mutation is present in less than 40% of lung cancer patients and in less than 2% of all cancer types (6). This indicates that this strategy to identify common mutations might not be an efficient way to identify universal cancer drivers. However, functional genomics and biochemistry studies have revealed a list of genes that perform similar functions across different cancer types (8-10). For example, TP53 has been identified as a cancer suppressor for an array of cancer types (11), suggesting that TP53 is a universal cancer regulator. Given the fact that more than 63,000 annotated genes, including protein-coding and noncoding RNAs, exist in the human genome (12), it is reasonable to hypothesize that a certain number of genes functionally serve as universal cancer regulators.
The identification of universal cancer regulators faces challenges. First, cancer genome abnormalities result from a combination of numerous factors, such as genetic heterogeneity, tissue type and a variety of epidemiological variables (13-16). Because of variations in these factors, the factor combination is complex in the human population and the identification of the causal relationship between cancer phenotype and primary factors is difficult. This poses difficulty in uncovering a conserved pattern of genomic changes that is consistent in all cancer types. Second, the current algorithms employed to identify cancer drivers have failed to comprise the complex factor combination. Conventionally, individual metrics (e.g. p-value derived from various analyses) have been employed to identify a cancer regulator (17), but it is unlikely for individual metrics to account for the majority of genome activation variations. The individual metrics obviously lead to biased results. Recent studies have employed several algorithms together to avoid biases from individual metrics (18). This certainly improves the variance interpretation, but it is uncertain if these algorithms are complementary or overlap and how much actual variances they can account for. Thus, all current results may be seriously biased.
In this present study, we systematically identified variance distributions in all cancers and found principal components that account for the majority of variances. Then, we developed algorithms to comprise all factors and most variances into a Cox proportional-hazards regression model (coxph), Furthermore, we also inserted stability-selection into the coxph to increase accuracy. These algorithms enabled us to identify universal cancer regulators, independent from of condition and cancer type. Our results revealed noncoding RNAs as primary cancer regulators, both as inducers and repressors. Strikingly, RP11−335K5.2, an antisense RNA, served as a universal cancer driver independent of any factor and condition. This finding advances our understanding of the fundamental drivers of tumorigenesis.
The Deadliest Cancerous Regulators
In order to identify the most important universal regulators of cancers, we need a gene regulation database that included all types of cancers and a software that can accurately infer the most important regulators. The Cancer Genome Atlas (TCGA) provides such a database, in which RNAseq and clinical data from all popular cancer types (36 types of cancers) are publicly available. Regulators inferred from this wide spectrum of cancer types should represent all cancers. We developed a software named ISURVIVAL (19) that can accurately infer an important regulator from its expression value and corresponding clinical variables, time and death status. ISURVIVAL inserts stability-selection into the conventional Cox proportional-hazards regression model (coxph), and the ISURVIVAL models increase inference accuracy. In contrast, the conventional coxph model lacks random sampling, leading to low accuracy.
We downloaded all available clinical and RNAseq samples (11,574) from a total of 36 cancer types. After applying quality control and filtering out samples that do not have clinical data matching RNAseq data, we obtained 9,457 samples, including 8,825 cancer and 632 normal samples. We applied ISURVIVAL model 1 (19) to estimate the significance of each gene from a cancer data matrix (8,825 samples) containing clinical data (time and status) and gene expression data. With a cutoff absolute coefficient >1 and a p-value<0.001, we selected 428 genes as the top most important regulators of all cancers. These 428 regulators were ranked on the basis of their absolute coefficients (coef). A regulator with a higher coefficient was more important in regulating cancer death. Details were deposited in our online database (20).
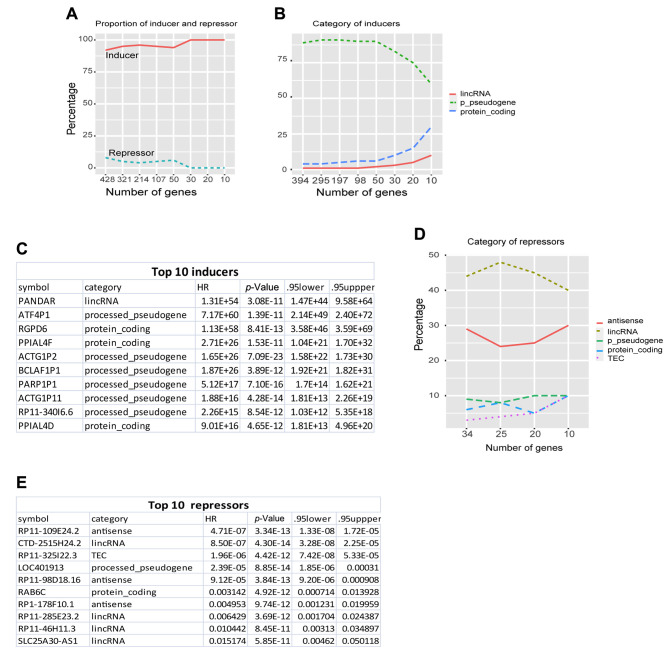
To further understand the composition of these 428 regulators, we calculated the proportion of inducers and repressors and their gene annotated categories. Among 428 regulators, 394 (92%) were cancer inducers (coef > 1, HR>2.72), and only 34 (<8%) were cancer repressors (coef< -1, HR< 0.37) (Figure 1A). Furthermore, 100% of the top 30 regulators were cancer inducers. Interestingly, among 394 inducers, more than 80% were processed-pseudogenes (p-pseudogenes) (Figure 1B). The top 10 inducers included 6 (60%) p-pseudogenes (Figure 1B and C). Among cancer suppressors, 40% and 30% were lincRNAs and antisense RNA, respectively (Figure 1D and E). This suggested that noncoding RNAs are the most important cancer regulators and pseudogenes are primary inducers. This is consistent with our recent study on cancer mechanisms revealing pseudogenes as the core drivers for all types of cancer (21).
Figure 1. The deadliest cancerous regulators. A, Proportion (%) of inducers and repressors of the most important cancer regulators. B, gene categories of the most important inducers. For clear illustration, only top gene categories with abundance of >10% are shown here and thereafter in this study. C, top 10 inducer list. D, gene categories of the top most important suppressors. E, top 10 suppressors list.
Activation Variability in Most Important Cancer Regulators
Genes that are consistently activated across all cancer types are of great interest because they can help to interpret the common mechanisms of all cancers and provide basic guidelines for therapy. To examine the activation consistency of the most important regulators identified above, we investigated whether they consistently function as inducers (positive activation, coef >0) or repressors (negative activation, coef<0) across different cancer types. After computing the coefficient of each gene in each cancer type, we found no consistence, but rather a high variability in regulator activation. From the top 10 inducers and suppressors identified above (Figure 1), only <60% and <67% of inducers and suppressors were, respectively, activated as inducers (coef>0) and repressors (coef<0) across different cancer types (Figure 2A, upper and bottom panel). For example, the top 1 inducer PANDAR (Figure 1C), did not always function as an inducer, and served as an inducer in only 57% of cancer types and as a suppressor in the rest.
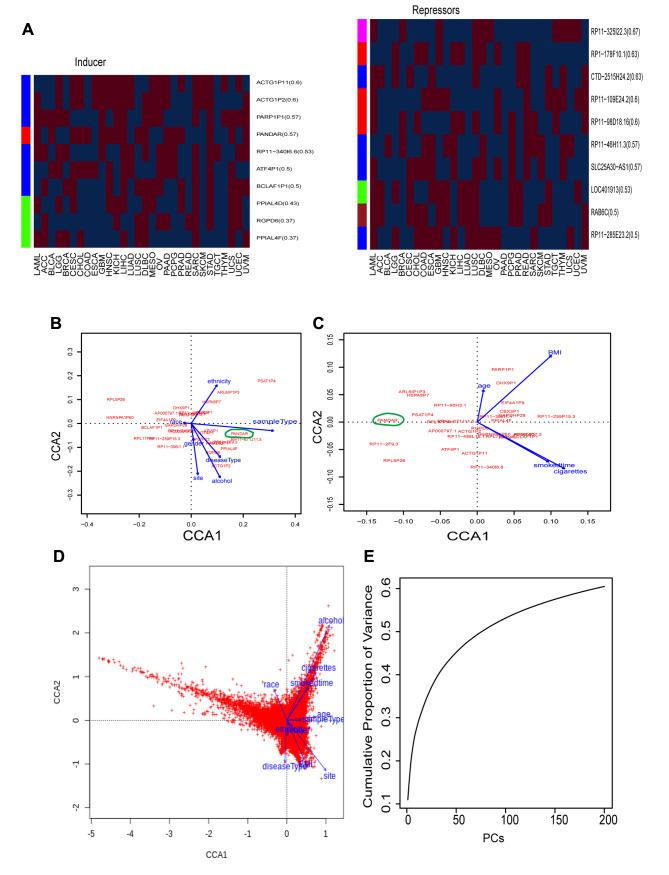
Figure 2. Personal clinic variables contributing to variations of cancerous gene activation. A, gene coefficient heatmap of the top 10 inducers (left panel) and suppressors (right panel) across 30 individual cancer types. Each row denotes a gene and each column a cancer type. For illustration proposes, red color denotes inducers (coef>0) and blue repressors (coef <0), while white colors as none (coefficient=0). The number following gene symbol represents the frequency (%) of this gene in 30 cancer types. For example, in the inducer panel (left panel), ACTG1P11(0.6) means it is an inducer occurring in 60% cancer types, 0.6=18 (red as inducer)/30 (total cancer types). Left bar colors represent gene categories. In the inducer panel (left), blue: processedpseudogenes (p-pseudogene, thereafter), red: lincRNA, green: proteincoding. In the suppressor panel (right), pink: TEC, red: antisense, blue: linRNA, green: p-pseudogene, brown: protein-coding gene (protein, thereafer). B, Canonical correspondence analysis (CCA) plot of the top 30 regulators and 7-character clinic variables from all cancer samples. C, CCA plot of the top 30 regulators and 4 digital clinic variables from all cancer samples. D, CCA plot of all genes and total 11 clinic variables from all cancer samples. Cancer genome was clearly seperated into 3 clusters based on clinic variables. E, the cumulative variance represented by each principal component (PC), from PC1 to PC200, derived from principal component analysis (PCA) of expression of all genes and all cancer samples.
Factors Contributing to the Variability of Regulator Activation
To understand the factors contributing to regulator activation variability, we systematically examined the relationship between clinical variables and gene activation by employing canonical correspondence analysis (CCA), which is a multivariate analysis to elucidate the multiple variable relationships between a variable matrix (e.g. a gene matrix) and another variable matrix (e.g. a clinical variable matrix). For clear illustration, we first run CCA on the top 30 regulators and clinical variables, including character and digital variables. In a CCA plot, each clinical variable was plotted as a vector, which carries both the amount of variance (line length) and the direction (line arrow). Among the 7-character variables, sample type and alcohol had the highest impact, the longest lines (Figure 2B), while smoking and BMI as the digital variables accounted for most of variances (Figure 2C). Interestingly, we found that PANDAR was positively correlated with sample type and negatively with race (Figure 2B), and it was significantly higher in primary tumors (p=5.8e-9, t-test) and Asian populations (p=0.00019). However, PANDAR did not seem to respond very well to cancer type and tissue (Figure 2B). Yet cancer type was classified based on tissue types, and this explained its high activation variability (only 57% consistence) in current classification system-disease type (Figure 2A upper panel). This also indicated PANDAR as a condition-dependent inducer instead of a universal cancer inducer.
The observation above encouraged us to extend our CCA analysis to all genes and all 11 clinical variables. The whole cancer genome was systematically separated into three clear sections (Figure 2D). The first was positively affected by alcohol and smoking, the second by site, BMI and disease type, and the third by unidentified factors. Astonishingly, alcohol served as the strongest factor altering cancer genome activation, even stronger than smoking and BMI (Figure 2D). This indicated that cancer genome activation is not only based on tissue as practiced by the current classification system, but also, importantly, other epidemiological variables.
Personal epidemiological variables vary, and factors contributing to cancer genome activation seem countless in the human population. To understand the systematic variance distribution caused by these unclear factors and personal variables, we performed principal component analysis (PCA) on genome expression of all cancers. Plotting the principal components (PCs) against the accumulated variance, we found that the first three PCs contributed to only 16% variance. PC1, PC2, and PC3 accounted only for 10%, 4%, 2% variance, respectively (Figure 2E). This suggested that any single metric only represents around 10% variance and the combination of the first three PCs, PC1-3, do not represent the overall variance very well as is the case of conventional practices; they normally represent >50% variance. This may explain why the current scheme based on combined individual metrics has failed so far to identify a universal cancer driver. Furthermore, the accumulated variance did not reach 50% until PC80 and was barely stable at 60% when PCs reached 200 (Figure 2E). This suggested that the number of factors contributing to cancer genome activation is much larger than expected and it is unlikely for any algorithm to uncover a universal cancer driver without integrating the variances of 200 PCs.
Noncoding RNAs Function as the Most Important Universal Drivers for all Cancers
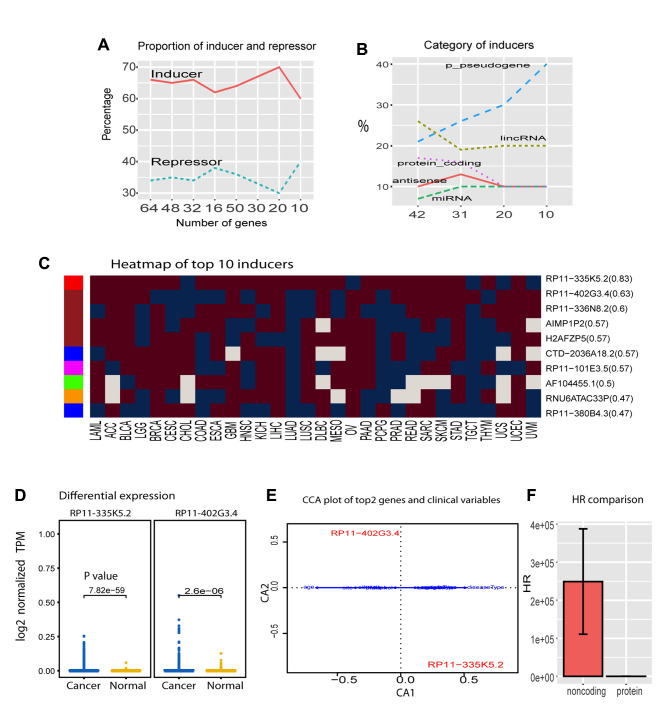
After learning the sources of complex variance for cancer genome activation, we developed the algorithm ISURVIVAL model 2 to comprise the majority of systems of variance to identify the most important drivers universal for all cancers (19). ISURVIVAL model 2 includes the systematic clinical variables and the first 200 PCs as covariates. To make an accurate estimation, we also inserted stability-selection with bootstrap random sampling into coxph model in ISURVIVAL model 2 (19). Applying ISURVIVAL model 2 to all RNAseqs, we found 64 significant regulators (p<0.05 and HR>1.1 or HR<0.9) deposited in (20). The 64 regulators contained 42 (65.6%) inducers (HR>1.1) and 22 (34.4%) repressors (HR<0.9) (Figure 3A). Among the inducers, noncoding RNAs dominated all profilings, accounting for more than 80% of the population. Especially p-pseudogenes represented 40% of the top 10, and only 1 protein encoding gene was within the top 10 (Figure 3B). As expected, these noncoding inducers were consistently activated in most cancer types (Figure 3C). The overall percentage of inducer activation was significantly higher than the previous one without incorporating personal clinic variables and PCs shown in Figure 2A (p=0.03151 for top 20 inducers, t-test). Strikingly, the top 1 regulator, antisense RP11-335k5.2, was activated in 83% of cancer types, and the pseudogenes RP11−402G3.4 also served as an inducer in >63% cancer types (Figure 3C). RP11-335k5.2 was also significantly activated between all cancer samples (8,825 samples) and all normal samples (632 samples) (Figure 3D). Furthermore, RP11-335k5.2 activation did not depend on any epidemiological variables according to our algorithm (Figure 3E), indicating RP11-335k5.2 as the most important universal cancer driver. This also suggested ISURVIVAL model 2 as a reliable algorithm to identify universal drivers.
Figure 3. Universal regulators of cancers. A, inducer and repressor proportion in total universal regulators of cancers. B, Gene categories of universal inducers for all cancers. C, coefficient heatmap of top 10 universal inducers in 30 cancer types. Left bar colors denote gene categories, red: antisense, brown: p-pseudogene, blue:lincRNA, pink: protein, green: miRNA, yellow: snRNA. D, differential expression of top 2 inducers between cancer and normal. E, CCA plot of top 2 inducers and clinical variables. F, HR comparison of noncoding RNAs and protein in top 42 inducers.
Consistently, RP11-335k5.2 was also shown, in our cancer network, to have a frequency score [total hits/total trails (22)] >0.95 and interact with CHRNB4 (cholinergic receptor nicotinic beta 4 subunit) (23). Frequency score normally represents universality; the high score of RP11-335k5.2 suggests that it is a universal inducer for all types of cancer. In contrast, TP53 has been widely reported as a universal regulator, but TP53 was not in our top 428 regulator list [detailed in (20)]. Consistently, TP53 does not show up in the cancer network with a frequency score >0.95, but only in the network with lower frequency score=0.9 (23), indicating that it is a regulator in most cases but does not have the universality of RP11-335k5.2.
To further understand the causal difference between noncoding RNAs and genes encoding proteins, we compared the HR between them in the 42 inducers selected above for all cancers. Noncoding RNA inducers had significant higher HR than protein encoding inducers (Figure 3F). These inducers were independent from any cancer type and clinical variable, suggesting noncoding RNAs as the most important universal drivers for all types of cancer.
All universal repressors were also noncoding RNAs, including p-pseudogenes, lincRNAs, TEC, antisense and sense_intronic. Taken together, these results suggested noncoding RNAs as the universal regulators for all cancers.
Noncoding RNAs Dominate the Most Important Hubs in Cancer Regulatory Network
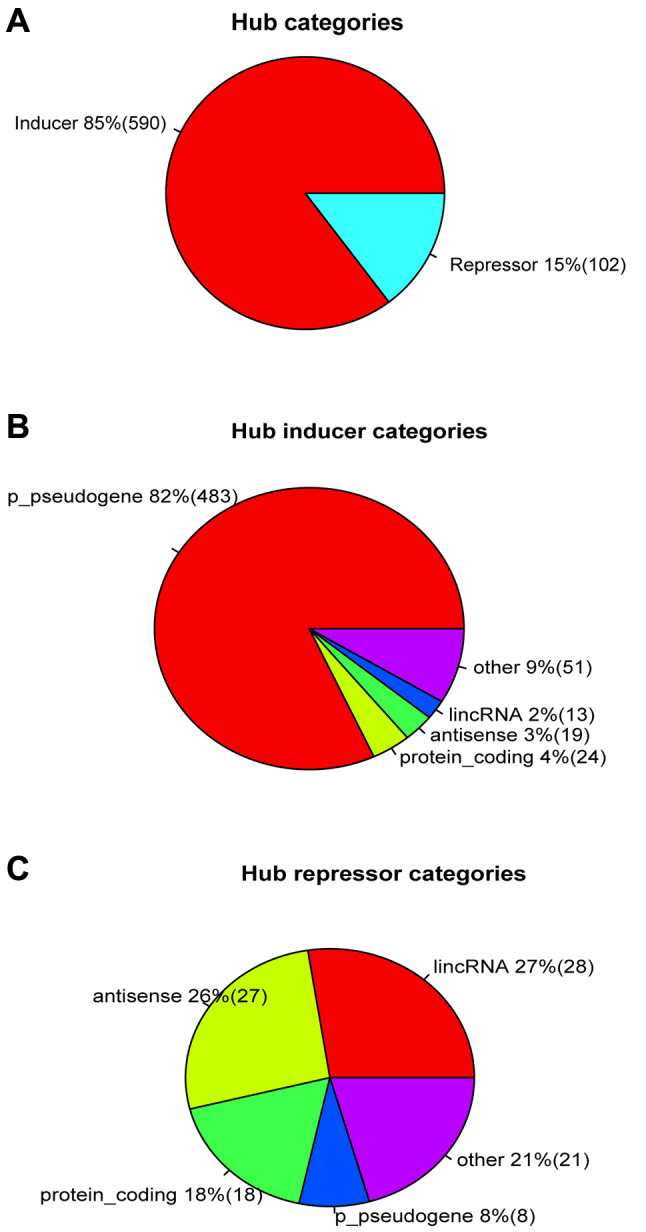
Network hubs play crucial functional roles in a regulatory network. We investigated the most important hubs in the cancer regulatory network established by us in our parallel study of the mechanism (21). We computed the network centrality to identify the top hubs for all types of cancer and then filtered these hubs with survival data (p<0.000001 and HR<0.5 or HR>1.5) to obtain the most important hubs, resulting in a total of 692 deadliest hubs. Among these 692 hubs, more than 85% (590 hubs) were inducers and only 15% (102 hub) were repressors (Figure 4A). Among the 590 inducers, 82% (483 hubs) were p_pseudogenes (Figure 4B), and only 4% protein coding genes. Thus, noncoding RNAs represented a total of 96% of inducers. Moreover, noncoding RNAs also comprised 82% of the strongest repressors, including lincRNAs, antisense, and protein encoding genes only accounted for 18% repressors (Figure 4C). These results further suggested noncoding RNAs, especially pseudogenes, as the most important drivers for all types of cancer.
Figure 4. Noncoding RNAs served as the primary hubs in cancer regulatory network. A, inducer and repressor proportion in network hubs. B, gene category proportion in inducer hubs. C, gene categories of repressor hubs.
Noncoding RNAs as Cancer Biomarkers
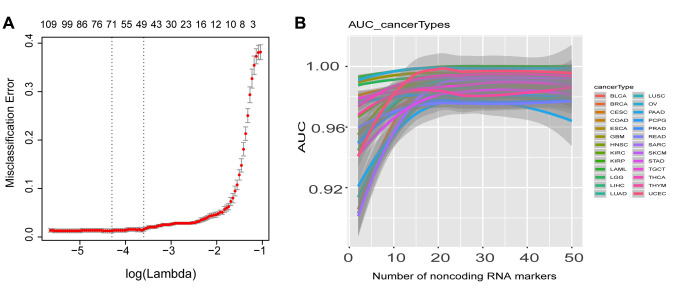
We next examined whether noncoding RNAs can be used as biomarkers to discriminate cancer and normal samples. We treated a total of 632 normal samples from the TCGA project as the control group and then used this normal group to discriminate cancer samples of each cancer type, and employed elastic-net with stability-selection to select biomarkers for each cancer type. Typically, around 50 noncoding RNAs could discriminate cancer from normal with misclassification error around 0 (Figure 5A). We used the top 50 noncoding RNAs in each cancer type as biomarkers to calculate the discrimination accuracy by computing AUC (area under the curve) of ROC (receiver operating characteristics). Beginning with the top 2 biomarkers, the AUC of each cancer type reached >90%, and with the top 20 biomarkers, the AUC reached a stable state (>96%) for all cancer types (Figure 5B). This indicated that noncoding RNAs can be used as biomarkers to discriminate cancer types.
Figure 5. Noncoding RNAs function as biomarkers to discriminate 30 cancer types. A, a typical cross-validation curve of noncoding RNAs as biomarkers. When around 50 noncoding RNAs were selected, the mean cross-validated error reached minimum, and this corresponding lambda was selected as lambda.min for biomarker selection for all 30 cancer types. B, AUC discriminating 30 cancer types when noncoding RNAs were used as biomarkers. When 10 noncoding RNAs were used as biomarkers, AUCs for most cancer types did not reach stable states, but when 20 noncoding RNAs were selected as biomarkers, all AUCs were stable, with AUC>0.96.
Discussion
This study revealed the most important universal regulators for all types of cancers. The universal regulators have been widely studied and various strategies have been developed for their identification. Most recent efforts have focused on identifying common mutations across cancers, but such mutations have been found with <2% consensus in cancers (6). Here, we found that RP11−335K5.2 was consistently induced in >83% of all cancer types regardless of any other condition. We have also found that RP11−335K5.2 also functioned as an inducer in a systems regulatory network (21), suggesting that it is a systems cancer universal driver. RP11−335K5.2 is more universal than TP53, which has been widely reported as a universal suppressor. TP53 did not show up on our top regulator list and our parallel mechanism study (21), indicating that TP53 is a regulator for most cases but it is not as universal as was previously thought. In addition, recent epigenetic sequencing technology has provided a powerful strategy to identify noncoding RNAs as universal regulators (24,25). For example, PANDAR has been found as a regulator in several cancer types. However, our data showed that PANDAR served as an inducer only in 57% of cancer types, and PANDAR is a condition-dependent regulator; it depends on sample type and race, suggesting that PANDAR is not a universal cancer driver. Taken together, these results suggested that the real universal cancer drivers have not been identified with the exemption of RP11−335K5.2 that was uncovered here.
Numerous studies have been performed on the biology of noncoding RNAs, which are associated with cancers through different suspectedness (26-28). Mutations in pseudogenes have also been identified in cancer (29), and a few pseudogenes have been recognized as cancer regulators (30). However, the current studies are biased to specific conditions and the systems role of noncoding RNAs on cancer genome remains unclear. This study and our systems network study revealed a clearer picture of the roles of noncoding RNAs in cancers, especially that of pseudogenes, which function as the most important cancer drivers (21), instead of protein-encoding genes as conventionally thought. Although we do not know the specific functions of RP11−335K5.2, our results, indicating that noncoding RNAs function as cancer drivers, advance our understanding of the mechanisms of tumorigenesis.
Noncoding RNAs comprise a significant fraction of the human genome and function as a flexible and energy-saving class of molecules. We are starting to recognize their importance in the genome, but understanding of their biological roles is still in an infant stage (30). The present study revealed that noncoding RNAs are the most important cancer drivers and suppressors, opens a field exploring their biological potentials in various physiological conditions as well as in therapy.
The failure of current studies to uncover universal cancer drivers results from the complexity of cancer phenotype, which is contributed by numerous factors, not only DNA sequences, but also complex epidemiological variables. These complex factors result in genome activation variation and measurement variability. Understanding the variance distribution that derives from these complex factors is essential for capturing the universal core from such complex data. However, current studies usually ignore this variance and only use individual metrics to identify cancer drivers. Our results showed only 10% variance for PC1. This 10% should be the maximum variance for any individual metric. The individual metrics could lose 90% variance, leading to biased results. To avoid biases resulting from the individual metrics strategy, recent studies combined several metrics to select cancer regulators (18). Combining metrics can slightly improve estimation, but the accumulated variance of PC1-3 only reached 16%. Therefore, improvement from the combination strategy is limited. This can be overcome by using 80 non-overlapped and complementary single metrics, in which variance could reach 50%, as shown in our data, but these 80 metrics are usually unavailable, and it is hard to develop such metrics. Therefore, all cancer drivers resulting from current strategies should still be biased and be condition-dependent. To solve this problem, we systematically studied the variation of genome activation, learned the variance distribution and then developed the algorithm ISURVIVAL model 2 to systematically combine clinical variables and 200 PCs with >60% variance plus stability-selection. This algorithm takes into account most of the variance in cancer genome activation and can produce unbiased universal drivers for all cancers.
Conflicts of Interest
No competing interests exist regarding this study.
Authors’ Contributions
A.W. designed this project, developed algorithms and software, analyses, wrote and edited this manuscript. R.H designed this project and edited this manuscript.
Acknowledgments
The Authors deeply thank Dr. Paul J Ride for reading and editing this manuscript. This work was supported by University of California Riverside initial funding.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol Poznan Pol. 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howard TP, Vazquez F, Tsherniak A, Hong AL, Rinne M, Aguirre AJ, Boehm JS, Hahn WC. Functional genomic characterization of cancer genomes. Cold Spring Harb Symp Quant Biol. 2016;81:237–246. doi: 10.1101/sqb.2016.81.031070. [DOI] [PubMed] [Google Scholar]
- 4.Calabrese C, Davidson NR, Demircioğlu D, Fonseca NA, He Y, Kahles A, Lehmann K-V, Liu F, Shiraishi Y, Soulette CM, Urban L, Greger L, Li S, Liu D, Perry MD, Xiang Q, Zhang F, Zhang J, Bailey P, Erkek S, Hoadley KA, Hou Y, Huska MR, Kilpinen H, Korbel JO, Marin MG, Markowski J, Nandi T, Pan-Hammarström Q, Pedamallu CS, Siebert R, Stark SG, Su H, Tan P, Waszak SM, Yung C, Zhu S, Awadalla P, Creighton CJ, Meyerson M, Ouellette BFF, Wu K, Yang H, Brazma A, Brooks AN, Göke J, Rätsch G, Schwarz RF, Stegle O, Zhang Z. Genomic basis for RNA alterations in cancer. Nature. 2020;578:129–136. doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, Satpathy AT, Mumbach MR, Hoadley KA, Robertson AG, Sheffield NC, Felau I, Castro MAA, Berman BP, Staudt LM, Zenklusen JC, Laird PW, Curtis C, Cancer Genome Atlas Analysis Network , Greenleaf WJ, Chang HY. The chromatin accessibility landscape of primary human cancers. Science. 2018;362:eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, Harsha B, Hathaway C, Jupe SC, Kok CY, Noble K, Ponting L, Ramshaw CC, Rye CE, Speedy HE, Stefancsik R, Thompson SL, Wang S, Ward S, Campbell PJ, Forbes SA. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Cancer Genome Atlas Research Network , Hu H. An Integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 9.Moya IM, Castaldo SA, Mooter LV den, Soheily S, Sansores-Garcia L, Jacobs J, Mannaerts I, Xie J, Verboven E, Hillen H, Algueró-Nadal A, Karaman R, Haele MV, Kowalczyk W, Waegeneer MD, Verhulst S, Karras P, Huffel L van, Zender L, Marine J-C, Roskams T, Johnson R, Aerts S, Grunsven LA van, Halder G. Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science. 2019;366:1029–1034. doi: 10.1126/science.aaw9886. [DOI] [PubMed] [Google Scholar]
- 10.Haddadi N, Lin Y, Travis G, Simpson AM, Nassif NT, McGowan EM. PTEN/PTENP1: “Regulating the regulator of RTK-dependent PI3K/Akt signalling”, new targets for cancer therapy. Mol Cancer. 2018;17:37. doi: 10.1186/s12943-018-0803-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 12.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 14.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 16.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. 2014;157:776–783. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, Ng PK-S, Jeong KJ, Cao S, Wang Z, Gao J, Gao Q, Wang F, Liu EM, Mularoni L, Rubio-Perez C, Nagarajan N, Cortés-Ciriano I, Zhou DC, Liang W-W, Hess JM, Yellapantula VD, Tamborero D, Gonzalez-Perez A, Suphavilai C, Ko JY, Khurana E, Park PJ, Van Allen EM, Liang H, MC3 Working Group, Cancer Genome Atlas Research Network , Lawrence MS, Godzik A, Lopez-Bigas N, Stuart J, Wheeler D, Getz G, Chen K, Lazar AJ, Mills GB, Karchin R, Ding L. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, Hornshøj H, Hess JM, Juul RI, Lin Z, Feuerbach L, Sabarinathan R, Madsen T, Kim J, Mularoni L, Shuai S, Lanzós A, Herrmann C, Maruvka YE, Shen C, Amin SB, Bandopadhayay P, Bertl J, Boroevich KA, Busanovich J, Carlevaro-Fita J, Chakravarty D, Chan CWY, Craft D, Dhingra P, Diamanti K, Fonseca NA, Gonzalez-Perez A, Guo Q, Hamilton MP, Haradhvala NJ, Hong C, Isaev K, Johnson TA, Juul M, Kahles A, Kahraman A, Kim Y, Komorowski J, Kumar K, Kumar S, Lee D, Lehmann K-V, Li Y, Liu EM, Lochovsky L, Park K, Pich O, Roberts ND, Saksena G, Schumacher SE, Sidiropoulos N, Sieverling L, Sinnott-Armstrong N, Stewart C, Tamborero D, Tubio JMC, Umer HM, Uusküla-Reimand L, Wadelius C, Wadi L, Yao X, Zhang C-Z, Zhang J, Haber JE, Hobolth A, Imielinski M, Kellis M, Lawrence MS, Mering C von, Nakagawa H, Raphael BJ, Rubin MA, Sander C, Stein LD, Stuart JM, Tsunoda T, Wheeler DA, Johnson R, Reimand J, Gerstein M, Khurana E, Campbell PJ, López-Bigas N, Weischenfeldt J, Beroukhim R, Martincorena I, Pedersen JS, Getz G. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578:102–111. doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang A. ISURVIVAL. Available at: combai.org/software/survival/:1, 2019. [Last accessed October 16, 2020]
- 20.Wang A. Deadliest Cancer Database. Available at: https://www.combai.org/database/deadliest/ [Last accessed October 16,2020]
- 21.Wang A, Hai R. Big-data analysis unearths the general regulatory regime in normal human genome and cancer. BioRxiv. 2019 doi: 10.1101/791970. [DOI] [Google Scholar]
- 22.Wang A, Hai R. FINET: Fast Inferring NETwork. BioRxiv. 2020 doi: 10.1101/733683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang A. Human cancer regulatory network. Available at:https://combai.org/network/cancer/ [Last accessed October 16, 2020]
- 24.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 25.Probert C, Curtis C. A role for chromatin regulatory dynamics in breast cancer evolution. Nat Med. 2018;24:1309–1311. doi: 10.1038/s41591-018-0182-8. [DOI] [PubMed] [Google Scholar]
- 26.Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin T, Hou P-F, Meng S, Chen F, Jiang T, Li M-L, Shi M-L, Liu J-J, Zheng J-N, Bai J. Emerging roles of p53 related lncRNAs in cancer progression: a systematic review. Int J Biol Sci. 2019;15:1287–1298. doi: 10.7150/ijbs.33218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooke SL, Shlien A, Marshall J, Pipinikas CP, Martincorena I, Tubio JMC, Li Y, Menzies A, Mudie L, Ramakrishna M, Yates L, Davies H, Bolli N, Bignell GR, Tarpey PS, Behjati S, Nik-Zainal S, Papaemmanuil E, Teixeira VH, Raine K, O’Meara S, Dodoran MS, Teague JW, Butler AP, Iacobuzio-Donahue C, Santarius T, Grundy RG, Malkin D, Greaves M, Munshi N, Flanagan AM, Bowtell D, Martin S, Larsimont D, Reis-Filho JS, Boussioutas A, Taylor JA, Hayes ND, Janes SM, Futreal PA, Stratton MR, McDermott U, Campbell PJ, ICGC Breast Cancer Group. Processed pseudogenes acquired somatically during cancer development. Nat Commun. 2014;5:3644. doi: 10.1038/ncomms4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature1298. [DOI] [PMC free article] [PubMed] [Google Scholar]