Abstract
Staphylococcus aureus is a common cause of invasive and life-threatening infections that are often multidrug resistant. To develop novel treatment approaches, a detailed understanding of the complex host–pathogen interactions during infection is essential. This is particularly true for the molecular processes that govern the formation of tissue abscesses, as these heterogeneous structures are important contributors to staphylococcal pathogenicity. To fully characterize the developmental process leading to mature abscesses, temporal and spatial analytical approaches are required. Spatially targeted proteomic technologies such as microliquid extraction surface analysis offer insight into complex biological systems including detection of bacterial proteins and their abundance in the host environment. By analyzing the proteomic constituents of different abscess regions across the course of infection, we defined the immune response and bacterial contribution to abscess development through spatial and temporal proteomic assessment. The information gathered was mapped to biochemical pathways to characterize the metabolic processes and immune strategies employed by the host. These data provide insights into the physiological state of bacteria within abscesses and elucidate pathogenic processes at the host–pathogen interface.
Keywords: abscess formation, host–pathogen interface, microLESA, proteomics, Staphylococcus aureus
Graphical Abstract
Staphylococcus aureus is one of the leading causes of bloodstream infections worldwide.1 In the United States alone, this bacterium is responsible for more than 600,000 hospitalizations annually,2 and patients with S. aureus bacteremia have a 30-day mortality rate of 20%.3 S. aureus bacteremia often spreads to other body sites, leading to the formation of abscesses, most commonly affecting the liver, kidneys, brain, and heart tissues.4
The formation of organ abscesses is a critical strategy to ensure S. aureus survival within the host.5 Abscesses offer a temporary refuge for S. aureus, allowing the enclosed bacteria to resist the actions of the immune system, thereby securing persistence within host tissues.6,7 In a murine model of systemic infection, the formation of soft tissue abscesses follows distinct phases (stages I–IV),5 driven by the recruitment of immune cells (e.g., neutrophils) ~2 days post infection (dpi)7 and the development of a bacterial nidus in the center of the abscess ~4–5 dpi (staphylococcal abscess colony, SAC).7 The developing SAC is surrounded by necrotic tissue, a fibrin pseudocapsule,7,8 and an outer microcolony-associated meshwork.6 At the end of abscess development (~15–30 dpi), the persistent and increasingly larger lesions rupture and release bacteria that seed new abscesses or cause secondary infections.7
Abscess formation requires the involvement of both host and bacterial factors. Several staphylococcal proteins, including both coagulases Coa and vWbp, are essential for fibrin pseudocapsule formation and abscess development.9 Additionally, a few other S. aureus proteins associated with virulence (e.g., Emp, Eap, Hla, IsdA, IsdB, SdrC, SrtA, Spa) have been found to be required for abscess formation.6–8,10,11 The majority of these data describing bacterial contributions to abscess development were generated from histological stains or characterization of isogenic S. aureus mutants and their ability to persist within host tissues. However, unbiased studies that aim to assess the bacterial abscess proteome are sparse. This is similarly true for proteinaceous host factors in proximity to the abscess. While it is established that specific cell types (e.g., fibroblasts and neutrophils12) or immune proteins (e.g., the metal scavenging protein calprotectin13) play essential roles during the immune response to S. aureus, we lack a detailed understanding of specific cellular processes involved in the host response to tissue colonization by S. aureus. Although a previous study from our laboratory addressed some of these shortcomings by assessing the proteinaceous composition of kidneys infected with S. aureus,14 the current study also examines the temporal and spatial aspects of abscess development.
Various proteomic technologies offer insight into complex biological systems, including the interplay between host and pathogen during infection.15,16 Recent technological advances have enabled pairing of spatially targeted surface sampling with high-performance mass spectrometry for molecular analysis of tissue. Imaging mass spectrometry (IMS) technology, such as matrix-assisted laser desorption/ionization (MALDI) IMS, offers the unique combination of high molecular specificity and high spatial resolution imaging.17–19 However, the sensitivity of MALDI IMS for analysis of large proteins (>30 kDa) can be limited due to low ionization efficiency of proteins from tissue and the poor transmission efficiency of high mass-to-charge ratio (m/z) ions.19,20 The identification of proteins with MALDI IMS is also challenging due to inefficient fragmentation of low charge state species. To facilitate this process, we have investigated the utility of complementary surface sampling technologies for discovery-based proteomic studies.
Traditional liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) of peptides derived from proteolytic digestion provides the greatest proteomic sensitivity.21 LC-MS/MS requires liquid samples, usually through homogenization of tissue, limiting spatial information from the sample of interest. To address this, we recently introduced a spatially targeted, bottom-up proteomics workflow used in analysis of S. aureus kidney infection.22 Specific foci were targeted using picoliter-sized droplets of trypsin protease, and the resulting proteolytic peptides were sampled using liquid extraction surface analysis (LESA). The entire process, termed microLESA, is histologically guided using autofluorescence microscopy. Herein, we expand on this work using microLESA to analyze regions from the abscess community (SAC), host–pathogen interface surrounding the community, and regions of cortical tissue of S. aureus infected kidneys. Spatial and temporal proteomic changes were examined by sampling different time points, allowing us to follow responses of both the pathogen and the immune system over the course of infection. By studying three defined regions, and their dynamics over the course of the infection, changes in both host and pathogen can be observed across the organ.
RESULTS AND DISCUSSION
Identification of Bacterial and Host Proteins in Infected Tissue.
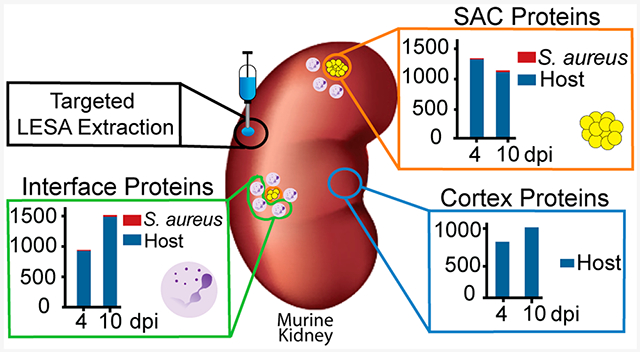
To explore the role of host and bacterial proteins throughout the development of staphylococcal organ abscesses, we analyzed proteins in the abscesses and surrounding areas over time. We focused on kidneys as one of the most commonly utilized model systems for staphylococcal abscess formation. Samples were extracted from one of three defined regions (SAC, interface, and cortex; shown in Figure 1A) at 4 and 10 dpi (Figure 1B). These extractions were guided by fluorescence microscopy images (transgenic fluorophore and autofluorescence), which highlights the fluorescent bacterial communities and overall tissue morphology that allowed for differentiation between SAC and adjacent regions. The two time points were selected to ensure sufficient abscess size for extraction, as at 4 dpi, abscesses are consistently large enough for micoLESA extraction, and at 10 dpi, the infection has greatly progressed. We hypothesized that the unique spatial and temporal proteomic analysis would discern (i) S. aureus physiology and production of virulence determinants within the abscess microenvironment (SAC), (ii) onset of the immune response including infection-mediated influx and action of immune cells (interface), as well as (iii) organ-wide responses to infection (cortex). A total of 2399 proteins were identified across all three regions of interest (ROI) and two time points (Tables S1 and S2) with an average of 1153 proteins per time point and ROI (Figure 1B, Tables S1 and S2). Of the proteins detected, 32 were of bacterial origin (Table S2). We identified an additional 30 bacterial proteins that were present in only one set of serial sections or one biological replicate and are not included in Figure 1B due to lower confidence (Table S3). This variability in detection could result from low protein abundance or heterogeneity among abscesses, a known challenge when studying abscess formation.23,24 Nevertheless, we conclude the identification of these proteins is a strength of the method, as bacterial proteins from S. aureus infections are difficult to detect and measure due to ion suppression effects from abundant host proteins within tissues. Since we spatially targeted the abscess region, the inherent “chemical noise” from the highly abundant host proteins was reduced, greatly improving the sensitivity for the bacterial proteins. Improving the coverage of the bacterial proteome detectable within the tissue microenvironment provided a more complete description of how S. aureus molecular machinery contributes to abscess formation and progression.
Figure 1.
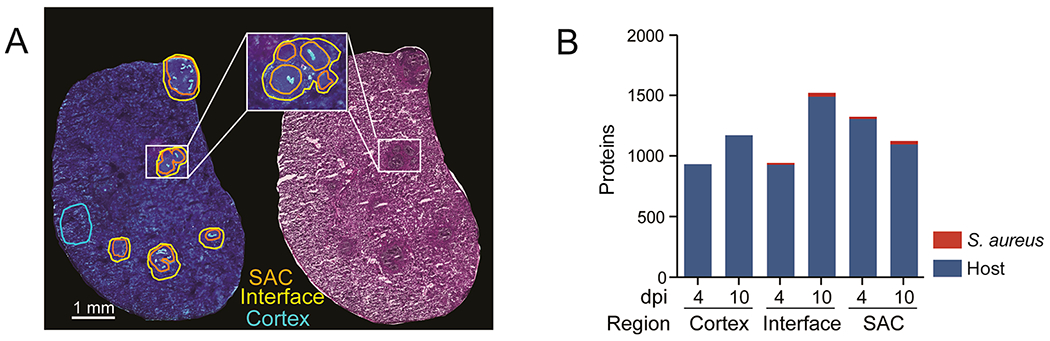
microLESA sampling strategy and results. (A) Schematic of regions selected for collection shown using autofluorescence and H&E. (B) Overview of bacterial and host proteins identified at different time points and across different regions within infected renal tissue. Total host proteins: 2367. Total bacterial proteins: 32.
Bacterial Factors.
Detection of High Abundance Proteins Related to Translation.
The microLESA workflow detected 32 bacterial proteins in the defined SAC and interface regions, several of which are involved in the translation process (Figure 2). These include ribosomal proteins (50S ribosomal proteins L7/L12 [RplL] and L17 [RplQ]), a tRNA-ligase (ThrS) as well as elongation factors Tu (Tuf)25 and Ts (Tsf).26 Several of these intracellular highly abundant proteins were detected in the SAC region and also in the abscess interface (Figure 2). While we cannot exclude that a small fraction of the bacterial population resides outside of the observable SAC or the potential for these proteins to “leak” from the SAC into adjacent regions during the sample preparation process, cell death or autolysin (Atl)-mediated lysis of a staphylococcal subpopulation may cause the release of bacterial factors into the abscess microenvironment.27 It was previously shown that decreased autolysis by S. aureus results in a moderate decrease in renal bacterial numbers in a murine model of systemic infection at 7 dpi,28 supporting the notion that Atl plays a role during spread to or colonization of this organ. While our data do not allow us to finally conclude why we detect cellular bacterial proteins outside of the SAC, it is intriguing to speculate that (auto)lytic processes are important during abscess formation, potentially aiding infection through the release of intracellular factors.
Figure 2.
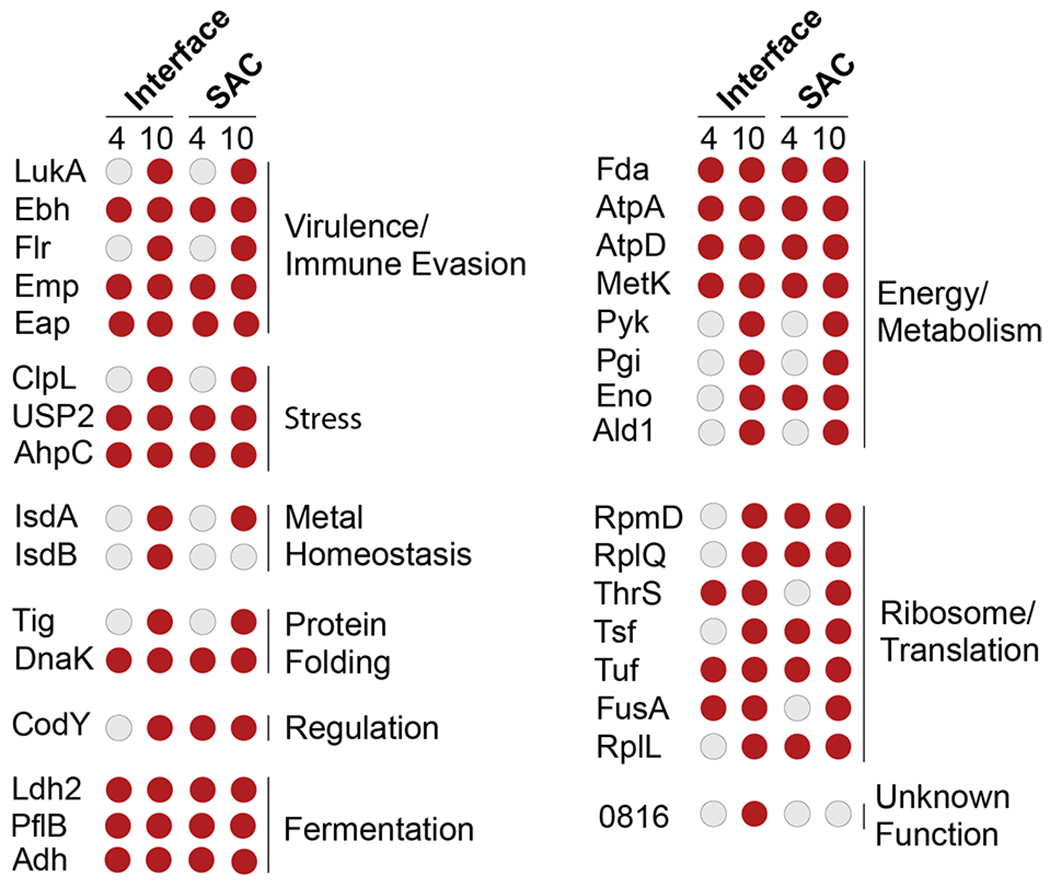
Overview of S. aureus proteins identified in the different abscess regions. Staphylococcal proteins were found in the interface and SAC of renal abscesses at 4 or 10 dpi. Red circles denote proteins that were present at the specific time point/region, while gray circles depict the absence of a specific protein. General functional categories for proteins and protein groups are shown. Further information concerning proteins in this list can be found in S2. 0816: SAUSA300_0816; 1656: SAUSA300_1656.
S. aureus Heme Metabolism Proteins in the Abscess.
In response to the presence of S. aureus in tissue, the host immune system initiates a variety of antibacterial strategies. Nutritional immunity is one such approach in which the immune system limits bacterial access to essential micronutrients (e.g., transition metals) to hinder bacterial growth and stall infection progression.29 Host-imposed Fe-starvation within the abscess was recently shown by our group using in vivo imaging and IMS strategies.23,24 Bacterial pathogens have evolved a number of mechanisms to overcome this Fe limitation.29 To ensure sufficient levels of the Fe-containing cofactor heme, S. aureus imports host heme through the action of the Isd heme uptake system.30 Additionally, heme can be synthesized de novo via a coproporphyrin-dependent bacterial heme biosynthesis pathway31,32. In our analysis, we observed components of the Isd system (IsdA and IsdB: Figure 2, Table S2 as well as ChdC (identified with lower confidence, Table S3, a member of the heme biosynthetic pathway, to be present within the abscess.33 Identification of proteins involved in heme uptake as well as de novo heme synthesis suggests that (i) S. aureus within renal abscesses is indeed heme starved and (ii) that multiple strategies are employed to overcome this limitation. These proteins were only detected at 10 dpi (Figure 2), indicating that metal starvation and corresponding bacterial responses are likely more abundant at later time points during infection.
Bacterial Factors Related to Protein Stress.
Other host-imposed stresses, including production of reactive oxygen species (ROS), reactive nitrogen species (RNS), and elevated temperatures during fever, can damage the bacterial protein pool.34–37 To protect the staphylococcal proteins from host-derived stresses, bacterial inducible heat shock proteins (Hsps)38 assist with protein folding and proteostasis.39 Although this study cannot distinguish between basal and stress-induced expression of such factors, the detection of the major bacterial Hsp DnaK at both the 4 dpi and 10 dpi time points (Figure 2, Table S2) is in accordance with the established roles of Hsps during stress and contact with the host. DnaK is not only involved in maintenance of cellular protein pools, but also plays a role in the folding of de novo synthesized proteins, e.g. when adapting to changing environmental conditions.40 This function is performed in concert with other chaperones, such as a ribosome-associated intracellular peptidyl prolyl cis/trans isomerase (PPIase),41 referred to as Trigger factor (TF, Tig),42 detected in the SAC at 10 dpi (Figure 2, Table S2). The cooperative nature of these two proteins during stress conditions is demonstrated by both dnak and tig mutants being viable under laboratory conditions, but a DnaK/TF double mutant being synthetic lethality at temperatures above 30 °C. This lethality is likely due to increased protein misfolding under these conditions.42 In line with the synergism nature of these proteins, we also found TF to be associated with the SAC at 10 dpi (Figure 2, Table S2). To our knowledge, the importance of DnaK and TF in S. aureus virulence has not been investigated in vivo, and it is intriguing to speculate about the importance of these systems during abscess development. The roles of TF are of particular interest because another S. aureus PPIase, PpiB, was previously shown to impact S. aureus virulence through a potential function in secretion of nucleolytic and hemolytic proteins.43
S. aureus Metabolism Is Shaped by the Host Environment.
S. aureus can circumvent cellular pathways that have been disrupted by macrophage-derived nitrosative stress41,44 or hypoxia at sites of infection.45 Specifically, fermentative pathways46 can be employed during glucose catabolism under hypoxia or if oxidative phosphorylation is impaired due to the damage of terminal oxidases by radical nitric oxide.47
Fermentation of pyruvate to lactate by S. aureus is facilitated by different lactate dehydrogenases (i.e., Ldh1, Ldh2, Ddh).46 We detected Ldh2 in the SAC and interface at 4 and 10 dpi (Figure 2, Table S2), and Ldh1, a nitric oxide-inducible lactate dehydrogenase, in the abscesses at 4 and 10 dpi, as well as in the interface at 10 dpi (Table S3). It was previously shown that loss of Idh1 increases staphylococcal sensitivity to nitrosative stress and decreases the ability to form renal abscesses.46 Furthermore, the additional loss of ldh2 was found to augment the latter phenotype,46 highlighting the importance of fermentative metabolism for S. aureus pathogenicity. In addition to maintaining the cells ability to generate energy under conditions encountered in the abscess, the creation of lactate as a byproduct of fermentation was recently shown to aid in staphylococcal immune evasion.48 Briefly, bacterial-derived lactate causes alterations of gene expression in host immune cells, e.g. stimulates the production of the anti-inflammatory cytokine Il-10, allowing for persistence in host tissue. These exciting findings further emphasize how bacterial metabolism shapes to its behavior as a pathogen and ultimately its interaction with the immune system.
Activity of metabolic pathways in S. aureus is dependent on environmental conditions and controlled by a large number of transcriptional regulators.49 Analogously, various regulatory proteins govern pathogenesis of the bacterium.50 Because the production of the vast array of virulence factors encoded by S. aureus is energetically costly, cellular nutritional status and virulence are intimately linked, e.g. through the action of transcriptional regulators that sense and respond to alterations in nutrient availability.51 A prime example of such a connection is the global transcriptional regulator CodY.52,53 This bifunctional regulator senses the availability of branched chain amino acids54,55 and GTP56 and responds by controlling a large number of metabolic and virulence-related traits. We detected CodY, as only one of two bona fide bacterial transcriptional regulators in our data set, at 4 and 10 dpi in the SAC and at 10 dpi in the interface (Figure 2, Table S2). While physical presence of the protein itself is not indicative of its regulatory state, the detection of CodY serves as reminder that S. aureus pathophysiology is directly affected by the conditions encountered in the abscess microenvironment and that sensing of these stimuli guides virulence of the bacterium within the abscess.
S. aureus Secreted Proteins.
In addition to the intracellular factors discussed thus far, we identified several staphylococcal proteins that are actively secreted into the abscess environment. Two adhesins belonging to the group of secretable expanded repertoire adhesive molecules (SERAMs)57 were detected: the extracellular matrix binding protein (Emp)58,59 and the extracellular adhesion protein (Eap).60 Both proteins were among the most consistently observed molecules in the SAC as well as in the interface (Figure 2, Table S2). Of note, our data set was manually screened for the presence of Eap as the encoding sequence is found in the USA300 LAC genome, but it is not annotated as a protein. Using this approach, we identified peptides for Eap, indicating production of this protein within host tissue. The presence of Emp and Eap is concurrent with their roles in abscess formation and maintenance.61 Detection of both proteins in all samples at 4 and 10 dpi supports the notion that these factors are abundant and important in both the early and late stages of abscess development.7
The action of neutrophils is essential for the immune system to clear staphylococcal infection. Neutrophils are recruited to the site of infection after the recognition of host or pathogen derived factors, including C5a, bacterial formylated peptides, or antimicrobial peptides. Corresponding receptors for these signals are the neutrophil receptor C5aR, the formyl peptide receptor (FPR), and the formyl peptide receptor-like-1 (FPRL1).62,63 To decrease the negative impact of neutrophil recruitment, S. aureus secretes chemotaxis inhibitory proteins that limit the infiltration of immune cells to the site of infection. These secreted factors bind to neutrophil receptors, therefore blocking the recognition of infection-related signal molecules and ultimately preventing neutrophil recruitment. Staphylococcal antichemotactic factors include, among others,64 the chemotaxis inhibitory protein of S. aureus (CHIPS) that antagonizes C5aR and FPR65 and the FPRL1 inhibitory protein (FLIPr) that blocks the FPRL1 and FPR receptors66. Both proteins were identified by our analysis (FLIPr: high confidence, Figure 2, Table S2; CHIPS: lower confidence, Table S3). Notably, we were only able to detect CHIPS and FLIPr at 10 dpi, suggesting that these factors are expressed or accumulate at advanced stages of abscess development. To our knowledge, this is the first time that production of these chemotaxis inhibitory proteins has been detected by an unbiased in vivo screen.
Another staphylococcal strategy to minimize the impact of the recruited immune cells is the secretion of pore-forming toxins.67 Upon secretion by S. aureus, these proteins insert into and disrupt the plasma membrane of host cells (e.g., neutrophils), ultimately leading to immune cell death. We identified both components of the bicomponent leukotoxin LukAB68,69 (LukA: high confidence, LukB: lower confidence) at 10 dpi (Figure 2, Tables S2 and S3). Presence of these proteins was recently correlated to abscess formation, additionally validating our method.68 LukB, as well as several other proteins identified by our microLESA approach (i.e., Chp, Emp, and Eap) are expressed under the control of the major virulence regulatory system SaeRS,70 whose activity can be modulated in response to host-derived signals (e.g., Zn-bound calprotectin or human neutrophil peptides).71,72 These results indicate that the SaeRS system may be active during adaptation to the abscess microenvironment and further highlight the interconnected nature of the host–pathogen interface, where the actions of host and bacteria are inseparably intertwined.
Host-Derived Factors.
Spatial and Temporal Changes in the Host Proteome.
The majority of proteins identified in this study were of host origin. The large number of murine proteins detected (2368) allowed for the probing of the relationships between different tissue regions as well as the determination of the proteomic changes within regions over time. At 4 dpi, the majority of identified proteins (676) were common among all three regions. This suggests that 4 days may not be sufficient for the full immune response to be observed and proteinaceously distinct abscess regions to form. The most unique region at this time point is the SAC (Figure 3). The SAC region also displayed the least changes over the course of infection, where the vast majority of proteins in the region were present at both time points (964, Figure 3B). This indicates that once an abscess community has been formed, the makeup of this region appears to remain fairly stable. At 4 dpi, the interface and SAC show high degrees of similarity with an overlap of 211 proteins. As the infection progresses this overlapping region becomes one of the most prominent groups represented in our data set (635) after proteins unique to the cortex (657), while the overlap between all three regions is less than at 4 dpi (441 vs 676). This indicates that as the infection continues, the site of infection (SAC and interface) and the cortex grow increasingly more distinct. As depicted in Figure 3B, only half of the proteins detected in the cortex were found at 4 and 10 dpi, which is in stark contrast to the trend seen in the SAC. These large-scale changes in the cortex suggest organ-wide effects of infection by S. aureus. We hypothesize that this increase in unique proteins in the cortex is likely due to the resolution of the immune response, in regions that are not in direct contact with the pathogen. This hypothesis is discussed further in the host response section regarding metabolism.
Figure 3.
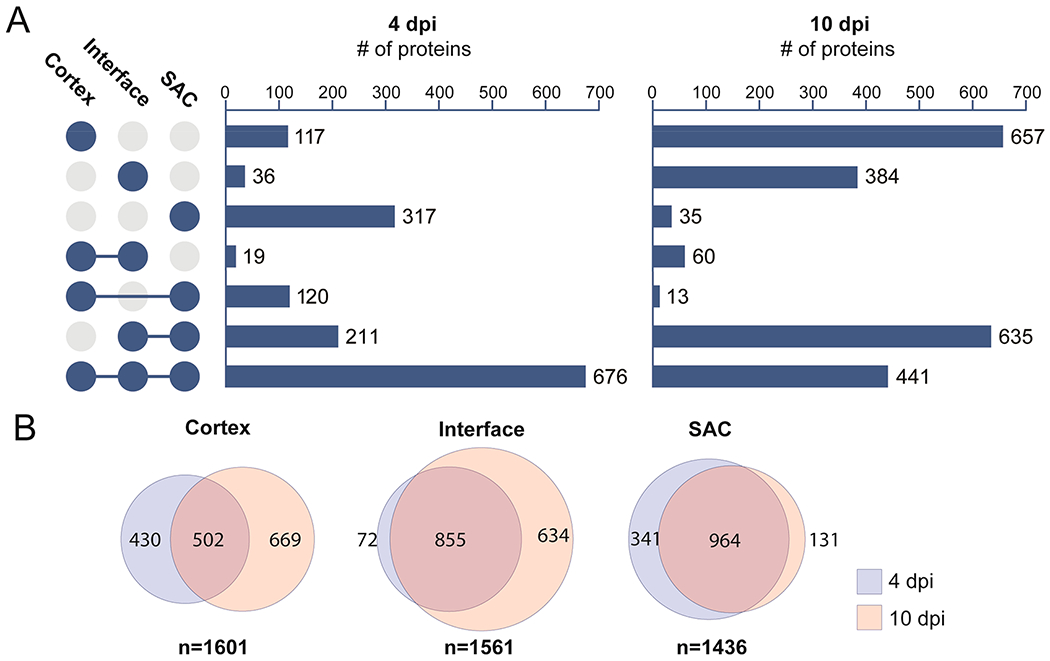
Comparison of the proteome from the different regions in and around staphylococcal tissue abscesses. Data are combined host and bacterial proteins. (A) UpSet plot displaying unique and shared proteins identified from the three sampled locations. (B) Comparison of the proteome from individual regions over the course of infections (4 vs 10 dpi).
Among the investigated regions, the most notable trends are in the interface. The number of proteins detected only in the interface showed dramatic change over the course of the experiment, increasing greater than 10-fold from 4 dpi (36 proteins) to 10 dpi (384 proteins) as shown in Figure 3. Additionally, many of the proteins found in the SAC at 4 dpi are shared with the interface by 10 dpi (635 proteins), suggesting that the interface becomes a major site of the competition between host and pathogen. The finding that many proteins were uniquely detected at the interface suggests that we are able to assess the proteome of this region without unwanted contamination from neighboring regions. If the host proteins were largely shared between the two regions, then unintended cross-contamination between the SAC and the interface would be a concern. This was not observed, however, in this data set. These findings also support the observation of bacterial cytosolic proteins detected at the interface indeed is biologically relevant and not merely an artifact of sample acquisition. Our results indicate that the interface is a unique and deeply informative tissue environment for understanding staphylococcal pathogenesis.
Immune Cell Distributions.
Similar to the rich biology seen from our analysis of bacterial proteins, thousands of identified murine proteins characterize the host response to S. aureus infection. A summary of all identified host-derived proteins, including information about localization and time of identification, can be found in Table S1. Consistent with a predicted presence of immune cells in the abscess, the panleukocyte marker Receptor-type tyrosine-protein phosphatase C (Ptprc or CD45) is present in the SAC and interface at 4 and 10 dpi (Figure 4). Curiously, other proteins thought to be specifically expressed by cells of hematopoietic lineages do not follow the same pattern as CD45. Lymphocyte-specific protein 1 (Lsp1) is also found in the cortical regions of the kidney both 4 and 10 dpi, while dedicator of cytokinesis protein 2 (Dock2) is only found in the abscess and interface 10 dpi (Figure 4). This suggests subtle spatial and temporal differences in the leukocyte populations during infection.
Figure 4.
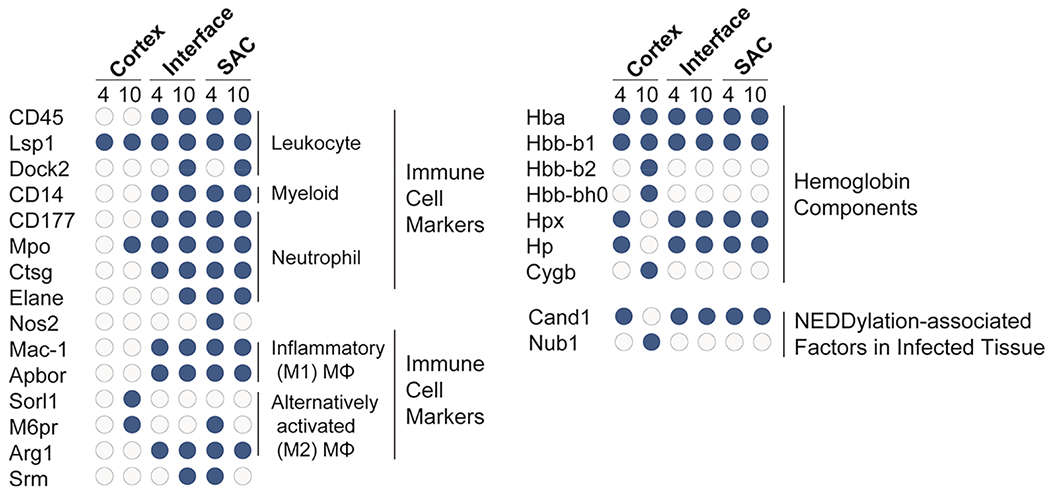
Spatiotemporal distribution of immune cell markers, hemoglobin components, and NEDDylation-associated factors in infected tissue. Host immune proteins were found in the interface and SAC of renal abscesses at 4 and 10 dpi. Blue circles denote proteins that were present at the specified time point/region, while gray circles depict the absence of a specific protein.
During abscess formation, neutrophils are recruited to the site of infection in high numbers.73–75 We identified the myeloid cell marker CD14 and neutrophil marker CD177 within the abscess and interface (Figure 4). Coinciding with the neutrophil surface markers, multiple neutrophil-specific antimicrobial factors are also found in the abscess and interface regions including myeloperoxidase (Mpo), cathepsin G (Ctsg), and neutrophil elastase (Elane) (Figure 4). In addition, neutrophils use NADPH oxidase to generate high levels of ROS in response to S. aureus, and an abundance of NADPH oxidase proteins are also specifically within the abscess and interface. The inducible nitric oxide synthase (Nos2), however, is only found in the abscess 4 dpi (Figure 4). While it is not possible to fully exclude that Nos2 is present in low concentrations below our limit of detection at this later time point, Nos2 was reliably detected in nearly all samples at the 4 dpi time point. This suggests that even if Nos2 is not absent from the abscess at 10 dpi, the concentration decreases over time.
Relative to neutrophils, macrophages make up a smaller percentage of the immune cells in the abscess.76 Nevertheless, the presence of integrin alpha-M (Itgam or Mac-1) indicates that activated inflammatory macrophages are found in the abscess and interface (Figure 4). The expression of apolipoprotein B receptor (Apobr) by macrophages is critical to combating S. aureus, as it suppresses activation of the Agr system, a major component of the staphylococcal regulatory landscape76 (Figure 4). Consistent with an integral role in combating S. aureus, Apobr is present both within the abscess and interface at 4 and 10 dpi. The presence of sortilin-related receptor (Sorl1), and mannose-6-phosphate receptor (M6pr) at 10 dpi in the cortex suggests the presence of alternatively activated M2-like macrophages involved in tissue repair and remodeling outside of the abscess (Figure 4). The classical M2-like macrophage marker arginase-1 (Arg1) is not detected in the cortex and is present ubiquitously in the abscess and interface (Figure 4). Despite Arg1 being linked to M2 polarization, Arg1 is only produced by a quarter of all M2-like macrophages77 and is furthermore up-regulated in inflammatory M1-like macrophages.78,79 Arg1 is necessary for the production of spermine and spermidine, which are uniquely toxic to most methicillin-resistant S. aureus (MRSA) strains and integral to killing S. aureus during the tissue resolution phase of skin infections.80 The presence of spermidine synthase (Srm) in the abscess at 4 dpi suggests spermine and spermidine production may play a critical antimicrobial function within abscesses in the kidney81 (Figure 4). These data suggest that during S. aureus infection of kidneys, Arg1 may be present in M1 macrophages in the abscess and might not be a reliable marker for the M2 macrophages associated with tissue repair in the cortex 10 dpi.
Heme Distribution.
Heme acts as a critical source of iron for S. aureus at the site of infection.82 While hemoglobin subunit alpha (Hba) and beta-1 (Hbb-b1) are ubiquitously present during infection, hemoglobin subunit beta-2 (Hbb-b2) and beta-H0 (Hbb-b0) are only found in the cortex at 10 dpi (Figure 4). Importantly, the host can restrict free heme and hemoglobin from S. aureus through binding and sequestration by hemopexin (Hpx)83 and haptoglobin (Hp),84 respectively. Both Hpx and Hp were found in the abscess, interface, and cortex at 4 dpi, but not in the cortex at 10 dpi (Figure 4). This suggests that while free heme and hemoglobin may be present at the site of infection in the kidney, the simultaneous presence of Hpx and Hp may render heme and hemoglobin biologically unavailable to S. aureus. The presence of host factors that limit heme availability to S. aureus also serves as an explanation for the previously discussed presence of different staphylococcal proteins aimed to counteract heme limitation during infection (i.e., IsdA, IsdB, and ChdC) (Figure 2). Another heme-containing protein, cytoglobin (Cygb), is exclusively present in the cortex at 10 dpi. Cygb contributes to oxygen diffusion for collagen synthesis during wound healing,85 regulating nitric oxide levels86,87 and detoxifying reactive oxygen species88 (Figure 4). Cygb mirrors the presence of M2 macrophages in the cortex at 10 dpi, that suggests Cygb plays a role in tissue repair and abscess resolution during S. aureus infection.
Host Signaling.
The power of microLESA is not just in confirming the presence or absence of proteins, but in allowing for unbiased observations about the regulatory states of the cells. NEDDylation and ubiquitylation are post-translational modifications that regulate protein degradation by the proteasome, thereby influencing signal transduction,89–94 inflammasome activation,95–97 autophagy,98,99 and cell death.100,101 The cullin-associated NEDD8-dissociated protein 1 (Cand1) regulates NEDD8 activity by sterically inhibiting the assembly of cullin-RING ubiquitin ligases and preventing NEDDylation,102 while the NEDD8 ultimate buster 1 (Nub1) specifically recruits NEDD8 to the proteasome for degradation.103,104 In the abscess, interface (both at 4 and 10 dpi), and cortex (at 4 dpi) Cand1 was present and Nub1 absent, suggesting that NEDDylation is being regulated by Cand1 (Figure 4). However, in the cortex at 10 dpi, the phenotype reversed with the presence of Nub1, and absence of Cand1, suggesting that NEDDylation is impaired by the Nub1-mediated degradation of NEDD8. The reduction of total NEDD8 protein would have significant implications in multiple signal transduction pathways, including NFκB and HIF1α, and suggests the signaling environment within the cortex at 10 dpi is unique. Enzymes necessary for ubiquitination and proteasomal degradation of proteins, including the ubiquitin activating enzymes (E1) that catalyze the first step in the ubiquitination reaction, and ubiquitin ligases (E3) that catalyze the transfer of ubiquitin from the E2 enzyme to the protein substrate, show similar spatial and temporal patterns. Because E1 and E3 enzymes interact directly with the protein substrate, it is possible that varying complexes of E1, E2, and E3 enzymes exhibit unique activities to exert diverse biological functions (Table S1).
Host Metabolism: Glycolysis and Glucogenesis.
Not only does signal transduction vary spatially and temporally, but the metabolic niche changes at different renal locations when comparing 4 and 10 dpi (Figure 4 and S2). We utilized a systems biology pathway analysis tool, SIMONE, to visualize the interactions between proteins of interest. These proteins were determined using external pathway mapping tools and entered into Reactome,105 and the resulting list of proteins associated with metabolism were input into SIMONE106 as seed proteins. This tool constructs networks using the MAGINE framework,107 which derives protein connection information from multiple databases (Figure S2). By uncovering how these proteins are connected, pathways can be predicted from spatiotemporal proteomics data. These pathways were summarized and combined in Figure 5. While we observed alterations in abundance of various proteins related to different metabolic processes (discussed below), the enzymes necessary for glycolysis are generally present during infection in the abscess, interface, and cortex, suggesting glucose conversion to pyruvate. Many of the enzymes necessary for the pentose phosphate pathway that runs parallel to glycolysis and enzymes necessary to breakdown fructose that feed into glycolysis are not detected in the cortex at 10 dpi. However, the enzyme bisphosphoglycerate mutase (Bpgm) is solely present in the cortex. This enzyme is necessary to form 2,3-bisphosphoglycerate from the glycolysis intermediate 1,3-bisphophoglycerate. The absence of Bpgm in the abscess and interface is consistent with a hypoxic environment in the abscess, as 2,3-bisphosphoglycerate binds hemoglobin at a high affinity and causes a conformational change resulting in the release of oxygen108,109 (Figure 5 and S2).
Figure 5.
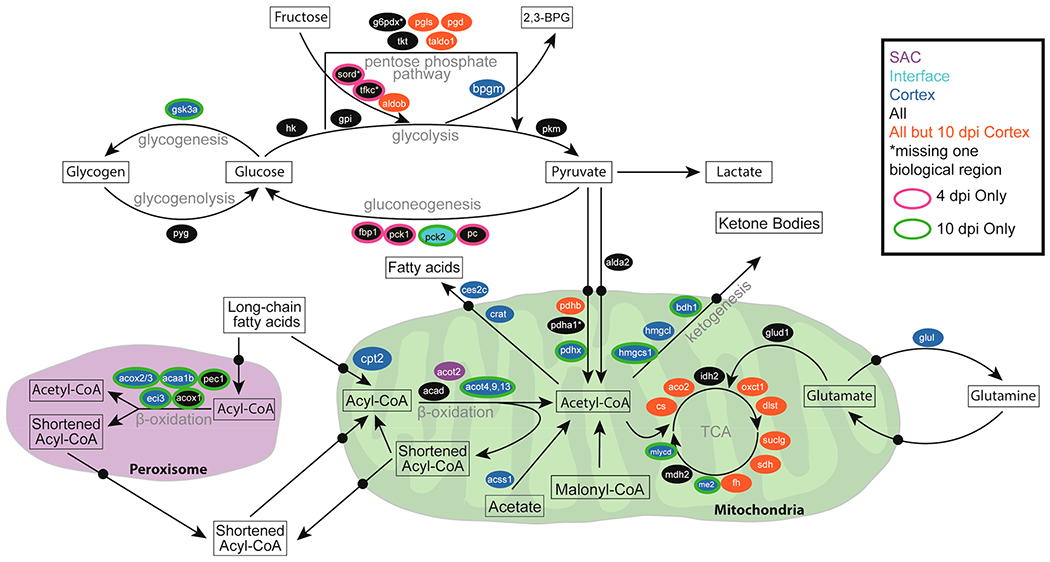
Overview of spatiotemporal distribution of proteins involved in central metabolism. Arrows denote metabolic pathways; ovals indicate genes, and colors indicate time point and region where proteins were detected.
The enzymes necessary for host gluconeogenesis, a process that converts noncarbohydrate substrates into glucose, were only detected in the abscess and cortex at 4 dpi, suggesting that gluconeogenesis may not occur late in infection (Figure 5 and S2). The enzymes necessary for glycogenolysis are present in the abscess, interface, and cortex; however, glycogen synthase kinase-3 alpha (Gsk3a), an enzyme that contributes to glycogenesis, is not detected in the abscess and interface (Figure 5 and S2). The lack of enzymes for glucose formation and long-term storage in the form of glycogen and the presence of enzymes necessary for the breakdown of glycogen into glucose in the abscess and interface are consistent with high levels of glycolysis during inflammation (reviewed in ref 110). The presence of Gsk3a exclusively in the cortex at 10 dpi suggests a decreased metabolic demand for glucose and possible glycogen formation. In addition, Gsk3a plays a central role in regulating the transition between pro-inflammatory and immune-suppressive response to S. aureus by controlling cytokine production.111 These results suggest a specific role for Gsk3a in altering the metabolic and cytokine landscape of the cortex 10 dpi during S. aureus infection of the kidney.
Host Metabolism: Metabolism in the Abscess.
While the metabolic enzymes present in the abscess and interface suggest the formation of pyruvate, the complex necessary for conversion of pyruvate into acetyl coenzyme A (acetyl-CoA) in the mitochondria may not be fully formed. Pyruvate dehydrogenase protein X component (Pdhx) tethers the E3 dimers to the E2 core of the pyruvate dehydrogenate complex112,113 (Figure 5 and S2). Therefore, lacking detectable Pdhx in the abscess and interface suggests that the pyruvate dehydrogenase complex would not be functional and that conversion of pyruvate into acetyl-CoA in the mitochondria would be impaired. Pyruvate can also be converted into acetyl-CoA in the cytosol,114 but the acetyl-coenzyme A synthetase (Acss2), which is necessary for transport into the mitochondria, is also not detected within the abscess and interface (Figure 5 and S2). Instead, the ubiquitous expression of the lactate dehydrogenase (Ldha and Ldhb) suggests that in the abscess and interface, the resulting pyruvate from glycolysis is being converted to lactate, consistent with anaerobic glycolysis in the hypoxic environment of the abscess (Figure 5 and S2).
Many of the enzymes associated with the tricarboxylic acid (TCA) cycle are present in the abscess and interface during infection, but pyruvate does not seem to be fueling downstream oxidative phosphorylation and ATP generation due to the lack of Pdhx and Acss2 (Figure 5 and S2). Many of the enzymes necessary for β-oxidation in the mitochondria are present in the abscess and interface, and the resulting formation of acetyl-CoA could fuel the TCA cycle (reviewed in ref 115). However, long-chain fatty acids (LCFAs) would likely not be able to serve as the carbon source for the TCA cycle. The O-palmitoyltransferase 2 (Cpt2), catalyzes the formation of palmitoyl-CoA from palmitoylcarnitine, a necessary step for LCFAs prior to β-oxidation (reviewed in ref 115). Thus, the lack of Cpt2 suggests that LCFAs cannot undergo β-oxidation in the mitochondria and therefore are not feeding into the TCA cycle in the abscess and interface (Figure 5 and S2). This does not exclude the possibility that short-chain fatty acids (SCFAs), which passively gain access to the mitochondria, could be fueling the TCA cycle. Carnitine O-acetyltransferase (Crat) blunts acetyl-CoA from fueling the TCA cycle by forming acetyl-carnitine for transport into the cytosol, and once in the cytosol, acylcarnitine hydrolase (Ces2c) liberates fatty acids from l-carnitine. Both Crat and Ces2c are absent in the in the abscess and interface supporting the possibility that SCFAs undergoing β-oxidation could serve as a carbon source for the TCA cycle (Figure 5 and S2). Glutamine may also act as an alternative carbon source for the TCA cycle.116 Glutamate dehydrogenase 1 (Glud1) catalyzes the oxidative deamination of glutamate into α-ketoglutarate, which feeds into the TCA cycle. The presence of Glud1 and absence of glutamine synthase (Glul), which converts glutamate into glutamine, in the abscess and interface suggests that glutamine may also be fueling the TCA cycle in the abscess environment (Figure 5 and S2).
Host Metabolism: Metabolism in the Cortex.
By contrast, in the cortex at 10 dpi, many of the enzymes necessary to run the TCA cycle are not detected. Only isocitrate dehydrogenase (Idh2), NAD-dependent malic enzyme (Me2), and malate dehydrogenase (Mdh2) are detected (Figure 5 and S2), suggesting that in the cortex at 10 dpi, the full TCA cycle may not be used. The cortex at 10 dpi is the only region to contain all the acyl-CoA dehydrogenases as well as almost exclusive presence of the enzymes necessary for β-oxidation in the peroxisome. Unlike the abscess and interface, Cpt2, is detected in the cortex suggesting that β-oxidation of LCFAs may occur, but the presence of Ces2c and Crat suggest that the resulting acetyl-CoA is shunted out of the mitochondria rather than feeding into the TCA cycle. M2 macrophages in the tissue surrounding the abscess (Figure 5 and S2) are necessary for the conversion to the resolution phase by cleaning up apoptotic cells and cellular debris, and are consistent with previous studies assessing abscess biology in the skin and soft tissue.80 Cellular debris contains high concentrations of lipids, thereby requiring β-oxidation for its degradation. The high presence of lipids in resolving damaged tissue outside the abscess could account for the increased presence of proteins necessary for β-oxidation.
Acetyl-coenzyme A synthetase 2-like (Acss1) and malonyl-CoA decarboxylase (Mlycd) are also present in the cortex 10 days postinfection (Figure 5 and S2). These enzymes shunt mitochondrial acetate and malonyl-CoA away from lipid synthesis and toward the formation of acetyl-CoA. The combined activity of β-oxidation, Acss1, and Mlycd with a limited presence of TCA cycle enzymes could cause an accumulation of acetyl-CoA. The presence of Crat and Cesc2c may relieve some of the acetyl-CoA burden by exporting acetyl-CoA as free fatty acids into the cytosol. Alternatively, many of the enzymes necessary for ketogenesis are present in the cortex at 10 dpi suggesting that the abundance of acetyl-CoA could also be converted to ketone bodies (Figure 5 and S2).
A byproduct of β-oxidation is oxidative stress, and consistent with increased β-oxidation, the cortex at 10 dpi is the only region to express the full array of glutathione S-transferases, synthase, reductase, and peroxidase to combat oxidative stress (Table S1). Since oxidative phosphorylation from the TCA cycle can create oxidative stress, this could explain the absence of TCA cycle enzymes in the cortex at 10 dpi (Figure 5 and S2). Finally, many of the downstream enzymes for β-oxidation in the mitochondria and peroxisome are not detected in the cortex at 10 dpi that are present at 4 days or in the abscess and interface (Figure 5 and S2). This suggests that while β-oxidation of LCFAs is occurring in the cortex at 10 dpi, the lipids are not fully broken down. In addition, acyl-coenzyme A thioesterases (Acot) are thought to maintain a sufficient CoA pool for β-oxidation by terminating β-oxidation after a set number of cycles (reviewed in ref 117). The specific presence of Acot4 in the peroxisome and Acot9 and Acot13 in the mitochondria could result in the formation of specific lipid products that is important in maintaining β-oxidation in high lipid environments to avoid oxidative stress (Figure 5 and S2). The metabolic environment in the cortex at 10 dpi is consistent with an immune response that has altered to a resolving phase that is cleaning up lipid-dense apoptotic cells and cellular debris following inflammation.
CONCLUSION
We performed spatially and temporally targeted proteomics, aided by a robust systems biology data processing workflow, to molecularly investigate staphylococcal abscess formation and development. This approach is suited for identification of bacterial proteins and enables characterization of the host proteome during infection. Such a comprehensive assessment of the abscess proteome allows us to molecularly characterize the host–pathogen interface over time. By pairing information about the spatiotemporal distribution of bacterial proteins with data defining the host proteome, we can elucidate how the pathogen and host shape the abscess (micro)environment and how, in turn, both parties react to these biomolecular changes.
The current study was specifically designed to assess (i) the proteomic changes between early and late phase of infection within each of the three distinct regions as well as (ii) the cross-comparison of the proteomes of the three regions throughout the course of staphylococcal disease. Using our microLESA workflow, we characterized the metabolic niche at the site of infection. Our findings indicate the influx of immune cells (i.e., neutrophils and different macrophage populations) while also elucidating specific metabolic processes employed by these host cell populations. The action of macrophages was not limited to the SAC and interface. We also detected markers for M2 macrophages in the renal cortex, suggesting a role in tissue repair and remodeling. In contrast, multiple neutrophil-specific antimicrobial factors (e.g., Mpo, Ctsg, Elane) were found to be expressed in proximity to the staphylococcal abscess. We found S. aureus produces CHIPS and FLIPr, which antagonize the host neutrophil receptors to prevent neutrophilic entrance into the abscess. Staphylococcal leukotoxins (LukAB) produced during abscess formation were also identified, again aiding in immune evasion and persistence in the host tissue. Indicative for the onset of nutritional immunity, we observed bacterial heme uptake and biosynthetic proteins present in the abscess. This limitation could be explained by the presence of host proteins that restrict the availability of heme for S. aureus (Hpx and Hp), particularly in the SAC and interface.
We describe several other additional mechanisms of how host and pathogen directly affect each other during infection and how both sides counteract the challenges presented to them. These data highlight the powerful nature of our experimental setup and offer insights into the processes at the host–pathogen interface, beyond the specific examples discussed here. The ability to elucidate when, where, and how pathogens and the immune system interact during abscess formation and disease progression is an invaluable resource to identify potential points of intervention when developing new antistaphylococcal therapeutic strategies.
Although microLESA is a powerful technology that provides spatial context to proteomic analysis, as with any analytical approach, the method has inherent limitations. These challenges are primarily associated with (i) the trade-off between spatial fidelity and molecular depth of coverage and (ii) ensuring sampling consistency when probing and comparing molecular features from distinct cell types and tissue regions. In our study, we identified 57 staphylococcal proteins alongside a myriad of host proteins. The bacterial proteins detected are largely consistent with those reported in S. aureus literature and relevant to understanding the pathophysiological state of the bacterium during infection. The number of bacterial proteins we detected, though, is relatively small when compared to the thousands of proteins in the staphylococcal proteome that went undetected. This is not unexpected as sensitivity and depth of coverage are typically sacrificed for improved spatial resolution due to fewer molecules being sampled from smaller foci. Still, the ability to characterize many staphylococcal proteins within the developing abscess in vivo is a significant advancement.
A second inherent challenge of microLESA is sampling consistency due to potential variation in extraction and ionization efficiency when probing different tissue regions. This is often driven by differences in molecular composition at each sampling location. To ensure accuracy of data generation and interpretation, only proteins reliably detected in the majority of replicates should be included in downstream analysis, as outlined in the Methods section of this paper and utilized for these analyses. Additionally, comparisons between sampling locations should be performed conservatively. Here, we based our conclusions on global changes in biological pathways rather than individual proteins to arrive at robust and reproducible trends in the data set. Further, the use of a systems biology pathway analysis workflow allowed us to focus on specific biological processes affected by the proximity to the site of infection. The use of such pathway analysis tools and a focus on global data trends is an efficient method for mining microLESA data and would be effective for any spatial targeted proteomics study. With these considerations in mind, microLESA has tremendous potential to be the cornerstone of future investigations attempting to spatially resolve the proteinaceous composition of complex tissues, including low abundance proteins or those difficult to ionize with other strategies.
METHODS
All sample preparation was completed using the method previously described by Ryan et al.22 Pathway analysis was conducted using a similar workflow described previously by Gutierrez and colleagues.106 Additional information on methods is included in the Supporting Information.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the NIH National Institute of Allergy and Infectious Diseases (R01AI138581 awarded to E.P.S and J.M.S. and R01AI069233 and R01AI073843 awarded to E.P.S.) and NIH National Institute of General Medical Sciences (2P41 GM103391-07 awarded to R.M.C.). A.W. is supported by the American Heart Association (18POST33990262) and the NIH National Institute of Environmental Health Sciences (T32ES007028). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ABBREVIATIONS USED
- dpi
days post infection
- SAC
staphylococcal abscess community
- LESA
liquid extraction surface analysis
- MALDI
matrix-assisted laser desorption/ionization
- IMS
imaging mass spectrometry
- LC-MS/MS
liquid chromatography tandem mass spectrometry
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00647.
Detailed methods; Figure S1, examples of microLESA from different regions, including the locations of microLESA sampling (colored dots) on autofluorescence images of infected kidneys (10 dpi); Figure S2, spatiotemporal distribution of metabolic proteins during infection (PDF)
Table S1, list of host-derived proteins found by presence at time point and biological region, including platform used for identification (PDF)
S2, list of bacterial proteins detected and their localizations based on stringent search criteria from Protalizer and MaxQuant (PDF)
Table S3, list of bacterial proteins detected and their localizations with lower identification confidence (PDF)
Accession Codes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE118 partner repository with the data set identifier PXD019920.
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.0c00647
Contributor Information
Emma R. Guiberson, Mass Spectrometry Research Center and Department of Chemistry, Vanderbilt University, Nashville, Tennessee 37203, United States.
Andy Weiss, Department of Pathology, Microbiology, and Immunology, Vanderbilt University School of Medicine, Nashville, Tennessee 37203, United States.
Daniel J. Ryan, Mass Spectrometry Research Center and Department of Chemistry, Vanderbilt University, Nashville, Tennessee 37203, United States.
Andrew J. Monteith, Department of Pathology, Microbiology, and Immunology, Vanderbilt University School of Medicine, Nashville, Tennessee 37203, United States
Kavya Sharman, Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37203, United States.
Danielle B. Gutierrez, Mass Spectrometry Research Center, Vanderbilt University, Nashville, Tennessee 37203, United States.
William J. Perry, Mass Spectrometry Research Center and Department of Chemistry, Vanderbilt University, Nashville, Tennessee 37203, United States
Richard M. Caprioli, Mass Spectrometry Research Center, Department of Chemistry, Department of Biochemistry, Department of Medicine, and Department of Pharmacology, Vanderbilt University, Nashville, Tennessee 37203, United States
Eric P. Skaar, Department of Pathology, Microbiology, and Immunology, Vanderbilt University School of Medicine, Nashville, Tennessee 37203, United States
Jeffrey M. Spraggins, Mass Spectrometry Research Center, Department of Chemistry, and Department of Biochemistry, Vanderbilt University, Nashville, Tennessee 37203, United States.
REFERENCES
- (1).Laupland KB (2013) Defining the epidemiology of bloodstream infections: the ‘gold standard’ of population-based assessment. Epidemiol. Infect 141 (10), 2149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Klein EY, Jiang W, Mojica N, Tseng KK, McNeill R, Cosgrove SE, and Perl TM (2018) National Costs Associated With Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Hospitalizations in the United States, 2010–2014. Clin. Infect. Dis 68 (1), 22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, and Gosbell IB (2012) Predictors of mortality in Staphylococcus aureus Bacteremia. Clin Microbiol Rev. 25 (2), 362–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fowler VG Jr., Olsen MK, Corey GR, Woods CW, Cabell CH, Reller LB, Cheng AC, Dudley T, and Oddone EZ (2003) Clinical identifiers of complicated Staphylococcus aureus bacteremia. Arch. Intern. Med 163 (17), 2066–72. [DOI] [PubMed] [Google Scholar]
- (5).Cheng AG, DeDent AC, Schneewind O, and Missiakas D (2011) A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 19 (5), 225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Guggenberger C, Wolz C, Morrissey JA, and Heesemann J (2012) Two distinct coagulase-dependent barriers protect Staphylococcus aureus from neutrophils in a three dimensional in vitro infection model. PLoS Pathog. 8 (1), No. e1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, and Missiakas DM (2009) Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23 (10), 3393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lam GT, Sweeney FJ Jr, Witmer CM, and Wise RI (1963) Abscess-Forming Factor(S) Produced by Staphylococcus Aureus. Ii. Abscess Formation and Immunity by a Staphylococcus and Its Mutants. J. Bacteriol 86, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, and Schneewind O (2010) Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog. 6 (8), No. e1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, and He C (2006) An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol 2 (11), 591–5. [DOI] [PubMed] [Google Scholar]
- (11).Rauch S, DeDent AC, Kim HK, Bubeck Wardenburg J, Missiakas DM, and Schneewind O (2012) Abscess formation and alpha-hemolysin induced toxicity in a mouse model of Staphylococcus aureus peritoneal infection. Infect. Immun 80 (10), 3721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kobayashi SD, Malachowa N, and DeLeo FR (2015) Pathogenesis of Staphylococcus aureus abscesses. Am. J. Pathol 185 (6), 1518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, and Skaar EP (2008) Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319 (5865), 962–5. [DOI] [PubMed] [Google Scholar]
- (14).Attia AS, Cassat JE, Aranmolate SO, Zimmerman LJ, Boyd KL, and Skaar EP (2013) Analysis of the Staphylococcus aureus abscess proteome identifies antimicrobial host proteins and bacterial stress responses at the host-pathogen interface. Pathog. Dis 69 (1), 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang CG, Chromy BA, and McCutchen-Maloney SL (2005) Host-pathogen interactions: a proteomic view. Expert Rev. Proteomics 2 (2), 187–202. [DOI] [PubMed] [Google Scholar]
- (16).Schmidt F, and Volker U (2011) Proteome analysis of host-pathogen interactions: Investigation of pathogen responses to the host cell environment. Proteomics 11 (15), 3203–11. [DOI] [PubMed] [Google Scholar]
- (17).Buchberger AR, DeLaney K, Johnson J, and Li L (2018) Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem 90 (1), 240–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Shariatgorji M, Svenningsson P, and Andren PE (2014) Mass spectrometry imaging, an emerging technology in neuropsychopharmacology. Neuropsychopharmacology 39 (1), 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Spraggins JM, Rizzo DG, Moore JL, Noto MJ, Skaar EP, and Caprioli RM (2016) Next-generation technologies for spatial proteomics: Integrating ultra-high speed MALDI-TOF and high mass resolution MALDI FTICR imaging mass spectrometry for protein analysis. Proteomics 16 (11–12), 1678–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Liu Z, and Schey KL (2008) Fragmentation of multiply-charged intact protein ions using MALDI TOF-TOF mass spectrometry. J. Am. Soc. Mass Spectrom 19 (2), 231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zhang Y, Fonslow BR, Shan B, Baek MC, and Yates JR 3rd (2013) Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 113 (4), 2343–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ryan DJ, Patterson NH, Putnam NE, Wilde AD, Weiss A, Perry WJ, Cassat JE, Skaar EP, Caprioli RM, and Spraggins JM (2019) MicroLESA: Integrating Autofluorescence Microscopy, In Situ Micro-Digestions, and Liquid Extraction Surface Analysis for High Spatial Resolution Targeted Proteomic Studies. Anal. Chem 91 (12), 7578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cassat JE, Moore JL, Wilson KJ, Stark Z, Prentice BM, Van de Plas R, Perry WJ, Zhang Y, Virostko J, Colvin DC, Rose KL, Judd AM, Reyzer ML, Spraggins JM, Grunenwald CM, Gore JC, Caprioli RM, and Skaar EP (2018) Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl. Med 10 (432), eaan6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Perry WJ, Spraggins JM, Sheldon JR, Grunenwald CM, Heinrichs DE, Cassat JE, Skaar EP, and Caprioli RM (2019) Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc. Natl. Acad. Sci. U. S. A 116 (44), 21980–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Widjaja M, Harvey KL, Hagemann L, Berry IJ, Jarocki VM, Raymond BBA, Tacchi JL, Grundel A, Steele JR, Padula MP, Charles IG, Dumke R, and Djordjevic SP (2017) Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci. Rep 7 (1), 11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Miller DL, and Weissbach H (1970) Interactions between the elongation factors: the displacement of GPD from the TU-GDP complex by factor Ts. Biochem. Biophys. Res. Commun 38 (6), 1016–22. [DOI] [PubMed] [Google Scholar]
- (27).Pasztor L, Ziebandt AK, Nega M, Schlag M, Haase S, Franz-Wachtel M, Madlung J, Nordheim A, Heinrichs DE, and Gotz F (2010) Staphylococcal Major Autolysin (Atl) Is Involved in Excretion of Cytoplasmic Proteins. J. Biol. Chem 285 (47), 36794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).McCarthy H, Waters EM, Bose JL, Foster S, Bayles KW, O’Neill E, Fey PD, and O’Gara JP (2016) The major autolysin is redundant for Staphylococcus aureus USA300 LAC JE2 virulence in a murine device-related infection model. FEMS Microbiol Lett. 363 (9), fnw087. [DOI] [PubMed] [Google Scholar]
- (29).Hood MI, and Skaar EP (2012) Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol 10 (8), 525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Skaar EP, and Schneewind O (2004) Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6 (4), 390–7. [DOI] [PubMed] [Google Scholar]
- (31).Dailey HA, Gerdes S, Dailey TA, Burch JS, and Phillips JD (2015) Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc. Natl. Acad. Sci. U. S. A 112 (7), 2210–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lobo SA, Scott A, Videira MA, Winpenny D, Gardner M, Palmer MJ, Schroeder S, Lawrence AD, Parkinson T, Warren MJ, and Saraiva LM (2015) Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol. Microbiol 97 (3), 472–87. [DOI] [PubMed] [Google Scholar]
- (33).Mayfield JA, Hammer ND, Kurker RC, Chen TK, Ojha S, Skaar EP, and DuBois JL (2013) The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. J. Biol. Chem 288 (32), 23488–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Fang FC (1999) Nitric oxide and infection, pp xxv–517, Kluwer Academic/Plenum Publishers, New York. [Google Scholar]
- (35).Guerra FE, Borgogna TR, Patel DM, Sward EW, and Voyich JM (2017) Epic Immune Battles of History: Neutrophils vs. Staphylococcus aureus. Front. Cell. Infect. Microbiol 7, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Stryjewski ME, Kanafani ZA, Chu VH, Pappas PA, Harding T, Drew LA, Benjamin DK, Reller LB, Lee BA, Corey GR, and Fowler VG (2009) Staphylococcus Aureus Bacteremia Among Patients with Health Care-associated Fever. Am. J. Med 122 (3), 281–U116. [DOI] [PubMed] [Google Scholar]
- (37).Ezraty B, Gennaris A, Barras F, and Collet JF (2017) Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol 15 (7), 385–96. [DOI] [PubMed] [Google Scholar]
- (38).Henderson B, Allan E, and Coates AR (2006) Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun 74 (7), 3693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, and Chamberlain NR (2007) Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology 153 (9), 3162–73. [DOI] [PubMed] [Google Scholar]
- (40).Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, and Hartl FU (2012) DnaK Functions as a Central Hub in the E. coli Chaperone Network. Cell Rep. 1 (3), 251–64. [DOI] [PubMed] [Google Scholar]
- (41).Liu CP, Zhou QM, Fan DJ, and Zhou JM (2010) PPIase domain of trigger factor acts as auxiliary chaperone site to assist the folding of protein substrates bound to the crevice of trigger factor. Int. J. Biochem. Cell Biol 42 (6), 890–901. [DOI] [PubMed] [Google Scholar]
- (42).Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, and Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400 (6745), 693–6. [DOI] [PubMed] [Google Scholar]
- (43).Keogh RA, Zapf RL, Wiemels RE, Wittekind MA, and Carroll RK (2018) The Intracellular Cyclophilin PpiB Contributes to the Virulence of Staphylococcus aureus Independently of Its Peptidyl-Prolyl cis/trans Isomerase Activity. Infect. Immun 86 (11), 1 DOI: 10.1128/IAI.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sasaki S, Miura T, Nishikawa S, Yamada K, Hirasue M, and Nakane A (1998) Protective role of nitric oxide in Staphylococcus aureus infection in mice. Infect. Immun 66 (3), 1017–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Vitko NP, Spahich NA, and Richardson AR (2015) Glycolytic Dependency of High-Level Nitric Oxide Resistance and Virulence in Staphylococcus aureus. mBio 6 (2), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Richardson AR, Libby SJ, and Fang FC (2008) A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319 (5870), 1672–6. [DOI] [PubMed] [Google Scholar]
- (47).Brown GC, McBride AG, Fox EJ, McNaught KS, and Borutaite V (1997) Nitric oxide and oxygen metabolism. Biochem. Soc. Trans 25 (3), 901–4. [DOI] [PubMed] [Google Scholar]
- (48).Heim CE, Bosch ME, Yamada KJ, Aldrich AL, Chaudhari SS, Klinkebiel D, Gries CM, Alqarzaee AA, Li Y, Thomas VC, Seto E, Karpf AR, and Kielian T (2020) Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol 5, 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ibarra JA, Perez-Rueda E, Carroll RK, and Shaw LN (2013) Global analysis of transcriptional regulators in Staphylococcus aureus. BMC Genomics 14, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Jenul C, and Horswill AR (2018) Regulation of Staphylococcus aureus Virulence. Microbiol Spectr. 6 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Somerville GA, and Proctor RA (2009) At the Crossroads of Bacterial Metabolism and Virulence Factor Synthesis in Staphylococci. Microbiol. Mol. Biol. Rev 73 (2), 233–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, and Sonenshein AL (2008) Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol 190 (7), 2257–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, and Brinsmade SR (2016) A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol. Microbiol 101 (3), 495–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kaiser JC, King AN, Grigg JC, Sheldon JR, Edgell DR, Murphy MEP, Brinsmade SR, and Heinrichs DE (2018) Repression of branched-chain amino acid synthesis in Staphylococcus aureus is mediated by isoleucine via CodY, and by a leucine-rich. PLoS Genet. 14 (1), e1007159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, and Wolz C (2009) CodY in Staphylococcus aureus: a Regulatory Link between Metabolism and Virulence Gene Expression. J. Bacteriol 191 (9), 2953–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Han AR, Kang HR, Son J, Kwon DH, Kim S, Lee WC, Song HK, Song MJ, and Hwang KY (2016) The structure of the pleiotropic transcription regulator CodY provides insight into its GTP-sensing mechanism. Nucleic Acids Res. 44 (19), 9483–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Chavakis T, Wiechmann K, Preissner KT, and Herrmann M (2005) Staphylococcus aureus interactions with the endothelium: the role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb. Haemostasis 94 (2), 278–85. [DOI] [PubMed] [Google Scholar]
- (58).Hussain M, Becker K, von Eiff C, Schrenzel J, Peters G, and Herrmann M (2001) Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacterid 183 (23), 6778–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Geraci J, Neubauer S, Pollath C, Hansen U, Rizzo F, Krafft C, Westermann M, Hussain M, Peters G, Pletz MW, Loffler B, Makarewicz O, and Tuchscherr L (2017) The Staphylococcus aureus extracellular matrix protein (Emp) has a fibrous structure and binds to different extracellular matrices. Sci. Rep 7 (1), 13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Harraghy N, Hussain M, Haggar A, Chavakis T, Sinha B, Herrmann M, and Flock JI (2003) The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology 149 (10), 2701–7. [DOI] [PubMed] [Google Scholar]
- (61).Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, and Missiakas DM (2009) Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23 (10), 3393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Rollins TE, and Springer MS (1985) Identification of the polymorphonuclear leukocyte C5a receptor. J. Biol. Chem 260 (12), 7157–7160. [PubMed] [Google Scholar]
- (63).Le Y, Murphy PM, and Wang JM (2002) Formyl-peptide receptors revisited. Trends Immunol. 23 (11), 541–8. [DOI] [PubMed] [Google Scholar]
- (64).Teng T-S, Ji A-L, Ji X-Y, and Li Y-Z (2017) Neutrophils and immunity: from bactericidal action to being conquered. J. Immunol. Res 2017, 9671604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Postma B, Poppelier MJ, van Galen JC, Prossnitz ER, van Strijp JA, de Haas CJ, and van Kessel KP (2004) Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol 172 (11), 6994–7001. [DOI] [PubMed] [Google Scholar]
- (66).Prat C, Bestebroer J, de Haas CJ, van Strijp JA, and van Kessel KP (2006) A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J. Immunol 177 (11), 8017–26. [DOI] [PubMed] [Google Scholar]
- (67).Reyes-Robles T, and Torres VJ (2016) Staphylococcus aureus Pore-Forming Toxins. Curr. Top. Microbiol. Immunol 409, 121–44. [DOI] [PubMed] [Google Scholar]
- (68).Dumont AL, Nygaard TK, Watkins RL, Smith A, Kozhaya L, Kreiswirth BN, Shopsin B, Unutmaz D, Voyich JM, and Torres VJ (2011) Characterization of a new cytotoxin that contributes to Staphylococcus aureus pathogenesis. Mol. Microbiol 79 (3), 814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ventura CL, Malachowa N, Hammer CH, Nardone GA, Robinson MA, Kobayashi SD, and DeLeo FR (2010) Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5 (7), No. e11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Liu Q, Yeo WS, and Bae T (2016) The SaeRS Two-Component System of Staphylococcus aureus. Genes 7 (10), 81. [Google Scholar]
- (71).Geiger T, Goerke C, Mainiero M, Kraus D, and Wolz C (2008) The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol 190 (10), 3419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Cho H, Jeong DW, Liu Q, Yeo WS, Vogl T, Skaar EP, Chazin WJ, and Bae T (2015) Calprotectin Increases the Activity of the SaeRS Two Component System and Murine Mortality during Staphylococcus aureus Infections. PLoS Pathog. 11 (7), No. e1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Kim MH, Granick JL, Kwok C, Walker NJ, Borjesson DL, Curry FR, Miller LS, and Simon SI (2011) Neutrophil survival and c-kit(+)-progenitor proliferation in Staphylococcus aureus-infected skin wounds promote resolution. Blood 117 (12), 3343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Spinardi JR, Berea R, Orioli PA, Gabriele MM, Navarini A, Marques MT, Neto MN, and Mimica MJ (2017) Enterococcus spp. and S. aureus colonization in neutropenic febrile children with cancer. GERMS 7 (2), 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Verdrengh M, and Tarkowski A (1997) Role of neutrophils in experimental septicemia and septic arthritis induced by Staphylococcus aureus. Infect. Immun 65 (7), 2517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Thammavongsa V, Missiakas DM, and Schneewind O (2013) Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science 342 (6160), 863–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado J. d. D., Popovich PG, Partida-Sanchez S, and Guerau-de-Arellano M (2015) Novel Markers to Delineate Murine M1 and M2Macrophages. PLoS One 10 (12), No. e0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Raes G, Noel W, Beschin A, Brys L, de Baetselier P, and Hassanzadeh GH (2002) FIZZ1 and Ym as tools to discriminate between differentially activated macrophages. Dev. Immunol 9 (3), 151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, Konig T, Schleicher U, Koo MS, Kaplan G, Fitzgerald KA, Tuomanen EI, Orme IM, Kanneganti TD, Bogdan C, Wynn TA, and Murray PJ (2008) Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol 9 (12), 1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Thurlow LR, Joshi GS, and Richardson AR (2018) Peroxisome Proliferator-Activated Receptor gamma Is Essential for the Resolution of Staphylococcus aureus Skin Infections. Cell Host Microbe 24 (2), 261–70. [DOI] [PubMed] [Google Scholar]
- (81).Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, and Richardson AR (2013) Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13 (1), 100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Skaar EP, Humayun M, Bae T, DeBord KL, and Schneewind O (2004) Iron-source preference of Staphylococcus aureus infections. Science 305 (5690), 1626–8. [DOI] [PubMed] [Google Scholar]
- (83).Torres VJ, Pishchany G, Humayun M, Schneewind O, and Skaar EP (2006) Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol 188 (24), 8421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Mikkelsen JH, Runager K, and Andersen CBF (2020) The human protein haptoglobin inhibits IsdH-mediated heme-sequestering by Staphylococcus aureus. J. Biol. Chem 295 (7), 1781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Burmester T, Gerlach F, and Hankeln T (2007) Regulation and role of neuroglobin and cytoglobin under hypoxia. Adv. Exp. Med. Biol 618, 169–80. [DOI] [PubMed] [Google Scholar]
- (86).Gardner PR, Gardner AM, Brashear WT, Suzuki T, Hvitved AN, Setchell KD, and Olson JS (2006) Hemoglobins dioxygenate nitric oxide with high fidelity. J. Inorg. Biochem 100 (4), 542–50. [DOI] [PubMed] [Google Scholar]
- (87).Gardner AM, Cook MR, and Gardner PR (2010) Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in rat hepatocytes. J. Biol. Chem 285 (31), 23850–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Xu R, Harrison PM, Chen M, Li L, Tsui TY, Fung PC, Cheung PT, Wang G, Li H, Diao Y, Krissansen GW, Xu S and Farzaneh F (2006) Cytoglobin overexpression protects against damage-induced fibrosis. Mol. Ther 13 (6), 1093–100. [DOI] [PubMed] [Google Scholar]
- (89).Amir RE, Iwai K, and Ciechanover A (2002) The NEDD8 pathway is essential for SCF(beta -TrCP)-mediated ubiquitination and processing of the NF-kappa B precursor p105. J. Biol. Chem 277 (26), 23253–9. [DOI] [PubMed] [Google Scholar]
- (90).Ehrentraut SF, Kominsky DJ, Glover LE, Campbell EL, Kelly CJ, Bowers BE, Bayless AJ, and Colgan SP (2013) Central role for endothelial human deneddylase-1/SENP8 in fine-tuning the vascular inflammatory response. J. Immunol 190 (1), 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Sufan RI, and Ohh M (2006) Role of the NEDD8 modification of Cul2 in the sequential activation of ECV complex. Neoplasia. 8 (11), 956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Curtis VF, Ehrentraut SF, Campbell EL, Glover LE, Bayless A, Kelly CJ, Kominsky DJ, and Colgan SP (2015) Stabilization of HIF through inhibition of Cullin-2 neddylation is protective in mucosal inflammatory responses. FASEB J. 29 (1), 208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Bonita DP, Miyake S, Lupher ML Jr, Langdon WY, and Band H (1997) Phosphotyrosine binding domain-dependent upregulation of the platelet-derived growth factor receptor alpha signaling cascade by transforming mutants of Cbl: implications for Cbl’s function and oncogenicity. Mol. Cell. Biol 17 (8), 4597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, and Yarden Y (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12 (23), 3663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Palazon-Riquelme P, Worboys JD, Green J, Valera A, Martin-Sanchez F, Pellegrini C, Brough D, and Lopez-Castejon G (2018) USP7 and USP47 deubiquitinases regulate NLRP3 inflammasome activation. EMBO Rep. 19 (10), e44766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Chui AJ, Okondo MC, Rao SD, Gai K, Griswold AR, Johnson DC, Ball DP, Taabazuing CY, Orth EL, Vittimberga BA, and Bachovchin DA (2019) N-terminal degradation activates the NLRP1B inflammasome. Science 364 (6435), 82–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Sandstrom A, Mitchell PS, Goers L, Mu EW, Lesser CF, and Vance R E. (2019) Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364 (6435), eaau1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, and Cecconi F (2013) mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol 15 (4), 406–16. [DOI] [PubMed] [Google Scholar]
- (99).Shi CS, and Kehrl JH (2010) TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signaling 3 (123), No. ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Roy C, Brown DL, Little JE, Valentine BK, Walker PR, Sikorska M, Leblanc J, and Chaly N (1992) The topoisomerase II inhibitor teniposide (VM-26) induces apoptosis in unstimulated mature murine lymphocytes. Exp. Cell Res 200 (2), 416–24. [DOI] [PubMed] [Google Scholar]
- (101).Delic J, Morange M, and Magdelenat H (1993) Ubiquitin pathway involvement in human lymphocyte gamma-irradiation-induced apoptosis. Mol. Cell. Biol 13 (8), 4875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Liu J, Furukawa M, Matsumoto T, and Xiong Y (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10 (6), 1511–8. [DOI] [PubMed] [Google Scholar]
- (103).Kamitani T, Kito K, Fukuda-Kamitani T, and Yeh ET (2001) Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem 276 (49), 46655–60. [DOI] [PubMed] [Google Scholar]
- (104).Kito K, Yeh ET, and Kamitani T (2001) NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J. Biol. Chem 276 (23), 20603–9. [DOI] [PubMed] [Google Scholar]
- (105).Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, Loney F, May B, Milacic M, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai T, Weiser J, Wu G, Stein L, Hermjakob H, and D’Eustachio P (2019) The reactome pathway knowledgebase. Nucleic Acids Res. 48 (D1), D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Gutierrez DB, Gant-Branum RL, Romer CE, Farrow MA, Allen JL, Dahal N, Nei YW, Codreanu SG, Jordan AT, Palmer LD, Sherrod SD, McLean JA, Skaar EP, Norris JL, and Caprioli RM (2018) An Integrated, High-Throughput Strategy for Multiomic Systems Level Analysis. J. Proteome Res 17 (10), 3396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Pino JC, Lubbock ALR, Harris LA, Gutierrez DB, Farrow MA, Muszynski N, Norris JL, Caprioli RM, Wikswo JP, and Lopez CF (2020) A computational framework to explore cellular response mechanisms from multi-omics datasets. bioRxiv, 1. [Google Scholar]
- (108).Benesch R, and Benesch RE (1967) The effect of organic phosphates from the human erythrocyte on the allosteric properties of hemoglobin. Biochem. Biophys. Res. Commun 26 (2), 162–7. [DOI] [PubMed] [Google Scholar]
- (109).Benesch R, Benesch RE, and Yu CI (1968) Reciprocal binding of oxygen and diphosphoglycerate by human hemoglobin. Proc. Natl. Acad. Sci. U. S. A 59 (2), 526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).O’Neill LA, and Pearce EJ (2016) Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med 213 (1), 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Silva-Garcia O, Rico-Mata R, Maldonado-Pichardo MC, Bravo-Patino A, Valdez-Alarcon JJ, Aguirre-Gonzalez J, and Baizabal-Aguirre VM (2018) Glycogen Synthase Kinase 3alpha Is the Main Isoform That Regulates the Transcription Factors Nuclear Factor-Kappa B and cAMP Response Element Binding in Bovine Endothelial Cells Infected with Staphylococcus aureus. Front. Immunol 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Powers-Greenwood SL, Rahmatullah M, Radke GA, and Roche TE (1989) Separation of protein X from the dihydrolipoyl transacetylase component of the mammalian pyruvate dehydrogenase complex and function of protein X. J. Biol. Chem 264 (7), 3655–7. [PubMed] [Google Scholar]
- (113).Lawson JE, Behal RH, and Reed LJ (1991) Disruption and mutagenesis of the Saccharomyces cerevisiae PDX1 gene encoding the protein X component of the pyruvate dehydrogenase complex. Biochemistry 30 (11), 2834–9. [DOI] [PubMed] [Google Scholar]
- (114).Liu X, Cooper DE, Cluntun AA, Warmoes MO, Zhao S, Reid MA, Liu J, Lund PJ, Lopes M, Garcia BA, Wellen KE, Kirsch DG, and Locasale JW (2018) Acetate Production from Glucose and Coupling to Mitochondrial Metabolism in Mammals. Cell 175 (2), 502–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Houten SM, and Wanders RJ (2010) A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J. Inherited Metab. Dis 33 (5), 469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, Rutter J, Merritt ME, and DeBerardinis RJ (2014) Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56 (3), 414–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Tillander V, Alexson SEH, and Cohen DE (2017) Deactivating Fatty Acids: Acyl-CoA Thioesterase-Mediated Control of Lipid Metabolism. Trends Endocrinol. Metab 28 (7), 473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, and Vizcaino JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


