Abstract
The final steps in the Renin-Angiotensin-Aldosterone signaling System (RAAS) involve binding of the corticosteroid hormone, aldosterone to its mineralocorticoid receptor (MR). The bound MR interacts with response elements to induce or repress the transcription of aldosterone-regulated genes. Along with the classic genomic targets of aldosterone that alter mRNA and protein expression, aldosterone also regulates the expression of non-coding RNAs (ncRNAs). Short ncRNAs termed microRNAs (miRs) have been shown to play a role in transducing aldosterone’s actions via MR signaling. The role of miRs in homeostatic regulation of aldosterone signaling, and the potential for aldosterone-regulated miRs to act as feedback regulators of MR have been recently reported. In this review, the role of miRs in RAAS signaling and feedback regulation of MR in kidney epithelial cells will be discussed.
Keywords: non-coding RNA, ncRNA, microRNA, miR, mineralocorticoid receptor, MR, aldosterone, RAAS
1. Introduction to non-coding RNAs
Sequencing of the human genome demonstrated that over 95% of the human genome is composed of DNA that does not code for proteins, with transcribed and non-translated RNA sequences far exceeding the protein coding regions (1,2). Studies subsequently demonstrated that far from being “junk” DNA the non-coding RNA (ncRNA) served numerous regulatory roles. NcRNA has been categorized into small and long ncRNA based on the length of the RNA transcript. Long non-coding RNAs (lncRNAs) are defined as RNA transcripts, without protein-coding potential, greater than 200 nucleotides long (3), and small ncRNA are less than 200 nucleotides long. The lncRNAs can circularize when the transcribed ncRNA is spliced onto itself to form a closed circular RNA structure (4,5). Reports have yet to emerge describing a role for lncRNAs in the regulation of or by the mineralocorticoid receptor so this review will focus on the short ncRNAs. Several species of small ncRNA have been identified in eukaryotes (6). The short ncRNAs are classified by their length (20–30 nucleotides) as well as their association with the Argonaute family of proteins (7). The inhibitory small ncRNAs are classed as microRNAs (miRs), small interfering RNA (siRNA) and PIWI-interacting RNA (piRNA) all of which selectively silence genetic transcripts (7,8).
2. MicroRNA background
Of the small ncRNAs, miRs constitute the predominant group and are important post-transcriptional regulators of gene expression (7). MiRs are typically 18–25 nucleotides in length, are widely expressed in plants and animals. MiRs bind predominantly to the 3’untranslated regions (UTRs) of messenger RNAs (mRNAs) to induce translational suppression or mRNA degradation (9–11). The major role of miRs is therefore to suppress protein expression by degrading target mRNA. Exceptions to this inhibitory pathway are known(7,12–16), but the most common function of miRs is to act as negative regulators of proteins, including in signaling pathways (see below). The function of miRs was first noted with their discovery in Caenorhabditis elegans in the 1990’s (17). In these studies, miRs were shown to be critical in the posttranslational control of gene expression associated with C.elegans development (18). In humans, about 2000 bona fide miRs have been identified. It is estimated that >60% of the protein-coding genes contain a conserved miR-binding site (19,20). This, combined with the number of non-conserved binding sites, and interactions with other small ncRNAs makes it likely that the majority of protein-coding genes will be subject to some form of ncRNA regulation. The biogenesis of miRs is tightly controlled and while this topic has been extensively reviewed (see for example;(21–27)) it will be briefly discussed to illustrate how miRs may be regulated by hormonal signaling.
3. MicroRNA synthesis
MiRs are transcribed as precursor forms in the nucleus. MiR precursors typically form a stem-loop secondary RNA structure. Sequences producing this stem-loop structure are encoded throughout the genome, embedded within protein coding genes (introns and exons) or encoded as stand-alone sequences. There is a diversity of strategies that transcribe miRs (detailed in numerous reviews including (7,28–33). However, when transcribed the ncRNAs adopt a characteristic stem-loop structure that needs to be further processed to produce the mature ~23nt miR. MiRs can be transcribed by a unique promoter (34–36) or embedded within genes to share the gene’s promoter (37,38). This allows for independent regulation of miR expression for stand-alone miRs. Alternatively, changes in miR expression are linked to the gene in which the miR is embedded. MiRs can be produced as a single stem-loop structure or be found within a cluster of stem-loop sequences encoded in close genomic proximity (39,40).
MiRs sequences are transcribed by RNA polymerase II into primary miRs (pri-miRs) (22,41,42). PrimiRs fold into the stem-loop hairpin structures, but are longer (>70nt) than mature miRs and contain a polyadenylated tail (7,43,44). The pri-miRs are trimmed in the nucleus in a complex containing the polymerase III enzyme Drosha, to produce a precursor miR (pre-miR) of approximately 70nt in length (27,42,45). Pre-miRs are exported to cytoplasm by Exportin-5 (46,47). In the cytoplasm the pre-miRs are further trimmed by the enzyme Dicer. Processing by Dicer is by structural rather than sequence recognition, with the loop of the pri-miR loaded into a cleavage pocket in the enzyme. The elongated tail is removed, and the loop clipped to produce a double-stranded RNA 22 or 23nt long (7,48). Due to the fact the Dicer cleaves at a set distance from the loop, mature miRs are, for the most part, identical in length. This mature miR is loaded onto Argonaute-2 (AGO2) containing complex. AGO2 is an endonuclease and together with the miR forms the central component of a complex of proteins that recognize and bind to target RNA sequences. The complex is guided by the miR to bind to complementary sequences on the 3’-UTR of target mRNA. Here the complex sterically hinders RNA polymerases to inhibit protein translation or Argonaute cleaves the mRNA to result in its degradation (16). The miR/protein assembly with AGO2 has been termed the RNA-induced silencing complex (RISC) (49–54). The net result of miR action is a reduction in mRNA translation either by translational inhibition or mRNA degradation. The major action of miRs is therefore to suppress protein production and steady-state levels.
4. RAAS cascade
The renin-angiotensin-aldosterone-signaling system (RAAS) is a well-established hormonal cascade that leads to release of the mineralocorticoid steroid hormone aldosterone in response to decreased plasma sodium (Na+), increased circulating potassium (K+), or decreased effective circulating volume (55). RAAS constitutes a critical homeostatic feedback mechanism that coordinates the activity of renal and vascular tissues to regulate blood volume and contribute to maintain blood pressure (55–57). The mineralocorticoid steroid hormone, aldosterone is the final constituent RAAS signaling cascade (58–61). Aldosterone is produced in the zona glomerulosa cells of the adrenal gland in response to renin (62–65). By binding to the mineralocorticoid receptor (MR) aldosterone induces the transcription of proteins that function together to establish a coordinated cellular genomic response (66,67). The specificity of aldosterone signaling in most aldosterone-sensitive tissues is protected by the action of 11β-hydroxysteroid dehydrogenase type 2 (11β-HSD2) which converts cortisol to cortisone to regulate access to the MR (68,69). A major action of aldosterone signaling is to increase Na+ reabsorption from glomerular filtrate. In the kidney, aldosterone modulates the expression of proteins responsible for increasing Na+ and water reabsorption in the distal nephron, leading to volume expansion. Homeostatic feedback following aldosterone stimulation reverses the normal signaling cascade as the cues to release aldosterone are diminished when plasma Na+ and volume are restored (70–72).
5. The Mineralocorticoid Receptor
The MR is a family member of the steroid nuclear receptors which includes receptors for progesterone, androgen, estrogen and glucocorticoids (73–75). As the name suggests, the MR responds to mineralocorticoid hormones including aldosterone. The receptor is capable of binding to other steroid hormones like glucocorticoids, primary amongst these being cortisol. Evolutionarily the MR signaling pathway involving aldosterone is first observed in the lungfish (Neoceratodus forsteri) and lobed-finned fish (76). While a MR analogue is present in cartilaginous fish, aldosterone is not found, indicating that this receptor is activated by other ligands. However, both aldosterone and the MR are represented in tetrapods suggesting the importance of aldosterone signaling in adaptation to living on land (76). Indeed, the central role for aldosterone in the maintenance of sodium (and by association water) homeostasis has been understood for many years and has been extensively reviewed elsewhere (see for example (55,56,60,65,69,77)). The main tissue targets for aldosterone/MR signaling to alter sodium transport are the distal kidney nephron epithelial cells, colon, sweat and salivary glands, where binding of aldosterone to MR induces an increase in sodium reabsorption with corresponding increases in the secretion of potassium and protons (66,67,78). MR is also found in cardiovascular tissues like the endothelial and smooth muscle cells of the vasculature and the heart, but it is not necessarily associated with Na+/K+/H+ transport here.
Unbound, MR is found in the cell cytosol held in an inactive state by binding of chaperones (like the 90-kD heat shock protein) that prevent nuclear localization and transcriptional activity(74,78). On ligand binding, MR undergoes a confirmation change that dislocates these chaperones and allows dimerization of the receptor. The dimer translocates to nucleus and binds to hormonal response elements to initiate DNA transcription. In concert with co-activators or repressors the active MR can then direct the transcription or inhibition of MR targeted genes (78). Target MR induced genes include the serum and glucocorticoid induced kinase (SGK1), subunits of the epithelial sodium channel (ENaC), the MR gene itself (NR3C2), epidermal growth factor receptor (EGFR), insulin-like growth factor 1 receptor (IGF1R) and angiotensin II receptor 1 (AGTR1) (73,74). The main impact of MR activation is to increase sodium reabsorption primarily via SGK1 and an increase in ENaC number in epithelia cells of the kidney distal nephron. This action of aldosterone has been well documented. However, the mechanisms that reverse this signaling pathway or keep excessive aldosterone activation in check are less well characterized
6. RAAS signaling disfunction
There are known cellular mechanisms that reverse the action of long-term aldosterone exposure and keep the signaling cascade in check by altering MR expression. Studies have demonstrated that aldosterone stimulation results in post-translational modifications to the MR that result in its inactivation and degradation (79,80). It was also recently demonstrated that mRNA expression of MR was reduced in response to long term aldosterone stimulation (81). A decrease in MR would diminish the ability to elicit a full aldosterone stimulation and protect the cells from aldosterone excess. The observation that MR RNA expression was decreased in mice placed on extended (7-day) low Na+ diets to induce aldosterone signaling, suggested the possibility of a post-transcriptional regulation of the mRNA message, and this task is ideally suited to miR actions.
A failure to reverse aldosterone signaling can result in pathophysiological disease states. For example, inappropriately elevated levels of aldosterone lead to the development of hypertension (82–84). Primary aldosteronism (PA) is characterized by excessive secretion of aldosterone and results in hypertension, electrolyte abnormalities like hypokalemic alkalosis, and cardiovascular disfunction (84–87). PA is the most common form of secondary hypertension and is seen in approximately 20% of patients with resistant hypertension (56,77,88). PA patients have a higher incidence of cardiovascular abnormalities and suffer greater morbidity and mortality compared to non-PA patients exhibiting similar levels of hypertension (89–91). Excessive aldosterone release leads to a chronic upregulation of the cellular signaling pathways that act to reabsorb Na+ (92). This drives water uptake from the kidney distal nephron to expand plasma volume. The cellular mechanism to keep aldosterone signaling in check are therefore essential to prevent pathophysiological disease states from emerging, particularly if long-term aldosterone signaling is not repressed. Leveraging the negative feedback controls that keep MR signaling in check may be a novel mechanism in disease prevention especially in the case of chronic conditions like long-term hypertension. MiRs appear to fill a negative feedback a roll.
7. MicroRNAs and the mineralocorticoid receptor
It is evident that miRs can exert regulation over MR signaling, altering expression of both the receptor and 11β-HSD2 which protects aldosterone signaling. Two reports have implicated miRs-124 and −135a in the regulation of MR expression. In the first report both miRs were able to downregulate the expression of a MR 3’-UTR luciferase reporter construct (93). In a separate study by Mannironi et al.(94), miR-135a and miR-124 binding to the MR 3’-UTR were also confirmed using luciferase reporter constructs. In addition, protein expression of MR was decreased by the overexpression of each miR and increased when these miRs were inhibited using antisense oligonucleotides. MiR-20a was shown to be enriched in the cortical collecting duct where it reduced the expression of 11β-HSD2 in rat models (95). Aldosterone also modulates the expression of miRs. Studies in cultured mouse kidney cortical collecting duct (CCD) cells demonstrated that aldosterone, acting through MR, modifies miR expression (96). This resulted in both a decrease and increase in several miRs (97). MiR regulation occurs in other aldosterone-sensitive cells, like vascular smooth muscle cells (98) where an upregulation in miR expression is noted after aldosterone stimulation. MiRs have been shown to target constituents of the RAAS cascade. Production of signaling hormones, expression of receptors and signaling proteins are all impacted by changes in miR expression (99–105).
8. MicroRNA’s role in aldosterone signaling in the kidney
When aldosterone levels were increased in the kidney in vivo, mouse miR-192 was shown to be inhibited (106). A validated target of miR-192 was the serine-threonine kinase, with no lysine (WNK1), specifically the long form (L-WNK1). A reduction in miR-192 expression resulted in an increase in L-WNK1 levels (106). L-WNK1 is known to be an important regulator of both K+ and Na+ transport and miR regulation by aldosterone represents an additional layer of control in fine tuning ion transport in the kidney (107–111).
Regulation of renal outer medullary potassium (ROMK) channels by miR-194 was observed when dietary K+ was increased in rodent models. An increase in dietary K+ typically results in an increase in aldosterone release and miR-194 was regulated by alterations in K+ diet (112). The authors describe a scaffold and regulatory protein, intersectin-1 that was regulated by miR-194. Decreasing intersectin-1 expression increased ROMK surface expression by delaying retrieval of the channel from the plasma membrane. As a result, an increase in channel surface residency increased K+ transport. An earlier study by the same group identified that miR-802 was similarly regulated by altered K+ diet. In this case the target protein was caveolin-1 (113). The phenotype would be similar for both miRs. A high K+ diet would increase aldosterone production, induced miR-802 expression which in turn inhibited caveolin-1 to reduced ROMK internalization. The change in dietary K+ provides two separate mechanisms to regulate K+ transport via changes in separate miRs, with similar outcomes.
Na+ regulation by miRs has been reported mainly by altering the function of the epithelial sodium channel (ENaC). In these studies, both a cortical collecting duct (mCCD) cell line treated with aldosterone and mice placed on a low Na+ diet to induce aldosterone release, produced parallel changes in miR expression (96). A cohort of miRs were described to be both upregulated and repressed after 24 hr aldosterone stimulation (in the mCCD cells) or on extended Na+-deficient diets in the CCD cells isolated from mouse kidney (96). The downregulated miR-335–3p targeted a membrane scaffold protein, ankyrin 3 (Ank3). The mechanism of ENaC regulation by Ank3 was elucidated in a follow-up study (114). Increasing Ank3 expression accelerated the delivery of ENaC to the apical surface of mCCD cells. The result of increasing ENaC abundance at the membrane surface was a net increase in Na+ reabsorption across the epithelial cells, a reported action of aldosterone in these cells (see Figure 1).
Figure 1:
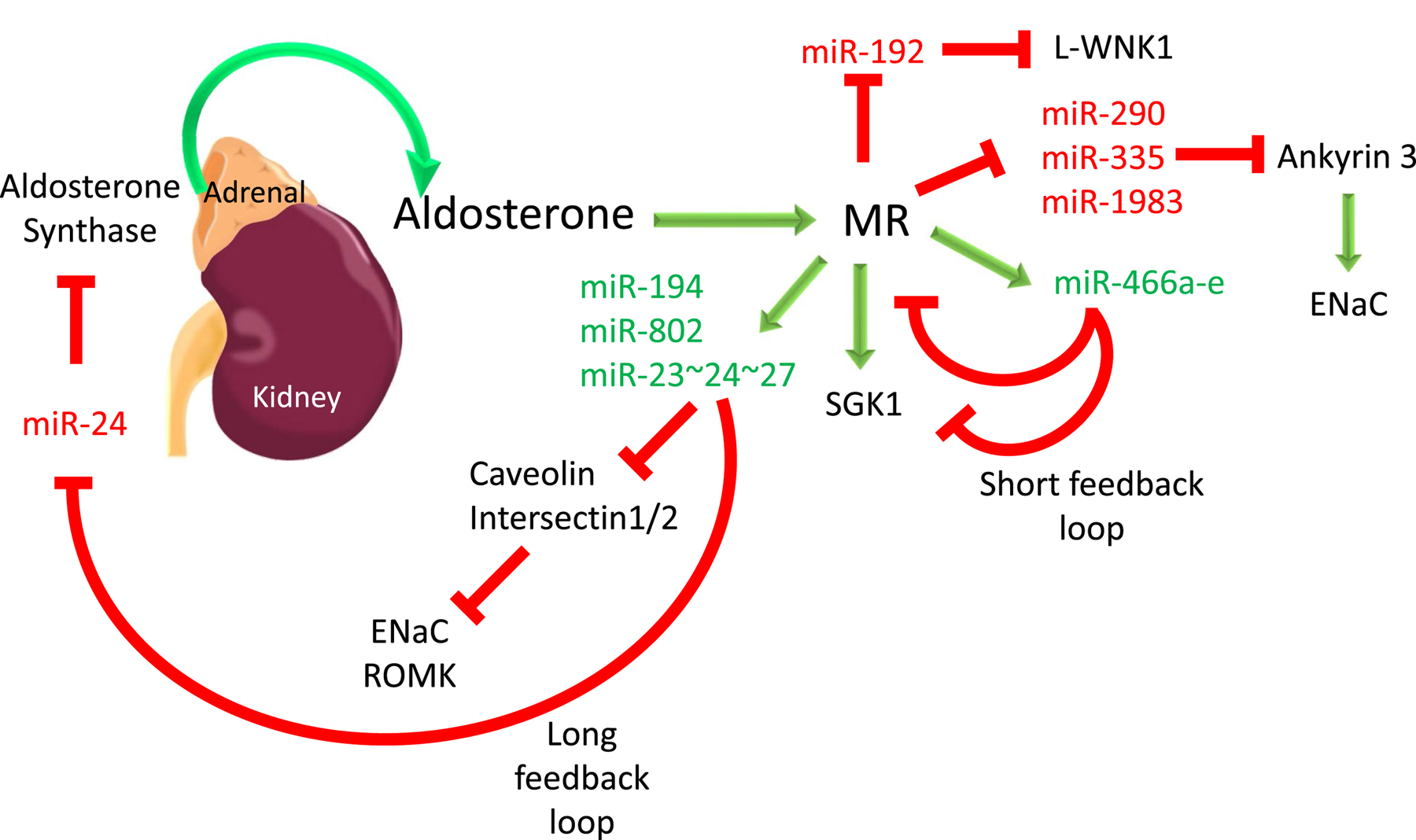
A schematic figure outlining the miR signaling pathways established by aldosterone stimulation through the MR. Red lines represent repression and green arrows stimulation of expression. It is likely that several of the miRs induced by aldosterone, feedback to negatively regulate parts of the RAAS signaling pathway (long feedback loop). There is evidence for localized feedback with miR-466 repressing the expression of MR and SGK1 in the same cells that are stimulated by aldosterone (short feedback loop). The miRs and targets depicted in this figure are discussed in the text.
In addition to the downregulation of miRs, aldosterone has been shown to increase miR expression in the same CCD cells in response to aldosterone stimulation. For ENaC regulation, the miR cluster mmu-miR-23~24~27, was significantly upregulated by aldosterone, along with several other miRs (97). These clusters were previously reported to be enriched in the kidney (115). Alteration in the expression of these miRs has been reported in numerous disease states, such as cardiac hypertrophy (116) or interstitial lung disease (117). The induction of these miRs by aldosterone in the kidney CCD was confirmed in both an mCCD cell line, and in isolated CCD cells from mice placed on low Na+ diets (97).
This increase in all family members was likely due to an MRE upstream of the clusters, although this has yet to be validated. A predicted target of these induced miRs was intersectin-2 (Itsn2). Similar to the reports for ROMK and the regulation of K+ transport, when Itsn2 protein was decreased, an increase in ENaC-mediated Na+ transport was observed. The 3’-UTR of Itsn2 was shown to be a target of these upregulated miRs, and a reduction in Itsn2 protein levels increased Na+ transport via ENaC. When Itsn2 was exogenously upregulated Na+ transport was diminished. The mechanism of action is likely similar to that reported for ROMK (See Figure 1).
It should be noted that MR/aldosterone induced miR changes are not the only miRs known to regulate ENaC. Regulation of ENaC by other miRs has been reported. For example changes in miR-16 in alveolar epithelial cells and miR-263a in enterocytes downregulates the expression ENaC subunits (118). Conversely, the activity of ENaC was shown to alter the regulation of miR-101 and miR-199 in endometrial cells (119). However, the contextual nature of MR-induced miR regulation in the kidney fine-tunes Na+ homeostasis in line with the function of aldosterone.
9. MiRs in feedback regulation of MR signaling
There have been few studies investigating the role of miRs to feedback and regulate MR and/or the the RAAS pathway. This regulation seems possible. For example, miR-24 is induced by aldosterone in MR-sensitive cells including epithelial cells of the distal nephron (97), and this miR negatively regulates aldosterone production by aldosterone synthase (120). Similarly, miR-27a targets the angiotensin converting enzyme. This miR- is part of the miR-23~24~27 cluster that is induced by aldosterone. Induction of the miR-23~24~27 family by aldosterone could therefore in theory decrease expression of two enzymes critical in the production of aldosterone (Long feedback loop, Figure 1). However, this long-range regulation has not been investigated or reported. A more localized negative feedback loop has been reported by our group. In a study by Ozbaki-Yagan et al., the upregulation of miR induced by long-term exposure to aldosterone was investigated (121). A miR cluster, miR-466a-e was identified that increased expression after days (>3days) of aldosterone exposure in principal cells of the mouse cortical collecting duct (mCCD) both in vitro and in vivo. Unlike earlier studies investigating changes in miR expression after 24hr aldosterone stimulation, these miRs did not show a significant upregulation at 24hrs, but steadily accumulated over time so that by 3 days their cellular expression was significantly raised, and they remained elevated for extended periods. The predicted and validated targets of these miRs were both MR and SGK1. By inhibiting these miRs, we demonstrated that not only was the expression of MR and SGK1 significantly greater than in control cells, the sensitivity of the mCCD cells to aldosterone was increased (121). This provides the first evidence for a localized feedback loop established by MR signaling (short feedback loop, Figure 1). The ability for aldosterone-stimulated miRs to target both the receptor and downstream effector of aldosterone protects the cells from excessive signaling that prolonged aldosterone would produce. This could be viewed as a form of aldosterone escape, desensitizing cells to long-term aldosterone stimulation. Of interest, additional miRs were identified in the samples stimulated by extended aldosterone exposure that hint at additional roles for miRs in regulation of MR signaling and these warrant additional investigation.
10. Summary
There is a diversity of responses to aldosterone signaling that varies in tissue and cell type, and between males and females (58,122,123), that remains under-investigated. The impact of extended aldosterone stimulation on miR regulation, Na+ homeostasis and blood pressure are relevant to human conditions where cardiac diseases often take years to manifest. The role of miRs in aldosterone signaling and as feedback regulators of aldosterone is beginning to emerge. Early evidence supports the idea that the induction of miRs by aldosterone, via the MR leads to both local and distant feedback loops that target MR, or components of the RAAS cascade to damp down long-term aldosterone signaling. In addition, the short-term miR regulation setup by MR signaling facilitates signaling responses, namely increases in Na+ transport, required for aldosterone action in the epithelial cells of the distal nephron.
It is therefore apparent that ncRNAs are vital components of aldosterone signaling via the MR. These ncRNAs need to be further investigated both under normal physiological conditions and in pathophysiological states. They are certain to be important points of intervention to regulate normal homeostasis and their anomalous regulation likely contributes to disease states.
Acknowledgements
Thanks goes to current and previous members of the lab who worked on aspects of ncRNA regulation of and by aldosterone. We acknowledge the invaluable collaboration with Dr. Jaqueline Ho’s lab who has assisted with so much of the in vivo work related to this research.
Funding: The work presented in this review (MBB) is supported by funding from the NIH NIDDK DK102843.
Abbreviations:
- MR
Mineralocorticoid receptor
- miR
MicroRNA
- mCCD
Mouse cortical collecting duct
- ncRNA
Non-coding RNA
- RAAS
Renin-Angiotensin-Aldosterone signaling System
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gardiner K (1995) Human genome organization. Curr Opin Genet Dev 5, 315–322 [DOI] [PubMed] [Google Scholar]
- 2.Zuckerkandl E (1997) Junk DNA and sectorial gene repression. Gene 205, 323–343 [DOI] [PubMed] [Google Scholar]
- 3.Paraskevopoulou MD, and Hatzigeorgiou AG (2016) Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol 1402, 271–286 [DOI] [PubMed] [Google Scholar]
- 4.Thomson DW, and Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17, 272–283 [DOI] [PubMed] [Google Scholar]
- 5.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, and Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 [DOI] [PubMed] [Google Scholar]
- 6.Eddy SR (2001) Non-coding RNA genes and the modern RNA world. Nat Rev Genet 2, 919–929 [DOI] [PubMed] [Google Scholar]
- 7.Ha M, and Kim VN (2014) Regulation of microRNA biogenesis. Nature reviews. Molecular cell biology 15, 509–524 [DOI] [PubMed] [Google Scholar]
- 8.Ishizu H, Siomi H, and Siomi MC (2012) Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 26, 2361–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, and Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906 [DOI] [PubMed] [Google Scholar]
- 10.Zeng Y, and Cullen BR (2003) Sequence requirements for micro RNA processing and function in human cells. RNA 9, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennecke J, Hipfner DR, Stark A, Russell RB, and Cohen SM (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 [DOI] [PubMed] [Google Scholar]
- 12.Yang JS, and Lai EC (2010) Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle 9, 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim VN, Han J, and Siomi MC (2009) Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 14.Flynt AS, Greimann JC, Chung WJ, Lima CD, and Lai EC (2010) MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Molecular cell 38, 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chua JH, Armugam A, and Jeyaseelan K (2009) MicroRNAs: biogenesis, function and applications. Curr Opin Mol Ther 11, 189–199 [PubMed] [Google Scholar]
- 16.Miyoshi K, Miyoshi T, and Siomi H (2010) Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Molecular genetics and genomics : MGG 284, 95–103 [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Feinbaum RL, and Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 18.Wightman B, Ha I, and Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 [DOI] [PubMed] [Google Scholar]
- 19.Friedman RC, Farh KK, Burge CB, and Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome research 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lujambio A, and Lowe SW (2012) The microcosmos of cancer. Nature 482, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP, and Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5, 396–400 [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 23.Murchison EP, and Hannon GJ (2004) miRNAs on the move: miRNA biogenesis and the RNAi machinery. Current opinion in cell biology 16, 223–229 [DOI] [PubMed] [Google Scholar]
- 24.He L, and Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 25.Sontheimer EJ, and Carthew RW (2005) Silence from within: endogenous siRNAs and miRNAs. Cell 122, 9–12 [DOI] [PubMed] [Google Scholar]
- 26.Filipowicz W, Jaskiewicz L, Kolb FA, and Pillai RS (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Current opinion in structural biology 15, 331–341 [DOI] [PubMed] [Google Scholar]
- 27.Kim VN (2005) Small RNAs: classification, biogenesis, and function. Molecules and cells 19, 1–15 [PubMed] [Google Scholar]
- 28.Bhatt K, Mi QS, and Dong Z (2011) microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. American journal of physiology. Renal physiology 300, F602–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF, and Jeyaseelan K (2012) Role of microRNAs in kidney homeostasis and disease. Kidney international 81, 617–627 [DOI] [PubMed] [Google Scholar]
- 30.Wei Q, Mi QS, and Dong Z (2013) The regulation and function of microRNAs in kidney diseases. IUBMB life 65, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trionfini P, Benigni A, and Remuzzi G (2015) MicroRNAs in kidney physiology and disease. Nature reviews. Nephrology 11, 23–33 [DOI] [PubMed] [Google Scholar]
- 32.Kim YK, and Kim VN (2007) Processing of intronic microRNAs. The EMBO journal 26, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhang X, and Jin H (2010) Multiple distinct small RNAs originate from the same microRNA precursors. Genome biology 11, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, and Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JY, Kim S, Hwang do W, Jeong JM, Chung JK, Lee MC, and Lee DS (2008) Development of a dual-luciferase reporter system for in vivo visualization of MicroRNA biogenesis and posttranscriptional regulation. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 49, 285–294 [DOI] [PubMed] [Google Scholar]
- 36.Breving K, and Esquela-Kerscher A (2010) The complexities of microRNA regulation: mirandering around the rules. The international journal of biochemistry & cell biology 42, 1316–1329 [DOI] [PubMed] [Google Scholar]
- 37.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, and Olson EN (2007) An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proceedings of the National Academy of Sciences of the United States of America 104, 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinske LC, Galante PA, Kuo WP, and Ohno-Machado L (2010) A potential role for intragenic miRNAs on their hosts’ interactome. BMC genomics 11, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chhabra R, Dubey R, and Saini N (2010) Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24–2 cluster and its implication in human diseases. Molecular cancer 9, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janga SC, and Vallabhaneni S (2011) MicroRNAs as Post-Transcriptional Machines and their Interplay with Cellular Networks. Advances in experimental medicine and biology 722, 59–74 [DOI] [PubMed] [Google Scholar]
- 41.Lee Y, Jeon K, Lee JT, Kim S, and Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. The EMBO journal 21, 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, and Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 43.Han J, Lee Y, Yeom KH, Kim YK, Jin H, and Kim VN (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18, 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y, and Kim VN (2007) In vitro and in vivo assays for the activity of Drosha complex. Methods in enzymology 427, 89–106 [DOI] [PubMed] [Google Scholar]
- 45.Ying SY, Chang DC, Miller JD, and Lin SL (2006) The microRNA: overview of the RNA gene that modulates gene functions. Methods Mol Biol 342, 1–18 [DOI] [PubMed] [Google Scholar]
- 46.Lund E, Guttinger S, Calado A, Dahlberg JE, and Kutay U (2004) Nuclear export of microRNA precursors. Science 303, 95–98 [DOI] [PubMed] [Google Scholar]
- 47.Kim VN (2004) MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends in cell biology 14, 156–159 [DOI] [PubMed] [Google Scholar]
- 48.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, and Kim VN (2011) Dicer recognizes the 5’ end of RNA for efficient and accurate processing. Nature 475, 201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sontheimer EJ, and Carthew RW (2004) Molecular biology. Argonaute journeys into the heart of RISC. Science 305, 1409–1410 [DOI] [PubMed] [Google Scholar]
- 50.Vaucheret H, Vazquez F, Crete P, and Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18, 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamura K, Ishizuka A, Siomi H, and Siomi MC (2004) Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev 18, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siomi MC, Tsukumo H, Ishizuka A, Nagami T, and Siomi H (2005) A potential link between transgene silencing and poly(A) tails. RNA 11, 1004–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perron MP, and Provost P (2009) Protein components of the microRNA pathway and human diseases. Methods Mol Biol 487, 369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choe J, Cho H, Lee HC, and Kim YK (2010) microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO reports 11, 380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibrahim HN, Rosenberg ME, and Hostetter TH (1997) Role of the renin-angiotensinaldosterone system in the progression of renal disease: a critical review. Seminars in nephrology 17, 431–440 [PubMed] [Google Scholar]
- 56.Campese VM, and Park J (2006) The kidney and hypertension: over 70 years of research. Journal of nephrology 19, 691–698 [PubMed] [Google Scholar]
- 57.Hsueh WA, and Wyne K (2011) Renin-Angiotensin-aldosterone system in diabetes and hypertension. J Clin Hypertens (Greenwich) 13, 224–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komukai K, Mochizuki S, and Yoshimura M (2010) Gender and the renin-angiotensinaldosterone system. Fundamental & clinical pharmacology 24, 687–698 [DOI] [PubMed] [Google Scholar]
- 59.Jansen PM, Danser AH, Imholz BP, and van den Meiracker AH (2009) Aldosterone-receptor antagonism in hypertension. Journal of hypertension 27, 680–691 [DOI] [PubMed] [Google Scholar]
- 60.Engeli S (2006) Role of the renin-angiotensin- aldosterone system in the metabolic syndrome. Contributions to nephrology 151, 122–134 [DOI] [PubMed] [Google Scholar]
- 61.Lakkis J, Lu WX, and Weir MR (2003) RAAS escape: a real clinical entity that may be important in the progression of cardiovascular and renal disease. Current hypertension reports 5, 408–417 [DOI] [PubMed] [Google Scholar]
- 62.Willenberg HS, Schinner S, and Ansurudeen I (2008) New mechanisms to control aldosterone synthesis. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme 40, 435–441 [DOI] [PubMed] [Google Scholar]
- 63.Hattangady NG, Olala LO, Bollag WB, and Rainey WE (2012) Acute and chronic regulation of aldosterone production. Molecular and cellular endocrinology 350, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nogueira EF, Bollag WB, and Rainey WE (2009) Angiotensin II regulation of adrenocortical gene transcription. Molecular and cellular endocrinology 302, 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spat A, and Hunyady L (2004) Control of aldosterone secretion: a model for convergence in cellular signaling pathways. Physiol Rev 84, 489–539 [DOI] [PubMed] [Google Scholar]
- 66.Berger S, Bleich M, Schmid W, Greger R, and Schutz G (2000) Mineralocorticoid receptor knockout mice: lessons on Na+ metabolism. Kidney international 57, 1295–1298 [DOI] [PubMed] [Google Scholar]
- 67.Fejes-Toth G, Pearce D, and Naray-Fejes-Toth A (1998) Subcellular localization of mineralocorticoid receptor in living cells: effects of receptor agonist and antagonists. PNAS 95, 2973–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Odermatt A, and Kratschmar DV (2012) Tissue-specific modulation of mineralocorticoid receptor function by 11beta-hydroxysteroid dehydrogenases: an overview. Molecular and cellular endocrinology 350, 168–186 [DOI] [PubMed] [Google Scholar]
- 69.Funder JW, Pearce PT, Smith R, and Smith AI (1988) Mineralocorticoid action: target tissue specificity is enzyme, not receptor, mediated. Science 242, 583–585 [DOI] [PubMed] [Google Scholar]
- 70.Strazzullo P, Galletti F, and Barba G (2003) Altered renal handling of sodium in human hypertension: short review of the evidence. Hypertension 41, 1000–1005 [DOI] [PubMed] [Google Scholar]
- 71.Meneton P (2000) Comparative roles of the renal apical sodium transport systems in blood pressure control. Journal of the American Society of Nephrology : JASN 11 Suppl 16, S135–139 [PubMed] [Google Scholar]
- 72.Antes LM, Kujubu DA, and Fernandez PC (1998) Hypokalemia and the pathology of ion transport molecules. Seminars in nephrology 18, 31–459459287 [Google Scholar]
- 73.Fuller PJ, Yang J, and Young MJ (2017) 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Coregulators as mediators of mineralocorticoid receptor signalling diversity. J Endocrinol 234, T23–T34 [DOI] [PubMed] [Google Scholar]
- 74.Ong GS, and Young MJ (2017) Mineralocorticoid regulation of cell function: the role of rapid signalling and gene transcription pathways. J Mol Endocrinol 58, R33–R57 [DOI] [PubMed] [Google Scholar]
- 75.Rogerson FM, Brennan FE, and Fuller PJ (2004) Mineralocorticoid receptor binding, structure and function. Molecular and cellular endocrinology 217, 203–212 [DOI] [PubMed] [Google Scholar]
- 76.Rossier BC, Baker ME, and Studer RA (2015) Epithelial sodium transport and its control by aldosterone: the story of our internal environment revisited. Physiol Rev 95, 297–340 [DOI] [PubMed] [Google Scholar]
- 77.Pimenta E, and Calhoun DA (2007) Resistant hypertension and aldosteronism. Current hypertension reports 9, 353–359 [DOI] [PubMed] [Google Scholar]
- 78.Yang J, and Young MJ (2009) The mineralocorticoid receptor and its coregulators. J Mol Endocrinol 43, 53–64 [DOI] [PubMed] [Google Scholar]
- 79.Shibata S, Rinehart J, Zhang J, Moeckel G, Castaneda-Bueno M, Stiegler AL, Boggon TJ, Gamba G, and Lifton RP (2013) Mineralocorticoid receptor phosphorylation regulates ligand binding and renal response to volume depletion and hyperkalemia. Cell metabolism 18, 660–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faresse N, Vitagliano JJ, and Staub O (2012) Differential ubiquitylation of the mineralocorticoid receptor is regulated by phosphorylation. FASEB J 26, 4373–4382 [DOI] [PubMed] [Google Scholar]
- 81.Poulsen SB, Limbutara K, Fenton RA, Pisitkun T, and Christensen BM (2018) RNA sequencing of kidney distal tubule cells reveals multiple mediators of chronic aldosterone action. Physiol Genomics 50, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tomaschitz A, Pilz S, Ritz E, Obermayer-Pietsch B, and Pieber TR (2010) Aldosterone and arterial hypertension. Nature reviews. Endocrinology 6, 83–93 [DOI] [PubMed] [Google Scholar]
- 83.Sowers JR, Whaley-Connell A, and Epstein M (2009) Narrative review: the emerging clinical implications of the role of aldosterone in the metabolic syndrome and resistant hypertension. Annals of internal medicine 150, 776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rossi GP, Sechi LA, Giacchetti G, Ronconi V, Strazzullo P, and Funder JW (2008) Primary aldosteronism: cardiovascular, renal and metabolic implications. Trends in endocrinology and metabolism: TEM 19, 88–90 [DOI] [PubMed] [Google Scholar]
- 85.Beuschlein F (2010) Animal models of primary aldosteronism. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme 42, 446–449 [DOI] [PubMed] [Google Scholar]
- 86.Nagata K (2008) Mineralocorticoid antagonism and cardiac hypertrophy. Current hypertension reports 10, 216–221 [DOI] [PubMed] [Google Scholar]
- 87.Trewet CL, and Ernst ME (2008) Resistant hypertension: identifying causes and optimizing treatment regimens. Southern medical journal 101, 166–173 [DOI] [PubMed] [Google Scholar]
- 88.Singer GM, and Setaro JF (2008) Secondary hypertension: obesity and the metabolic syndrome. J Clin Hypertens (Greenwich) 10, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reincke M, Fischer E, Gerum S, Merkle K, Schulz S, Pallauf A, Quinkler M, Hanslik G, Lang K, Hahner S, Allolio B, Meisinger C, Holle R, Beuschlein F, Bidlingmaier M, Endres S, and German Conn’s Registry-Else Kroner-Fresenius-Hyperaldosteronism, R. (2012) Observational study mortality in treated primary aldosteronism: the German Conn’s registry. Hypertension 60, 618–624 [DOI] [PubMed] [Google Scholar]
- 90.Monticone S, D’Ascenzo F, Moretti C, Williams TA, Veglio F, Gaita F, and Mulatero P (2018) Cardiovascular events and target organ damage in primary aldosteronism compared with essential hypertension: a systematic review and meta-analysis. Lancet Diabetes Endocrinol 6, 41–50 [DOI] [PubMed] [Google Scholar]
- 91.Monticone S, Burrello J, Tizzani D, Bertello C, Viola A, Buffolo F, Gabetti L, Mengozzi G, Williams TA, Rabbia F, Veglio F, and Mulatero P (2017) Prevalence and Clinical Manifestations of Primary Aldosteronism Encountered in Primary Care Practice. J Am Coll Cardiol 69, 1811–1820 [DOI] [PubMed] [Google Scholar]
- 92.Chrissobolis S (2017) Vascular Consequences of Aldosterone Excess and Mineralocorticoid Receptor Antagonism. Curr Hypertens Rev 13, 46–56 [DOI] [PubMed] [Google Scholar]
- 93.Sober S, Laan M, and Annilo T (2010) MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochemical and biophysical research communications 391, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mannironi C, Camon J, De Vito F, Biundo A, De Stefano ME, Persiconi I, Bozzoni I, Fragapane P, Mele A, and Presutti C (2013) Acute stress alters amygdala microRNA miR-135a and miR-124 expression: inferences for corticosteroid dependent stress response. PLoS One 8, e73385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rezaei M, Andrieu T, Neuenschwander S, Bruggmann R, Mordasini D, Frey FJ, Vogt B, and Frey BM (2014) Regulation of 11beta-hydroxysteroid dehydrogenase type 2 by microRNA. Hypertension 64, 860–866 [DOI] [PubMed] [Google Scholar]
- 96.Edinger RS, Coronnello C, Bodnar AJ, Labarca M, Bhalla V, LaFramboise WA, Benos PV, Ho J, Johnson JP, and Butterworth MB (2014) Aldosterone regulates microRNAs in the cortical collecting duct to alter sodium transport. Journal of the American Society of Nephrology : JASN 25, 2445–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu X, Edinger RS, Klemens CA, Phua YL, Bodnar AJ, LaFramboise WA, Ho J, and Butterworth MB (2017) A MicroRNA Cluster miR-23–24–27 Is Upregulated by Aldosterone in the Distal Kidney Nephron Where it Alters Sodium Transport. J Cell Physiol 232, 1306–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bretschneider M, Busch B, Mueller D, Nolze A, Schreier B, Gekle M, and Grossmann C (2016) Activated mineralocorticoid receptor regulates micro-RNA-29b in vascular smooth muscle cells. FASEB J 30, 1610–1622 [DOI] [PubMed] [Google Scholar]
- 99.Ilatovskaya DV, Levchenko V, Pavlov TS, Isaeva E, Klemens CA, Johnson J, Liu P, Kriegel AJ, and Staruschenko A (2019) Salt-deficient diet exacerbates cystogenesis in ARPKD via epithelial sodium channel (ENaC). EBioMedicine 40, 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Butterworth MB (2018) Role of microRNAs in aldosterone signaling. Curr Opin Nephrol Hypertens 27, 390–394 [DOI] [PubMed] [Google Scholar]
- 101.Parreira RC, Lacerda LHG, Vasconcellos R, Lima SS, Santos AK, Fontana V, Sandrim VC, and Resende RR (2017) Decoding resistant hypertension signalling pathways. Clin Sci (Lond) 131, 2813–2834 [DOI] [PubMed] [Google Scholar]
- 102.Pacurari M, and Tchounwou PB (2015) Role of MicroRNAs in Renin-Angiotensin-Aldosterone System-Mediated Cardiovascular Inflammation and Remodeling. Int J Inflam 2015, 101527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen LJ, Xu R, Yu HM, Chang Q, and Zhong JC (2015) The ACE2/Apelin Signaling, MicroRNAs, and Hypertension. Int J Hypertens 2015, 896861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Batkai S, and Thum T (2012) MicroRNAs in hypertension: mechanisms and therapeutic targets. Current hypertension reports 14, 79–87 [DOI] [PubMed] [Google Scholar]
- 105.Nossent AY, Hansen JL, Doggen C, Quax PH, Sheikh SP, and Rosendaal FR (2011) SNPs in microRNA binding sites in 3’-UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. American journal of hypertension 24, 999–1006 [DOI] [PubMed] [Google Scholar]
- 106.Elvira-Matelot E, Zhou XO, Farman N, Beaurain G, Henrion-Caude A, Hadchouel J, and Jeunemaitre X (2010) Regulation of WNK1 expression by miR-192 and aldosterone. Journal of the American Society of Nephrology : JASN 21, 1724–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lang F, Capasso G, Schwab M, and Waldegger S (2005) Renal tubular transport and the genetic basis of hypertensive disease. Clinical and experimental nephrology 9, 91–99 [DOI] [PubMed] [Google Scholar]
- 108.Yang CL, Zhu X, Wang Z, Subramanya AR, and Ellison DH (2005) Mechanisms of WNK1 and WNK4 interaction in the regulation of thiazide-sensitive NaCl cotransport. The Journal of clinical investigation 115, 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Subramanya AR, Yang CL, McCormick JA, and Ellison DH (2006) WNK kinases regulate sodium chloride and potassium transport by the aldosterone-sensitive distal nephron. Kidney international 70, 630–634 [DOI] [PubMed] [Google Scholar]
- 110.Wade JB, Fang L, Liu J, Li D, Yang CL, Subramanya AR, Maouyo D, Mason A, Ellison DH, and Welling PA (2006) WNK1 kinase isoform switch regulates renal potassium excretion. Proceedings of the National Academy of Sciences of the United States of America 103, 8558–8563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang CL, and Kuo E (2007) Mechanisms of disease: WNK-ing at the mechanism of salt-sensitive hypertension. Nature clinical practice. Nephrology 3, 623–630 [DOI] [PubMed] [Google Scholar]
- 112.Lin DH, Yue P, Zhang C, and Wang WH (2014) MicroRNA-194 (miR-194) regulates ROMK channel activity by targeting intersectin 1. American journal of physiology. Renal physiology 306, F53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin DH, Yue P, Pan C, Sun P, and Wang WH (2011) MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol 22, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klemens CA, Edinger RS, Kightlinger L, Liu X, and Butterworth MB (2017) Ankyrin G Expression Regulates Apical Delivery of the Epithelial Sodium Channel (ENaC). J Biol Chem 292, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tian Z, Greene AS, Pietrusz JL, Matus IR, and Liang M (2008) MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome research 18, 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, and Li PF (2009) miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proceedings of the National Academy of Sciences of the United States of America 106, 12103–12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hunter MP, Russo A, and O’Bryan JP (2013) Emerging Roles for Intersectin (ITSN) in Regulating Signaling and Disease Pathways. International journal of molecular sciences 14, 7829–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tamarapu Parthasarathy P, Galam L, Huynh B, Yunus A, Abuelenen T, Castillo A, Kollongod Ramanathan G, Cox R Jr., and Kolliputi N (2012) MicroRNA 16 modulates epithelial sodium channel in human alveolar epithelial cells. Biochemical and biophysical research communications 426, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sun X, Ruan YC, Guo J, Chen H, Tsang LL, Zhang X, Jiang X, and Chan HC (2014) Regulation of miR-101/miR-199a-3p by the epithelial sodium channel during embryo implantation: involvement of CREB phosphorylation. Reproduction 148, 559–568 [DOI] [PubMed] [Google Scholar]
- 120.Robertson S, MacKenzie SM, Alvarez-Madrazo S, Diver LA, Lin J, Stewart PM, Fraser R, Connell JM, and Davies E (2013) MicroRNA-24 is a novel regulator of aldosterone and cortisol production in the human adrenal cortex. Hypertension 62, 572–578 [DOI] [PubMed] [Google Scholar]
- 121.Ozbaki-Yagan N, Liu X, Bodnar AJ, Ho J, and Butterworth MB (2020) Aldosterone-induced microRNAs act as feedback regulators of mineralocorticoid receptor signaling in kidney epithelia. FASEB J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yoshida H, Rosano G, Shimizu M, Mochizuki S, and Yoshimura M (2011) Gender differences in the effects of angiotensin receptor blockers on cardiovascular disease. Current pharmaceutical design 17, 1090–1094 [DOI] [PubMed] [Google Scholar]
- 123.Miller JA, Anacta LA, and Cattran DC (1999) Impact of gender on the renal response to angiotensin II. Kidney international 55, 278–285 [DOI] [PubMed] [Google Scholar]


