Abstract
The coronavirus disease 2019 (COVID-19) pandemic has claimed the lives of over one million people worldwide. The causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a member of the Coronaviridae family of viruses that can cause respiratory infections of varying severity. The cellular host factors and pathways co-opted during SARS-CoV-2 and related coronavirus life cycles remain ill defined. To address this gap, we performed genome-scale CRISPR knockout screens during infection by SARS-CoV-2 and three seasonal coronaviruses (HCoV-OC43, HCoV-NL63, and HCoV-229E). These screens uncovered host factors and pathways with pan-coronavirus and virus-specific functional roles, including major dependency on glycosaminoglycan biosynthesis, sterol regulatory element-binding protein (SREBP) signaling, bone morphogenetic protein (BMP) signaling, and glycosylphosphatidylinositol biosynthesis, as well as a requirement for several poorly characterized proteins. We identified an absolute requirement for the VMP1, TMEM41, and TMEM64 (VTT) domain-containing protein transmembrane protein 41B (TMEM41B) for infection by SARS-CoV-2 and three seasonal coronaviruses. This human coronavirus host factor compendium represents a rich resource to develop new therapeutic strategies for acute COVID-19 and potential future coronavirus pandemics.
Keywords: COVID-19, SARS-CoV-2, coronavirus, CRISPR, genetic screens, TMEM41B, host factors, HCoV-229E, HCoV-OC43, HCoV-NL63
Graphical Abstract
Schneider et al. conducted parallel genome-wide CRISPR knockout screens with SARS-CoV-2 and three seasonal coronaviruses to identify pan-coronavirus and virus-specific host factor requirements. They identified an interconnected network of host factors required by these four viruses and validated TMEM41B as a pan-coronavirus host factor required for a post-entry step in the coronavirus life cycle.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic, has claimed the lives of more than 1.4 million people worldwide in less than a year (Zhou et al., 2020; Zhu et al., 2020a; https://coronavirus.jhu.edu/map.html). SARS-CoV-2 is a beta-CoV in the Coronaviridae family, which is composed of enveloped positive-sense RNA viruses with large (>30 kb) genomes that can infect a variety of vertebrate hosts (Cui et al., 2019). Seasonal human CoVs (HCoVs), such as the beta-CoV OC43, as well as the alpha-CoVs NL63 and 229E can cause mild to moderate upper respiratory tract infections with cold-like symptoms in humans (Cui et al., 2019). In stark contrast, highly pathogenic beta-CoVs have been responsible for multiple deadly outbreaks in the 21st century, including SARS-CoV (2003), Middle East respiratory syndrome coronavirus (MERS-CoV; 2012), and SARS-CoV-2 (2019) (Cui et al., 2019). The spread of SARS-CoV and MERS-CoV was contained in part because of their comparatively low transmissibility (Cui et al., 2019). However, SARS-CoV-2 spreads more readily and remains largely uncontrolled across the globe, presenting an urgent health crisis.
A complete understanding of the host factors and pathways co-opted by SARS-CoV-2 and other CoVs for execution of their life cycles could contribute to development of therapies to treat COVID-19 and increase preparedness for potential future outbreaks. Large-scale forward genetic approaches based on RNA interference, insertional mutagenesis, and CRISPR have proven to be powerful for identifying host factors required for infection by different viruses (reviewed in Puschnik et al., 2017). Here we performed parallel genome-scale CRISPR-Cas9 knockout screens to generate an extensive functional catalog of host factors required for infection by SARS-CoV-2 and three seasonal CoVs (HCoV-OC43, HCoV-NL63, and HCoV-229E). We identified multiple genes and pathways with pan-CoV and virus-specific functional roles, including factors involved in glycosaminoglycan (GAG) biosynthesis, sterol regulatory element-binding protein (SREBP) signaling, bone morphogenetic protein (BMP) signaling, and glycosylphosphatidylinositol (GPI) biosynthesis, as well as several poorly characterized proteins, such as transmembrane protein 41B (TMEM41B). We show that the VMP1, TMEM41, and TMEM64 (VTT) domain-containing protein TMEM41B is a critical host factor required for infection by SARS-CoV-2, HCoV-OC43, HCoV-NL63, and HCoV-229E as well as several flaviviruses of high interest to public health (Hoffmann et al., 2020a [this issue of Cell]), nominating TMEM41B as a broad-spectrum RNA virus liability and potential high-priority target for future drug development efforts.
Results
Genome-wide CRISPR Screens Identify Host Factors Required for SARS-CoV-2 Infection
We set out to develop an extensive catalog of human host factors required for infection by SARS-CoV-2 and three seasonal CoVs (HCoV-OC43, HCoV-NL63, and HCoV-229E) using pooled CRISPR-Cas9 genetic screening (Cui et al., 2019; Figure 1 A). Our screens used the Brunello genome-wide library, which is composed of 76,441 single guide RNAs (sgRNAs) targeting 19,114 human genes (Doench et al., 2016). We screened Cas9-expressing Huh-7.5 hepatoma cells (Huh-7.5-Cas9), which endogenously express the SARS-CoV-2 cellular receptor, angiotensin-converting enzyme 2 (ACE2), as well as transmembrane serine protease 2 (TMPRSS2), a key mediator of SARS-CoV-2 entry (Hoffmann et al., 2020). We recently showed that Huh-7.5-Cas9 cells are permissive to infection by SARS-CoV-2, HCoV-OC43, HCoV-NL63, and HCoV-229E and that they are a robust system for CRISPR-based genetic screening (Hoffmann et al., 2020b).
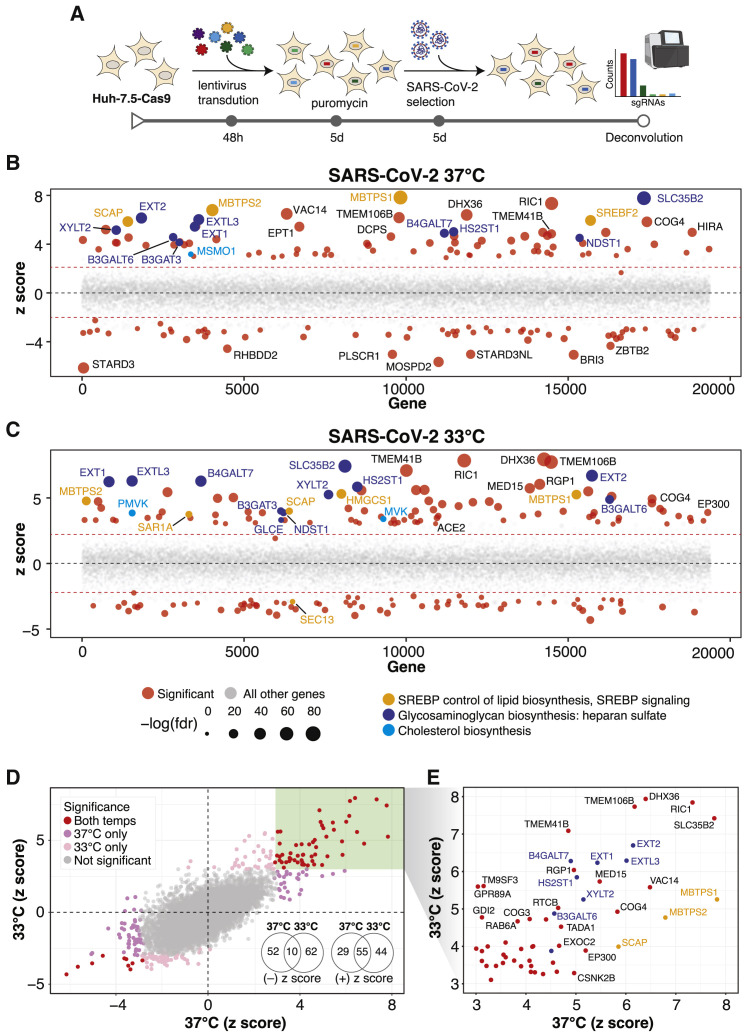
Figure 1.
Genome-wide CRISPR Screens Identify Host Factors Required for SARS-CoV-2 Infection
(A) Genome-wide CRISPR screening workflow. Cas9-expressing Huh-7.5 cells are transduced with the Brunello genome-wide CRISPR library, selected with puromycin, and infected with SARS-CoV-2 or one of three seasonal CoVs (HCoV-OC43, HCoV-NL63, or HCoV-229E). Surviving cells and mock controls are then harvested, and sgRNA abundance is determined using next-generation sequencing.
(B) Bubble plot of data from SARS-CoV-2 screens at 37°C. Red lines denote z = ±2.
(C) Bubble plot of data from SARS-CoV-2 screens at 33°C. Red lines denote z = ±2.
(D) Scatterplot comparing Z scores from (B) and (C) for SARS-CoV-2 screens at 37°C and 33°C, respectively.
(E) Subset of significantly enriched genes from SARS-CoV-2 screens at 37°C and 33°C.
We performed a series of parallel genetic screens by transducing Huh-7.5-Cas9 cells with the Brunello library followed by antibiotic selection and expansion for 7 days to ensure CRISPR-based knockout of host factor genes prior to CoV infection. In this context, cells expressing sgRNAs targeting genes required for virus infection or virus-induced death should survive, whereas those expressing neutral sgRNAs or sgRNAs targeting genes irrelevant to infection are expected to deplete. Similarly, cells expressing sgRNAs targeting essential genes with no roles in virus infection or virus-induced death are expected to deplete under mock-infected (uninfected) and virus-infected conditions. SARS-CoV-2 screens were performed in triplicate at two physiologically relevant temperatures, 33°C and 37°C, to mimic the temperatures of the upper and lower airways, respectively (V’kovski et al., 2020). Surviving cells were harvested 5 days post-infection and subjected to genomic DNA extraction and screen deconvolution using high-throughput sequencing.
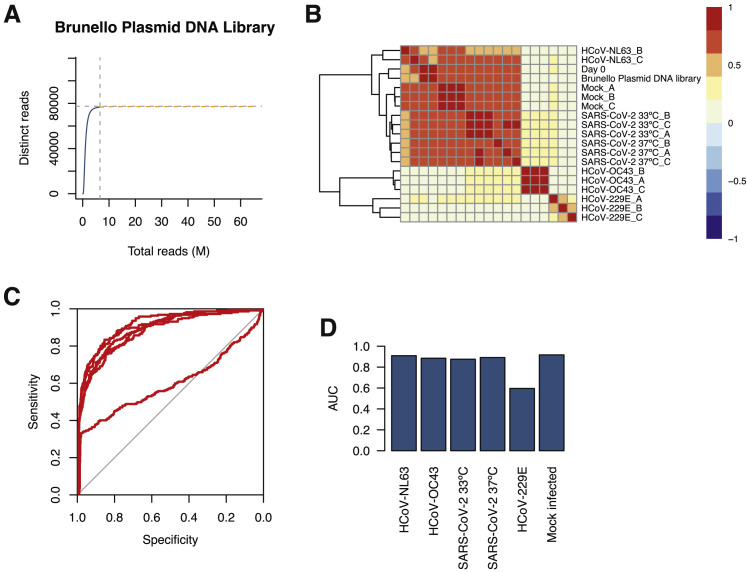
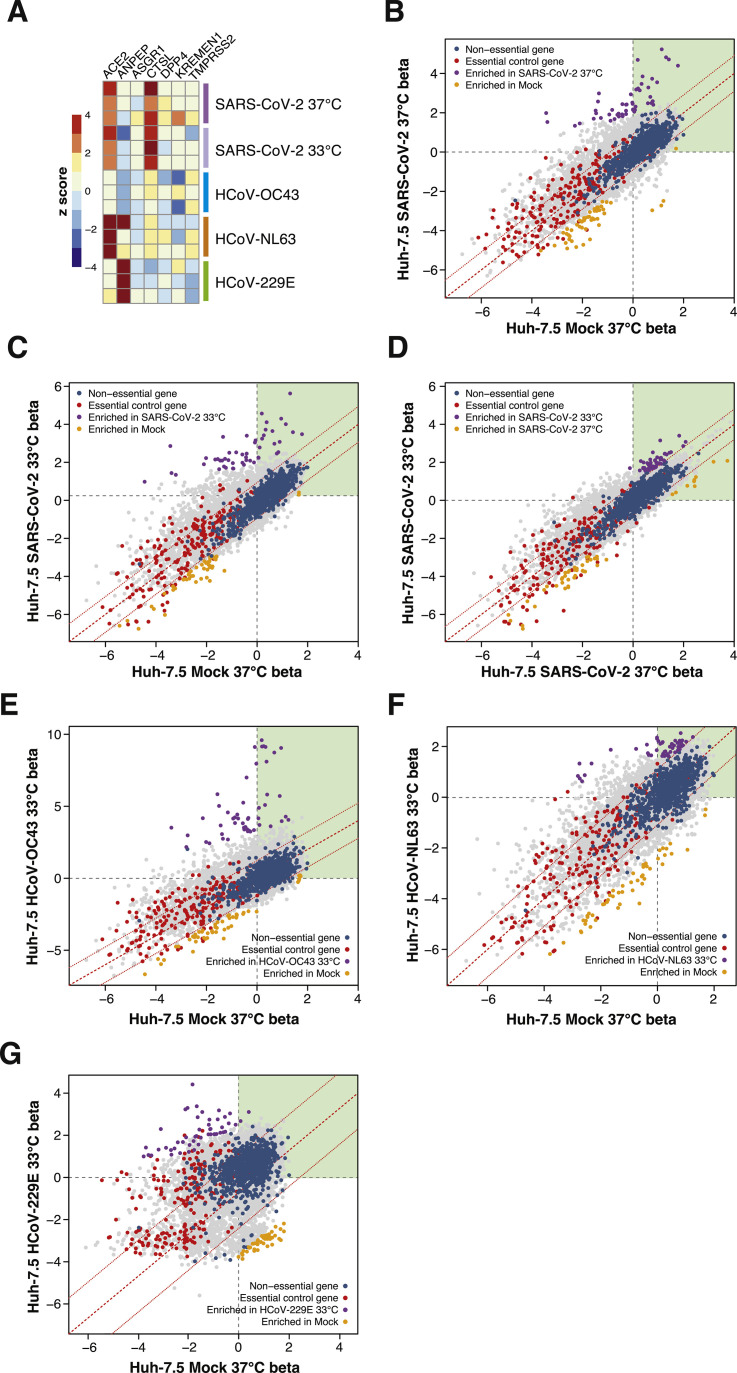
Several quality control (QC) metrics demonstrated excellent technical performance across all screens and biological replicates (Figure S1 ). First, we confirmed that 76,160 of 76,441 of sgRNAs (99.6%) from the Brunello library were recovered from the plasmid preparation and that all screen libraries were sequenced to saturation (Figure S1A). Second, pairwise correlation analyses demonstrated that biological replicates from each genetic screen clustered together with high correlation coefficients (Figure S1B). Third, receiver operating characteristic (ROC) curves generated based on the fitness effects of disruption of previously defined neutral and essential genes from the Brunello library confirmed robust gene disruption in the cell pools (Figures S1C and S1D). The HCoV-229E screen, although successful, was particularly stringent, resulting in a lower area under the curve (AUC) relative to the other screens in this study. As described in our recent study (Hoffmann et al., 2020b), we performed a Z score analysis and computed a gene essentiality (beta) score using a published maximum likelihood estimation (MLE) algorithm (Li et al., 2014). The gene essentiality analysis allowed us to stratify candidate host factor targets based on their effects on cellular fitness under mock-infected conditions followed by identification of high-confidence gene hits in virus-infected cells. Specifically, genes with beta scores similar to essential genes could affect cell survival in the presence or absence of infection and may be confounded by effects on cellular fitness. Conversely, genes with beta scores similar to neutral sgRNAs are predicted to affect cell survival only during viral infection and are more likely to be true positives. As expected, the screens identified the SARS-CoV-2 receptor ACE2 and the well-known host factor cathepsin L (CTSL) (Hoffmann et al., 2020; Letko et al., 2020; Yeager et al., 1992; Figure S2 A). The MERS-CoV receptor DPP4 (Earnest et al., 2017; Wang et al., 2013) and the putative SARS-CoV-2 receptors KREMEN1 and ASGR1 did not score in any screen (Gu et al., 2020). Our analysis identified 146 and 171 genes that significantly influenced SARS-CoV-2-induced cell death at 37°C and 33°C, respectively (false discovery rate [FDR] < 0.05) (summarized in Figures 1B–1E, Figures S2B and S2C, and Tables S1A and S1B). A total of 84 (37°C) and 99 (33°C) genes scored as candidate host factors that may facilitate SARS-CoV-2 infection (Z score > 0, FDR < 0.05). Conversely, 62 (37°C) and 72 (33°C) genes scored as candidate antiviral host factors (Z score < 0; FDR < 0.05). As expected, neutral and essential gene-targeting sgRNAs scored similarly across mock and SARS-CoV-2 conditions (blue and red symbols in Figures S2B and S2C, respectively). Integrating the 33°C and 37°C SARS-CoV-2 screening datasets allowed us to obtain a clearer picture of candidate temperature-specific host factors that either support or antagonize SARS-CoV-2 viral infection (Figures 1D and 1E; Figure S2D). These results demonstrate that the human genome encodes a catalog of host factors that functionally contribute to the SARS-CoV-2 life cycle.
Figure S1.
Screen QC Data, Related to Figures 1 and 2
(A) Species accumulation curve for unique reads with ≥ 10 counts.
(B) Heatmap for correlation coefficients between samples.
(C) ROC curves for each screen.
(D) Area under the curve (AUC) for each ROC curve in (C).
Figure S2.
Analysis of Established and Putative CoV Host Factors and Gene-wise Fitness Scores for SARS-CoV-2 and 3 Seasonal CoVs, Related to Figures 1 and 2
(A) Heatmap of z-scores for known and putative coronavirus host factors. (B-D) Genewise fitness beta scatterplots comparing SARS-CoV-2 at 37°C versus mock (B), SARS-CoV-2 at 33°C versus mock (C), and SARS-CoV-2 at 33°C versus SARS-CoV-2 at 37°C (D). Non-targeting controls and essential control genes are highlighted in blue and red, respectively. (E) HCoV-OC43 versus mock infected. (F) HCoV-NL63 versus mock infected. (G) HCoV-229E versus mock infected.
Parallel Genome-wide CRISPR Screening against Multiple HCoVs Uncovers Host Factor Networks with Pan-CoV and Virus-Specific Functional Roles
Viruses within the same family often require the same host factors to complete their respective life cycles (Dimitrov, 2004). Nevertheless, there are several examples of closely related viruses with discrete host factor requirements. For example, SARS-CoV-2 and HCoV-NL63 engage ACE2 as a cellular receptor, whereas HCoV-229E uses membrane alanyl aminopeptidase (ANPEP), and HCoV-OC43 has no known essential proteinaceous receptor (Cui et al., 2019; Forni et al., 2017). Beyond attachment factors and receptors, closely related viruses can also exploit distinct components of intracellular pathways in a virus-specific manner. A comprehensive functional understanding of the commonalities and differences among CoVs and other virus families could pave the way for specific and general antiviral therapies. Toward this goal, we expanded our functional genomics efforts to develop an extensive functional catalog of human host factors required for infection by members of the Coronaviridae family, including two alpha-CoVs (HCoV-NL63 and HCoV-229E) and one additional beta-CoV (HCoV-OC43).
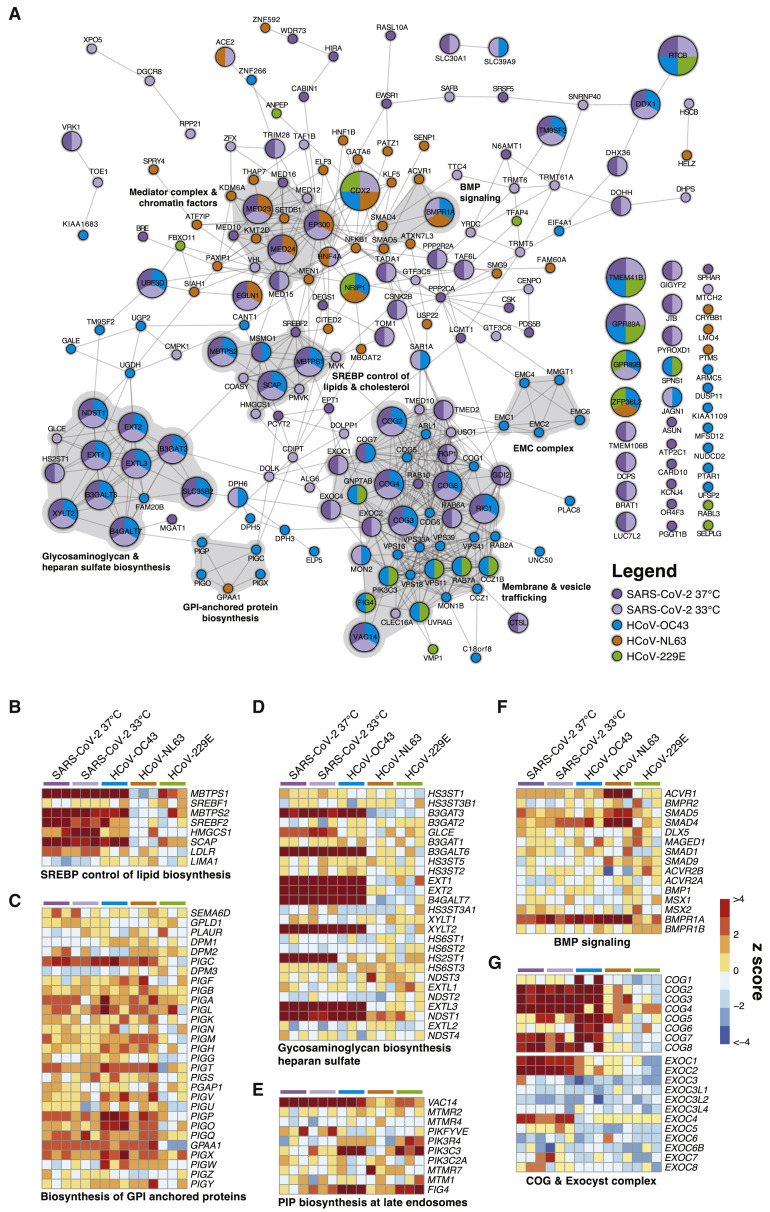
The results of these screens are shown in Figures 2 A–2C; Figure S2; Tables S1C–S1E. An integrative analysis that also includes the two SARS-CoV-2 screens described above is shown in Figure 2D. These screens identified numerous CoV-specific and pan-CoV host factors that appear to play critical roles during infection by each of these viruses. This extensive network of human host factors functionally implicates numerous cellular pathways, as shown in Figure 3 A. We present a selection of comparative analyses below that highlight pan-CoV and virus-specific host factors through the lens of SARS-CoV-2.
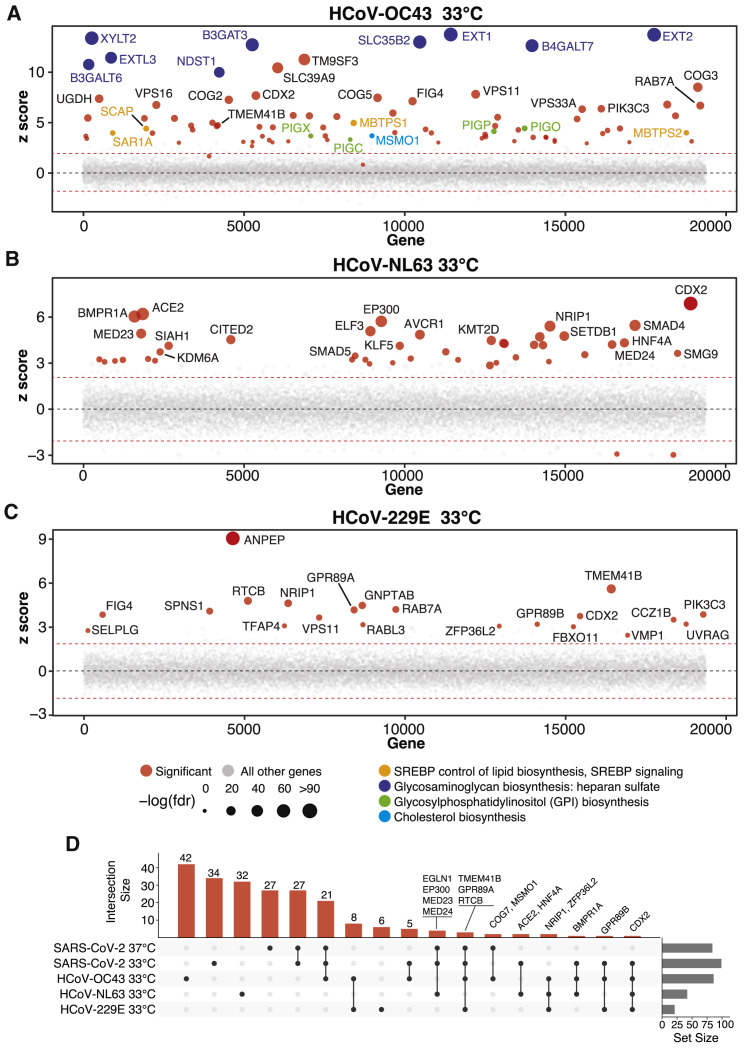
Figure 2.
Parallel Genome-wide CRISPR Screening against Multiple HCoVs Uncovers Host Factors and Pathways with Pan-CoV and Virus-Specific Functional Roles
(A) Bubble plot of data from HCoV-OC43 screens at 33°C. Red lines denote z = ±2.
(B) Bubble plot of data from HCoV-NL63 screens at 33°C. Red lines denote z = ±2.
(C) Bubble plot of data from HCoV-229E screens at 33°C. Red lines denote z = ±2.
(D) UpSet plot showing enriched hits overlapping in screens across all four viruses. Select genes for enriched sgRNAs are indicated.
Figure 3.
CoVs Co-opt an Extensive Network of Human Proteins and Pathways to Complete Their Life Cycles
(A) Network analysis of human CoV host factors for all significant screening hits using the STRING-db protein:protein interaction network. Nodes are subdivided by number of virus screens by color and size for which the node was significant. Highly interconnected and functionally related genes are further highlighted in gray.
(B–G) Comparative pathway-focused heatmaps showing enriched and depleted sgRNAs across all CRISPR screens.
SARS-CoV-2 and HCoV-OC43 (Beta-CoVs)
The beta-CoVs, SARS-CoV-2 and HCoV-OC43, co-opt an overlapping set of host factors to carry out their life cycles. These include genes involved in pathways related to GAG biosynthesis (e.g., heparan sulfate) and transport, such as EXT1, EXT2, EXTL3, B3GALT6, B3GAT3, B4GALT7, SLC35B2, XYLT2, HS2ST1, and NDST1 (Aikawa et al., 2001; Bai et al., 2001; Casanova et al., 2008; Cuellar et al., 2007; Kitagawa et al., 1998; Kreuger and Kjellén, 2012; Lind et al., 1998; Okajima et al., 1999; Pönighaus et al., 2007). We also identified multiple factors that regulate intracellular protein trafficking, processing, and sorting through the cis-oligomeric Golgi (COG) complex, including COG2, COG3, COG4, COG7, and COG8 (Blackburn et al., 2019; Smith and Lupashin, 2008) (Figures 3A, 3D, 3G). Consistent with the role of heparan sulfate as an attachment factor for multiple viruses, the heparan sulfate biosynthesis pathway has been implicated previously as a critical host pathway for several viruses and virus families, including herpes simplex virus (O’Donnell and Shukla, 2008), human papillomavirus (Giroglou et al., 2001; Joyce et al., 1999), respiratory syncytial virus (Bourgeois et al., 1998; Escribano-Romero et al., 2004; Feldman et al., 2000; Hallak et al., 2000; Harris and Werling, 2003; Karger et al., 2001; Krusat and Streckert, 1997; Martínez and Melero, 2000; Techaarpornkul et al., 2002), adenoviruses (Dechecchi et al., 2000, 2001), hepatitis C virus (Xu et al., 2015), dengue and Zika virus (Cruz-Oliveira et al., 2015; Marceau et al., 2016; Savidis et al., 2016), West Nile virus (Perera-Lecoin et al., 2013), Rift Valley fever virus (Riblett et al., 2015), Eastern equine encephalitis virus (Gardner et al., 2011), and HIV (Ibrahim et al., 1999), among others. These studies support the role of heparan sulfate proteoglycans and other GAGs as common mediators of binding and entry for many viruses. Indeed, recent cellular and biochemical evidence suggests that SARS-CoV-2 exploits heparan sulfate proteoglycans cooperatively with ACE2 to bind to and gain entry into cells (Clausen et al., 2020). Given that no protein has been identified as a cellular receptor for HCoV-OC43 entry, it is possible that this beta-CoV engages one or more GAGs to invade target cells.
Another set of factors in common between the SARS-CoV-2 and HCoV-OC43 screens was related to cholesterol homeostasis, particularly as related to SREBP cleavage-activating protein (SCAP)-mediated cholesterol sensing and the SREBP pathway (Figures 3A and 3B; Figures S3 A and S3B). Indeed, genes known to be functionally involved in sensing and biosynthesis of cholesterol, such as SCAP, SREBF2 (but not SREBF1), MBTPS1, MBTPS2, and SAR1A, were among the top enriched genes for these two viruses (Figures 3A and 3B). These results are consistent with our recent discovery that SARS-CoV-2 and other CoVs require SCAP, NPC2, and EMC1 to carry out their life cycles (Hoffmann et al., 2020b) and extend these findings to further elaborate the essential regulatory components of SREBP signaling. We also note a significant reliance of HCoV-OC43 on factors involved in synthesis of GPI-anchored proteins (Figures 3A and 3C). Collectively, these results nominate factors involved in GAG biosynthesis and transport, intracellular protein trafficking, processing, and sorting, and cholesterol homeostasis as potential targetable factors to inhibit SARS-CoV-2 and HCoV-OC43.
Figure S3.
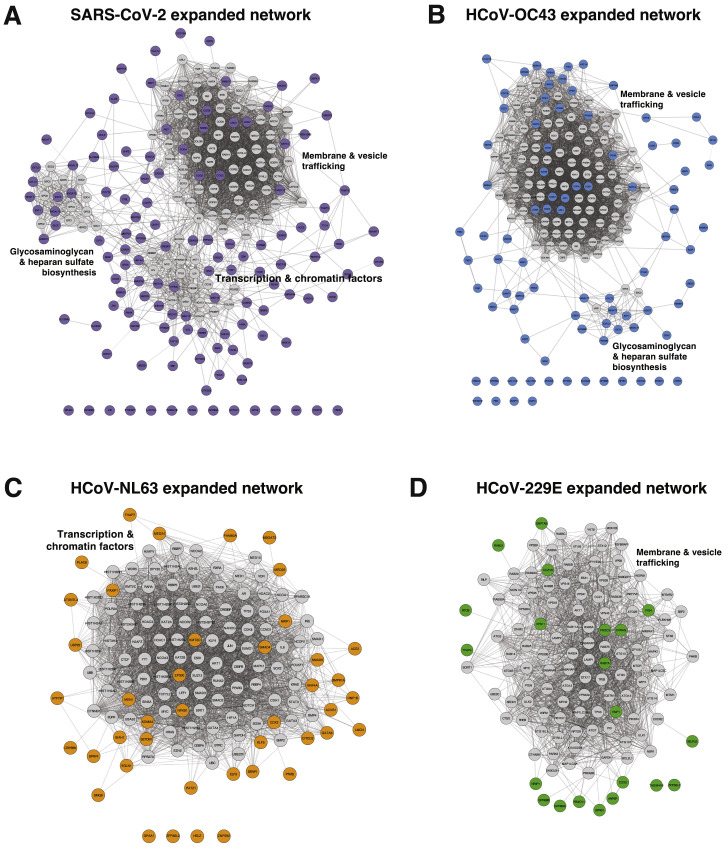
Expanded CoV-Specific Networks, Related to Figure 3
(A–D) Network diagram of all coronavirus screen hits and the next 100 adjacent interactors, shown in gray. Broad functional categories of highly interconnected gene neighborhoods are indicated. Diagrams are for SARS-CoV-2 (A), HCoV-OC43 (B), HCoV-NL63 (C) and HCoV-229E (D).
SARS-CoV-2 Host Factors
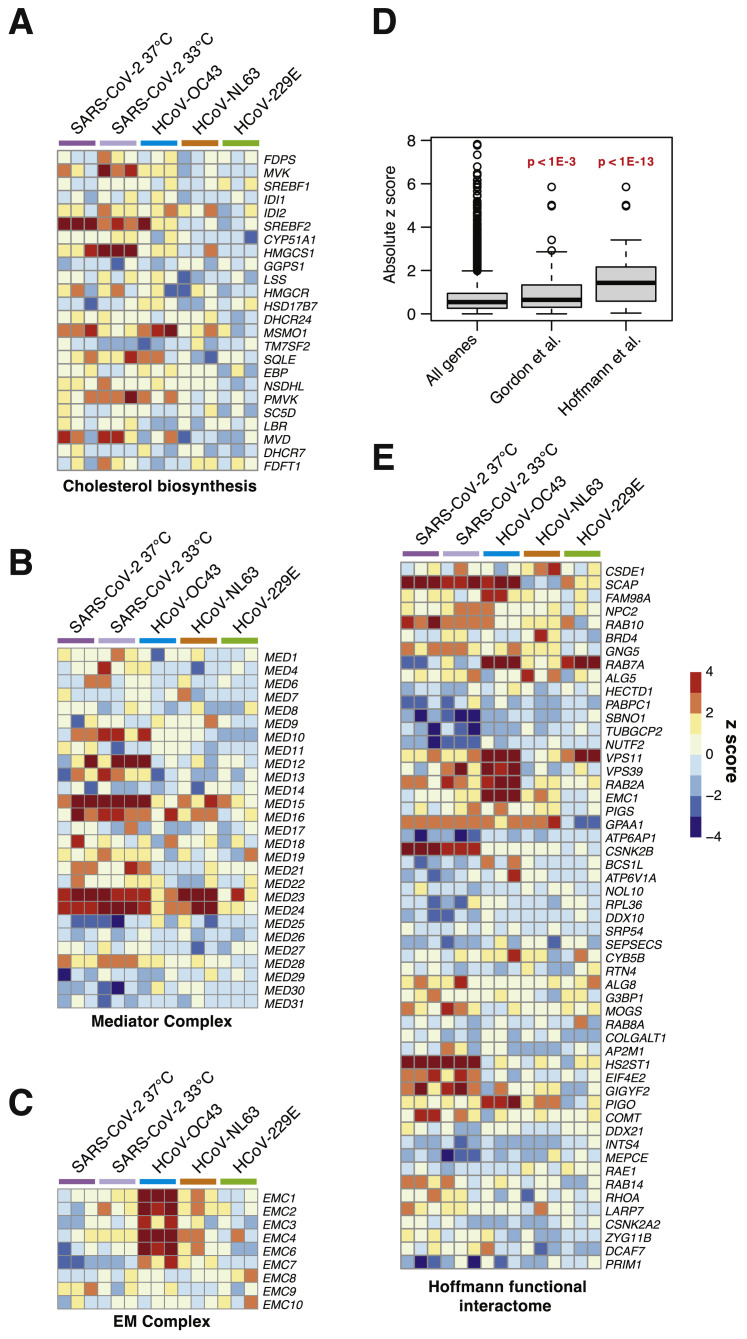
We also identified host factors and pathways that appear to be required for SARS-CoV-2 infection but less so for other CoVs tested. Genes in the mevalonate pathway, which is regulated by SREBP and is responsible for converting mevalonate into sterol isoprenoids, such as cholesterol (Buhaescu and Izzedine, 2007; Goldstein and Brown, 1990), were among the top scoring hits (Figures 3A and 3B; Figures S3 A and S4A). Multiple sgRNAs targeting 3-hydroxy-3-methylglutaryl-coenzyme A (CoA) synthase 1 (HMGCS1), which catalyzes conversion of HMG-CoA to mevalonic acid; mevalonate kinase (MVK), which catalyzes phosphorylation of mevalonic acid into phosphomevalonate; and phosphomevalonate kinase (PMVK), which converts phosphomevalonate to mevalonate 5-diphosphate were significantly enriched in SARS-CoV-2 screens, although to a lesser extent than SREBP signaling (Figures 3A and 3B; Figure S4A). These results suggest that factors and intermediates of the mevalonate pathway, which are known to play important roles in post-translational modification of many proteins involved in key processes, such as intracellular signaling and protein glycosylation (Buhaescu and Izzedine, 2007; Goldstein and Brown, 1990), are important for the SARS-CoV-2 life cycle.
Figure S4.
Additional Enriched Pathways and Subset Analysis of a High-Confidence SARS-CoV-2 Protein:Protein Interactome, Related to Figure 3
(A) Heatmap of z-scores for the cholesterol biosynthesis gene set.
(B) Heatmap of z-scores for the mediator complex gene set.
(C) Heatmap of z-scores for the endoplasmic reticulum membrane complex gene set.
(D) Subset analysis of absolute z-scores for SARS-CoV-2 screen at 37°C for the Gordon et al. high-confidence interactome and the Hoffmann et al., functional interactome. Whiskers represent the range, bar represents median, box represents first and third quartiles. Significance tests are two-sided Wilcoxon rank sum test.
(E) Heatmap of z-scores from functional interactome hits in Hoffmann et al. subsetted from genome-scale CRISPR screens.
Another set of top-scoring genes in SARS-CoV-2 screens encode multiple subunits of the exocyst complex, which regulates tethering of secretory vesicles to the plasma membrane and their subsequent SNARE-mediated membrane fusion and exocytosis (Martin-Urdiroz et al., 2016; Wu and Guo, 2015; Figure 3G). Multiple sgRNAs targeting EXOC1, EXOC2, and EXOC4 were significantly enriched, suggesting a critical role of these factors in mediating SARS-CoV-2 infection. The mammalian exocyst complex is known to interact with Rab GTPases to coordinate intracellular trafficking (Babbey et al., 2010; Mei and Guo, 2018). Indeed, sgRNAs targeting RAB6A and RAB10 were also among the most significantly enriched hits in SARS-CoV-2 screens (Figure 3), a finding that is consistent with our recent work identifying RAB10 as an important host factor for SARS-CoV-2 (Hoffmann et al., 2020b). In addition, we observed that RIC1 and RGP1, which together encode a guanine nucleotide exchange factor complex for RAB6, and Rab guanosine diphosphate dissociation inhibitor beta (GDI2), were significant hits. These results suggest that SARS-CoV-2 relies on specific intracellular host factors and complexes that govern intracellular transport.
Another complex that appears to play an essential role in the SARS-CoV-2 life cycle is the Mediator complex (Figure 3; Figure S4B). The mammalian Mediator is an evolutionarily conserved protein complex composed of at least 28 subunits that regulates transcription by functionally connecting general transcription factors with the core transcriptional machinery (Allen and Taatjes, 2015). The Mediator subunits MED10, MED12, MED15, MED16, MED23, and MED24 were among the top-scoring genes in SARS-CoV-2 screens, suggesting a critical role of this complex during infection and death by this virus (Figure 3). Intriguingly, a non-overlapping set of Mediator subunits was recently implicated in HIV-1 replication, including MED6, MED7, MED11, MED14, MED21, MED26, MED27, MED28, and MED30 (Ruiz et al., 2014), suggesting that different viruses might have specific requirements for members of this complex during transcription and replication of their genomes.
Beyond well-characterized pathways that were represented by multiple components, we also identified factors with less understood network-level connections (Figure 3A). These include the endoplasmic reticulum membrane protein complex (EMC) (Figure S4C), the DEAH-box helicases DHX36 and DHX38, the Golgin family GOLGA6L1 and GOLGA8O, the general transcription factor IIIC subunits GTF3C5 and GTF3C6, the tRNA methyltransferases TRMT5 and TRMT6, the G protein-coupled receptors GPR89A and GPR89B, the transmembrane P24-trafficking proteins TMED2 and TMED10, and genes involved in phosphatidylethanolamine biosynthesis, such as PCYT2 and EPT1, among others (Figure 3A; Table S1). Further mining of SARS-CoV-2 host factor networks could expand the repertoire of potential targetable factors to treat COVID-19.
Given the genome-scale depth of these screening data, we sought to determine whether interactome-focused networks were significantly enriched in a genome-wide context. A recent SARS-CoV-2 CRISPR screen in African green monkey VeroE6 cells (Wei et al., 2020) failed to detect significant enrichment from a SARS-CoV-2-human protein-protein network derived from immunoprecipitation (IP)-mass spectrometry (MS) (Gordon et al., 2020). We recently tested genes from this protein-protein interactome with a focused CRISPR screen and assigned functional relevance to putative interactors (Hoffmann et al., 2020b). We subsetted Z scores from the full interactome in Gordon et al. (2020) and detected a modest but significant enrichment for hits in a genome-wide context (Figure S4D). Upon subsetting the functionally relevant members from our focused screen in Hoffmann et al., 2020b we observed a striking increase in the degree of enrichment (Figure S4D). The subsetted heatmap of hits in Hoffmann et al. mapped onto genome-scale data largely cross-validated many but not all of our prior findings (Figure S4E). These results demonstrate the power of focused CRISPR screens to complement the breadth of genome-wide efforts.
HCoV-NL63 and HCoV-229E (Alpha-CoVs)
Interestingly, the catalog of host factors essential for infection by the alpha-CoVs HCoV-NL63 and HCoV-229E is substantially different than that of beta-CoVs (Figures 2 and 3; Figures S3C and S3D), suggesting that alpha- and beta-CoVs rely on different pathways to carry out their life cycles. Both alpha-CoV screens successfully identified known cognate virus receptors, as evidenced by robust enrichment of sgRNAs targeting ACE2 (HCoV-NL63) and ANPEP (HCoV-229E) (Hoffmann et al., 2020; Letko et al., 2020; Yeager et al., 1992; Figures 2 and 3). Intriguingly, HCoV-NL63 seems to rely on a core set of host chromatin regulators with known functional interactions, including EP300, KDM6A (also known as UTX), KMT2D (also known as MLL4), MED23, MED24, MEN1, PAXIP1, and SETDB1. This raises the tantalizing possibility that HCoV-NL63 co-opts the well established UTX-MLL4-EP300 enhancer remodeling network (which also contains PAXIP1) (Wang et al., 2017) to reprogram the host transcriptome for successful infection (Figure 3A, orange nodes). In addition, we observed a requirement for factors involved in BMP signaling with the HCoV-NL63-specific factors SMAD4, SMAD5, ACVR1, and BMPR1A (Figure 3F). Collectively, our results demonstrate that even closely related viruses, such as the alpha-CoVs HCoV-NL63 and HCoV-229E, may employ different host factor pathways during infection, which may in part be linked to their different receptor usage.
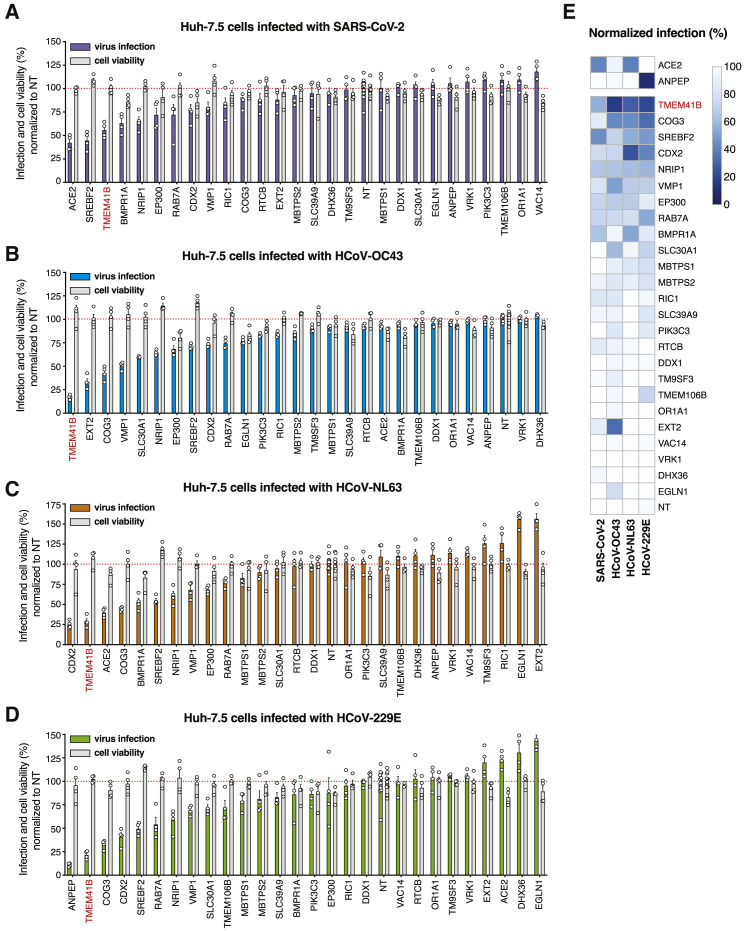
Orthogonal Validation of Candidate CoV Host Factors
As shown in Figure 3 and described above, many of the genes enriched in each virus screen converged on specific pathways and protein complexes. Given the size of this network, we performed targeted validation experiments on a representative number of putative pan-CoV and virus-specific host factors (n = 27 genes) using arrayed CRISPR and direct viral antigen staining as an orthogonal measurement of infection. To do so, we transfected Huh-7.5-Cas9 cells with crRNA:tracrRNA in a 96-well format where each well contained four unique crRNAs per targeted gene. We incubated the cells for 4 days post-transfection to allow time for genome editing and protein depletion before infecting cells with each of the four CoVs. The results from these validation experiments are displayed in Figure 4 . Known receptors—ANPEP for HCoV-229E and ACE2 for SARS-CoV-2 and HCoV-NL63—were among the genes that displayed the strongest requirement for infection, validating our assay (Figures 4A and 4C–4E). Disruption of other genes, including SREBF2 (SREBP signaling), CDX2 (a transcription factor known to be regulated by BMP signaling), and COG3 (from the COG complex), among others, reduced infection by all four CoVs tested. Strikingly, these data also highlighted TMEM41B as a candidate host factor whose disruption appeared to block infection by all CoVs similar to the levels achieved by ablation of cognate viral receptors (Figure 4E). Overall, as highlighted in Figures 4A–4D and depicted in the heatmap presented in Figure 4E, TMEM41B scored among the top three required genes for all CoVs tested. Based on these results, we chose to investigate TMEM41B in greater depth.
Figure 4.
Validation of High-Confidence Coronaviridae Host Factors
(A) Candidate validation in Huh-7.5 cells with SARS-CoV-2 infection at 33°C.
(B) Candidate validation in Huh-7.5 cells with HCoV-OC43 infection at 33°C.
(C) Candidate validation in Huh-7.5 cells with HCoV-NL63 at 33°C.
(D) Candidate validation in Huh-7.5 cells with HCoV-229E at 37°C.
(E) Heatmap representation of data from (A)–(D). The SARS-CoV-2 and HCoV-NL-63 receptor (ACE2) and the HCoV-229E receptor (ANPEP) are shown separately.
TMEM41B Is a Pan-CoV Host Factor
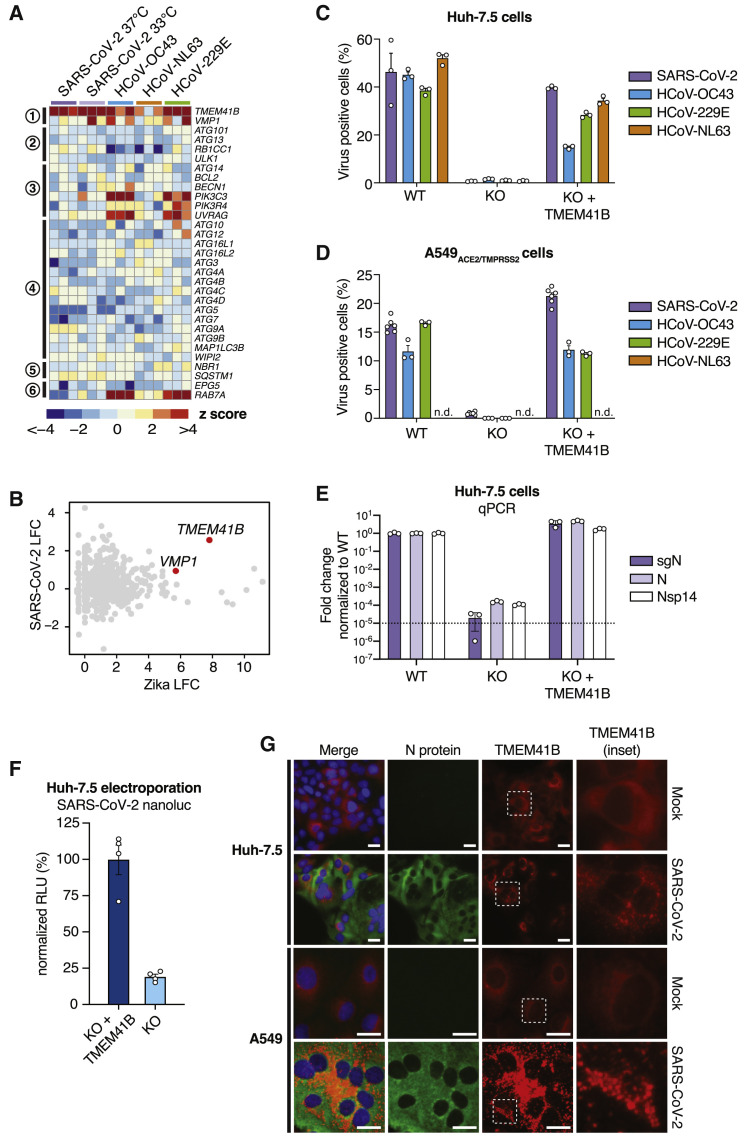
TMEM41B is a poorly understood endoplasmic reticulum (ER)-localized transmembrane protein that was previously found to be required for synaptic transmission in motor circuit neurons (Imlach et al., 2012, Lotti et al., 2012) and has been recently implicated in the autophagy pathway (Moretti et al., 2018; Morita et al., 2018; Shoemaker et al., 2019). Specifically, TMEM41B deficiency has been shown to lead to accumulation of ATG proteins, blocking the autophagy pathway at the early step of autophagosome formation (Morita et al., 2018). In addition, TMEM41B deficiency has been shown to trigger the abnormal accumulation of intracellular lipid droplets. These phenotypes have been linked to the function of another autophagy factor, vacuole membrane protein 1 (VMP1), which shares a rare and characteristic VTT domain (Morita et al., 2019). Interestingly, multiple sgRNAs targeting VMP1 were significantly enriched in the HCoV-229E screen (Figure 2C), and VMP1 was also validated in our orthogonal arrayed CRISPR experiments (Figure 4). These findings prompted us to inspect the autophagy network across all CoV screens in more detail.
TMEM41B was the only autophagy-related gene that scored as a significant hit across multiple CoV screens, and only a handful of genes involved in the nucleation and tethering steps scored for HCoV-OC43 and HCoV-229E (Figure 5 A). This was a striking finding given that, in related work, we also identified TMEM41B as a critical host factor required for infection by numerous members of the Flaviviridae family (Hoffmann et al., 2020a). To illustrate this, we compared Zika virus and SARS-CoV-2 screens and found that TMEM41B was the top-scoring host factor critical for infection by both viruses (Figure 5B; Hoffmann et al., 2020a). Indeed, similar to the pan-flavivirus requirement for TMEM41B, genetic deletion of TMEM41B in Huh-7.5 and A549ACE2/TMPRSS2 cells strongly impaired infection by all four CoVs tested, and infectivity was restored upon reconstitution with TMEM41B cDNA (Figures 5C–5E).
Figure 5.
TMEM41B Is a Pan-CoV Host Factor
(A) Heatmap of genes from the autophagy pathway, ordered sequentially by function in the autophagy cascade.
(B) Comparative analysis of Zika virus and SARS-CoV-2 screens by log2 fold change (LFC).
(C) CoV infectivity assay in parental Huh-7.5 cells, clonal TMEM41B knockout cells, and TMEM41B knockout cells reconstituted with TMEM41B cDNA.
(D) CoV infectivity assay in parental A549ACE2/TMPRSS2 cells, clonal TMEM41B knockout cells, and TMEM41B knockout cells reconstituted with TMEM41B cDNA.
(E) Quantitative PCR analysis of the levels of three SARS-CoV-2 RNA transcripts in parental Huh-7.5 cells, clonal TMEM41B knockout cells, and TMEM41B knockout cells reconstituted with TMEM41B cDNA. Error bars represent standard deviation (SD).
(F) Measurement of SARS-CoV-2 replication in Huh-7.5 TMEM41B knockout cells and TMEM41B knockout cells reconstituted with TMEM41B cDNA electroporated with RNA of SARS-CoV-2 expressing NanoLuc. Measurements were performed 18 h post-electroporation and are expressed in relative luciferase units (RLUs) normalized to reconstituted knockout cells.
(G) Fluorescence microscopy images of SARS-CoV-2 immunostaining (green), tagRFP-TMEM41B (red), and DNA (DAPI) in parental Huh-7.5 and A549ACE2/TMPRSS2 cells, clonal TMEM41B knockout cells, and TMEM41B knockout cells reconstituted with TMEM41B cDNA. Scale bars, 30 μm.
Based on TMEM41B studies in the flavivirus replication cycle (Hoffmann et al., 2020a), we proposed that TMEM41B is required post-entry to facilitate the ER membrane remodeling necessary to form replication organelles. Like flaviviruses, CoVs also remodel ER membranes to establish membrane-protected viral RNA replication complexes (Knoops et al., 2008; Snijder et al., 2020). It is possible that TMEM41B plays a similar role in the CoV life cycle. To begin to test this hypothesis, we next utilized a SARS-CoV-2 reporter virus expressing nanoluciferase (NanoLuc) to determine whether TMEM41B was required after virus entry. To do so, we electroporated Huh-7.5 TMEM41B knockout and TMEM41B reconstituted knockout cells with SARS-CoV-2 RNA encoding NanoLuc in place of ORF7a and quantified luciferase activity 18 h post-electroporation. As shown in Figure 5F, we observed a more than 80% reduction in luciferase activity in cells lacking TMEM41B. Although TMEM41B may also have a role in CoV entry, this result demonstrates that TMEM41B is required for SARS-CoV-2 replication even when the entry step is bypassed.
We found previously that TMEM41B relocalizes from a diffuse reticular-like pattern consistent with ER localization to large aggregates upon flavivirus infection (Hoffmann et al., 2020a). To determine whether TMEM41B also changes subcellular localization upon infection with CoVs, we reconstituted TMEM41B knockout Huh-7.5 and A549ACE2/TMPRSS2 cells with a red fluorescent protein (RFP)-tagged TMEM41B construct followed by SARS-CoV-2 infection. Interestingly, we found that TMEM41B sub-cellular localization changed dramatically in both cell lines 24 h after infection (Figure 5G). SARS-CoV-2-infected cells were characterized by distinct TMEM41B cytosolic aggregates and positive immunostaining for the SARS-CoV-2 nucleoprotein (Figure 5G). These data establish TMEM41B as a critical pan-CoV host factor.
Discussion
The full complement of human proteins and pathways required for infection by SARS-CoV-2 remains poorly defined. A more complete understanding of the cellular and molecular mechanisms that are co-opted by CoVs could catalyze drug development efforts to combat the ongoing COVID-19 pandemic and prepare for potential future CoV outbreaks. We performed parallel genome-scale CRISPR-Cas9 knockout screens to generate an extensive functional catalog of host factors required for infection by SARS-CoV-2 and three seasonal CoVs (HCoV-OC43, HCoV-NL63, and HCoV-229E).
This catalog contains multiple host factors and pathways that play critical pan-CoV and virus-specific functional roles. For instance, we identified a requirement for factors involved in GAG biosynthesis, modification, and transport as potential targetable factors to inhibit SARS-CoV-2 and HCoV-OC43. We also identified proteins that mediate cholesterol homeostasis via SREBP signaling as key host factors for infection by SARS-CoV-2 and HCoV-OC43. These results agree with and substantially extend results from our recent functional interactome study, demonstrating that SARS-CoV-2 and other CoVs require SCAP, NPC2, and EMC1 (Hoffmann et al., 2020b). These results are also consistent with work on other viruses, including Ebola virus (Carette et al., 2011) and hantavirus (Kleinfelter et al., 2015). We also identified key enzymes from the mevalonate pathway. Given that statins block mevalonate production by inhibiting HMG-CoA reductase and have been associated recently with improved outcomes among COVID-19 patients (Fajgenbaum and Rader, 2020; Zhang et al., 2020), we speculate that pharmacological modulation of this pathway could be a strategy for treating COVID-19. Interestingly, we also identified STARD3, STARD3NL, and the key STARD3 interactor MOSPD2 as negative regulators of SARS-CoV-2 infection. STARD3 mediates ER-to-endosome cholesterol transport. One study linked STARD3 repression to genetic obesity, positing a role for lipid export (Soffientini et al., 2014). Given that obesity is a major risk factor for COVID-19, further studies are warranted to determine whether STARD3 repression plays a role in SARS-CoV-2 pathogenesis.
Another set of critical SARS-CoV-2 host factors are Rab GTPases and Rab GTPase regulatory proteins that regulate intracellular transport, tethering, and exocytosis of secretory vesicles. These results support our recent nomination of RAB10 as a putative SARS-CoV-2 host factor (Hoffmann et al., 2020b) and suggest that different CoVs might have differential requirements for intracellular transport proteins.
Despite the many common host factor requirements identified in our SARS-CoV-2 screens at 33°C and 37°C, we also identified host factors that appear to be differentially required at these temperatures. It will be interesting to determine whether these differences are reproducible in more relevant model systems (e.g., animal models) and how such differences may influence tissue tropism and disease pathogenesis.
Our data also suggest that alpha-CoVs and beta-CoVs differentially co-opt a number of host factors. For instance, HCoV-NL63 seems to be particularly dependent on chromatin regulators with known functional interactions. By integrating our Coronaviridae screening data, we identified TMEM41B as a critical host factor for infection by SARS-CoV-2, HCoV-OC43, HCoV-NL63, and HCoV-229E. TMEM41B is a poorly understood ER-localized transmembrane protein that has been shown recently by three independent groups to regulate autophagy in conjunction with VMP1 (Moretti et al., 2018; Morita et al., 2018; Shoemaker et al., 2019). Strikingly, TMEM41B was the only gene implicated in autophagy that scored as a significant hit across the SARS-CoV-2, HCoV-OC43, and HCoV-229E CRISPR screens, and it was subsequently validated as a cofactor for HCoV-NL63 as well. This suggests a putative autophagy-independent role for TMEM41B as a pan-CoV replication factor. Moreover, our related functional and mechanistic work showed that TMEM41B is also required for infection by more than 10 diverse flaviviruses (Hoffmann et al., 2020a). Thus, TMEM41B is a critical host factor required for infection by all of the CoVs tested in our study as well as several other viruses of high public health interest and, therefore, is an attractive target for further investigation.
CoV host factor discovery and validation is an active area of research, with multiple studies appearing in press and on preprint servers in recent months (Baggen et al., 2020; Daniloski et al., 2020; Heaton et al., 2020; Hoffmann et al., 2020b; Wang et al., 2020; Wei et al., 2020; Zhu et al., 2020b). One group performed genome-wide CRISPR screens in the African green monkey kidney cell line VeroE6 and reported strong dependency on ACE2 and CTSL, consistent with our study (Wei et al., 2020). Another group performed a similar screen in ACE2-overexpressing A549 cells and identified serine/arginine-rich protein-specific kinase 1 (SRPK1) as a single dominant hit unique to their study (Heaton et al., 2020). Zhu et al. (2020b) also performed a screen using A549-ACE2 cells; however, in contrast to Heaton et al. (2020), they identified ACE2, CTSL, and elements of the retromer COMMD/CCDC22/CCDC93 (CCC), Wiskott-Aldrich syndrome protein and SCAR homolog (WASH), and actin-related proteins-2/3 (Arp2/3) complexes but not SRPK1. More recently, a series of screens using Huh-7.5.1 cells overexpressing ACE2 and TMPRSS2 identified four statistically significant host factors, including SCAP, validating our recent study (Wang et al., 2020; Hoffmann et al., 2020b). The other three factors (TMEM106B, VAC14, and ACE2) were also identified in the present study. Another group performed SARS-CoV-2 and HCoV-229E screens using Huh-7 cells and highlighted TMEM106B, similar to Wang et al. (2020) and our study, as well as TMEM41B, the focus of this study and our accompanying manuscript describing TMEM41B as a pan-flavivirus host factor (Baggen et al., 2020; Hoffmann et al., 2020a). Most recently, Daniloski et al. (2020) performed screens in A549-ACE2 cells and validated eight genes via secondary screening, including ACE2 and CTSL. Overall, our study identified 128 high-confidence SARS-CoV-2 host factors— the largest catalog reported to date—by performing genome-scale CRISPR screens in cells that did not require ectopic ACE2 expression.
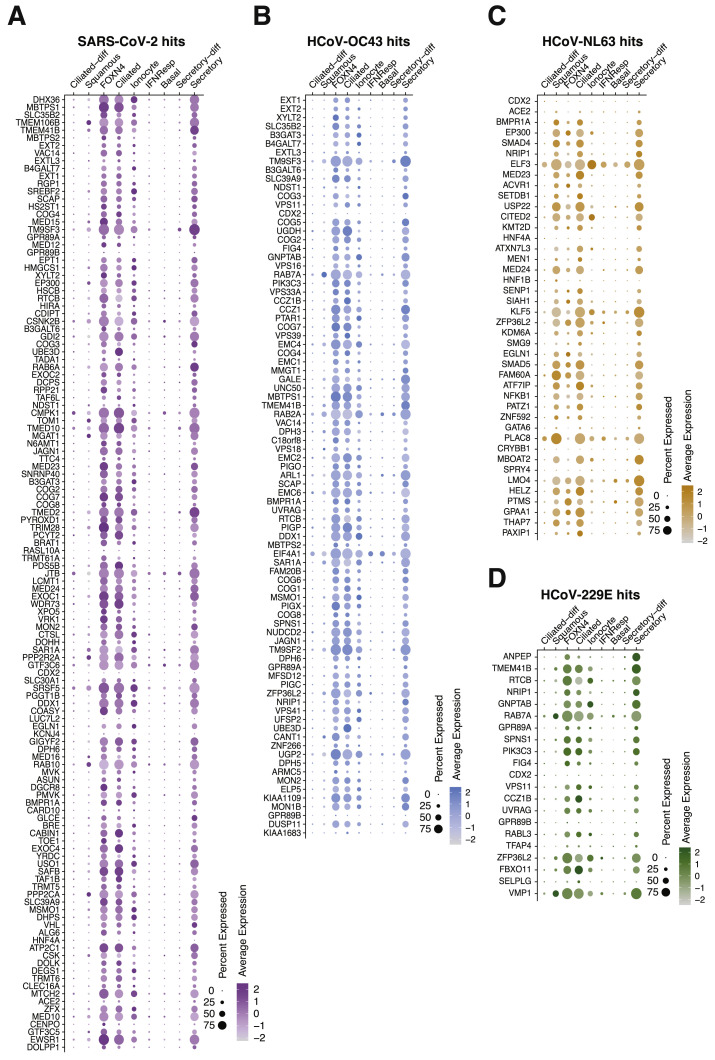
The results of this study should be interpreted within the context of its limitations. First, pooled CRISPR screens may not identify functionally redundant or buffering genes (Ewen-Campen et al., 2017). Second, Huh-7.5 cells were chosen based on their infectivity by multiple CoVs, but they are not airway cells. Nevertheless, the present study and recent work have demonstrated that hits in Huh-7 cells translate to human cells of lung origin (Baggen et al., 2020). Furthermore, as shown in Figure S5 , the majority of genes identified here are expressed in human cells and tissues known to be infected by SARS-CoV-2. Last, our current experimental system is limited to assessing survival and is therefore best suited to identify host factors required for virus replication rather than virus restriction factors. Furthermore, pooled cell survival assays are not well suited to interrogate host factors required for virus egress from cells or identify genes that play important roles in immune modulation and pathogenesis.
Figure S5.
A Majority of Functional CoV Host Factors Are Expressed in the Airway, Related to Figures 1, 2, and 3
(A–D) scRNaseq expression dotplot diagrams from select cells in the airway of the average expression and the percent of cells expressing coronavirus host factors for SARS-CoV-2 (A), HCoV-OC43 (B), HCoV-NL63 (C) and HCoV-229E (D). Rows for each diagram are ordered top to bottom from highest to lowest z-score. Data are from Chua et al. (2020).
Strengths of our study include internal consistency among individual screens and across four independent CoVs. Furthermore, our cell line platform is permissive to all four CoVs screened without modification. Notably, we found that ACE2 overexpression rendered cells permissive to SARS-CoV-2, but it also promoted syncytia, resulting in massive multinucleated cells. Syncytia is likely caused by ectopic ACE2 overexpression, causing cells to fuse when they are adjacent to infected cells producing SARS-CoV-2 Spike protein. Syncytia is a major technical and biological limitation for pooled CRISPR screens because SARS-CoV-2-resistant cells that would normally survive infection may be killed by their infected neighbors, limiting the number of host factors that can be discovered and biasing the results toward genes that alter cell surface ACE2 levels.
In summary, we identified complex interconnected networks of CoV host factors and pathways that are essential for virus infection, nominating hundreds of host proteins that represent liabilities for SARS-CoV-2 and potential opportunities for therapeutic intervention. This represents an extensive functional catalog of host factors required for infection by SARS-CoV-2 and three seasonal CoVs (HCoV-229E, HCoV-NL63, and HCoV-OC43), providing a larger context in which to interpret ongoing and future large-scale CRISPR studies. Future efforts will focus on dissecting the complex interplay between virus and host and direct medicinal chemistry and drug-repurposing resources toward the most chemically tractable targets.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit polyclonal anti-SARS-CoV-2 nucleoprotein | GeneTex, Inc. | Cat#GTX135357; RRID: AB_2868464 |
| mouse monoclonal anti-dsRNA antibody J2 | Scicons | Cat#10010500; RRID: AB_2651015 |
| goat polyclonal anti-rabbit IgG conjugated to AF594 | ThermoFisher Scientific, Inc. | Cat#A-11012; RRID: AB_141359 |
| goat polyclonal anti-mouse IgG conjugated to AF488 | ThermoFisher Scientific, Inc. | Cat#A-11001; RRID: AB_2534069 |
| goat polyclonal anti-rabbit IgG conjugated to AF488 | ThermoFisher Scientific, Inc. | Cat#A-11008; RRID: AB_143165 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 (strain: WA1/2020) GenBank: MT246667.1 |
BEI Resources | Cat#NR-52281 |
| HCoV-NL63 GenBank: AY567487 |
BEI Resources | Cat#NR-470 |
| HCoV-OC43 | ZeptoMetrix | Cat#0810024CF |
| HCoV-229E | Laboratory of Volker Thiel | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tri-reagent | Zymo research | Cat#R2050 |
| TRIzol Reagent | ThermoFisher Scientific | Cat#15596026 |
| DharmaFECT-4 | ThermoFisher Scientific | Cat#NC1411281 |
| Hoechst 33342 solution | ThermoFisher Scientific, Inc. | Cat#62249; CAS: 23491-52-3 |
| BsmBI Restriction Enzyme | New England Biolabs | Cat#R0739L |
| Anti-Reverse Cap Analog (ARCA) | NEB | Cat#S1411S |
| Critical Commercial Assays | ||
| Superscript III First-Strand Synthesis System Kit | Invitrogen | Cat#18080051 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Cat#A25742 |
| iScribe T7 High Yield RNA Synthesis kit | NEB | Cat#E2040S |
| Nano-Glo® Luciferase Assay System | Promega | Cat#N1120 |
| Monarch Genomic DNA Purification kit | New England Biolabs | Cat#T3010L |
| Direct-zol RNA Miniprep Plus kit | Zymo Research | Cat#R2072 |
| RNeasy mini kit | QIAGEN | Cat#74014 |
| Monarch® RNA Cleanup Kit | NEB | Cat#T2050L |
| Deposited Data | ||
| Raw NGS reads and raw counts | This study | GEO: GSE162038 |
| Screening pipeline output | This study | https://doi.org/10.17632/7bd5bhmhmz.1 |
| Experimental Models: Cell Lines | ||
| Human: Lenti-X 293T (embryonic kidney epithelial) | Takara Bio Inc. | Cat. #632180; RRID: CVCL_4401 |
| Human: Huh-7.5 (hepatocyte) | Laboratory of Charles M. Rice | RRID: CVCL_7927 |
| Human: A549 (lung epithelial) | ATCC | Cat. #CCL-185; RRID: CVCL_0023 |
| Oligonucleotides | ||
| SARS-CoV-2 subgenomic N forward: 5′-GTTTATACCTTCCCAGGTAACAAACC-3′ | This study | N/A |
| SARS-CoV-2 subgenomic N reverse: 5′-GTAGAAATACCATCTTGGACTGAGATC-3′ | This study | N/A |
| RPS11 forward: 5′-GCCGAGACTATCT GCACTAC-3′ |
This study | N/A |
| RPS11 reverse: 5′-ATGTCCAGCCTC AGAACTTC-3′ |
This study | N/A |
| SARS-CoV-2 N forward: 5′-TAATC AGACAAGGAACTGATTA-3′ |
Chu et al., 2020 | N/A |
| SARS-CoV-2 N reverse: 5′-CGAAG GTGTGACTTCCATG-3′ |
Chu et al., 2020 | N/A |
| SARS-CoV-2 Nsp14 forward: 5′-TGG GGYTTTACRGGTAACCT-3′ |
Chu et al., 2020 | N/A |
| SARS-CoV-2 Nsp14 reverse: 5′-AACRCGCTTAACAAAGCACTC-3′ |
Chu et al., 2020 | N/A |
| Recombinant DNA | ||
| Plasmid: LentiCas9-Blast | Sanjana et al., 2014 | Addgene: Cat#52962; RRID: Addgene_52962 |
| Plasmid: LentiCRISPRv2 | Sanjana et al., 2014 | Addgene: Cat#52961; RRID: Addgene_52961 |
| Plasmid: HIV-1 Gag-Pol | This study | N/A |
| Plasmid: VSV-G | This study | N/A |
| Human CRISPR Brunello library | Doench et al., 2016 | Addgene: Cat#73178-LV RRID: Addgene_73178 |
| Plasmid: pSCRPSY_TMPRSS2-2A-NeoR_ACE2 | This study | N/A |
| Software and Algorithms | ||
| ImageXpress Micro XLS | Molecular Devices | |
| Prism | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK) | Li et al., 2014 | https://sourceforge.net/p/mageck/wiki/Home/ |
| Cytoscape | Shannon et al., 2003 | https://cytoscape.org/ |
| Seurat | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| Other | ||
| Revolve inverted microscope | ECHO | https://discover-echo.com/revolve |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John T. Poirier (John.Poirier@nyulangone.org).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Data supporting the findings of this study are reported in Figures S1–S6 and Tables S1-S2. The accession number for the CRISPR screen sequencing data reported in this paper is GEO: GSE162038. Networks are available on NDEx. All reagents and materials generated in this study will be available to the scientific community through Addgene and/or MTAs.
Experimental Model and Subject Details
Cell culture
Lenti-X 293T cells (H. sapiens; sex: female) (Takara, cat. #632180), Huh-7.5 cells (H. sapiens; sex: male) (Blight et al., 2002), and A549 cells (H. sapiens; sex: male) (ATCC®, cat. #CCL-185) were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM, Fisher Scientific, cat. #11995065) supplemented with 0.1 mM nonessential amino acids (NEAA, Fisher Scientific, cat. #11140076) and 10% hyclone fetal bovine serum (FBS, HyClone Laboratories, Lot. #AUJ35777). All cell lines have tested negative for contamination with mycoplasma.
Production and titration of coronavirus stocks
SARS-CoV-2 (strain: USA-WA1/2020) and HCoV-NL63 were obtained from BEI Resources (NR-52281 and NR-470). HCoV-OC43 was obtained from ZeptoMetrix (cat. #0810024CF) and HCoV-229E was generously provided by Volker Thiel (University of Bern). All viruses were amplified at 33°C in Huh-7.5 cells to generate a P1 stock. To generate working stocks, Huh-7.5 cells were infected at a multiplicity of infection (MOI) of 0.01 plaque forming unit (PFU)/cell (SARS-CoV-2, HCoV-NL63, HCoV-OC43) and 0.1 PFU/cell (HCoV-229E) and incubated at 33°C until virus-induced CPE was observed. Supernatants were subsequently harvested, clarified by centrifugation (3,000 g × 10 min) at 4 dpi (HCoV-229E), 6 dpi (SARS-CoV-2, HCoV-OC43) and 10 dpi (HCoV-NL63), and aliquots stored at −80°C.
Viral titers were measured on Huh-7.5 cells by standard plaque assay. Briefly, 500 μL of serial 10-fold virus dilutions in Opti-MEM were used to infect 4 × 105 cells seeded the day prior into wells of a 6-well plate. After 90 min adsorption, the virus inoculum was removed, and cells were overlaid with DMEM containing 10% FBS with 1.2% microcrystalline cellulose (Avicel). Cells were incubated for 4 days (HCoV-229E), 5 days (SARS-CoV-2, HCoV-OC43) and 6 days (HCoV-NL63) at 33°C, followed by fixation with 7% formaldehyde and crystal violet staining for plaque enumeration. All SARS-CoV-2 experiments were performed in a biosafety level 3 laboratory.
To confirm the identity of the viruses, RNA from 200 μl of each viral stock was purified by adding 800 μl TRIzol Reagent (ThermoFisher Scientific, cat. #15596026) plus 200 μl chloroform then centrifuged at 12,000 g x 5 min. The upper aqueous phase was moved to a new tube and an equal volume of isopropanol was added. This was then added to an RNeasy mini kit column (QIAGEN, cat. #74014) and further purified following the manufacturer’s instructions. Viral stocks were confirmed via next generation sequencing at the NYU Genome Technology Center using an Illumina stranded TruSeq kit and omitting the polyA selection step. Libraries were then sequenced by MiSeq Micro (2 × 250 bp paired end reads).
Method Details
Plasmids and sgRNA cloning
To generate stable Cas9-expressing cell lines, we used lentiCas9-Blast (Addgene, cat. #52962). To express sgRNAs, we used lentiGuidePurov2, a variant of lentiGuide-Puro (Addgene, cat. #52963) that contains an improved sgRNA scaffold based on Chen et al. (2013). For sgRNA cloning, lentiGuidePurov2 was linearized with BsmBI (NEB) and ligated with BsmBI-compatible annealed and phosphorylated oligos encoding sgRNAs using high concentration T4 DNA ligase (NEB). HIV-1 Gag-Pol and VSV-G plasmid sequences are available upon request.
Arrayed validation of gene candidates using CRISPR knockdown
A total of 4 × 103 Huh-7.5-Cas9 cells per well were seeded on 96-well plates in quadruplicate, co-transfected with tracRNA and crRNAs targeting individual genes (Dharmacon, Inc.) 2-4 hours post-seeding using DharmaFECT-4 (ThermoFisher Scientific, cat. #NC1411281) according to the manufacturer’s protocol. Cells were incubated for 4 days at 37°C followed by infection with different coronaviruses under optimal conditions for each virus by directly applying 50 ul of virus inoculum to each well at the following MOI’s: SARS-CoV-2: 500 PFU/well, HCoV-OC43: 15,000 PFU/well, HCoV-229E: 150 PFU/well and HCoV-NL63: 100 PFU/well. SARS-CoV-2 infected plates were incubated for 48 hours at 33°C, HCoV-229E-infected plates for 48 hours at 37°C, and HCoV-OC43- and HCoV-NL63-infected plates for 72 hours at 33°C. Cells were then fixed by adding an equal volume of 7% formaldehyde to the wells, followed by permeabilization with 0.1% Triton X-100 for 10 min. After extensive washing, cells were incubated for 1 hour at room temperature with a blocking solution of 5% goat serum in PBS (Jackson ImmunoResearch, cat. #005–000-121). To stain SARS-CoV-2 infected cells, a rabbit polyclonal anti-SARS-CoV-2 nucleocapsid antibody (GeneTex, cat. #GTX135357) was added to the cells at 1:1,000 dilution in blocking solution. The mouse monoclonal anti-dsRNA antibody J2 (Scicons, cat. #10010500) was used at a 1:1,000 dilution to stain cells infected with HCoV-229E, HCoV-NL63 and HCoV-OC43. After overnight incubation at 4°C, cells were washed and stained with secondary antibodies at a 1:2,000 dilution: goat anti-rabbit AlexaFluor 594 (Life Technologies, cat. #A-11012) and goat anti-mouse AlexaFluor 488 (Life Technologies, cat. #A-11001). Nuclei were stained with Hoechst 33342 (ThermoFisher Scientific, cat. #62249) at 1 μg/ml. Images were acquired with a fluorescence microscope and analyzed using ImageXpress Micro XLS (Molecular Devices, Sunnyvale, CA). We included non-targeting crRNAs (Dharmacon, Inc.) as controls for all of these experiments.
Generation of A549ACE2/TMPRSS2 cells
To render A549 cells permissive to SARS-CoV-2 infection, we delivered human ACE2 and TMPRSS2 cDNA to cells by lentivirus transduction with pSCRPSY_TMPRSS2-2A-NeoR_ACE2, a modified SCRPSY vector (GenBank: KT368137.1). We generated the pSCRPSY_TMPRSS2-2A-NeoR_ACE2 lentiviral construct by cloning the ACE2 open reading frame into the multiple cloning site and by replacing the PAC (puromycin acetyl transferase) 2A (stop-start/skip from FMDV) tagRFP (red fluorescent protein) cassette with a TMRPSS2-2A-NeoR (neomycin phosphotransferase II; NPT II) cassette. The pSCRPSY_TMPRSS2-2A-NeoR_ACE2 plasmid sequence is available upon request.
Infection of TMEM41B knockout cells with SARS-CoV-2, HCoV-OC43, HCoV-NL63, and HCoV-229E
Huh-7.5 and A549 TMEM41B knockout cells (KO) and their reconstituted (tagRFP-TMEM41B) counterpart were generated as described in (Hoffmann et al., 2020a). To facilitate infection of A549 cells with SARS-CoV-2, cells were stably reconstituted to express ACE2 and TMPRSS2 as described above. The day prior to infection, parental Huh-7.5 and A549ACE2/TMPRSS2 WT, clonal TMEM41B KO and reconstituted KO cells were seeded into 96-well plates at different densities relative to time of fixation e.g., 1 × 104, 7.5 × 103 and 5 × 103 cells/well for a 24, 48, and 72 hours post infection time point, respectively. Cells were infected with the different coronaviruses under optimal conditions for each virus by directly applying 50 uL of virus inoculum to each well (n = 3-6) at the following MOIs for Huh-7.5 cells: SARS-CoV-2: 0.05 PFU/cell, HCoV-OC43: 2 PFU/cell, HCoV-229E: 0.15 PFU/cell and HCoV-NL63: 0.05 PFU/cell, and incubated for 24 hours at 37°C (HCoV-229E), 48 hours at 33°C (SARS-CoV-2), and 72 hours at 33°C (HCoV-OC43 and HCoV-NL63). A549ACE2/TMPRSS2 cells were infected under the following conditions and MOIs: SARS-CoV-2: 0.05 PFU/cell, HCoV-OC43: 2 PFU/cell, HCoV-229E: 0.1 PFU/cell and HCoV-NL63: 0.03 PFU/cell, and incubated for 24 hours at 37°C (SARS-CoV-2) and 72 hours at 33°C (HCoV-OC43, HCoV-229E and HCoV-NL63). Cells were subsequently fixed and stained as described above. Both secondary antibodies goat anti-rabbit and goat anti-mouse were conjugated to AlexaFluor 488 (Life Technologies: cat. #A-11008 and cat. #A-11001) to allow imaging tagRFP-TMEM41B. Images for quantification of virus infection and cell viability were acquired with a fluorescence microscope and analyzed using ImageXpress Micro XLS (Molecular Devices, Sunnyvale, CA). Images for assessment of tagRFP-TMEM41B subcellular localization were obtained using a Revolve inverted microscope (Echo, San Diego, CA).
qPCR of SARS-CoV infected cells
Parental Huh-7.5 WT, clonal TMEM41B KO and TMEM41B-reconstituted KO cells were seeded into 24-well plates in triplicate at 5 × 104 cells/well. The next day, cells were washed once with OptiMEM and infected with SARS-CoV-2 (MOI = 0.35 PFU/cell) diluted in OptiMEM and supplemented with 1 μg/ml TPCK-treated trypsin (Sigma-Aldrich, cat. #T1426) increasing the rate of infection. After an 1 hour incubation period at 37°C, the inoculum was removed, cells were washed three times with OptiMEM to remove residual virus before adding back regular DMEM medium. After incubating for 24 hours at 37°C, supernatants were aspirated, cells were washed three times with PBS and subsequently lysed in 250 μl Tri-reagent (Zymo, cat. #R2050) per well. RNA was extracted using the Direct-zol RNA Miniprep Plus kit (Zymo Research, cat. #R2072) according to the manufacturer’s protocol, followed by reverse transcription into cDNA using random hexamer primers with the Superscript III First-Strand Synthesis System Kit (Invitrogen, cat. #18080051) following the manufacturer’s instructions. Gene expression was quantified by qRT-PCR using PowerUp SYBR Green Master Mix (Applied Biosystems, cat. #A25742) and gene-specific primers for RPS11 (forward: 5′-GCCGAGACTATCTGCACTAC-3′ and reverse: 5′-ATGTCCAGCCTCAGAACTTC-3′) and SARS-CoV-2 subgenomic N (Leader forward: 5′-GTTTATACCTTCCCAGGTAACAAACC-3′ and N reverse: 5′-GTAGAAATACCATCTTGGACTGAGATC-3′). SARS-CoV-2 primers targeting genomic N (forward: 5′-TAATCAGACAAGGAACTGATTA-3′ and reverse: 5′-CGAAGGTGTGACTTCCATG-3′) and Nsp14 regions (forward: 5′-TGGGGYTTTACRGGTAACCT-3′ and reverse: 5′-AACRCGCTTAACAAAGCACTC-3′) are from Chu et al. (2020). The following PCR conditions were used: 50°C for 2 min and 95°C for 2 min (initial denaturation); 45 cycles 95°C for 1 s, 60°C for 30 s (PCR); followed by 95°C for 15 s, 65°C for 10 s, a slow increase to 95°C (0.07°C/sec) for a melt curve. The data were analyzed by melt curve analysis for product specificity as well as ΔΔCT analysis for fold changes (after normalization to housekeeping genes) and graphed using Prism 8 (GraphPad).
SARS-CoV-2 mNeon-NanoLuc reporter virus assay
The SARS-CoV-2 mNeon-NanoLuc reporter virus was constructed by yeast transformation-associated recombination (TAR) and cloned into a pCC1-BAC-HIS3 vector, as described in Thao et al. (2020). Monomeric NeonGreen (mNeon) fused to nanoluciferase (NanoLuc) (Hall et al., 2012) was cloned in place of ORF7a. DNA Template for in-vitro transcription was prepared as in Thao et al. (2020) with modifications, digested with EagI enzyme (NEB), and cleaned with phenol-chloroform-isoamylalcohol (SigmaAldrich, cat. #77617) followed by ethanol precipitation. RNA was synthesized from 1 μg linear DNA template using HiScribe T7 High Yield RNA Synthesis kit (NEB, cat. #E2040S) and Anti-Reverse Cap Analog (ARCA) (NEB, cat. #S1411S) at a GTP:ARCA ratio of 1:2.8. After in vitro transcription, RNA was treated with Ambion DNase I (ThermoFisher Scientific, cat. #AM2222) and cleaned using Monarch® RNA Cleanup Kit (NEB, cat. #T2050L). 5 μg SARS-CoV2 mNeon-NanoLuc RNA and 2 μg SARS-CoV2 N RNA were co-electroporated into 6 × 106 Huh-7.5 TMEM41B KO and TMEM41B-reconstituted KO cells in a 2-mm cuvette (BTX, cat. #45-0125) using a BTX ElectroSquare Porator ECM 830 (710 V, 99 μs, five pulses). Electroporated cells were incubated at room temperature for 10 min prior to resuspension in DMEM medium and seeded into 24-well plates at 1.5 × 105 cells per well. At 18 hours post electroporation, cells were washed with 1 mL PBS, lysed with diluted 5x Passive Lysis buffer (Promega, cat. #E1941) and processed using the Nano-Glo® Luciferase Assay System (Promega, cat. #N1120) and the Fluostar Omega microplate reader (BMG Labtech).
CRISPR-Cas9 genetic screening
Huh-7.5-Cas9 cells were generated by lentiviral transduction of lentiCas9-Blast (Addgene, cat. #52962) followed by selection and expansion in the presence of 5 μg/ml blasticidin. The human CRISPR Brunello library (Doench et al., 2016) was obtained through Addgene as a ready-to-use lentiviral pooled library at a titer ≥ 1 × 107 TU/mL (Addgene, cat. #73178-LV). To deliver the Brunello sgRNA library, 2.04 × 108 Huh-7.5-Cas9 cells were transduced by spinoculation at 1,000 g x 1 h in media containing 4 μg/ml polybrene (Millipore, cat. #TR-1003-G) and 20 mM HEPES (GIBCO, cat. #15630080) at a MOI = 0.21 to achieve ~560-fold overrepresentation of each sgRNA. Cells were spinoculated at 3 × 106 cells/well in 12-well plates in 1.5 mL final volume. Six hours post transduction, cells were trypsinized and transferred to T175 flasks at 7 × 106 cells/flask. Two days later, media was replaced with fresh media containing 1.5 μg/ml puromycin and cells were expanded for five additional days prior to seeding for coronavirus infection. Huh-7.5-Cas9 cells transduced with the Brunello sgRNA library were seeded in p150 plates at 4.5 × 106 cells/plate with two plates per replicate (e.g., 9 × 106 cells) and three replicates for each condition (mock, HCoV-229E, HCoV-NL63, HCoV-OC43).
For biosafety reasons, SARS-CoV-2 infection was performed in T175 screw top flasks. For infections at 37°C, we seeded cells at 5 × 106 cells per flask and used two flasks (e.g., 1 × 107 cells) per replicate. For infections at 33°C, we seeded cells at 6.7 × 106 cells per flask and used three flasks (e.g., 2 × 107 cells) per replicate. Both SARS-CoV-2 screens were performed in triplicate. The following day, the media was removed and viruses diluted in 10 ml/plate OptiMEM were added to cells. The inocula of HCoV-229E, HCoV-NL63 and SARS-CoV-2 were supplemented with 1 μg/ml TPCK-treated trypsin (Sigma-Aldrich, cat. #T1426) increasing the rate of infection. After two hours on a plate rocker at 37°C, 10 ml/plate media was added and plates were moved to 5% CO2 incubators set to 33°C (HCoV-229E, HCoV-NL63, HCoV-OC43, and SARS-CoV-2) or 37°C (SARS-CoV-2). Coronavirus screens were performed at the following MOIs in PFU/cell: HCoV-229E = 0.05 at 33°C; HCoV-NL63 = 0.01 at 33°C; HCoV-OC43 = 1 at 33°C; SARS-CoV-2 = 0.01 at 33°C and 0.1 at 37°C. Mock cells cultured at both temperatures were passaged every 3-4 days and re-seeded at 4.5 × 106 cells/plate with two plates per replicate. Media was changed on virus infected plates as needed to remove cellular debris. We optimized infection conditions empirically in an attempt to achieve robust selection and recovery of 1.5 × 106 cells per replicate at the experimental endpoint for virus infected cells and 1.5 × 107 for mock infected cells. Mock cells and cells that survived coronavirus infection were harvested approximately one to two weeks post infection.
Genomic DNA (gDNA) was isolated via ammonium acetate salt precipitation if greater than 1.5 × 106 cells were recovered or using the Monarch Genomic DNA Purification kit (NEB) if fewer per the manufacturer’s instructions. gDNA concentrations were quantitated via UV spectroscopy and normalized to 250 ng/μl with 10 mM Tris. The library was amplified from gDNA by a two-stage PCR approach. For PCR1 amplification, gDNA samples were divided into 50 ul PCR reactions. Each well consisted of 25 μl of NEB Q5 High-Fidelity 2X Master Mix, 2.5 ul of 10 μM forward primer Nuc-PCR1_Nextera-Fwd Mix, 2.5 ul of 10 μM reverse primer Nuc-PCR1_Nextera-Rev Mix and 20 μl of gDNA (5 μg each reaction). PCR1 cycling settings: initial 30 s denaturation at 98°C; then 10 s at 98°C, 30 s at 65°C, 30 s at 72°C for 25 cycles; followed by 2 min extension at 72°C. PCR1 samples were cleaned up by isopropanol precipitation, and normalized to 20 ng/μl. Each PCR2 reaction consisted of 25 μl of NEB Q5 High-Fidelity 2X Master Mix, 2.5 μl 10 μM Common_PCR2_Fwd primer, and 2.5 ul of 10 μM reverse i7 indexing primer. PCR2 cycling settings: initial 30 s at 98°C; then 10 s at 98°C, 30 s at 65°C, 30 s at 72°C for 13 cycles. PCR products were again purified by SPRI, pooled and sequenced on an Illumina NextSeq 500 at the NYU Genome Technology Center using standard Nextera sequencing primers and 75 cycles.
Quantification and Statistical Analysis
Statistical details of experiments can be found in the figure legends, including the statistical tests used. In all figures, center represents the mean and error bars represent standard error of the mean (SEM), unless otherwise noted in the figure legend. Where non-parametric significance tests are indicated, the data was not tested for normality. p < 0.05 was considered statistically significant. Generation of plots and statistical analyses were performed using the R statistical computing environment or Prism 8 (GraphPad).
Analysis of CRISPR-Cas9 genetic screen data
FASTQ files were processed and trimmed to retrieve sgRNA target sequences followed by enumeration of sgRNAs in the reference sgRNA library file using MAGeCK (Li et al., 2014). MAGeCK was also used to determine gene essentiality (beta) using its maximum likelihood estimation (MLE) algorithm. Z-scores for visualization in the form of heatmaps were computed using the following approach: for each condition, the log2 fold change with respect to the initial condition was computed. A natural cubic spline with 4 degrees of freedom was fit to each pair of infected and control cells and residuals were extracted. To obtain gene-wise data, the mean residuals for each group of sgRNAs was calculated, a z-score was computed, and a p value was determined using a 2-sided normal distribution test. P values were combined across screens using Fisher’s sumlog approach and corrected for multiple testing using the method of Benjamini & Hochberg.
Functional clustering and network analysis of screening data
High confidence CRISPR hits with FDR cutoffs below 0.05 were extracted for functional clustering and network building. Briefly, enriched pathways were identified from the NIH NCATS BioPlanet database (Huang et al., 2019), which aggregates currates pathways from multiple sources, using competitive gene set testing of z-scores in pre-ranked mode (Wu and Smyth, 2012). For construction of the network in Figure 3, significant CRISPR hits from any virus were searched using the STRING database (https://string-db.org; Szklarczyk et al., 2019) using default parameters and imported into Cytoscape (Shannon et al., 2003). Overlapping hits per virus were calculated and subsequently depicted as pie charts per node in Adobe Illustrator. For virus specific networks in Figure S2, significant CRISPR hits per virus and the next adjacent 100 interactors were extracted and graphed in Cytoscape.
Analysis of scRNaseq data
For scRNaseq analysis, Seurat objects were downloaded from figshare: (https://doi.org/10.6084/m9.figshare.12436517.v2; Chua et al., 2020). Dotplots for select cell identities and for all hgh confidence CRISPR hits per virus were plotted using the DotPlot function in Seurat (Stuart et al., 2019).
Acknowledgments
This work was initiated and conducted under unusual circumstances. As New York City and much of the world were sheltering in place to reduce the spread of SARS-CoV-2, all of the authors here were sustained during the shutdown by generous funding intended for related and unrelated work. During this time, we were fortunate to obtain funding from government and charitable agencies that allowed this COVID-19 work to continue. For funding directly related to these COVID-19 efforts, we thank the G. Harold and Leila Y. Mathers Charitable Foundation and the BAWD Foundation for their generous awards. We also thank Fast Grants (https://fastgrants.org), a part of Emergent Ventures at the Mercatus Center, George Mason University. Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award R01AI091707. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We further received funding for COVID-19 related work from an administrative supplement to U19AI111825. The authors were also supported by non-COVID-19 funding by the following awards, foundations, and charitable trusts: R01CA190261, R01CA213448, U01CA213359, R01AI143295, R01AI150275, R01AI124690, R01AI116943, P01AI138938, P30CA008748, P30CA016087, R03AI141855, R21AI142010, W81XWH1910409, EMBO fellowship ALTF 380-2018, F32AI133910, the Robertson Foundation, and an Agilent Technologies Thought Leader Award. S.W.L. is the Geoffrey Beene Chair of Cancer Biology and a Howard Hughes Medical Institute Investigator. F.J.S.-R. is an HHMI Hanna Gray Fellow and was partially supported by an MSKCC Translational Research Oncology Training Fellowship (NIH T32-CA160001). We also thank the NYU Langone Health Genome Technology Center. We also wish to thank Rodrigo Romero (MSKCC) for assistance with Revolve fluorescence microscopy and Aileen O’Connell, Santa Maria Pecoraro Di Vittorio, Glen Santiago, Mary Ellen Castillo, Arnella Webson, and Sonia Shirley for outstanding administrative and technical support.
Author Contributions
Conceptualization, W.M.S., H.-H.H., J.M.L., F.J.S.-R., A.A.L., and J.T.P.; Methodology, W.M.S., H.-H.H., J.M.L., F.J.S.-R., A.A.L., and J.T.P.;Formal Analysis, J.M.L. and J.T.P.; Investigation, W.M.S., H.-H.H., A.A.L., A.W.A., J.L.P., E.M., I.R.-L., A.F.S., and J.T.P.; Resources, S.W.L., C.M.R., and J.T.P.; Data Curation, J.M.L., J.L.P., and J.T.P.; Supervision, C.M.R. and J.T.P.; Visualization, J.M.L., F.J.S.-R., and J.T.P.; Writing – Original Draft, F.J.S.-R., J.M.L., and J.T.P.; Writing – Review & Editing, W.M.S., J.M.L., H.-H.H., F.J.S.-R., J.L.P., I.R.-L., M.R.M., C.M.R., and J.T.P.; Project Administration, A.W.A., A.P., and M.R.M.; Funding Acquisition, S.W.L., C.M.R., and J.T.P.
Declaration of Interests
S.W.L. is an advisor for and has equity in the following biotechnology companies: ORIC Pharmaceuticals, Faeth Therapeutics, Blueprint Medicines, Geras Bio, Mirimus Inc., PMV Pharmaceuticals, and Constellation Pharmaceuticals. C.M.R. is a founder of Apath LLC; a Scientific Advisory Board member of Imvaq Therapeutics; Vir Biotechnology, and Arbutus Biopharma; and an advisor for Regulus Therapeutics and Pfizer.
Published: December 9, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.12.006.
Supplemental Information
Table S1A: Huh-7.5 37°C SARS-CoV-2 z-scores. Table S1B: Huh-7.5 33°C SARS-CoV-2 z-scores. Table S1C: Huh-7.5 33°C HCoV-OC43 z-scores. Table S1D: Huh-7.5 33°C HCoV-NL63 z-scores. Table S1E: Huh-7.5 33°C HCoV-229E z-scores.
Table S2A: Primers used to prepare Illumina-compatible libraries for high-throughput sequencing of CRISPR screens. Table S2B: Full sequence of sgRNA expression lentiviral vectors constructed and used in this study.
References
- Aikawa J., Grobe K., Tsujimoto M., Esko J.D. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J. Biol. Chem. 2001;276:5876–5882. doi: 10.1074/jbc.M009606200. [DOI] [PubMed] [Google Scholar]
- Allen B.L., Taatjes D.J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbey C.M., Bacallao R.L., Dunn K.W. Rab10 associates with primary cilia and the exocyst complex in renal epithelial cells. Am. J. Physiol. Renal Physiol. 2010;299:F495–F506. doi: 10.1152/ajprenal.00198.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen J., Persoons L., Jansen S., Vanstreels E., Jacquemyn M., Jochmans D., Neyts J., Dallmeier K., Maes P., Daelemans D. Identification of TMEM106B as proviral host factor for SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.09.28.316281. [DOI] [Google Scholar]
- Bai X., Zhou D., Brown J.R., Crawford B.E., Hennet T., Esko J.D. Biosynthesis of the linkage region of glycosaminoglycans: cloning and activity of galactosyltransferase II, the sixth member of the beta 1,3-galactosyltransferase family (beta 3GalT6) J. Biol. Chem. 2001;276:48189–48195. doi: 10.1074/jbc.M107339200. [DOI] [PubMed] [Google Scholar]
- Blackburn J.B., D’Souza Z., Lupashin V.V. Maintaining order: COG complex controls Golgi trafficking, processing, and sorting. FEBS Lett. 2019;593:2466–2487. doi: 10.1002/1873-3468.13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight K.J., McKeating J.A., Rice C.M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois C., Bour J.B., Lidholt K., Gauthray C., Pothier P. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J. Virol. 1998;72:7221–7227. doi: 10.1128/jvi.72.9.7221-7227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhaescu I., Izzedine H. Mevalonate pathway: a review of clinical and therapeutical implications. Clin. Biochem. 2007;40:575–584. doi: 10.1016/j.clinbiochem.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.C., Kuhn J., Kleesiek K., Götting C. Heterologous expression and biochemical characterization of soluble human xylosyltransferase II. Biochem. Biophys. Res. Commun. 2008;365:678–684. doi: 10.1016/j.bbrc.2007.10.206. [DOI] [PubMed] [Google Scholar]
- Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:P1043–P1057. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Oliveira C., Freire J.M., Conceição T.M., Higa L.M., Castanho M.A., Da Poian A.T. Receptors and routes of dengue virus entry into the host cells. FEMS Microbiol. Rev. 2015;39:155–170. doi: 10.1093/femsre/fuu004. [DOI] [PubMed] [Google Scholar]
- Cuellar K., Chuong H., Hubbell S.M., Hinsdale M.E. Biosynthesis of chondroitin and heparan sulfate in chinese hamster ovary cells depends on xylosyltransferase II. J. Biol. Chem. 2007;282:5195–5200. doi: 10.1074/jbc.M611048200. [DOI] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T.X., Wessels H.H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell. 2020 doi: 10.1016/j.cell.2020.10.030. Published online October 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechecchi M.C., Tamanini A., Bonizzato A., Cabrini G. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology. 2000;268:382–390. doi: 10.1006/viro.1999.0171. [DOI] [PubMed] [Google Scholar]
- Dechecchi M.C., Melotti P., Bonizzato A., Santacatterina M., Chilosi M., Cabrini G. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 2001;75:8772–8780. doi: 10.1128/JVI.75.18.8772-8780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov D.S. Virus entry: molecular mechanisms and biomedical applications. Nat. Rev. Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Hantak M.P., Li K., McCray P.B., Jr., Perlman S., Gallagher T. The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog. 2017;13:e1006546. doi: 10.1371/journal.ppat.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Romero E., Rawling J., García-Barreno B., Melero J.A. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J. Virol. 2004;78:3524–3532. doi: 10.1128/JVI.78.7.3524-3532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B., Mohr S.E., Hu Y., Perrimon N. Accessing the Phenotype Gap: Enabling Systematic Investigation of Paralog Functional Complexity with CRISPR. Dev. Cell. 2017;43:6–9. doi: 10.1016/j.devcel.2017.09.020. [DOI] [PubMed] [Google Scholar]
- Fajgenbaum D.C., Rader D.J. Teaching Old Drugs New Tricks: Statins for COVID-19? Cell Metab. 2020;32:145–147. doi: 10.1016/j.cmet.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman S.A., Audet S., Beeler J.A. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 2000;74:6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner C.L., Ebel G.D., Ryman K.D., Klimstra W.B. Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci. USA. 2011;108:16026–16031. doi: 10.1073/pnas.1110617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroglou T., Florin L., Schäfer F., Streeck R.E., Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J.L., Brown M.S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Cao J., Zhang X., Gao H., Wang Y., Wang J., Zhang J., Shen G., Jiang X., Yang J., et al. Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv. 2020 doi: 10.1101/2020.09.09.287508. [DOI] [Google Scholar]
- Hall M.P., Unch J., Binkowski B.F., Valley M.P., Butler B.L., Wood M.G., Otto P., Zimmerman K., Vidugiris G., Machleidt T., et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallak L.K., Collins P.L., Knudson W., Peeples M.E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000;271:264–275. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- Harris J., Werling D. Binding and entry of respiratory syncytial virus into host cells and initiation of the innate immune response. Cell. Microbiol. 2003;5:671–680. doi: 10.1046/j.1462-5822.2003.00313.x. [DOI] [PubMed] [Google Scholar]
- Heaton B.E., Trimarco J.D., Hamele C.E., Harding A.T., Tata A., Zhu X., Tata P.R., Smith C.M., Heaton N.S. SRSF protein kinases 1 and 2 are essential host factors for human coronaviruses including SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.14.251207. [DOI] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.-H., Schneider W.M., Rozen-Gagnon K., Miles L.A., Schuster F., Razooky B., Jacobson E., Wu X., Yi S., Rudin C.M., et al. TMEM41B is a pan-flavivirus host factor. Cell. 2020;184 doi: 10.1016/j.cell.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.-H., Sánchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Grishagin I., Wang Y., Zhao T., Greene J., Obenauer J.C., Ngan D., Nguyen D.T., Guha R., Jadhav A., et al. The NCATS BioPlanet - An Integrated Platform for Exploring the Universe of Cellular Signaling Pathways for Toxicology, Systems Biology, and Chemical Genomics. Front. Pharmacol. 2019;10:445. doi: 10.3389/fphar.2019.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim J., Griffin P., Coombe D.R., Rider C.C., James W. Cell-surface heparan sulfate facilitates human immunodeficiency virus Type 1 entry into some cell lines but not primary lymphocytes. Virus Res. 1999;60:159–169. doi: 10.1016/s0168-1702(99)00018-0. [DOI] [PubMed] [Google Scholar]
- Imlach W.L., Beck E.S., Choi B.J., Lotti F., Pellizzoni L., McCabe B.D. SMN is required for sensory-motor circuit function in Drosophila. Cell. 2012;151:427–439. doi: 10.1016/j.cell.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce J.G., Tung J.S., Przysiecki C.T., Cook J.C., Lehman E.D., Sands J.A., Jansen K.U., Keller P.M. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 1999;274:5810–5822. doi: 10.1074/jbc.274.9.5810. [DOI] [PubMed] [Google Scholar]
- Karger A., Schmidt U., Buchholz U.J. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 2001;82:631–640. doi: 10.1099/0022-1317-82-3-631. [DOI] [PubMed] [Google Scholar]
- Kitagawa H., Tone Y., Tamura J., Neumann K.W., Ogawa T., Oka S., Kawasaki T., Sugahara K. Molecular cloning and expression of glucuronyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 1998;273:6615–6618. doi: 10.1074/jbc.273.12.6615. [DOI] [PubMed] [Google Scholar]
- Kleinfelter L.M., Jangra R.K., Jae L.T., Herbert A.S., Mittler E., Stiles K.M., Wirchnianski A.S., Kielian M., Brummelkamp T.R., Dye J.M., Chandran K. Haploid Genetic Screen Reveals a Profound and Direct Dependence on Cholesterol for Hantavirus Membrane Fusion. MBio. 2015;6:e00801. doi: 10.1128/mBio.00801-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger J., Kjellén L. Heparan sulfate biosynthesis: regulation and variability. J. Histochem. Cytochem. 2012;60:898–907. doi: 10.1369/0022155412464972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusat T., Streckert H.J. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 1997;142:1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind T., Tufaro F., McCormick C., Lindahl U., Lidholt K. The putative tumor suppressors EXT1 and EXT2 are glycosyltransferases required for the biosynthesis of heparan sulfate. J. Biol. Chem. 1998;273:26265–26268. doi: 10.1074/jbc.273.41.26265. [DOI] [PubMed] [Google Scholar]
- Lotti F., Imlach W.L., Saieva L., Beck E.S., Hao L.T., Li D.K., Jiao W., Mentis G.Z., Beattie C.E., McCabe B.D., Pellizzoni L. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau C.D., Puschnik A.S., Majzoub K., Ooi Y.S., Brewer S.M., Fuchs G., Swaminathan K., Mata M.A., Elias J.E., Sarnow P., Carette J.E. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Urdiroz M., Deeks M.J., Horton C.G., Dawe H.R., Jourdain I. The Exocyst Complex in Health and Disease. Front. Cell Dev. Biol. 2016;4:24. doi: 10.3389/fcell.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I., Melero J.A. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 2000;81:2715–2722. doi: 10.1099/0022-1317-81-11-2715. [DOI] [PubMed] [Google Scholar]
- Mei K., Guo W. The exocyst complex. Curr. Biol. 2018;28:R922–R925. doi: 10.1016/j.cub.2018.06.042. [DOI] [PubMed] [Google Scholar]
- Moretti F., Bergman P., Dodgson S., Marcellin D., Claerr I., Goodwin J.M., DeJesus R., Kang Z., Antczak C., Begue D., et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 2018;19:e45889. doi: 10.15252/embr.201845889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Hama Y., Izume T., Tamura N., Ueno T., Yamashita Y., Sakamaki Y., Mimura K., Morishita H., Shihoya W., et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 2018;217:3817–3828. doi: 10.1083/jcb.201804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Hama Y., Mizushima N. TMEM41B functions with VMP1 in autophagosome formation. Autophagy. 2019;15:922–923. doi: 10.1080/15548627.2019.1582952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell C.D., Shukla D. The Importance of Heparan Sulfate in Herpesvirus Infection. Virol. Sin. 2008;23:383–393. doi: 10.1007/s12250-008-2992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T., Yoshida K., Kondo T., Furukawa K. Human homolog of Caenorhabditis elegans sqv-3 gene is galactosyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J. Biol. Chem. 1999;274:22915–22918. doi: 10.1074/jbc.274.33.22915. [DOI] [PubMed] [Google Scholar]
- Perera-Lecoin M., Meertens L., Carnec X., Amara A. Flavivirus entry receptors: an update. Viruses. 2013;6:69–88. doi: 10.3390/v6010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pönighaus C., Ambrosius M., Casanova J.C., Prante C., Kuhn J., Esko J.D., Kleesiek K., Götting C. Human xylosyltransferase II is involved in the biosynthesis of the uniform tetrasaccharide linkage region in chondroitin sulfate and heparan sulfate proteoglycans. J. Biol. Chem. 2007;282:5201–5206. doi: 10.1074/jbc.M611665200. [DOI] [PubMed] [Google Scholar]
- Puschnik A.S., Majzoub K., Ooi Y.S., Carette J.E. A CRISPR toolbox to study virus-host interactions. Nat. Rev. Microbiol. 2017;15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riblett A.M., Blomen V.A., Jae L.T., Altamura L.A., Doms R.W., Brummelkamp T.R., Wojcechowskyj J.A. A Haploid Genetic Screen Identifies Heparan Sulfate Proteoglycans Supporting Rift Valley Fever Virus Infection. J. Virol. 2015;90:1414–1423. doi: 10.1128/JVI.02055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Pauls E., Badia R., Riveira-Muñoz E., Clotet B., Ballana E., Esté J.A. Characterization of the influence of mediator complex in HIV-1 transcription. J. Biol. Chem. 2014;289:27665–27676. doi: 10.1074/jbc.M114.570341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidis G., McDougall W.M., Meraner P., Perreira J.M., Portmann J.M., Trincucci G., John S.P., Aker A.M., Renzette N., Robbins D.R., et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep. 2016;16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C.J., Huang T.Q., Weir N.R., Polyakov N.J., Schultz S.W., Denic V. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019;17:e2007044. doi: 10.1371/journal.pbio.2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.D., Lupashin V.V. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffientini U., Caridis A.M., Dolan S., Graham A. Intracellular cholesterol transporters and modulation of hepatic lipid metabolism: Implications for diabetic dyslipidaemia and steatosis. Biochim. Biophys. Acta. 2014;1842:1372–1382. doi: 10.1016/j.bbalip.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Techaarpornkul S., Collins P.L., Peeples M.E. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology. 2002;294:296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- Thao T.T.N., Labroussaa F., Ebert N., Jores J., Thiel V. In-Yeast Assembly of Coronavirus Infectious cDNA Clones Using a Synthetic Genomics Pipeline. Methods Mol. Biol. 2020;2203:167–184. doi: 10.1007/978-1-0716-0900-2_13. [DOI] [PubMed] [Google Scholar]
- V’kovski P., Gultom M., Steiner S., Kelly J., Russeil J., Mangeat B., Cora E., Pezoldt J., Holwerda M., Kratzel A., et al. Disparate temperature-dependent virus – host dynamics for SARS-CoV-2 and SARS-CoV in the human respiratory epithelium. bioRxiv. 2020 doi: 10.1101/2020.04.27.062315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.P., Tang Z., Chen C.W., Shimada M., Koche R.P., Wang L.H., Nakadai T., Chramiec A., Krivtsov A.V., Armstrong S.A., et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Mol. Cell. 2017;67:308–321.e6. doi: 10.1016/j.molcel.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K., Hayashi J.M., Carlson-Stevermer J., Oki J., Holden K., Krogan N.J., et al. Functional genomic screens identify human host factors for SARS-CoV-2 and common cold coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.09.24.312298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell. 2020 doi: 10.1016/j.cell.2020.10.028. Published online October 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Guo W. The Exocyst at a Glance. J. Cell Sci. 2015;128:2957–2964. doi: 10.1242/jcs.156398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Smyth G.K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Martinez P., Séron K., Luo G., Allain F., Dubuisson J., Belouzard S. Characterization of hepatitis C virus interaction with heparan sulfate proteoglycans. J. Virol. 2015;89:3846–3858. doi: 10.1128/JVI.03647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-J., Qin J.-J., Cheng X., Shen L., Zhao Y.-C., Yuan Y., Lei F., Chen M.-M., Yang H., Bai L., et al. In-Hospital Use of Statins Is Associated with a Reduced Risk of Mortality among Individuals with COVID-19. Cell Metab. 2020;32:176–187.e4. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., et al. The S1/S2 boundary of SARS-CoV-2 spike protein modulates cell entry pathways and transmission. bioRxiv. 2020 doi: 10.1101/2020.08.25.266775. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1A: Huh-7.5 37°C SARS-CoV-2 z-scores. Table S1B: Huh-7.5 33°C SARS-CoV-2 z-scores. Table S1C: Huh-7.5 33°C HCoV-OC43 z-scores. Table S1D: Huh-7.5 33°C HCoV-NL63 z-scores. Table S1E: Huh-7.5 33°C HCoV-229E z-scores.
Table S2A: Primers used to prepare Illumina-compatible libraries for high-throughput sequencing of CRISPR screens. Table S2B: Full sequence of sgRNA expression lentiviral vectors constructed and used in this study.
Data Availability Statement
Data supporting the findings of this study are reported in Figures S1–S6 and Tables S1-S2. The accession number for the CRISPR screen sequencing data reported in this paper is GEO: GSE162038. Networks are available on NDEx. All reagents and materials generated in this study will be available to the scientific community through Addgene and/or MTAs.