Abstract
The evolving threat of antibiotic resistance development in pathogenic bacteria necessitates the continued cultivation of new technologies and agents to mitigate associated negative health impacts globally. It is no surprise that infection prevention and control are cited by the Centers for Disease Control and Prevention (CDC) as two routes for combating this dangerous trend. One technology that has gained great research interest is antimicrobial photodynamic inactivation of bacteria, or APDI. This technique permits controllable activation of antimicrobial effects by combining specific light excitation with the photodynamic properties of a photosensitizer; when activated, the photosensitizer generates reactive oxygen species (ROS) from molecular oxygen via either a Type I (electron transfer) or Type II (energy transfer) pathway. These species subsequently inflict oxidative damage on nearby bacteria, resulting in suppressed growth and cell death. To date, small molecule photosensitizers have been developed, yet the scalability of these as widespread sterilization agents is limited due to complex and costly synthetic procedures. Herein we report the use of brominated carbon nanodots (BrCND) as new photosensitizers for APDI. These combustion byproducts are easily and inexpensively collected; incorporation of bromine into the nanodot permits photosensitization effects that are not inherent to the carbon nanodot structure alone—a consequence of triplet character gained by the heavy atom effect. BrCND demonstrate both Type I and Type II photosensitization under UV-A irradiation, and furthermore are shown to have significant antimicrobial effects against both Gram-negative Escherichia coli and Gram-positive Staphylococcus aureus and Listeria monocytogenes as well. A mechanism of “dark” toxicity is additionally reported; the pH-triggered release of reactive nitrogen species is detected from a carbon nanodot structure for the first time. The results described present the BrCND structure as a competitive new antimicrobial agent for controllable sterilization of bacteria.
Keywords: photodynamic inactivation, carbon nanodots, heavy atom effect, photosensitization, reactive oxygen species, reactive nitrogen species, antibiotic resistance, phosphorescence
Graphical Abstract
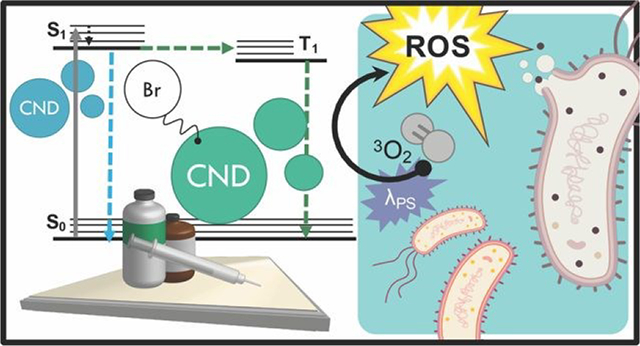
Light-responsive antimicrobial activity is achieved from tuning carbon nanodot structures via bromination, a direct result of the heavy atom effect.
1.0. INTRODUCTION
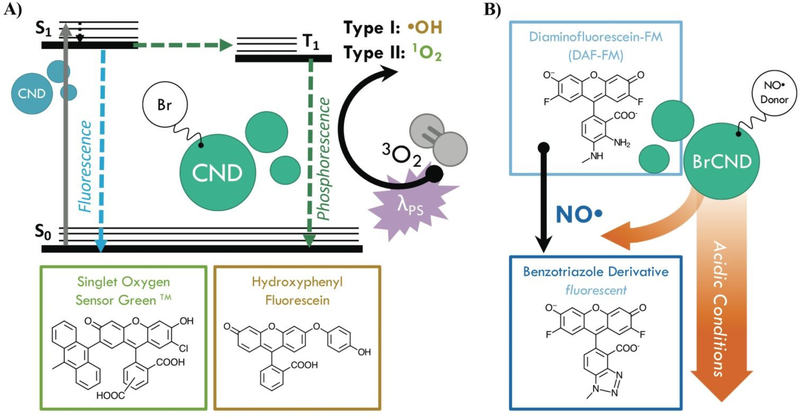
Infection from antibiotic resistant bacteria is not a new threat, yet it is one that is continually growing and demands urgency of response. Antibiotics, which have been the core preventative tactic and treatment strategy against bacterial infections for many decades, require lengthy timelines and rigorous classification before they are available for public use; as such, researchers have increasingly begun to investigate alternative technologies to mitigate the global crisis. Included in this is a focus on prevention, whereby the overall negative health effects from resistant bacteria can be attenuated by simply reducing the rate of infections within the population. This is particularly important for high-risk environments; two examples are hospitals, where those exposed are particularly susceptible, and airports, which are hubs for global transit, although these are not the only areas in which a highly efficient sterilization material would be beneficial. Although numerous agents for this purpose have been developed to date,1, 2 controllable antimicrobial mechanisms are desirable to prevent unwanted resistance development to sterilization procedures or negative environmental impacts. One sterilization technique that has seen expanded interest for this reason is the antimicrobial photodynamic inactivation of bacteria, or APDI.3 This process combines a specific light source with a respective photosensitizing agent (photosensitizer); when excited, the photosensitizer interacts with molecular oxygen to produce reactive oxygen species (ROS), which then inflict oxidative damage upon nearby bacteria—ultimately resulting in cell death.3, 4 Photosensitizers function via either Type I (radical electron transfer) or Type II (energy transfer) mechanisms to generate ROS from molecular oxygen.4 Although numerous photosensitizers have been developed, many of these are small molecules which have complex and expensive synthetic requirements.5 Carbon nanodots, or quasi-spherical nanoparticles from many-layered oxidized graphene sheets, present a promising alternative.6 These particles are actually a combustion byproduct and are simply, rapidly, and inexpensively collected from low-heat, or sooting, flames.7, 8 Further, these particles are frequently reported to resist photodegradation,6 and in our laboratory have been historically stable for use over several years, suggesting advantageous properties in terms of shelf-life in application. Although primarily researched for fluorescence applications such as diagnostics,9–11 carbon nanodots have received heightened interest as antimicrobial agents in recent years, with reports in the literature investigating their intrinsic antimicrobial effects (both photodynamic and otherwise)12–18 and their potential for synergistic toxicity with antibiotics.19–23 Regarding their use for APDI, We have demonstrated recently that the composition of these particles may be tuned to gain luminescence properties characteristic of ROS photosensitizers.24, 25 Namely, incorporation of bromine into the carbon nanodots for a “brominated carbon nanodot” structure (BrCND) permits efficient spin-orbit coupling and subsequent phosphorescence detection,24, 25 as illustrated in Scheme 1a.
Scheme 1.
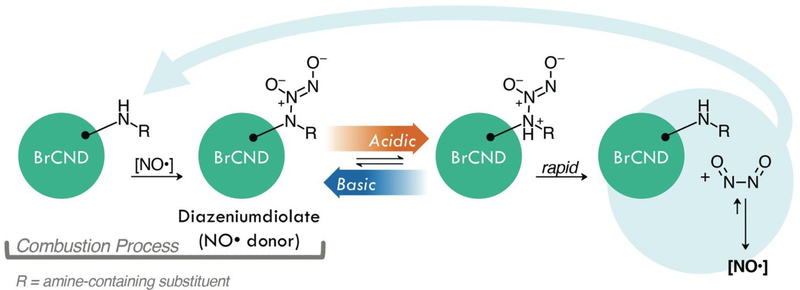
Diagrams demonstrating the use of brominated carbon nanodots (BrCND) as antimicrobial agents. A) BrCND as reactive oxygen species photosensitizers. Carbon nanodots (CND) alone are only fluorescent, as shown by the Jablonski diagram. Incorporation of bromine facilitates the heavy atom effect and phosphorescence from the triplet excited (T1) state. This excited state may also generate reactive oxygen species via a Type I or Type II photosensitization pathway; products of this reaction may be detected by fluorescent probes such as Singlet Oxygen Sensor Green (SOSGTM, 1O2), or hydroxyphenyl fluorescein (HPF, •OH). B) BrCND as donors of nitric oxide under acidic cycled conditions. Products of this reaction may be detected by the fluorescence-on probe diaminofluorescein-FM (DAF-FM).
This result was predicted, as incorporation of atoms such as bromine into small molecules has been a long-standing strategy for achieving phosphorescence from fluorophores, in a phenomenon known as the heavy atom effect.26 Triplet character is desired for ROS generation, as triplet-triplet interactions are favorable between ground state molecular oxygen (3O2) and triplet excited agents. Additionally, the long-lived (psec-msec) lifetime of triplet excited states improve the probability that electron transfer may occur between oxygen and the agent; this is far less likely for fluorescent species due to rapid (nsec) radiative decay.4, 27 A recent study by Zhang et al has linked the photodynamic antimicrobial effects of carbon dots against Gram-negative bacteria Escherichia coli and Salmonella to their phosphorescent character, citing nitrogen content in the dots as the source of phosphorescence tuning; further, the authors demonstrated how carbon dot structures could exhibit photosensitization efficacy competitive even to photosensitizers such as phloxine B and rose bengal.28 For phosphorescent carbon dots, however, the photodynamic toxicity of the structures against Gram-positive bacteria, such as Staphylococcus aureus and particularly Listeria monocytogenes, has received less attention. It is important to examine both Gram-positive and - negative bacterium when proposing a new broad-spectrum photodynamic antimicrobial agent, as both Gram-types exhibit different susceptibilities to APDI and indeed the varying reactive species generated by this process. The outer membrane of Gram-negative bacteria, for example, is known to lower cell permeability for certain photosensitizers, reducing the effects of APDI by blocking access to the plasma membrane and cytoplasm.29 Herein we show that the BrCND structures are in fact able to generate ROS via both Type I and Type II photosensitization mechanisms, employing the fluorescence-on probes Singlet Oxygen Sensor Green™ and hydroxyphenyl fluorescein to detect singlet oxygen (1O2, Type II) and hydroxyl radical (•OH, Type I) respectively (Scheme S1a). The efficacy of this novel photosensitizer is further examined under practical considerations, demonstrated by growth inhibition reported for both Gram-positive Staphylococcus aureus and Listeria monocytogenes and Gram-negative E. coli. The Centers for Disease Control and Prevention identifies both E. coli and S. aureus (particularly methicillin-resistant S. aureus or MRSA) as threats in the 2019 “Antibiotic Resistance Threats in the United States” report;30 L. monocytogenes is a common food-borne pathogen with emerging accounts of antibiotic resistance in recent years.31 We additionally observe and identify an unexpected secondary, “dark” toxicity mechanism from BrCND that functions to inhibit bacterial colony growth in the absence of photodynamic processes. Using the fluorescence-on probe diaminofluorescein-FM (DAF-FM, Scheme 1b), the pH-triggered release of reactive nitrogen species (namely nitric oxide, NO•) is detected for the first time from a carbon nanodot structure. The results presented herein expand the potential of carbon nanodot structures as controllable antimicrobial agents for future materials development and sterilization against antibiotic resistant bacteria.
2.0. EXPERIMENTAL METHODS
2.1. Synthesis of Brominated Carbon Nanodots and Solution Preparation.
Brominated carbon nanodots (BrCND) were collected according to a previously published procedure from our lab;24 key characterization data for these structures are provided in the ESI, Appendix B. In brief, 5M hydrobromic acid (HBr, Acros Organics) was added to a glass impinger. A vacuum was applied for six hours over the impinger, with a hosing line running to a collection funnel positioned over a low-heat flame. Previously reported data has shown burn duration to affect signal strength by modulating concentration;24 the 6-hour period was thus chosen to achieve a sufficiently concentrated sample for subsequent dilutions and analysis. To collect non-bromine-containing carbon nanodots, deionized water was used in place of hydrobromic acid. The maximum concentration of bromide ion (assuming no incorporation of bromide into the carbon dot during synthesis “[Br-]max”), was calculated from the final and initial sample volumes. It is important to note here that previous studies whereby non-bromine-containing carbon dots were first collected then refluxed with hydrobromic acid exhibited phosphorescence only after 6-hour reflux times, with only weak emission achieved.24 These data are also given in the ESI, Appendix B. These results suggest that bromination of the structures occurs during synthesis, and that phosphorescence is not simply a consequence of bromide ion diffusing in the carbon dot solution. Accordingly, no dialysis steps were performed to remove the excess bromide.
To achieve specific pH solutions of varying BrCND concentrations, different ratios of deionized water to raw BrCND solutions were added to trisodium citrate (~0.17M, Sigma Aldrich) and the initial pH tested using an Accumet® Basic AB15 benchtop pH meter. Adjustments to the desired pH were made using 10M hydrobromic acid or sodium hydroxide until the reading was stable over several minutes. Control solutions were prepared using only deionized water, sodium citrate, and hydrobromic acid. Approximate bromide concentrations were determined from the hydrobromic acid and BrCND aliquot volumes and the final solution volume. It should be noted that this bromide concentration value for any solution containing BrCND is only approximate, as the true concentration of free bromide in the initial sample was unknown. For these, [Br]max is reported.
2.2. Spectroscopic and Physical Characterization.
All absorption and fluorescence measurements were conducted in a quartz cuvette. Absorption readings were performed on an Agilent Technologies Cary 60 UV-Vis spectrophotometer with Cary WinUV Scan application software; fluorescence measurements were completed using a FluoroMax®−4P spectrophotometer. Spectra were extracted and plotted, with signal responses reported as “percent signal changes” (ΔF) according to Equation 1,
| (Equation 1) |
where F is the fluorescence intensity recorded at a particular emission wavelength, “pre” denotes the pre-exposure measurement, “post” denotes the post-exposure measurement, and λmin − λmax encompass the detected emission wavelength range.
Dynamic light scattering and zeta potential measurements were performed using a Malvern Zetasizer Nano-ZS; the latter were collected at pH 3.5 with μM salt concentrations. Gel electrophoresis was conducted using a Bio Rad PowerPac HC (100 V, 40 min) and a 1.75% Certified™ Molecular Biology Agarose (Bio Rad) gel prepared with 1% TBE buffer (Fisher Bioreagents®). Sample aliquots at 50 μL were run. Gels were imaged on a Bio Rad Gel Doc™ EZ Imager using Ethidium Bromide settings.
2.3. Fluorescence Detection of Reactive Oxygen Species.
Singlet Oxygen Sensor Green™ (SOSG™), Hydroxyl Phenyl Fluorescein (HPF), and Diaminofluorescein-FM (DAF-FM) were purchased from Invitrogen® and were prepared as stock probe solutions according to the manufacturer recommendations. The following procedure describes the technique used for reactive oxygen species (ROS) detection using both SOSG™ and HPF, as both probes are fluorescein-based and therefore behave similarly. To a pH ~3 BrCND solution, a small (<5% total solution volume) aliquot of sodium hydroxide was added to achieve a pH of >12; an aliquot of stock probe was added to achieve a 4.8 μM solution of the probe, and the initial (“pre”) probe fluorescence measurement was obtained. An additional aliquot of stock probe was added to achieve a 95 μM solution of the probe. Fluorescence (λex = 473 nm, slit widths = 2 nm) measurements of the pre-exposure solutions were performed. The pH was then adjusted back to ~3 using hydrochloric acid (HCl, Acros Organics) and was exposed for four minutes to ultraviolet (UV) light using an Entela Blak-Ray® Long Wave Ultraviolet lamp (Model B 100 AP/R, λmax = 365 nm, “exposed”); exposure powers were recorded using a ThorLabs PM100D power meter and energy densities (J•cm−2) were calculated from exposure times and sample surface area approximations. For “dark” conditions, the sample was covered for the exposure interval. For gas-purged conditions, a steady stream of oxygen or argon (Airgas, Inc.) was bubbled through the solution for 1-minute prior to exposure; when complete, the sample was capped then exposed. For “air purged” samples, bubbling was conducted from the laboratory air valve. Between fluorescence readings the samples were purged with nitrogen (2-minutes) to normalize the dissolved gas content for fluorescent readings. If not specifically indicated, the sample was not purged and therefore contains atmospheric levels of dissolved oxygen. For the “post” exposure fluorescence reading, sodium hydroxide (<5%) was again added to the sample to restore the pH to >13.
To conduct the pH cycling experiments for nitric oxide detection using DAF-FM, the BrCND solutions were first adjusted to pH 3.0 following the procedure described in section 2.1, including a control containing no BrCND that was buffered to the same pH, concentration of trisodium citrate, and concentration of bromide ion. BrCND solutions used were diluted significantly from their original prepared concentrations using the buffered control; the absorption of BrCND at 365 nm was approximately zero. The pH of each sample was then adjusted to 12–12.5 with NaOH and confirmed using colorimetric pH test strips; initial fluorescence (λex = 473 nm, slit widths = 1 nm) and absorption measurements were recorded for the BrCND prior to DAF-FM addition. The probe was then added (final concentration = 2.7 μM), mixed, and fluorescence/absorption immediately recorded. The pH was adjusted to ~2.5 using a small (<10% by volume) aliquot of HCl. The sample then underwent a 4-minute “exposure” period at room temperature under either UV-irradiated or dark conditions. After exposure, the pH was returned to basic pH (12–12.5) using a small aliquot of NaOH, and the final fluorescence/absorption measurements were recorded. For the dilution control, both HCl/NaOH aliquots (excluding the initial NaOH addition) were replaced by deionized water; accordingly, the pH of the sample was 12–12.5 for the entire cycling procedure.
2.4. Bacterial Growth and Sample Preparation.
Strains of Escherichia coli and Staphylococcus aureus were cultured overnight on Luria-Bertani (“LB,” for E. coli and S. aureus) agar plates prepared in-house. Listeria monocytogenes was cultured either on Blood Sheep Agar (Fisher Scientific) plates or brain heart infusion (“BHI”) plates that had been prepared in-house. Single colonies were then suspended in DI water immediately before an experiment was performed such that the solution optical density was between 0.11–0.12 a.u. at 600 nm (108 CFU/mL). Depending on the strain and experimental conditions, subsequent serial dilutions were performed into DI water for the optimal experimental concentration of bacteria.
2.5. Antimicrobial Control Methods.
For each strain, effects of UV exposure, pH variation, and bromide ion concentration were examined. In the case of exposure, different zones of UV power under the exposure source were determined using a ThorLabs PM100D power meter. The bacterial samples were added to a 96-well plate positioned in these zones, with addition times noted for each sample. Aliquots were removed after the exposure time period and were added to phosphate buffered saline solution (PBS, Fisher Bioreagents®) in light-sensitive centrifuge tubes. Once all samples had been collected, 10 μL aliquots of each sample, and subsequent tenfold serial dilutions, were plated and placed in the incubator overnight. This same procedure was performed for both the pH (range: 2–6) and bromide concentration (0–5M, deionized water, neutral pH) experiments in the absence of UV exposure. In all cases, the initial bacterial solution (described in section 2.4) was serially diluted into the experimental solutions prior to the exposure window (ESI Scheme S1). As a final control, bromide concentration effects were determined under the photosensitization experimental parameters (pH 3–4, UV exposure). Bromide solutions were prepared according to the procedure described for the control solutions in section 2.1, adjusting the overall bromide concentrations using sodium bromide (Sigma Aldrich). All plated experiments were photographed, and colonies counted after incubation overnight. For detailed solvent descriptions for bacterial experiments, the reader is referred to the ESI Table S1.
2.6. Antimicrobial Photodynamic Inactivation of Bacteria: Methods.
Bacterial solutions were prepared according to section 2.4. Brominated carbon nanodot and control solutions were prepared according to section 2.1 (additional details in the ESI Table S1) and were added to individual light-sensitive centrifuge tubes. UV power zones (3 mW) for a 96-well plate positioned under the exposure source were determined as described in section 2.5. The experiment was timed, with aliquots of the initial bacterial solution being added at regular intervals to each experimental solution tested. After the exposure window, aliquots from each sample were removed and transferred to PBS to restore near-neutral pH conditions. Once all samples had been collected, at least 2x tenfold serial dilutions (optimized for countable colony formation) of each sample were performed into PBS; 10 μL aliquots of each dilution were plated and the bacteria permitted to grow overnight. For a diagram of the procedure, see supporting Scheme S1. All plates were photographed and those which were countable were analyzed for colony formation either manually or using the Colony Counter plugin for ImageJ or the Promega Colony Counter application for iPhone. In some cases, high density estimates were performed for samples with crowded growth. For more information on these procedures, the reader is referred to Appendix A in the ESI. From the colony counting data, quantities of relative viability (R) and growth inhibition by UV (IUV) are reported, calculated from equations 2 and 3 respectively:
| (Equation 2) |
| (Equation 3) |
where A denotes a particular absorption value (at 365 nm) for the BrCND sample for which the count was obtained and n indicates a non-zero concentration of BrCND. Additionally, normalized IUV values were obtained by dividing IUV (%) values of n samples by the IUV,n=0 (%) value.
3.0. RESULTS AND DISCUSSION.
3.1. Type II Photosensitization by Brominated Carbon Nanodots: 1O2.
In order to assess if the brominated carbon nanodots would perform as a photosensitizer, we first examined the ability of these particles to generate singlet oxygen, which is a product of Type II photosensitization. This ROS, as mentioned previously, is generated when BrCND are in the excited triplet state and dissolved molecular oxygen is present in solution. The cumulative singlet oxygen generated by a particular agent may be monitored over an exposure time period using the fluorescence-on probe, Singlet Oxygen Sensor Green™. Prior to singlet oxygen detection, this fluorescein-based probe has a low fluorescence quantum yield due to quenching from intramolecular photoinduced electron transfer (PET). Upon reacting irreversibly with singlet oxygen, the probe forms a new endoperoxide (SOSG™-EP) structure that does not undergo PET quenching. As a result, the fluorescence quantum yield increases significantly, and the detection of singlet oxygen concentrations are confirmed. Because the reaction favors product formation, the final fluorescence measurement reflects the relative concentration of singlet oxygen generated by the system. Using this probe, singlet oxygen generation from BrCND was examined. First, atmospheric concentrations of dissolved oxygen were considered (“air purge”) as reported in Figure 1.
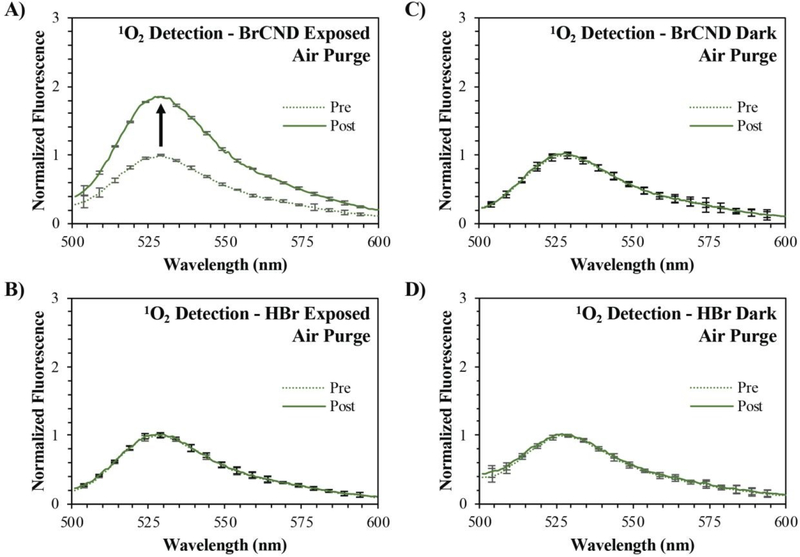
Figure 1.
Normalized fluorescence spectra of Singlet Oxygen Sensor Green (“SOSG,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, ~0.5 J•cm−2) under air-purged conditions. Fluorescence spectra are reported for SOSG UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. Reported spectra are the average of three analyzed solutions from one sample trial.
Comparing the “pre” and “post” exposure intensities for the control samples containing no BrCND (“HBr,” Fig. 1 b/d) it is apparent that there are no notable issues with probe photostability following 4-minutes of UV irradiation. No signal change is also reported for the non-irradiated BrCND sample (Fig. 1c), which is expected under the mechanism of photosensitization. When the sample is irradiated, conversely, the fluorescence intensity of the SOSG™ “post” exposure measurement has now increased relative to the initial intensity (Fig. 1a). These results indicate that the BrCND are in fact acting as a singlet oxygen photosensitizer under atmospheric conditions.
Singlet oxygen generation from the BrCND was further analyzed under different concentrations of dissolved oxygen, as shown in Figure 2.
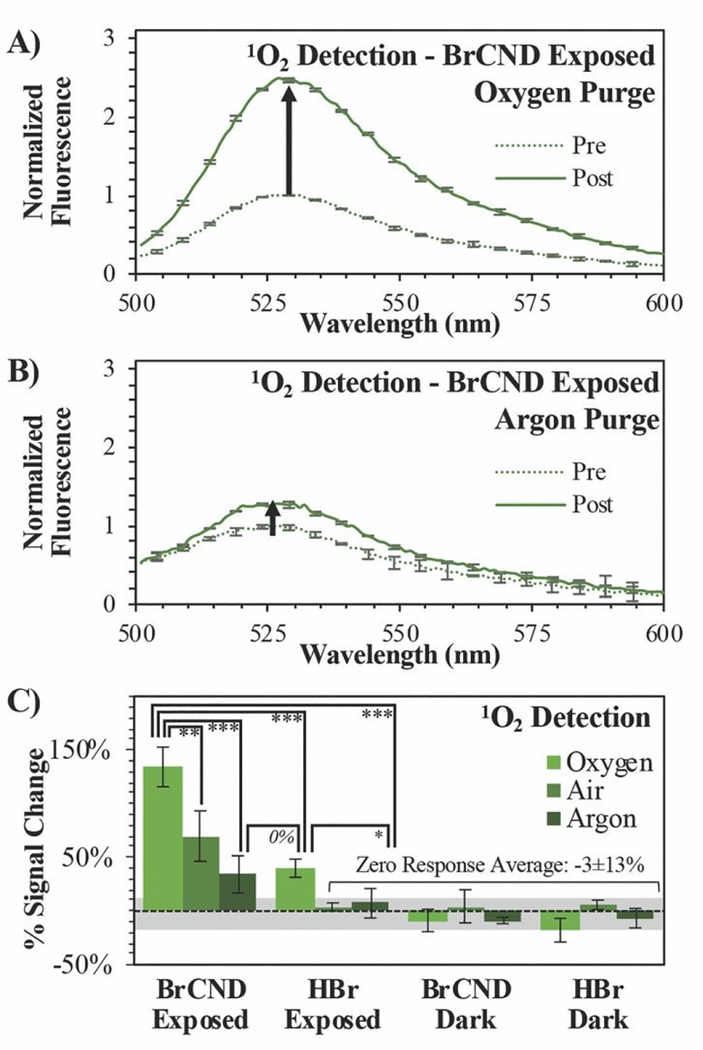
Figure 2.
Singlet Oxygen Sensor Green (“SOSG,” λexcitation = 473 nm) detection of singlet oxygen (1O2) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, ~0.5 J•cm−2). Normalized fluorescence spectra are reported for SOSG under A) oxygen and B) argon purging conditions. Reported spectra are the average of three analyzed solutions from one sample trial. C) Percent signal changes are reported for these spectra and for control samples (ESI figures S1–4) under all oxygen concentration conditions with statistical analysis reported. Values are the average of n ≥ 3 trials for each condition, with error from standard deviation reported. *p < 0.10, **p < 0.05, ***p < 0.01.
Rather than conducting the exposure period at atmospheric concentrations of oxygen, the samples were purged prior to exposure either with oxygen or argon gas to enrich or deplete the dissolved oxygen concentration in solution respectively (spectra shown in the ESI, Fig. S1/S2). It is difficult to estimate the exact concentrations of dissolved oxygen in these solutions, as salt concentration is known to have a deleterious effect on oxygen solubility in solution. As the atmosphere is only ~21% oxygen, we assume that the solution is not oxygen saturated (maximum solubility of O2 in pure water ~1.3 mM). In this case, as the partial pressure of each respective gas is increased via purging, so does the mole fraction of said gas increase in the solution. This is known as Raoult’s Law and is the basis for our purging experiments. As the partial pressure of oxygen increases in the sample during purging, the mole fraction similarly will increase for dissolved oxygen above that which is present under atmospheric conditions. Purging with argon, conversely, will decrease the partial pressure of oxygen and thereby reduce its concentration in solution to some degree. Examining the exposed BrCND samples, the fluorescence intensity from the reacted probe is indeed much higher for the oxygen-purged system than that reported for either the air- or argon-purged conditions (Fig. 1a, 2a/b); the percent signal change (increase) associated with singlet oxygen generation from BrCND is in fact proportional to the overall concentration of molecular oxygen in solution and is statistically higher than the reported controls (“zero response average”), as shown by Fig. 2b. Regarding the BrCND/argon system, a non-zero probe response is observed. This is attributable to trace oxygen concentrations in the oxygen-depleted system; molecular oxygen is therefore limiting—but not absent—from the overall reaction scheme, yielding low signal responses after singlet oxygen photosensitization from BrCND. Interestingly, a statistically comparable result is observed for the “HBr”/oxygen system, despite the absence of BrCND; however, it is key to note in this case that the signal detected for the HBr/oxygen exposed system represents an oxygen-enriched environment. Previous literature has demonstrated that SOSG™ is actually able to a small degree to behave as a singlet oxygen photosensitizer itself as well as a detection probe.32–34 It is likely therefore that in such an oxygen-rich system, singlet oxygen is instead being produced via UV photosensitization from SOSG™. While this is likely at play for all oxygen enriched measurements, the oxygen-purging condition for exposed BrCND yields a signal change that is statistically and significantly higher than that from HBr exposed sample (ESI Fig. S3; further statistical analysis of the various purging conditions and experimental versus control samples may be found in the ESI Fig. S4). These results confirm that BrCND are behaving as a photosensitizer for singlet oxygen, in a Type II photosensitization mechanism.
3.2. Type I Photosensitization by Brominated Carbon Nanodots.
Type II photosensitization is favorable largely due to the regeneration of the initial photosensitizer after the formation of singlet oxygen;4 however, it is likely that a photosensitizer will not proceed solely by this mechanism. Alternatively, the agent may participate in radical chemistry, or Type I photosensitization. By this route, an excited photosensitizer will undergo electron transfer steps to form the highly-reactive superoxide anion radical. Subsequently, downstream ROS such as peroxides and hydroxyl radical (•OH) can be generated.4 To characterize the potential of the BrCND to behave as Type I photosensitizers, we probed the generation of hydroxyl radical by the BrCND. Similar to the detection of singlet oxygen, a fluorescence-on probe was employed to detect the species. The probe used is hydroxyphenyl fluorescein (HPF); prior to reacting with •OH, the probe has a low quantum yield. Upon reacting, p-benzoquinone is released, and the substituent is replaced by a hydroxyl group thereby restoring the classic structure of fluorescein and its fluorescence intensity. It should be noted also that HPF is sensitive, albeit less so, to peroxynitrite in addition to hydroxyl radical; this species is only formed if nitric oxide is available to react with superoxide anion radical. While we did not initially expect nitric oxide to be generated by BrCND structures, this will be addressed in later sections. Herein, we assign the signal from HPF to •OH for simplicity; yet in either case, detection of •OH or peroxynitrite from photosensitization does in fact confirm Type I photosensitization.
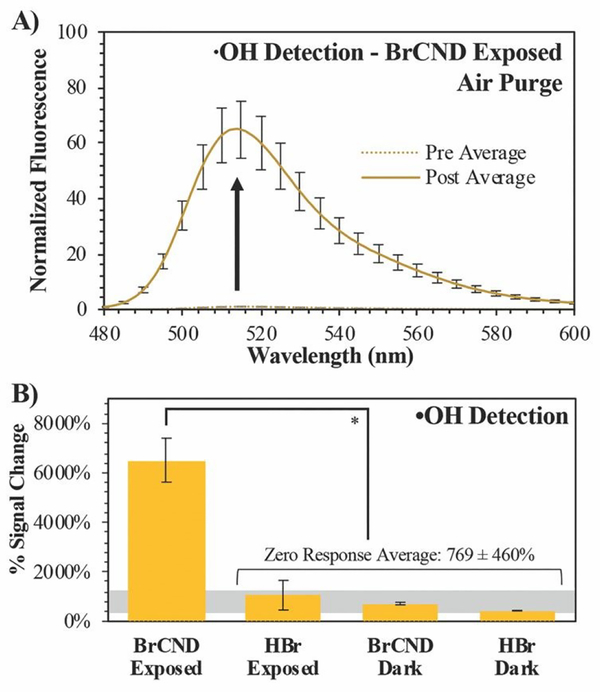
This was further investigated under atmospheric concentrations of molecular oxygen; the normalized spectra for the photosensitized system are reported in Figure 3a, with control spectra available in the ESI, Figure S5. Calculated percent signal changes (signal responses) for all conditions are presented in Figure 3b. Unlike the spectra for SOSG™, there is a substantial percent signal increase associated with each of the control samples for HPF. This could be the result of a few potential factors, including general stability of the probe, stability under UV exposure or pH cycling, or reactions with solvent species (see ESI Table S1), to name a few. Nonetheless, the photosensitization system containing BrCND is marked by a signal change over 6-fold greater than those detected for the control conditions, indicating that hydroxyl radicals are generated as a result of photosensitization from BrCND. These results have implications for other reactive oxygen species as well, since superoxide anion radical may instead be generated from single electron transfer as a predecessor to downstream to •OH or peroxynitrite in Type I photosensitization.35, 36
Figure 3.
Detection of hydroxyl radical (•OH) using hydroxyphenyl fluorescein (HPF). A) Normalized fluorescence spectra (λexcitation = 473 nm) of HPF before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, 1 J•cm−2) under air-purged conditions. Reported spectra are the average of three analyzed solutions from one sample trial. B) Percent signal changes are reported for these spectra and for control samples (ESI figure S5) with statistical analysis reported. Values are the average of n ≥ 3 trials for each condition, with error from standard deviation. *p < 0.001.
In cellular environments, all ROS produced downstream as a consequence of photosensitization may inflict cellular damage, permitting a Type I photosensitizer to be potentially very powerful in the antimicrobial photodynamic inactivation of bacteria.
3.3. Antimicrobial Photodynamic Inactivation of Bacteria by Brominated Carbon Nanodots: Time and Concentration Dependence of Antimicrobial Activity.
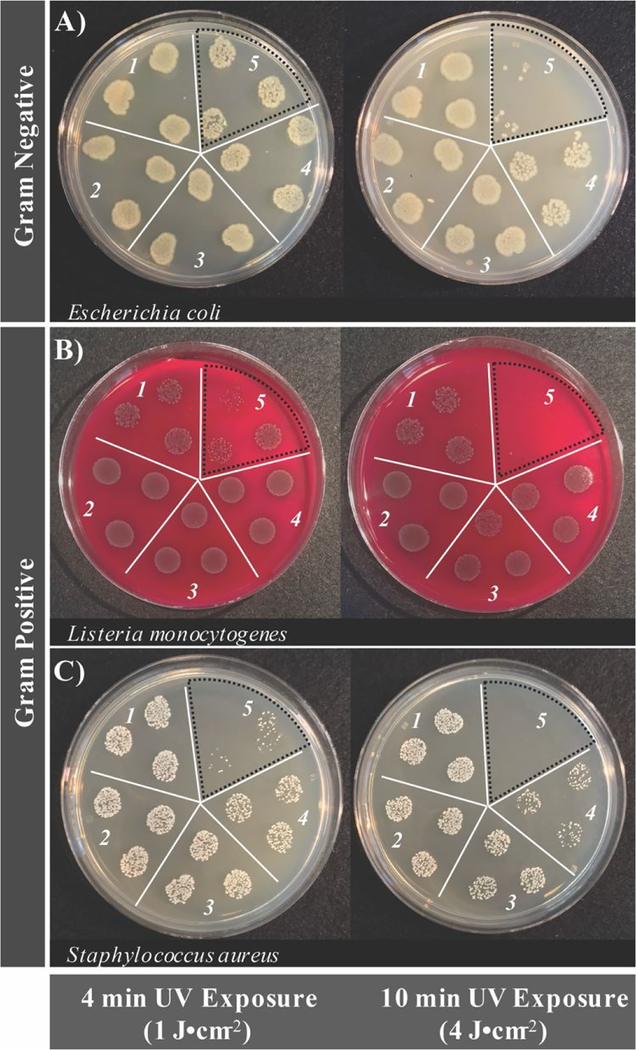
Although a photosensitizer may generate reactive oxygen species, this factor alone is insufficient to state definitively if an agent will be a potent photosensitizer for APDI. This is largely due to the adaptive features of pathogenic organisms. ROS are an endogenous feature of biological systems and will only induce cell death if present in sufficient concentrations, which involves overwhelming the biological pathways that are in place to mitigate oxidative stress.37 Accordingly, the antimicrobial activity of BrCND was investigated, as shown in Figure 4 (plotted counts reported in the ESI Figure S6). Additional control experiments to establish the experimental parameters (UV power distribution, pH tolerance, bromide salt concentration tolerance) are included for each bacterium in the ESI Figures S7–S12. The results shown in Figure 4 depict the bacterial colony growth for samples of E. coli, L. monocytogenes, and S. aureus after photosensitization for both 4- and 10-minute exposure periods, followed by plating and overnight incubation. For all 4-minute control solutions (Fig. 4, left, selection 1–4) there is no clear difference in growth patterns; this stands in contrast to the photosensitized BrCND sample (Fig. 4, left, selection 5), which remarkably exhibits decreased colony growth for all three bacteria. This is consistent with the ROS generation studies from sections 3.1 and 3.2, where only the photosensitized samples yielded singlet oxygen or hydroxyl radical. Furthermore, with 10-minutes of UV exposure the growth of each bacterium is further decreased, achieving minimal to no colony formation visible for each (Fig. 4, right, selection 5). It should be noted that the initial concentrations of each bacterial solution and brominated carbon dot solution for these experiments were not equal, and instead were optimized to demonstrate the time-dependent anti-microbial effects of the BrCND photosensitizer. This is particularly important when considering the antibacterial capabilities of these structures. As shown in the ESI Figure S6, at least a 2 log decrease in viability is reported for E. coli and L. monocytogenes after 10-minutes of exposure (although indeed some of the n = 3 sample trials resulted in eradication, Fig. 4a/b), while a 5 log decrease is reported for S. aureus. The potential then remains for complete eradication of all bacteria using more concentrated brominated carbon dot samples and/or longer exposure times. In each case, samples with complete growth inhibition were achievable, bolstering the potential of these compounds as future commercial antimicrobial agents. For this to be feasible, the various mechanisms of toxicity from the BrCND must be understood.
Figure 4.
Real-color photographs of bacterial growth inhibition from photosensitization of brominated carbon nanodots as a function of different exposure energy densities (pH 3.2 ± 0.2, λexposure = 365 nm, 3 mW). Strains tested include A) Escherichia coli, B) Listeria monocytogenes, and C) Staphylococcus aureus. Labels correspond to the following conditions: 1) DI water only, 2) HBr control with no UV exposure, 3) HBr control with UV exposure, 4) brominated carbon nanodots with no UV exposure, and 5) brominated carbon nanodots with UV exposure. Note: concentrations of brominated carbon nanodots are variable between bacterial strains. Bromine dot photosensitized sections are indicated by a black dotted line.
The difference in growth patterns observed for the photosensitized BrCND samples versus the controls were expected. Unexpectedly, growth patterns for the 10-minute dark BrCND samples (Fig. 4, right, selection 4) also did not compare to the other control conditions, particularly for E. coli and S. aureus, unlike what was observed at 4-minutes of exposure. Although growth inactivation was not as pronounced as is reported for the photosensitized sample (Fig. 4, selection 5), there is still a notable effect.
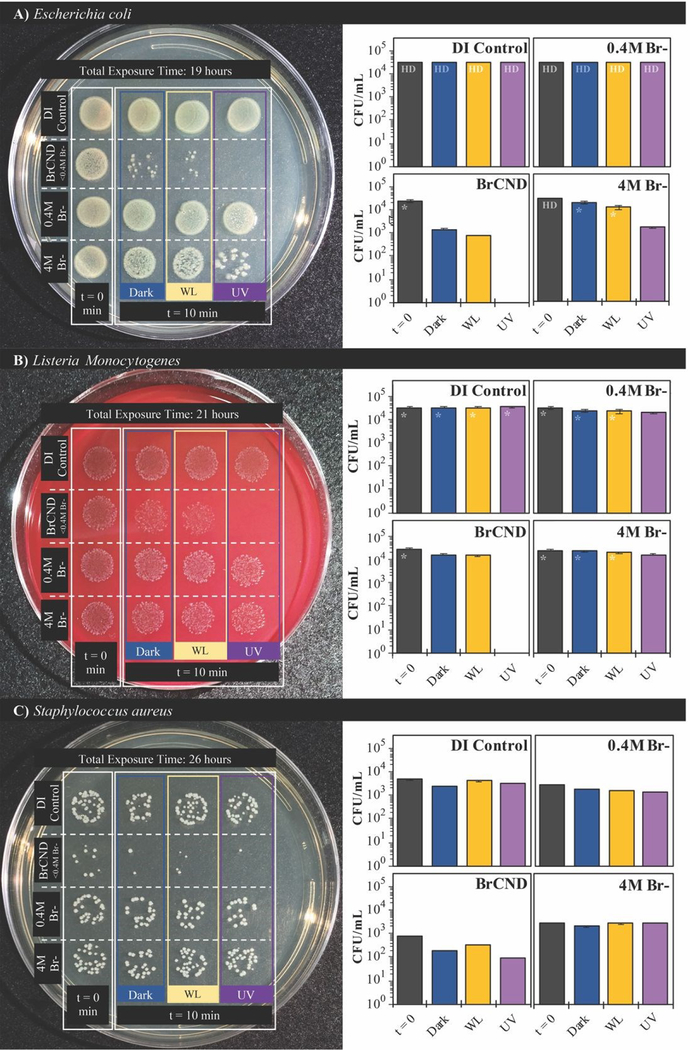
We subsequently considered the possibility that ambient room light may be causing low levels of photosensitization, as brominated carbon nanodots and indeed carbon nanodots in general are known to have broad absorption spectra.6, 24 Accordingly, the photosensitization effects from white light exposure were investigated compared to dark and UV-exposed antimicrobial effects and are reported in Figure 5. As a control, an additional sample, “t=0,” is included, where the bacterial solutions were mixed into the experimental solvents and were then immediately aliquoted into the preparation sample (PBS), restoring the bacteria to near-neutral pH and lowering the concentration of BrCND by an order of magnitude prior to incubation. For all other samples, this aliquoting process was completed at the end of the exposure period, “t=10,” prior to plating and incubation, as is true for all bacterial experiments reported herein. It becomes clear from the controls that neither exposure source nor varied bromide concentration has a deleterious effect on bacterial colony growth for either of the Gram-positive bacteria studied, L. monocytogenes (Fig. 5b) or S. aureus (Fig. 5c). E. coli does see some impact on growth resulting from the combination of light exposure and high salt concentrations (4 M); it should be noted, however, that the maximum concentration of free bromide contained within the BrCND experimental samples is <0.4M following sample collection and solution preparation, assuming zero incorporation of bromide into the nanodot structure itself during collection. This assumption is indeed conservative, given that previous studies have indicated the importance of bromide incorporation with the nanodot structure in order to achieve the heavy atom effect and subsequently triplet character.24 The actual concentration of free bromide is instead estimated to be much lower (<<0.4M), but nonetheless the 0.4M bromide solution shown in Figure 5 is suitable to control for this aspect. For all bacteria including E. coll, diminished growth is not observed for the 0.4 M concentration. Accordingly, the growth inhibition displayed by the BrCND for all bacteria is indeed attributable to the brominated carbon nanodots themselves. Upon closer examination of the data, each bacterium sees some diminished growth for the dark conditions, which is only further exacerbated with UV exposure, as observed previously. The use of white light even at much higher exposure energy densities than the UV source, alternatively, does not yield effects substantially different than those observed under dark conditions, demonstrating the superiority of UV wavelengths for BrCND photosensitization and subsequent APDI. These results further do not support the previously stated notion that ambient room light may be producing apparent “dark toxicity;” therefore, an alternate explanation is needed, as will be discussed in more detail in section 3.5.
Figure 5.
Growth of A) Escherichia coli, B) Listeria monocytogenes, and C) Staphylococcus aureus with exposure to brominated carbon nanodots (“BrCND”) at pH 3.5 under UV (λexposure = 365 nm, 40 J•cm−2) and white light (λmax = 572 nm, 300 J•cm−2) exposure conditions versus dark exposure to BrCND. Real-color photographs of labeled plates after overnight bacterial incubation (left) and corresponding colony counts for each sample condition (right). Error is from counts by 3x individuals to reduce bias in counting. Colony growth too dense for adequate counting is indicated as “HD”; high density estimates are indicated by “*”.
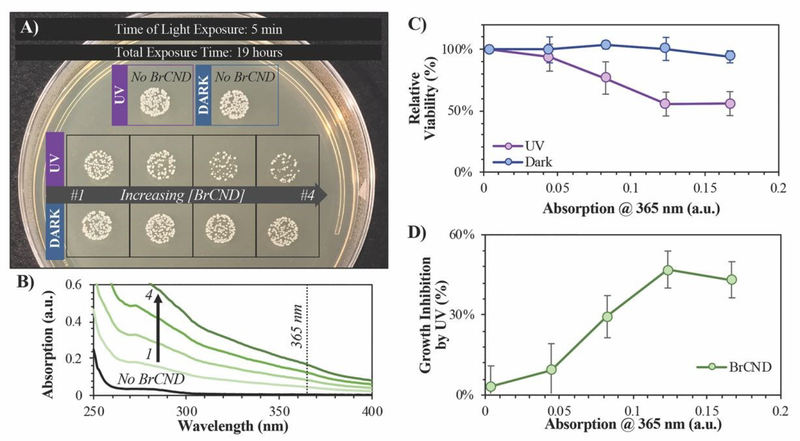
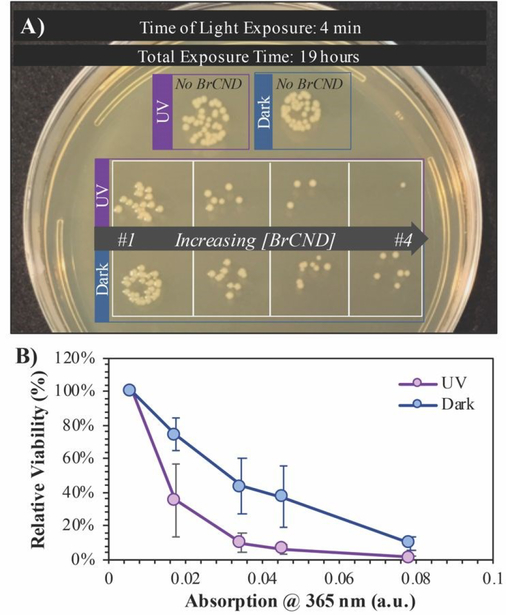
In order to better elucidate the relationship of BrCND to photosensitization, and to also observe dark toxicity effects, we subsequently investigated the antimicrobial impact of varying BrCND concentration—and therefore sample absorption at 365 nm—within the system for both dark and UV-exposed conditions, as shown for S. aureus in Figure 6 (all plated trials and corresponding counts are given in the ESI Figure S13). From the plated samples, colony counts were determined. From these values, viability of the bacterial sample was calculated relative to the “No BrCND” control; ultimately, we were also able to calculate and subsequently report the percentage of growth inhibition attributable to the BrCND photosensitization mechanism. Using these parameters, there is no significant toxicity observed from BrCND under dark conditions for S. aureus. With UV exposure, however, the antimicrobial efficacy from BrCND as a photosensitizer is evident and is established to be a concentration-dependent effect, consistent with the mechanism of APDI. It is important to note here that the formation of reactive bromine species may also be playing a role in the photodynamic antibacterial effect, as the antibacterial response has been shown to improve for other photosensitizing agents upon incorporation of sodium bromide.38 In our case, free bromide is indeed present in the solution (see ESI Table S1), although it should be noted that for all samples the concentration of free bromide is ~0.4 M, thereby reducing variability from this potential mechanism between samples.
Figure 6.
Viability of Staphylococcus aureus after 5-minute exposure to brominated carbon nanodot (“BrCND”) solutions of varying concentrations. Bacterial samples were both kept in dark (no light) and photosensitization (λexposure = 365 nm or “UV”, 3.0 ± 0.1 mW, 2 J•cm−2) conditions at a pH of 3.0. A) Real-color photograph of S. aureus growth after samples were adjusted to neutral pH and incubated overnight. Photo is representative of n = 3 trials. B) Absorption spectra of each BrCND solution (1–4). Black line is the absorption of the control solution. C) Relative viability of dark versus UV-exposed samples. D) Growth inhibition due to UV photosensitization for each solution absorption at the photosensitization wavelength. Error is from the standard deviation of n = 3 trials.
Upon photosensitization and the generation of Type I ROS from BrCND, the following could result:
| (Reaction 1) |
This radical cation in turn could react with free bromide similar to what was suggested by Wu et al for a titanium dioxide photocatalyst, resulting in the generation of hypobromite following bromide oxidation.38 This potential mechanism is undoubtedly possible for the brominated carbon nanodot photosensitizers as described here, and will likely be the focus of future work.
3.4. Antimicrobial Photodynamic Inactivation of Bacteria by Brominated Carbon Nanodots: Photosensitization Efficiency and Physical Properties.
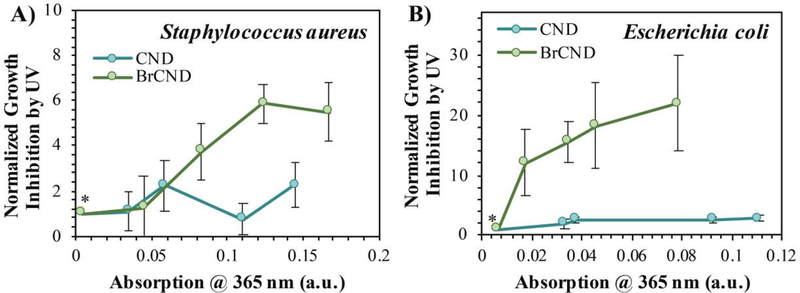
As mentioned previously, the incorporation of bromide into the nanodot structure is a crucial component to achieve strong triplet character, which can also lead to strong photosensitization of ROS. For our methods, phosphorescence from carbon nanodots was only observed upon incorporation of heavy atoms—including bromide—and was not observed for carbon dots collected into water.24 It then follows that the BrCND should exhibit superior antimicrobial properties from UV photosensitization as compared to carbon nanodots (CND) alone. Accordingly, we repeated the experimental design from Figure 6, substituting CND samples for the BrCND, but otherwise keeping all other components consistent. Growth inhibition from UV, or essentially the APDI efficacy, is plotted in Figure 7 for BrCND versus CND structures (plate photos and counts are given in the ESI Figures S13–S16).
Figure 7.
Normalized growth inhibition of bacteria by UV (λexposure = 365 nm, pH = 3.0) when exposed with brominated carbon nanodot (“BrCND”) or carbon nanodot (“CND”) solutions of varying absorption intensities (due to sample concentration differences). Bacterial strains include A) Staphylococcus aureus (2 J•cm−2) and B) Escherichia coli (1 J•cm−2). Growth inhibition values were normalized against the “0” nanodot condition (*) = 1. Error is propagated from the standard deviation of n ≥ 3 trials.
These are reported for both Gram-positive S. aureus (Fig. 7a) and Gram-negative E.coli (Fig. 7b). In the case of either bacterium, CND do not exhibit strong photosensitization effects, especially in comparison to the BrCND. As BrCND concentration, and therefore solution absorption, increases there is instead a marked increase in growth inhibition from UV exposure that is not observed for the CND samples. It follows then that the antimicrobial photodynamic inactivation of bacteria from carbon nanodot structures is enhanced by the incorporation of heavy atoms such as bromide, akin to the effects observed for phosphorescence from these species in comparison.24
It is interesting to note with the BrCND that such strong photodynamic antimicrobial character is observed, as both analysis of zeta potential and gel electrophoresis confirm that the particles are predominantly negatively charged, although some positive species are present in solution (ESI, Appendix B). In the development of APDI photosensitizers, it is often desirable to employ an agent with sufficiently cationic characteristics, due to the negative surface charge of both Gram-negative and -positive bacteria.5, 39 Taking advantage of the attractive electrostatic interactions between photosensitizer and bacterium, the agent can localize at the membrane surface, thereby reducing the distance that any ROS must diffuse before inflicting oxidative damage on the bacterium rather than the photosensitizer itself. Using an anionic agent such as BrCND, conversely, it would be expected that the bacteria and particles would repel one another, reducing the antimicrobial efficacy of the BrCND as a photosensitizer. Interestingly, this need not be the case. There are instances in the literature, perhaps counterintuitively, that demonstrate aggregation of negatively charged bacteria with negatively charged particles.40 The combined system, for example between E. coli and graphene oxide, has been shown to exhibit a reduced overall zeta potential; as this value approaches zero, the particulate matter in solution becomes unstable and prone to aggregation.40 This is a possibility also for the BrCND system, as zeta potential measurements are distributed near to zero (ESI, Appendix B). In a system where negatively charged particles may nonetheless localize with bacteria, the challenge of ROS lifetime and diffusion is addressed, and photosensitization can be effective, as demonstrated in Figure 7 for both S. aureus and E. coli.
3.5. Dark Toxicity of Carbon Nanodots and Reactive Nitrogen Species Generation.
Although no dark toxicity effects are reported for S. aureus in Figure 6, previously discussed data herein does in fact demonstrate growth inhibition where no photosensitization mechanism is at play. This is illustrated clearly by the BrCND concentration-dependent growth of E. coli, reported in Figure 8 (additional data in ESI Fig. S17). The UV exposed samples exhibit a decrease in growth (Fig. 8a), and therefore relative viability (Fig. 8b), consistent with the APDI mechanism observed also for S. aureus (Fig. 6/7). Yet obvious differences are observed in the “dark” condition, where no photosensitization occurs. Even under these conditions, there is a significant impact of BrCND concentration on viability, which is only further enhanced by the added photosensitization mechanism (Fig. 8b). This observation is consistent with reports mentioned earlier, which incorporate the “dark” toxicity from outer membrane disruption to improve the antibacterial efficacy of small-molecule photosensitizers in Gram-negative bacteria.29, 41 Given the surface charges reported from zeta analysis (ESI, Appendix B) of BrCND, it is possible that some of these structures carry a polycationic charge capable of E. coli membrane disruption, similar to what has been reported for other carbon dot structures.17,18
Figure 8.
Viability of Escherichia coli after 4-minute exposure to brominated carbon nanodot (“BrCND”) solutions of varying concentrations. Bacterial samples were both kept in dark (no light) and photosensitization (λexposure = 365 nm or “UV”, 3.0 ± 0.2 mW, 1 J•cm−2) conditions at a pH of 3.0. A) Real-color photograph of E. coli growth after samples were adjusted to neutral pH and incubated overnight. Photo is representative of n = 3 trials. B) Relative viability of dark versus UV-exposed samples.
This is an intriguing possibility and will likely be the focus of a future report. In the context of oxidative stress from reactive species, we considered that additional reactive species may be present that were not a product of photosensitization. Reactive nitrogen species, for example, produce different antimicrobial responses in E. coli and S. aureus due to different susceptibilities to nitrosative damage;42, 43 adaptability to stress from nitric oxide (NO•), for example, is a studied feature of S. aureus in the literature,43 although broadly speaking nitric oxide itself is only weakly antibacterial. In fact, “dark” toxicity from a NO• precursor could be due to a number of downstream reactive species, generated by oxidation of NO• by dissolved oxygen, as detailed in the following reactions:44
| (Reaction 2) |
| (Reaction 3) |
| (Reaction 4) |
Of course, growth of bacterial colonies is highly dependent on a number of factors beyond a single reactive species or mechanism, so direct comparison between the two bacteria is challenging at best; however, these observations indeed triggered interest in potential NO• release from BrCND. Furthermore, the potential contribution from NO• was not discountable particularly in light of the HPF response. As mentioned previously, HPF is sensitive to •OH as well as peroxynitrite, which itself has antibacterial character. This species may also be generated by Type I photosensitization, for example via the following reaction pathway,45
| (Reaction 5) |
and thereby could feasibly play some role in the antibacterial activity observed under photosensitization conditions if NO• is generated. Peroxynitrite, however, is only generated when both superoxide anion radical and NO• are available to react; yet, NO• is not a common product of photosensitization. If indeed present, there necessitated an alternative generation mechanism. We noted that some key small-molecule NO• donor structures are pH dependent, with NO• release occurring only in more acidic environments.46 This bolstered our thinking that such a species may possibly be generated by the BrCND particles.
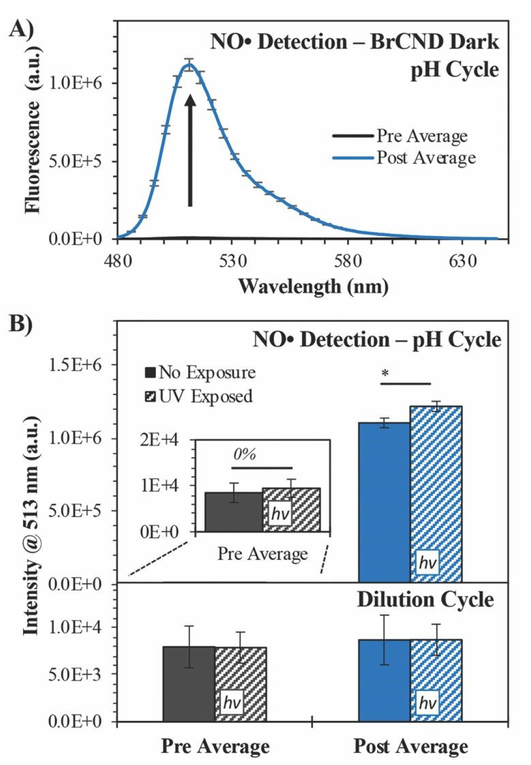
In order to test this, we employed the fluorescence-on probe Diaminofluorescein-FM (DAF-FM), the structure for which may be viewed in Scheme 1. The probe was incorporated into the BrCND solutions and underwent the pH cycling procedures described in section 2.3, with the added “dilution cycle” control where all aliquots following the initial pH adjustment to basic (pH = 12–12.5) were deionized water. As such, for this control, the sample remained at basic pH for the entirety of the experiment including the exposure period. The results of this are reported in Figure 9 (for additional spectra see the ESI Figure S18).
Figure 9.
Detection of nitric oxide (NO•) using Diaminofluorescein-FM (DAF-FM). A) Fluorescence spectra (λexcitation = 473 nm) of DAF-FM before (“pre average,” maximum intensity ≈ 104) and after (“post average”) exposure to brominated carbon nanodots (“BrCND,” pH ≈ 2.5) under dark conditions. Reported spectra are the average of three trials. B) Fluorescence intensity values both “pre” and “post” exposure conditions with BrCND (λexposure = 365 nm, 0.56 ± 0.04 mW, 0.1 J•cm−2). Values are reported for both top - pH cycled and bottom - dilution cycled (pH ≈ 12) conditions. Values are the average of n = 3 trials for each condition, with error from standard deviation. *p < 0.05.
For the dilution cycle, no significant change in signal was detected in the “post” exposure measurement as compared to the “pre” exposure conditions (Fig. 9b); this is the case for both the dark and UV-exposed systems, indicating that UV exposure alone has no notable impact on the probe fluorescence properties. When comparing the dilution versus pH cycle results, it becomes apparent that NO• generation here is pH-dependent. When the sample is cycled under acidic conditions, the fluorescence of DAF-FM increases substantially, indicating the generation of NO• and downstream species.47 This is true not just under UV-exposed conditions, which would be expected for a photosensitization mechanism, but notably under dark conditions as well. The signal is only slightly increased by photosensitization. This observation may be accounted for by considering that the presence of oxygen radicals opens other pathways for NO• reactions. Given that the mechanism for NO• sensing by DAF-FM requires the formation of an intermediate,47 it is possible that the presence of ROS reagents leads to a change in formation rate (and subsequently net concentration) of the intermediates over the exposure period. This would yield a different net response from the probe to NO• concentrations generated by the BrCND under UV-exposed, pH cycling conditions.
Although the mechanism of this generation from BrCND indeed requires more extensive analysis to elucidate, we suggest a potential pathway by which the BrCND may produce NO• (Scheme 2).
Scheme 2.
Graphical representation of one possible mechanism for acid-mediated nitric oxide (NO•) donation characteristics from a diazeniumdiolate form of brominated carbon nanodots (BrCND). For this schematic, NO• is generated by BrCND after acid cycling. The products generated restore the original structure. In the absence of competing pathways, NO• may react with the BrCND to restore the diazeniumdiolate.
Diazeniumdiolate structures are well-studied NO• donors, which release NO• in a pH-dependent manner as is observed for BrCND. These structures can be generated from amines upon reaction with NO•,46 which is particularly important when considering the formation of the carbon nanodot structures as combustion byproducts. NO• is also a known product from combustion in atmospheric conditions,48 and therefore is likely present during the nanodot synthesis. It proves difficult to elucidate specific information regarding functional groups from the FTIR spectra of BrCND structures, which are largely unstructured due to great variation in sample composition; however, it indeed seems possible that such a reaction as presented in Scheme 2 may occur during synthesis. Post-collection under this scheme, acidic environments permit the rapid release of NO• concentrations, restoring the original amine structure which may re-generate the diazeniumdiolate if reacted with generated NO•.46 As an alternative donor mechanism for reactive nitrogen species, nitroalkane substituents with adjacent aliphatic carbons may pyrolyze under basic to acidic cycling conditions to release nitrous acid; a common example of this in organic chemistry is known as the Nef reaction performed with the commercially available Oxone® reagent.49 In sufficiently high concentrations, aqueous nitrous acid can react to produce N2O3,50 which has been proposed as the key intermediate in the reaction mechanism of DAF-FM47 and is itself an acute oxidizer and is therefore highly toxic.51, 52 Extensive investigation must be conducted to understand the exact mechanism of pH-dependent generation of reactive nitrogen species from brominated carbon nanodots, yet it is clear that these particles do exhibit the unique capacity for contributing significant antimicrobial properties both from photo-dependent reactive oxygen species and pH-dependent reactive nitrogen species generation mechanisms.
4.0. CONCLUSION
Herein we report the generation of reactive oxygen and nitrogen species from brominated carbon nanodots. The BrCND, first described recently by our lab,24 are effective ROS photosensitizers by both Type I and Type II photosensation mechanisms. Under UVA (365 nm) exposure, the BrCND generate singlet oxygen in both oxygen-rich (oxygen purged) and oxygen-depleted (argon purged) solutions, in a manner consistent with the Type II photosensitization mechanism. Relative singlet oxygen yields are reported for each system using the fluorescence-on probe Singlet Oxygen Sensor Green™, demonstrating the oxygen concentration dependence of the system for the formation of singlet oxygen. Type I photosensitization of oxygen-derived radicals, such as hydroxyl radical, is also confirmed from BrCND using the fluorescence-on probe hydroxyphenyl fluorescein (HPF). Furthermore, the efficacy of the BrCND as APDI photosensitizing agents was investigated using both Gram-negative and -positive microbes including Escherichia coli, Staphylococcus aureus, and Listeria monocytognes. For all bacteria, photosensitization of the BrCND resulted in suppressed colony growth, consistent with APDI. Photosensitization effects from non-bromine-containing carbon nanodots were also compared and displayed minimal to no UV-dependent toxicity; this result is consistent with previous reports, in which triplet character was observed only for the brominated carbon nanodots compared to nanodots alone.24 As such, improved ROS photosensitization is also a consequence of the heavy atom effect in this case. The overall antimicrobial effects of BrCND further can be adjusted by varying the bacterial concentration during exposure, the concentration of BrCND, and the duration of UV exposure. Interestingly, dark toxicity effects from the BrCND were observed in some cases, which could not be attributed to activation from ambient room light exposure. This prompted the investigation into an additional antimicrobial mechanism from BrCND: the pH-triggered release of reactive nitrogen species. Nitric oxide was released from BrCND as a result of pH cycling (basic ➔ acidic ➔ basic), both under dark and UV-exposed conditions. The fluorescence-on probe DAF-FM was used in the detection of this species. Two potential sources of this NO• donating character are discussed, including the possibility of forming diazeniumdiolate groups or nitroalkane substituents at the surface of BrCND structures during combustion-based collection; these groups each may undergo chemical alteration during a pH cycle and release reactive nitrogen species, and therefore are cited as potential sources of the DAF-FM signal response. The findings described herein set the foundation for future incorporation and application of BrCND as antimicrobial materials. Featuring the combination of an inexpensive and rapid collection procedure with pH- and light-driven antimicrobial properties, these structures present a scalable solution to combating the widespread global threat of infection from antibiotic resistant bacteria.
Supplementary Material
Electronic Supplementary Information (ESI).
APPENDIX A: Calculating High Density Estimates of Colony Formation in Antimicrobial Experiments.
APPENDIX B: Characterization of Brominated Carbon Nanodots
APPENDIX C: Supporting Figures
Scheme S1. Experimental schematic for the bacterial experiment design used to assess the antimicrobial efficacy of brominated carbon nanodots (BrCND) under varying exposure conditions.
Table S1. Experimental Solvents Used for the Antimicrobial Efficacy of Brominated Carbon Nanodot Bacterial Experiments and Controls.
Figure S1. Normalized fluorescence spectra of Singlet Oxygen Sensor Green (“SOSG,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, ~0.5 J•cm−2) under oxygen-purged conditions. Fluorescence spectra are reported for SOSG UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. Reported spectra are the average of three analyzed solutions from one sample trial.
Figure S2. Normalized fluorescence spectra of Singlet Oxygen Sensor Green (“SOSG,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, ~0.5 J•cm−2) under argon-purged conditions. Fluorescence spectra are reported for SOSG UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. B/C) Reported spectra are the average of three analyzed solutions from one sample trial. A/D) Spectra are the average of two solutions from one sample trial.
Figure S3. Statistical analysis of percent signal change values determined for singlet oxygen generation (1O2) by photosensitization from brominated carbon nanodots (pH = 3, λexposure = 365 nm, ~0.5 J•cm−2) compared to control conditions. Values are reported for A) oxygen, B) air, and C) argon purged systems. Values are the average of a minimum of three trials for each condition, with error from standard deviation reported. *1.00 > p > 0.10, **p = 0.10, ***p = 0.05, ****p ≤ 0.01.
Figure S4. Statistical analysis of percent signal change values determined for singlet oxygen generation (1O2) by photosensitization from A) UV-exposed brominated carbon nanodots (“BrCND,” pH = 3, λexposure = 365 nm, ~0.5 J•cm−2) compared to control conditions including B) unexposed (dark) BrCND, C) exposed hydrobromic acid (”HBr”) solution, and D) unexposed HBr. Values are the average of a minimum of three trials for each condition, with error from standard deviation reported. *1.00 < p ≤ 0.20, **p = 0.10, ***p = 0.05, ****p ≤ 0.02.
Figure S5. Normalized fluorescence spectra of Hydrophenyl Fluorescein (“HPF,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, 1 J•cm−2) under air-purged conditions. Fluorescence spectra are reported for HPF UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. Reported spectra are the average of a minimum of three independent trials.
Figure S6. Results of bacterial growth inhibition from photosensitization of brominated carbon nanodots (pH 3.2 ± 0.2, λexposure = 365 nm), presented in the main text figure 4. Strains tested include A) Escherichia coli, B) Listeria monocytogenes, and C) Staphylococcus aureus. Labels correspond accordingly: “DI” - deionized water only, pH 5.5; “BrCND(D)” - brominated carbon nanodots adjusted to pH 3.2, no UV exposure; “BrCND(UV)” - brominated carbon nanodots adjusted to pH 3.2 with exposure. Note: concentrations of BrCND are variable between bacterial strains, but not between energy densities. Colony growth too dense for adequate counting is indicated as “HD” and is estimated by the maximum colony count obtained in n = 3 trials; high density estimates are indicated by “*”. Error is from standard deviation of n = 3 trials.
Figure S7. Escherichia coli control experiments for inactivation by brominated carbon nanodot photosensitization. A) Effects of UV (λex = 365 nm) exposure on bacteria alone at 10- and 25-mW exposure powers (power measured at 400 nm) for 4- and 10-minute exposure intervals. B) Effects of exposure solution pH on bacteria alone for a pH range of 2 to 6. C) Effects of bromide ion concentration in exposure solution for bacteria alone for concentrations ranging from 0- to 5-M.
Figure S8. Escherichia coli growth dependence on variable concentrations of bromide ion at pH = 3.5. A) Photograph of E. coli growth on LB agar after 10 minute exposure under various conditions followed by incubation. B) Colony counts for each sample shown in (A). Error is from counts by 3x individuals to reduce bias in manual counting. Colony growth too dense for adequate counting is indicated as “HD” and is estimated by the maximum colony count obtained in n = 3 trials; high density estimates are indicated by “*”.
Figure S9. Listeria monocytogenes control experiments for inactivation by brominated carbon nanodot photosensitization. A) Effects of UV (λex = 365 nm) exposure on bacteria alone at 10- and 25-mW exposure powers (power measured at 400 nm) for 4- and 10-minute exposure intervals. B) Effects of exposure solution pH on bacteria alone for a pH range of 2 to 6. C) Effects of bromide ion concentration in exposure solution for bacteria alone for concentrations ranging from 0- to 5-M.
Figure S10. Listeria monocytogenes growth dependence on variable concentrations of bromide ion at pH = 3.5. A) Photograph of L. monocytogenes growth after 10 minute exposure under various conditions followed by incubation. B) Colony counts for each sample shown in (A). Error is from counts by 3x individuals to reduce bias in manual counting.
Figure S11. Staphylococcus aureus growth control experiments for inactivation by brominated carbon nanodot photosensitization. A) Effects of UV (λex = 365 nm) exposure on bacteria alone at 10- and 25-mW exposure powers (power measured at 400 nm) for 4- and 10-minute exposure intervals. B) Effects of exposure solution pH on bacteria alone for a pH range of 2 to 6. C) Effects of bromide ion concentration in exposure solution for bacteria alone for concentrations ranging from 0- to 5-M.
Figure S12. Staphylococcus aureus growth dependence on variable concentrations of bromide ion at pH = 3.5. A) Photograph of S. aureus growth on LB agar after 10 minute exposure under various conditions followed by incubation. B) Colony counts for each sample shown in (A). Error is from counts by 3x individuals to reduce bias in manual counting.
Figure S13. Real-color photographs and corresponding antibacterial activity of Staphylococcus aureus exposed to bromine dot (“BrCND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.1 mW, 2 J•cm−2) conditions. The data here is all the experimental data for the n = 3 trials (“T”) used in further analysis, presented in figures 6 and 7 of the main text. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - second log dilution, 104 CFU/mL.
Figure S14. Real-color photographs and corresponding antibacterial activity of Staphylococcus aureus exposed to carbon dot (“CND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.1 mW, 2 J•cm−2) conditions. The data here is all the experimental data for the n = 4 trials (“T”) used in further analysis, presented in figure 7 of the main text. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - second log dilution, 104 CFU/mL.
Figure S15. Real-color photographs and corresponding antibacterial activity of Escherichia coli exposed to bromine dot (“BrCND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.2 mW, 1 J•cm−2) conditions. The data here is all the experimental data for the n = 3 trials (“T”) used in further analysis, presented in figures 7 and 8 of the main text, and ESI figure S17. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - second log dilution, 104 CFU/mL.
Figure S16. Real-color photographs and corresponding antibacterial activity of Escherichia coli exposed to carbon dot (“CND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.2 mW, 1 J•cm−2) conditions. The data here is all the experimental data for the n = 3 trials (“T”) used in further analysis, presented in figure 7 of the main text. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - se8ond log dilution, 104 CFU/mL.
Figure S17. Viability of Escherichia coli after 4-minute exposure to brominated carbon nanodot (“BrCND”) solutions of varying concentrations. Bacterial samples were both kept in dark (no light) and photosensitization (λexposure = 365 nm or “UV”, 3.0 ± 0.2 mW, 1 J•cm−2) conditions at a pH of 3.0. A) Growth inhibition due to UV photosensitization for each solution absorption at the photosensitization wavelength. Error is from the standard deviation of n = 3 trials. B) Absorption spectra of each BrCND solution (1–4, Fig. 7–8). Black line is the absorption of the control solution. Inset. Magnified absorption window at the photosensitization wavelength.
Figure S18. Fluorescence spectra of Diaminofluorescein-FM (“DAF-FM,” λexcitation = 473 nm) before (“pre”) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, 0.56 ± 0.04 mW, 0.1 J•cm−2). Fluorescence spectra are reported for DAF-FM pH cycled under A) dark and B) UV-exposed conditions and dilution cycled (pH 12–12.5) for C) dark and D) UV-exposed conditions. Reported spectra are the average of n = 3 independent trials.
ACKNOWLEGEMENT
This work was supported by the National Science Foundation Graduate Research Fellowship Program (2018262827) and the HHS/NIH/National Institute of General Medical Sciences (NIGMS) through the Chemistry/Biology Interface Program at the University of Maryland Baltimore County (5T32GM066706). The authors also acknowledge the Institute of Fluorescence (IoF) as well as the Department of Chemistry and Biochemistry at the University of Maryland Baltimore County (UMBC) as sources of internal funding. The authors would additionally like to thank the Daniel lab at UMBC for providing instrumentation and assistance with dynamic light scattering and zeta potential measurements.
ABBREVIATIONS:
- APDI
Antimicrobial Photodynamic Inactivation (of bacteria)
- BrCND
Brominated Carbon Nanodots (brominated dots)
- CND
Carbon Nanodots
- ROS
Reactive Oxygen Species
- 1O2
Singlet Oxygen
- 3O2
Ground State (triplet) Oxygen
- •OH
Hydroxyl Radical
- NO•
Nitric Oxide
- SOSG™
Singlet Oxygen Sensor Green™
- HPF
Hydroxyphenyl Fluorescein
- DAF-FM
Diaminofluorescein-FM
Footnotes
Conflicts of Interest.
There are no conflicts of interest to declare.
REFERENCES
- 1.Lansdown ABG, Silver in Healthcare: Its Antimicrobial Efficacy and Safety in Use, Royal Society of Chemistry, Cambridge, 2010. [Google Scholar]
- 2.Bakthavatchalu S and Noel G, in Frontiers in Clinical Drug Research - Anti Infectives, ed. Rahman A.- u., Bentham Science Publishers Ltd, Sharjah, 2017, vol. 3, ch. 5, pp. 187–218. [Google Scholar]
- 3.Hamblin MR, Current Opinion in Microbiology, 2016, 33, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mróz P and Hamblin MR, Advances in Photodynamic Therapy: Basic, Translational, and Clinical, Artech House, Inc, Boston, 2008. [Google Scholar]
- 5.Ghorbani J, Rahban D, Aghamiri S, Teymouri A and Bahador A, Laser Therapy, 2018, 27, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Namdari P, Negahdari B and Eatemadi A, Biomedicine & Pharmacotherapy, 2017, 87, 209–222. [DOI] [PubMed] [Google Scholar]
- 7.Schmitz R, PhD Thesis, University of Maryland Baltimore County, 2016. [Google Scholar]
- 8.Zhang H, Liang J, Liu J, Chen S, Zhang H, Tian Z, Cai Y, Wang P, Ye Y and Liang C, RSC Advances, 2016, 6, 8456–8460. [Google Scholar]
- 9.Yuan F, Li S, Fan Z, Meng X, Fan L and Yang S, Nano Today, 2016, 11, 565–586. [Google Scholar]
- 10.Wang J and Qiu J, Journal of Materials Science, 2016, 51, 4728–4738. [Google Scholar]
- 11.Roy P, Chen P-C, Periasamy AP, Chen Y-N and Chang H-T, Materials Today, 2015, 18, 447–458. [Google Scholar]
- 12.Du FF, Shuang SM, Guo ZH, Gong XJ, Dong C, Xian M and Yang ZH, Talanta, 2020, 206, 120243 8p. [DOI] [PubMed] [Google Scholar]
- 13.Al-Jumaili A, Alancherry S, Bazaka K and Jacob MV, Materials (1996–1944), 2017, 10, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang JJ, Gao G, Zhang XD, Ma YH, Chen XK and Wu FG, Carbon, 2019, 146, 827–839. [Google Scholar]
- 15.Al Awak MM, Wang P, Wang S, Tang Y, Sun Y-P and Yang L, RSC Advances, 2017, 7, 30177–30184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Z, Zhao C.-x., Li Y.-y. and Yang Y.-l., Applied Microbiology and Biotechnology, 2019, 103, 4585–4593. [DOI] [PubMed] [Google Scholar]
- 17.Abu Rabe DI, Al Awak MM, Yang F, Okonjo PA, Dong X, Teisl LR, Wang P, Tang Y, Pan N, Sun Y-P and Yang L, International Journal of Nanomedicine, 2019, 14, 2655–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meziani MJ, Dong XL, Zhu L, Jones LP, LeCroy GE, Yang F, Wang SY, Wang P, Zhao YP, Yang LJ, Tripp RA and Sun YP, ACS Applied Materials & Interfaces, 2016, 8, 10761–10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JJ, Lu SY, Tang QL, Zhang K, Yu WX, Sun HC and Yang B, Nanoscale, 2017, 9, 7135–7142. [DOI] [PubMed] [Google Scholar]
- 20.Jijie R, Barras A, Bouckaert J, Dumitrascu N, Szunerits S and Boukherroub R, Colloids and Surfaces B-Biointerfaces, 2018, 170, 347–354. [DOI] [PubMed] [Google Scholar]
- 21.Thakur M, Pandey S, Mewada A, Patil V, Khade M, Goshi E and Sharon M, Journal of Drug Delivery, 2014, 2014, 282193 9p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandal S, Prasad SR, Mandal D and Das P, ACS Applied Materials & Interfaces, 2019, 11, 33273–33284. [DOI] [PubMed] [Google Scholar]
- 23.Sidhu JS, Mayank T. Pandiyan, Kaur N and Singh N, Chemistryselect, 2017, 2, 9277–9283. [Google Scholar]
- 24.Knoblauch R, Bui B, Raza A and Geddes CD, Physical Chemistry Chemical Physics, 2018, 20, 15518–15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knoblauch R, Ra E and Geddes CD, Physical Chemistry Chemical Physics: PCCP, 2019, 21, 1254–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turro NJ, Modern molecular photochemistry, Mill Valley, Calif. : University Science Books, c1991, 1991. [Google Scholar]
- 27.Lakowicz JR, Principles of fluorescence spectroscopy, Springer, New York, 3rd edn., 2006. [Google Scholar]
- 28.Zhang J, Lu X, Tang D, Wu S, Hou X, Liu J and Wu P, ACS Applied Materials and Interfaces, 2018, 10, 40808–40814. [DOI] [PubMed] [Google Scholar]
- 29.Sperandio FF, Huang Y-Y and Hamblin MR, Recent patents on anti-infective drug discovery, 2013, 8, 108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.“CDC Antibiotic Resistance Threats in the United States 2019” Report, https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf, (accessed March 2020).
- 31.Olaimat AN, Al-Holy MA, Shahbaz HM, Al-Nabulsi AA, Abu Ghoush MH, Osaili TM, Ayyash MM and Holley RA, Comprehensive Reviews in Food Science and Food Safety, 2018, 17, 1277–1292. [DOI] [PubMed] [Google Scholar]
- 32.Ragas X, Jimenez-Banzo A, Sanchez-Garcia D, Batllori X and Nonell S, Chemical Communications, 2009, 2920–2922. [DOI] [PubMed] [Google Scholar]
- 33.Marazzi M, Besancenot V, Gattuso H, Lassalle H-P, Grandemange S and Monari A, Journal of Physical Chemistry B, 2017, 121, 7586–7592. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Fujitsuka M and Majima T, Journal of Physical Chemistry B, 2013, 117, 13985–13992. [DOI] [PubMed] [Google Scholar]
- 35.Bowler RP, Barnes PJ and Crapo JD, COPD: Journal of Chronic Obstructive Pulmonary Disease, 2004, 1, 255–277. [DOI] [PubMed] [Google Scholar]
- 36.Gao Z, Yang DZ, Wan Y and Yang YL, Analytical and Bioanalytical Chemistry, 2020, 412, 871–880. [DOI] [PubMed] [Google Scholar]
- 37.Kashmiri ZN and Mankar SA, International Journal of Current Microbiology and Applied Sciences, 2014, 3, 34–40. [Google Scholar]
- 38.Wu X, Huang Y-Y, Kushida Y, Bhayana B and Hamblin MR, Free Radical Biology and Medicine, 2016, 95, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tim M, Journal of Photochemistry & Photobiology, B: Biology, 2015, 150, 2–10. [DOI] [PubMed] [Google Scholar]
- 40.Gusev A, Zakharova O, Vasyukova I, Muratov DS, Rybkin I, Bratashov D, Lapanje A, Il’inikh I, Kolesnikov E and Kuznetsov D, Materials Science & Engineering C, 2019, 99, 275–281. [DOI] [PubMed] [Google Scholar]
- 41.Hamblin MR, O Donnell DA, Murthy N, Rajagopalan K, Michaud N, Sherwood ME and Hasan T, Journal of Antimicrobial Chemotherapy, 2002, 49, 941–951. [DOI] [PubMed] [Google Scholar]
- 42.Meng Q, Yin J, Jin M and Gao H, Applied and Environmental Microbiology, 2018, 84, e00559–00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grosser MR, Weiss A, Shaw LN and Richardson AR, 2016, 198, 2043–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis JI, Jon MF, Jeannete MG, Norma ER and Russell EB, PNAS, 1993, 90, 8103–8107.7690141 [Google Scholar]
- 45.Radi R, PNAS, 2018, 115, 5839–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hrabie JA and Keefer LK, Chemical Reviews, 2002, 102, 1135–1154. [DOI] [PubMed] [Google Scholar]
- 47.Namin SM, Nofallah S, Joshi MS, Kavallieratos K and Tsoukias NM, Nitric Oxide, 2013, 28, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altshuller AP, Journal of the Air Pollution Control Association, 1956, 6, 97–100. [DOI] [PubMed] [Google Scholar]
- 49.Ceccherelli P, Curini M, Marcotullio MC, Epifano F and Rosati O, Synthetic Communications, 1998, 28, 3057–3064. [Google Scholar]
- 50.Rosadiuk KA and Bohle DS, European Journal of Inorganic Chemistry, 2017, 2017, 5461–5465. [Google Scholar]
- 51.Liaudet L, Soriano FG and Szabo C, Critical Care Medicine, 2000, 28, N37–N52. [DOI] [PubMed] [Google Scholar]
- 52.Gunaydin H and Houk KN, Chemical Research in Toxicology, 2009, 22, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electronic Supplementary Information (ESI).
APPENDIX A: Calculating High Density Estimates of Colony Formation in Antimicrobial Experiments.
APPENDIX B: Characterization of Brominated Carbon Nanodots
APPENDIX C: Supporting Figures
Scheme S1. Experimental schematic for the bacterial experiment design used to assess the antimicrobial efficacy of brominated carbon nanodots (BrCND) under varying exposure conditions.
Table S1. Experimental Solvents Used for the Antimicrobial Efficacy of Brominated Carbon Nanodot Bacterial Experiments and Controls.
Figure S1. Normalized fluorescence spectra of Singlet Oxygen Sensor Green (“SOSG,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, ~0.5 J•cm−2) under oxygen-purged conditions. Fluorescence spectra are reported for SOSG UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. Reported spectra are the average of three analyzed solutions from one sample trial.
Figure S2. Normalized fluorescence spectra of Singlet Oxygen Sensor Green (“SOSG,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, ~0.5 J•cm−2) under argon-purged conditions. Fluorescence spectra are reported for SOSG UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. B/C) Reported spectra are the average of three analyzed solutions from one sample trial. A/D) Spectra are the average of two solutions from one sample trial.
Figure S3. Statistical analysis of percent signal change values determined for singlet oxygen generation (1O2) by photosensitization from brominated carbon nanodots (pH = 3, λexposure = 365 nm, ~0.5 J•cm−2) compared to control conditions. Values are reported for A) oxygen, B) air, and C) argon purged systems. Values are the average of a minimum of three trials for each condition, with error from standard deviation reported. *1.00 > p > 0.10, **p = 0.10, ***p = 0.05, ****p ≤ 0.01.
Figure S4. Statistical analysis of percent signal change values determined for singlet oxygen generation (1O2) by photosensitization from A) UV-exposed brominated carbon nanodots (“BrCND,” pH = 3, λexposure = 365 nm, ~0.5 J•cm−2) compared to control conditions including B) unexposed (dark) BrCND, C) exposed hydrobromic acid (”HBr”) solution, and D) unexposed HBr. Values are the average of a minimum of three trials for each condition, with error from standard deviation reported. *1.00 < p ≤ 0.20, **p = 0.10, ***p = 0.05, ****p ≤ 0.02.
Figure S5. Normalized fluorescence spectra of Hydrophenyl Fluorescein (“HPF,” λexcitation = 473 nm) before (“pre,” maximum intensity = 1) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, 1 J•cm−2) under air-purged conditions. Fluorescence spectra are reported for HPF UV-exposed with A) BrCND and B) hydrobromic acid (“HBr,” pH = 3.0) control, and under dark conditions (no exposure) for C) BrCND and D) HBr control solution. Reported spectra are the average of a minimum of three independent trials.
Figure S6. Results of bacterial growth inhibition from photosensitization of brominated carbon nanodots (pH 3.2 ± 0.2, λexposure = 365 nm), presented in the main text figure 4. Strains tested include A) Escherichia coli, B) Listeria monocytogenes, and C) Staphylococcus aureus. Labels correspond accordingly: “DI” - deionized water only, pH 5.5; “BrCND(D)” - brominated carbon nanodots adjusted to pH 3.2, no UV exposure; “BrCND(UV)” - brominated carbon nanodots adjusted to pH 3.2 with exposure. Note: concentrations of BrCND are variable between bacterial strains, but not between energy densities. Colony growth too dense for adequate counting is indicated as “HD” and is estimated by the maximum colony count obtained in n = 3 trials; high density estimates are indicated by “*”. Error is from standard deviation of n = 3 trials.
Figure S7. Escherichia coli control experiments for inactivation by brominated carbon nanodot photosensitization. A) Effects of UV (λex = 365 nm) exposure on bacteria alone at 10- and 25-mW exposure powers (power measured at 400 nm) for 4- and 10-minute exposure intervals. B) Effects of exposure solution pH on bacteria alone for a pH range of 2 to 6. C) Effects of bromide ion concentration in exposure solution for bacteria alone for concentrations ranging from 0- to 5-M.
Figure S8. Escherichia coli growth dependence on variable concentrations of bromide ion at pH = 3.5. A) Photograph of E. coli growth on LB agar after 10 minute exposure under various conditions followed by incubation. B) Colony counts for each sample shown in (A). Error is from counts by 3x individuals to reduce bias in manual counting. Colony growth too dense for adequate counting is indicated as “HD” and is estimated by the maximum colony count obtained in n = 3 trials; high density estimates are indicated by “*”.
Figure S9. Listeria monocytogenes control experiments for inactivation by brominated carbon nanodot photosensitization. A) Effects of UV (λex = 365 nm) exposure on bacteria alone at 10- and 25-mW exposure powers (power measured at 400 nm) for 4- and 10-minute exposure intervals. B) Effects of exposure solution pH on bacteria alone for a pH range of 2 to 6. C) Effects of bromide ion concentration in exposure solution for bacteria alone for concentrations ranging from 0- to 5-M.
Figure S10. Listeria monocytogenes growth dependence on variable concentrations of bromide ion at pH = 3.5. A) Photograph of L. monocytogenes growth after 10 minute exposure under various conditions followed by incubation. B) Colony counts for each sample shown in (A). Error is from counts by 3x individuals to reduce bias in manual counting.
Figure S11. Staphylococcus aureus growth control experiments for inactivation by brominated carbon nanodot photosensitization. A) Effects of UV (λex = 365 nm) exposure on bacteria alone at 10- and 25-mW exposure powers (power measured at 400 nm) for 4- and 10-minute exposure intervals. B) Effects of exposure solution pH on bacteria alone for a pH range of 2 to 6. C) Effects of bromide ion concentration in exposure solution for bacteria alone for concentrations ranging from 0- to 5-M.
Figure S12. Staphylococcus aureus growth dependence on variable concentrations of bromide ion at pH = 3.5. A) Photograph of S. aureus growth on LB agar after 10 minute exposure under various conditions followed by incubation. B) Colony counts for each sample shown in (A). Error is from counts by 3x individuals to reduce bias in manual counting.
Figure S13. Real-color photographs and corresponding antibacterial activity of Staphylococcus aureus exposed to bromine dot (“BrCND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.1 mW, 2 J•cm−2) conditions. The data here is all the experimental data for the n = 3 trials (“T”) used in further analysis, presented in figures 6 and 7 of the main text. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - second log dilution, 104 CFU/mL.
Figure S14. Real-color photographs and corresponding antibacterial activity of Staphylococcus aureus exposed to carbon dot (“CND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.1 mW, 2 J•cm−2) conditions. The data here is all the experimental data for the n = 4 trials (“T”) used in further analysis, presented in figure 7 of the main text. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - second log dilution, 104 CFU/mL.
Figure S15. Real-color photographs and corresponding antibacterial activity of Escherichia coli exposed to bromine dot (“BrCND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.2 mW, 1 J•cm−2) conditions. The data here is all the experimental data for the n = 3 trials (“T”) used in further analysis, presented in figures 7 and 8 of the main text, and ESI figure S17. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - second log dilution, 104 CFU/mL.
Figure S16. Real-color photographs and corresponding antibacterial activity of Escherichia coli exposed to carbon dot (“CND”) solutions of varying concentrations (dilution ratios) under either no-light (“dark”) or UV-exposed (“UV,” λexposure = 365 nm, 3.0 ± 0.2 mW, 1 J•cm−2) conditions. The data here is all the experimental data for the n = 3 trials (“T”) used in further analysis, presented in figure 7 of the main text. Left - 106 CFU/mL sample; Middle - first log dilution, 105 CFU/mL; Right - se8ond log dilution, 104 CFU/mL.
Figure S17. Viability of Escherichia coli after 4-minute exposure to brominated carbon nanodot (“BrCND”) solutions of varying concentrations. Bacterial samples were both kept in dark (no light) and photosensitization (λexposure = 365 nm or “UV”, 3.0 ± 0.2 mW, 1 J•cm−2) conditions at a pH of 3.0. A) Growth inhibition due to UV photosensitization for each solution absorption at the photosensitization wavelength. Error is from the standard deviation of n = 3 trials. B) Absorption spectra of each BrCND solution (1–4, Fig. 7–8). Black line is the absorption of the control solution. Inset. Magnified absorption window at the photosensitization wavelength.
Figure S18. Fluorescence spectra of Diaminofluorescein-FM (“DAF-FM,” λexcitation = 473 nm) before (“pre”) and after (“post”) exposure with brominated carbon nanodots (“BrCND,” pH = 3.0, λexposure = 365 nm, 0.56 ± 0.04 mW, 0.1 J•cm−2). Fluorescence spectra are reported for DAF-FM pH cycled under A) dark and B) UV-exposed conditions and dilution cycled (pH 12–12.5) for C) dark and D) UV-exposed conditions. Reported spectra are the average of n = 3 independent trials.