Summary
While cell transplantation can rescue motor defects in Parkinson’s disease (PD) models, whether and how grafts functionally repair damaged neural circuitry in the adult brain is not known. We transplanted hESC-derived midbrain dopamine (mDA) or cortical glutamate neurons into the substantia nigra or striatum of a mouse PD model and found extensive graft integration with host circuitry. Axonal pathfinding towards the dorsal striatum was determined by the identity of the grafted neurons and anatomical presynaptic inputs were largely dependent on graft location, while inhibitory vs. excitatory input was dictated by identity of grafted neurons. hESC-derived mDA neurons display A9 characteristics and restore functionality of the reconstructed nigrostriatal circuit to mediate improvements in motor function. These results indicate similarity in cell type-specific pre- and post-synaptic integration between transplant-reconstructed circuit and endogenous neural networks, highlighting the capacity of hPSC-derived neuron subtypes for specific circuit repair and functional restoration in the adult brain.
Graphical Abstract
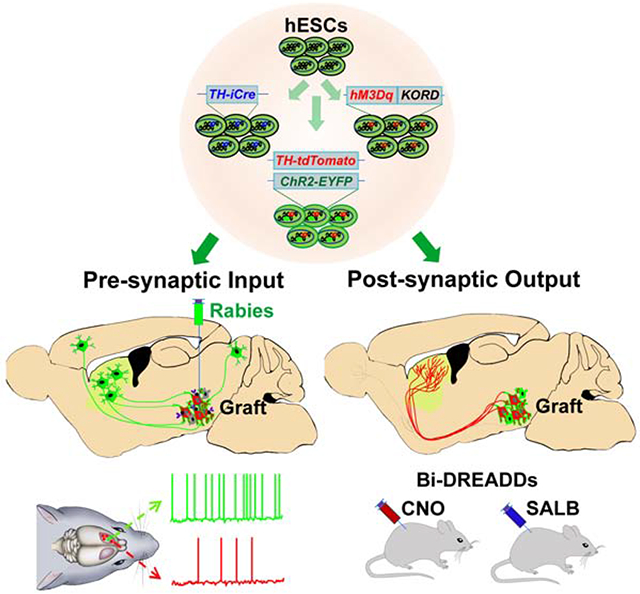
eTOC Blurb
Xiong et al. found that the genetically labeled hESC-derived midbrain dopamine (mDA) neurons, following transplantation into the brain of Parkinson’s disease mice, repair the nigra-striatal circuit anatomically and functionally. Furthermore, functional innervation into the graft depends on the cell identity, highlighting the necessity of correct cell types for cell therapy.
Introduction
The adult mammalian brain has a limited capacity to regenerate after injury or diseases. Transplantation of neural cells to replace the lost neuronal function (cell therapy) holds promise for neurological disorders (Steinbeck and Studer, 2015). For cell therapy to succeed, grafted neuronal progenitors need to differentiate into specific types of neurons, which send out axons through the mature CNS environment to find and make functional synaptic connections with their target. Furthermore, grafted neurons should receive proper inputs from host neurons so that the activity of the grafted cells is appropriately regulated (Rossi and Cattaneo, 2002). Together, the grafted cells replace the lost neurons and restore the circuit function.
Substantial efforts have been made to repair damaged neuronal circuitry by transplanting neurons into their home location (homotopic transplantation) in the adult brain. Embryonic cortex neurons, transplanted into the cerebral cortex of adult mice, extend long-range projections to and form synapses with neurons in the cortex, thalamus, and spinal cord (Falkner et al., 2016; Fricker-Gates et al., 2002; Gaillard et al., 2007). When the embryonic ventral mesencephalic (VM) tissues are grafted to the substantia nigra (SN) of 6-OHDA lesioned Parkinson’s disease (PD) animals, a portion of axons grow into the striatum (Gaillard et al., 2009; Grealish et al., 2014; Isacson and Deacon, 1996; Thompson et al., 2008; Wictorin et al., 1992). Similarly, cortical (Palma-Tortosa et al., 2020; Tornero et al., 2013) or mDA progenitor cells (Adler et al., 2019; Cardoso et al., 2018; Grealish et al., 2014), derived from human embryonic stem cells (hESCs) or induced pluripotent stem cells (iPSCs), following homotopic transplantation into the cortex or SN of model mice, respectively, can project axons to distal brain regions including their cognate areas and synapse with host neurons. These findings highlight the capacity for grafted immature neuronal cells to send out long axons in the adult brain, raising the prospect of neural progenitor transplantation therapy for neurological conditions.
Axonal growth in the adult brain is necessary for regeneration but the specificity of axonal projection is essential for proper functional restoration of a damaged circuit (Isacson and Deacon, 1997; Rossi and Cattaneo, 2002). The projection patterns of grafted neurons are often examined by using species-specific antibodies (Adler et al., 2019; Grealish et al., 2014; Wictorin et al., 1992), or fluorescent labeling mediated by plasmids or viral particles (Falkner et al., 2016; Tornero et al., 2013). Such methods label all the transplanted cells that are almost always heterogeneous. By staining for the human neuronal cell adhesion molecule (hNCAM) to label transplanted cells, previous studies showed that hESC-derived mDA neurons, grafted to the SN, project to the ventral and dorsal striatum. However, a significant portion of fibers project to the medial septum (Adler et al., 2019; Grealish et al., 2014) which is usually not or less innervated by endogenous mDA neurons (Poulin et al., 2018). These phenomena raise the question of whether the fibers to different brain regions are from different grafted cell types and what determines the specificity of axonal projection and target-finding.
Grafted cells also need to receive appropriate pre-synaptic inputs for proper circuit function. It is recently reported that transplanted neurons receive presynaptic inputs from host brain regions based on rabies virus tracing (Cardoso et al., 2018; Falkner et al., 2016; Grealish et al., 2015; Linaro et al., 2019) and the input is dependent upon the graft location (Adler et al., 2019; Doerr et al., 2017). However, in these studies, the glycoprotein and TVA (EnvA envelope glycoprotein for viral infection) were expressed in all types of transplanted neurons; hence, the presynaptic neurons traced are a collection of inputs to all cell types in the graft. It is not known whether a specific neuron type will establish specific presynaptic inputs and what determines the source of their inputs. More importantly, it is unknown if the anatomical connections are functional and what determines the nature of functional inputs.
By transplanting genetically labeled mDA and forebrain glutamate (Glu) neurons, derived from hESCs, into the SN or striatum of PD model mice, we found that the nigrally grafted mDA, but not Glu, neurons predominantly projected to the dorsal striatum. Rabies-mediated tracing identified inputs to the nigrally grafted human mDA neurons similar to those to endogenous mDA neurons. These inputs became functional over the 3-6-month period with the mDA neurons receiving more inhibitory but less excitatory innervations than non-mDA neurons. The animals transplanted with mDA neurons recovered from motor deficits, which was abrogated or enhanced by regulating the activity of the nigrally grafted mDA neurons.
RESULTS
Grafted human mDA and Glu neurons project to differential targets.
During development, targeted axonal projection is often determined by the intrinsic property of the cells (Goldberg, 2003; Stoeckli, 2018). To address if cell intrinsic property also determines target-finding in the adult brain, we transplanted hESC-derived mDA or forebrain Glu neuron progenitors into the midbrain of the PD model mice. The mDA or Glu neuron progenitors were differentiated from hESCs according to our protocols (Li et al., 2009; Xi et al., 2012). At day 32 of mDA neuron differentiation (the day for transplantation), most of the progenitors expressed the floor plate and midbrain markers, CORIN, FOXA2, LMX1A and EN1 (Figures S1A and S1C). By day 42, 69% of total cells or 84% of TUJ1+ neurons were positive for tyrosine hydroxylase (TH) as well as EN1, FOXA2, LMX1A, and NURR1 (Figures S1D and S1F), suggesting an mDA neuronal identity. Furthermore, most of the TH+ neurons co-expressed GIRK2 (>85%), a marker relatively enriched in A9 mDA neurons, but fewer for Calbindin (CALB) (15%), a marker expressed in A10 mDA neurons (Figures S1D and S1F). Thus, most of the TH+ neurons carry A9 mDA neuron characteristics. Meanwhile, at day 32 of Glu neuron differentiation, most cells expressed dorsal forebrain marker PAX6 and forebrain marker FOXG1, demonstrating their dorsal forebrain progenitor identity (Figures S1B and S1C). By day 42, 82% of total cells became positive for vGLUT1, and most of these neurons expressed CTIP2 (>80%) or TBR1 (>85%), suggesting a forebrain layer 5/6 cortical glutamate neuron identity (Figures S1E and S1F).
We then transplanted the mDA (Figure 1A) or Glu progenitors (Figure 1B) into the SN of PD mice. Six months following transplantation, grafts were present in all the grafted animals. Serial sagittal sections of the mDA neuron-transplanted brain revealed that most of the hNCAM+ fibers distributed in the caudate putamen (CPu) (Figures 1A and 1D), the area mainly innervated by A9 mDA neurons (Bjorklund and Dunnett, 2007). Quantification of hNCAM+ fiber density from three representative sagittal planes (Figure 1C) showed that 72% in plane L2.16, 62% in plane L1.44, and 45% in plane L0.72 of the total hNCAM+ fibers were distributed in the CPu (Figure 1E). Fewer hNCAM+ fibers were detected in the olfactory tubercle (Tu, 17-20%) and accumbens nucleus (Acb, 9-16%), areas targeted by A10 mDA neurons. The remaining areas, including the amygdala and cortex, accounted for less than 15% of the total fibers (Figure 1E). Within the striatum, 72% and 87% of human fibers projected to the dorsal striatum (CPu) in plane L0.72 and L1.44 respectively (Figure 1F). The distribution pattern of the hNCAM+ fibers, especially within CPu, is similar to that of endogenous TH+ fibers in normal animals (Figures 1D and S1G).
Figure 1. Axonal projection of nigrally transplanted human neurons.
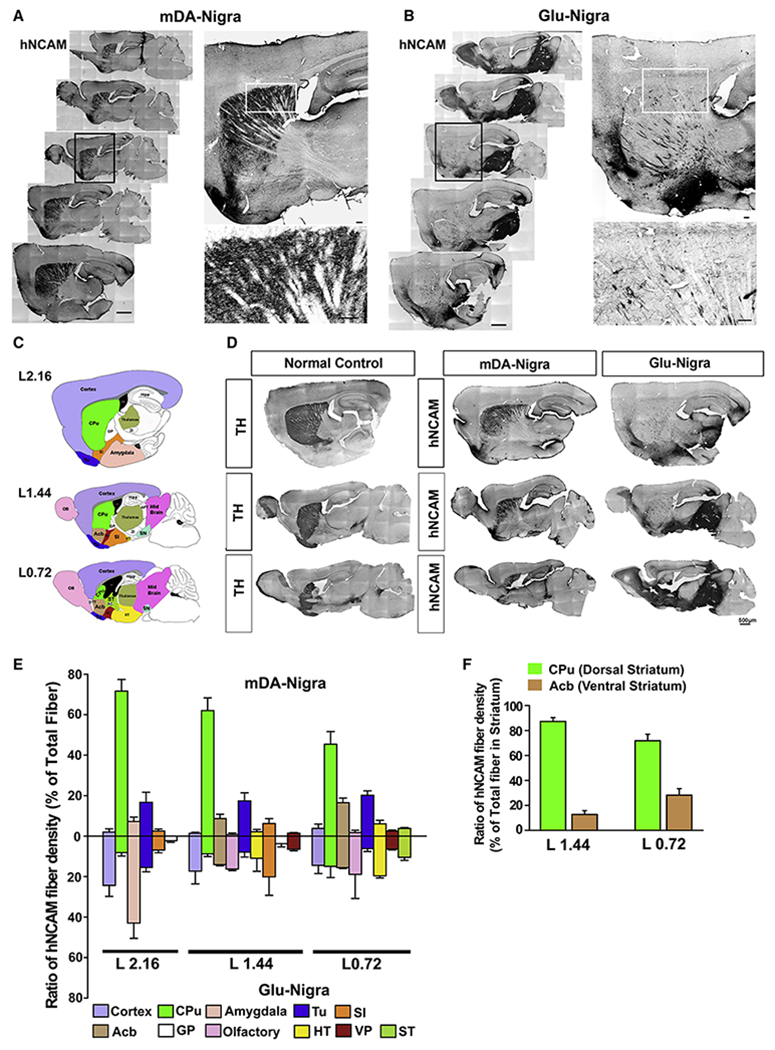
(A-B) immunohistochemistry of sagittal sections for hNCAM from nigral graft with mDA (A) or Glu neurons (B), Scale bar = 250 μm. The black boxed areas are amplified at the upper right. The white boxed areas are amplified at the lower right. Scale bar = 25 μm for the amplified images. (C) Schematic sagittal diagram of anatomical structures in three representative planes. See Table S1 for abbreviations. (D) Immunostaining for TH in wild type mice (left panel) or hNCAM in PD mice transplanted with mDA neurons (middle panel) or Glu neurons (right panel) at corresponding sagittal planes. (E) Quantification of the regional distribution of hNCAM+ fibers in different areas. n = 8 for mDA neuron group, n = 6 for Glu neuron group. (F) Relative distribution of hNCAM+ fibers in the dorsal (CPu) and ventral striatum (Acb) from the two representative planes of mDA-transplanted mice.
See also Figure S1.
In contrast, the Glu neurons extended axons locally, filling the entire midbrain (Figure 1B). They also sent out axons over a long distance, but mostly to the amygdala, olfactory bulb (OB), substantia innominata (SI), and the cerebral cortex, with only a small portion of hNCAM+ fibers (2-8%) detected in the CPu (Figures 1B, 1D, and 1E).
Together, these results indicate that the grafted human neural progenitors differentiate to respective neuronal types and project axons to differential brain regions.
Grafted human mDA neurons project through their cognate path
Examination from serial sagittal sections (Figure 2A) revealed that the grafted mDA neurons extended axons in the rostral direction along the medial forebrain bundle (MFB) in a well-defined and fasciculated tract (Figure 2B, L1.08 and red arrowheads in 2C). Laterally, they were seen to extend through the SI (Figure 2B, L1.44) and amygdala (Figure 2B, L2.04). Rostrally, the hNCAM+ fibers penetrated the Acb (Figures 2B–2E) where a large portion of hNCAM+ fibers ascended into the CPu along the boundary between cortex and striatum (Figures 2A and red arrows in 2D). A few hNCAM+ fibers were detected in the cortex (blue arrowheads in Figures 2D and 2F). These results suggest that most of the axonal projection from grafted mDA neurons follows the endogenous nigra-striatal pathway (Voorn et al., 1988).
Figure 2. Axonal projection pathways of nigrally transplanted neurons.
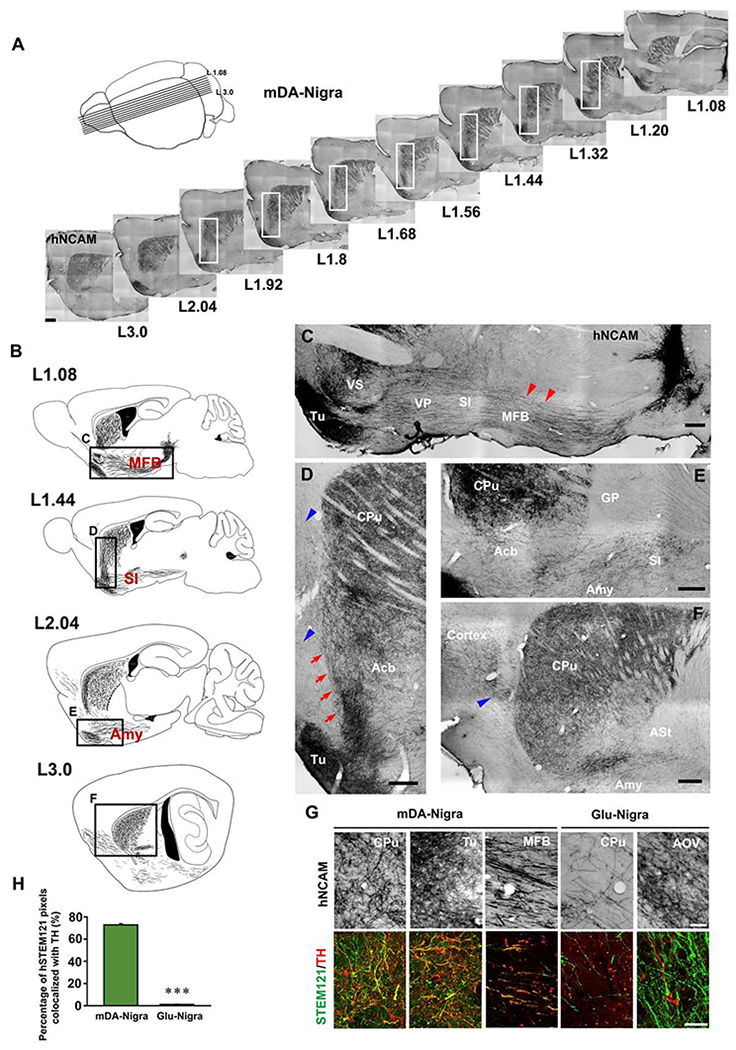
(A) A schematic of the approximate medial-lateral planes and the corresponding serial sagittal sections immunostained for hNCAM from the mouse brain transplanted with mDA neurons. (B) Neurolucida drawing of hNCAM+ axonal projection at different planes of sagittal sections. The boxed areas are magnified in (C-F). (C-F) High magnification illustrates the axonal projection pathways and territory. Scale bar = 250 μm. The red arrowheads in (C) indicate the hNCAM+ axons in MFB. The red arrows in (D) indicate ascending hNCAM+ axons. (F) Distribution of hNCAM+ fibers in the lateral striatum. The blue arrowheads in (D) and (F) indicate hNCAM+ fibers in the cortex. See Table S1 for abbreviations. (G) The morphology of hNCAM+ fibers and co-labeling of human STEM121 and TH in fibers derived from mDA or Glu neuron graft in different host brain regions. Scale bar = 50 μm for the upper panel, 25 μm for the lower panel. (H) Quantification of the percentage of human STEM121 pixels colocalized with TH in CPu of mice transplanted with mDA or Glu neurons. Data are represented as mean ± SEM. Student-t test. ***p < 0.001.
See also Figure S2.
In the dorsal/lateral striatum, the dense hNCAM+ fibers exhibited elaborately ramified, fine-beaded terminal networks (Figures 2D–2F and S2A). The ramification of the axons was also observed in Tu (Figures 2C and 2D), but not in the Acb, amygdala, and MFB (Figure S2B), suggesting target-specific axonal branching. Most of the human fibers, stained by STEM121, were positive for TH (Figures 2G and 2H), demonstrating the dopaminergic identity. Ramification of hNCAM+ fibers from Glu neuron graft was observed in the OB and cortex (Figure S2C).
Immunohistochemistry analysis of mDA graft showed that 68% of the grafted cells co-expressed TH and human nuclei (hN), and most of the TH+ cells also expressed GIRK2 as well as FOXA2 and LMX1A (Figures S2D–S2F), suggestive of A9 mDA identity. The human specific synaptophysin (hSyn) punctae distributed along the TH+ fibers in CPu, and some localized on the GABA+ cell body (Figure S2G). In contrast, few hSyn+ punctae were observed in the forebrain neuron grafted group and hardly any of them were localized on the GABA+ cell body (Figure S2H). These results suggest synaptic connections between grafted human mDA neurons and target cells in the host brain.
Genetic labeling reveals specific axonal innervation by human mDA neurons
To elucidate the specific axonal innervation by grafted mDA neurons, we created a TH reporter hESC line with tdTomato expression recapitulating that of the endogenous TH gene (see Method Details). We further knocked in a ChR2-EYFP fusion protein expression cassette into the AAVS1 locus to enable specifically labelling and manipulating transplanted human cells (Figures 3A, 3B, S3A and S3B). The final hESCs, named TH-tdTomato/AAVS1-ChR2-EYFP hESCs, constitutively expressed ChR2-EYFP during the entire mDA neuron differentiation process whereas tdTomato was expressed only at the later stages of mDA neuron differentiation (Figure S3C) and exclusively in the TH+ mDA neurons (Figures 3C and S3D), highlighting the specific reporting of TH+ cells.
Figure 3. Axonal projection and electrophysiological properties of genetically labeled human mDA neurons.
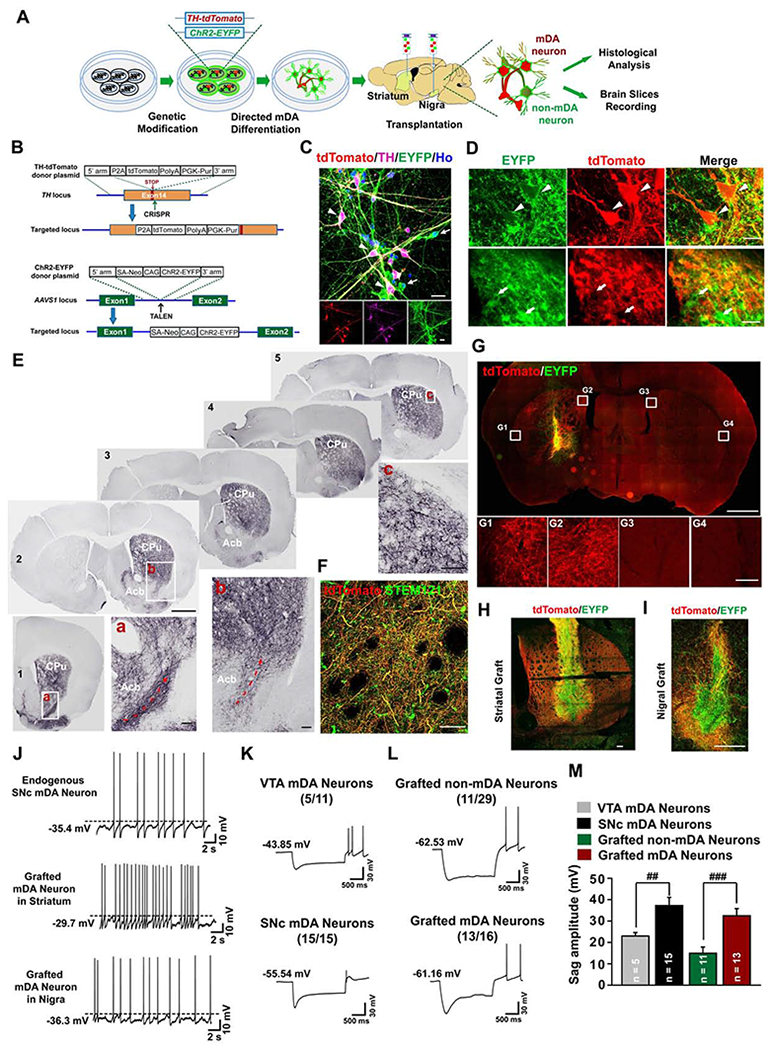
(A) The strategy for visualization and electrophysiological recording of the grafted human mDA or non-mDA neurons. (B) The strategy for generation of TH-tdTomato/ChR2-EYFP dual locus knock-in hESC line (TH-tdTomato/AAVS1-ChR2-EYFP hESCs). (C) Immunostaining of day 42 cultures derived from the above hESCs shows co-expression of tdTomato and EYFP in TH+ neurons (white arrowheads), and expression of EYFP but not tdTomato in TH− neurons (white arrows). Scale bar = 20 μm. (D) Immunohistochemistry images show that nigral graft with the transgenic human mDA neurons contains both tdTomato+/EYFP+ mDA neurons (white arrowheads) and tdTomato−/EYFP+ non-mDA neuronal cells (white arrows). (E) Serial coronal sections immunostained for tdTomato from the PD mouse brain with nigral graft, Scale bar = 1 mm. Boxed areas are magnified as indicated, Scale bar = 0.1 mm. Dotted red arrows indicate ascending axons from rostral-ventral part of the striatum. (F) Co-labeling of human STEM121 and tdTomato. Scale bar = 50 μm. (G-I) Coronal sections in the graft site immunostained for tdTomato and EYFP from the PD mouse brain with striatal (G and H) or nigral (I) graft. Boxed areas in (G) are magnified below. The image in (I) is a composite from two separate images for the upper part and lower part of the same graft. Scale bar = 1 mm, upper panel in (G). Scale bar = 100 μm lower panel in (G). Scale bar = 250 μm in (H and I). (J) Typical traces of spontaneous action potentials (sAPs) in endogenous SNc mDA neurons from mDA neuron reporter mice (DAT-Cre/Ai9), or striatally or nigrally grafted human mDA neurons at 3 months after transplantation. (K-M) Typical traces of voltage sag measurement from endogenous medial VTA or SNc mDA neurons in mDA neuron reporter mice (K), or striatally grafted human non-mDA neurons or mDA neurons at 6 months after transplantation (L). The number in parenthesis represents the number of neurons displaying sag components among recorded cells. The sag amplitude was plotted in (M). The sample number for statistics is indicated in the column. Data are represented as mean ± SEM. Student-t test, ##p < 0.01, ###p < 0.001.
See also Figures S3 and S4.
We then transplanted the mDA progenitors derived from TH-tdTomato/AAVS1-ChR2-EYFP hESCs into the SN or striatum of PD model mice (Figure 3A). Six months after transplantation, EYFP+ grafts were present in all the grafted animals (Figure 3D), and tdTomato was only expressed in the TH+ mDA neurons but not non-mDA neurons, e.g. 5-HT+ serotonin neurons (Figures S3E and S3F). Serial coronal sections of the brain with the SN graft showed that most of the human mDA neuron projections distributed in the CPu, where fibers formed dense and ramified networks (Figures 3E, panel b and panel c). Only a small proportion of mDA neuron projections were present in the Acb (Figure 3E, Slices 1-3). The rostral ascending projection path of mDA neuron axons were identified in these coronal sections (Figure 3E, Slice 1-2 panel a and b, dotted red arrows). The tdTomato+ projections were positive for STEM121 (Figure 3F), confirming their human identity. These results verify the specific axonal path-finding and targeting from grafted human mDA neurons shown by hNCAM staining (Figures 1 and 2). In the striatal graft, tdTomato+ mDA neuron fibers occupied the entire CPu and most of the fibers were confined to the CPu (Figures 3G, 3H, and S3G). In the nigral graft, the tdTomato+ fibers displayed abundant dendritic outgrowth but limited to the graft surrounding (Figure 3I), suggesting cell- and target-specific ramification of human mDA dendrites and axons. In addition, the hESC-derived DA neurons were distributed in the periphery of both the striatal and nigral grafts (Figures 3H and 3I), a phenomenon strikingly similar to that of human fetal grafts in PD patients (Mendez et al., 2005; Mendez et al., 2008).
The cellular features (Figures S1D, S2D, and S2F) and the specific projection pattern (Figures 1–3) suggest that our mDA neurons resemble those in the SNc. Taking advantage of the genetic reporter, we performed whole-cell patch-clamp recording on the mDA neurons (EYFP+/tdTomato+) and non-mDA neurons (EYFP+/tdTomato−) (Figure S3H). We found that both human mDA neurons and non-mDA neurons in the striatal or nigral grafts displayed current- or blue light-induced action potentials (APs) and spontaneous APs (sAPs) by 3 months after transplantation (Figures 3J and S4A–S4F), suggesting functional maturation of grafted human neurons. Importantly, the sAPs of human mDA neurons grafted in both striatum and nigra displayed regular discharge at a low spiking rate (0.87 ± 0.20 Hz for striatal group; 0.83 ± 0.15 Hz for nigral group) and slow sub-threshold oscillatory potentials (0.29 ± 0.06 Hz for striatal group; 0.26 ± 0.13 Hz for nigral group) (Figures 3J, S4G, and S4H). Prominent afterhyperpolarization (AHP) was observed in the sAP of human mDA neurons 6 months after transplantation (Figures S4I and S4J). These physiological features are consistent with characteristics of endogenous SNc (A9) mDA neurons (Figure 3J) (Guzman et al., 2009; Lammel et al., 2008; Nedergaard et al., 1993). In addition, grafted mDA neurons displayed a higher membrane capacitance (Cm) and a trend of lower neuronal input resistance (Rm) when compared with non-mDA neurons in the striatal or nigral graft 6 months after transplantation (Figures S4K and S4L). Furthermore, endogenous SNc mDA neurons are characterized by sag potentials in response to a hyperpolarizing current injection (Evans et al., 2017; Lammel et al., 2008; Neuhoff et al., 2002). Using mDA neuron reporter mice (DAT-Cre/Ai9), we found that all of the recorded SNc neurons displayed a typical sag component in the subthreshold range (37.2 ± 3.8 mV, n=15/15) (Figures 3K and 3M). However, only 5 out of 11 endogenous medial VTA mDA neurons showed sag component, the amplitude of which was much smaller (23.0 ± 1.6 mV, n=5/11) (Figures 3K and 3M). Interestingly, the grafted human mDA neurons in the striatum displayed a pronounced sag component (32.5±3.3 mV, n=13/16). In contrast, only 11 out of 29 grafted non-mDA neurons showed sag component with a much smaller amplitude (14.9 ± 2.9 mV, n=11/29) (Figures 3L and 3M). Together, these results indicate that grafted human mDA neurons have functional characteristics of A9 mDA neurons.
Anatomical synaptic inputs to human neurons are associated with transplant sites
Rabies-mediated tracing has been used to track the anatomical inputs onto the transplanted cells in the PD model (Adler et al., 2019; Cardoso et al., 2018; Grealish et al., 2015). To reveal the presynaptic inputs specifically to the grafted human mDA neurons, we combined the Cre-loxP gene expression system with rabies-mediated trans-synaptic tracing (Figure 4A). We created a TH-iCre hESC line, allowing Cre recombinase expression directed to TH-expressing cells without disrupting endogenous TH expression (Figures 4B, S5A and S5B). The specificity of this system was evidenced by the exclusive mCherry expression in TH+ mDA neurons following infection of the TH-iCre hESC-derived neuronal culture with lentivirus expressing Cre-dependent mCherry (Figures S5C and S5D).
Figure 4. Rabies-mediated tracing of inputs to genetically labeled human mDA neurons.
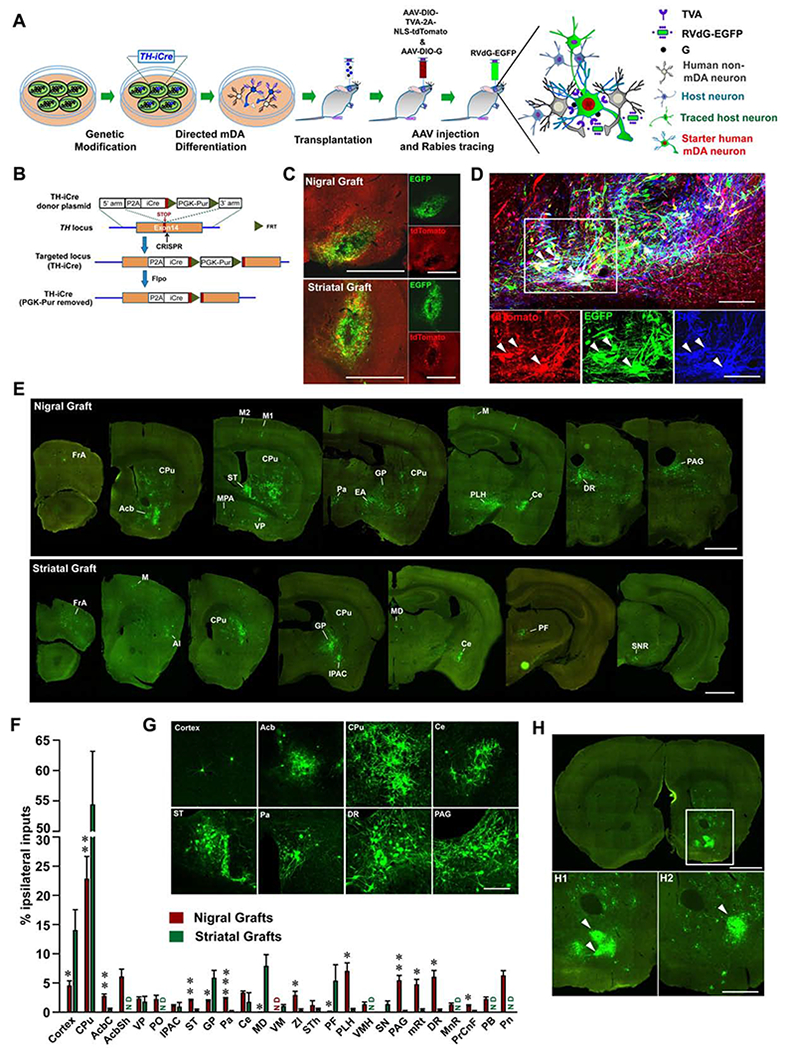
(A) The strategy for tracing inputs to genetically labeled human mDA neurons transplanted into the SN or striatum of PD mice. (B) Schematic diagram showing the generation of TH-iCre hESC line. (C) EGFP and tdTomato expressing neurons in the graft site. Scale bar = 1 mm. (D) Immunohistochemistry images show the expression of tdTomato, EGFP and TH in neurons at the SN graft site. The white arrowheads indicate co-expression of EGFP and tdTomato in TH+ neurons. Scale bar = 100 μm. (E) Serial coronal sections show distribution of traced host neurons (EGFP+/tdTomato−) to nigrally or striatally grafted human mDA neurons. Only the side ipsilateral to the graft is shown. Scale bar = 1 mm. (F) Quantification of ipsilaterally labeled inputs to nigrally or striatally grafted human mDA neurons, shown as a percentage of all ipsilateral inputs. Data are represented as mean ± SEM. Only the brain regions with the average input percentage > 1% to either nigral or striatal graft are shown. n = 3 for striatal graft, n = 5 for nigral graft. ND, not detected. Student-t test, * p < 0.05, **p < 0.01, ***p < 0.001. (G) Magnified images of labeled input neurons to nigrally grafted mDA neurons in different host brain regions. Scale bar = 200 μm. See Table S1 for abbreviations. (H) Coronal section shows labeled input neurons to nigrally grafted mDA neurons in Acb. Boxed area is magnified below (H1). Example distribution of input neurons from another animal is shown (H2). White arrowheads indicate patch-like distribution of labeled input neurons. Scale bar = 1 mm for upper image; Scale bar = 0.5 mm for lower images.
See also Figure S5.
Five months following the transplantation of the TH-iCre mDA progenitors into the nigra or striatum of PD mice, AAV expressing a Cre-dependent TVA and NLS-tdTomato (AAV-DIO-TVA-2A-NLS-tdTomato) as well as AAV expressing a Cre-dependent rabies glycoprotein (G) (AAV-DIO-G) were co-injected into the graft sites (Figure 4A). One month later, the EnvA-pseudotyped and G-deleted rabies virus expressing EGFP (RVdG-EGFP) was injected into the graft sites (Figure 4A). Since only grafted human mDA neurons express Cre recombinase, the expression of TVA, tdTomato and G will be restricted to the human mDA neurons, not non-mDA neurons. Indeed, tdTomato was expressed only in TH+ human mDA neurons (Figure S5E). As the RVdG-EGFP only infects TVA-expressing (tdToamto+) human mDA neurons; the grafted starter human mDA neurons will co-express EGFP and tdTomato. Co-expression of G in the grafted human mDA neurons enabled RVdG-EGFP to spread trans-synaptically to their presynaptic partners (Wickersham et al., 2007); hence, the host presynaptic neurons will express EGFP only (Figure 4A).
The starter neurons (EGFP+/tdTomato+) were only found in the human grafts and they were TH positive (Figures 4C and 4D). Trans-synaptically labeled host neurons (EGFP+/ tdTomato−) were readily detected in the brain regions away from the graft (Figure 4E). In the nigrally grafted brain, the most abundantly labelled host neurons were found in the striatum including CPu and Acb (Figures 4E–4G). The labeled host neurons in the CPu expressed GABA and DARPP32, suggesting striatal medium spiny neurons (Figure S5F). In the Acb, the labeled host neurons formed patches, which was consistently observed in different samples (Figure 4H). In the cortical area, CTIP2+ and SATB2+ cortical neurons were found to project to the nigral human mDA neurons (Figures 4E–4G and S5F). In the hypothalamic area, peduncular part of lateral hypothalamus (PLH) and the paraventricular hypothalamic nucleus (Pa) projected strongly to the human mDA neurons in SN. Densely labelled host neurons were also found in the bed nucleus of stria terminalis (ST) and the central amygdala nucleus (Ce), but not other amygdala regions (Figures 4E–4G). In addition, scattered neurons were observed in the globus pallidus (GP), ventral pallidum (VP), and extended amygdala (EA). In more caudal regions, the dorsal raphe (DR), periaqueductal gray (PAG) and pontine reticular nucleus (Pn) contained large number of labeled neurons (Figures 4E–4G). We identified 5-HT+ serotonin neurons in DR projecting to the human mDA neurons in SN (Figure S5F). The distribution of presynaptic inputs to the nigrally grafted human mDA neurons was strikingly similar to that onto the endogenous SNc mDA neurons in DAT-Cre mice which express Cre in dopamine neurons (Figures S5G and S5H), and the patch-like distribution of inputs to the endogenous mDA neurons was also observed in the Acb (Figure S5I).
In the brain with the striatal graft, dense presynaptic inputs were found in the CPu, but not Acb. More inputs were observed in the GP and cortical areas when compared with those to nigrally grafted human mDA neurons. Parafascicular thalamic nucleus (PF) and mediodorsal thalamic nucleus (MD) preferentially projected to striatally grafted human mDA neurons. The labeled host neurons were also found in Ce and substantia nigra reticular part (SNR) in the brain with striatal graft. Few labeled neurons were found in the more caudal brain regions such as PAG, DR, PB, and Pn (Figures 4E, 4F and S5J).
Thus, human mDA neurons grafted in the striatum and nigra receive inputs from largely different brain regions, suggesting location-dependent presynaptic inputs. The nigrally grafted human mDA neurons receive extensive inputs from similar brain regions as endogenous mDA neurons do (Watabe-Uchida et al., 2012).
The identity of grafted neuron determines its functional input characteristics
Electrophysiological recording showed that few sEPSCs and sIPSCs (spontaneous excitatory and inhibitory postsynaptic currents) were detected in human mDA and non-mDA neurons in the striatal or nigral grafts 3 months post transplantation (Figures 5A–5D). Strikingly, the mean frequency, but not the amplitude, of both sIPSC and sEPSC from mDA and non-mDA neurons in the striatal or nigral grafts were significantly increased at 6 months after transplantation (Figures 5A–5D). No differences in sEPSC or sIPSCs rise time or decay time between grafted non-mDA and mDA neurons were observed (Figures S6A–S6F). These results demonstrate that the functional inputs are established during the 3-6-month period, which is not affected by graft locations or neuronal types.
Figure 5. Electrophysiological properties of inputs to human mDA or non-mDA neurons.
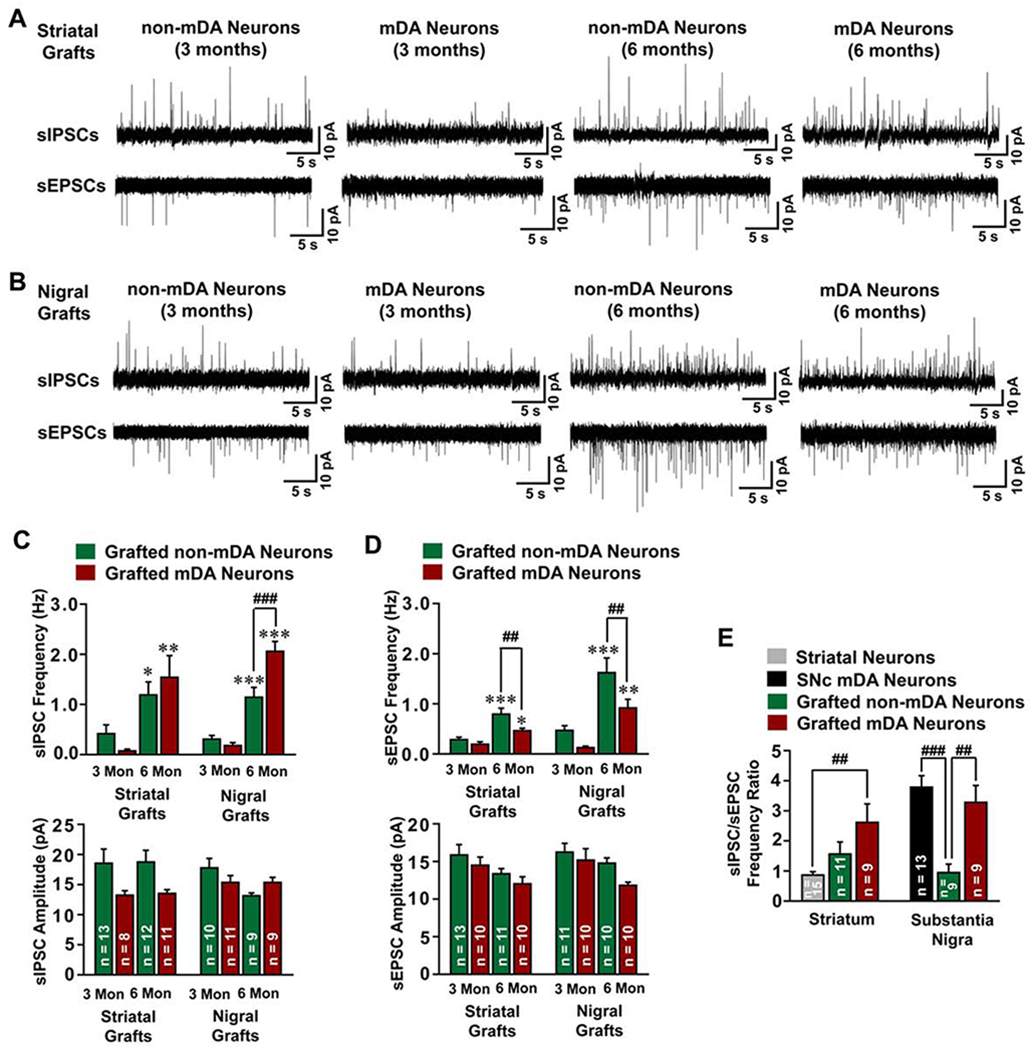
(A and B) Typical traces of sEPSCs and sIPSCs in striatally (A) or nigrally (B) grafted human mDA or non-mDA neurons at 3 or 6 months after transplantation. (C and D) The frequency and amplitude of sIPSCs (C) and sEPSCs (D) were plotted. Data are represented as mean ± SEM. One-way ANOVA followed by Holm-Sidak post hoc test. * p < 0.05, **p < 0.01, ***p < 0.001, comparison between 3-month and 6-month grafted mDA or non-DA neurons. ##p < 0.01, ###p < 0.001, comparison between grafted mDA and non-mDA neurons. (E) The sIPSC/sEPSC ratio in endogenous striatal or SNc neurons in wild type SCID mice, or non-mDA or mDA neurons in striatum or nigra at 6 months after transplantation were plotted. Data are represented as mean ± SEM. One-way ANOVA followed by Holm-Sidak post hoc test. ##p < 0.01, ###p < 0.001. The sample number for statistics is indicated in the column.
See also Figure S6.
Interestingly, the sIPSC frequency in mDA neurons was higher than that in non-mDA neurons in the nigral grafts (Figure 5C). In contrast, the sEPSC frequency in mDA neurons was significantly lower than that in non-mDA neurons in both striatal and nigral grafts (Figure 5D). By calculating the sIPSC/sEPSC ratio, we found that nigrally grafted human mDA neurons receive more inhibitory inputs with a sIPSC/sEPSC ratio of 3.28, a pattern similar to that of endogenous mDA neurons, whereas the human non-mDA neurons received almost equal inhibitory/excitatory inputs, with a sIPSC/sEPSC ratio of 0.95 (Figures 5E and S6G). A trend of higher sIPSC/sEPSC ratio (2.61) of human mDA neurons when compared with non-mDA neurons was also observed in the striatal grafts (Figure 5E) whereas the endogenous striatum neurons received more excitatory inputs with a sIPSC/sEPSC ratios of 0.86 (Figures 5E and S6H). These results suggest that the identity of the grafted neurons, rather than the graft site, determines the inhibitory and excitatory input characteristics of the grafted neurons.
Transplantation of mDA but not Glu neurons corrects motor deficits in PD mice
The functional effect of the nigral grafts was assessed by amphetamine-induced rotation, rotarod test and the cylinder test before and every 4 weeks after grafting (Figure 6A). PD mice with the striatal graft began to recover at 3 months and recovered 4 months post-transplantation in amphetamine-induced rotation. Over time the mice gradually exhibited overcompensation by rotating to the contralateral side (Figure 6B). Similar overcorrected phenotypes were observed from other studies (Chen et al., 2016; Kirkeby et al., 2012; Kirkeby et al., 2017). Functional recovery was also observed in PD mice with nigral mDA graft 4-5 months later, however, no overcompensation was detected up to 6 months (p<0.001) (Figure 6B). In contrast, those received nigral Glu neurons or ACSF (controls) did not show any sign of recovery in amphetamine-induced rotation (p>0.05) (Figure 6B).
Figure 6. Behavioral consequence of transplanted animals.
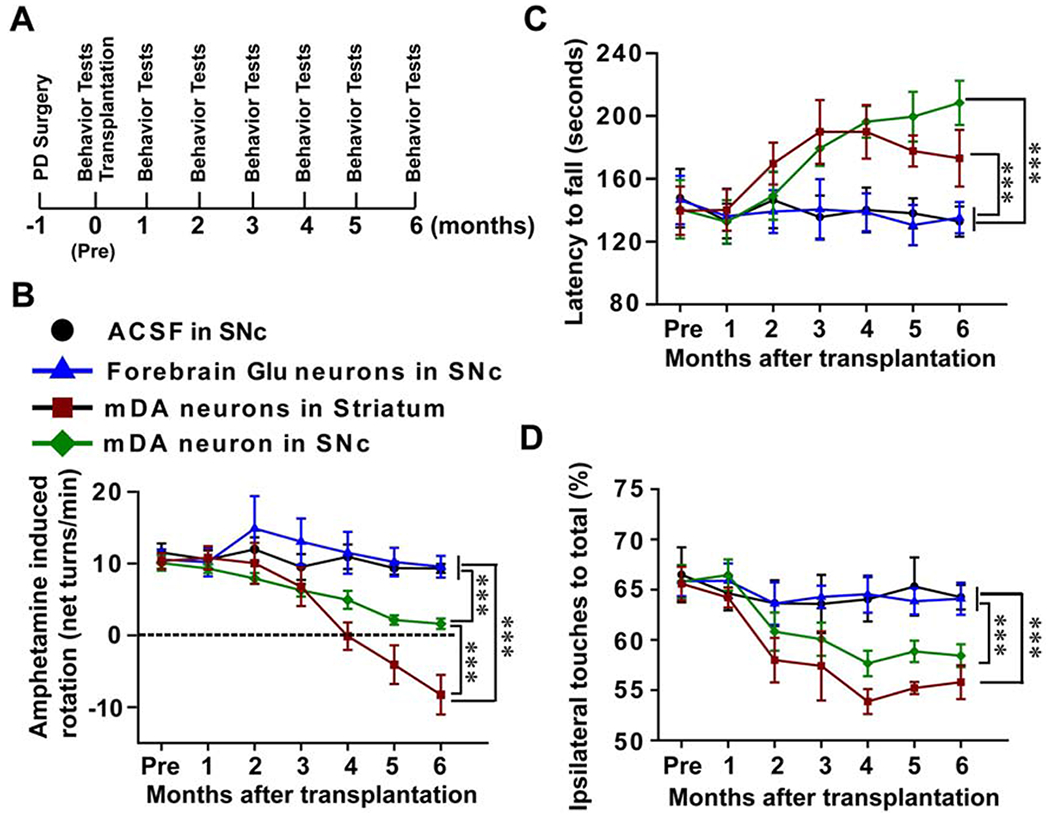
(A) The experimental process of the establishment of the animal model, transplantation and behavioral analysis. These animals were tested monthly using behavior tests including Amphetamine-induced rotation, Rotarod test and Cylinder test. (B) Amphetamine-induced rotation behavior changes over the 6 months post-transplantation. (C) Rotarod test shows the changes in latency to fall before and after transplantation. (D) Cylinder test shows the preferential ipsilateral touches before and after transplantation. In all three behavioral tests, n = 11 in nigral mDA group, n = 8 in nigral Glu group, n = 8 in nigral ACSF group, n = 8 in striatal mDA group. Data are presented as mean ± SEM. Two-way ANOVA followed by Holm-Sidak test. ***, p<0.001.
In the rotarod test, which is used to assess motor coordination and balance, and does not depend on pharmacological stimulation of the dopaminergic system, the latency to fall was significantly increased over time in PD mice that received nigral or striatal transplantation of human mDA neurons (p<0.001), but not in those received nigral Glu neurons or ACSF (p>0.05, Figure 6C).
In the cylinder test, a measure of forelimb akinesia, all the groups presented preferential ipsilateral touches after 6-OHDA lesion. The ipsilateral touching preferences were significantly reduced (close to 50%) 4 months after nigral or striatal transplantation of human mDA-rich neurons (p<0.001), but not the nigral Glu neurons or ACSF (p>0.05, Figure 6D).
Motor recovery depends on the functional reconstruction of the nigra-striatal circuit
To determine whether the behavioral recovery of the PD model animals depends on the reconstructed nigra-striatal circuit, we transplanted mDA neurons that were derived from hESCs with a “bi-directional switch” (Figure 7A). The hESC line, named Bi-DREADD-hESCs, was established by inserting in the AAVS1 locus two DREADDs (Designer Receptors Exclusively Activated by Designer Drugs), an excitatory hM3Dq activated by CNO (Alexander et al., 2009) and an inhibitory KORD activated by Salvinorin B (SALB) (Vardy et al., 2015) (Figures 7B, S7A and S7B). hESCs with mCherry expression cassette knocked-in the AAVS1 locus (mCherry-hESCs) were included as a control (Figures 7B, S7A and S7B). The expression of transgenes was readily detected in the neurons derived from Bi-DREADD-hESCs or mCherry-hESCs in culture or after transplantation (Figures 7C, 7D, and S7C–S7G). Six months post transplantation, the motor recovery was observed in both the Bi-DREADD group and the mCherry (control) group, evidenced by reduced amphetamine-induced rotation and reduced ipsilateral touching preferences (p<0.05) (Figures 7E–7G). Using cylinder test and spontaneous rotation test which do not require stimulation of DA release by amphetamine, we found that CNO (1 mg/kg) treatment further decreased the preferential ipsilateral touches (p<0.05) (Figure 7H) whereas SALB (5 mg/kg) treatment increased the ipsilateral touches in the same animals with the Bi-DREADD graft (p<0.01) (Figure 7H). CNO or SALB treatment had no effect on the ipsilateral touches in mice receiving mCherry-expressing graft (p>0.05) (Figure 7H). In the spontaneous rotation test, the vehicle treated mice did not show obvious motor balance changes. However, CNO treatment significantly induced more contralateral than ipsilateral rotations (p<0.01) in mice transplanted with the Bi-DREADD cells (Figure 7I). This is better shown by the ipsilateral rotation ratio from 48.49 ± 8.10 to 34.83 ± 6.23 (p<0.05) (Figure. 7J). In contrast, SALB significantly increased ipsilateral net rotations (p<0.05), or an ipsilateral rotation ratio of 58.42 ± 9.91 from 48.49 ± 8.10 (p<0.01) (Figure 7J). The mCherry mice did not show obvious changes with or without CNO or SALB treatment (p>0.05) (Figures 7I and 7J). These results suggest that the recovery of forelimb akinesia and asymmetric rotation depends on the graft activity.
Figure 7. Bi-directional control of transplanted PD mice.
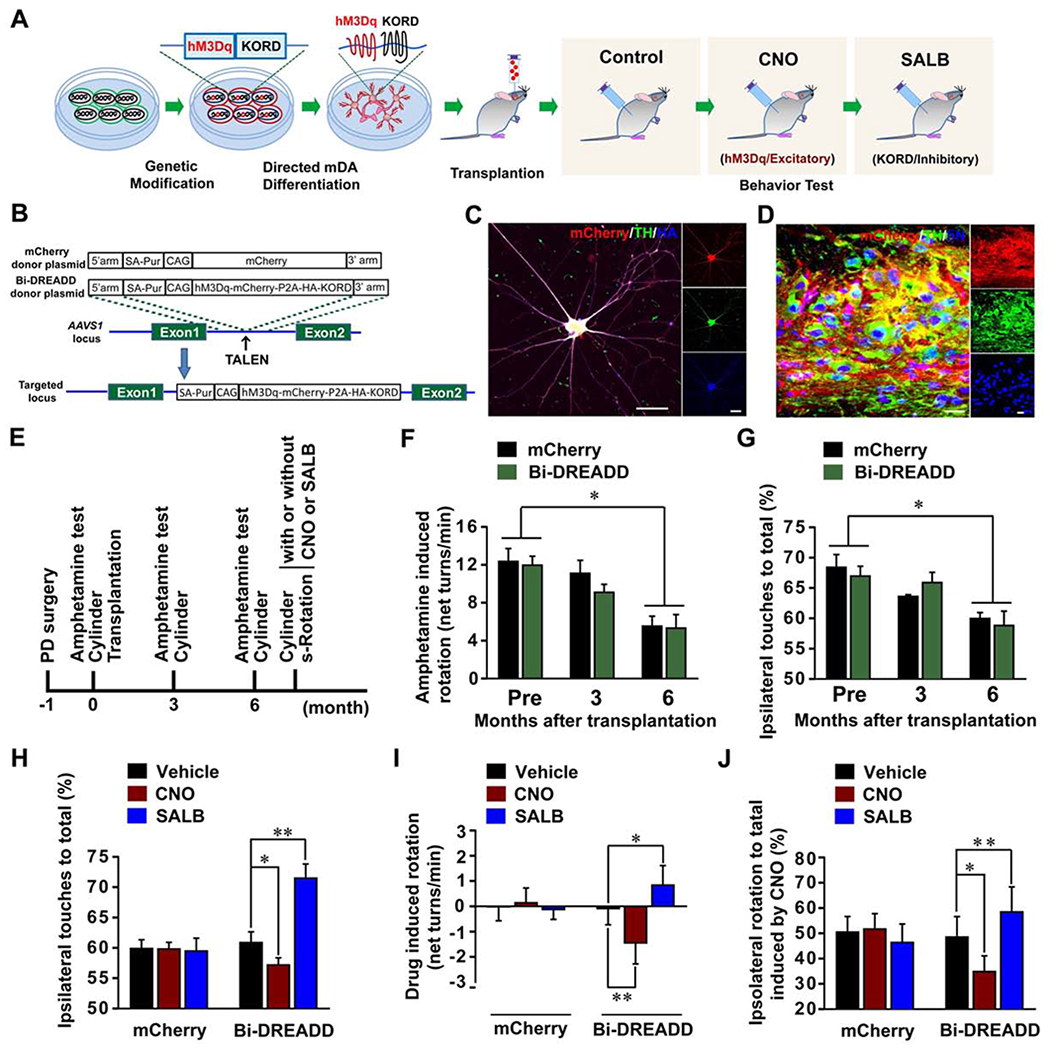
(A) The strategy for bi-directional regulation of human mDA neuron transplants in PD mice. (B) The strategy for generation of mCherry or Bi-DREADD hESC lines. (C) Immunostaining shows co-expression of TH, hM3Dq-mcherry, and HA-tagged KORD in mDA neurons at day 42 of differentiation from Bi-DREADD hESCs. Scale bar = 50 μm. (D) Immunohistochemistry images show that the nigrally grafted mDA neurons from Bi-DREADD hESCs co-expressed human nuclei (hN), mCherry and TH. Scale bar = 20 μm. (E) The experimental process of animal model, transplantation, and behavioral analysis. S-Rotation, spontaneous rotation. (F and G) Amphetamine-induced rotation and Cylinder test show the changes of the rotation behavior (F) or preferential ipsilateral touches (G). (H) Cylinder test shows the changes of preferential ipsilateral touches of PD mice after treatment with vehicle, CNO or SALB. (I and J) Spontaneous rotation test shows CNO- or SALB-induced changes in net ipsilateral rotations (I) and preferential ipsilateral rotations (J). Data are presented as mean ± SEM. Two-way RM ANOVA followed by Tukey’s post hoc test in (F) and G), or Paired t-test in (H), (I), (J). In all behavioral tests, n=6 in each group. **, p<0.01. *, p<0.05.
See also Figure S7.
Discussion
In this study, we developed genetic labeling strategies to precisely map the projection from and synaptic inputs to the grafted human mDA neurons in a mouse model of PD. We found that human mDA neurons, transplanted in the nigra, specifically projected to the dorsal striatum. The grafted mDA neurons receive synaptic inputs, revealed by rabies-mediated tracing, in a pattern strikingly similar to those to the endogenous mDA neurons. Electrophysiological recording revealed predominantly inhibitory inputs to the grafted mDA neurons, which appears dependent on the cell identity but not the transplant site. With the pre- and post-synaptic integration, the homotopically grafted human mDA neurons rescue the motor deficits of PD mice in a manner dependent on graft activity. These findings reveal a cell type-dependent functional circuit integration by transplanted neurons, highlighting the prospect of specialized neuronal types from stem cells for repairing the neural circuit to treat neurological conditions.
What determines the targeted projection by grafted neurons in the mature brain remains unknown. By transplanting two types of projection neurons, mDA and Glu neurons, into the SN of PD mice, we found that both neuronal types project axons over a long distance but to different targets via distinct routes with the majority axons of the grafted mDA neurons target CPu. This is further verified by using the TH reporter cells, showing near exclusive projection to the dorsal (CPu) but not ventral striatum (Acb). Since CPu is the main target of SNc (A9) mDA neurons (Bjorklund and Dunnett, 2007; Joel and Weiner, 2000), our finding suggests that our human mDA neurons are mostly A9-like cells. Indeed, our cellular characterization and, in particular, our electrophysiological recording confirm the A9 identity of the human mDA neurons. This interpretation suggests that many of the axonal projection to other brain regions, seen in the previous studies (Adler et al., 2019; Grealish et al., 2014), may be coming from non-mDA neurons. Together, these results strongly suggest that the path-finding and target projection are largely determined by the identity of the grafted neurons.
Correct synaptic inputs into the grafted neurons are also critical for restoration of lost function. The rabies tracing combined with the Cre-loxP system (TH-iCre) in our study enables identification of monosynaptic inputs to grafted human mDA neurons specifically, which revealed an interesting pattern. By looking at the graft in general, the anatomical synaptic inputs seem associated with the graft sites even though both the nigrally and striatally grafted mDA neurons receive inputs from overlapping areas like the dorsal striatum, similar to the observation made by Parmar and colleagues (Adler et al., 2019). However, the striking similarity between the inputs to the nigrally grafted human mDA neurons and endogenous mDA neurons, suggests that the cell identity plays a role in dictating the synaptic inputs. This is displayed more clearly at the functional level. The grafted mDA neurons receive more inhibitory but less excitatory inputs when compared to non-mDA neurons, whether the cells are transplanted into the striatum or nigra, suggesting that the identity of the grafted neurons dictates the functional synaptic inputs.
With the pre- and post-synaptic integration by transplanted mDA neurons, it is natural to believe that the reconstructed nigra-striatal circuit contributes to motor recovery of the PD mice. It is shown that human mDA neurons, transplanted into the striatum, functionally connect with striatal neurons and contribute to animal behavioral recovery using optogenetic and chemogenetic tools (Chen et al., 2016; Steinbeck et al., 2015). In the current study, the use of Bi-DREADD strategy, which enables excitation and inhibition on the same grafted cells, demonstrates clearly that the reconstructed nigra-striatal circuit is functional, underlying the behavioral recovery of PD mice.
Taken together, with the precise genetic labeling and functional measurement, we have revealed that the restoration of a neural circuit by transplanted cells in the mature brain, including path-finding, targeting specificity, and functional input establishment, is largely determined by the intrinsic property of the grafted neurons. Because of that, it becomes critical to transplant highly enriched, appropriately fated neural progenitors in order to achieve reconstruction of specific circuits for therapeutic outcomes. Most of the major neural cell types can now be efficiently generated from hESCs (Tao and Zhang, 2016). Hence, cell-based therapy for treating neurological conditions is realistic.
Limitations of the Study
Our study revealed a mechanism by which grafted human mDA neurons repair the nigra-striatal circuit anatomically and functionally in the mouse brain. Given the substantially (10X) longer distance between the substantia nigra and the striatum in the human brain, studies using animals that are more relevant to humans such as non-human primates are needed to further assess the potentials of hESC-derived neurons to repair specific circuit, and the methodology to speed up the axonal growth from the grafted DA neurons is desired. Technically, we identified the grafted DA neurons by genetically targeting the TH locus, which, however, may not be sufficiently specific. Some of the low TH-expressing cells might not be true DA neurons due to the high sensitivity of the Cre-LoxP system. Combinatorial targeting with additional DA gene(s) may be necessary to precisely label DA neurons or even DA neuron subtypes.
STAR ★Methods
Resource Availability
Lead Contact
Further information and requests for sources should be directed to, and will be fulfilled by, the Lead Contact, Yuejun Chen (yuejunchen@ion.ac.cn).
Materials Availability
Plasmids and hESC lines generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study. The datasets supporting this study are available from the lead contact, Dr. Yuejun Chen (yuejunchen@ion.ac.cn) upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture
H9 hESCs and reporter H9 hESCs were cultured on a feeder of irradiated mouse embryonic fibroblasts (MEFs) in the ESC medium consisting of Dulbecco’s Modified Eagle’s medium/Nutrient Mixture F-12 (DMEM/F-12), 1× Non-Essential Amino Acids (NEAA), 0.5× Glutamax, 0.1 mM 2-Mercaptoethanol, and 4 ng/ml FGF-2. Cells were fed daily with ESC medium and passaged weekly by Dispase II.
Generation of Midbrain Dopamine Neurons and Forebrain Glutamate Neurons
Induction of the midbrain dopaminergic progenitors was carried out based on protocol described previously (Xi et al., 2012) with modification. Briefly, hESCs (1 day after passaging) on MEF feeder layer were cultured in the neural induction medium (NIM) (DMEM/F-12, 1× NEAA, 1× N2 supplement) supplemented with SB431542 (10 μM) and DMH-1 (2 μM). To pattern the differentiating cells to the midbrain FP progenitors, SHH (C25II, 500 ng/ml) and CHIR99021 (0.4 μM) were added to the cultures from day 1 till day 7. On day 7, individual colonies of neuroepithelial cells were gently blown off with a pipette and replated on mouse embryonic fibroblast feeder in the NIM containing SAG (2 μM) and SHH (100 ng/ml) and CHIR99021 (0.4 μM) for additional 6 days (D7-12). On day 12, CHIR99021 was removed, SHH was reduced to 20 ng/ml, SAG (0.5 μM) and FGF8b (100 ng/ml) was added to the culture to expand the progenitors in suspension till day19. Then, 20 ng/ml SHH and 20 ng/ml FGF8b were kept in the neural induction medium till transplantation at day 32. For in vitro analysis, the neurospheres were dissociated by incubation in Accutase at 37°C for 3-5 minutes on day 32 and plating onto glass coverslips that were coated with Matrigel. Cells were fed with Neural differentiation medium (Neurobasal™ Medium, 1× N2 supplement, 1× B27 supplement) (NDM) supplemented with brain-derived neurotrophic factor (BDNF, 10 ng/ml), glial cell line derived neurotrophic factor (GDNF, 10 ng/ml), ascorbic acid (AA, 200 μM), cAMP (1 μM), transforming growth factor β3 (1 ng/ml) and Compound E (0.1 μM).
Induction of forebrain glutamate neurons from hESCs was described before (Zhang et al., 2001). Briefly, H9 hESC colonies were cultured for 1 week with daily ES medium change. Then, hESC colonies were detached from the feeder layer and grown in the ES medium for 4 days to help form cell aggregates. For neural induction, cell aggregates were cultured in flasks fed with NIM supplemented with 2 μM SB431542 and 2 μM DMH-1 for 3 days. The ESC aggregates were then adhered to vitronectin coated 6-well plates in the presence of NIM till formation of neural tube-like rosettes at around day 16. The rosettes were gently blown off by a 1-ml pipette and suspended in the same medium for another 10 days. Then, progenitors were digested into small spheres with Accutase for 4 min at day 26. After an additional 1 day culture in flask with NIM, small spheres were collected and transplanted into animal models or seeded and matured in NDM supplemented with BDNF (10 ng/ml), GDNF(10 ng/ml), AA (200 μM), cAMP (1 μM), IGF1 (10 ng/ml) on glass coverslips for another 1 week for immunofluorescence staining.
PD model and Cell Transplantation
All animal experiments were conducted according to a protocol approved by the animal care and use committee at University of Wisconsin-Madison, or the Institute of Neuroscience, CAS Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences. The surgical procedure for producing the PD model was performed in SCID mice as described (Chen et al., 2016). Briefly, adult SCID mice (8-12 weeks) were anesthetized with 1%–2% isoflurane mixed in oxygen. 1 μl 6-OHDA (3 mg/ml, in saline with 1% ascorbic acid) was directly injected into the left substantial nigra (anterior-posterior [AP] = − 2.9 mm, lateral [L] = −1.1 mm, vertical [V] = −4.5 mm, from skull). Animals with amphetamine-induced rotation at >6/min over 1.5 h period were selected for cell transplantation 4 weeks after 6-OHDA-lesion surgery. Animals were randomly grouped and transplanted with Glutamatergic, dopaminergic progenitors, or artificial cerebral spinal fluid (ACSF) (control). 5×104 cells were resuspended in 1 μl ACSF containing Rock inhibitor (0.5 μM), B27, 20 ng/ml BDNF and injected into the left nigra ( [AP] = − 2.9 mm, lateral [L] = −1.1 mm, vertical [V] = −4.4 mm, from skull) or left striatum (AP = +0.6 mm, L = −1.8 mm, V = −3.2 mm, from dura).
METHOD DETAILS
Donor Plasmid Construction
TALEN pair, Human codon-optimized Streptococcus pyogenes wild-type Cas9 (Cas9-2A-GFP), Cas9 nickase (Cas9D10A-2A-GFP), and pCAG-Flpo was obtained from Addgene (plasmid #52342, plasmid #52341, plasmid #44719, plasmid #44720, plasmid #60662) (Ding et al., 2013; Qian et al., 2014; Xue et al., 2014). PL652 donor plasmid vector containing a FRT-flanked PGK-puromycin expression cassette was constructed by replacing the loxP sequence in the PL552 (#68407) (Chen et al., 2015) with the FRT sequence. To generate the TH-iCre donor plasmid, DNA fragments with left or right homology arm were PCR amplified from the genomic DNA immediately upstream or downstream of the STOP codon of TH gene. The DNA fragment with iCre gene fused to P2A sequence was PCR amplified from pDIRE (Addgene plasmid #80945) (Osterwalder et al., 2010). P2A sequences were included in the PCR primers. These three fragments were then cloned into the multiple cloning sites of plasmid PL652. To generate TH-tdTomato donor plasmid, the DNA fragment with tdTomato gene fused to P2A was PCR amplified from pAAV-FLEX-ArchT-tdTomato (Addgene plasmid #28305) (Han et al., 2011). P2A sequences were included in the PCR primers. The DNA fragment with hGH polyA signal sequences was PCR amplified from AAVS1-pur-CAG-EGFP plasmid (Addgene plasmid #80945) (Chen et al., 2016). The DNA fragments with left or right homology arm for TH-iCre donor plasmid, DNA fragment containing P2A-tdTomato sequences, and DNA fragments containing hGH polyA signal sequences were cloned into the multiple cloning sites of plasmid PL552. To generate AAVS1-neo-CAG-ChR2-EYFP donor plasmid, we replaced the FlpeERT2 gene in the AAVS1-Neo-CAG-Flpe-ERT2 plasmid (Addgene plasmid #68460) (Chen et al., 2016) with ChR2-EYFP gene which is PCR amplified from pAAV-hSyn-hChR2 (H134R)-EYFP (Addgene plasmid #26973). To generate the AAVS1-pur-CAG-Bi-DREADD donor plasmid (AAVS1-pur-CAG-hM3Dq-mcherry-P2A-HA-KORD) or AAVS1-pur-CAG-mCherry donor plasmid, we amplified mCherry or hM3Dq-mCherry from AAVS1-pur-CAG-hM3Dq-mCherry (Addgene plasmid #80948) (Chen et al., 2016), HA-KORD from pAAV-hSyn-dF-HA-KORD-IRES-mCitrine (Addgene plasmid #65417) (Vardy et al., 2015). The two DREADD genes, hM3Dq-mcherry and HA-KORD, were linked by P2A peptide (hM3Dq-mcherry-P2A-HA-KORD) to ensure simultaneous expression of these two genes in the same cells. The hM3Dq-mcherry-P2A-HA-KORD or mCherry gene was inserted into the AAVS1-pur-CAG-EGFP to replace EGFP. SA-Neo, splice acceptor sequence followed by T2A self-cleaving peptide sequence, and then the neomycin resistance gene. CAG, synthetic CAGGS promoter with the actin enhancer and the cytomegalovirus early promoter.
Electroporation and Generation of Reporter hESC Lines
H9 hESCs were pretreated with Rho-kinase (ROCK) inhibitor for 6-8 h (0.5 mM). Cells were then digested by TrypLE™ Express Enzyme, dispersed into single cells, and electroporated with appropriate combination of Cas9 or TALEN plasmids, sgRNA and donor plasmids in 500 μl of electroporation buffer (5 mM KCl, 5 mM MgCl2, 15 mM HEPES, 102.94 mM Na2HPO4, and 47.06 mM NaH2PO4, pH = 7.2) using the Gene Pulser Xcell System (Bio-Rad) at 250 V, 500 mF in a 0.4 cm cuvette (Phenix Research Products). Cells were then seeded on MEF feeder layer in 6-well plate in MEF-conditioned ESC medium with ROCK inhibitor. Cells were fed daily with MEF-conditioned ESC medium. 72 hours later, puromycin (0.5 μg/ml) or G418 (50-100 μg/ml) were added into MEF-conditioned ESC medium for selection for two weeks. After selection, cells were pretreated with ROCK inhibitor for 6-8 h, and then individual clones were picked up. Genomic PCR was applied to examine the integration of the transgene.
For generation of TH-iCre knock-in hESC line, the cassette containing a P2A peptide sequence-linked codon-improved Cre recombinase (iCre) gene with a STOP codon and then FRT-flanked PGK-Pur sequence (PGK promoter-driven puromycin-resistance gene followed by polyA signal) was introduced to immediately upstream of the STOP codon of the endogenous TH gene of H9 ESCs by CRISPR. FRT-flanked PGK-Pur was then removed by transient expression of Flpo. For generation of TH-tdTomato/AAVS1-ChR2-EYFP hESC line, the cassette containing a P2A peptide sequence-linked tdTomato gene with STOP codon followed by polyA sequence and then PGK-Pur sequence was introduced to immediate upstream of the STOP codon of the endogenous TH gene of H9 ESCs by CRISPR, and then the ChR2 expression cassette was knocked into the AAVS1 locus by TALEN. For generation of Bi-DREADD or mCherry hESCs. The hM3Dq-mCherry-P2A-HA-KORD or mCherry expression cassette was inserted into the AAVS1 locus of H9 hESCs using TALEN.
Whole-Cell Patch-Clamp and Brain Slice Recording
Coronal brain slices (350 μm thick) at the level of the forebrain or the midbrain were prepared from recovered animals at 3 and 6 months post-transplantation using a vibratome (Leica VT1200S) in ice-cold cutting solution (100 mM glucose, 75 mM NaCl, 26 mM NaHCO3, 2.5 mM KCl, 2 mM MgCl2-6H20, 1.25 mM NaH2PO4-6H20, and 0.7 mM CaCl2). The slices were transferred to the recording artificial cerebrospinal fluid (ACSF, 124 mM NaCl, 4.4 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 25 mM NaHCO3, 1 mM NaH2PO4, and 10 mM glucose) saturated with 95% O2/5% CO2. Voltage and current signals were recorded by Axon 700B amplifier (Axon). The recording electrodes (3–5 MΩ) were filled with a solution containing 112 mM Cs-Gluconate, 5 mM TEA-Cl, 3.7 mM NaCl, 0.2 mM EGTA, 10 mM HEPES, 2 mM MgATP, 0.3 mM Na3GTP and 5 mM QX-314 (adjusted to PH 7.2 with CsOH) for spontaneous excitatory post-synaptic current (sEPSC) and spontaneous inhibitory post-synaptic current (sIPSC) recording. For sEPSC or sIPSC recording, cells were voltage clamped at −60 mV or 0 mV, respectively. The initial access resistance was monitored throughout the experiment, ranging from 15-30 MΩ. Cells with the access resistance changed > 15% were discarded. Data were filtered at 1 kHz and digitized at 10 kHz. Action potentials (APs) in response to the blue light stimulation (473 nm, frequency 5 Hz, intensity 10 mM/mm2) or the depolarizing currents (0-100 pA, step 10 pA, duration 2s) were recorded in current clamp mood. Voltage sag measurement was conducted under current clamp mode by injection of −90 pA or −120 pA currents into the grafted or endogenous mDA neurons, respectively. Transplanted mDA neurons and non-mDA neurons were identified by their tdTomato fluorescence in the EYFP-positive graft.
Viral Injections and Rabies Tracing Experiments
For rabies tracing experiments, 200 nl AAV expressing Cre-dependent TVA and tdTomato with nuclear location signal linked through a 2A peptide (AAV2/9-Ef1a-DIO-TVA-2A-NLS-tdTomato, titer 1.29×1012 genome copies (gc)/ml), or 200 nl AAV expressing Cre-dependent Rabies Glycoprotein (AAV2/9-Ef1a-DIO-G, titer 1.29×1012 gc/ml) were co-injected into the graft site (For nigral graft: AP = −2.9 mm, L = −1.1 mm, V = −4.4 mm, from skull; For striatum graft: AP = +0.6 mm, L = −1.8 mm, V = −3.2 mm, from dura) of PD mice 5 months after transplantation. 3 weeks later, EnVA-pseudotyped, rabies G deleted, EGFP-expressing rabies virus (RVdG-EGFP, 400 nl, titer 2×108 pfu/ml) was injected into the same site for trans-synaptic labelling. One week later, the mice were sacrificed for histological analysis. For endogenous mDA neuron, viruses were injected to the SNc (AP = −2.9 mm, L = −1.1 mm, V = −4.5 mm, from skull) of DAT-Cre/Ai9 mice. After fixation, the brain was sectioned (30 μm thick) with a freezing microtome. All coronal sections (1:4 series) without staining were imaged with a fluorescence microscope (Olympus VS120). The locations of labeled neurons and the outlines of brain areas were manually labeled using Photoshop according to the Mouse Brain in Stereotaxic Coordinates (Third Edition). Some sections underwent immunostaining to elucidate cell identity.
Tissue Preparation and Immunohistochemistry
Animals were sacrificed with an overdose of pentobarbital (250 mg/kg, i.p.) and perfused with saline followed by 4% ice-cold phosphate-buffered paraformaldehyde (PFA). The brains were removed and immersed sequentially in 20% and 30% sucrose until sunk. Serial sagittal (0.12 to 3.12 mm from medial to Lateral) or coronal (1.42 to −0.10 mm from the Bregma) sections were cut on a freezing microtome (Leica SM2010R) at a thickness of 30 μm and stored at −20°C in a cryoprotectant solution. Free-floating sections were incubated with a primary antibody in 4°C for 1-2 nights, and then the unbound primary antibodies were removed. For DAB staining, sections were incubated with corresponding biotinylated secondary antibodies for 1h followed by avidin-biotin peroxidase for 1h at room temperature. Immunoreactivity was visualized with DAB staining kit. The sections were then dehydrated with ethanol, permeabilized in xylene, and mounted in neutral resin. For fluorescent immunolabeling, sections were incubated with corresponding fluorescent secondary antibodies for 1h at room temperature. Then sections were mounted by Fluoromount-G.
Lentivirus Production
Lentiviruses were generated in 293T cells by transfecting packaging and backbone plasmids using calcium phosphate/DNA coprecipitation method. 293T cell were cultured in Dulbecco’s MEM (DMEM) containing 10% FBS. The supernatant containing the viral particles was collected 72 hours after transfection, and concentrated by ultracentrifugation at 27000 rpm for 2 hours at 4°C. The viral particles were then resuspended in DPBS.
Imaging and Cellular Quantification
To quantify the population of EN1, FOXA2, LMX1A, NURR1, GIRK2, and TUJ-1 expressing cells among total TH or TH to total (Hoechst-labeled) cells, at least five randomly chosen images from coverslips were counted with ImageJ software. Data were replicated three times and were expressed as mean ± SEM. For measuring the human fiber density in the brain slices, images were captured by a Nikon TE600 microscope. The optical density of human fibers in different areas of the mouse brain was measured by image processing and analysis system (Image Pro Plus 5.1 software). Data were shown as optical density in different areas. For TH, GIRK2, LMX1A, human nuclei (hN) and FOXA2 staining, the graft was outlined and captured by a 60x objective with Nikon A1R-Si laser-scanning confocal microscope (Nikon, Tokyo, Japan) or a fluorescence microscope (Olympus VS120). Single or double stained cells were counted manually with Image J. Data were presented as ratio of TH-, LMX1A-, FOXA2- to total hN or GIRK2/TH/hN to TH/hN cells. All data are expressed as means ± S.E.M.
Behavioral Tests
Rotation Test
Amphetamine-induced rotations were tested before transplantation and every month till 6 months after transplantation. Rotation was recorded by a video camera for 1.5 h, 5-10 min after peritoneally amphetamine (2 mg/ml in saline, 5 mg/kg) injection. Data were presented as the average net number of rotations per minute during 90 min.
For spontaneous rotation test, animals were recorded for 60 min after injection of CNO (1.2 mg/kg) for 20 min, SALB (5 mg/kg) for 5 min, or saline for 20 min.
Cylinder Test
Individual animal was placed in a glass cylinder and recorded by a camera for 3 min. The ipsilateral and contralateral paw touches to the wall of the cylinder were counted. The data were expressed as the percentage of ipsilateral touches to total touches. For drug treatment, animals were treated by CNO (1.2 mg/kg) for 20 min, SALB (5 mg/kg) for 5 min, or saline for 20 min before Cylinder test.
Rotarod Test
An accelerating Rotarod (Med Associates Instruments) was used to test motor coordination. All animals were pre-trained for two days in order to reach a stable performance. On day-1, mice were trained on a rotating rod that accelerated from 2 per minute (rpm) to 20 rpm in a period of 300 s for three times. On day-2, mice were trained on rod accelerated from 3 rpm to 30 rpm twice, and from 4 rpm to 40 rpm once, in a period of 300 s. The test was performed from the third day on a rotating rod that accelerated from 4 rpm to 40 rpm in a period of 300 s. The period of time the mouse stayed on the rod was monitored. The average duration from three repeated tests of each animal was used for data analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
SPSS software was used for statistical analysis. In all studies, data were analyzed by Student-t test, Paired t test, two-way ANOVA followed by Holm-Sidak test, Two-way RM ANOVA followed by Tukey’s post hoc test, or One-way ANOVA followed by Holm-Sidak test. Statistical significance was determined at P < 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-human nuclei | Millipore | Cat#MAB1281; RRID: AB_94090 |
| Mouse monoclonal anti-STEM121 | Takara Bio Inc | Cat#AB-121-U-050; RRID: AB_2632385 |
| Mouse monoclonal anti-EN1 | DSHB | Cat#4G11; RRID: AB_528219 |
| Goat polyclonal anti-FOXA2 | Santa Cruz Biotechnology | Cat#sc-6554; RRID: AB_2262810 |
| Mouse monoclonal anti-synaptophysin | Novus | Cat#NBP1-19222; RRID: AB_1643135 |
| Rabbit polyclonal anti-Calbindin D-28k | Swant | Cat#CB38; RRID: AB_2721225 |
| Rabbit polyclonal anti-LMX-1 | Millipore | Cat#AB10533; RRID: AB_10805970 |
| Mouse monoclonal anti-beta-Tubulin III | Sigma-Aldrich | Cat#T8660; RRID: AB_477590 |
| Rabbit polyclonal anti-Tyrosine Hydroxylase | Pel-Freez Biologicals | Cat#P40101; RRID: AB_2313713 |
| Mouse monoclonal anti-Tyrosine Hydroxylase | Sigma-Aldrich | Cat#T1299; RRID: AB_477560 |
| Mouse monoclonal anti-Human NCAM | Santa Cruz Biotechnology | Cat#sc-106; RRID: AB_627128 |
| Rabbit polyclonal anti-FOXG1 | Abcam | Cat#ab18259; RRID: AB_732415 |
| Rabbit polyclonal anti-TBR1 | Abcam | Cat#ab31940; RRID: AB_2200219 |
| Rat monoclonal anti-CTIP2 | Abcam | Cat#ab18465; RRID: AB_2064130 |
| Rabbit monoclonal anti-SATB2 | Abcam | Cat#ab92446; RRID: AB_10563678 |
| Rabbit polyclonal anti-NURR1 | Santa Cruz Biotechnology | Cat#sc-990; RRID: AB_2298676 |
| Rat monoclonal anti-Corin | R&D | Cat#MAB2209; RRID: AB_2082224 |
| Rabbit polyclonal anti-VGLUT1 | Synaptic Systems | Cat#135303; RRID: AB_887875 |
| Mouse monoclonal anti-PAX6 | DSHB | Cat#pax6; RRID: AB_528427 |
| Rabbit polyclonal anti-GABA | Sigma-Aldrich | Cat#A2052; RRID: AB_477652 |
| Rabbit monoclonal anti-DARPP32 | Abcam | Cat#ab40801; RRID: AB_731843 |
| Rabbit polyclonal anti-Serotonin (5-HT) | Sigma-Aldrich | Cat#S5545; RRID: AB_477522 |
| Goat polyclonal anti-GIRK2 | Abcam | Cat#ab65096; RRID: AB_1139732 |
| Rabbit polyclonal anti-HA | Sigma-Aldrich | Cat#H6908; RRID: AB_260070 |
| Rat monoclonal anti-mCherry | Thermo Fisher Scientific | Cat#M11217; RRID: AB_2536611 |
| Rat monoclonal anti-GFP | Nacalai Tesque | Cat#04404-84; RRID: AB_10013361 |
| Goat polyclonal anti-OTX2 | R&D | Cat#AF1979; RRID: AB_2157172 |
| Goat polyclonal anti-tdTomato | SICGEN | Cat#AB8181-200; RRID: AB_2722750 |
| Bacterial and Virus Strains | ||
| Lentiviral particles expressing Cre-dependent mCherry | This paper | N/A |
| AAV2/9-Ef1a-DIO-TVA-2A-NLS-tdTomato | BrainVTA | Cat#PT-0149 |
| AAV2/9-Ef1a-DIO-G | BrainVTA | Cat#PT-0023 |
| RVdG-EGFP | BrainVTA | Cat#R01001 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SB431542 | Stemgent | Cat#04-0010 |
| DMH-1 | Tocris | Cat#4126 |
| CHIR99021 | Tocris | Cat#4423 |
| bFGF | PeproTech | Cat#100-18B |
| FGF8b | PeproTech | Cat#100-25 |
| SAG | Calbiochem | Cat#566660 |
| BDNF | Peprotech | Cat#450-02 |
| GDNF | Peprotech | Cat#450-10 |
| ascorbic acid | Sigma-Aldrich | Cat#A4544 |
| TGF β3 | R&D | Cat#243-B3-010 |
| Compound E | Calbiochem | Cat#565790 |
| IGF1 | Peprotech | Cat#100-11 |
| cAMP | Sigma-Aldrich | Cat#A9501 |
| Puromycin | Invivogen | Cat#ant-pr-1 |
| G418 | Invivogen | Cat#ant-gn-1 |
| Clozapine N-oxide | Enzo Life Sciences | Cat#BML-NS105-0025 |
| Salvinorin B | Tocris | Cat#5611 |
| Rho kinase Inhibitor | Calbiochem | Cat#555550 |
| 6-Hydroxydopamine hydrochloride (6-OHDA) | Sigma-Aldrich | Cat#H4381 |
| Matrigel | Corning | Cat#354230 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| Fluoromount-G | SouthernBiotech | Cat#0100-01 |
| Hoechst | Life Technologies | Cat#33342 |
| StemPro™ Accutase™ Cell Dissociation Reagent | GIBCO | Cat#A1110501 |
| N-2 supplement (100x) | GIBCO | Cat#17502048 |
| B-27 supplement (50x), minus vitamin A | GIBCO | Cat#12587010 |
| B-27 supplement (50x) | GIBCO | Cat#17504044 |
| GlutaMAX™ supplement | GIBCO | Cat#35050061 |
| MEM Non-Essential Amino Acids Solution, 100X | GIBCO | Cat#11140050 |
| Neurobasal™ Medium | GIBCO | Cat#21103049 |
| DPBS, no calcium, no magnesium | GIBCO | Cat#14190144 |
| DMEM/F-12, HEPES | GIBCO | Cat#11330032 |
| DMEM, high glucose | GIBCO | Cat#11965092 |
| KnockOut™ Serum Replacement | GIBCO | Cat#10828028 |
| TrypLE™ Express Enzyme (1X), no phenol red | GIBCO | Cat#12604021 |
| Dispase II, powder | GIBCO | Cat#17105041 |
| TrypLE™ Express Enzyme (1X), no phenol red | GIBCO | Cat#12604021 |
| Certified Fetal Bovine Serum (FBS) | Biological Industries | Cat#04-002-1A |
| Critical Commercial Assays | ||
| VECTASTAUN Elite ABC Kit, Rabbit IgG | Vector Laboratories | Cat#PK6101 |
| VECTASTAUN Elite ABC Kit, mouse IgG | Vector Laboratories | Cat#PK6102 |
| Experimental Models: Cell Lines | ||
| H9 hESCs | WiCell | Cat#WA09 |
| H9 TH-tdTomato/AAVS1-ChR2-EYFP | This paper | N/A |
| H9 TH-iCre | This paper | N/A |
| H9 AAVS1-mCherry | This paper | N/A |
| H9 AAVS1-Bi-DREADDs | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: CB17.Cg-PrkdcscidLystbg-J/Crl (SCID Beige) | Charles river | Cat# CRL:250; RRID:IMSR_CRL:250 |
| Mouse: B6.SJL-Slc6a3tm1.1(cre)Bkmn/J | The Jackson Laboratory | JAX: 006660; RRID: IMSR_JAX:006660 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX: 007909; RRID: IMSR_JAX:007909 |
| Oligonucleotides | ||
| sgRNA: human TH GAGGTTGGGAAGGGCCCTCA | This paper | N/A |
| sgRNA: human TH CCTGCACTGTCCCGGAGCTC | This paper | N/A |
| Recombinant DNA | ||
| hAAVS1 TALEN Left | Addgene | Addgene Plasmid #52341; RRID: Addgene_52341 |
| hAAVS1 TALEN Right | Addgene | Addgene Plasmid #52342; RRID: Addgene_52342 |
| pCas9_GFP | Addgene | Addgene Plasmid #44719; RRID: Addgene_44719 |
| pCas9D10A_GFP | Addgene | Addgene Plasmid #44720; RRID: Addgene_44720 |
| pCAG-Flpo | Addgene | Addgene Plasmid #60662; RRID: Addgene_60662 |
| pDIRE | Addgene | Addgene Plasmid #26745; RRID: Addgene_26745 |
| PL552 | Addgene | Addgene Plasmid #68407; RRID: Addgene_68407 |
| pAAV-FLEX-ArchT-tdTomato | Addgene | Addgene Plasmid #28305; RRID: Addgene_28305 |
| AAVS1-Pur-CAG-EGFP | Addgene | Addgene Plasmid #80945; RRID: Addgene_80945 |
| AAVS1-Neo-CAG-Flpe-ERT2 | Addgene | Addgene Plasmid #68460; RRID: Addgene_68460 |
| pAAV-hSyn-hChR2(H134R)-EYFP | Addgene | Addgene Plasmid #26973; RRID: Addgene_26973 |
| AAVS1-Pur-CAG-hM3Dq-mCherry | Addgene | Addgene Plasmid #80948; RRID: Addgene_80948 |
| pAAV-hSyn-dF-HA-KORD-IRES-mCitrine | Addgene | Addgene Plasmid #65417; RRID: Addgene_65417 |
| PL652 | This paper | N/A |
| AAVS1-pur-CAG-Bi-DREADD | This paper | N/A |
| AAVS1-pur-CAG-mCherry | This paper | N/A |
| Th-iCre | This paper | N/A |
| Th-tdTomato | This paper | N/A |
| AAVS1-Neo-CAG-ChR2-EYFP | This paper | N/A |
| Lenti-Ubi-DIO-mCherry | This paper | N/A |
| Software and Algorithms | ||
| Photoshop CS2 | Adobe | https://www.adobe.com/products/photoshop.html; RRID:SCR_014199 |
| Illustrator CS2 | Adobe | http://www.adobe.com/products/illustrator.html; RRID:SCR_010279 |
| SigmaPlot 11.0 | SigmaPlot | http://www.sigmaplot.com/products/sigmaplot/; RRID:SCR_003210 |
| Neurolucida | MBF Bioscience | http://www.mbfbioscience.com/neurolucida; RRID:SCR_001775 |
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/; RRID: SCR_003070 |
| Image-Pro Plus 5.1 | Media Cybernetics | https://www.mediacy.com/imageproplus; RRID:SCR_007369 |
Highlights.
Human ESC-derived mDA neurons exhibit A9-like DA neuron characteristics
Functional synaptic inputs depend on the grafted neuronal type but not graft site
Human mDA neurons repair the nigra-striatal circuit with precision
Functionally repaired nigra-striatal circuit restores the animal behavior
ACKNOWLEDGMENTS
We thank Dr. A. Bhattacharyya and Y. Dong for helpful comments on the manuscript. This study was supported in part by the National Key Research and Development Program of China (2018YFA0108000), Strategic Priority Research Program of the Chinese Academy of Sciences( XDA16010310) , Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), NIH-NINDS (NS096282, NS076352, NS086604), NIH-NIMH (MH099587, MH100031), the Bleser Family Foundation, the Busta Foundation, and the NICHD (U54 HD090256), the National Medical Research Council of Singapore (MOH-000212, MOH-000207), the National Natural Science Foundation of China (31771137, 31722024, 81974174, 3170060187), Thousand Young Talents Program, Shanghai Natural Science Foundation (17ZR1448500), Shanghai Rising-Star Program (17QA1400600), Shanghai Pujiang Program (17PJ1410200). Su-Chun Zhang is a cofounder of BrainXell, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests
REFERENCES
- Adler AF, Cardoso T, Nolbrant S, Mattsson B, Hoban DB, Jarl U, Wahlestedt JN, Grealish S, Bjorklund A, and Parmar M (2019). hESC-Derived Dopaminergic Transplants Integrate into Basal Ganglia Circuitry in a Preclinical Model of Parkinson’s Disease. Cell Rep 28, 3462–3473 e3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. (2009). Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 63, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund A, and Dunnett SB (2007). Dopamine neuron systems in the brain: an update. Trends Neurosci 30, 194–202. [DOI] [PubMed] [Google Scholar]
- Cardoso T, Adler AF, Mattsson B, Hoban DB, Nolbrant S, Wahlestedt JN, Kirkeby A, Grealish S, Bjorklund A, and Parmar M (2018). Target-specific forebrain projections and appropriate synaptic inputs of hESC-derived dopamine neurons grafted to the midbrain of parkinsonian rats. J Comp Neurol 526, 2133–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cao J, Xiong M, Petersen AJ, Dong Y, Tao Y, Huang CT, Du Z, and Zhang SC (2015). Engineering Human Stem Cell Lines with Inducible Gene Knockout using CRISPR/Cas9. Cell Stem Cell 17, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xiong M, Dong Y, Haberman A, Cao J, Liu H, Zhou W, and Zhang SC (2016). Chemical Control of Grafted Human PSC-Derived Neurons in a Mouse Model of Parkinson’s Disease. Cell Stem Cell 18, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, and Musunuru K (2013). Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr J, Schwarz MK, Wiedermann D, Leinhaas A, Jakobs A, Schloen F, Schwarz I, Diedenhofen M, Braun NC, Koch P, et al. (2017). Whole-brain 3D mapping of human neural transplant innervation. Nature communications 8, 14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RC, Zhu M, and Khaliq ZM (2017). Dopamine Inhibition Differentially Controls Excitability of Substantia Nigra Dopamine Neuron Subpopulations through T-Type Calcium Channels. J Neurosci 37, 3704–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner S, Grade S, Dimou L, Conzelmann KK, Bonhoeffer T, Gotz M, and Hubener M (2016). Transplanted embryonic neurons integrate into adult neocortical circuits. Nature 539, 248–253. [DOI] [PubMed] [Google Scholar]
- Fricker-Gates RA, Shin JJ, Tai CC, Catapano LA, and Macklis JD (2002). Late-stage immature neocortical neurons reconstruct interhemispheric connections and form synaptic contacts with increased efficiency in adult mouse cortex undergoing targeted neurodegeneration. J Neurosci 22, 4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard A, Decressac M, Frappe I, Fernagut PO, Prestoz L, Besnard S, and Jaber M (2009). Anatomical and functional reconstruction of the nigrostriatal pathway by intranigral transplants. Neurobiol Dis 35, 477–488. [DOI] [PubMed] [Google Scholar]
- Gaillard A, Prestoz L, Dumartin B, Cantereau A, Morel F, Roger M, and Jaber M (2007). Reestablishment of damaged adult motor pathways by grafted embryonic cortical neurons. Nat Neurosci 10, 1294–1299. [DOI] [PubMed] [Google Scholar]
- Goldberg JL (2003). How does an axon grow? Genes & development 17, 941–958. [DOI] [PubMed] [Google Scholar]
- Grealish S, Diguet E, Kirkeby A, Mattsson B, Heuer A, Bramoulle Y, Van Camp N, Perrier AL, Hantraye P, Bjorklund A, et al. (2014). Human ESC-derived dopamine neurons show similar preclinical efficacy and potency to fetal neurons when grafted in a rat model of Parkinson’s disease. Cell Stem Cell 15, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealish S, Heuer A, Cardoso T, Kirkeby A, Jonsson M, Johansson J, Bjorklund A, Jakobsson J, and Parmar M (2015). Monosynaptic Tracing using Modified Rabies Virus Reveals Early and Extensive Circuit Integration of Human Embryonic Stem Cell-Derived Neurons. Stem cell reports 4, 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman JN, Sanchez-Padilla J, Chan CS, and Surmeier DJ (2009). Robust pacemaking in substantia nigra dopaminergic neurons. J Neurosci 29, 11011–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chow BY, Zhou H, Klapoetke NC, Chuong A, Rajimehr R, Yang A, Baratta MV, Winkle J, Desimone R, et al. (2011). A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Frontiers in systems neuroscience 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isacson O, and Deacon T (1997). Neural transplantation studies reveal the brain’s capacity for continuous reconstruction. Trends Neurosci 20, 477–482. [DOI] [PubMed] [Google Scholar]
- Isacson O, and Deacon TW (1996). Specific axon guidance factors persist in the adult brain as demonstrated by pig neuroblasts transplanted to the rat. Neuroscience 75, 827–837. [DOI] [PubMed] [Google Scholar]
- Joel D, and Weiner I (2000). The connections of the dopaminergic system with the striatum in rats and primates: an analysis with respect to the functional and compartmental organization of the striatum. Neuroscience 96, 451–474. [DOI] [PubMed] [Google Scholar]
- Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, Lindvall O, and Parmar M (2012). Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep 1, 703–714. [DOI] [PubMed] [Google Scholar]
- Kirkeby A, Nolbrant S, Tiklova K, Heuer A, Kee N, Cardoso T, Ottosson DR, Lelos MJ, Rifes P, Dunnett SB, et al. (2017). Predictive Markers Guide Differentiation to Improve Graft Outcome in Clinical Translation of hESC-Based Therapy for Parkinson’s Disease. Cell Stem Cell 20, 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Hackel O, Jones I, Liss B, and Roeper J (2008). Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron 57, 760–773. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, and Zhang SC (2009). Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development 136, 4055–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linaro D, Vermaercke B, Iwata R, Ramaswamy A, Libe-Philippot B, Boubakar L, Davis BA, Wierda K, Davie K, Poovathingal S, et al. (2019). Xenotransplanted Human Cortical Neurons Reveal Species-Specific Development and Functional Integration into Mouse Visual Circuits. Neuron 104, 972–986 e976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez I, Sanchez-Pernaute R, Cooper O, Vinuela A, Ferrari D, Bjorklund L, Dagher A, and Isacson O (2005). Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson’s disease. Brain 128, 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez I, Vinuela A, Astradsson A, Mukhida K, Hallett P, Robertson H, Tierney T, Holness R, Dagher A, Trojanowski JQ, et al. (2008). Dopamine neurons implanted into people with Parkinson’s disease survive without pathology for 14 years. Nat Med 14, 507–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard S, Flatman JA, and Engberg I (1993). Nifedipine- and omega-conotoxin-sensitive Ca2+ conductances in guinea-pig substantia nigra pars compacta neurones. The Journal of physiology 466, 727–747. [PMC free article] [PubMed] [Google Scholar]
- Neuhoff H, Neu A, Liss B, and Roeper J (2002). I(h) channels contribute to the different functional properties of identified dopaminergic subpopulations in the midbrain. J Neurosci 22, 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, and Lopez-Rios J (2010). Dual RMCE for efficient re-engineering of mouse mutant alleles. Nature methods 7, 893–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Tortosa S, Tornero D, Gronning Hansen M, Monni E, Hajy M, Kartsivadze S, Aktay S, Tsupykov O, Parmar M, Deisseroth K, et al. (2020). Activity in grafted human iPS cell-derived cortical neurons integrated in stroke-injured rat brain regulates motor behavior. Proc Natl Acad Sci U S A 117, 9094–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin JF, Caronia G, Hofer C, Cui Q, Helm B, Ramakrishnan C, Chan CS, Dombeck DA, Deisseroth K, and Awatramani R (2018). Mapping projections of molecularly defined dopamine neuron subtypes using intersectional genetic approaches. Nat Neurosci 21, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian K, Huang CT, Chen H, Blackbourn L.W.t., Chen Y, Cao J, Yao L, Sauvey C, Du Z, and Zhang SC (2014). A simple and efficient system for regulating gene expression in human pluripotent stem cells and derivatives. Stem Cells 32, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, and Cattaneo E (2002). Opinion: neural stem cell therapy for neurological diseases: dreams and reality. Nat Rev Neurosci 3, 401–409. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck JA, Choi SJ, Mrejeru A, Ganat Y, Deisseroth K, Sulzer D, Mosharov EV, and Studer L (2015). Optogenetics enables functional analysis of human embryonic stem cell-derived grafts in a Parkinson’s disease model. Nat Biotechnol 33, 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbeck JA, and Studer L (2015). Moving stem cells to the clinic: potential and limitations for brain repair. Neuron 86, 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckli ET (2018). Understanding axon guidance: are we nearly there yet? Development 145. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Kirik D, and Bjorklund A (2008). Non-dopaminergic neurons in ventral mesencephalic transplants make widespread axonal connections in the host brain. Exp Neurol 213, 220–228. [DOI] [PubMed] [Google Scholar]
- Tornero D, Wattananit S, Gronning Madsen M, Koch P, Wood J, Tatarishvili J, Mine Y, Ge R, Monni E, Devaraju K, et al. (2013). Human induced pluripotent stem cell-derived cortical neurons integrate in stroke-injured cortex and improve functional recovery. Brain 136, 3561–3577. [DOI] [PubMed] [Google Scholar]
- Vardy E, Robinson JE, Li C, Olsen RHJ, DiBerto JF, Giguere PM, Sassano FM, Huang XP, Zhu H, Urban DJ, et al. (2015). A New DREADD Facilitates the Multiplexed Chemogenetic Interrogation of Behavior. Neuron 86, 936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorn P, Kalsbeek A, Jorritsma-Byham B, and Groenewegen HJ (1988). The pre- and postnatal development of the dopaminergic cell groups in the ventral mesencephalon and the dopaminergic innervation of the striatum of the rat. Neuroscience 25, 857–887. [DOI] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, and Uchida N (2012). Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74, 858–873. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, and Callaway EM (2007). Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature methods 4, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wictorin K, Brundin P, Sauer H, Lindvall O, and Bjorklund A (1992). Long distance directed axonal growth from human dopaminergic mesencephalic neuroblasts implanted along the nigrostriatal pathway in 6-hydroxydopamine lesioned adult rats. J Comp Neurol 323, 475–494. [DOI] [PubMed] [Google Scholar]
- Xi J, Liu Y, Liu H, Chen H, Emborg ME, and Zhang SC (2012). Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells 30, 1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Atallah BV, and Scanziani M (2014). Equalizing excitation-inhibition ratios across visual cortical neurons. Nature 511, 596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brustle O, and Thomson JA (2001). In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19, 1129–1133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. The datasets supporting this study are available from the lead contact, Dr. Yuejun Chen (yuejunchen@ion.ac.cn) upon request.


