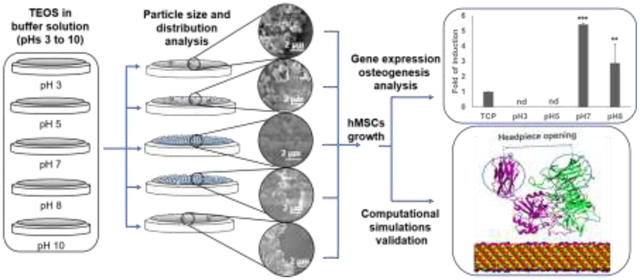
Abstract
Understanding the properties and behavior of biomineralized protein-based materials at the organic-inorganic interface is critical to optimize the performance of such materials for biomedical applications. To that end, this work investigates biomineralized protein-based films with applications for bone regeneration. These films were generated using a chimeric protein fusing the consensus repeat derived from the spider Nephila clavipes major ampullate dragline silk with the silica-promoting peptide R5 derived from the Cylindrotheca fusiformis silaffin gene. The effect of pH on the size of silica nanoparticles during their biomineralization on silk films was investigated, as well as the potential impact of nanoparticle size on the differentiation of human mesenchymal stem cells (hMSCs) into osteoblasts. To that end, induction of the integrin αV subunit and the osteogenic markers Runx2 transcription factor and Bone Sialoprotein (BSP) was followed. The results indicated that pH values of 7–8 during biomineralization maximized the coverage of the film surface by silica nanoparticles yielding nanoparticles ranging 200–500 nm and showing enhanced osteoinduction in gene expression analysis. Lower (3–5) or high (10) pH values led to lower biomineralization and poor coverage of the protein surfaces, showing reduced osteoinduction. Molecular dynamics simulations confirmed the activation of the integrin αVβ3 in contact with silica nanoparticles, correlating with the experimental data on the induction of osteogenic markers. This work sheds light on the optimal conditions for the development of fit-for-purpose biomaterial designs for bone regeneration, while the agreement between experimental and computational results shows the potential of computational methods to predict the expression of osteogenic markers for biomaterials.
Keywords: Biomaterial, Spider silk fusion, Biosilicification, Silica nanoparticle sizes, Multiscale modeling
Graphical Abstract
1. Introduction
Biomaterials for bone regeneration aim at promoting the regeneration of damaged bone. Their performance depends on various factors, such as porosity, surface properties or mechanical properties [1]. These parameters affect the speed of differentiation of stem cells into osteoblasts, which ultimately should lead to the growth of new bone tissue [2]. There are several types of biomaterials that can be grafted in damaged areas of the body to trigger bone regeneration. Among them, protein-based biopolymers have shown good biocompatibility, as well as high mechanical strength, while being extremely lightweight [3–5], which are sought-after properties for bone regeneration biomaterials.
Protein-based biopolymers are typically harvested from natural sources (e.g., silkworm cocoons, animal tissue) [6,7], but technological advances in bioprocess engineering, metabolic engineering and molecular and synthetic biology are making it increasingly possible to also produce them in fermentative processes using recombinant DNA technology [8]. Biosynthesized protein-based biopolymers present several advantages over chemically synthesized polymers [8–10]: (i) biosynthesis leads to significantly higher yields and molecular weights than solid-phase peptide synthesis; (ii) they present monodisperse, complex and well-defined chain sequences and interactions; (iii) cells can confer protein polymers with post-translational modifications difficult to realize via chemical synthesis; and (iv) the physicochemical and biological properties of biosynthesized proteins can be fine-tuned through molecular design and sequence modification, incorporating diverse functional domains to create de novo chimeric proteins.
One of such protein-based biopolymers is spider silk, which is actively researched for the synthesis of biomaterials in the form of nanoparticles, fibers, thin films or hydrogels, showing promising results for various biomedical applications (e.g., drug delivery, biosensing, tissue regeneration) [11–14]. Furthermore, silk biomaterials can be biomineralized with osteoinductive inorganic materials (e.g., silica, hydroxyapatite, bioactive glass, tricalcium phosphate) [1] to produce composite materials that promote the differentiation of stem cells into osteoblasts [15–20]. These biomineralization processes can be facilitated by the use of the R5 peptide, which promotes the precipitation of silica [21,22]. To that end, silk proteins provide structural support, controlling the mechanical properties of the biomaterial, whereas the R5 peptide biomineralizes the solvent exposed surface of the biomaterial with silica to enhance osteoinductivity.
In previous studies, it was shown that fusing the silk-like consensus repeat derived from the spider Nephila clavipes’ major ampullate dragline silk with the silica-promoting peptide R5 peptide derived from the silaffin gene of the diatom Cylindrotheca fusiformis [23] did not inhibit the biomineralizing ability of the R5 peptide [24]. The cytocompatibility of these types of fusion proteins was also demonstrated, as well as their ability to trigger differentiation of human mesenchymal stem cells (hMSCs) into osteoblasts [19,20,22]. By combining experimental work with computational simulations, folded structures for silk-silica fusion proteins were predicted [25], identifying also intracellular pathways activated after the adhesion of hMSCs to silk-silica scaffolds due to the activation of the αVβ3 integrin [20]. This integrin is a cell adhesion receptor, and its extracellular domains interact with the extracellular matrix to trigger the intracellular mitogen-activated protein kinase (MAPK) pathways, which was shown to be involved in important cellular processes, such as embryogenesis, tissue development, angiogenesis, or immune systems [26–33].
In the present work, the structure-property relationships of silk-silica composite biomaterials were studied. Silk-silica films produced using chimeric silk proteins with the R5 peptide were biosilicified at different pHs to generate coatings with silica nanoparticles of different sizes, following the Stöber process [24,34–36]. The aim was to elucidate the effect of the pH on silica nanoparticle size during the biosilicification process, and the subsequent impact of the resulting silk-silica composite biomaterials on the osteoinduction in hMSCs. The expression profile of several relevant genes identified in previous work [20] was analyzed, and compared with molecular dynamics (MD) simulations, to study the structure-function relationships for silk-silica biomaterials at the silica-organic interface [37]. This experimental-computational approach identified silica nanoparticles of 200 nm (formed at pH 7) as more effective in triggering differentiation of hMSC into osteoblasts. Beyond providing a detailed study of these biomaterials for bone regeneration, this work also demonstrated the ability of MD simulations to predict biological outcomes of a biomaterial, in this case expression of gene markers related to osteoinduction in hMSCs.
2. Materials and methods
2.1. Recombinant production of silk-silica fusion protein
The spider silk fusion protein used in this study was constructed using a 15-unit consensus repeat from the major ampullate spidroin 1 (MaSp1) protein of N. clavipes spider silk [(SGRGGLGGQGAGAAAAAGGAGQGGYGGLGSQGT)15, molecular weight of approximately 40 kDa]. Previous publications by the authors determined that the N-terminal region was the optimal location for the R5 silica-inducing domain to maximize the biomineralization of this composite biomaterial [18]. Thus, the R5 silica-promoting peptide (SSKKSGSYSGSKGSKRRIL) was appended at the N-terminal of the silk repeat. A histidine tag (ch) was located at the C-terminal, yielding a fusion protein with the sequence R5–15mer-ch. The construct was cloned into commercially available pET30a(+) vector (Novagen, San Diego, CA, USA), as described previously [38]. The recombinant construct was expressed in Escherichia coli strain BL21 Star (DE3) (Invitrogen, Grand Island, NY, USA) and purified as described previously [39]. Briefly, the recombinant strain was grown overnight at 37°C in Lysogeny Broth (LB) medium. Afterwards, a seeding culture was transferred to yeast extract medium and cultured at 37°C and pH 6.8 using a New Brunswick BioFlo 3000 fermentor (New Brunswick Scientific, Edison, NJ, USA). When the optical density OD600 reached approximately 0.8, expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Sigma-Aldrich, St. Louis, MO, USA). Purification was performed using immobilized metal affinity chromatography. The protein was dialyzed against water using a Slide-A-Lyzer cassettes (Pierce, Rockford, IL, USA) with a molecular weight cut-off of 10 kDa and the protein solution was then lyophilized. The purity of the expressed protein was verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Colloidal Blue staining.
2.2. Preparation of silk-chimera films
Lyophilized recombinant R5–15mer-ch silk-silica fusion protein, containing the R5 silica-promoting peptide at the N-terminal, followed by the 15 silk-unit repetition and a histidine tag at the C-terminal, was dissolved at a final concentration of 2% (w/v) in ultrapure water overnight at 4°C. For their characterization via Scanning Electronic Microscopy (SEM), circle cover glasses (Fisher Scientific, Waltham, MA) (R=12 mm) were used as substrate, on which 30 μL aliquots of the protein solution were deposited. For cell culture, 400 μL of dissolved protein was deposited onto each well in a 6-well plate. The resulting films were air-dried overnight in a fume hood. Then, the films were subjected to water vapor annealing using an isotemp vacuum oven for 24 h at room temperature [22,39] to induce the formation of β-sheets. Finally, the films were air-dried overnight in a fume hood at room temperature.
2.3. Biosilicification reaction on fusion protein films
To induce silica precipitation, R5–15mer-ch protein films were treated with pre-hydrolyzed tetraethyl orthosilicate (TEOS) in 100 mM bis-tris propane/citric acid buffer at pHs ranging from 3 to 10, according to protocols previously described [22,25,39]. For the SEM material characterization, a total of 1 mL of 30 mM TEOS at different pHs was added into each well containing a cover glass of a 12-well plate to cover the film formed in the cover glass for 1, 2 or 5 h at room temperature. For cell culture, 4 mL of 30 mM TEOS at different pHs were added into each well of a 6-well plate to cover the film formed in the well for 1h at room temperature. The films were then washed twice with distilled water and air-dried overnight inside a fume hood.
2.4. Scanning electron microscopy (SEM)
The silica deposition, nanoparticle size and particle size distribution of silica on the films was analyzed by SEM. Imaging was performed on a Zeiss EVO-10MA microscope (Zeiss, Oberkochen, Germany) at 5 kV accelerating voltage. Prior to imaging, samples were thoroughly air-dried in a fume hood for 24 h and coated with ~10 nm gold using a SC7620 sputter coater (Quorum Technologies, UK). Three images at each pH were analyzed using ImageJ software (NIH, Bethesda, USA) to obtain the particle size distribution.
2.5. Cell culture and film seeding
Films for cell culture were sterilized using ethylene oxide for 16 h at 4°C [40] and stored aseptically until seeding. hMSCs were obtained and cultured as described previously [41]. hMSCs were isolated from fresh bone marrow aspirates (Lonza, Basel, Switzerland), cultured in Dulbecco’s Modified Eagle Medium (supplemented with 10% fetal bovine serum, 0.1 mM non-essential amino acids, 1 ng/mL bFGF, 1% antibiotic/antimycotic) and seeded at passage 2 [41]. Cells were seeded at a density of 5,000 cells per cm2 in hMSC medium for 3 h, allowing the cells to adhere to the surface. Then, the medium was changed to osteogenic medium StemPro Osteogenesis Differentiation Kit (Gibco, Life Technologies, Grand Island, NY, USA). All cell cultures were performed in an incubator maintained at 37°C and 5% CO2. Cell growth and shape were monitored using a phase-contrast light microscope (Carl Zeiss, Jena, Germany).
2.6. RNA isolation and quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR)
Total RNA, DNA and proteins were extracted after 24 h cultures as previously described [20]. The SurePrep RNA/DNA/Protein Purification kit (Thermo Scientific, Carlsbad, CA, USA) was used to extract total RNA, DNA and proteins. After RNA isolation, RNA samples were DNase I-treated according to the manufacturer’s instructions (Ambion, Carlsbad, CA, USA). The concentration and purity of the RNA samples were assessed using a Nanodrop™ 2000 (Thermo Scientific, Carlsbad, CA, USA). The Transcriptor First Strand cDNA Synthesis kit (Roche, Indianapolis, IN, USA) was used to synthesize total cDNA in 20-μL reactions containing 100 ng of total RNA, 1 μM concentration of each dNTP, 10 units of reverse transcriptase, 20 units of Protector RNase Inhibitor, and 60 μM random hexamers; samples were then incubated at 25°C for 10 min and then at 55°C for 30 min following the manufacturer′s protocol. Reactions were terminated by incubation at 85°C for 5 min. The cDNA samples were analyzed for gene expression relative to the GAPDH housekeeping gene using LightCycler® 480 SYBR Green I Master (Roche, Indianapolis, IN, USA) in a Stratagene Mx3000 qPCR system (Agilent, Santa Clara, CA, USA). PCR amplifications were carried out with one denaturation cycle (95°C for 10 min), followed by 50 cycles of amplification (95°C for 10 s, 60°C for 10 s, and 72°C for 10 s). After amplification, melting curves were generated to confirm amplification of a single product. To amplify transcripts from GAPDH the primers F 5′-ACCCAGAAGACTGTGGATGG −3′ and R 5′-CAGTGAGCTTCCCGTTCAG-3′were used; to amplify integrin αV subunit primers F 5′ AATCTTCCAATTGAGGATATCAC 3′ and R 5′-AAAACAGCCAGTAGCAACAAT-3′ were used; to amplify Runx2 transcripts primers F 5′-ATGCTTCATTCGCCTCAC-3′and R 5′-ACTGCTTGCAGCCTTAAAT-3′ were used; to amplify collagen type I (Col) transcripts primers F 5′-GTGCGATGACGTGATCTGTGA-3′ and R 5′-CGGTGGTTTCTTGGTCGGT-3′ were used; to amplify BSP transcripts primers F 5′-ATGGCCTGTGCTTTCTCAATG-3′and R 5′-GGATAAAAGTAGGCATGCTTG-3′ were used. Expression of these genes was compared to control samples cultured on tissue culture plastic (TCP). For each sample, the Ct value was defined as the cycle number at which the amplification of each target gene was in the linear range of the reaction. Relative expression levels for each gene were calculated by normalizing to the Ct value of GAPDH housekeeping gene and to the normalized level of the control sample (2−ΔΔCt) [42]. The analysis was performed in three technical replicates from three biological samples.
2.7. Statistical analysis
Statistical analyses were performed by analysis of variance (ANOVA) to determine significant differences between means. The Student–Newman–Keuls (SNK) test was used to assess significant differences between pH values. All data was analyzed using SigmaPlot software. Statistical significance was evaluated at P<0.001 (***), P<0.01 (**) and P<0.05 (*).
2.8. Computational methods
Q2/Q3 ionized silica surfaces with 13.4×13.7×2.7 nm in size and 6.9 silanol groups per nm2 were used as models for Stöber-type silica nanoparticles with diameters of 200 nm or more [43]. Nanoparticles with 200 nm in diameter were simulated using Q2/Q3 surfaces with 18% ionization of their silanol groups, corresponding to ca. 1.2 siloxide groups per nm2. This surface model was available as part of the documentation of the Interface Force Field from the Heinz group [37]. Nanoparticles with 500 nm in diameter were simulated using Q2/Q3 surfaces with 30% ionization (corresponding to ca. 2.0 siloxide groups per nm2) [43], by modifying surfaces with 18% ionization using the software Materials Studio, rearranging the atomic charges accordingly. The procedure to modify surface charges and atomic composition on silica surfaces to attain a 30% ionization of silanol groups is shown in Fig. S1. This procedure involved the modification of the charges in the silicon atom and the oxygen atom that was deprotonated. Moreover, the hydrogen atom was replaced by a sodium atom.
The crystal structure of the extracellular subunit of the integrin αVβ3 was obtained from the RCSB protein data bank (PDB) with PDB ID 1L5G [44]. Only the β propeller domain from the α leg, and the hybrid and βA domains from the β leg were simulated. The βA domain contained divalent metal cations, and the metal ion dependent adhesion site (MIDAS) was coordinated with Mg2+. The other two cation binding sites were coordinated with Ca2+ [20]. The protein structure at physiological pH was placed 5 Å above the silica surface, and the system was fully solvated in TIP3P water molecules. To mimic physiological conditions and neutralize the system charge, a NaCl concentration of 0.15 M was applied. The system dimensions were only allowed to change in the vertical direction with respect to the silica surface, keeping the xy plane dimensions fixed. The ShakeH algorithm was applied to all the bonds in which hydrogen atoms were involved. The solvation box was made sufficiently large in the vertical direction with respect to the silica surface to avoid spurious effects of self-interactions in periodic images for the silica-integrin system.
Fully atomistic molecular dynamic simulations were performed using the software NAMD 2.12 [45], using the Chemistry at HARvard Macromolecular Mechanics (CHARMM) [46] force field for the solvated protein system and the Interface Force Field [37] for the silica surfaces. 3D periodic boundary conditions were applied. For each system, the enhanced sampling method termed temperature intervals with global exchange of replicas with hybrid potential energy approximation and solvent shell algorithm (TIGER2hs) was used [47]. This method comprised of several replicas undergoing repeating heating-sampling-quenching cycles in explicit solvent at different temperatures. This was followed by the evaluation of the potential energies of the replicas after quenching to the baseline temperature (300 K), in order to select structures for the ensemble of accepted states for the system. The energy evaluations were performed using a hybrid system composed of implicit solvent and an explicit solvation water layer around the protein. This represented a major difference with conventional replica exchange molecular dynamics (REMD), where water fluctuations can severely influence the determination of energies, thus impacting the exchange between replicas.
Each cycle consisted of 2000 timesteps (4 ps) of heating, 38,000 timesteps of sampling (76 ps), and 15000 timesteps of quenching (30 ps) to allow the adjustment of the potential energies to the baseline temperature (300 K). The equations of motions were integrated using a time step of 2 fs. 7 replicas were run in explicit solvent, covering a temperature range from 300 to 500 K. An additional replica was run beside the explicit solvent replica for exchange decisions after each quenching step, using a hybrid solvent that comprised of a combination of explicit TIP3P water molecules and Generalized Born (GB) implicit solvent. The calculations in hybrid solvent used two explicit solvation shells around the integrin, comprised of 1500 water molecules. A cutoff distance of 12 Å was applied for electrostatic and van der Waals interactions of the explicit solvent replicas, with a switch distance of 9 Å to avoid hard cuts. The electrostatic Coulombic interactions were calculated using particle mesh Ewald method with a grid spacing of 1 Å. Langevin thermostat and barostat were used to control temperature and pressure (1 bar) in the system. For the calculations with a hybrid solvent system, a cutoff of 18 Å was applied, with a switch distance of 16 Å. Each replica was run for 45 ns, including sampling and quenching. Thus, the total simulation time was 315 ns.
The output of the simulations was analyzed using in-house TCL and bash scripts. The Seaborn library from Python was used for data visualization. The atomic structures were visualized using visual molecular dynamics (VMD) graphics software [48]. The root mean square deviation (RMSD) for the ensemble of selected states during the simulation was analyzed for the integrin backbone using the RMSD Trajectory Tool from VMD. The headpiece opening of the αVβ3 integrin was calculated as the separation between the centers of mass of the β propeller domain from the α leg of the integrin (residues α1–438) and the hybrid domain in the β leg of the integrin (residues β55–108 and β353–432) [49,50] (Fig. S3). This headpiece opening distance was obtained from the ensemble of accepted states in the last 2.5 ns of simulation per replica. The electrostatic and van der Waals interaction energies between the integrin and the silica surface were obtained using the NAMD Energy plugin from VMD. The electrostatic potential map of the integrin was calculated with the Adaptive Poisson-Boltzmann Solver (APBS) [51] as implemented in the software Chimera. Atomic charges were assigned according to the CHARMM forcefield with PDB2PQR [52,53].
3. Results
3.1. pH effect on biosilicification, silica nanoparticle size and particle size distribution
Successful expression of the R5–15mer-ch fusion protein (Fig. 1a) is shown in Fig. 1b, and this chimeric protein was used to produce silk-silica film biomaterials. The effect of pH on the resulting silica nanoparticle was examined; the size was successfully tuned during the biosilicification process by varying the pH (Fig. 1c), agreeing with previous reports [22,24,36]. The SEM images showed that using a buffer at pH 3 during biosilicification resulted in a low surface coverage (around 10%) of aggregates and nanoparticles of silica (Fig. 2a), with nanoparticle sizes ranging widely (between 50 and 950 nm). The average nanoparticle size was 80 nm (Fig. 2b and c), which represented more than 65% of the nanoparticles present. At pH 5, the surface coverage increased up to approximately 50% (Fig. 2a), and the nanoparticle sizes ranged between 50 and 1200 nm. The mean centered at 50 nm (Fig. 2b and c) represented more than 55% of the nanoparticles deposited on the surface, while the mean at 950 nm represented 5% of the nanoparticles. Increasing the pH to 7 increased the surface coverage up to ca. 90% (Fig. 2a), with nanoparticle sizes ranging between 50 and 1,000 nm and a peak centered at 200 nm (Fig. 2b and c) that represented more than 63% of the nanoparticles on the film. At pH 8, the surface coverage decreased to approximately 70% (Fig. 2a). In this case, the nanoparticle sizes ranged between 250 and 1,200 nm with a peak at 500 nm and a second peak at 1,000 nm (Fig. 2b and c), representing more than 50 and 25% respectively of the nanoparticles identified on the surface. Finally, at pH 10 a major reduction in surface coverage was observed, leading to just ca. 5% coverage with aggregates of silica deposited on the films. The nanoparticle size ranged between <1 and 25 nm, with a peak at 25 nm (Fig. 2b and c) representing more than 50% of the nanoparticles on the film. The results at pH 10 were similar to those at pH 3, and thus from this point onwards only the data at pH 3 was considered. Extending the biosilicification at pH 5 for 2 and 5 h did not result in an increase in surface coverage (Fig. S3). Thus, only short biosilicification times (1 h) were further considered in this study.
Figure 1 -.
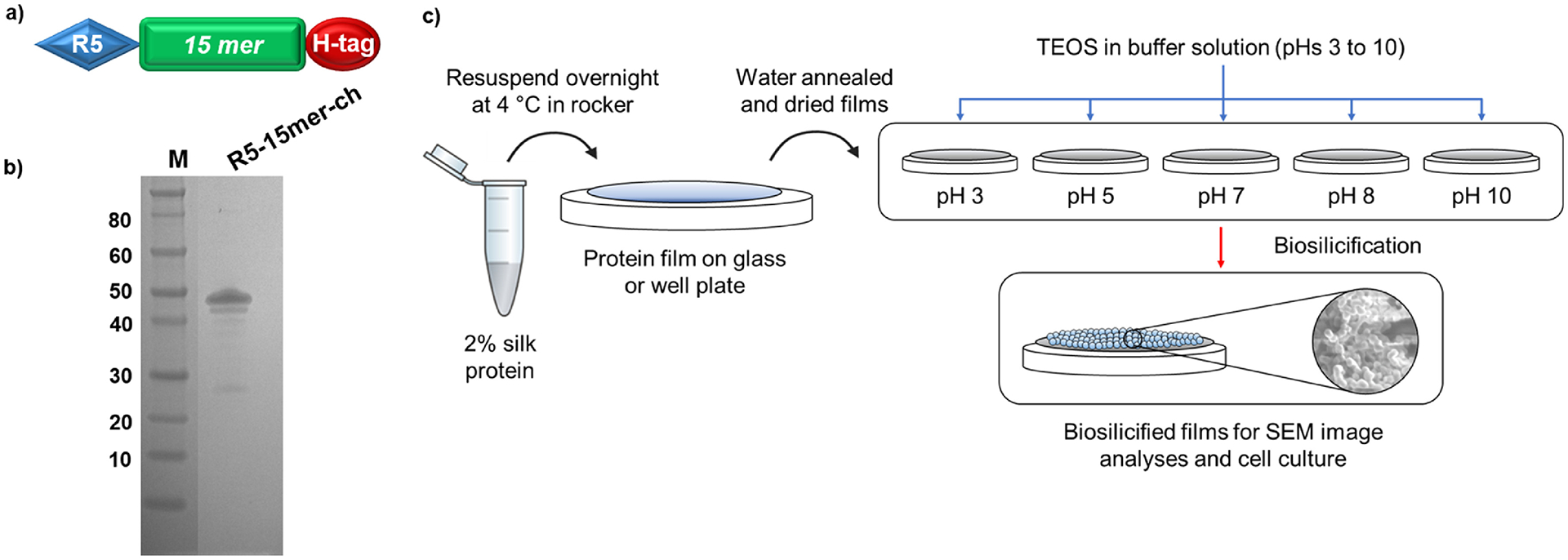
a) Schematic representation of the design of silk-silica chimeric proteins; R5 domain (blue block) was added to a spider silk 15mer (green block) at the N-terminal end, and a His-tag (H-tag, red block) was added C-terminal end. b) SDS-PAGE of purified silk-silica chimeric R5–15mer-ch (~43 kDa) proteins, run on the 4%−12% Bis-Tris acrylamide gel stained with Simple Blue dye. Marker (M) sizes are indicated on the left. c) Schematic of material preparation: the protein was dissolved in water, casted in cover glass or plastic well plate, water annealed and dried; then biosilicification was induced using silica precursor in buffer. Different pH values are tested to create films covered with silica nanoparticles of different sizes.
Figure 2 –
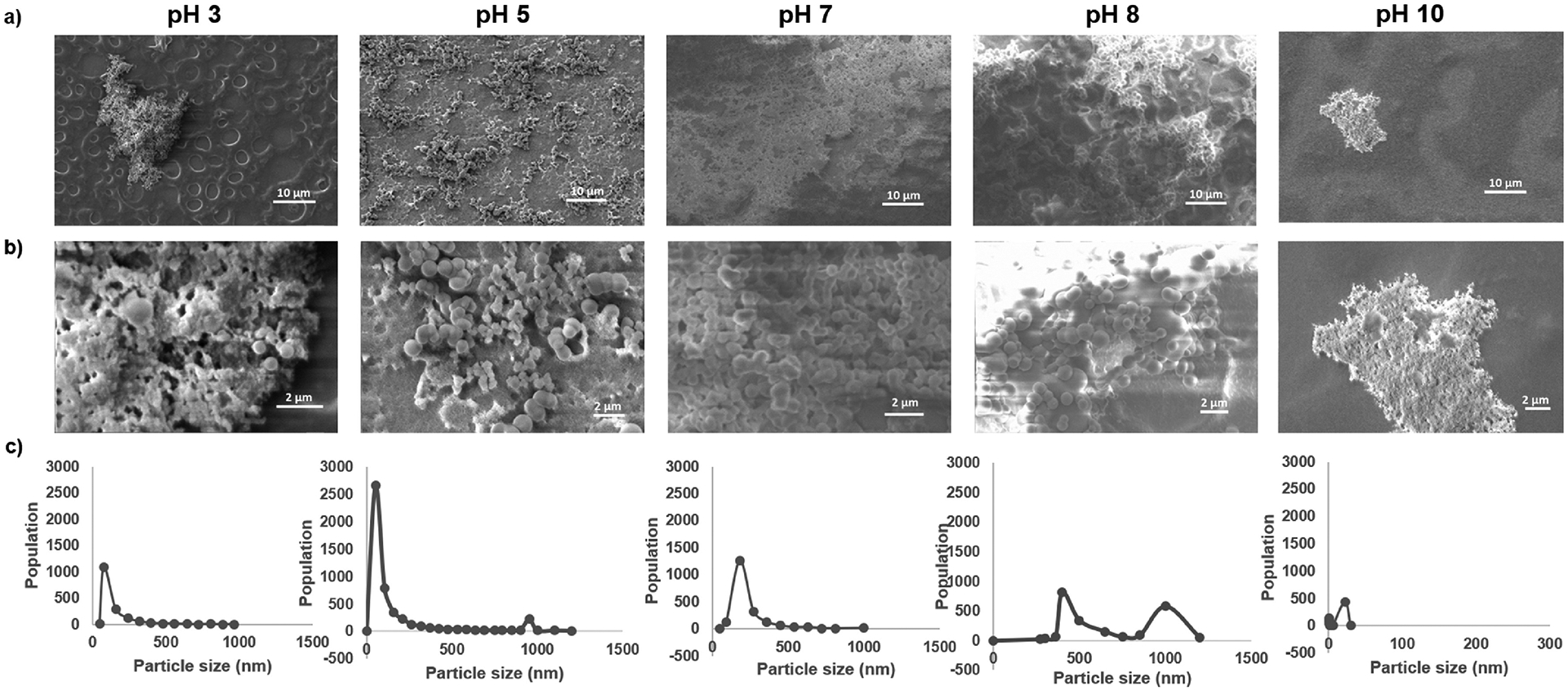
Analysis of silica nanoparticle deposition and distribution at different pH values. a) Representative SEM images showing the surface coverage of silica nanoparticles deposited after biosilicification at different pH, and b) representative magnifications used for nanoparticle size quantification. c) Quantification of silica particle size distribution after biosilicification at different pHs. The graph shows the quantification of three different images.
3.2. Silica nanoparticle size effect on osteogenic induction in vitro.
Gene expression of selected key markers, identified in previous work by the authors [20], was analyzed to study the effect of different silica nanoparticle sizes on osteoinduction. These components included the integrin αV subunit and the osteogenic markers Runx2 transcription factor and Bone Sialoprotein (BSP), as well as an earlier osteogenic marker, collagen type I (ColI). The level of gene expression of the markers 24 h and 7 days after seeding was monitored by quantitative PCR (qPCR) and compared to control cells seeded on tissue culture plastic (TCP). After 24 h, cells seeded on biosilicified films at pH 3 or 5 had no detectable (n.d.) levels of expression of markers or the integrin subunit, similarly to TCP (Fig. 3a–d). Conversely, the cells seeded on the biosilicified films at pH 7 or 8 showed increased induction of gene expression of the αV integrin subunit, Runx2 transcription factor and ColI marker (Fig. 3a–c). The expression of BSP also displayed increased induction in the samples biosilicified at pH 7, but not at pH 8 (Fig. 3d). Notably, the induction observed was always higher for the samples biosilicified at pH 7 than at pH 8 (Fig. 3a–d). After 7 days, as observed after 24 h, cells seeded on biosilicified films at pH 3 or 5 had no detectable (n.d.) levels of expression of markers or the integrin subunit (Fig. 3e–h). Cells seeded on the biosilicified films at pH 7 or 8 showed increased induction of gene expression for all the markers analyzed; this expression was always higher for samples biosilicified at pH 7 (Fig. 3e–h).
Figure 3 -.
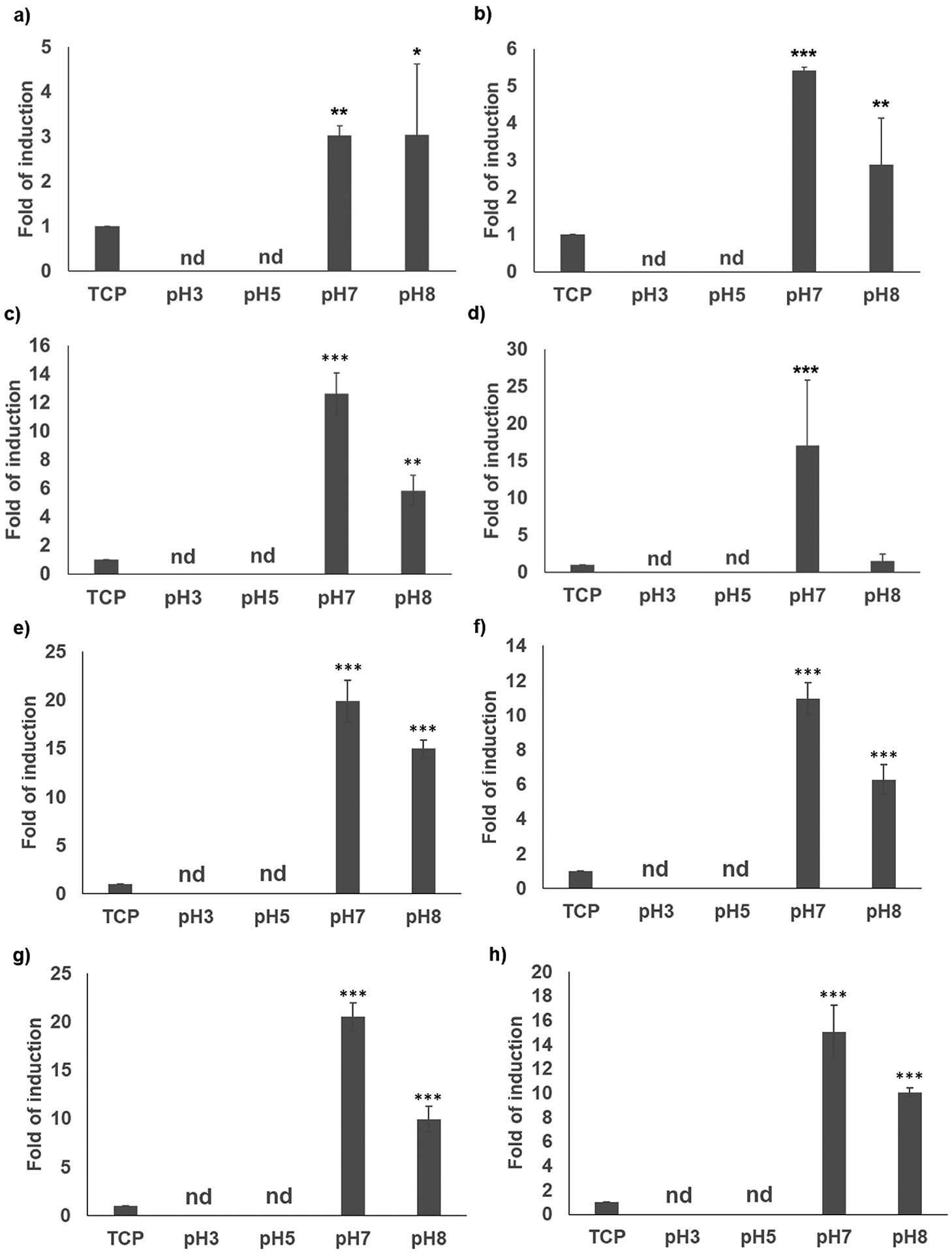
Gene expression of αV integrin subunit (a,e), Runx2 (b,f), collagen I (c,g), and BSP (d,h) markers of hMSCs seeded for 24 h (a-d) or 7 days (e-h) on tissue culture plastic (TPC), and R5–15mer-ch films silicified at different pHs. “nd” stands for not detected. Data represents mean ± SE (n=3) *: p<0.05; **: p <0.01; ***: p <0.001.
3.3. Computational simulations
The number of accepted states per cycle for the baseline ensemble was 1.5–1.6 for the different simulations herewith reported. The root mean square deviation (RMSD) of the backbone of the integrin was calculated for each accepted state produced during the simulation (Fig. 4a), showing a decreasing rate of change as the simulation progressed. The integrin displayed a larger RMSD when in contact with silica nanoparticles. The root mean-square fluctuations (RMSF) for each residue revealed that the hybrid domain exhibited the highest flexibility (Fig. 4b–c), which also agreed with the RMSD for each domain revealed a higher mobility for the hybrid domain (Fig. S4).
Figure 4 –
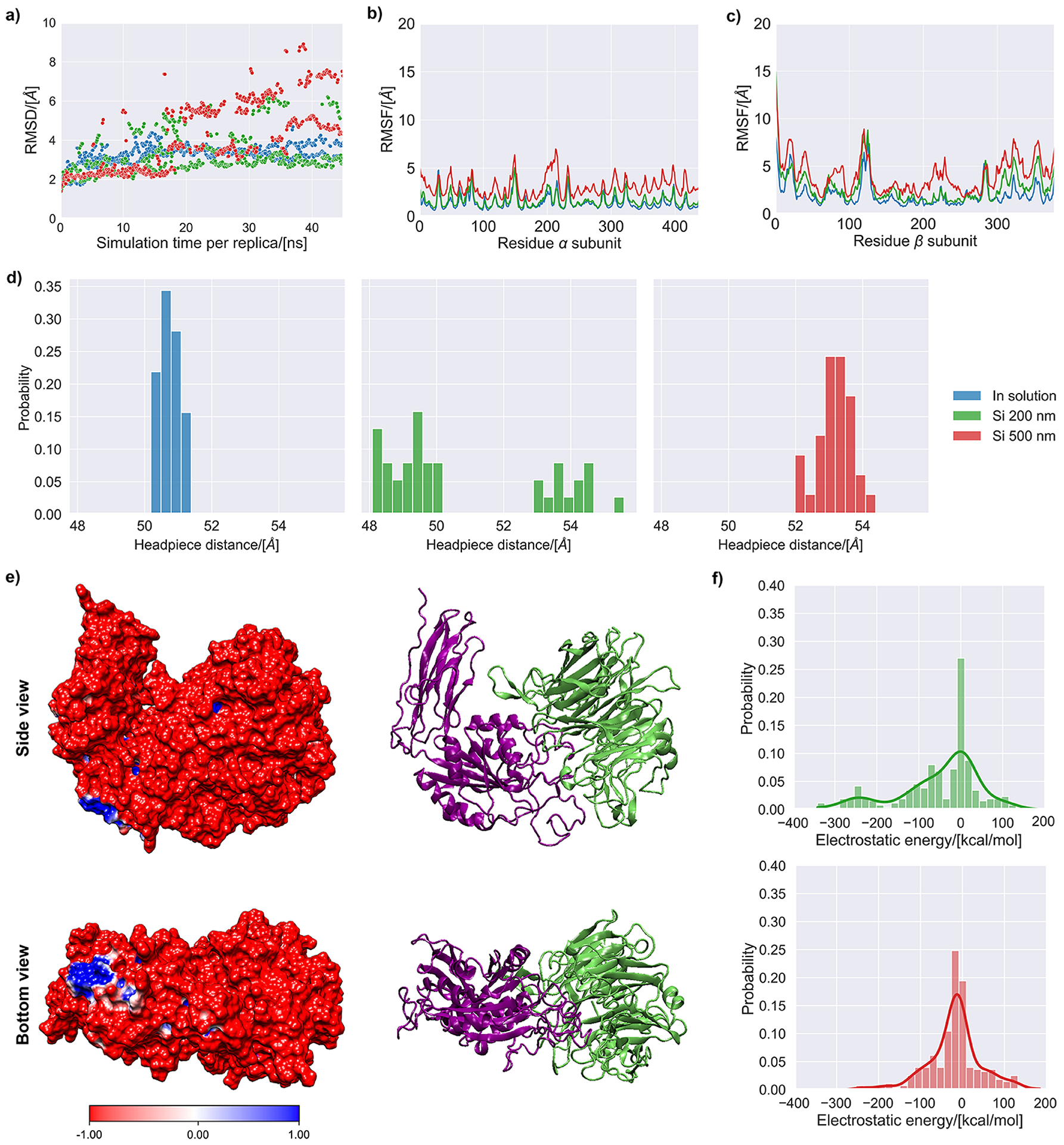
MD simulation results of the αVβ3 integrin subunit in aqueous solution (0.15 M NaCl) and in contact with silica surfaces of 200 and 500 nm. (a) Root mean square deviation of the backbone of the αVβ3 integrin for the accepted states during the simulation. (b) Root mean square fluctuations (RMSF) of the backbone of the α chain for the accepted states during the simulation. (c) Root mean square fluctuations (RMSF) of the backbone of the β chain for the accepted states during the simulation. (d) Histograms representing the headpiece distance for the accepted states collected during the last 2.5 ns of simulation. (e) Side and bottom views of the αVβ3 integrin’s molecular electrostatic potential map, with cartoon representation shown next to it for reference (lime color=α subunit, purple color=β subunit). (f) Histograms of the electrostatic and van der Waals energies for the interaction of the αVβ3 integrin with the silica surfaces representing nanoparticles of 200 and 500 nm. The continuous probability density curve is plotted the KDE method in the Seaborn library from Python.
The ensemble of accepted states for the integrin, collected over the last 2.5 ns of sampling time, was used to average the headpiece opening distance of the integrin subunit. Simulations of the integrin in contact with silica nanoparticles yielded structures in the ensemble of accepted states with a headpiece opening higher than in solution (Fig. 4d). For silica nanoparticles of 500 nm, one cluster of structures was identified. These structures had a headpiece distance that was on average ca. 2.5 Å larger than for the integrin in solution. In the case of silica nanoparticles of 200 nm, two structural subpopulations were identified. One of them had a headpiece opening with a similar value to that of the integrin in solution, while the other resembled the integrin in contact with silica nanoparticles of 500 nm.
The subunits of the αVβ3 integrin used in these simulations had a net negative electric charge number of −27. This was confirmed by its molecular electrostatic maps (Fig. 4e), where regions of lower electrostatic potential (red) were predominant and only a small region of higher electrostatic potential (blue) was found in the bottom part of the integrin. To assess the impact of the charges of the integrin and the silica surfaces, the non-bonded energies between the integrin and the silica surface were calculated for the ensemble of accepted states (Fig. 4f). The dominant non-bonded interaction was electrostatic in nature, being around one order of magnitude higher than the van de Waals interactions (Fig. S5). The electrostatic interactions in the accepted states for the simulation with nanoparticles of 200 nm displayed more negative values than with nanoparticles of 500 nm, indicating more favorable electrostatic interactions for nanoparticles of 200 nm.
4. Discussion
The design and use of chimeric fusion proteins containing silk and silica-forming domains in the synthesis of silk–silica nanocomposites has been extensively studied [19,20,22,24,25,39,54], as well as the effect of the pH on silica nanoparticle size during silicification via Stöber process [24,35,36]. Nevertheless, there has been no extensive study of the effect of pH on the biosilicification of protein-based films or the osteoinductive potential of the resulting films. To shed light on these questions, an experimental-computational approach was applied in this work.
R5–15mer-ch chimera proteins were used to generate films that were biosilicified at pH values from 3 to 10. A pH 3 led to a low surface coverage (around 10%) of dispersed amorphous structures containing nanoparticles with an average size of 80 nm. This agreed with previous work at the same pH that showed the presence of silica aggregates on silk structures with poor control of size and shape and low yield, with a predominant deposition of silica in the form of small nanoparticles [24]. At pH 5, two different subpopulations of nanoparticles of 50 and 950 nm were observed. Similar data was reported in a previous work with the same chimeric proteins, where two subpopulations of nanoparticles of 100 and 1,400 nm were generated at pH 5 [22]. When increasing the pH to 7, an distribution of silica nanoparticle sizes centered at 200 nm was obtained, being this size half the value of the average nanoparticle size of 400 nm previously reported for the same conditions [22,25]. At pH 8, again two subpopulations were detected, consisting of particles of 500 and 1,000 nm in diameter. Similar values were reported in previous work for silica particles in solution, leading to particles of 800 to 2,000 nm at this pH [24]. Finally, at pH 10, dispersed aggregates with nanoparticles of 25 nm were observed in the present study, which agreed with data from solution assays, where significantly reduced yields and dispersed structures containing few spheres were observed at pH >9 [24]. The discrepancy in particle size values between the present study and previous work may be due to the differences in experimental sampling: while measurements in this work were derived from measurements directly on silk films, earlier studies were obtained from measurements of particles in a protein solution. Nonetheless, the changes in sizes in the present study followed the same trends as previous work.
Induction of osteogenenic markers was evaluated via qPCR. For films biosilicified at pH 3 and 5, no detectable levels were found. This could be most likely linked to the poor surface coverage obtained after biosilicification. Due to this poor coverage, integrins in the hMSCs were sensing silk protein instead of silica. To that end, it was previously demonstrated that chimeric films without silica were not inductive or even repressive of the expression of integrin αV subunit, Runx2 transcription factor and osteogenic markers [20]. Conversely, films biosilicified at pH 7, with an average silica nanoparticle size of 200 nm and the highest surface silica coverage achieved in this work (approximately 90%), showed induction of all markers both after 24 h or 7 days of growth, agreeing with previous findings at the same pH [20]. The films biosilicified at pH 8, with an average silica nanoparticle size of 500 nm and lower coverage than films biosilicified at pH 7, showed upregulated gene expression of αV integrin subunits and Runx2 transcription factor and ColI markers (albeit lower than at pH 7) but not BSP after 24 h of cell growth, whereas all the markers were induced after 7 days of cell growth. Data reported elsewhere showed comparable results for films biosilicified using different concentrations of protein and leading to the precipitation of nanoparticles between <200 nm and 500 nm [19]. In that study, films with average nanoparticle sizes of 500 nm showed induction of just Alkaline Phosphatase (ALP), whereas films with silica nanoparticles <200 nm and a better surface coverage showed induction of ALP, BSP and ColI markers. It should be noted that the reduced surface coverage at pH 8 in the present study (average nanoparticle size of 500 nm) could also have a negative impact on the osteogenic induction because less cells will be in contact with silica, similarly to surfaces generated at pH 3 and 5. Moreover, after a short period of time only early osteogenic markers are induced (like Runx2 or ColI), whereas BSP -a later marker- would require more time for induction.
Fully atomistic MD simulations were used to study the onset of the extracellular domain of the αVβ3 integrin activation in contact with silica. Silica nanoparticles with diameters of 200 and 500 nm were simulated, corresponding to the main peaks in the particle size distributions obtained during biosilicification at pH values of 7 and 8, respectively. Previous REMD simulations revealed that αVβ3 integrin became activated in contact with silk-silica biomaterials due to the presence of silica, while silk did not affect the integrin folded state [20]. A similar simulation set-up was applied in the present work, albeit using the TIGER2hs method, which is expected to improve the efficiency of the sampling of the integrin conformation.
The larger values for the integrin RMSD obtained in the simulations with silica indicate that the presence of silica notably affects the integrin conformation. This was caused by changes in the relative position of the three domains simulated, which maintained a compact conformation. Their secondary structure remained stable throughout the simulation (Fig. S6), agreeing with earlier conformational data for intact, full-length human αIIbβ3 integrin obtained using cryo-EM [55]. The hybrid domain exhibited the highest mobility during the simulations, as shown by its RMFS, which matched previous data suggesting that the hybrid domain swings out during the onset of integrin activation [50].
Several studies proposed that increased headpiece openings can be interpreted as a sign of integrin activation [49,56,57]. Thus, the headpiece opening was monitored during MD simulations in this work. The results for the headpiece opening agreed with the experimental data about the expression of genes related to the differentiation of hMSCs into osteoblasts in biomineralized films with silica nanoparticles at pH 7 and 8. The ensemble of accepted states for the structure of the integrin during the last 2.5 ns of simulation exhibited structures with a higher headpiece opening than for simulations in solution, both for silica nanoparticles of 200 and 500 nm.
As mentioned in Section 2.8, nanoparticles with 200 nm in diameter had 18% of their surface silanol groups ionized (ca. 1.2 siloxide groups per nm2). This ionization increased for nanoparticles with 500 nm in diameter (30% ionization, ca. 2.0 siloxide groups per nm2). Thus, the surface representing nanoparticles of 500 nm was richer in fixed negative charges due to a ca. 50% higher content of O− atoms in siloxide groups. The enhanced osteoinduction observed experimentally with nanoparticles of 200 nm (pH 7) could be thus linked to the more energetically favorable electrostatic interactions between the negatively charged integrin and the less negatively-charged surface of silica nanoparticles of 200 nm (Fig. 4g).
This work combined experimental data (including nanoparticle measurements and in vitro osteoinduction) with MD simulations to shed light on the control of the silica nanoparticle size during biomineralization, as well as on the effect of silica nanoparticle size on the osteoinductive potential of silk-silica composite biomaterials. Taken together, the results suggest that the osteogenic induction ability of biosilicified films depends on both silica nanoparticle size (governing the cell-silica interactions) and film coverage (controlling the number of cells in contact with the inducing material, silica in this case). These results deepened the understanding of the structure-property relationships of such biomaterials, while highlighting the potential of MD simulations to predict and explain the biological outcomes (in this case, expression of gene markers related to osteoinduction in hMSCs) of biosilicified silk biomaterials.
5. Conclusions
Biomineralized silk-silica films were produced at different pH values, using a chimeric protein consisting of the consensus domain of spider Nephila clavipes major ampullate dragline silk and the silica-promoting R5 peptide. Using pH values of 7–8 during biomineralization maximized the coverage of the film surface by silica nanoparticles, as well as the size of those nanoparticles. The gene expression profile of cells growing on the resulting biomineralized silk-silica films revealed an enhanced osteoinduction for films biomineralized at pH 7, predominantly covered with silica nanoparticles of 200 nm. Lower (3–5) or high (10) pH values led to lower biomineralization and poor coverage of the protein surfaces. At pH 8 large silica nanoparticles (>500 nm) were formed, showing a reduced osteoinduction. MD simulations demonstrated that the presence of silica nanoparticles of 200 and 500 nm led to a larger headpiece opening of the αVβ3 integrin extracellular subunit. Moreover, the slightly enhanced activation observed experimentally for nanoparticles of 200 nm could be caused by the reduced electrostatic repulsion between the negatively charged integrin and the surface of silica nanoparticles of 200 nm, as shown by the more favorable electrostatic interaction observed during MD simulations. Overall, these results provide guidelines for the optimization of the synthesis of biomineralized silk-silica biomaterials, while revealing MD as a valuable tool to predict their biological outcome (i.e., expression of gene markers of osteoinduction).
Supplementary Material
Statement of significance.
The ability of biomineralized materials to induce hMSCs differentiation for bone tissue regeneration applications was analyzed. Biomaterials were created using a recombinant protein formed by the consensus repeat derived from the spider Nephila clavipes major ampullate dragline silk and the silica-promoting peptide R5 derived from the Cylindrotheca fusiformis silaffin gene. A combination of computational and experimental techniques revealed the optimal conditions for the synthesis of biomineralized silk-silica films with enhanced expression of markers related to bone regeneration.
Acknowledgments
We acknowledge the NIH (U01EB014976, R01AR068048) for support of this work. Computational simulations were performed on the MIT Engaging Cluster and the Extreme Science and Engineering Discovery Environment (XSEDE) which is supported by the National Science Foundation grant number TG-MSS090007.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Bobbert FSL, Zadpoor AA, Effects of bone substitute architecture and surface properties on cell response, angiogenesis, and structure of new bone, J. Mater. Chem. B 5 (2017) 6175–6192. 10.1039/c7tb00741h. [DOI] [PubMed] [Google Scholar]
- [2].Klosterhoff BS, Nagaraja S, Dedania JJ, Guldberg RE, Willett NJ, Material and Mechanobiological Considerations for Bone Regeneration, in: Mater Devices Bone Disord., Elsevier Inc., 2017: pp. 197–264. 10.1016/B978-0-12-802792-9.00005-7. [DOI] [Google Scholar]
- [3].Bryksin AV, Brown AC, Baksh MM, Finn MG, Barker TH, Learning from nature - Novel synthetic biology approaches for biomaterial design, Acta Biomater. 10 (2014) 1761–1769. 10.1016/j.actbio.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhou Z, Zhang S, Cao Y, Marelli B, Xia X, Tao TH, Engineering the Future of Silk Materials through Advanced Manufacturing, Adv. Mater 30 (2018) 1706983 10.1002/adma.201706983. [DOI] [PubMed] [Google Scholar]
- [5].Farokhi M, Mottaghitalab F, Samani S, Shokrgozar MA, Kundu SC, Reis RL, Fatahi Y, Kaplan DL, Silk fibroin/hydroxyapatite composites for bone tissue engineering, Biotechnol. Adv 36 (2018) 68–91. 10.1016/j.biotechadv.2017.10.001. [DOI] [PubMed] [Google Scholar]
- [6].Ling S, Kaplan DL, Buehler MJ, Nanofibrils in nature and materials engineering, Nat. Rev. Mater 3 (2018) 1–15. 10.1038/natrevmats.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Schnepp Z, Biopolymers as a Flexible Resource for Nanochemistry, Angew. Chemie Int. Ed 52 (2013) 1096–1108. 10.1002/anie.201206943. [DOI] [PubMed] [Google Scholar]
- [8].Yang YJ, Holmberg AL, Olsen BD, Artificially Engineered Protein Polymers, Annu. Rev. Chem. Biomol. Eng 8 (2017) 549–575. 10.1146/annurevchembioeng-060816-101620. [DOI] [PubMed] [Google Scholar]
- [9].Frandsen JL, Ghandehari H, Recombinant protein-based polymers for advanced drug delivery, Chem. Soc. Rev 41 (2012) 2696–2706. 10.1039/c2cs15303c. [DOI] [PubMed] [Google Scholar]
- [10].Price R, Poursaid A, Ghandehari H, Controlled release from recombinant polymers, J. Control. Release 190 (2014) 304–313. 10.1016/j.jconrel.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huang W, Ling S, Li C, Omenetto FG, Kaplan DL, Silkworm silk-based materials and devices generated using bio-nanotechnology, Chem. Soc. Rev 47 (2018) 6486–6504. 10.1039/c8cs00187a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Numata K, Kaplan DL, Silk-based delivery systems of bioactive molecules, Adv. Drug Deliv. Rev 62 (2010) 1497–1508. 10.1016/j.addr.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL, Spider silks and their applications, Trends Biotechnol. 26 (2008) 244–251. 10.1016/j.tibtech.2008.02.006. [DOI] [PubMed] [Google Scholar]
- [14].An B, Tang-Schomer MD, Huang W, He J, Jones JA, Lewis RV, Kaplan DL, Physical and biological regulation of neuron regenerative growth andnetwork formation on recombinant dragline silks, Biomaterials. 48 (2015) 137–146. 10.1016/j.biomaterials.2015.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Au AY, Au RY, Demko JL, McLaughlin RM, Eves BE, Frondoza CG, Consil® bioactive glass particles enhance osteoblast proliferation and selectively modulate cell signaling pathways in vitro, J. Biomed. Mater. Res. - Part A 94 (2010) 380–388. 10.1002/jbm.a.32668. [DOI] [PubMed] [Google Scholar]
- [16].Lu ZF, Zreiqat H, Beta-tricalcium phosphate exerts osteoconductivity through α2β1 integrin and down-stream MAPK/ERK signaling pathway, Biochem. Biophys. Res. Commun 394 (2010) 323–329. 10.1016/j.bbrc.2010.02.178. [DOI] [PubMed] [Google Scholar]
- [17].Shie MY, Ding SJ, Integrin binding and MAPK signal pathways in primary cell responses to surface chemistry of calcium silicate cements, Biomaterials. 34 (2013) 6589–6606. 10.1016/j.biomaterials.2013.05.075. [DOI] [PubMed] [Google Scholar]
- [18].Chen X, Wang J, Chen Y, Cai H, Yang X, Zhu X, Fan Y, Zhang X, Roles of calcium phosphate-mediated integrin expression and MAPK signaling pathways in the osteoblastic differentiation of mesenchymal stem cells, J. Mater. Chem. B 4 (2016) 2280–2289. 10.1039/c6tb00349d. [DOI] [PubMed] [Google Scholar]
- [19].Mieszawska AJ, Nadkarni LD, Perry CC, Kaplan DL, Nanoscale control of silica particle formation via silk-silica fusion proteins for bone regeneration, Chem. Mater 22 (2010) 5780–5785. 10.1021/cm101940u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martín-Moldes Z, Ebrahimi D, Plowright R, Dinjaski N, Perry CC, Buehler MJ, Kaplan DL, Intracellular Pathways Involved in Bone Regeneration Triggered by Recombinant Silk–Silica Chimeras, Adv. Funct. Mater 28 (2018). 10.1002/adfm.201702570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tajuddin I, Voelcker N, Mitchell J, Silica nanostructure formation from synthetic R5 peptide, in: Voelcker NH (Ed.), Smart Mater. IV, SPIE, 2006: p. 64130Q 10.1117/12.695946. [DOI] [Google Scholar]
- [22].Plowright R, Dinjaski N, Zhou S, Belton DJ, Kaplan DL, Perry CC, Influence of silk-silica fusion protein design on silica condensation in vitro and cellular calcification, RSC Adv. 6 (2016) 21776–21788. 10.1039/c6ra03706b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Knecht MR, Wright DW, Functional analysis of the biomimetic silica precipitating activity of the R5 peptide from Cylindrotheca fusiformis, Chem. Commun 9 (2003) 3038–3039. 10.1039/b309074d. [DOI] [PubMed] [Google Scholar]
- [24].Belton DJ, Mieszawska AJ, Currie HA, Kaplan DL, Perry CC, Silk-silica composites from genetically engineered chimeric proteins: Materials properties correlate with silica condensation rate and colloidal stability of the proteins in aqueous solution, Langmuir. 28 (2012) 4373–4381. 10.1021/la205084z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dinjaski N, Ebrahimi D, Ling S, Shah S, Buehler MJ, Kaplan DL, Integrated Modeling and Experimental Approaches to Control Silica Modification of Design Silk-Based Biomaterials, ACS Biomater. Sci. Eng 3 (2017) 2877–2888. 10.1021/acsbiomaterials.6b00236. [DOI] [PubMed] [Google Scholar]
- [26].Avraamides CJ, Garmy-Susini B, Varner JA, Integrins in angiogenesis and lymphangiogenesis, Nat. Rev. Cancer 8 (2008) 604–617. 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Moreno-Layseca P, Streuli CH, Signalling pathways linking integrins with cell cycle progression, Matrix Biol. 34 (2014) 144–153. 10.1016/j.matbio.2013.10.011. [DOI] [PubMed] [Google Scholar]
- [28].M.C.J.A.M.H. Humphries JD, Signal transduction via integrin adhesion complexes, Curr. Opin. Cell Biol 56 (2018) 14–21. [DOI] [PubMed] [Google Scholar]
- [29].Kechagia JZ, Ivaska J, Roca-Cusachs P, Integrins as biomechanical sensors of the microenvironment, Nat. Rev. Mol. Cell Biol 20 (2019) 457–473. 10.1038/s41580-019-0134-2. [DOI] [PubMed] [Google Scholar]
- [30].Pan L, Zhao Y, Yuan Z, Qin G, Research advances on structure and biological functions of integrins, Springerplus. 5 (2016) 1094 10.1186/s40064-016-2502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Greenblatt MB, Shim J-H, Glimcher LH, Mitogen-Activated Protein Kinase Pathways in Osteoblasts, Annu. Rev. Cell Dev. Biol 29 (2013) 63–79. 10.1146/annurev-cellbio-101512-122347. [DOI] [PubMed] [Google Scholar]
- [32].Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L, Rusu A, Irimie A, Atanasov AG, Slaby O, Ionescu C, Berindan-Neagoe I, A comprehensive review on MAPK: A promising therapeutic target in cancer, Cancers (Basel). 11 (2019). 10.3390/cancers11101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Geest CR, Coffer PJ, MAPK signaling pathways in the regulation of hematopoiesis, J. Leukoc. Biol 86 (2009) 237–250. 10.1189/jlb.0209097. [DOI] [PubMed] [Google Scholar]
- [34].Stöber W, Fink A, Bohn E, Controlled growth of monodisperse silica spheres in the micron size range, J. Colloid Interface Sci 26 (1968) 62–69. 10.1016/0021-9797(68)90272-5. [DOI] [Google Scholar]
- [35].Kobayashi M, Juillerat F, Galletto P, Bowen P, Borkovec M, Aggregation and charging of colloidal silica particles: Effect of particle size, Langmuir. 21 (2005) 5761–5769. 10.1021/la046829z. [DOI] [PubMed] [Google Scholar]
- [36].Gorrepati EA, Wongthahan P, Raha S, Fogler HS, Silica precipitation in acidic solutions: Mechanism, pH effect, and salt effect, Langmuir. 26 (2010) 10467–10474. 10.1021/la904685x. [DOI] [PubMed] [Google Scholar]
- [37].Heinz H, Lin TJ, Kishore Mishra R, Emami FS, Thermodynamically consistent force fields for the assembly of inorganic, organic, and biological nanostructures: The INTERFACE force field, Langmuir. 29 (2013) 1754–1765. 10.1021/la3038846. [DOI] [PubMed] [Google Scholar]
- [38].Rabotyagova OS, Cebe P, Kaplan DL, Self-assembly of genetically engineered spider silk block copolymers, Biomacromolecules. 10 (2009) 229–236. 10.1021/bm800930x. [DOI] [PubMed] [Google Scholar]
- [39].Zhou S, Huang W, Belton DJ, Simmons LO, Perry CC, Wang X, Kaplan DL, Control of silicification by genetically engineered fusion proteins: Silk-silica binding peptides, Acta Biomater. 15 (2015) 173–180. 10.1016/j.actbio.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mauney JR, Blumberg J, Pirun M, Volloch V, Vunjak-Novakovic G, Kaplan DL, Osteogenic Differentiation of Human Bone Marrow Stromal Cells on Partially Demineralized Bone Scaffolds in Vitro, in: Tissue Eng, 2004: pp. 81–92. 10.1089/107632704322791727. [DOI] [PubMed] [Google Scholar]
- [41].Altman GH, Horan RL, Lu HH, Moreau J, Martin I, Richmond JC, Kaplan DL, Silk matrix for tissue engineered anterior cruciate ligaments, Biomaterials. 23 (2002) 4131–4141. 10.1016/S0142-9612(02)00156-4. [DOI] [PubMed] [Google Scholar]
- [42].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method, Methods. 25 (2001) 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [43].Emami FS, Puddu V, Berry RJ, Varshney V, Patwardhan SV, Perry CC, Heinz H, Prediction of specific biomolecule adsorption on silica surfaces as a function of pH and particle size, Chem. Mater 26 (2014) 5725–5734. 10.1021/cm5026987. [DOI] [Google Scholar]
- [44].Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA, Crystal structure of the extracettutar segment of integrin αVβ3, Science (80-.) 294 (2001) 339–345. 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K, Scalable molecular dynamics with NAMD, J. Comput. Chem 26 (2005) 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M, CHARMM: The biomolecular simulation program, J. Comput. Chem 30 (2009) 1545–1614. 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Geist N, Kulke M, Schulig L, Link A, Langel W, Replica-Based Protein Structure Sampling Methods II: Advanced Hybrid Solvent TIGER2hs, J. Phys. Chem. B 123 (2019) 5995–6006. 10.1021/acs.jpcb.9b03134. [DOI] [PubMed] [Google Scholar]
- [48].Humphrey W, Dalke A, Schulten K, VMD: Visual molecular dynamics, J. Mol. Graph 14 (1996) 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [49].Paradise RK, Lauffenburger DA, Van Vliet KJ, Acidic Extracellular pH Promotes Activation of Integrin αvβ3, PLoS One. 6 (2011) e15746 10.1371/journal.pone.0015746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhu J, Zhu J, Springer TA, Complete integrin headpiece opening in eight steps, J. Cell Biol 201 (2013) 1053–1068. 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: Application to microtubules and the ribosome, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 10037–10041. 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA, PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations, Nucleic Acids Res. (2007). 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA, PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations., Nucleic Acids Res. 32 (2004) W665–7. 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Foo CWP, Patwardhan SV, Belton DJ, Kitchel B, Anastasiades D, Huang J, Naik RR, Perry CC, Kaplan DL, Novel nanocomposites from spider silk-silica fusion (chimeric) proteins, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 9428–9433. 10.1073/pnas.0601096103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xu XP, Kim E, Swift M, Smith JW, Volkmann N, Hanein D, Three-Dimensional Structures of Full-Length, Membrane-Embedded Human αiIbβ3 Integrin Complexes, Biophys. J 110 (2016) 798–809. 10.1016/j.bpj.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Takagi J, Petre BM, Walz T, Springer TA, Global conformational earrangements in integrin extracellular domains in outside-in and inside-out signaling, Cell. 110 (2002) 599–611. 10.1016/S0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- [57].Puklin-Faucher E, Gao M, Schulten K, Vogel V, How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation, J. Cell Biol 175 (2006) 349–360. 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


