Abstract
Background/Aim: Prognosis of advanced stages of laryngeal squamous cell carcinoma (LSCC) remains poor. To clarify therapeutic targets and improve survival rate, identification of new specific and prognostic biomarkers of LSCC is required. The study aimed to evaluate the impact of IL-10:rs1800871, rs1800872, rs1800896 single nucleotide polymorphisms (SNPs), and IL-10 serum levels on LSCC development and determine associations of selected SNPs with patient survival rate. Patients and Methods: A total of 300 LSCC patients and 533 controls were included in the study. Genotyping was carried out using RT-PCR; IL-10 serum levels were analyzed by ELISA. Results: Significant associations were identified between IL-10 rs1800871 variants and advanced stage of LSCC patient group in the codominant, recessive and additive models (OR=0.473, p=0.027; OR=0.510, p=0.040; and OR=0.733; p=0.037). Significant variants of IL-10 rs1800872 were determined in the codominant, recessive and additive models (OR=0.473, p=0.027; OR=0.510, p=0.040; and OR=0.733, p=0.037). The distribution of IL-10 SNPs genotypes did not impact LSCC patient survival rate (respectively, p=0.952; p=0.952; p=0.991). Conclusion: IL-10:rs1800871 and rs1800872 SNPs are associated with advanced stage of LSCC. The genotypic distribution of IL-10 SNPs does not influence the survival rate of LSCC patients.
Keywords: Laryngeal squamous cell carcinoma, IL-10, rs1800871, rs1800872, rs1800896, gene polymorphism, serum, levels, survival rate
Laryngeal squamous cell carcinoma (LSCC) represents the largest subgroup of head and neck squamous cell carcinoma (HNSCC) and is one of the most common tumors in the respiratory system (1). In all respiratory tract malignancies, LSCC ranks second in terms of the mortality rate (2). LSCC is more commonly diagnosed in men than women (3). According to Global Cancer Observatory data, 154,977 new male and 22,445 female cases were registered in 2018, demonstrating almost seven-times higher incidence in the male population (4). According to racial disparities, younger age African Americans have a higher incidence and mortality compared to Caucasians (1). Furthermore, race is a prognostic factor for 5-year overall survival for white, black, Hispanic, and Asian patients; 60.6%, 52.7%, 59.5% and, 65.7%, respectively (5). Sophisticated diagnostics tools (video laryngostroboscopy, flexible endoscopy, contact endoscopy), surgical (endolaryngeal laser approach, partial laryngectomy, total laryngectomy) and/or nonsurgical options (advanced radiotherapy and/or chemotherapy) and multidisciplinary team-based decision making are used to choose the most suitable treatment modality for LSCC. However, LSCC patient survival rate of 1-, 3-, 5-, and 10- years have not significantly improved and remain relatively low: 81%, 62%, 53%, and 38%, respectively (6). The low survival rate, especially at advanced stages, might be due to little worrying symptoms such as hoarseness, which delay the diagnosis of LSCC and allow progression of the disease resulting in laryngeal stenosis with dyspnoea and stridor. Also, a lack of screening programs (endoscopic laryngeal examination performed by an otorhinolaryngologist is the first diagnostic tool in LSCC suspicion) in LSCC development indicates the necessity for further research to determine new specific blood- or tissue-based biomarkers to earlier diagnose this disease (7,8). To identify new biological immunomarkers and evaluate their correlation with the morphological and clinical manifestations of LSCC, recent interest has focused on the role of single nucleotide polymorphisms (SNP) in carcinogenesis (9-11).
Interleukin-10 (IL-10) is a pleiotropic cytokine that is originally called cytokine synthesis inhibitory factor (CSIF) and has structural similarities to interferon-gamma (INFγ) (12,13). In humans, IL-10 is encoded by the IL-10 gene and located on chromosome 1 (1q31-1q32) (14). Various cells can produce IL-10, including macrophages, mast cells, T cells regulatory subsets, dendritic cells, B cells, a cluster of differentiation (CD) 4+ and CD8+ T cells, and natural killer (NK) cells (12,15,16). The receptor of IL-10 is a two-receptor complex composed of two copies each of IL-10 specific chain – IL10R1 and IL10R2 proteins (17). Activated IL-10R complex induces signal transducer and activator of transcription 3 (STAT3) signaling via phosphorylation of the cytoplasmic tails of IL-10R1 and IL10R2 by Janus Kinase 1 (JAK1) and Tyrosine kinase 2 (Tyk2) (12,17). In unstimulated tissues, the expression of IL-10 is deficient, suggesting the importance of IL-10 for anti-inflammatory functions and indicating that provoking factors as massive inflammation are required for observation of elevated IL-10 levels (18).
IL-10 is known as an immunosuppressive cytokine that stimulates proliferation and metastasis of tumor cells (19). However, the role of IL-10 in carcinogenesis remains controversial and considered to be paradoxical (12,20,21). Various studies have demonstrated a positive correlation between IL-10 levels in serum and tumor tissues and a poor prognosis in melanoma, lung cancer, T/NK cells lymphomas, and HNSCC (22-26). Previous studies also suggested that tumor cells can produce IL-10 by themselves, and IL-10 might be a potential biomarker for IL-10-targeted therapy leading to the conclusion that antagonists of IL-10 might be used as a novel treatment of cancer patients (22-25,27,28). In contrast, the IL-10 anti-tumor effect is also demonstrated in murine tumor models where rapid tumor rejection that increased secretion of IL-10 was identified (12). Moreover, it is suggested that IL-10 plays a critical role in memory development and the function of appropriate CD8 T cells (29). Regarding the IL-10 role in anti-tumor immune responses, it can be presumed that the transcription activity of IL-10 or IL-10 protein structure could be modified by genetic polymorphisms, which could provide susceptibility to different oncological disorders involving HNSCC (30,31).
Several studies indicating the role of IL-10: rs1800872, rs1800871, rs1800896 polymorphisms in HNSCC have already been published (32-35). However, there is still a lack of data analyzing the associations between these polymorphisms and the pure group of LSCC patients and the impact on patients’ survival rate.
The objective of this study was to evaluate the impact of IL-10: rs1800871, rs1800871, and rs1800896 SNPs and IL-10 serum levels on LSCC development and to determine possible associations of selected SNPs with the survival rate of LSCC patients.
Patients and Methods
The present case-control study was carried out at the Department of Otorhinolaryngology, Lithuanian University of Health Sciences (LUHS), Kaunas, Lithuania, and the Laboratory of Ophthalmology, Neuroscience Institute, LUHS, Kaunas, Lithuania.
Ethics Statement. The study protocol was approved by the Kaunas Regional Ethics Committee for Biomedical Research, LUHS (authorization number BE-2-37). An Informed Consent Form, in accordance with the Declaration of Helsinki, was obtained from all subjects involved in the study.
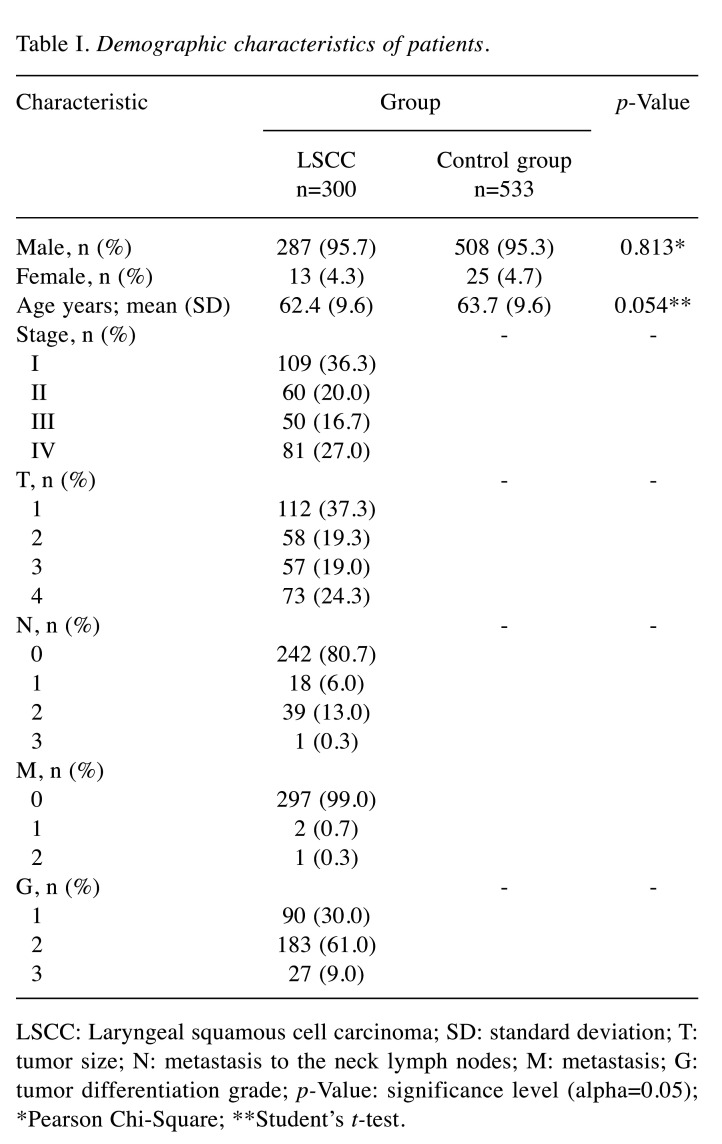
Study population. A total of 833 subjects were selected for the present study: 300 patients with LSCC and 533 healthy controls as a reference group. The demographic characteristics of LSCC patients and healthy control subjects are presented in Table I.
Table I. Demographic characteristics of patients.
LSCC: Laryngeal squamous cell carcinoma; SD: standard deviation; T: tumor size; N: metastasis to the neck lymph nodes; M: metastasis; G: tumor differentiation grade; p-Value: significance level (alpha=0.05); *Pearson Chi-Square; **Student’s t-test.
Selection of study population
LSCC group. A detailed otorhinolaringological examination (flexible endoscopy and/ or video laryngostroboscopy, neck palpation) was performed for all LSCC patients at the Out-patient Office of the Department of Otorhinolaryngology, LUHS. Peripheral venous blood samples collection was accomplished prior to preparation for general anaesthesia. Direct microlaryngoscopy with biopsy was performed for all patients. The diagnosis of LSCC was histologically confirmed at the Department of Pathology, LUHS. Clinical data were obtained by review of patients’ case records and personal interviews. The final diagnosis of LSCC was based on clinical data and the results of histological examination and laryngeal and neck CT or MRI data. The staging of LSCC was accomplished according to Guidelines for Head and Neck Cancers classification, Version 2.2020 accepted by National Comprehensive Cancer Network (NCCN) (36). Patients diagnosed with another type and localization of oncological disease, acute or chronic infectious diseases, as well as individuals using psychomotor suppressants and anti-epileptic drugs and persons younger than 18 years old, were rejected from this study.
Healthy controls. Patients who had an otorhinolaryngologist’s consultation at the Out-patient Office of the Department of Otorhinolaryngology, LUHS, and were selected for surgical treatment (nasal bones reposition, septoplasty, rhinoseptoplasty, tympanoplasty, ossiculoplasty, tympanostomy, uvulopalatopharyngoplasty, or radiofrequency thermoablation) were involved in the present study as healthy control group subjects. The collection of peripheral venous blood samples was accomplished from the same catheter to induce general anaesthesia. Patients who attended the family doctor’s consultation for the general check-up and had a collected peripheral blood sample were also included in this study. All the patients with the diagnosed oncologic disease, acute or chronic infectious diseases, individuals using psychomotor suppressants and anti-epileptic drugs, and persons younger than 18 years old were rejected from this study.
Subjects in the healthy control group were adjusted by age (p=0.054) and sex (p=0.813) according to demographic characteristics of the LSCC group (Table I).
SNP selection. The pathogenesis of LSCC on the molecular level remains unrevealed. According to published data, environmental carcinogens (37-42) can induce damage to DNA and cause genomic irregularity. Therefore, further studies based on genetic alterations are required.
Recently, the controversial role of IL-10 suggested that IL-10 might be a significant biomarker for IL-10 targeted treatment for oncological patients (28). In accordance with these findings, for the present study, we selected IL-10: rs1800872, rs1800871, rs1800896 SNPs to determine possible associations with LSCC development. Selected IL-10 SNPs are the most frequently analyzed in the scientific literature and are strongly related to malignancies (43-45).
IL-10: rs1800872, rs1800871, rs1800896. A few IL-10 promoter polymorphic sites have been characterized with determination of three polymorphisms from transcription initiation site at positions -819 (rs1800871), -592 (rs1800872) and -1082 (rs1800896) (-1082) (46). These IL-10 promoter polymorphisms have been the most widely investigated in various diseases: asthma, cardiovascular diseases, diabetic nephropathy, systemic lupus erythematosus, rheumatoid arthritis, chronic periodontitis, Alzheimer’s disease (47-53). Furthermore, these polymorphisms are strongly associated with various serum levels of IL-10 in vivo (54). Moreover, these three SNPs’ significant role was demonstrated in the carcinogenesis of lung, thyroid, gastric, cervical, prostate cancer, and oral squamous cell carcinoma (55-60).
A few studies analyzing IL-10: rs1800872, rs1800871, rs1800896 associations with HNSCC have been published (32-35,60). However, there is no data in the literature analyzing the association of these SNPs with the development of LSCC in a pure group of LSCC patients and the impact of these SNPs on the survival rate of LSCC patients.
DNA extraction, genotyping, and analysis of serum IL-10 levels. The DNA extraction and analysis of IL-10: rs1800872, rs1800871, rs1800896 gene polymorphisms were performed at the Laboratory of Ophthalmology, Neuroscience Institute, LUHS.
The DNA was extracted from peripheral venous blood samples (leucocytes) collected in 200 μl test-tubes utilizing the silica-based membrane technology using a genomic DNA extraction kit (GeneJET Genomic DNA Purification Kit, Thermo Fisher Scientific, Vilnius, Lithuania), according to manufacturer’s recommendations. The genotyping of IL-10: rs1800872, rs1800871, rs1800896 was carried out using the real-time polymerase chain reaction (PCR) method. Identification of all single-nucleotide polymorphisms (SNPs) was performed using TaqMan® Genotyping assays (Thermo Fisher Scientific, Inc, Pleasanton, USA). The genotyping was performed using a “StepOnePlus” real-time PCR quantification system (Thermo Fisher Scientific, Singapore). Data of individual genotypes were received using the Allelic Discrimination program during the real-time PCR.
IL-10 serum levels were evaluated in 20 control subjects and 20 LSCC patients. The assay was performed using the Invitrogen ELISA Kit (Cat. No. KHC0101) for human IL-10, standard curve sensibility range=0-500 pg/ml, sensitivity <1 pg/ml, following the manufacturer’s instructions, and analyzed on the Multiskan FC Microplate Photometer (Thermo Scientific, Waltham, MA) at 450 nm.
Quality control of genotyping. The repetitive analysis of 5% randomly chosen initial samples was accomplished for selected SNPs to confirm the same rate of genotypes from initial and repetitive genotyping.
Survival rate. The LSCC group data about the mortality rate, including the survival period after diagnosis of LSCC, were collected from the Lithuanian State Register of Death Cases and Their Causes, Institute of Hygiene, Vilnius.
Statistical analysis. The data about the study participants’ demographic characteristics are presented as absolute numbers with percentages in brackets and compared between the control group subjects and LSCC group using the Chi-square test. The frequencies of all SNPs genotypes and alleles are presented in percentages.
The Analysis of Hardy-Weinberg using a chi-square test was performed in the control group to compare the analyzed and expected frequencies of IL-10: rs1800872, rs1800871, rs1800896 SNPs. Chi-square test was used to compare the distribution of IL-10: rs1800872, rs1800871, rs1800896 SNPs in the LSCC, and control groups. Binomial logistic regression analysis with an adjusted odds ratio (OR) and its 95% confidence interval (95% CI) was utilized to evaluate the influence of IL-10: rs1800872, rs1800871, rs1800896 genotypes and alleles on LSCC development and the risk prediction for LSCC patients with these polymorphisms. The binomial logistic regression analysis results are represented as genetic models: codominant, dominant, recessive, overdominant, and additive. Akaike Information Criterion (AIC) was used for the best genetic model selection. Only statistically significant variables are presented in the tables.
Statistical analysis was performed using the SPSS/W 20.0 software (Statistical Package for the Social Sciences for Windows, Inc., Chicago, Illinois, USA). The findings were considered statistically significant when p<0.05.
Results
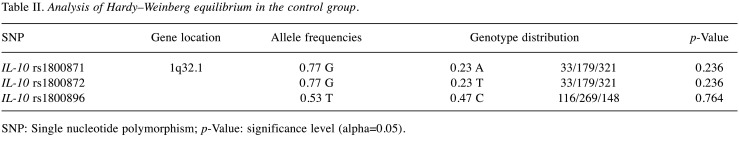
Hardy-Weinberg equilibrium (HWE) test was carried out between the control group subjects. The results demonstrated that genotypes of SNPs in the control group did not deviate from HWE (p>0.05) (Table II).
Table II. Analysis of Hardy–Weinberg equilibrium in the control group.
SNP: Single nucleotide polymorphism; p-Value: significance level (alpha=0.05).
Association between IL-10 SNPs and LSCC. In the present study, we performed genotyping of IL-10: rs1800872, rs1800871, and rs1800896 SNPs in 833 subjects (300 LSCC patients and 533 control group subjects) and studied possible associations between selected SNPs and LSCC development. Table III lists the frequencies of selected SNPs genotype profiles between the control group and LSCC patients. However, we did not find any statistically significant differences between the control group and LSCC patients (Table III).
Table III. Frequencies of genotypes and alleles of IL-10 rs1800871, rs1800872, and rs1800896 in the control and LSCC group.
LSCC: Laryngeal squamous cell carcinoma; p-Value: significance level (alpha=0.05).
Interestingly, we noticed that genotypes at position -819 (rs1800871) were changed synchronously with -592 (rs1800872) position. To evaluate the impact of IL-10 rs1800871, rs1800872, and rs1800896 on the LSCC development, a binomial logistic regression was applied. However, our results did not show any statistically significant results.
Genotypic and allelic distribution according to the development of LSCC by stages. To analyze the possible genotypic differences to the progress of LSCC, we divided the LSCC group into two subgroups: the early-stage LSCC group (composed of I and II stages LSCC patients) and the advanced-stage LSCC group (formed of III and IV stages LSCC patients) and compared them with control group subjects and between each other.
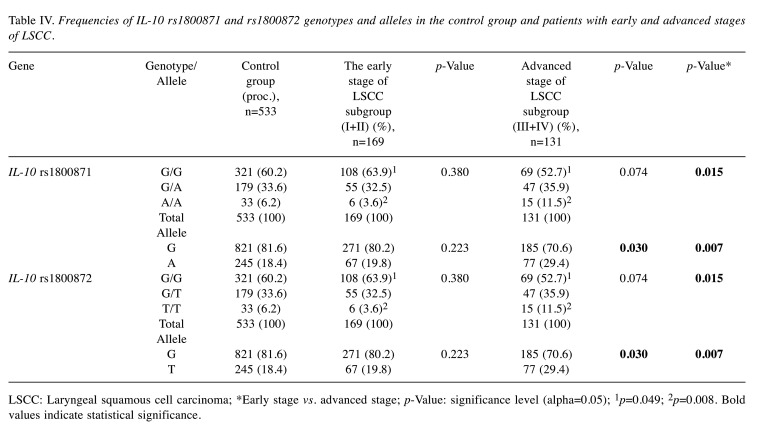
We found statistically significant differences in the allele distribution of IL-10 rs1800871 and rs1800872 in the advanced-stage LSCC subgroup compared to the control group: the G allele at rs1800871 was less frequent in the LSCC group than in controls, while the A allele was statistically significantly more frequent in the advanced stage of LSCC subgroup than in the control group (accordingly: 70.6% vs. 81.6% and 29.4% vs. 18.4%; p=0.030). Respectively, the G allele at 1800872 was less common in the advanced-stage LSCC subgroup than in controls, although the T allele at rs1800872 was statistically significantly more frequent in the advanced stage of LSCC subgroup than in the control group (accordingly: 29.4% vs.18.4%; p=0.030) (Table IV).
Table IV. Frequencies of IL-10 rs1800871 and rs1800872 genotypes and alleles in the control group and patients with early and advanced stages of LSCC.
LSCC: Laryngeal squamous cell carcinoma; *Early stage vs. advanced stage; p-Value: significance level (alpha=0.05); 1p=0.049; 2p=0.008. Bold values indicate statistical significance.
SNPs analysis between the early and advanced stages of LSCC revealed statistically significant differences in the genotypic distribution of IL-10 rs1800871 and rs1800872. We found out that the GG genotype at rs1800871 was more frequent in the early stage compared to the advanced stage of the LSCC subgroups (63.9% vs. 52.7%; p=0.049), while the AA genotype was less frequent in the early stage compared to the advanced stage of the LSCC subgroups (3.6% vs. 11.5%; p=0.008). Also, the G allele at rs1800871 was more frequent in the early stage LSCC subgroup compared to the advanced stage of LSCC patients (80.2% vs. 70.6%; p=0.007). The rs1800872 GG genotype was more frequent in the early stage compared to the advanced stage of the LSCC subgroup (63.9% vs. 52.7%; p=0.049), while the TT genotype was less frequent in the early stage compared to the advanced stage of the LSCC subgroup (3.6% vs. 11.5%, p=0.008). Also, the G allele at rs1800872 was more frequent in the early stage of LSCC comparing to the advanced stage of LSCC patients (80.2% vs. 70.6%, p=0.007) (Table IV).
Binomial logistic regression analysis was performed to evaluate the impact of selected IL-10 SNPs on LSCC development in the disease’s advanced stage. It revealed that IL-10 rs1800871 AA genotype is associated with about 0.5-fold decreased odds of LSCC development in the codominant and recessive models (OR=0.473; 95% CI=0.244-0.918; p=0.027 and OR=0.510; 95% CI=0.268-0.971; p=0.040, respectively). Also, each allele A is associated with 0.7-fold decreased odds of LSCC development in the additive model (OR=0.733; 95% CI=0.548-0.982; p=0.037) (Table V).
Table V. Binomial logistic regression analysis of IL-10 rs1800871 and rs1800872 in the control group and patients with the advanced stage of LSCC.
OR: Odds ratio; CI: confidence interval; p-Value: significance level (alpha=0.05); AIC: Akaike information criterion. Bold values indicate statistical significance.
Correspondingly, it was identified that the TT genotype at IL-10 rs1800872 was associated with about 0.5-fold decreased odds of LSCC development in the codominant and recessive models (OR=0.473; 95% CI=0.244-0.918; p=0.027 and OR=0.510; 95% CI=0.268-0.971; p=0.040). Equally, each allele T at IL-10 rs1800872 was associated with 0.7-fold decreased odds of LSCC development in the additive model (OR=0.733; 95% CI=0.548-0.982; p=0.037) (Table V).
Genotypic and allelic distribution according to the metastasis to the neck lymph nodes. To better understand the development of LSCC, we divided LSCC patients group into two subgroups: patients with no metastasis to the neck lymph nodes (NMNLN) subgroup and patients with metastasis to the neck lymph nodes (WMNLN) subgroup. The subgroups were compared with the control group separately and between each other. After statistical analysis, it was revealed that the GG genotype at rs1800871 was less frequent in the WMNLN subgroup than in control subjects (44.8% vs. 60.2%; p=0.024), while the AA genotype was more frequent in the WMNLN subgroup than in control subjects (17.2% vs. 6.2%; p=0.002). The GG genotype at rs1800872 was less frequent in the WMNLN subgroup than in control subjects (44.8% vs. 60.2%; p=0.024), while the TT genotype was more frequent in the WMNLN subgroup than in control subjects (17.2% vs. 6.2%; p=0.002). Also, the allele G in both SNPs (rs1800871 and rs1800872) was less frequent in the WMNLN subgroup than in the control group (63.8% vs. 81.6%; p=0.002 and 63.8% vs. 81.6%; p=0.002, respectively). The T allele at rs1800872 was more frequent in the WMNLN subgroup than in the control group (36.2% vs. 18.4%; p=0.002) (Table VI).
Table VI. Frequencies of IL-10 rs1800871 and rs1800872 genotypes and alleles in the control group and LSCC patients with no metastasis and with metastasis to the neck lymph nodes.
*No metastasis to neck lymph nodes vs. metastasis to neck lymph nodes; p-Value: significance level (alpha=0.05); 1p=0.024; 2p=0.002; 3p=0.015; 4p=0.001. Bold values indicate statistical significance.
Comparing the NMNLN and WMNLN subgroups between each other, we identified that the genotypes and alleles at rs1800871, and respectively at rs1800872, were more frequent in the NMNLN subgroup than in the WMNLN subgroup (p=0.001).
The GG genotype at rs1800871 was less frequent in the WMNLN subgroup than in the NMNLN subgroup (44.8% vs. 62.4%; p=0.015), while the AA genotype was more frequent (17.2% vs. 4.5%; p=0.001). Correspondingly, the GG genotype at rs1800872 was less frequent in the WMNLN subgroup than in the NMNLN subgroup (44.8% vs. 62.4%; p=0.015), while the TT genotype was more frequent (17.2% vs. 4.5%; p=0.001). Also, the G allele in both SNPs (rs1800871 and rs1800872) were less frequent in the WMNLN subgroup than in the NMNLN subgroup (63.8% vs. 78.9%; p=0.001) (Table VI).
After binomial logistic regression analysis, the statistically significant differences in the impact of IL-10 rs1800871 and rs1800872 on the WMNLN patients’ subgroup were revealed. It was clarified that genotype AA at IL-10 rs1800871 was associated with 3.7-fold increased odds of LSCC development in the WMNLN subgroup under the codominant model (OR=3.741; 95% CI=1.660-8.432; p=0.001) and 3.2- fold increased odds under the recessive model (OR=3.224; 95% CI=1.495-6.949; p=0.003). Both genotypes (AG+AA) under the dominant model were associated with 1.8-fold increased odds of LSCC with the WMNLN subgroup (OR=1.805; 95% CI=1.042-3.2137; p=0.035). Each allele A was associated with 1.8-fold increased odds of LSCC development in the WMNLN subgroup under the additive model (OR=1.799; 95% CI=1.211-2.672; p=0.004) (Table VII).
Table VII. Binomial logistic regression analysis of IL-10 rs1800871 and rs1800872 in the control group and LSCC patients with no metastasis and with metastasis to the neck lymph nodes.
OR: Odds ratio; CI: confidence interval; p-Value: significance level (alpha=0.05); AIC: Akaike information criterion. Bold values indicate statistical significance.
Correspondingly, it was noticed that IL-10 rs1800872 TT genotype was associated with 3.7-fold increased odds of LSCC development in the WMNLN subgroup patients under the codominant model (OR=3.741; 95% CI=1.660-8.432; p=0.001) and 3.2-fold increased odds under the recessive model (OR=3.224; 95% CI=1.495-6.949; p=0.003). Both genotypes (TG+TT) under the dominant model were associated with 1.8-fold increased odds of LSCC with the WMNLN subgroup (OR=1.805; 95% CI=1.042-3.2137; p=0.035). Each allele T at IL-10 rs1800871 was associated with 1.8-fold increased odds of LSCC development in the WMNLN subgroup under the additive model (OR=1.799; 95 % CI=1.211-2.672; p=0.004) (Table VII).
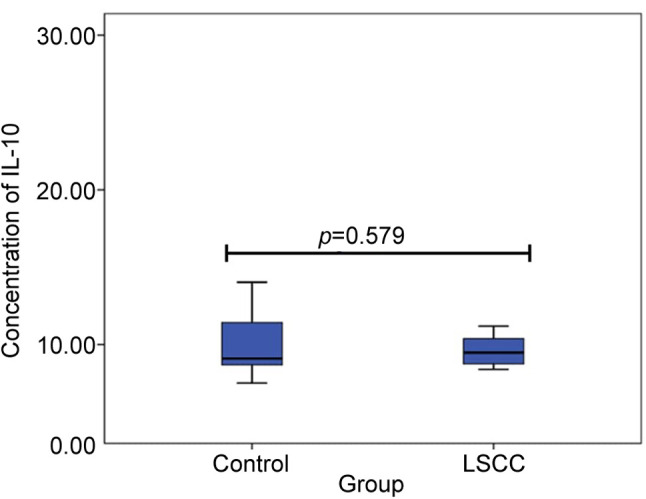
Serum concentrations of IL-10 in control and LSCC group. IL10 serum concentrations were measured in 20 subjects of the control group and 20 patients of the LSCC group. Observed variations in serum IL-10 concentrations in the patients with LSCC and the control group are presented in Figure 1. No statistically significant differences of IL-10 concentration between the control and LSCC groups (10.3 pg/ml ±15.99 vs. 14.6 pg/ml ±2.61; p=0.579) were found. Also, we performed a comparison of the genotypic distribution of serum levels of IL-10 between the control and LSCC groups. No statistically significant results were revealed.
Figure 1. Serum IL10 concentrations in controls and patients with LSCC.
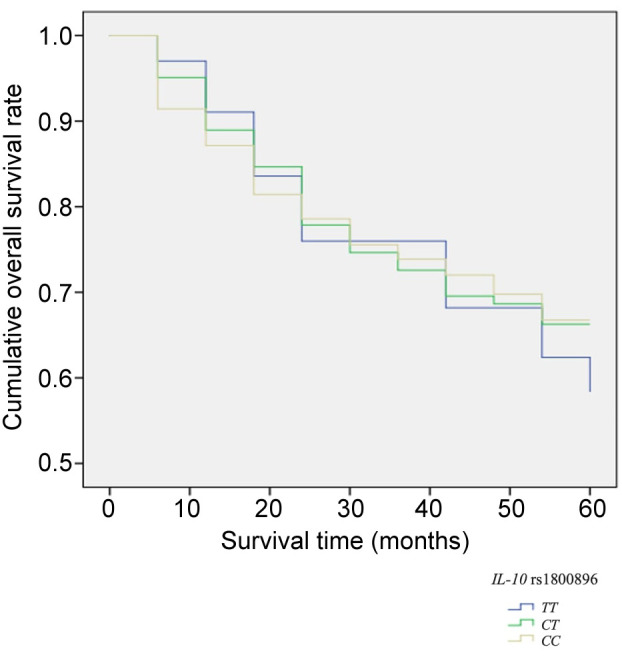
Association between SNPs and five-year survival rate. The five-year survival rate of selected 300 LSCC patients was 65%. Analyzing LSCC patients’ five-year survival rate and the genotype distribution, we did not find any statistically significant differences between the genotypic distribution of selected SNPs and the survival rate of the LSCC patients (Figure 2 and Figure 3).
Figure 2. Five-year survival rate according to the distribution of IL-10 rs1800871 and IL-10 rs1800872 genotypes.

Figure 3. Five-year survival rate according to the distribution of IL-10 rs1800896 genotypes.
Discussion
Recent studies suggested that polymorphisms in pro- and anti-inflammatory cytokines promote different diseases, including carcinogenesis (34,61). It has been indicated that immune cytokines such as IL-10 and inflammation play an essential role during carcinogenesis by inducing angiogenesis, immune suppression, and cell metastasis (62,63). According to that, the molecular effect of IL-10 gene promoter SNPs is not precisely understood, and the results remain conflicting (17,34). For this study, we selected three IL-10: rs1800871, rs1800872, and rs1800896 SNPs to identify their possible role in LSCC development and determine associations of genetic distribution of selected SNPs and patients’ survival rate; IL-10 serum levels were measured as well. To the best of for knowledge, this is the first study analyzing the impact of before mentioned SNPs on the survival rate of LSCC patients.
The immunosuppressive characteristics of IL-10, corresponding with the high levels of IL-10 identified in tumor tissues are well determined in the literature (20). In tumor cells, IL-10 inhibits the expression of major histocompatibility complex (MHC) class II in the antigen-presenting cell, MHC class I expression, and natural killer group 2D (NKG2D) ligand expression (12,64). Conditions as carcinogenesis trigger upregulation of NKG2D, inducing the recognition of the NKG2D receptor (20). This results in the activation of signaling pathways, leading to stimulation of antitumor functions of NK cells (65). Moreover, blockage of NKG2D pathways presents increased susceptibility of induced malignant transformations of mice (66). Hence, IL-10 induced inhibition of NKG2D ligand expression influences tumor immune surveillance (64). However, during the down-regulation of MHC class I expression, it stimulates human leukocyte antigen-G (HLA-G) molecules expression that is usually observed in pathological mechanisms, as carcinogenesis (20). HLA-G is associated with tumor immune escape, metastasis, and poor prognosis (67). Referring to the critical role of IL-10 in immune response regulation in malignant proceses, a huge interest has been focused on the identification of naturally occurring gene polymorphisms in IL-10 and flanking regions (17). It is suggested that alterations in IL-10 promoter activity might be one of the mechanisms provoking IL-10 production modifications (68). However, the molecular effects of IL-10 rs1800871, rs1800872, and rs1800896 polymorphisms genotypic distribution in malignancy pathogenesis remain not completely understood (17). According to previous studies and focusing on histological aspects that LSCC belongs to squamous cell carcinomas, significant associations between IL-10 rs1800871, rs1800872, and rs1800896 polymorphisms with lung, thyroid, gastric, cervical, and prostate cancer are identified (55,57-59).
Several studies announced that IL-10 has pro-tumorigenic and anti-tumorigenic effects (12,22-26). For this reason, numerous studies evaluating the association between polymorphisms of IL-10 as potential biomarkers and different types of cancer have been reported. From all IL-10 promoter polymorphisms, the rs1800896 (-1082A/G) is the one most widely studied. A meta-analysis by Khan et al. analyzed twenty-three case-control relevant studies of IL-10 promoter-1082 A>G (rs1800896) polymorphism significance in the increased risk of carcinogenesis involving 4753 cases (576 cases of HNSCC) and 6,086 control subjects. It concluded that IL-10 promoter-1082 polymorphism is not associated under a heterozygous (GG vs. AG) model with cancer developing risk (32). However, in a meta-analysis conducted by Niu et al., a systematic review of 9 relevant studies including 2,258 cases of only HNSCC and 2,887 control subjects (623 cases of nasopharyngeal carcinoma, 1,357 – oral SCC, and 278 cases of all HNSCC without specific location) was performed and suggested that IL-10 -1082A>G (rs1800896) polymorphism might be associated with increased risk of HNSCC development (34). In the present study, we identified argumentative results: all genotypes of IL-10 rs1800896 (TT, TC, and CC) were very similar in the LSCC and control groups (23.3 vs. 27.8%, 54.3 vs. 50.5%, and 22.3 vs. 21,8%; p=0.364) demonstrating that there was no statistically significant difference between IL-10 rs1800896 in both investigated groups.
The impact of IL-10 promoter polymorphisms on vocal fold leukoplakia and LSCC was investigated by Zhou et al. (35). The study involved 146 patients with LSCC (97.3% male and 2.7% female) and 119 control subjects (95.8% male and 4.2% female) and revealed that the -592A/C (rs1800872), -819T/C (rs1800871) and -1082A/G (rs1800896) polymorphisms and elevated IL-10 plasma levels are associated with an increased risk of LSCC and vocal fold leukoplakia development. Moreover, higher IL-10 plasma levels and higher OR values of selected SNPs were identified in the advanced stages of LSCC patients and the patients with metastasis to the neck lymph nodes. In the present study, our LSCC group was two times larger compared to Zhou et al. study and was composed of 300 patients. Furthermore, our control group was more than four-times larger than in the previously mentioned study and was formed of 533 subjects adjusted by age and sex. Nevertheless, we did not identify any statistically significant differences according to genotype distribution after the binomial logistic regression analysis was accomplished. However, results of our study indicated associations of IL-10 rs1800871 and IL-10 rs1800872 variants with the advanced stage of LSCC subgroup patients under the codominant, recessive, and additive models (p=0.027, p=0.040, and p=0.037, respectively) and with patients who had metastasis to the neck lymph nodes under the codominant, recessive, dominant and additive models (p=0.001, p=0.003, p=0.035, and p=0.004, respectively).
Zhou et al., in their study, mentioned that the sample size of subjects might be the main limitation of their research, suggesting that increasing the study population could lead to more statistically reliable results (35). Our study results suggest that IL-10 rs1800871 and rs1800872 polymorphisms play a significant role only in the advanced stages of LSCC development compared to healthy controls and comparing them to the early stage of the LSCC group. Hence, it leads us to the presumption that LSCC might develop slower in patients who carry the AA genotype at rs1800871 and the TT at rs1800872 with about 0.5-fold decreased odds of LSCC development under the codominant and recessive (p=0.027, p=0.040 and p=0.027, p=0.040, respectively) models. Also, each allele A at rs1800871 and as well each T allele at rs1800872 is associated with 0.7-fold decreased odds of LSCC development under the additive model (p=0.037 and p=0.037, respectively).
Makni et al. investigated possible IL-10 promoter gene variants in 194 HNSCC patients – 137 nasopharyngeal cancer (NPC), 57 LSCC and in 263 control subjects and found out that all three SNPs were significant in NPC development, but only IL-10 rs1800871 and AT haplotype may be used for identification of higher risk of LSCC (33). In this study, the synchronous changing of genotypes at position -819 (rs1800871) with -592 (rs1800872) position was not identified. However, this was noticed both in our study and in the study designed by Zhou et al. (35). Also, in the study performed by Makni et al. the dominant sex in the control group was female: 171 (65.8%) female from 263 control subjects; respectively 6 (10.5%) females from 57 LSCC patients. In the present study, we selected and adjusted both investigated groups for age and sex (LSCC group: 95.7% males and 4.3% females; control group: 95.3% males and 4.7% females; p=0.813), with a reference that reported predominant sex of LSCC is male (3,33).
In the literature, huge interest has been given on elevated IL-10 plasma levels as a significant marker and adverse prognostic factor in HNSCC (35,69,70). Increased IL-10 serum levels of HNSCC patients have been observed at advanced stages of the disease (19.3±1.1.2 pg/ml, n=29 vs. 9.1±0.9 pg/ml, n=31, p<0.0005) and in patients with nodal metastasis (19.1±1.6 pg/ml, n=29 vs. 9.4±1.4 pg/ml, n=31) (26). However, some researchers suggest that serum cytokine levels cannot be used as potential biomarker in LSCC (71). In the present study, the IL-10 plasma level was slightly lower in LSCC patients than in control subjects; however, no statistically significant differences were observed (10.3±15.99 pg/ml, n=20 vs. 14.6±2.61 pg/ml, n=20; p=0.579). The median IL-10 plasma level in HNSCC patients was also lower comparing to the control group in the study carried out by Makni et al. 2.25 pg/ml vs. 4.65 pg/ml; p=0.34) (33).
Regarding the poor prognosis of HNSCC and urgently needed prognostic biomarkers to diagnose these disorders earlier, the present study was the first to investigate possible associations of IL-10: rs1800871, rs1800872, rs1800896 SNPs with LSCC patients’ survival rate. As a result, we identified that the distribution of selected SNPs genotypes had no significant impact on LSCC patients’ survival rate (respectively, p=0.952; p=0.952, and p=0.991).
This study’s strength was the inclusion of a large study population (833 subjects in total), pure LSCC patients group, and precise selection of investigated groups – both groups were adjusted for age and sex.
Generally, the results of the present study are in concordance with the data of the literature. Some discrepancies between our results and literature data can be possibly explained by the significant diversity of malignances traditionally united under the umbrella of the HNSCC term because this term includes malignant tumors of different localizations (oral, pharyngeal, laryngeal regions) as well as of different etiology, biological and clinical behavior. Therefore, pooling all cancer types together may mask the possible significant associations of IL-10: rs1800871, rs1800872, and rs1800896 with individual cancer types. In the present study, a pure cohort of LSCC patients was investigated. Results revealed no statistically significant differences in the distribution of IL-10: rs1800871, rs1800872, rs1800896 variants between the LSCC patients and control groups. Presumably, these findings reflect the biological and clinical peculiarities of the LSCC tumor.
To the best of our knowledge, this is the first report that associates the role of IL-10: rs1800871, rs1800872, rs1800896 SNPs, and development of LSCC in large, pure and homogenous LSCC patients cohort and age- and sex-matched control subjects. This particuliarity allowed us to accomplish a precise analysis of associations between selected SNPs and LSCC development, with a concrete tumor in one anatomical region of head and neck. It is well known from clinical practice that LSCC (notably, the glottis cancer) is characterized as less aggressive in accordance with local spreading and low metastatic rate compared to other HNSCC (72,73). Thus, not identified discrepancies of IL-10: rs1800871, rs1800872, rs1800896 variants distribution between this low aggressive LSCC patients’ and sex-matched control subjects is comprehensible. However, a statistically significant association between IL-10 rs1800871 and rs1800872 gene polymorphisms and LSCC in patients with the advanced stage of LSCC and patients with metastasis to the neck lymph nodes suggests the importance of these SNPs in the development of this peculiar tumor.
Certain limitations of the present study, as involvement of environmental factors should be deliberated; smoking and alcohol consumption habits were not investigated. However, this is a targeted task for future investigation.
Conclusion
The present study indicated a significant association between IL-10 rs1800871 and rs1800872 gene polymorphisms with an increased risk for LSCC development in advanced stages of LSCC and patients with metastasis to the neck lymph nodes. These results might be useful for selecting candidate prognostic factors in identifying and elaborating novel targeted therapies and immunotherapy strategies and could improve LSCC patients’ survival rate. However, further studies involving risk factors such as smoking and alcohol consumption are necessary to define these SNPs’ role in LSCC carcinogenesis.
Conflicts of Interest
The Authors declare no potential conflicts of interest associated with this study.
Authors’ Contributions
R.L., G.G., A.V. and V.U. designed the research; V.U., V.L. and A.P. collected patients’ samples; A.P., G.G. and A.V. performed research; R.L., G.G., A.V. and A.P. analyzed data; and A.P.; R.L.; V.L. and V.U. wrote the paper.
References
- 1.Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Wang N, Lv H, Huang M. Impact of gender on survival in patients with laryngeal squamous cell carcinoma: A propensity score matching analysis. Int J Clin Exp Pathol. 2020;13(3):573–58. [PMC free article] [PubMed] [Google Scholar]
- 4.The Global Cancer Observatory 2018 Estimated cancer incidence, mortality and prevalence Wordwide in 2018. Lyon, International Agency for Research on Cancer, 2018. Available at: https://gco.iarc.fr/today/data/factsheets/cancers/14-Larynxfact-sheet.pdf [Last accessed on 15th November 2020]
- 5.Shin JY, Truong MT. Racial disparities in laryngeal cancer treatment and outcome: A population-based analysis of 24,069 patients. Laryngoscope. 2015;125(7):1667–1674. doi: 10.1002/lary.25212. [DOI] [PubMed] [Google Scholar]
- 6.Daneshi N, Fararouei M, Mohammadianpanah M, Zare-Bandamiri M, Parvin S, Dianatinasab M. Effects of different treatment strategies and tumor stage on survival of patients with advanced laryngeal carcinoma: A 15-year cohort study. J Cancer Epidemiol. 2018;2018:e9678097. doi: 10.1155/2018/9678097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gogarty DS, Shuman A, O’Sullivan EM, Sheahan P, Kinsella J, Timon C, O’Neill JP. Conceiving a national head and neck cancer screening programme. J Laryngol Otol. 2016;130(1):8–14. doi: 10.1017/S0022215115003084. [DOI] [PubMed] [Google Scholar]
- 8.Oloomi M, Moazzezy N, Bouzari S. Comparing blood versus tissue-based biomarkers expression in breast cancer patients. Heliyon. 2020;6(4):e03728. doi: 10.1016/j.heliyon.2020.e03728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skalski M, Ustaszewski A, Jaskiewicz K, Kiwerska K, Wierzbicka M, Klimza H, Grenman R, Giefing M. Single nucleotide polymorphism rs11614913 associated with CC genotype in miR-196a2 is overrepresented in laryngeal squamous cell carcinoma, but not salivary gland tumors in Polish population. J Appl Genet. 2018;59(3):301–304. doi: 10.1007/s13353-018-0445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv Y, Jia C, Jiang A, Zhang H, Wang Y, Liu F, Yang L, Sun Y, Song W. Analysis of association between MGMT and p53 gene single nucleotide polymorphisms and laryngeal cancer. Anticancer Res. 2017;37(8):4399–4403. doi: 10.21873/anticanres.11834. [DOI] [PubMed] [Google Scholar]
- 11.Insodaite R, Liutkevicius V, Uloza V, Solovejute R, Smalinskiene A. Association between MMP8 gene polymorphisms and laryngeal squamous cell carcinoma. Anticancer Res. 2020;40(4):2003–2009. doi: 10.21873/anticanres.14156. [DOI] [PubMed] [Google Scholar]
- 12.Mannino MH, Zhu Z, Xiao H, Bai Q, Wakefield MR, Fang Y. The paradoxical role of IL-10 in immunity and cancer. Cancer Lett. 2015;367(2):103–107. doi: 10.1016/j.canlet.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Oft M. IL-10: Master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol Res. 2014;2(3):194–199. doi: 10.1158/2326-6066.CIR-13-0214. [DOI] [PubMed] [Google Scholar]
- 14.Eskdale J, Kube D, Tesch H, Gallagher G. Mapping of the human IL10 gene and further characterization of the 5’ flanking sequence. Immunogenetics. 1997;46(2):120–128. doi: 10.1007/s002510050250. [DOI] [PubMed] [Google Scholar]
- 15.Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol Res. 2010;47(1-3):185–206. doi: 10.1007/s12026-009-8150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maynard CL, Weaver CT. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol Rev. 2008;226(1):219–233. doi: 10.1111/j.1600-065X.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosser D, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Mai J, Virtue A, Yin Y, Gong R, Sha X, Gutchigian S, Frish A, Hodge I, Jiang X, Wang H, Yang XF. IL-35 is a novel responsive anti-inflammatory cytokine – a new system of categorizing anti-inflammatory cytokines. PLoS One. 2012;7(3):e33628. doi: 10.1371/journal.pone.0033628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore KW, De Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 20.Costa Brandão BF, Brajão de Oliveira K. IL-10 in cancer: Just a classical immunosuppressive factor or also an immunostimulating one. AIMS Allergy Immunol. 2018;2(2):88–97. doi: 10.3934/allergy.2018.2.88. [DOI] [Google Scholar]
- 21.Mumm JB, Oft M. Pegylated IL-10 induces cancer immunity: The surprising role of IL-10 as a potent inducer of IFN-γ-mediated CD8+ T cell cytotoxicity prospects & overviews. BioEssays. 2013;35(7):623–663. doi: 10.1002/bies.201300004. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Daniel V, Maher DW, Hersey P. Production of IL-10 by melanoma cells: Examination of its role in immunosuppression mediated by melanoma. Int J Cancer. 1994;56(5):755–760. doi: 10.1002/ijc.2910560524. [DOI] [PubMed] [Google Scholar]
- 23.Nemunaitis J, Fong T, Shabe P, Martineau D, Ando D. Comparison of serum interleukin-10 (il-10) levels between normal volunteers and patients with advanced melanoma. Cancer Invest. 2001;19(3):239–247. doi: 10.1081/CNV-100102550. [DOI] [PubMed] [Google Scholar]
- 24.Li C, Li H, Jiang K, Li J, Gai X. TLR4 signaling pathway in mouse Lewis lung cancer cells promotes the expression of TGF-β1 and IL-10 and tumor cells migration. Biomed Mater Eng. 2014;24(1):869–875. doi: 10.3233/BME-130879. [DOI] [PubMed] [Google Scholar]
- 25.Boulland ML, Meignin V, Leroy-Viard K, Copie Bergman C, Brière J, Touitou R, Kanavaros P, Gaular P. Human interleukin-10 expression in T/natural killer-cell lymphomas: Association with anaplastic large cell lymphomas and nasal natural killer- cell lymphomas. Am J Pathol. 1998;153(4):1229–1237. doi: 10.1016/S0002-9440(10)65667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farzin M, Ghapanchi J, Tadbir AA. Serum level of interleukin-10 in patients with head and neck squamous cell carcinoma. Aust J Basic Appl Sci. 2012;6(8):282–286. [Google Scholar]
- 27.Gastl GA, Abrams JS, Nanus DM, Oosterkamp R, Silver J, Liu F, Chen M, Albino AP, Bander NH. Interleukin-10 production by human carcinoma cell lines and its relationship to interleukin-6 expression. Int J Cancer. 1993;55(1):96–101. doi: 10.1002/ijc.2910550118. [DOI] [PubMed] [Google Scholar]
- 28.Zhao S, Wu D, Wu P, Wang Z, Huang J. Serum IL-10 predicts worse outcome in cancer patients: A meta-analysis. PLoS One. 2015;10(10):1–15. doi: 10.1371/journal.pone.0139598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foulds KE, Rotte MJ, Seder RA. IL-10 is required for optimal CD8 T cell memory following Listeria monocytogenes infection. J Immunol. 2006;177(4):2565–2574. doi: 10.4049/jimmunol.177.4.2565. [DOI] [PubMed] [Google Scholar]
- 30.Hong P, Feng WY, Fu LH, Jin J, Fu JP. Associations between genetic polymorphisms in interleukin-10 and hematological oncology: Evidence from a meta-analysis. Cancer Biol Ther. 2020;21(4):372–378. doi: 10.1080/15384047.2019.1702404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mocellin S, Marincola FM, Young HA. Interleukin-10 and the immune response against cancer: A counterpoint. J Leukoc Biol. 2005;78(5):1043–1051. doi: 10.1189/jlb.0705358. [DOI] [PubMed] [Google Scholar]
- 32.Khan MT, Afzal S, Rehman AU, Zeb T. Interleukin 10 (IL-10) promoter-1082 and risk of cancer. Meta-analysis. Adv life Sci. 2015;2:67–73. [Google Scholar]
- 33.Makni L, Ben Hamda C, Al-Ansari AK, Souiai O, Gazouani E, Mezlini A, Almawi WY, Yacoubi-Loueslati B. Association of common il-10 promoter gene variants with the susceptibility to head and neck cancer in tunisia. Turkish J Med Sci. 2019;49(1):123–128. doi: 10.3906/sag-1805-21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu YM, Du XY, Cai HX, Zhang C, Yuan RX, Zeng XT, Luo J et. Increased risks between Interleukin-10 gene polymorphisms and haplotype and head and neck cancer: A meta-analysis. Sci Rep. 2015;5:1–10. doi: 10.1038/srep17149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Zhang D, Chen B, Li Q, Zhou L, Liu F, Chou KY, Tao L, Lu LM. Association of interleukin-10 promoter polymorphisms and corresponding plasma levels with susceptibility to laryngeal squamous cell carcinoma. Oncol Lett. 2014;7(5):1721–1727. doi: 10.3892/ol.2014.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister DG, Spencer S, Adelstein D, Adkins D, Anzai Y, Brizel DM, NCCN Working Group. Head and neck cancers, version 2.2020. J Natl Compr Cancer Netw. 2020;18(7):873–898. doi: 10.6004/jnccn.2020.0031. [DOI] [PubMed] [Google Scholar]
- 37.Stell PM, Mcgill T. Asbestos and laryngeal carcinoma. Lancet. 1973;302(7826):416–417. doi: 10.1016/S0140-6736(73)92275-7. [DOI] [PubMed] [Google Scholar]
- 38.Paget-Bailly S, Cyr D, Luce D. Occupational exposures and cancer of the larynx-systematic review and meta-analysis. J Occup Environ Med. 2012;4(1):71–84. doi: 10.1097/JOM.0b013e31823c1343. [DOI] [PubMed] [Google Scholar]
- 39.Di Maso M, Talamini R, Bosetti C, Montella M, Zucchetto A, Libra M, Negri E, Levi F, La Vecchia C, Franceschi S, Serraino D, Polesel J. Red meat and cancer risk in a network of case-control studies focusing on cooking practices. Ann Oncol. 2013;24(12):3107–3112. doi: 10.1093/annonc/mdt392. [DOI] [PubMed] [Google Scholar]
- 40.Gama RR, Carvalho AL, Filho AL, Scorsato AP, Mendoza López RV, Rautava J, Syrjänen S, Syrjänen K. Detection of human papillomavirus in laryngeal squamous cell carcinoma: Systematic review and meta-analysis. Laryngoscope. 2016;126(4):885–893. doi: 10.1002/lary.25738. [DOI] [PubMed] [Google Scholar]
- 41.Kuper H, Boffetta P, Adami HO. Tobacco use and cancer causation: Association by tumour type. J Intern Med. 2002;252(3):206–224. doi: 10.1046/j.1365-2796.2002.01022.x. [DOI] [PubMed] [Google Scholar]
- 42.Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7(2):149–156. doi: 10.1016/S1470-2045(06)70577-0. [DOI] [PubMed] [Google Scholar]
- 43.Moghimi M, Ahrar H, Karimi-Zarchi M, Aghili K, Marjansadat S, Zare-Shehneh M, Neamatzadeh H. Association of IL-10 rs1800871 and rs1800872 polymorphisms with breast cancer risk: A systematic review and meta-analysis. Asian Pacific J Cancer Prev. 2018;19(12):3353–3359. doi: 10.31557/APJCP.2018.19.12.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Yang F, Xu G, Zhong S. The roles of IL-6, IL-8 and IL-10 gene polymorphisms in gastric cancer: A meta-analysis. Cytokine. 2018;111:230–236. doi: 10.1016/j.cyto.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 45.Fei C, Yao XM, Sun Y, Gu XZ, Yu LQ, Lai X. Interleukin-10 polymorphisms associated with susceptibility to acute myeloid leukemia. Genet Mol Res. 2015;14(1):925–930. doi: 10.4238/2015.February.2.15. [DOI] [PubMed] [Google Scholar]
- 46.Gao J, Wei L, Fu R, Wei J, Niu D, Wang L, Ge H, Yu Q, Wang M, Liu X, Zhang W. Association of interleukin-10 polymorphisms (rs1800872, rs1800871, and rs1800896) with predisposition to IgA nephropathy in a Chinese Han population: A case-control study. Kidney Blood Press Res. 2017;42(1):89–98. doi: 10.1159/000471899. [DOI] [PubMed] [Google Scholar]
- 47.Huang ZY, Cheng BJ, Wan Y, Zhou C. Meta-analysis of the IL-10 promoter polymorphisms and pediatric asthma susceptibility. Genet Mol Res. 2016;15(2):1–16. doi: 10.4238/gmr.15028320. [DOI] [PubMed] [Google Scholar]
- 48.Tabrez S, Ali M, Jabir NR, Firoz CK, Ashraf GM, Hindawi S, Damanhouri GA, Alama MN. A putative association of interleukin-10 promoter polymorphisms with cardiovascular disease. IUBMB Life. 2017;69(7):522–527. doi: 10.1002/iub.1637. [DOI] [PubMed] [Google Scholar]
- 49.Naing C, Htet NH, Basavaraj AK, Nalliah S. An association between IL-10 promoter polymorphisms and diabetic nephropathy: A meta-analysis of case-control studies. J Diabetes Metab Disord. 2018;17(2):333–343. doi: 10.1007/s40200-018-0349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang TP, Lv TT, Xu SZ, Pan HF, Ye DQ. Association of interleukin-10 gene single nucleotide polymorphisms with rheumatoid arthritis in a Chinese population. . Postgrad Med J. 2018;94(1111):284–288. doi: 10.1136/postgradmedj-2017-135441. [DOI] [PubMed] [Google Scholar]
- 51.Mohammadi S, Saghaeian Jazi M, Zare Ebrahimabad M, Eghbalpour F, Abdolahi N, Tabarraei A, Yazdani Y. Interleukin 10 gene promoter polymorphisms (rs1800896, rs1800871 and rs1800872) and haplotypes are associated with the activity of systemic lupus erythematosus and IL10 levels in an Iranian population. Int J Immunogenet. 2019;46(1):20–30. doi: 10.1111/iji.12407. [DOI] [PubMed] [Google Scholar]
- 52.Emampanahi M, Masoudi Rad S, Saghaeian Jazi M, Mansour Samaei N, Behnampour N, Mohammadi S, Fakhari E. Association between interleukin-10 gene polymorphisms and severe chronic periodontitis. Oral Dis. 2019;25(6):1619–1626. doi: 10.1111/odi.13114. [DOI] [PubMed] [Google Scholar]
- 53.Vargas-Alarcón G, Juárez-Cedillo E, Martínez-Rodríguez N, Fragoso JM, García-Hernández N, Juárez-Cedillo T. Association of interleukin-10 polymorphisms with risk factors of Alzheimer’s disease and other dementias (SADEM study) Immunol Lett. 2016;177:47–52. doi: 10.1016/j.imlet.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol. 2012;32(1):23–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seifart C, Plagens A, Dempfle A, Clostermann U, Vogelmeier C, Von Wichert P, Seifart U. TNF-α, TNF-β, IL-6, and IL-10 polymorphisms in patients with lung cancer. Dis Markers. 2005;21(3):157–165. doi: 10.1155/2005/707131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erdogan M, Karadeniz M, Ozbek M, Ozgen AG, Berdeli A. Interleukin-10 gene polymorphism in patients with papillary thyroid cancer in Turkish population. J Endocrinol Invest. 2008;31 (9):750–754. doi: 10.1007/BF03349252. [DOI] [PubMed] [Google Scholar]
- 57.Xue H, Wang YC, Lin B, An J, Chen L, Chen J, Fang JY. A meta-analysis of interleukin-10 -592 promoter polymorphism associated with gastric cancer risk. PLoS One. 2012;7(7):e39868. doi: 10.1371/journal.pone.0039868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roh JW, Kim MH, Seo SS, Kim SH, Kim JW, Park NH, Song YS, Park SY, Kang SB, Lee HP. Interleukin-10 promoter polymorphisms and cervical cancer risk in Korean women. Cancer Lett. 2002;184(1):57–63. doi: 10.1016/S0304-3835(02)00193-3. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Song B, Bai X, Liu W, Li Z, Wang J, Zheng Y, Wang Z. Association of genetic polymorphisms in the interleukin-10 promoter with risk of prostate cancer in Chinese. BMC Cancer. 2010;10:e456. doi: 10.1186/1471-2407-10-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin L, Sturgis EM, Cao X, Song X, Salahuddin T, Wei Q, Li G. Interleukin-10 promoter variants predict HPV-positive tumors and survival of squamous cell carcinoma of the oropharynx. FASEB J. 2013;27(6):2496–2503. doi: 10.1096/fj.12-226803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Husain SR, Han J, Au P, Shannon K, Puri RK. Gene therapy for cancer: Regulatory considerations for approval. Cancer Gene Ther. 2015;22(12):554–563. doi: 10.1038/cgt.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okada F. Inflammation-related carcinogenesis: Current findings in epidemiological trends, causes and mechanisms. Yonago Acta Med. 2014;57(2):65–72. [PMC free article] [PubMed] [Google Scholar]
- 63.Landskron G, De La Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. doi: 10.1155/2014/149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gopal M. Role of cytokines in tumor immunity and immune tolerance to cancer. In: Cancer Immunology: A Translational Medicine Context. Rezaei N (ed.) New York, Springer. 2020:93–120. [Google Scholar]
- 65.Moretta L, Bottino C, Pende D, Castriconi R, Mingari MC, Moretta A. Surface NK receptors and their ligands on tumor cells. Semin Immunol. 2006;18(3):151–158. doi: 10.1016/j.smim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 66.Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med. 2005;202(5):583–588. doi: 10.1084/jem.20050994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noguchi K, Isogai M, Kuwada E. Detection of anti-HLA-F antibodies in Sera from cancer patients. Anticancer Res. 2004;24(5C):3387–3392. [PubMed] [Google Scholar]
- 68.Steinke JW, Barekzi E, Hagman J, Borish L. Functional analysis of −571 IL-10 promoter polymorphism reveals a repressor element controlled by Sp1. J Immunol. 2004;173(5):3215–3222. doi: 10.4049/jimmunol.173.5.3215. [DOI] [PubMed] [Google Scholar]
- 69.Eyigor M, Eyigor H, Osma U, Yilmaz MD, Erin N, Secul OT, Sezer C, Gultekin M, Koksoy S. Analysis of serum cytokine levels in larynx squamous cell carcinoma and dysplasia patients. Iran J Immunol. 2014;11(4):259–268. [PubMed] [Google Scholar]
- 70.Jebreel A, Mistry D, Loke D, Dunn G, Hough V, Oliver K, Stafford N, Greenman J. Investigation of interleukin 10, 12 and 18 levels in patients with head and neck cancer. J Laryngol Otol. 2007;121(3):246–252. doi: 10.1017/S0022215106002428. [DOI] [PubMed] [Google Scholar]
- 71.Sotirović J, Perić A, Vojvodić D, Baletić N, Zaletel I, Stanojević I, Erdoglija M, Milojević M. Serum cytokine profile of laryngeal squamous cell carcinoma patients. J Laryngol Otol. 2017;131(5):455–461. doi: 10.1017/S0022215117000573. [DOI] [PubMed] [Google Scholar]
- 72.Cadoni G, Giraldi L, Petrelli L, Pandolfini M, Paludetti G, Pastorino R, Leoncini E, Arzani D, Almadori G, Boccia S. Prognostic factors in head and neck cancer: A 10-year retrospetive analysis in a single-institutuon in Italy. Acta Otorhinolaryngol Ital. 2017;37(6):458–466. doi: 10.14639/0392-100X-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gatta G, Botta L, Sánchez MJ, Anderson LA, Pierannunzio D, Licitra L, EUROCARE Working Group. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur J Cancer. 2015;51(15):2130–2143. doi: 10.1016/j.ejca.2015.07.043. [DOI] [PubMed] [Google Scholar]