Abstract
Purpose:
Protein kinase A (PKA) subunit defects (in PRKAR1A and PRKACA) are known to contribute to adrenal tumor pathogenesis. We studied the PRKAR1B gene for any genetic changes in bilateral adrenocortical hyperplasia (BAH) and cortisol-producing adrenal adenomas (CPA).
Methods:
Exome sequencing and PRKAR1B copy number variant (CNV) analysis were performed in 74 patients with BAH and 21 with CPA. PKA activity was studied in tumors with defects; sequence variants were investigated in vitro.
Results:
Three PRKAR1B germline variants (p.I40V, p.A67V, p.A300T) were identified among 74 patients with BAH. PRKAR1B copy number gains (CNG) were found in 3 of 21 CPAs, one in a tumor carrying a somatic PRKACA “hot-spot” pathogenic variant p.L206R. CPAs bearing PRKAR1B CNGs showed higher PRKAR1B mRNA levels and reduced PKA activity. Baseline PKA activity was also decreased for p.A67V and p.A300T in vitro, and mutant PRKAR1β bound PRKACα in FRET recordings of co-transfected HEK293 cells stronger than normal.
Conclusion:
PRKAR1B is yet another PKA subunit that may potentially contribute to adrenal tumor formation. Its involvement in adrenocortical disease may be different from that of other subunits, because PRKAR1B variants and PRKAR1B CNG were associated with decreased (rather than increased) overall PKA activity in vitro.
Keywords: cyclic AMP, cortisol, adrenocortical hyperplasia, PRKAR1B gene
INTRODUCTION
Protein kinase A (PKA) is a tetrameric enzyme composed of a regulatory (R) dimer and two catalytic (C) subunits.1 There are four different R-subunits (RIα, RIβ, RIIα and RIIβ) and four C-subunits (Cα, Cβ, and Cγ, and PRKX) that act as serine threonine kinases.1 In 2000, defects of the PRKAR1A gene (MIM: 188830) coding for the RIα subunit and causing Carney complex (CNC [MIM: 160980]) were discovered. CNC is a rare multiple neoplasia syndrome inherited in an autosomal dominant manner.2,3 PRKAR1A pathogenic variants were also found in isolated primary pigmented nodular adrenocortical disease (PPNAD) and cortisol-producing adenomas (CPA) causing Cushing syndrome (CS).4,5 A single patient with CNC-associated tumors was found to have somatic mosaicism for a copy number gain (CNG) of the PRKACB gene coding the Cβ subunit (MIM: 176892).6 Recently, we described a somatic PRKACB sequencing variant in a CPA of a patient with CS.7 Defects in PRKACA, coding for the Cα subunit (MIM: 601639), were identified in isolated PPNAD, CPA, and other adrenocortical tumors8–11 and cardiac myxomas.12 PRKACA chromosomal rearrangements have been found in fibrolamellar hepatocellular carcinoma13 and in intraductal oncocytic papillary neoplasms of the pancreas and bile duct, along with PRKACB defects.14 The genomic profile of CPAs that did not have the common genetic defects was also investigated recently: 15,16 among the identified sequence abnormalities and copy number variants (CNV), chromosome 7p22 CNG that included the PRKAR1B gene, coding the R1β subunit (MIM: 176911), were the most frequent.16 In almost all cases, PRKAR1B CNG was seen in association with other defects that were thought to be drivers of the phenotype, such as for example PRKACA amplification (chromosome 19p13.3) or β-catenin (CTNNB1 [MIM: 116806]) activating pathogenic variants.16
The PRKAR1B subunit, less studied compared to PRKAR1A or PRKACA, is ubiquitously expressed, but at much lower levels than other PKA subunits with the exception of certain areas of the brain (https://v18.proteinatlas.org/ENSG00000188191-PRKAR1B/tissue).17 PRKAR1B is on chromosome 7p22, a region that has been implicated in primary hyperaldosteronism (PA) but not in CS,18 and in fusion sequences in various neoplastic conditions.19 The R1β PKA type-I (PKA-I) holoenzyme has a more than two-fold higher sensitivity to cyclic adenosine mono-phosphate (cAMP) than the R1α PKA-I which serves better neuronal transmission of cAMP signaling and associates with certain tissue-specific anchoring proteins.17,20
Following the identification of the first 7p22 CNGs in cortisol-producing lesions,15,16 we investigated patients with CS due to micronodular forms of bilateral adrenocortical hyperplasia (BAH) or CPA for PRKAR1B variants and 7p22 CNG. Our results confirmed PRKAR1B’s potential for being involved in the perturbations of cAMP signaling that give rise to adrenocortical disease.
MATERIALS AND METHODS
Ethics Statement
All patients were recruited under protocols for the study of PPNAD and other BAH, and CPA. Approval was obtained from the Institutional Review Boards of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) (1995–2011) and the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (2011–2020); the patient’s informed consent was obtained and the patient’s assent and/or parents’ consent as appropriate.
Clinical and DNA studies
Tumor samples were obtained from patients at surgery, as previously described.21 DNA was extracted from peripheral blood and fresh-frozen tissues, as previously described.4,7 Exome sequencing was conducted as we have described elsewhere.4,7,8 In brief, exome DNA was captured using the SureSelectXT Human All Exon version 4 Kit (Agilent, CA), following the manufacturer’s protocol, and then sequenced on a pair of SOLiD 5500xl flowchips (Thermo Fisher Scientific, MA). All patients had been screened for germline pathogenic variants of PRKAR1A, PDE11A (MIM: 604961), PRKACA, and related genes, as part of older studies;8,10,21 their peripheral DNA samples were negative for any variants in these genes, confirmed also by Sanger sequencing.
The PRKAR1B-coding and the flanking intronic sequences were amplified by PCR both in peripheral and tumor DNA and sequenced by the Sanger methodology, to confirm exome sequencing findings; both strands of the amplified products were directly sequenced with forward and reverse primers, as we have published elsewhere.7,8
CNV analysis through droplet digital polymerase chain reaction (ddPCR)
To quantify the DNA copy number of the PRKAR1B gene, we performed ddPCR using the FAM/MGB-labeled TaqMan probes for PRKAR1B (Hs04991422_cn, Hs04982006_cn, Hs04981093_cn) (Thermo Fisher Scientific, MA) and VIC/TAMRA-labeled TaqMan CNV RNAse P (#4403328) (Bio-Rad Laboratories, CA) as the internal control. This analysis was performed on DNA derived from adrenal tissues of the 21 patients with CPA. The ddPCR experiments were performed as follows: each 21-μL reaction mixture contained 2μL of DNA template (20 ng total), 2× ddPCR supermix for probes (no dUTP), and PRKAR1B and RNAse P probe assays. The assays were purchased as a 20× premix of probes and used at 1× concentration. After homogenization, the PCR reaction mixture and droplet generation oil for probes were loaded into an eight-channel droplet generator cartridge (Bio-Rad Laboratories, CA). The PCR reaction mixtures were partitioned into an emulsion of approximately 15,000 droplets (~1 nL per droplet) that were manually transferred to a 96-well PCR plate. The PCR plate was heat sealed and placed in a conventional thermal cycler, and PCR proceeded according to the manufacturer’s protocol. Following the PCR, the 96-well plate was loaded on a QX100 droplet reader (Bio-Rad Laboratories, CA). Analysis of the ddPCR data was performed with QuantaSoft software (Bio-Rad; version 1.7.4.0917, CA), which analyzes each droplet individually using a two-color detection system (set to detect FAM or VIC dyes). The absolute quantification of DNA is directly dependent on the number of accepted droplets (positive and negative) and the DNA quantity analyzed. The calculation of the 95% CI given by the Poisson law and the distribution of the CNV values according to our sample cohort and controls led us to consider a sample as duplicated if the CNV value was > 2.5 and Poisson CNV minimum value (CNVmin) (95% CI) > 2.0.
In silico analyses
Genetic variants were evaluated by MutationTaster (http://www.mutationtaster.org/), Polymorphism Phenotyping v2 algorithm tool (PolyPhen-2) (http://genetics.bwh.harvard.edu/pph2), SIFT (Sorting Tolerant From Intolerant) algorithm (http://sift.jcvi.org), Mutation Assessor (http://mutationassessor.org/r3) and FATHMM (Functional Analysis through Hidden Markov Models v2.3) (http://fathmm.biocompute.org.uk/), to predict the possible impact of the amino acid substitution on the structure and function of the corresponding proteins. Variants that were predicted as damaging by more than three in silico tools and also have a frequency in gnomAD less than 1% were selected for further studies.
In silico modeling
Structural images were prepared using the PyMOL software (www.pymol.org). The structure of the mouse full-length dimeric R1β:Cα (pdb: 4DIN)22 was used to display the PKA catalytic (Cα) and regulatory (R1β) subunit structures. To display the position of cAMP, an alignment of the cyclic nucleotide binding domain of R1α (pdb: 5KJX) was performed.23 For the visualization of a tetrameric R1β:Cα holoenzyme, an alignment with the tetrameric R2β(2):Cα(2) holoenzyme was used.24
DNA constructs and cell culture
The wild-type (WT) (NM_001164760.2) and 2 variant (p.A67V and p.A300T) PRKAR1B sequences were introduced into different expression vectors according to the method described elsewhere.7
Fluorescence resonance energy transfer (FRET) by acceptor photobleaching
HEK293 cells (ATCC) were seeded onto 12 well plates and left overnight to recover. The HEK293 cells were transfected with R1β-Venus and Cα-Cerulean vectors (1 μg each) and 9h later the cells were placed in low-serum medium (0.5% FBS) for 12 h before stimulations. Experiments were performed on the confocal microscope Zeiss LSM 880 Airyscan as described previously (Zeiss, NY).25 Cα-Cerulean was imaged with the 405-nm laser, R1β-Venus with the 514-nm laser. Using a custom region of interest (ROI), R1β-Venus in one cell was bleached with the 514-nm laser at 100% transmission until the overall intensity dropped to between 80% and 50% of pre-bleach values. The R1β-Cα interaction was calculated by the difference in Cerulean intensity pre-versus post-bleaching. In each experiment, 15–20 bleaches on different cells were performed. Results shown are the combined results of three separate experiments.
Protein kinase A (PKA) enzymatic activity assay
HEK293 cells (ATCC) were transfected with 1μg of PRKAR1B vector and left for 24h. Tissues were homogenized in freshly prepared lysis buffer (10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 1 mM dithiothreitol with 0.5 mM PMSF and protease inhibitor cocktail I (1:100; EMD Biosciences, CA). BCA assays were performed as per manufacturer’s protocol to determine the total protein concentrations of samples (Pierce) (Thermo Fisher Scientific, MA). Samples were diluted to 1 μg/μL and 10 μL of total protein was used for each reaction. PKA enzymatic assays were performed by kemptide assay, using 25 μM kemptide (Leu-Arg-Arg-Ala-Ser-Leu-Gly), as previously described with and without cAMP (5 μM).26 All reactions for basal and cAMP-stimulated (total) PKA activity were carried out in duplicate. Additionally, activity values for replicate reactions that were incubated in the presence of protein kinase inhibitor (PKI) (5 nM) were subtracted from activity values to account for non-specific kinase activity.
In vitro PKA activity assay in tumor cell lysates
PKA catalytic activity was measured with or without the addition of cAMP using the PepTag non-radioactive cAMP-dependent protein kinase assay (#V5340, Promega, WI) using Kemptide (LRRASLG) as previously described.26 Images of the gels were acquired with a gel documentation system (Herolab) and analyzed using the ImageJ software (http://rsbweb.nih.gov/ij).
Statistics
Data are presented as the mean ± SEM. Data normality was assessed by Shapiro-Wilk test and the appropriate statistical test was used. The significance was determined by one-way ANOVA with multiple comparisons in the PKA activity assay as the data were normally distributed. For the FRET by acceptor photobleaching experiment, data were not normally distributed and Mann-Whitney test was used. Statistical significance was set at P < 0.05 and analyses were carried out using the GraphPadPrism 6 (GraphPad®) software.
RESULTS
Clinical studies
We studied 74 patients with micronodular forms of BAH with an average age of onset of CS 11.7 years (the minimum age was 8 months and maximum 46 years) and 21 patients with CPA with an average age of onset of CS 40.4 years (minimum age 6 months and maximum 66 years ), all referred for the investigation of corticotropin (ACTH)-independent CS. These patients were recruited by only one center (National Institutes of Health, Bethesda, MD, USA) over 25 years (1995–2020).
A. Patients with germline PRKAR1B variants
Three patients among 74 with isolated micronodular adrenocortical disease (iMAD) and no signs of CNC were found to carry germline PRKAR1B variants. The first patient (CAR757.03) was a 4-year-old female patient with CS that underwent bilateral adrenalectomy (ADX) but eventually succumbed to complications of her disease; her case has been published.27 Her disease was initially categorized as PPNAD but upon further review she was reclassified as iMAD, since she had minimal pigmentation and no other signs of CNC.27,28 She was found to carry the c.118A>G (p.I40V) PRKAR1B variant. The second patient (CAR762.03) had been diagnosed with Beckwith-Wiedemann syndrome (BWS [MIM: 130650]) and was included in a case series of patients with BWS, adrenal lesions, and ACTH-independent CS that we reported elsewhere.29 She underwent ADX at the age of 8 months. There is no more clinical information on this patient at this time. She was found to carry the c.200C>T (p.A67V) PRKAR1B variant. The third patient (CAR521.01) carried the germline c.898G>A (p.A300T) PRKAR1B variant. She underwent ADX for CS at the age of 9 years; her histology was typical for iMAD without any findings of PPNAD.21,27 At the age of 16 years, the patient was diagnosed with liver focal nodular hyperplasia (FNH) (data not shown). At present, she is 28 years old and healthy on glucocorticoid and mineralocorticoid replacement without any signs of CNC or other tumors. In all cases, family DNA was not available to study the inheritance of these PRKAR1B variants.
B. Patients whose tumors carried somatic 7p.22 CNV containing the PRKAR1B gene
Three patients with CPA among 21 were found to have CNG of the chromosome 7p22 PRKAR1B gene-harboring locus in their tumors (14.3% of total) (Fig. 3c). None carried germline variants in any of the examined genes. The tumor DNA was fully sequenced in all cases, and 1 of the 3 tumors carried the L206R PRKACA “hot-spot” variant that we and others have described previously.8 The subject’s heavily pigmented CPA (ADT47.03) was included in a case series presented elsewhere.30
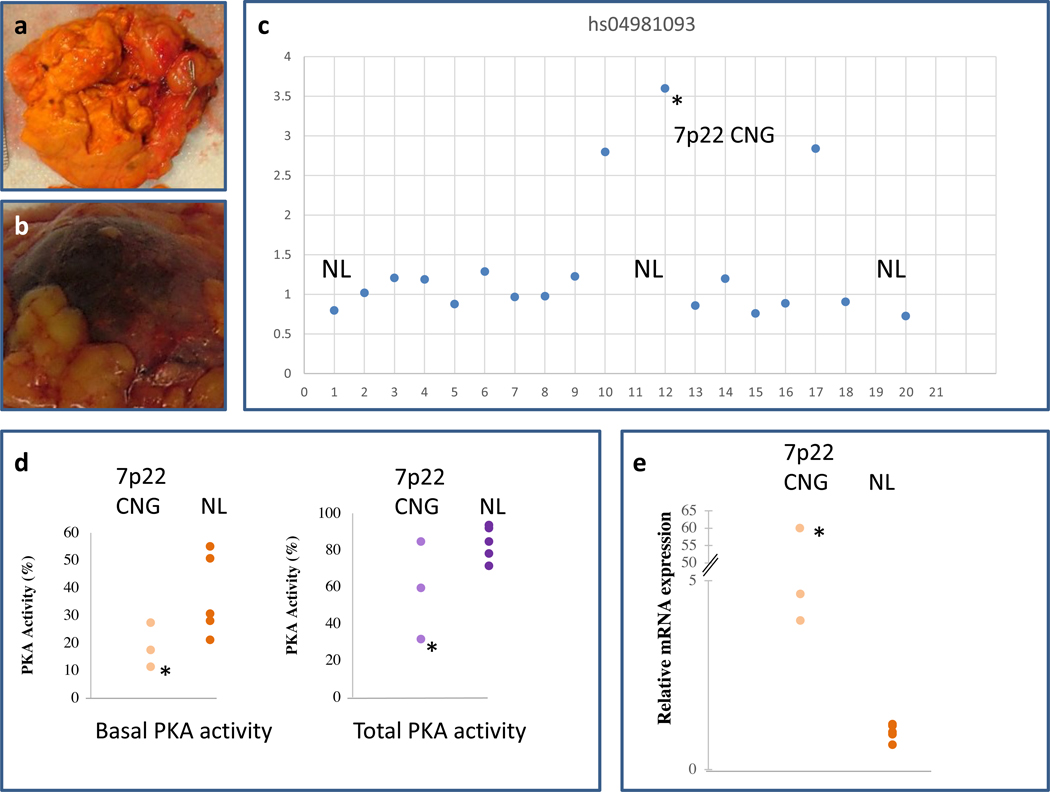
Fig. 3. PKA activity and PRKAR1B subunit expression in tumors with 7p22 CNG.
(a) Tumor sample ADT35.02. (b) Tumor sample ADT47.03. (c) Detection of 7p22 CNG, by the hs04981093 probe, in tumors ADT35.02, ADT47.03 and ADT183.02 at positions 10, 12 and 17 respectively. (d) PKA activity was measured in tumor cell lysates. The 3 tumors with 7p22 CNG had lower basal PKA activity (in absence of cAMP, left panel) but no differences in total PKA activity (in presence of cAMP, right panel) from NL CPA (N=5) that did not contain 7p22 CNG. (e) The 3 CPA with 7p22 CNG showed higher expression of the PRKAR1B mRNA compared to the 5 CPA that were genomically NL. NL stands for Normal and the asterisk (*) corresponds to tumor ADT47.03 as this is the CPA that also carried the L206R PRKACA “hot-spot” pathogenic variant.
Nothing unusual was identified in the histology of the other two CPA (ADT35.02 and ADT183.02). The clinical comparison for the two groups of patients is shown in Table S1. The mean age of presentation with ACTH-independent CS of the 3 patients with CPA and 7p22 CNG was 47.74 ± 3.35 years old; these patients were females. Among the remaining 18 patients, 15 and 3 were females and males respectively, with a mean age of 39.54 ± 5.03 years old. The biochemical data showed (Table S1) that the 3 patients with the CPA carrying 7p22 CNG had higher cortisol levels post high-dose dexamethasone suppression test (HDDST) (452.64 ± 0.00 versus 308.568 ± 57.13 nmol/L) and higher 17-hydroxysteroid (17-OHS) levels (15.00 ± 4.80 versus 8.88 ± 1.56 μg/g creatinine/24h) but similar 24h urinary free cortisol (UFC). The size of the tumors was also similar in the two groups. However, these differences were not statistically evaluated due to the small number of the tumors.
In silico analyses
Two of the identified PRKAR1B variants (p.A67V, p.A300T), the two rare ones, were in regions of the protein that are highly conserved evolutionarily (Table 1). The variant p.A67V was predicted as damaging for the function of the protein in 3 out of 5 in silico prediction tools used. And, the variant p.A300T was predicted as damaging by the 5 tools (Table S2). In the gnomAD database v2.0.1, the allele frequency for the p.I40V, p.A67V and p.A300T is 0.013545, 0.00019396 and 0.0000648, respectively (Table S2). None of the identified variants were found in the TCGA database of PRKAR1B sequence variation from a variety of human cancers (Table S3).
Table 1.
Multiple sequence alignment based on the PRKAR1B isoform NM_001164760.2
| Organism | Aminoacid Sequence | |||
|---|---|---|---|---|
| p.I40V | ||||
| variant sequence | ...YVQLHGIQQ VLKDCIVHLC | V | SKPER------------PMKFLRE---... | |
| Human | (Homo sapiens) | ...YVQLHGIQQ VLKDCIVHLC | I | SKPER------------PMKFLRE---... |
| Monkey | (Callithrix jacchus) | ...YVQLHGIQQVLKDCIVHLC | I | SKPER------------PMKFLRE---... |
| Rat | (Rattus norvegicus) | ...---------------------------QIMCV | V | SVCGEGMGWDPVEISQSHAE... |
| Chicken | (Gallus gallus) | ...YVQKHNIQQILKECIVNLC | I | AKPDR------------PMKFLRE---... |
| Frog | (Xenopus laevis) | ...YVQKYNIQQLLKECIVNLC | I | AKPDR------------PMKFLRE---... |
| Zebrafish | (Danio rerio) | ...FVQKHNIQQILKECIVNLC | I | AKPER------------PMKFLRE---... |
| p.A67V | ||||
| variant sequence | ...E------HFEKLEKEENRQIL | V | RQKSNSQSDSHDEEVSPTPP... | |
| Human | (Homo sapiens) | ...E------HFEKLEKEENRQIL | A | RQKSNSQSDSHDEEVSPTPP... |
| Monkey | (Callithrix jacchus) | ...E------HFEKLEKEENRQIL | A | RQKSNSQSDSHDEEVSPTPP... |
| Rat | (Rattus norvegicus) | ...SHAEGCAQTTGLEENRQIL | A | RQKSNSQCDSHDEEISPTPP... |
| Chicken | (Gallus gallus) | ...E------HFEKLEKEECKQIL | A | RQKSSSQSDSHDDEISPPPP... |
| Frog | (Xenopus laevis) | ...E------HFEKLEKEECKQIL | A | QQKSNSQSDSHDDEISPPPP... |
| Zebrafish | (Danio rerio) | ...E------HFEKLEKEECKQIM | A | RQKSNSQSDSHDDEVSPPPP... |
| p.A300T | ||||
| variant sequence | ...KIVVQGEPGDDFYIITEGT | T | SVLQRRSPNEEYVEVGRLGP... | |
| Human | (Homo sapiens) | ...KIVVQGEPGDDFYIITEGT | A | SVLQRRSPNEEYVEVGRLGP... |
| Monkey | (Callithrix jacchus) | ...KIVVQGEPGDDFYIITEGT | A | SVLQRRSPNEEYVEVGRLGP... |
| Rat | (Rattus norvegicus) | ...KIVVQGEPGDDFYIITEGT | A | SVLQRRSPNEEYVEVGRLGP... |
| Chicken | (Gallus gallus) | ...KIVVQGEPGDDFFIITEGT | A | SVLQRRSDNEEYVEVGRLGP... |
| Frog | (Xenopus laevis) | ...KIVVQGEPGDDFFIITEGT | A | SVLQRRSDNEEYVEVGRLGP... |
| Zebrafish | (Danio rerio) | ...KIVVQGEPGDDFFIITEGT | A | SVLQRRSDNEEYVEVGRLGP... |
In silico modeling
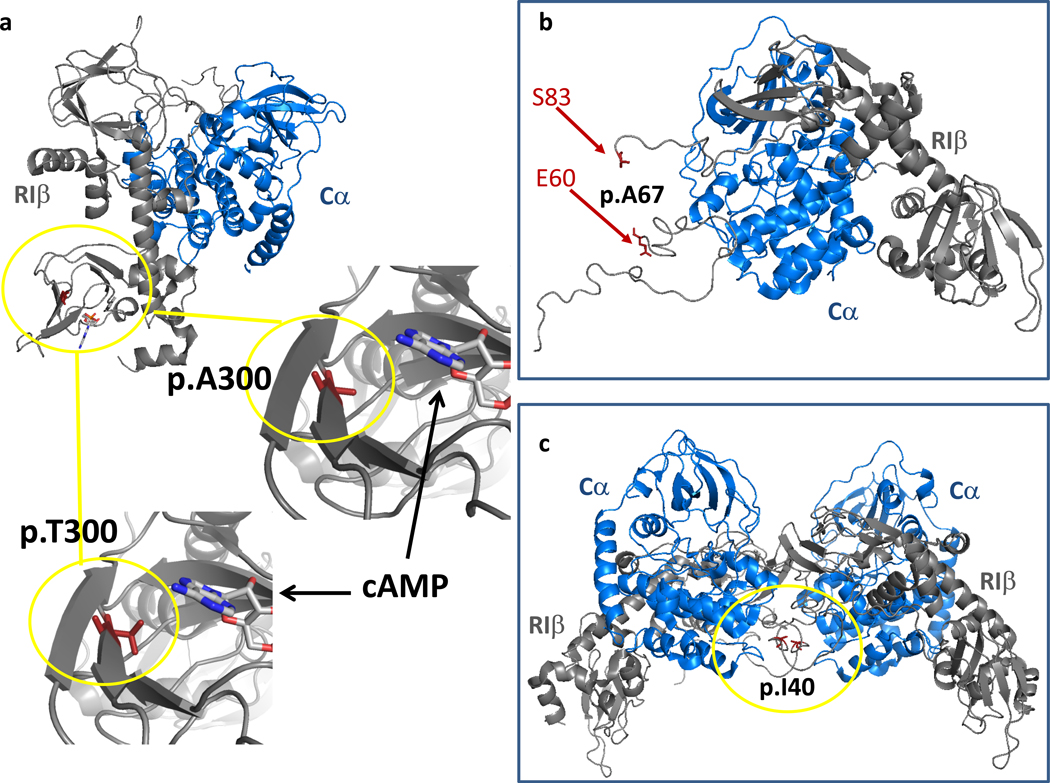
As shown in Fig. 1a, Alanine in position 300 is part of the cyclic nucleotide binding domain of R1β. Therefore, the p.A300T substitution may be affecting cAMP binding and, indirectly, the interaction of R1β with the, Cα subunit. As shown in the Fig. 1, threonine is bulkier compared to alanine and, thus, may interfere with cAMP binding, hindering the cAMP-dependent dissociation of the R1β - Cα complex. As depicted in Fig. 1b the Alanine in position 67 is part of the N-linker, a flexible region connecting the inhibitory sequence with the D/D domain for which there is little information on the possible effects on the R1β - Cα association or other functions.22 Mutation of the Isoleucine in position 40 (Fig. 1c) may interfere with holoenzyme formation or the association with PKA-binding proteins including “A-kinase anchoring proteins (AKAPs)” as predicted by Ilouz et al.22
Fig. 1. Location of p.A300T (a), p.A67V (b) and p.I40V (c) missense variants.
(a) Position of A300 in the R1β subunit (pdb: 4DIN). Zoomed in regions show A300 and its in silico replacement with T. cAMP is shown based on an alignment with the cyclic nucleotide binding domain of R1α (pdb: 5KJX). (b) Position of the flexible N-linker region, which is not resolved in the crystal structure and therefore does not reveal the position of A67. (c) Position of I40 in a tetrameric R1β-Cα complex based on an alignment of the dimeric R1β-Cα complex (pdb: 4DIN) to a tetrameric R2β-Cα holoenzyme (pdb: 3TNP). R1β and Cα subunits are depicted in grey and blue respectively.
In vitro expression studies and PKA enzymatic activity of PRKAR1B variants
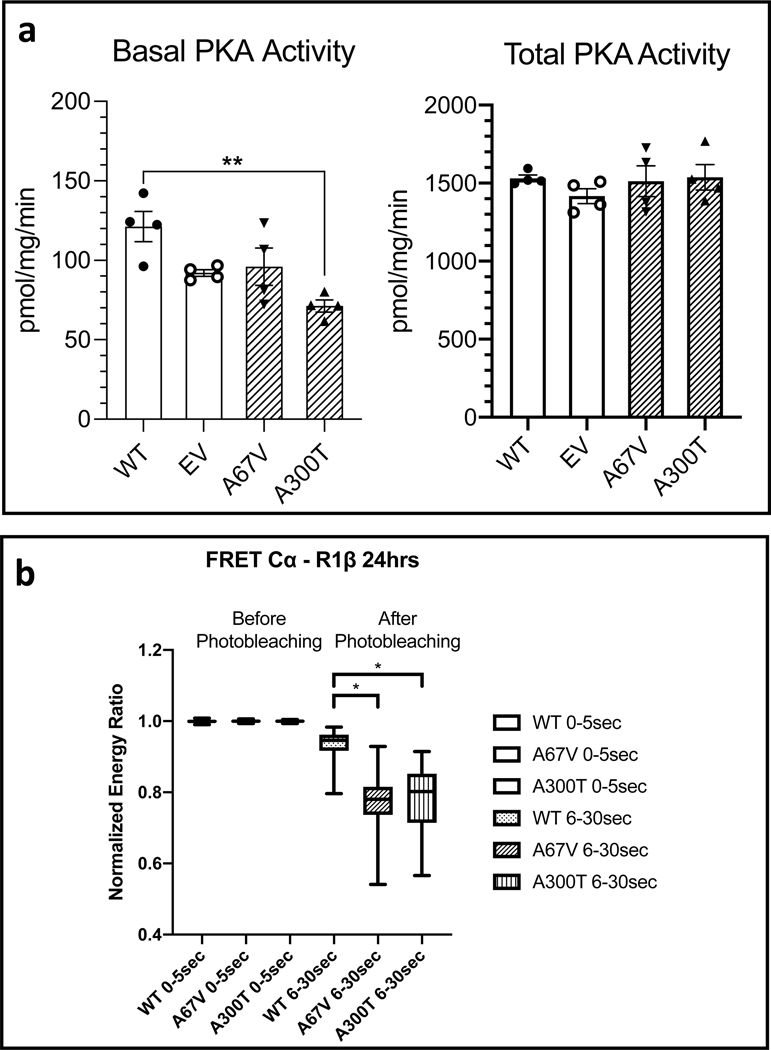
We focused our functional studies on the p.A67V and p.A300T variants only, as the p.I40V variant is located on an amino acid that is not conserved along the different species (Table 1) and is found relatively frequently in the general population; it was also predicted to be mostly benign by in silico analyses (Table S2). If this variant had any function it would most likely be related to AKAP-binding for which there is no readily available assay. As shown in Fig. 2a (left panel), when we expressed these two PRKAR1B variants (p.A67V and p.A300T) in HEK293 cells, they had decreased basal PKA activity although the data for the p.A67V-bearing construct failed to reach statistical significance. Both constructs behaved as the normal PRKAR1B did when stimulated with cAMP (Fig. 2a, right panel). Also, when performed fluorescence resonance energy transfer (FRET) studies, both constructs appeared to not release the main PKA catalytic subunit Cα (Fig. 2b) at an approximately equal rate; the difference from the WT R1β-Cα interaction was significant for both p.A67V and p.A300T variants.
Fig. 2. Effect of PRKAR1B variants on PKA activity and R1β-Cα interaction in HEK293 cells.
(a) PRKAR1B wild-type (WT), A67V and A300T variants or Empty Vector (EV) were expressed in HEK293 cells and PKA activity was measured. Both variants showed a lower basal PKA activity (in absence of cAMP, left panel) but only significantly different for A300T compared to the WT. Total PKA activity (in presence of cAMP, right panel) for both variants did not show any differences than the WT. Each experiment was done in duplicate. Data represent mean ± SEM and were analyzed by one-way ANOVA with multiple comparisons, **p < 0.01. (b) Results of Fluorescence resonance energy transfer (FRET) when R1β-Cα vectors were co-expressed in HEK293 cells. Energy transfer between R1β-Cα was measured before and after photobleaching (0–5 sec and 6–30 sec respectively). R1β harboring any of the two variants (p.A67V or p.A300T) appears to bind stronger to the catalytic PKA subunit. Results represent three combined separate experiments. Box plots show median values with upper and lower quartiles, whiskers represent minimal and maximal values. The different groups were compared by Mann-Whitney test, *p < 0.0001.
PKA activity and PKA subunit expression of CPA bearing chromosomal 7p22 gains
We measured PKA activity and PKA subunit expression in the tumors that had 7p22 CNG. Tumor ADT35.02 and ADT47.03 are depicted in Fig. 3a, b, respectively. These are the tumors in positions 10 and 12 in Fig. 3c which depicts just one of the three different probes that confirmed the presence of 7p22 CNG. Tumor ADT47.03 (heavily pigmented and shown in Fig. 3b is tumor 12 in the graph) is indicated throughout Fig. 3 with an asterisk (*), as this is the CPA that also carried the L206R PRKACA “hot-spot” pathogenic variant.30 The 3 tumors with 7p22 CNG had lower basal PKA activity but no differences in total PKA activity compared to the normal (NL) CPA (N=5) without 7p22 CNG (Fig. 3d). As expected from the 7p22 CNG containing the PRKAR1B gene, the 3 CPAs with 7p22 CNG had higher expression of the PRKAR1B mRNA compared to the 5 CPA that did not contain 7p22 CNG (Fig. 3e).
DISCUSSION
In this report, we identified both sequence variants and chromosomal amplification of the PRKAR1B gene in iMAD and CPA, respectively, associated with ACTH-independent CS. The study was prompted by a genome-wide search of CNV in CPA that identified 7p22 amplification in a small number of adenomas.15,16 To date, the only other human disease thought to be caused by a PRKAR1B defect is a rare form of a late-onset neurodegenerative disorder presenting with dementia or Parkinson’s syndrome that is characterized by abundant neuronal inclusions with immunoreactivity for intermediate filaments.31 A heterozygous c.149T>G (p.L50R) PRKAR1B missense substitution was found to segregate with the disease in 12 members of a large family. However, other studies failed to identify PRKAR1B defects in large numbers of patients with these rare forms of neurodegenerative disorders,32,33 and there were no functional studies of the p.L50R variant.
Two of the identified PRKAR1B variants (p.A67V, p.A300T) in our cohort were rare and in highly conserved parts of the protein (Table 1); in functional studies, they decreased basal PKA activity and were bound with higher affinity to the PKA catalytic subunit (Fig. 2a, b). The p.A67V variant is in the D/D domain which is hypothesized to be unique for its role in binding to the catalytic subunit in the R1β-Cα complex22 and the p.A300T variant may interfere with cAMP binding which is essential for the release of the catalytic subunit from the R1β-Cα complex (Fig. 1).
We also confirmed 7p22 CNG in 3 of 21 CPA (14.2%) with probes designed to specifically detect the PRKAR1B gene (Fig. 3c). This is a much higher percentage than what was reported previously; 15,16 however, the two techniques employed were different and we searched specifically for PRKAR1B amplification. A much larger study of our methodology needs to be completed to see how frequent these alterations are in CPAs, since our study was underpowered for determining the true rate of PRKAR1B-containing CNGs. Nevertheless, the amplification of the gene led to higher expression of the PRKAR1B mRNA which confirmed the genomic finding (Fig. 3e). Like in the previous study by Ronchi et al., in which 7p22 CNG were found in association mostly with CTNNB1 pathogenic variants16, in one of our subjects’ CPAs, PRKAR1B amplification was present along the L206R PRKACA “hot-spot” pathogenic variant.
In terms, of genotype-phenotype correlation, we did not see an obvious difference between CPAs that had PRKAR1B CNGs and those that did not (Table S1). We also did not see any differences among patients with BAH. The two groups of patients, BAH and CPAs differed in age, since BAHs in general occur much earlier in life. This is also expected for diseases that are due to germline defects versus those caused by somatic defects of the same gene, as seen in other conditions. Finally, in a much larger study of BAHs that is ongoing, we have not found any significant genetic or genomic changes that could account for additional phenotypic effects (data not shown).
The question that emerges of course is what is the functional significance of PRKAR1B CNG, if they are present alongside “driver” pathogenic variants such as CTNNB116 or PRKACA defects? The answer to that, in part, may be answered by the PKA enzymatic activity data for both PRKAR1B sequence variants found in our patients (Fig. 2a) and CPAs carrying PRKAR1B CNG (Fig. 3d). In all cases, PRKAR1B alterations led to decreases in basal PKA activity which is probably due to higher retention of the catalytic subunit by the R1β-Cα heterodimer in the PKA tetramer, as suggested by the FRET experiments (Fig. 2b). Additional studies of PRKAR1B activity in adrenocortical cell lines are needed to interpret our data, since what we presented here were results from HEK293 cells only. Nevertheless, it may be speculated that this tighter binding may be the case in tumors bearing PRKAR1B amplifications, the latter of which lead to an increase in available R1β protein. Such an increase in the amount of R1β in adrenocortical cells should not be overlooked as unimportant, both due to R1β’s role in mitochondria that are essential for steroidogenesis,22 as well as for its proven capacity to form heterodimers with R1α,17,20 a unique feature among PKA subunits.
Thus, one could speculate that the mechanism of tumorigenesis in BAHs (iMAD) and CPAs bearing PRKAR1B variants or CNG, respectively, is perturbation of PKA signaling in adrenocortical cells. This can be explained by disturbance of the relative ratios of PRKAR1A and PRKACA molecules that are available to the cAMP response, molecules with proven direct and causative involvement when mutated in other BAHs (PPNAD) and CPA, respectively.2,8 There is ample evidence that PRKAR1B can be altered when PRKAR1A is deficient in adrenocortical cells.34,35 Although Prkar1b-deficient mice did not have an obvious adrenocortical phenotype,36,37 most of their examination focused on neuronal abnormalities because of the preponderance of R1β expression in brain.17,22 Prkar1b-deficient mice had defective hippocampal long-term depression and depotentiation and diminished inflammation and nociceptive pain.36,37 However, PRKAR1B is not only expressed in the adrenal cortex and its levels are zone-specific,38 but also appears to be essential for steroidogenic differentiation and regulation of PKA signaling by histone methyltransferase EZH2.39 The latter data are an example of how altered PRKAR1B in Ezh2-deficient mouse adrenals can lead to PKA signaling defects resulting in phenotypes that one would typically associate with PRKAR1A or PRKACA; the fact is that any perturbation of PKA signaling can lead to adrenal abnormalities given the sensitive balance of functionally interacting pathways in adrenocortical zonation and tumorigenesis.39,40
In conclusion, this investigation reveals sequencing variants of the PRKAR1B PKA regulatory subunit in the germline of unrelated patients with iMAD and somatic PRKAR1B amplifications in CPA and confirms PRKAR1B’s potential for being involved in the perturbations of cAMP signaling that give rise to adrenocortical disease, although additional studies are needed for elucidation of R1β’s role in the pathogenesis of BAH and CPA leading to CS. Also, the data reveal possible new roles for R1β in disease pathogenesis; PRKAR1B should be included in the candidate genes for suspected PKA signaling defects in patients with phenotypes that overlap with those that carry PRKAR1A, PRKACA and PRKACB defects.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program, Eunice Kennedy Shriver National Institute of Child Health & Human Development, National Institutes of Health (project number Z1A 29 HD008920 to C.A.S) and DFG German Research Foundation (grant RO-5435/3-1 to C.L.R). K.B. was supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg. D.C. is supported by a Wellcome Trust Senior Research Fellowship (212313/Z/18/Z). The results of the identified somatic variants of PRKAR1B gene in different tissues here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
C.A.S. holds patent on the PRKAR1A, PDE11A and GPR101 genes and/or their function and his laboratory has received research funding from Pfizer Inc. F.R.F holds patent on the GPR101 gene and/or its function.
Footnotes
Conflict of interest statement
All other authors declare no competing interests.
REFERENCES
- 1.Calebiro D, Bathon K, Weigand I. Mechanisms of Aberrant PKA Activation by Calpha Subunit Mutations. Horm Metab Res. 2017;49(4):307–314. [DOI] [PubMed] [Google Scholar]
- 2.Kirschner LS, Carney JA, Pack SD, et al. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nature genetics. 2000;26(1):89–92. [DOI] [PubMed] [Google Scholar]
- 3.Horvath A, Bertherat J, Groussin L, et al. Mutations and polymorphisms in the gene encoding regulatory subunit type 1-alpha of protein kinase A (PRKAR1A): an update. Hum Mutat. 2010;31(4):369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertherat J, Horvath A, Groussin L, et al. Mutations in regulatory subunit type 1A of cyclic adenosine 5’-monophosphate-dependent protein kinase (PRKAR1A): phenotype analysis in 353 patients and 80 different genotypes. J Clin Endocrinol Metab. 2009;94(6):2085–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertherat J, Groussin L, Sandrini F, et al. Molecular and functional analysis of PRKAR1A and its locus (17q22–24) in sporadic adrenocortical tumors: 17q losses, somatic mutations, and protein kinase A expression and activity. Cancer research. 2003;63(17):5308–5319. [PubMed] [Google Scholar]
- 6.Forlino A, Vetro A, Garavelli L, et al. PRKACB and Carney complex. The New England journal of medicine. 2014;370(11):1065–1067. [DOI] [PubMed] [Google Scholar]
- 7.Espiard S, Knape MJ, Bathon K, et al. Activating PRKACB somatic mutation in cortisol-producing adenomas. JCI Insight. 2018;3(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beuschlein F, Fassnacht M, Assié G, et al. Constitutive activation of PKA catalytic subunit in adrenal Cushing’s syndrome. The New England journal of medicine. 2014;370(11):1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Dalmazi G, Kisker C, Calebiro D, et al. Novel somatic mutations in the catalytic subunit of the protein kinase A as a cause of adrenal Cushing’s syndrome: a European multicentric study. J Clin Endocrinol Metab. 2014;99(10):E2093–2100. [DOI] [PubMed] [Google Scholar]
- 10.Lodish MB, Yuan B, Levy I, et al. Germline PRKACA amplification causes variable phenotypes that may depend on the extent of the genomic defect: molecular mechanisms and clinical presentations. European journal of endocrinology. 2015;172(6):803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carney JA, Lyssikatos C, Lodish MB, Stratakis CA. Germline PRKACA amplification leads to Cushing syndrome caused by 3 adrenocortical pathologic phenotypes. Hum Pathol. 2015;46(1):40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng IC, Huang WJ, Jhuang YL, Chang YY, Hsu HP, Jeng YM. Microinsertions in PRKACA cause activation of the protein kinase A pathway in cardiac myxoma. The Journal of pathology. 2017;242(2):134–139. [DOI] [PubMed] [Google Scholar]
- 13.Honeyman JN, Simon EP, Robine N, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science (New York, NY). 2014;343(6174):1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singhi AD, Wood LD, Parks E, et al. Recurrent Rearrangements in PRKACA and PRKACB in Intraductal Oncocytic Papillary Neoplasms of the Pancreas and Bile Duct. Gastroenterology. 2020;158(3):573–582.e572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronchi CL, Sbiera S, Leich E, et al. Single nucleotide polymorphism array profiling of adrenocortical tumors--evidence for an adenoma carcinoma sequence? PLoS One. 2013;8(9):e73959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronchi CL, Di Dalmazi G, Faillot S, et al. Genetic Landscape of Sporadic Unilateral Adrenocortical Adenomas Without PRKACA p.Leu206Arg Mutation. The Journal of clinical endocrinology and metabolism. 2016;101(9):3526–3538. [DOI] [PubMed] [Google Scholar]
- 17.Solberg R, Taskén K, Wen W, et al. Human regulatory subunit RI beta of cAMP-dependent protein kinases: expression, holoenzyme formation and microinjection into living cells. Exp Cell Res. 1994;214(2):595–605. [DOI] [PubMed] [Google Scholar]
- 18.Elphinstone MS, Gordon RD, So A, Jeske YW, Stratakis CA, Stowasser M. Genomic structure of the human gene for protein kinase A regulatory subunit R1-beta (PRKAR1B) on 7p22: no evidence for mutations in familial hyperaldosteronism type II in a large affected kindred. Clin Endocrinol (Oxf). 2004;61(6):716–723. [DOI] [PubMed] [Google Scholar]
- 19.Charo LM, Burgoyne AM, Fanta PT, et al. A Novel PRKAR1B-BRAF Fusion in Gastrointestinal Stromal Tumor Guides Adjuvant Treatment Decision-Making During Pregnancy. J Natl Compr Canc Netw. 2018;16(3):238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson CR, Ruppelt A, Taskén K. A kinase anchoring protein (AKAP) interaction and dimerization of the RIalpha and RIbeta regulatory subunits of protein kinase a in vivo by the yeast two hybrid system. Journal of molecular biology. 2003;327(3):609–618. [DOI] [PubMed] [Google Scholar]
- 21.Horvath A, Boikos S, Giatzakis C, et al. A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nature genetics. 2006;38(7):794–800. [DOI] [PubMed] [Google Scholar]
- 22.Ilouz R, Bubis J, Wu J, et al. Localization and quaternary structure of the PKA RIβ holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(31):12443–12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz R, Moon EW, Kim JJ, et al. Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity. Biochem J. 2017;474(14):2389–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, Smith-Nguyen EV, Keshwani MM, Deal MS, Kornev AP, Taylor SS. Structure and allostery of the PKA RIIβ tetrameric holoenzyme. Science (New York, NY). 2012;335(6069):712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavrakis M, Lippincott-Schwartz J, Stratakis CA, Bossis I. Depletion of type IA regulatory subunit (RIalpha) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Human molecular genetics. 2006;15(19):2962–2971. [DOI] [PubMed] [Google Scholar]
- 26.Nesterova M, Yokozaki H, McDuffie E, Cho-Chung YS. Overexpression of RII beta regulatory subunit of protein kinase A in human colon carcinoma cell induces growth arrest and phenotypic changes that are abolished by site-directed mutation of RII beta. Eur J Biochem. 1996;235(3):486–494. [DOI] [PubMed] [Google Scholar]
- 27.Bartz SK, Karaviti LP, Brandt ML, et al. Residual manifestations of hypercortisolemia following surgical treatment in a patient with Cushing syndrome. Int J Pediatr Endocrinol. 2015;2015(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannah-Shmouni F, Stratakis CA. A Gene-Based Classification of Primary Adrenocortical Hyperplasias. Horm Metab Res. 2020;52(3):133–141. [DOI] [PubMed] [Google Scholar]
- 29.Carney JA, Ho J, Kitsuda K, Young WF Jr., Stratakis CA. Massive neonatal adrenal enlargement due to cytomegaly, persistence of the transient cortex, and hyperplasia of the permanent cortex: findings in Cushing syndrome associated with hemihypertrophy. The American journal of surgical pathology. 2012;36(10):1452–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angelousi A, Szarek E, Shram V, Kebebew E, Quezado M, Stratakis CA. Lipofuscin Accumulation in Cortisol-Producing Adenomas With and Without PRKACA Mutations. Horm Metab Res. 2017;49(10):786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong TH, Chiu WZ, Breedveld GJ, et al. PRKAR1B mutation associated with a new neurodegenerative disorder with unique pathology. Brain. 2014;137(Pt 5):1361–1373. [DOI] [PubMed] [Google Scholar]
- 32.Cohn-Hokke PE, Wong TH, Rizzu P, et al. Mutation frequency of PRKAR1B and the major familial dementia genes in a Dutch early onset dementia cohort. J Neurol. 2014;261(11):2085–2092. [DOI] [PubMed] [Google Scholar]
- 33.Pottier C, Baker M, Dickson DW, Rademakers R. PRKAR1B mutations are a rare cause of FUS negative neuronal intermediate filament inclusion disease. Brain. 2015;138(Pt 6):e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heyerdahl SL, Boikos S, Horvath A, Giatzakis C, Bossis I, Stratakis CA. Protein kinase A subunit expression is altered in Bloom syndrome fibroblasts and the BLM protein is increased in adrenocortical hyperplasias: inverse findings for BLM and PRKAR1A. Horm Metab Res. 2008;40(6):391–397. [DOI] [PubMed] [Google Scholar]
- 35.Robinson-White A, Meoli E, Stergiopoulos S, et al. PRKAR1A Mutations and protein kinase A interactions with other signaling pathways in the adrenal cortex. The Journal of clinical endocrinology and metabolism. 2006;91(6):2380–2388. [DOI] [PubMed] [Google Scholar]
- 36.Brandon EP, Zhuo M, Huang YY, et al. Hippocampal long-term depression and depotentiation are defective in mice carrying a targeted disruption of the gene encoding the RI beta subunit of cAMP-dependent protein kinase. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(19):8851–8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malmberg AB, Brandon EP, Idzerda RL, Liu H, McKnight GS, Basbaum AI. Diminished inflammation and nociceptive pain with preservation of neuropathic pain in mice with a targeted mutation of the type I regulatory subunit of cAMP-dependent protein kinase. J Neurosci. 1997;17(19):7462–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigand I, Ronchi CL, Rizk-Rabin M, et al. Differential expression of the protein kinase A subunits in normal adrenal glands and adrenocortical adenomas. Sci Rep. 2017;7(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathieu M, Drelon C, Rodriguez S, et al. Steroidogenic differentiation and PKA signaling are programmed by histone methyltransferase EZH2 in the adrenal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(52):E12265-e12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drelon C, Berthon A, Sahut-Barnola I, et al. PKA inhibits WNT signalling in adrenal cortex zonation and prevents malignant tumour development. Nat Commun. 2016;7:12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.