Abstract
Purpose:
Vascular malformations (VM) are primarily caused by somatic activating pathogenic variants in oncogenes. Targeted pharmacotherapies are emerging but require molecular diagnosis. Since variants are currently only detected in malformation tissue, patients may be ineligible for clinical trials prior to surgery. We hypothesized that cell-free DNA (cfDNA) could provide molecular diagnoses for patients with isolated VM.
Methods:
cfDNA was isolated from plasma or cyst fluid from patients with arteriovenous malformations (AVM), venous malformations (VeM), or lymphatic malformations (LM), and assayed for known pathogenic variants using droplet digital PCR (ddPCR). Cyst fluid cfDNA from an independent cohort of LM patients was prospectively screened for variants using a multiplex ddPCR assay.
Results:
Variants were detected in plasma cfDNA in patients with AVM (2/8) and VeM (1/3). Variants were detected in cyst fluid cfDNA (7/7) but not plasma (0/26) in LM patients. Prospective testing of cyst fluid cfDNA with multiplex ddPCR identified variants in LM patients who had never undergone surgery (4/5).
Conclusion:
Variants were detected in plasma from AVM and VeM patients, and in cyst fluid from patients with LM. These data support investigation of cfDNA-based molecular diagnostics for VM patients which may provide opportunities to initiate targeted pharmacotherapies without prior surgery.
Keywords: vascular malformations, cell-free DNA, PIK3CA, droplet digital PCR, multiplexing
Introduction
Vascular malformations (VM) are congenital malformations resulting from defective morphogenesis of arteries, veins, capillaries, and/or lymphatic vessels. Histologically they resemble disorganized and dilated blood/lymphatic vessels and are classified by dominant type of malformed vessel: capillary malformations (CM, MIM#163000), lymphatic malformations (LM), venous malformations (VeM), and arteriovenous malformations (AVM).1 LM, VeM, and AVM are associated with significant morbidity, causing pain, disfigurement, and functional impairment.2, 3 Current treatments are primarily invasive, including endovascular approaches (sclerotherapy, embolization) and open surgical resection.3–8
Although familial disorders exist, most VM are sporadic with the majority caused by post-zygotic, activating pathogenic variants in oncogenes within the PI3K-MTOR and RAS-MAPK pathways.9 Previous studies have shown clear genotype-phenotype demarcations: LM are associated with PIK3CA variants,10, 11 VeM with TEK and PIK3CA variants,12, 13 and AVM with MAP2K1, BRAF, and KRAS variants.14–16 These variants are restricted to a small fraction (1–20%) of cells within VM tissue and are generally not detected in DNA isolated from blood-cells or surrounding normal tissues (e.g. skin biopsies overlying the lesion).11 Consequently, molecular diagnosis currently requires surgically excised VM tissue. VM resection requires specialized surgical expertise, and associated risks include bleeding, infection, chyle leak, nerve injury, and poor wound healing. Several targeted therapies including sirolimus (MTOR inhibitor), alpelisib (PI3K inhibitor), and miransertib (AKT inhibitor) are currently being examined for efficacy in VM and overgrowth in clinical trials (clinicaltrials.gov: NCT03987152, NCT02638389, NCT04085653, NCT03094832).16–18 With the exception of sirolimus, which is currently used off-label for many VM despite mixed clinical evidence19–22, identification of a pathogenic variant must precede initiation of targeted therapies.
Over the last decade, liquid biopsies utilizing cell-free DNA (cfDNA) have gained prominence in cancer diagnosis, surveillance, and therapy.23, 24 Given the critical role mutant endothelial cells play in the development of VM, as well as their intimate contact with plasma and lymphatic fluid, we hypothesized that molecular diagnosis of VM could be achieved by analysis of cfDNA.
Here, we describe the successful molecular diagnosis of vascular malformations utilizing minimally invasive techniques. Highly sensitive targeted genotyping using droplet digital PCR (ddPCR) of plasma derived cfDNA from patients with extracranial AVM (MAP2K1-activating variants) and VeM (TEK-activating variants) detected known, tissue-based somatic variants. PIK3CA-activating variants were detected in cyst-fluid derived cfDNA but not in plasma cfDNA from patients with LM. Furthermore, we used a multiplex ddPCR assay to screen an independent cohort of LM patients who had never undergone surgery and detected PIK3CA-activating variants in the majority of samples at a diagnostic rate similar to tissue-based sequencing. These results demonstrate the feasibility of cfDNA as a diagnostic analyte for VM, providing an opportunity for patients with VM to initiate targeted medical therapies without the need for surgery.
Materials and Methods
Ethics Statement
This study was approved by the Institutional Review Board at Seattle Children’s Hospital. Written, informed consent was obtained for each individual in this study prior to sample and data collection. All individuals presented are de-identified.
Participants and sample collection
We included individuals with VM who had cyst fluid and/or plasma available for testing. Patients were designated as having isolated VM if there was no clinical evidence of multi-system disease such as congenital lipomatosis, overgrowth, vascular malformations, epidermal nevi, and scoliosis syndrome (CLOVES, MIM#612918), Klippel-Trenaunay syndrome (KTS, MIM%149000), Gorham-Stout disease (MIM123880), hereditary hemorrhagic telangiectasia (MIM#187300), blue rubber bleb nevus syndrome (MIM%11200), etc. Blood and cyst fluid were collected in either EDTA tubes or Cell-Free DNA BCT® tubes (“Streck tubes”, Streck Omaha, NE). Plasma and cyst fluid volumes were recorded. Cyst fluid was collected during surgery or sclerotherapy, or in clinic with ultrasound guidance. VM tissue samples were collected at time of clinically indicated surgical resection. Recorded clinical variables included age, sex, and location of VM.
Sample processing and DNA extraction
Whole blood and cyst fluid underwent centrifugation at 1,600g. Plasma was isolated from remaining blood, and cyst fluid supernatant separated from the cyst fluid pellet. Plasma and cyst fluid supernatant were then centrifuged at 16,000g for 15 minutes at 4 °C to remove any remaining debris. The resulting supernatants were isolated and stored at −80 °C along with the cyst fluid pellets. cfDNA extraction from plasma and cyst fluid supernatant was performed with QIAmp Circulating Nucleic Acid Kit (Qiagen, Venlo, Netherlands). Genomic DNA (gDNA) was extracted from affected tissue and cyst fluid pellets using PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA). Extracted cfDNA and gDNA were quantified with Qubit fluorometry (Thermo Fisher Scientific, Waltham, MA, USA). cfDNA was further analyzed on an Agilent 2200 Tapestation with High Sensitivity D1000 ScreenTape (Agilent, Santa Clara, CA, USA).
Singleplex ddPCR
For singleplex ddPCR experiments, all individuals had a variant detected from affected tissue by ddPCR or clinical sequencing. The following Bio-Rad-designed ddPCR assays (Bio-Rad, Hercules, CA) were used: MAP2K1 (NM_002755.3) p.Lys57Asn (c.171G>T, c.171G>C) and p.Gln56Pro (c.167A>C); BRAF (NM_004333.4) p.Val600Glu (c.1799T>A); PIK3CA (NM_006218.4) p.Glu542Lys (c.1624G>A), p.Glu545Lys (c.1633G>A), p.His1047Arg (c.3140A>G), and p.Gln546Lys (c.1636C>A); TEK (NM_000459.4) p.Leu914Phe (c.2740C>T). PCR was performed as previously described.11 All samples were run in quadruplicate with a maximum of 25 ng DNA per well or maximum volume 11 μL if cfDNA concentration was low. Each run included a no template control, wild-type (WT) gDNA control, WT cfDNA control, and variant gDNA control. Data were analyzed with QuantaSoft software (Bio-Rad, Hercules, CA). Samples were positive if the variant fluorescence was significantly different from the fluorescence of the WT control using 95% confidence intervals for total error (Figure S1). The total error is displayed by the QuantaSoft software and defined as the greater of either the technical error (Poisson error) or the empirical error (standard error of the mean).25 Variant allele fractions (VAFs) were calculated as the concentration of variant droplets out of the total concentration of droplets containing at least one copy of variant or WT DNA.
Multiplex ddPCR
A multiplex ddPCR assay to detect the four most common PIK3CA-activating variants was previously developed (see Supplemental Methods for details).26 Samples were run in quadruplicate and each run included a no template control, WT gDNA control, and WT cfDNA control. For positive controls, gene blocks of 170 bp were designed for each variant and associated WT sequence (Integrative DNA Technologies, Coralville, IA). Runs included four positive controls which consisted of a 10–25% dilution of variant gene block in WT gene block for each of the four variants. Data was analyzed with QuantaSoft software. VAFs were calculated as the concentration of variant droplets out of the total concentration of droplets containing DNA for the locus of interest. Interpretation was validated by three independent observers (KZ, DMJ, VD; Supplemental Methods).
Statistical Analysis
All statistical analyses were performed using R software.
Results
Subjects and plasma cfDNA characteristics
Plasma samples (n = 55) were obtained for 38 individuals: eight extracranial AVM patients (15 samples), three VeM patients (four samples), and 27 LM patients (36 samples). All individuals had a pathogenic variant previously detected in affected tissue (Table 1, Table S1). Forty-six of these plasma samples were collected into EDTA tubes between September 2012 and December 2019 and stored in a frozen biorepository (mean volume 1.1 mL, SD: 0.9). All AVM and VeM samples were collected in EDTA tubes and obtained from the frozen biorepository. Nine plasma samples, all from LM patients, were collected into Streck tubes (mean volume 3.7 mL, SD: 1.8). The average amount of DNA was 21.8 ng (SD: 20.7), excluding one EDTA-collected frozen sample that had >500 ng of DNA recovered. DNA recovery by volume was significantly lower from Streck tube samples (mean: 10.4 ng DNA/mL plasma, SD: 4.2) than from EDTA-collected frozen samples (mean: 21.5 ng DNA/mL plasma, SD: 12.5) likely indicating a larger amount of “contaminating” gDNA from white blood cells in the EDTA-collected frozen samples (mean difference: −11.1 ng DNA/mL plasma, 95% CI: −15.8, −6.42).
Table 1.
Somatic variants in plasma cfDNA from extracranial AVM and VeM.
| Plasma cfDNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Sex | Agea | VM Type | Location | Variant | Tissue VAF (%)b | Plasma volume (mL)c | VAF (%)b | Droplets (Var/WT) |
| LR16-173 | F | 15 | AVM | Ear |
MAP2K1 p.K57N (c.171G>T) |
8.2 | 3.3 0.9 0.5 |
0.4 NEG NEG |
21/4823 1/2151 0/1107 |
| LR17-049 | F | 18 | AVM | Face, temple |
MAP2K1 p.Q56P (c.167A>C) |
5.9 | 0.9 0.5 |
1.6 2.1 |
9/531 47/2096 |
| LR13-356 | M | 19 | AVM | Face, buccal |
MAP2K1 p.K57N (c.171G>T) |
7.2 | 0.9 | NEG | 2/1793 |
| LR17-050 | M | 1.4 | AVM | Anterior scalp |
BRAF p.V600E (c.1799T>A) |
21.2 | 0.4 | NEG | 1/810 |
| LR17-205 | M | 13 | AVM | Face, forehead |
MAP2K1 p.K57N (c.171G>T) |
5.2 | 0.6 1.3 |
NEG NEG | 0/38305 0/1437 |
| LR17-207 | M | 18 | AVM | Face, supraorbital |
MAP2K1 p.K57N (c.171G>T) |
1.0 | 3.0 | NEG | 0/2131 |
| LR17-208 | F | 11 | AVM | Hand |
MAP2K1 p.K57N (c.171G>C) |
13.7 | 1.3 0.6 0.9 |
NEG NEG NEG | 0/974 0/764 0/1337 |
| LR18-541 | M | 8 | AVM | Ear |
MAP2K1 p.K57N (c.171G>T) |
6.1 | 0.3 0.5 |
NEG NEG | 0/83 1/304 |
| LR18-542 | M | 4 | VeM | Face, buccal |
TEK p.L914F (c.2740C>T) |
3.3 | 0.8 | 1.6 | 21/1329 |
| LR16-024 | M | 8 | VeM | Tongue |
TEK p.L914F (c.2740C>T) |
7.5 | 4.0 | NEG | 1/1673 |
| LR17-197 | M | 3 | VeM | Face, buccal |
TEK p.L914F (c.2740C>T) |
8.0 | 2.3 0.7 |
NEG NEG |
0/1900 0/44 |
Abbreviations: AVM – arteriovenous malformation, NEG – no variant detected, VAF – variant allele fraction, Var – variant, VM – vascular malformation, VeM – venous malformation, WT – wild-type.
Age at earliest plasma collection in years.
VAF calculated using droplet concentrations and only reported for samples in which sample variant concentration was statistically different from WT cfDNA control variant concentration based on 95% total error confidence intervals.
Samples taken at separate time points when multiple volumes listed excepting LR18–541 who had two plasma samples from the same day.
MAP2K1 and TEK variants detected in plasma cfDNA from patients with AVM and VeM
Variants were detected in plasma cfDNA for two of the eight isolated extracranial AVM patients and one of the three isolated VeM patients (Table 1). In AVM patient LR16-173, the known MAP2K1 variant was detected in one of three independent plasma samples (Table 1). On further review it was noted that the one positive sample was collected on the day of surgery, while the two additional samples were collected after resection of the AVM. In AVM patient LR17-049 two independent plasma samples contained the MAP2K1 variant. Both samples were collected on the day of surgery, one at the initial resection and another at the time of repeat resection. One TEK variant was detected in a patient with VeM, LR18-542. The variant allele fractions (VAF) in plasma (range 0.4–2.1%) were lower than in tissues. Three of the remaining eight patients in whom a variant was not detected (LR17-050, LR17-208, LR18-541) had < 1500 wild-type droplets, indicating low DNA input and therefore decreased power to detect variants at very low VAF.25
PIK3CA variants not detected in plasma cfDNA from patients with isolated LM
We detected a PIK3CA variant (p.Glu545Lys) in a plasma sample from a single patient (LR14-285, Table S1) with an extensive LM in the setting of CLOVES syndrome. The remaining LM plasma samples came from patients with isolated LM and were negative for variants by ddPCR (Table S1). Two of the 35 negative LM plasma samples (LR16-266 and LR18-537) had three variant droplets detected, which would be reported as positive under less stringent criteria,26, 27 but did not meet our cutoff for positivity based on droplet concentration total error. However, it suggests that PIK3CA variants may be detected in the plasma of LM patients if larger amounts of cfDNA with less gDNA contamination were available for testing.
PIK3CA variants detected in cfDNA from cyst fluid in patients with isolated LM
LM have poor connectivity to the systemic circulation,28 so it is perhaps unsurprising that variants were not detected in plasma. However, LM have a cystic structure that permits direct sampling of lymphatic fluid. Seven LM patients with tissue detected PIK3CA pathogenic variants (six of which were also present in the plasma cohort above) had available cyst fluid samples (Table 2). Four cyst fluid samples (LR16-263, LR16-266, LR19-427, and LR19-474) were obtained during surgical excision of the LM. LR16-145 had cyst fluid aspirated by interventional radiology during transcutaneous catheter placement in LM to address acute respiratory distress. LR19-454 cyst fluid was collected in the operating room during seroma drainage one week following LM resection. Cyst fluid from LR19-545 was collected in clinic by needle aspiration using ultrasound guidance and local anesthetic. Cyst fluid volumes were higher than plasma (mean volume: 5.7 mL, SD: 3.1). Recovery of cfDNA ranged from 5.6 to 5920 ng DNA per mL of cyst fluid. Regardless of this variation, all cyst fluid cfDNA samples were positive for their respective PIK3CA variants with VAFs ranging from 0.13 to 7.9% (Table 2). Five of the seven cfDNA samples had a VAF equal to or greater than their corresponding tissue detected VAF. Cyst fluid pellet gDNA contained detectable variants in two of the seven samples (Table 2).
Table 2.
PIK3CA variants detected in LM cyst fluid cfDNA.
| Cyst fluid cfDNA | Cyst fluid pellet gDNA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Sex | Agea | Location | PIK3CA Variant | Tissue VAF (%)b | Cyst fluid volume (mL) | VAF (%)b | Droplets (Var/WT) | VAF (%)b | Droplets (Var/WT) |
| LR16-145c | M | 1.0 | Mediastinum | p.E542K (c.1624G>A) |
0.1 | 6.0 | 0.1 | 36/22993 | NEG | 2/10114 |
| LR16-263c | M | 2 mo | Tongue/FOM | p.Q546K (c.1636C>A) |
9.5 | 9.0 | 0.9 | 139/15053 | NEG | 1/7023 |
| LR16-266c | M | 1.4 | Neck | p.H1047R (c.3140A>G) |
0.5 | 3.0 | 1.4 | 18/1216 | NEG | 1/35 |
| LR19-427 | M | 14 | Parotid | p.E542K (c.1624G>A) |
1.1 | 3.3d | 3.7 | 207/5132 | NEG | 12/12430 |
| LR19-454 | F | 2.9 | FOM/neck | p.E542K (c.1624G>A) |
9.4 | 6.5d | 0.7 | 85/10378 | 0.1 | 11/16449 |
| LR19-474 | F | 2.2 | Neck | p.E545K (c.1633G>A) |
5.9 | 2.0d | 7.9 | 168/1939 | 11.9 | 44/324 |
| LR19-545 | F | 14 | Lower arm/hand | p.H1047R (c.3140A>G) |
2.2 | 10.2d | 3.2 | 156/4573 | NEG | 2/13624 |
Abbreviations: cfDNA – cell free DNA, mo – months old, mL – milliliters, NEG – no variant detected, VAF – variant allele fraction, Var– variant, WT – wild-type
Age at first cyst fluid collection, in years unless otherwise specified.
VAF calculated using droplet concentrations and only reported for samples in which sample variant concentration was statistically different from WT cfDNA control variant concentration based on 95% total error confidence intervals.
Patient previously reported.11
Sample collected in Streck tube.
Multiplex ddPCR in prospective LM cohort
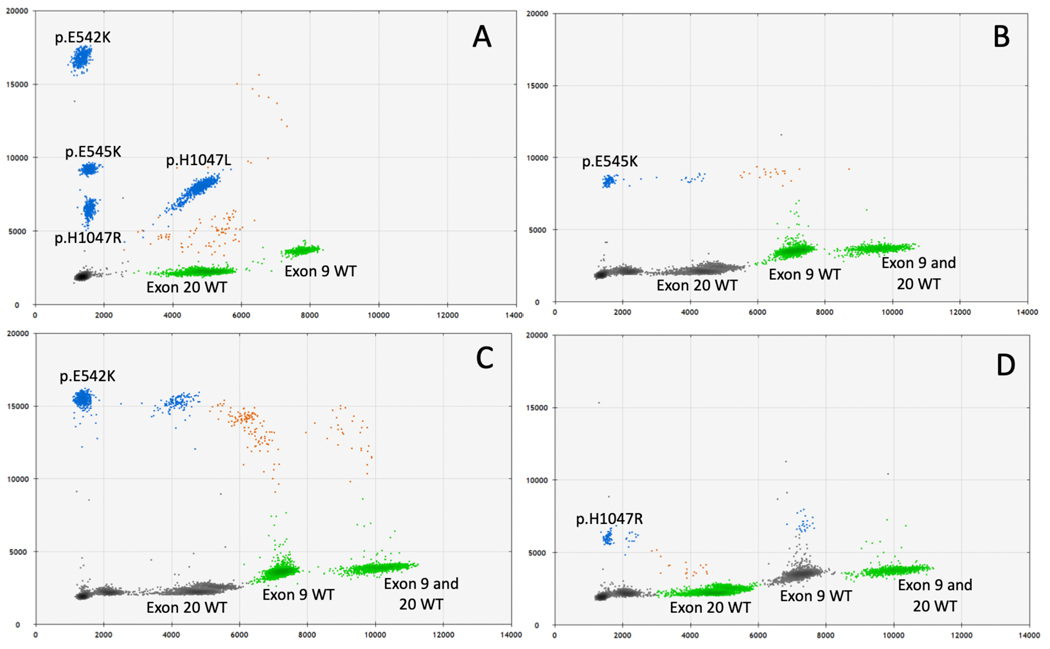
We next sought to determine if we could prospectively identify variants in LM patients who have not undergone surgical resection to obtain tissue for molecular diagnosis. Since the quantity of DNA obtained from cyst fluid can be low, a multiplex assay is ideal for prospective testing to maintain high sensitivity while maximizing use of low concentration samples. We utilized a previously published multiplex ddPCR assay that detects the four most common PIK3CA variants observed in cancer and LM (p.His1047Arg, p.Glu545Lys, p.Glu542Lys, and p.His1047Leu).26 We first tested this assay on synthetic DNA (“gene blocks”) and gDNA samples with known PIK3CA variants previously detected by ddPCR singleplex (Figure 1a, Figure S2). Both variant detection and VAF were reliably reproduced in the multiplex assay (Figure S2).
Figure 1.
PIK3CA multiplex ddPCR validation and performance with cyst fluid cfDNA. Positive controls for each of the four variants detected by multiplex ddPCR superimposed onto a single 2D fluorescence plot, HEX fluorescence on x-axis and FAM fluorescence on y-axis (QuantaSoft software), shows distinct separation of the four variant clusters and two corresponding wild-type (WT) clusters (A). The exon 9 WT cluster corresponds to p.E542K and p.E545K variant clusters while the exon 20 WT cluster corresponds to p.H1047R and p.H1047L clusters. The colors represent different fluorophores or combinations thereof: gray indicates droplets without DNA of interest, blue indicates droplets with variant DNA, green indicates droplets with WT DNA, and orange indicates both variant and WT DNA for the respective locus were present in the same droplet. Representative 2D-plots demonstrating ddPCR multiplex results from cyst fluid cfDNA for three individuals with isolated LM (B-D).
We then tested cyst fluid samples from five patients with isolated LM who had not undergone surgery and did not have tissue available for molecular diagnosis (Table 3). Four individuals had cyst fluid obtained by interventional radiology during sclerotherapy procedures, while one (LR19-481) had cyst fluid aspirated using ultrasound guidance while in the operating room for airway evaluation and gastrostomy tube placement. Using the multiplex assay to test cyst fluid cfDNA, three different PIK3CA variants were detected in four out of five individuals (80%) with VAF ranging from 0.2 – 6.7% (Table 3). The variants and VAF were confirmed using singleplex ddPCR (Table 3). Two individuals (LR16-265 and LR19-442) had the same PIK3CA variant detected in their corresponding cyst fluid pellet gDNA by multiplex ddPCR, though at lower VAF than cyst fluid cfDNA (Supplemental Table 2).
Table 3.
Prospective molecular diagnosis of LM cyst fluid cfDNA with multiplex ddPCR.
| Multiplex ddPCR | Singleplex ddPCR | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Sex | Agea | Location | Cyst fluid volume (mL) | PIK3CA Variant | VAF (%)b | Droplets (Var/WT) |
VAF (%)b | Droplets (Var/ WT) |
| LR16-265c | F | 11 | Axilla | 6.0 | p.H1047R (c.3140A>G) |
1.4 | 124/8172 | 1.4 | 170/10972 |
| LR19-442 | M | 6 | Retro-peritoneum | 9.0 | p.E545K (c.1633G>A) |
0.2 | 27/12098 | 0.2 | 20/11395 |
| LR19-443 | M | 1 mo | Axilla | 2.0 | NEG | NEG | - /30632 | - | - |
| LR19-446 | M | 9 mo | Chest wall | 5.0 | p.E542K (c.1624G>A) |
6.5 | 920/12116 | 6.7 | 856/10878 |
| LR19-481 | M | 9 mo | Neck, lower face | 6.5d | p.E545K (c.1633G>A) |
1.6 | 152/8910 | 1.5 | 162/9662 |
Abbreviations: cfDNA – cell free DNA, mo – months old, mL – milliliters, NEG – no variant detected, VAF – variant allele fraction, Var – variant, WT – wild-type
Age at first cyst fluid collection, in years unless otherwise specified.
VAF calculated using droplet concentrations and only reported for samples in which sample variant concentration was statistically different from WT cfDNA control variant concentration based on 95% total error confidence intervals.
Patient previously reported.11 No variant was found in previous publication when DNA was extracted from 18g needle biopsy of lesion tissue taken at the time of sclerotherapy treatment.
Sample collected in Streck tube.
Additional samples were collected from two individuals, one with isolated LM and another with KTS, during acute infections and were notably purulent. cfDNA isolation of the purulent samples yielded >100,000 ng DNA per mL of cyst fluid, indicating gross cfDNA contamination with white blood cell gDNA. Variants were not detected on multiplex ddPCR of DNA isolated from purulent samples (data not shown).
Discussion
We detected somatic activating variants in cfDNA from plasma and cyst fluid in three subtypes of vascular malformations: AVM, VeM, and LM. To our knowledge, this is the first demonstration of cfDNA based molecular diagnostics for isolated VM, and the first use of cyst fluid cfDNA as a diagnostic analyte.
Cell-free DNA enters the circulation through a variety of processes including necrosis, apoptosis, autophagy, and active secretion.29 Tumor-derived cfDNA is driven by rapid cell turnover in neoplastic conditions.30 While markers of proliferation have been documented in endothelial cells of AVM, cell turnover is not considerably increased in VM, making our ability to detect variants in plasma cfDNA surprising.31 VM develop due to postzygotic somatic mutations in the PI3K or RAS-MAPK pathways.9 Vascular endothelial cells are known to possess the somatic activating variants associated with VM and are hypothesized to be the driving cell type in VM developement,14, 32–35 A recent analysis of cfDNA methylation patterns in healthy individuals demonstrated that 10% of cfDNA was derived from vascular endothelial cells,36 thus our ability to detect variants in plasma from AVM and VeM patients may be due to contact between vascular endothelial cells and the systemic circulation. Overgrowth syndromes, such as CLOVES or facial infiltrating lipomatosis, are associated with the same activating pathogenic variants, however additional cell types beyond endothelial cells are affected.37 At this time it is unclear which cell type(s) may be contributing to the variants detected in plasma cfDNA. Additional studies such as cfDNA methylation analysis or nucleosome footprinting and longitudinal plasma collections before and after VM resection will be needed to address these questions.
We did not detect PIK3CA variants in plasma from patients with isolated LM (Table S1), which is consistent with LM anatomy as they have limited connections to the systemic circulation.28 We detected a PIK3CA variant in plasma from a single patient with an LM in the context of CLOVES syndrome. This is consistent with previous reports and may reflect a larger burden of PIK3CA mutant cells in CLOVES syndrome patients.27
Most isolated LM have a macrocystic or mixed cystic structure (cysts >2cm in diameter) that permits direct sampling of lymphatic fluid.4, 38 We took advantage of this unique compartment to assay lymphatic fluid directly for detection of PIK3CA variants. Every LM with a tissue detected variant had a detectable variant in cyst fluid cfDNA, in some cases at VAF higher than those detected from tissue samples (Table 2). Only a small fraction of cells in the LM possess the variant and there is significant intralesional heterogeneity, therefore multiple lesion samples may be required to establish a molecular diagnosis.11 Given our prior results suggesting a single LM sample may be insufficient for variant detection, we did not expect every cyst fluid sample to be variant positive. Interestingly, the cfDNA compartment was distinct from the pelleted material from which gDNA was extracted. In the retrospective cohort, only two of seven cyst fluid pellets were variant positive (Table 2). The superiority of cfDNA over pellet gDNA was also observed in a study of somatic mutation detection from pleural effusions in lung cancer.39 We hypothesize that the static nature of lymphatic fluid in LM, combined with its proximity to mutant lymphatic endothelial cells may result in an enrichment of variant PIK3CA molecules in cfDNA. Further work is needed to define the origin of variant PIK3CA molecules in cyst fluid cfDNA. Regardless, these results suggest that LM cyst fluid may be as or more sensitive an analyte than LM tissue for molecular diagnosis.
In addition to confirming known, tissue based PIK3CA variants, we demonstrated that cyst fluid cfDNA could be used as a first line diagnostic test, prior to surgery and any knowledge of the tissue-based variant. The variant spectrum in LM is narrow with four recurrent PIK3CA-activating variants accounting for >95% of identified variants.10, 11 Taking advantage of this narrow spectrum, we prospectively tested LM cyst fluid cfDNA for these four variants at once using a multiplex ddPCR assay previously developed for detection of PIK3CA variants in plasma cfDNA from individuals with breast cancer.26 We detected PIK3CA variants in four of five individuals with LM who have never undergone surgical resection (Table 3).
Most prior studies of VM genetics, and mosaic disease in general, have only included individuals with accessible cutaneous manifestations or lesions necessitating surgery, biasing analyses towards more severe presentations. For example, one-third of LM patients in a previous study of genotype-phenotype associations were high grade (de Serres stage IV or V), which contributes to only 15% of head and neck LM.11, 38, 40 Molecular diagnosis of individuals using cfDNA from either plasma or cyst fluid will provide opportunities to include a wider array of patients in future studies.
Historically VM were treated surgically, however the field is moving towards targeted pharmacotherapy for initial treatment. A PI3Kα inhibitor, alpelisib, recently demonstrated benefit in a cohort of 19 patients with PIK3CA-related overgrowth spectrum (PROS), of which LM is a subgroup.17 The efficacy of alpelisib and miransertib, another PI3K-AKT-MTOR pathway inhibitor, are currently being examined in patients with PROS who have a documented PIK3CA variant (clinicaltrials.gov: NCT04085653, NCT03094832. Before this study, surgical debulking was the only avenue for obtaining material for genetic diagnosis. Cell-free DNA based molecular diagnoses may provide opportunities for patients with disease not amenable to surgery, or not severe enough to warrant surgery, to receive targeted medical therapy for their specific variant. Furthermore, a cfDNA based approach may not only eliminate the need for debulking surgery prior to pharmacotherapy but may obviate the need for surgery altogether if targeted medical therapy proves effective.
The primary limitations of this study are small sample size and heterogeneity in plasma and cyst fluid collection. Small sample size is a constant challenge in the study of rare disease. We observed clear differences in DNA recovery based on plasma volumes and collection in either EDTA or Streck tubes. DNA concentration from EDTA-collected frozen plasma samples was higher than DNA concentration from Streck tubes, likely indicating gDNA contamination which reduces the sensitivity of variant detection assays. Many of these samples would be rejected by clinical lab quality control metrics, however we believe inclusion of all samples in this study strengthened our findings by demonstrating feasibility despite imperfect starting material. We hypothesize that larger sample volumes collected into Streck tubes with less contaminating gDNA will increase diagnostic yield in VeM and AVM. Future studies, including larger cohorts of patients with uniform sample collection, will be needed to define the true diagnostic sensitivity of cfDNA in VM patients.
Our criteria for determining positivity based on comparison of variant concentration confidence intervals are based on the strictest interpretation ddPCR results. Multiple groups have used the presence of ≥ 3 variant droplets as sufficient evidence for variant dection.26, 27 In this study we compared variant concentration to WT control variant concentration and only samples with non-overlapping total error confidence intervals were considered positive (Figure S1). We chose to accept a higher false negative rate to increase confidence when a positive result was observed. Although ddPCR is not currently in wide use in clinical laboratories for diagnosis of somatic variants, the platform is FDA approved for detection of BCR-ABL translocations in patients with chronic myeloid leukemia41, and we expect its usage to expand in clinical diagnostics for mosaic disorders like VM.
In conclusion, we detected somatic activating variants in cfDNA obtained from plasma and cyst fluid in patients with AVM, VeM, and LM. Multiplex ddPCR allowed for prospective molecular diagnosis from LM cyst fluid cfDNA in individuals without LM tissue resection. This represents an opportunity to initiate targeted medical therapy in VM patients prior to surgery. Furthermore, our results support future exploration of the use of cfDNA in diagnosis of non-neoplastic congenital disorders due to post-zygotic activating variants.
Supplementary Material
Acknowledgements
We thank the affected individuals, their families, and referring physicians for their important contribution to our ongoing work on these disorders. This study was funded by the US National Institutes of Health under NHLBI grants F32HL147398 (to K.Z.) and 1RO1HL103996 (to W.B.D.), a Seattle Children’s Hospital Guild Association Funding Focus Award (to J.A.P.), and Burroughs Wellcome Career Award for Medical Scientists 1014700 (to J.T.B.). We thank the Brotman Baty Institute for Precision Medicine for sharing their protocols and expertise on cfDNA isolation. We would also like to acknowledge the Seattle Children’s Vascular Anomalies Program and especially the interventional radiology team for their support in sample collection. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the funding sources.
Footnotes
Conflict of Interest Statement:
R. A. Bly: Co-founder EigenHealth, Inc, Consultant to SpiWay, LLC. Dr. Randall Bly holds a financial interest of ownership equity with Edus Health, Inc.
The remaining authors have declared that no conflict of interest exists.
Citations
- 1.Wassef M, Blei F, Adams D, et al. Vascular Anomalies Classification: Recommendations From the International Society for the Study of Vascular Anomalies. Pediatrics. 2015;136(1):e203–14. [DOI] [PubMed] [Google Scholar]
- 2.Bhatt N, Perakis H, Watts TL, Borders JC. Traumatic hemorrhage and rapid expansion of a cervical lymphatic malformation. Ear Nose Throat J. 2011;90(1):20–2. [DOI] [PubMed] [Google Scholar]
- 3.Liu AS, Mulliken JB, Zurakowski D, Fishman SJ, Greene AK. Extracranial arteriovenous malformations: natural progression and recurrence after treatment. Plastic and reconstructive surgery. 2010;125(4):1185–94. [DOI] [PubMed] [Google Scholar]
- 4.Adams MT, Saltzman B, Perkins JA. Head and neck lymphatic malformation treatment: a systematic review. Otolaryngol Head Neck Surg. 2012;147(4):627–39. [DOI] [PubMed] [Google Scholar]
- 5.Behravesh S, Yakes W, Gupta N, et al. Venous malformations: Clinical diagnosis and treatment. Cardiovascular Diagnosis and Therapy. 2016;6(6):557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elluru RG, Balakrishnan K, Padua HM. Lymphatic malformations: diagnosis and management. Semin Pediatr Surg. 2014;23(4):178–85. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert P, Dubois J, Giroux MF, Soulez G. New Treatment Approaches to Arteriovenous Malformations. Semin Intervent Radiol. 2017;34(3):258–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hage AN, Beecham Chick JF, Srinivasa RN, et al. Treatment of Venous Malformations: The Data, Where We Are, and How It Is Done. Techniques in Vascular and Interventional Radiology. 2018;21(2):45–54. [DOI] [PubMed] [Google Scholar]
- 9.Brouillard P, Vikkula M. Genetic causes of vascular malformations. Hum Mol Genet. 2007;16 Spec No. 2:R140–9. [DOI] [PubMed] [Google Scholar]
- 10.Luks VL, Kamitaki N, Vivero MP, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. The Journal of pediatrics. 2015;166(4):1048–54.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zenner K, Cheng CV, Jensen DM, et al. Genotype correlates with clinical severity in PIK3CA-associated lymphatic malformations. JCI insight. 2019;4(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limaye N, Wouters V, Uebelhoer M, et al. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat Genet. 2009;41(1):118–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Limaye N, Kangas J, Mendola A, et al. Somatic Activating PIK3CA Mutations Cause Venous Malformation. American journal of human genetics. 2015;97(6):914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Couto JA, Huang AY, Konczyk DJ, et al. Somatic MAP2K1 Mutations Are Associated with Extracranial Arteriovenous Malformation. American journal of human genetics. 2017;100(3):546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolaev SI, Vetiska S, Bonilla X, et al. Somatic Activating KRAS Mutations in Arteriovenous Malformations of the Brain. N Engl J Med. 2018;378(3):250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Olabi L, Polubothu S, Dowsett K, et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J Clin Invest. 2018;128(4):1496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venot Q, Blanc T, Rabia SH, et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558(7711):540–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lekwuttikarn R, Lim YH, Admani S, Choate KA, Teng JMC. Genotype-Guided Medical Treatment of an Arteriovenous Malformation in a Child. JAMA Dermatology. 2019;155(2):256–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams DM, Trenor CC 3rd, Hammill AM, et al. Efficacy and Safety of Sirolimus in the Treatment of Complicated Vascular Anomalies. Pediatrics. 2016;137(2):e20153257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker VER, Keppler-Noreuil KM, Faivre L, et al. Safety and efficacy of low-dose sirolimus in the PIK3CA-related overgrowth spectrum. Genet Med. 2019;21(5):1189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Triana P, Dore M, Cerezo VN, et al. Sirolimus in the Treatment of Vascular Anomalies. Eur J Pediatr Surg. 2017;27(1):86–90. [DOI] [PubMed] [Google Scholar]
- 22.Hammer J, Seront E, Duez S, et al. Sirolimus is efficacious in treatment for extensive and/or complex slow-flow vascular malformations: a monocentric prospective phase II study. Orphanet J Rare Dis. 2018;13(1):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nature reviews Cancer. 2017;17(4):223–38. [DOI] [PubMed] [Google Scholar]
- 24.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature reviews Cancer. 2011;11(6):426–37. [DOI] [PubMed] [Google Scholar]
- 25.BioRad. Droplet Digital PCR Applications Guide [Available from: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf. [Google Scholar]
- 26.Rowlands V, Rutkowski AJ, Meuser E, Carr TH, Harrington EA, Barrett JC. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci Rep. 2019;9(1):12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biderman Waberski M, Lindhurst M, Keppler-Noreuil KM, et al. Urine cell-free DNA is a biomarker for nephroblastomatosis or Wilms tumor in PIK3CA-related overgrowth spectrum (PROS). Genet Med. 2018;20(9):1077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whimster IW. The pathology of lymphangioma circumscriptum. Br J Dermatol. 1976;94(5):473–86. [DOI] [PubMed] [Google Scholar]
- 29.Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc. 2018;93(3):1649–83. [DOI] [PubMed] [Google Scholar]
- 30.Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PloS one. 2011;6(9):e23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meijer‐Jorna LB, van der Loos CM, de Boer OJ, van der Horst C. Microvascular proliferation in congenital vascular malformations of skin and soft tissue. J Clin Pathol. 602007. p. 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaser K, Dickie P, Neilson D, Osborn A, Dickie BH. Linkage of Metabolic Defects to Activated PIK3CA Alleles in Endothelial Cells Derived from Lymphatic Malformation. Lymphat Res Biol. 2018;16(1):43–55. [DOI] [PubMed] [Google Scholar]
- 33.Osborn AJ, Dickie P, Neilson DE, et al. Activating PIK3CA alleles and lymphangiogenic phenotype of lymphatic endothelial cells isolated from lymphatic malformations. Hum Mol Genet. 2015;24(4):926–38. [DOI] [PubMed] [Google Scholar]
- 34.Blesinger H, Kaulfus S, Aung T, et al. PIK3CA mutations are specifically localized to lymphatic endothelial cells of lymphatic malformations. PLoS ONE [Electronic Resource]. 2018;13(7):e0200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goss JA, Huang AY, Smith E, et al. Somatic mutations in intracranial arteriovenous malformations. PloS one. 2019;14(12):e0226852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moss J, Magenheim J, Neiman D, et al. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9(1):5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couto JA, Konczyk DJ, Vivero MP, et al. Somatic PIK3CA mutations are present in multiple tissues of facial infiltrating lipomatosis. Pediatr Res. 2017;82(5):850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balakrishnan K, Menezes MD, Chen BS, Magit AE, Perkins JA. Primary surgery vs primary sclerotherapy for head and neck lymphatic malformations. JAMA Otolaryngol Head Neck Surg. 2014;140(1):41–5. [DOI] [PubMed] [Google Scholar]
- 39.Xiang C, Huo M, Ma S, et al. Molecular Profiling for Supernatants and Matched Cell Pellets of Pleural Effusions in Non-Small-Cell Lung Cancer. J Mol Diagn. 2020;22(4):513–22. [DOI] [PubMed] [Google Scholar]
- 40.de Serres LM, Sie KC, Richardson MA. Lymphatic malformations of the head and neck. A proposal for staging. Arch Otolaryngol Head Neck Surg. 1995;121(5):577–82. [DOI] [PubMed] [Google Scholar]
- 41.Chung HJ, Hur M, Yoon S, et al. Performance Evaluation of the QXDx BCR-ABL %IS Droplet Digital PCR Assay. Ann Lab Med. 2020;40(1):72–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.