Abstract
GNA13, encoding one of the G protein alpha subunits of heterotrimeric G proteins that transduce signals of G protein-coupled receptors (GPCR), is frequently mutated in germinal center B-cell-like diffuse large B-cell lymphoma (GCB-DLBCL) with poor prognostic outcomes. Due to the “undruggable” nature of GNA13, targeted therapy for these patients is not available. In this study, we found that palmitoylation of GNA13 not only regulates its plasma membrane localization, but also regulates GNA13’s stability. It is essential for the tumor suppressor function of GNA13 in GCB-DLBCL cells. Interestingly, GNA13 negatively regulates BCL2 expression in GCB-DLBCL cells in a palmitoylation-dependent manner. Consistently, BCL2 inhibitors were found to be effective in killing GNA13-deficient GCB-DLBCL cells in a cell-based chemical screen. Furthermore, we demonstrate that inactivating GNA13 by targeting its palmitoylation enhanced the sensitivity of GCB-DLBCL to the BCL2 inhibitor. These studies indicate that the loss-of-function mutation of GNA13 is a biomarker for BCL2 inhibitor therapy of GCB-DLBCL and that GNA13 palmitoylation is a potential target for combination therapy with BCL2 inhibitors to treat GCB-DLBCL with wild-type GNA13.
Subject terms: B-cell lymphoma, B-cell lymphoma, Translational research
Introduction
GNA13 encodes one of the alpha subunits (GNA13/Gα13) of the heterotrimeric G proteins that transduce signals of G protein-coupled receptors (GPCR). It is expressed in various tissues, including lymphoid, vascular, and bone tissues in embryos and adults. Although GNA13 is classified into the Gα12/13 subfamily and highly homologous to GNA121, GNA13 has unique functions. It has been shown to play critical roles in localization of germinal center (GC) B cells2, angiogenesis3, female fertility4,5, bone homeostasis6, and platelet activation7,8.
Recurrent mutations in the GNA13 gene have been identified in multiple tumor types. As GNA13 activation can promote migration, invasion, and metastasis in pancreas, prostate, and ovarian cancer, it was originally classified as an oncogene9–11. However, loss-of-function mutations in GNA13 have recently been identified in diffuse large B-cell lymphoma (DLBCL)12–14, indicating that GNA13 may also function as a tumor suppressor. Consistent with this observation, GNA13-deficient mice develop GC B-cell-derived lymphoma2.
DLBCL is the most commonly diagnosed lymphoma and accounts for 25–35% of all B-cell non-Hodgkin lymphomas15. Based on the gene expression pattern and cell-of-origin, DLBCL is usually classified into two main subtypes, namely, GC B-cell-like (GCB) and activated B-cell-like (ABC) DLBCL16,17. Although nearly 60% of DLBCL patients can be cured by Rituximab plus chemotherapy-based standard treatment (R-CHOP), the rest may die due to therapy nonresponsiveness or disease relapse resulting from the complexity and heterogeneity of the disease13. Identifying valuable therapeutic targets for treating DLBCL remains an urgent need.
In the GC, B cells are strictly confined within follicles by the GPCR signaling, such as sphingosine-1-phosphate receptor S1PR2 and purinergic receptor P2RY8 signaling18–20. GNA13 was found to activate ARHGEF1-RHOA and subsequently inhibits the phosphoinositide 3-kinase (PI3K)/AKT pathway21. A recent CRISPR/Cas9-based screen in primary GC B cells showed that GNA13 depletion strikingly enhances cell survival and proliferation, indicating its major suppressive role in constraining GC B cells22. Consistent with this, over 18% of germinal center B-cell-like diffuse large B-cell lymphoma (GCB-DLBCL) patients harbor loss-of-function mutations or homozygous deletions in the GNA13 gene locus12–14. Additionally, some partners of GNA13, like S1PR2, P2RY8, ARHGEF1, and RHOA, are also frequently mutated or dysregulated in GCB-DLBCL, implying the critical role of this GPCR signaling in lymphomagenesis12,23,24.
Although GCB-DLBCL prognosis is generally more favorable than that of ABC-DLBCL, a recent comprehensive analysis of 1001 DLBCL patients revealed that GCB-DLBCL patients who harbor GNA13 mutations and also express high level of BCL2 have an extraordinarily high risk of poor outcomes25. However, no effective therapeutic strategy is available for this DLBCL subtype.
Post-translational protein modifications regulate protein function and can be used as therapeutic targets. S-palmitoylation involves palmitoyl acyltransferase (PAT)-mediated covalent lipid modification of cysteine side chains with the 16-carbon fatty acid, palmitate26,27. Palmitoylation regulates the membrane association, subcellular trafficking, stability, and function of proteins26. We previously showed that palmitoylation of NRAS is essential for its plasma membrane (PM) translocation, signal transduction, and leukemogenesis, both in vivo and in vitro28.
Palmitoylation is required for GNA13 to associate with the PM and the activation of Rho-dependent signaling29. Here, we show that palmitoylation of GNA13 also regulates its stability and is required for its tumor suppressor function in GCB-DLBCL cells. Interestingly, GNA13 negatively regulated BCL2 expression in GCB-DLBCL cells in a palmitoylation-dependent manner. Inactivating GNA13 by targeting its palmitoylation enhanced the sensitivity of GCB-DLBCL cells to the BCL2 inhibitors. Our studies suggested that GNA13 loss-of-function mutations may serve as a biomarker for BCL2 inhibitor-mediated precision therapy of DLBCL and that GNA13 palmitoylation may be a potential target for combination therapy with BCL2 inhibitors to treat DLBCL with wild-type (WT) GNA13.
Results
Palmitoylation regulates GNA13 protein stability
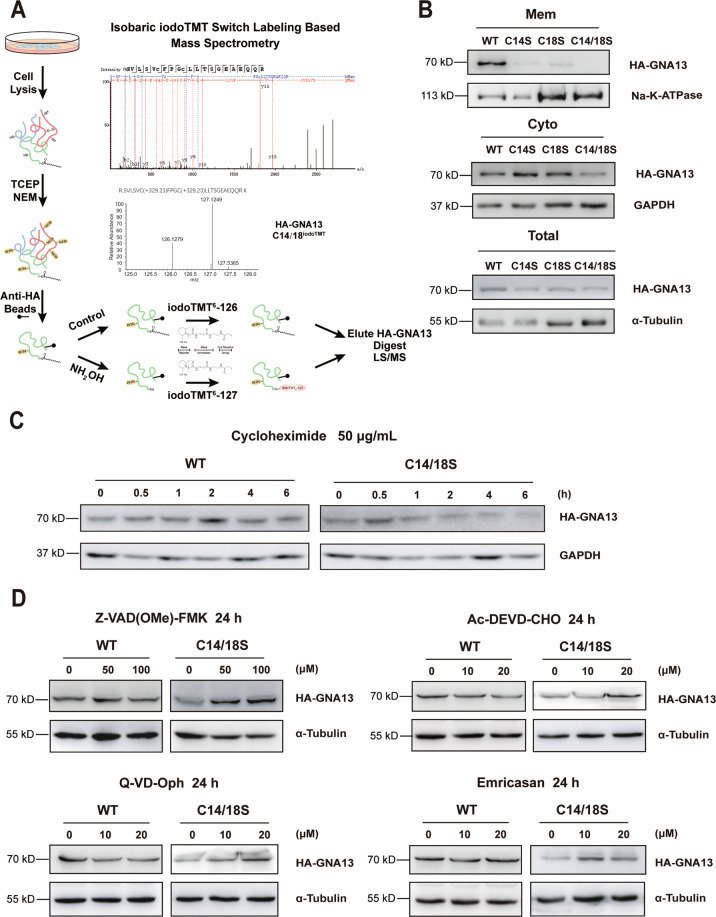
To elucidate the role of GNA13 palmitoylation in GCB-DLBCL, we first confirmed the palmitoylation sites in GNA13 employing isobaric iodoTMT switch labeling in HeLa cells stably expressing HA-tagged GNA13. The proteomics data showed that both cysteine 14 (C14) and 18 (C18) contained iodoTMT6-127, indicative of palmitoyl modifications (Fig. 1A). All other cysteines could be excluded as palmitoylation sites except for C236, because the tryptic peptide containing this residue could not be resolved by mass spectrometry owing to its small size. Similarly, a click chemistry-based, single-cell in situ proximity ligation assay (Supplementary Fig. S1A–C) showed that GNA13 was palmitoylated (red fluorescence) and that palmitoylation was almost abolished by the C14/18S double mutation. We further confirmed the above results using bioinformatic algorithms (CSS-PALM 4.030, MDD-PALM31) and an Acyl-RAC assay (Supplementary Fig. S1D, E). These results were consistent with previous findings29.
Fig. 1. Palmitoylation of GNA13 regulates its protein stability.
A The scheme of isobaric iodoTMT switch labeling-based mass spectrometry assay and results of MS/MS spectrum of palmitoylated peptide of GNA13. IodoTMT6-127 labeling on Cys14 and Cys18 (the two C in lowercase) of GNA13 peptide sequence was shown in peaks graph (upper panel). B Total, membrane (Mem) and cytosolic (Cyto) fractions of HeLa cells expressing HA-tagged WT GNA13, C14S, C18S, or C14/18S mutant of GNA13 were immunoblotted with an anti-HA antibody. α-Tubulin was used as a loading control for total cellular proteins, while Na-K-ATPase and GAPDH were used as markers of the membrane and cytosol, respectively. C HeLa cells overexpressing WT GNA13 or C14/18S mutant were incubated with cycloheximide (CHX) and analyzed by western blot at the indicated time points. D Protein levels of WT GNA13 and C14/18S mutant in HeLa cells treated with or without indicated caspase inhibitors for 24 h were detected by immunoblotting with an anti-HA antibody. α-Tubulin was used as a loading control.
Next, we characterized the PM localization of GNA13 by co-staining with the PM marker, Na-K ATPase (Supplementary Fig. S3A). A large fraction of WT GNA13 (GNA13WT) localized to the PM, whereas GNA13 harboring the single or double mutations in palmitoylation sites showed diffused cytoplasmic staining. Furthermore, 2-bromopalmitate, a pan-palmitoylation inhibitor, markedly inhibited the membrane localization of GFP-GNA13WT after a short treatment (Supplementary Fig. S3B). We further employed a biochemical method to isolate the PM and cytosolic fractions of the cell. Consistent with a previous report29, the data showed that the PM fraction of palmitoylation-deficient GNA13 mutants was considerably less than that of GNA13WT (Fig. 1B).
Notably, the expression level of the GNA13C14/18S mutant was lower than that of GNA13WT, as well as that of the single mutants (Fig. 1B), indicating that palmitoylation affected protein stability. A cycloheximide-based pulse-chase experiment revealed that the loss of palmitoylation accelerated GNA13 degradation compared with GNA13WT (Fig. 1C).
Palmitoylation can regulate protein stability by affecting critical proteolytic processes, such as those associated with the ubiquitin-proteasome32,33, autophagosome-lysosome34, and caspase systems35,36. To examine if palmitoylation regulates GNA13 protein stability through these systems, we tested whether the downregulation of the GNA13C14/18S mutant could be rescued by treatment with the proteasomal inhibitor MG132, the autophagy inhibitors HCQ and ULK-101, and several caspase inhibitors. Interestingly, the downregulation of GNA13C14/18S was marked reversed by exposure to the pan-caspase inhibitors Z-VAD(OMe)-FMK, Q-VD-Oph, and Emricasan, as well as by the group II caspase-specific inhibitor Ac-DEVD-CHO (Fig. 1D), indicating that the palmitoylation of GNA13 regulates its stability through the evasion of caspase-associated degradation.
Interestingly, the level of GNA13WT can be upregulated by MG-132 (Supplementary Fig. S2C), indicating that GNA13 is also subject to proteasome-mediated degradation. Meanwhile, protein levels of both GNA13WT and GNA13C14/18S can be moderately upregulated by autophagic inhibitors HCQ and ULK-101 (Supplementary Fig. S2D), suggesting that GNA13 can also be regulated by the autophagosome-lysosome system. Palmitoylation does not appear to affect these processes.
These data demonstrated that GNA13 palmitoylation regulates both its PM localization and its stability.
Palmitoylation of GNA13 is required for its tumor suppressor function
GNA13 is frequently mutated in GCB-DLBCL. Various sites are mutated throughout the GNA13 gene, consistent with their loss-of-function nature11–14. Through the whole genome sequencing data obtained from St. Jude Cloud37, we found that there were at least two patients harboring the GNA13 C14S mutation (Supplementary Fig. S3)38,39, suggesting that palmitoylation of GNA13 regulates its tumor suppression function.
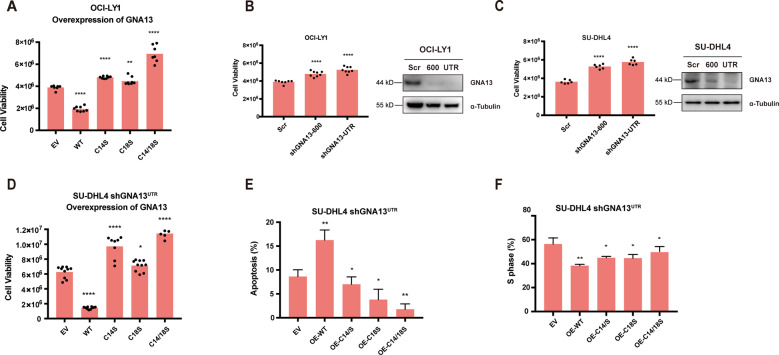
To examine the role of palmitoylation in GNA13’s tumor suppressor function, we compared the tumor suppressor activity of palmitoylation-deficient mutants of GNA13 to that of WT counterpart in two GCB-DLBCL cell lines. First, we transduced GNA13WT, GNA13C14S, GNA13C18S, and GNA13C14/18S into OCI-LY1, a GCB-DLBCL cell line harboring loss-of-function GNA13 mutations. Consistent with the tumor suppressor function of GNA13, we found that ectopic expression of GNA13WT markedly suppressed proliferation of OCI-LY1 cells (Fig. 2A). The three GNA13 palmitoylation mutants, on the other hand, did not inhibit proliferation of OCI-LY1 cells (Fig. 2A), indicating that palmitoylation of GNA13 is required for its tumor suppressor function.
Fig. 2. Palmitoylation of GNA13 is required for its tumor suppressor function.
A Viability of OCI-LY1 cells transfected with empty vector (EV), WT, C14S, C18S, or C14/18S GNA13 construct measured by CellTiter-Glo Luminescent Cell Viability Assay 48 h after plating. B, C Knocking down of the endogenous GNA13 in OCI-LY1 (B) or SU-DHL4 (C) cells by two GNA13-specific shRNAs, shGNA13-600 and shGNA13-UTR. Scrambled shRNA (Scr) was used as the negative control. Cell viability of each cell line was measured by CellTiter-Glo Luminescent Cell Viability Assay 48 h after plating. Protein levels of GNA13 in these cells were examined by western blot analysis. α-Tubulin was used as loading controls. D SU-DHL4 cells expressing shGNA13-UTR were transfected with EV, HA-tagged WT, C14S, C18S, or C14/18S mutant GNA13. Cell viability of each cell line was measured by CellTiter-Glo Luminescent Cell Viability Assay 48 h after plating. E Apoptosis assay of each SU-DHL4 cell line using Annexin V/PI staining was examined by flowcytometry 48 h after plating. OE, overexpression. F Cell-cycle analysis of each SU-DHL4 cell line by using BrdU/7-AAD staining. Percentage of the cells at S phase was shown as the proliferative portion of whole population.
Interestingly, the palmitoylation mutants of GNA13, particularly GNA13C14/18S, actually promoted proliferation of OCI-LY1 cells (Fig. 2A). This result suggests that OCI-LY1 cells retain partial GNA13 tumor suppressor functions, either the GNA13 mutant in OCI-LY1 cells retain partial tumor suppressor function or the presence of the wild-type allele of GNA13, and that palmitoylation mutants of GNA13 have a dominant negative effect. To test this possibility, we knocked down the endogenous GNA13 by introducing GNA13 shRNAs (Fig. 2B). We found that two independent GNA13 shRNAs, shGNA13600 and shGNA13UTR, both promoted proliferation of OCI-LY1 cells (Fig. 2B), supporting the assumption that OCI-LY1 cells retain partial GNA13 tumor suppressor functions.
Next, we further confirm the role of palmitoylation in GNA13’s tumor suppressor function in SU-DHL4, a GCB-DLBCL cell line with the WT GNA13. In this experiment, we first transfected SU-DHL4 cells with either scrambled or GNA13-specific shRNAs. The endogenous GNA13 expression was significantly reduced in SU-DHL4 cells transfected with specific shRNAs (Fig. 2C). More importantly, proliferation of SU-DHL4 cells transfected with GNA13 shRNAs was significantly increased as compared to SU-DHL4 cells transfected with control shRNA (Fig. 2C). We chose SU-DHL4 cell line stably expressing the GNA13-UTR shRNA (SU-DHL4-shGNA13UTR) for further experiments.
We then transduced GNA13WT, GNA13C14S, GNA13C18S, and GNA13C14/18S into SU-DHL4-shGNA13UTR cells. As expected, ectopic expression of GNA13WT markedly suppressed proliferation of SU-DHL4-shGNA13UTR cells (Fig. 2D). The three GNA13 palmitoylation mutants, on the contrary, did not inhibit proliferation of SU-DHL4-shGNA13UTR (Fig. 2D), further demonstrating that palmitoylation of GNA13 is required for its tumor suppressor function. The increased proliferation of SU-DHL4-shGNA13UTR expressing palmitoylation mutants of GNA13 compared to that transfected with the vector control may be a result of the dominant negative effect of palmitoylation mutants of GNA13 over the residual GNA13, as discussed above (Fig. 2A, D).
In addition, we found that SU-DHL4-shGNA13UTR expressing palmitoylation mutants of GNA13 exhibited a significant decrease in annexin V/PI positive apoptotic cell population compared to SU-DHL4-shGNA13UTR expressing the WT GNA13 (Fig. 2E and Supplementary Fig. S4A), indicating that palmitoylation of GNA13 is required for its pro-apoptotic function. We also found that the cells overexpressing GNA13WT showed a higher level of the cleaved Caspase3, which means the WT GNA13 could induce an active state of caspase3 (Supplementary Fig. S5). Consistently, a similar trend in cell proliferation could also been observed among these cells when we assessed the cell-cycle progression using a BrdU/7-AAD labeling assay (Fig. 2F and Supplementary Fig. S4B).
GNA13 negatively regulates BCL2 expression in GCB-DLBCL in a palmitoylation-dependent manner
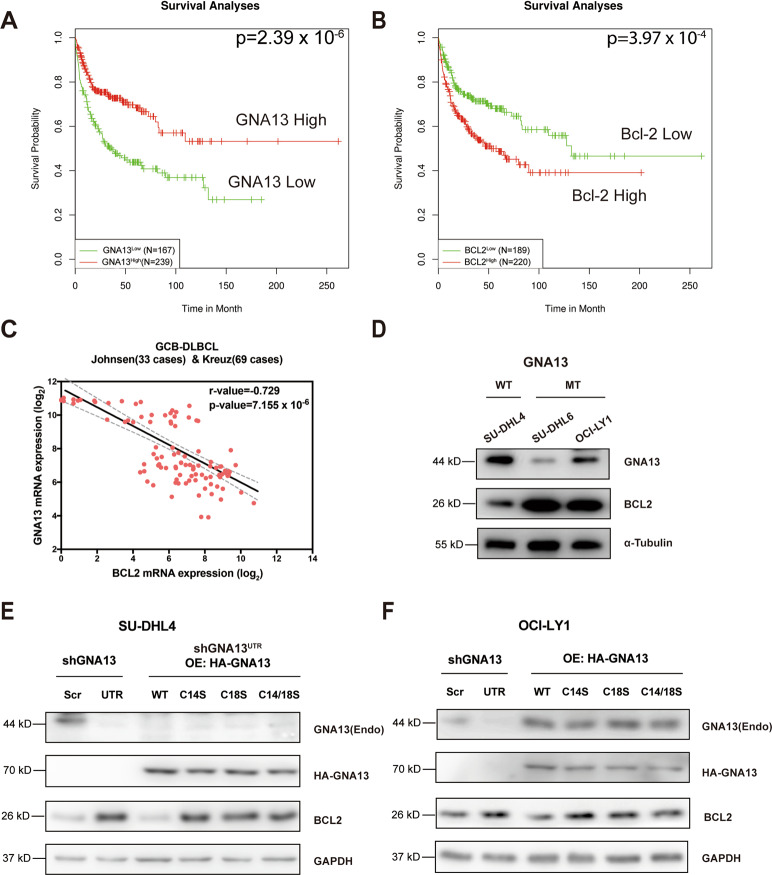
Intriguingly, the clinical data from PPISURV40 revealed that GNA13 and BCL2 expressions exhibited opposite prognostic effects on the survival of DLBCL patients (Fig. 3A, B). To gain insights into the mechanism by which GNA13 functions as a tumor suppressor in GCB-DLBCL, we analyzed RNA sequencing data of 102 GCB-DLBCL cases from public datasets (R2: Genomics Analysis and Visualization Platform, https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). We found that BCL2 expression and GNA13 expression are inversely correlated (Fig. 3C).
Fig. 3. GNA13 regulates the BCL2 expression in GCB-DLBCL cells in a palmitoylation-dependent manner.
A, B Relationship between survival of GCB-DLBCL and expression levels of GNA13 and BCL-2 was analyzed based on public datasets (PPISURV). (A) High-level expression of GNA13 is significantly associated with positive outcome of GCB-DLBCL. (B) Overexpression of BCL-2 is significantly associated with negative outcome of GCB-DLBCL. C Plot of GNA13 expression vs. BCL2 expression in 102 GCB-DLBCL cases (R2: Genomics Analysis and Visualization Platform, https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). The transcript levels of GNA13 and BCL2 are inversely related (r = −0.729, P = 7.155 × 10−6). D Western blot analysis of GNA13 and BCL2 expressions in SU-DHL4 cells, which harbor the WT GNA13, OCI-LY1, and SU-DHL6 cells, both of which harbor mutant GNA13. α-Tubulin was used as a loading control. E Western blot analysis of GNA13 and BCL2 expressions in SU-DHL4 cells expressing scrambled shRNA (Scr) or GNA13 knockdown shRNA (UTR) (left two lanes), and the SU-DHL4-shGNA13UTR cells ectopically overexpressing (OE) HA-tagged WT, C14S, C18S, or C14/18S mutant GNA13 (right four lanes). GAPDH was used as a loading control. F Western blot analysis of GNA13 and BCL2 expressions in OCI-LY1 cells expressing scrambled shRNA (Scr) or GNA13 knockdown shRNA (UTR) (left two lanes) and the OCI-LY1 cells ectopically expressing HA-tagged WT, C14S, C18S, or C14/18S mutant GNA13 (right four lanes). GAPDH was used as a loading control.
To check if the BCL2 expression is affected by the GNA13 activity, we examined BCL2 expression levels in GCB-DLBCL cell lines either with the WT GNA13 (SU-DHL4) or mutant GNA13 (OCI-LY1 and SU-DHL6). We found that the expression of BCL2 is significantly higher in cells with GNA13 mutant than the cells with WT GNA13 (Fig. 3D). These data suggest that GNA13 may exert its tumor suppressor function partially by regulating the BCL2 expression.
To test this hypothesis, we examined the expression level of BCL2 in SU-DHL4 cells vs. SU-DHL4-shGNA13UTR cells. We found that BCL2 expression was drastically elevated when GNA13 was knocked down (Fig. 3E), indicating that GNA13 negatively regulates the expression of BCL2. We then moved on to do a rescue experiment and found that ectopic expression WT GNA13 in SU-DHL4-shGNA13UTR inhibited the expression of BCL2 (Fig. 3E), confirming that GNA13 negatively regulates BCL2 expression level.
Consistent with the previous finding that palmitoylation of GNA13 is required for its tumor suppressor function, ectopic expression of palmitoylation mutants of GNA13 were found to be incapable of suppressing the expression of BCL2 in SU-DHL4-shGNA13UTR cells (Fig. 3E). Similar results were obtained in OCI-LY1 cells bearing a loss-of-function mutant of GNA13 (Fig. 3F). These data demonstrate that GNA13 is a negative regulator of BCL2 and that palmitoylation is required for this function of GNA13.
GNA13-deficient GCB-DLBCL cells are hypersensitive to the treatment with BCL2 inhibitors
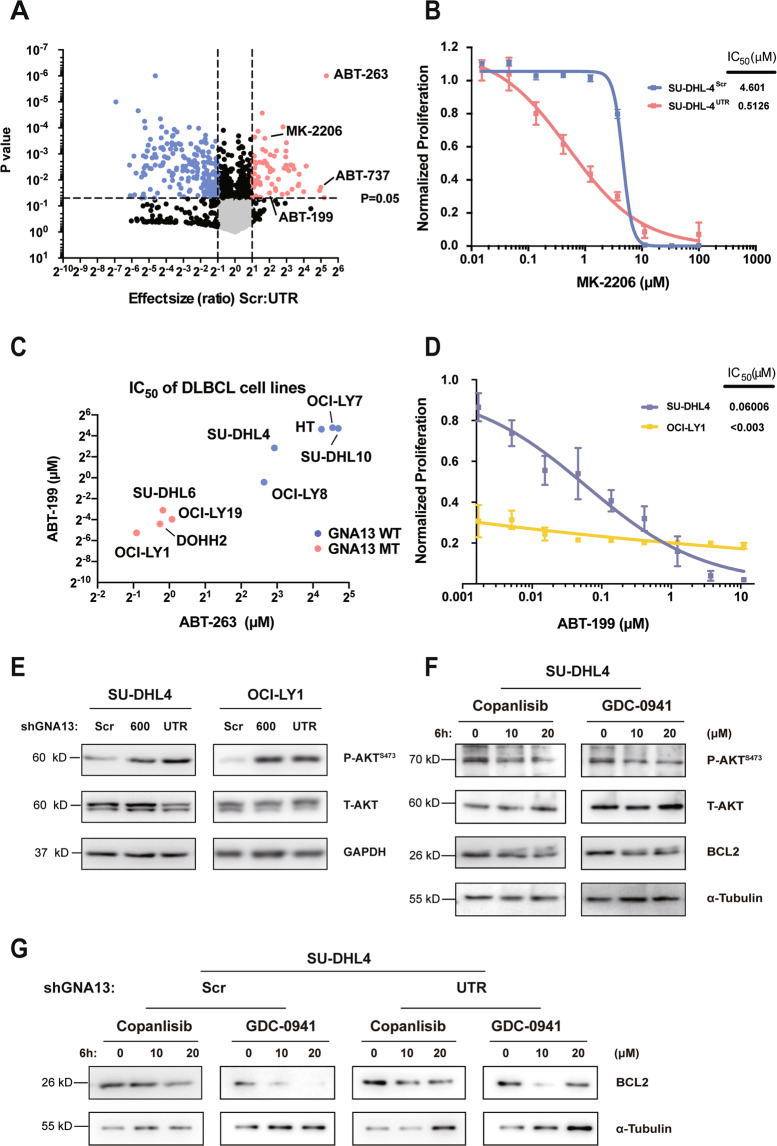
To find potential therapies for GNA13-deficient GCB-DLBCL, we carried out a cell-based drug screening using a chemical library comprising FDA-approved drugs and bioactive compounds with known targets. The SU-DHL4-shGNA13UTR described above was used as a model for GNA13-deficient GCB-DLBCL and the parental SU-DHL4 was used for the counter screening. We found that two BCL2 inhibitors, ABT-737 and ABT-263 (the first- and second-generation BCL2 inhibitors, respectively), exhibited the most significant efficacy in killing SU-DHL4-shGNA13UTR as compared to the SU-DHL4 control cells (Fig. 4A).
Fig. 4. GNA13-deficient GCB-DLBCL cells are hypersensitive to BCL2 inhibitors.
A Volcano plot of FDA-approved drugs and bioactive compounds with known targets on SU-DHL4-shGNA13Scr and SU-DHL4-shGNA13UTR cells. Drug effect size ratio between SU-DHL4-shGNA13Scr and SU-DHL4-shGNA13UTR vs. statistical significance (P value) were plotted. Red and blue points indicate drug identified as differentially inhibited between the two types of cells. B Dose–response curves for SU-DHL4-shGNA13Scr or SU-DHL4-shGNA13UTR treated with AKT inhibitor MK-2206. C IC50 of BCL2 inhibitors ABT-199 and ABT-263 in DLBCL cell lines with either wild-type (blue dots) or mutant (red dots) GNA13. D Dose–response curves for SU-DHL4 and OCI-LY1 cells treated with ABT-199. E SU-DHL4 or OCI-LY1 cells were transfected with constructs containing shGNA13-600, shGNA13-UTR, or a scrambled shRNA (Scr). Levels of total (T)-AKT and phosphorylated (P)-AKTS473 in these cells were examined by western blot analysis. GAPDH was used as loading controls. F Western blot analysis of BCL2, total (T)-AKT, and phosphorylated (P)-AKTS473 level in SU-DHL4 cells treated with PI3K inhibitors Copanlisib or GDC-0941 at indicated concentrations for 6 h, respectively. G Western blot analysis of BCL2 level in SU-DHL4-shGNA13Scr or SU-DHL4-shGNA13UTR cells treated with PI3K inhibitors Copanlisib or GDC-0941 at indicated concentrations for 6 h, respectively. α-Tubulin was used as a loading control.
It has been shown by us (Fig. 4E) and others2 that the PI3K-AKT signaling pathway is a well-known downstream target of GNA13. We found that inhibitors of the PI3K-AKT signaling pathway, such as MK-2206, can also effectively kill GNA13-deficient SU-DHL4 cells (Fig. 4A, B), demonstrating the validity of this screening.
Having found that GNA13-deficient SU-DHL4 cells are hypersensitive to the treatment with BCL2 inhibitors, we analyzed sensitivity of DLBCL cell lines to BCL2 inhibitors from published data41. Figure 4C shows that DLBCL cell lines with GNA13 mutations are more sensitive to the treatment with BCL2 inhibitors than those with WT GNA13. To further confirm that GCB-DLBCL cells with GNA13 mutations are hypersensitive to the treatment with BCL2 inhibitors, we compared the sensitivity of ABT-199 (also known as venetoclax, the third-generation BCL2 inhibitor41) in treating GCB-DLBCL cell lines either with the WT GNA13 (SU-DHL4) or mutant GNA13 (OCI-LY1). As shown in Fig. 4D, OCI-LY1 cells are much more susceptible to the treatment with ABT-199 than SU-DHL4 cells. Consistent with the function of GNA13 in suppressing the phosphoinositide 3-kinase (PI3K)/AKT pathway21, the AKT Serine 473 phosphorylation (P-AKTS473) level was elevated in SU-DHL4 cells transfected with GNA13 shRNAs (Fig. 4E). To test whether the high expression of BCL2 is correlated to the activation of PI3K-AKT pathway in DLBCL, we used the pan-PI3K inhibitor GDC-0941 and PI3Kα/δ inhibitor Copanlisib42 in treating WT SU-DHL4 cells. Both of the inhibitors could suppress the phosphorylation of AKTS473 and BCL2 protein expression accordingly (Fig. 4F). Likewise, in SU-DHL4-shGNA13UTR cells, the protein expression level of BCL2 was also drastically repressed upon PI3K-AKT pathway inhibition by the above two inhibitors (Fig. 4G), implying a high correlation between PI3K-AKT signaling and BCL2 anti-apoptosis pathway may exist in the GNA13-deficient background.
These data, together with the above finding that GNA13 negatively regulates the expression of BCL2, suggest that the loss-of-function mutation of GNA13 is a biomarker for the precision BCL2 inhibitor therapy for GCB-DLBCL.
Inactivation of GNA13 by targeting its palmitoylation sensitizes the GCB-DLBCL cells to BCL2 inhibitors
Although GCB-DLBCL with loss-of-function mutation of GNA13 could be treated effectively with BCL2 inhibitors, the majority of GCB-DLBCL patients harbor WT GNA13. As GNA13 negatively regulates BCL2 expression, the sensitivity of GCB-DLBCL to the treatment with BCL2 inhibitors, the effective therapy of GCB-DLBCL with WT GNA13 may be achieved by targeting both GNA13 and BCL2. Since our data show that palmitoylation of GNA13 is required for its function in regulating the BCL2 expression, inhibiting GNA13 palmitoylation may sensitize the GCB-DLBCL cells with WT GNA13 to BCL2 inhibitors.
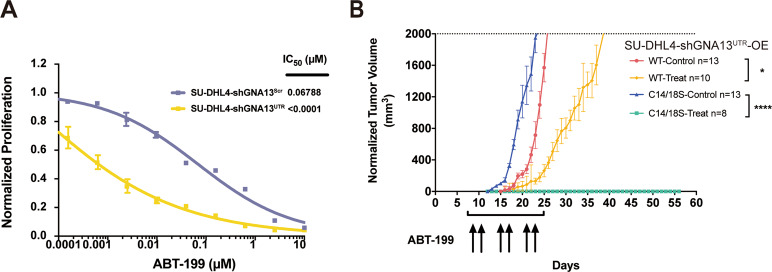
As a proof-of-concept experiment, we attempted to test the sensitivity of GCB-DLBCL cells bearing palmitoylation mutant of GNA13 to the treatment with BCL2 inhibitor. We first confirmed that SU-DHL4-shGNA13UTR cells, in which the WT GNA13 was knocked down with the specific GNA13 shRNA as described earlier, were more susceptible to the treatment with ABT-199 compared to the parental SU-DHL4 cells (Fig. 5A).
Fig. 5. GCB-DLBCL cells with palmitoylation-deficient GNA13 are hypersensitive to BCL2 inhibitor.
A Dose–response curves for SU-DHL4-shGNA13Scr and SU-DHL4-shGNA13UTR cells treated with ABT-199. B Tumor growth curves of SU-DHL4-shGNA13UTR cells either expressing the WT GNA13 (SU-DHL4-shGNA13UTR-OEWT) or the C14/C18S palmitoylation mutant of GNA13 (SU-DHL4-shGNA13UTR-OEC14/18S) in mice treated with either BCL2 inhibitor ABT-199 or vehicle. n, number.
To test the ABT-199 efficacy in vivo, we generated two xenograft models by serially transplanting SU-DHL4-shGNA13UTR-OEWT or SU-DHL4-shGNA13UTR-OEC14/18S cells into recipient NOD/SCID mice. Tertiary recipients transplanted with two different SU-DHL4 cells were randomly assigned to two cohorts that were orally administered 100 mg/kg ABT-199 or vehicle, respectively. As shown in Fig. 5B, SU-DHL4-shGNA13UTR-OEC14/18S tumors in mice treated with vehicle control (n = 13) grew faster than SU-DHL4-shGNA13UTR-OEWT tumors (n = 13), which is consistent with previous in vitro result in Fig. 2D. The ABT-199 treatment slowed down the growth of shGNA13UTR-OEWT tumors (n = 10), while the effect of the ABT-199 treatment was much more dramatic on SU-DHL4-shGNA13UTR-OEC14/18S tumors (n = 8) (Fig. 5B). At the end of this experiment (56 days), 8 out of 8 SU-DHL4-shGNA13UTR-OEC14/18S mice (100%) treated with the ABT-199 remained tumor-free. These data suggest that inhibition of palmitoylation of GNA13 is an effective therapeutic strategy for GCB-DLBCL in combination with the BCL2 inhibitor.
Discussion
In this study, we demonstrate that palmitoylation of GNA13 not only regulates its plasma membrane localization, but also regulates GNA13’s stability. It is essential for the tumor suppressor function of GNA13 in GCB-DLBCL cells. Interestingly, we found that GNA13 negatively regulates BCL2 expression in GCB-DLBCL in a palmitoylation-dependent manner. Consistently, we found that GCB-DLBCL with loss-of-function mutations of GNA13 are sensitive to the treatment with BCL2 inhibitors and that inactivating the WT GNA13 by targeting its palmitoylation dramatically enhanced the sensitivity of GCB-DLBCL to the BCL2 inhibitor. These results indicate that the loss-of-function mutation of GNA13 is a biomarker for the precision BCL2 inhibitor therapy for GCB-DLBCL and that GNA13 palmitoylation is a potential target for combination therapy with the BCL2 inhibitor to treat GCB-DLBCL with WT GNA13.
It has been shown that palmitoylation regulates protein stability26. We show here that palmitoylation of GNA13 does affect its stability (Fig. 1B). Our data show that downregulation of palmitoylation-deficient mutant GNA13C14/18S was rescued significantly by caspase inhibitors such as Z-VAD(OMe)-FMK (Fig. 1D), indicating that palmitoylation of GNA13 regulates its stability by inhibiting caspase-mediated protein degradation. It is possible that palmitoylation affects the conformation of GNA13, exposing the caspase cleavage sites. Alternatively, changed cellular localization due to lack of palmitoylation may subject GNA13 to the caspase-mediated protein degradation. These possibilities will be tested in the future.
Mutations of GNA13 have been found in about 18% in GCB-DLBCL, 13% Burkitt lymphoma (BL), and 15.6% Follicular Lymphoma (FL), respectively43–45, including multiple point mutations and truncated variants. In BL, several prevailing GNA13 point mutations, such as L184R, L197Q, and F245S, were shown to be loss-of-function mutations in terms of Gα13/RHOA activity in GPCR signaling46. Here, we identified two palmitoylation sites, C14 and C18, in the N-terminal GNA13 protein by using both genetic and biochemical approaches. Consistent with our finding that palmitoylation of GNA13 is required for its tumor suppressor function, at least two DLBCL patients were found to bear the GNA13 C14S mutation38,39. Our data further support the idea that GNA13 plays tumor suppressor function in germinal center B cells and loss-of-function mutations of GNA13 are involved in lymphomagenesis.
Venetoclax, the FDA-approved BCL2 inhibitor47,48, is being tested to extend its application in DLBCL49. Our study indicates that inactivation of GNA13 alone could lead to higher expression of BCL2 that eventually conferred higher drug sensitivity to BCL2 inhibitor, implying that DLBCL patients with inactive mutations of GNA13 are more sensitive to the BCL2 inhibitor treatment, providing a patient-stratification biomarker for the venetoclax therapy.
Inactivating a tumor suppressor may sound counterintuitive in fighting cancers. But it works as a strategy of synthetic lethality. A recent systematic drug sensitivity screening shows that loss of SETD2 sensitized tumor cells to CDK7 inhibitor, and BAP1 depletion confers vulnerability to inhibitor of DNMT150. Our work that inactivating GNA13 increases sensitivity of GCB-DLBCL cells to the BCL2 inhibitor may serve as a new therapeutic strategy for GCB-DLBCL.
S-palmitoylation is catalyzed by PATs. To date, at least 23 mammalian PATs have been identified51. The limited responsibilities of each PAT give hope that targeting specific PAT may be safe and effective in cancer therapies. It is important to identify which PAT is responsible for the palmitoylation modification of GNA13 and develop specific PAT inhibitors. Such agents would be effective for treating GCB-DLBCL with WT GNA13 in combination with the BCL2 inhibitor.
As mentioned in the Introduction, the role of GNA13 in tumorigenesis is cell-context dependent. It is not clear whether GNA13 also regulates the expression of BCL2 in tumors other than GCB-DLBCL and whether the therapeutic scenario for targeting palmitoylation of GNA13 in combination with the BCL2 inhibitor holds in other types of cancer. We will test the role of palmitoylation of GNA13 in other type of cancer in the future.
Materials and methods
Cell culture
The GCB-DLBCL cell lines SU-DHL6 and OCI-LY1 were kindly provided by Dr. Chenghua Yang, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences (CAS). The GCB-DLBCL cell line SU-DHL4 was obtained from Stem Cell Bank, CAS. All the GCB-DLBCL cell lines were cultured in RPMI 1640 (Basal Media, Shanghai, China) with 10% (v/v) FBS (Thermo, Waltham, MA, USA) in a humidified incubator at 37 °C under 5% CO2. The HeLa cells were bought from ATCC and cultured as previously described52. All cell lines were authenticated via STR profiling and periodically treated with Plasmocin (Invivogen, San Diego, CA, USA) to exclude mycoplasma contamination.
Cell proliferation/viability assay
Cell proliferation/viability were assessed using CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) as previously described52,53. Briefly, cells were seeded into 96-well cell plates (5000 cells/well) and supplemented with drugs at various concentrations. After 48 h of incubation, cells were lysed by CellTiter-Glo reagent and the resulting luminescence was measured using an Envision plate reader (PerkinElmer, Akron, OH, USA) after a 30-min incubation at room temperature.
Chemicals and antibodies
The chemical library containing FDA-approved drugs and bioactive compounds, BCL2 inhibitor navitoclax (ABT-263), AKT inhibitor MK-2206, and proteasome inhibitor MG-132 were purchased from Selleckchem (Houston, TX, USA). The BCL2 inhibitor venetoclax (ABT-199), the pan-caspase inhibitor Emricasan and Q-VD-Oph, the group II caspases inhibitor Ac-DEVD-CHO, the pan-PI3K inhibitor GDC-0941, and the PI3Kα/δ inhibitor Copanlisib were purchased from CSNpharm (Arlington Heights, IL, USA). The cycloheximide (CHX), autophagy inhibitors, Hydroxychloroquine sulfate (HCQ), ULK-101, and the pan-caspase inhibitor Z-VAD(OMe)-FMK were purchased from TargetMol (Wellesley Hills, MA, USA). For antibodies, the rabbit anti-GNA13 (ab128900), rabbit anti-Na-K-ATPase (ab76020), and rabbit anti-BCL2 (ab32124) antibodies were purchased from Abcam (Cambridge, UK). The rabbit anti-Caspase3 (9662), rabbit anti-phospho-AKT (Ser473(4060)), rabbit anti-pan-AKT (4691), rabbit anti-HA (3724), anti-Rabbit IgG (H + L), and F(ab′)2 Fragment (Alexa Fluor® 555 Conjugate) (4413) antibodies were purchased from Cell Signaling (Danvers, MA, USA). The HRP-conjugated mouse anti-GAPDH (HRP-60004) and HRP-conjugated mouse anti-α-Tubulin (HRP-66031) antibodies were bought from Proteintech (Rosemont, IL, USA). The goat anti-Biotin (B 3604) was purchased from Sigma Aldrich (St. Louis, MO, USA). The donkey anti-Goat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 (A-11056) was bought from Thermo.
Membrane and cytosolic protein isolation
Protein extractions from membrane and cytosolic part of cells were isolated as previously described54,55.
Western blot analysis
Western blot analysis was performed as previously described53. Briefly, cells were lysed in 1× sodium dodecyl sulfate (SDS) sample loading buffer, and then equal amounts of protein samples were loaded to polyacrylamide gel, transferred to nitrocellulose membrane, and then blotted with specific primary and secondary antibodies. Luminescence signals on membrane were detected with Immobilon Western HRP Substrate (Millipore, Darmstadt, Germany) and blots were imaged by the FluorChem Multiplex imaging system (ProteinSimple, San Jose, CA, USA).
Drug screening
Both SU-DHL4-Scr and SU-DHL4-UTR cells were seeded into 96-well plates at a density of 5000 cells/well. Individual chemicals from FDA-approved Drug Library were added into each well in duplicates by an Explorer automation workstation (Perkin Elmer) at the concentration of 2 μM. After 48 h of incubation, cell viability was measured using CellTiter-Glo (Promega) following the manufacturer’s instructions.
Statistical analysis
GraphPad Prism 7 and the Student’s t-test were used for data analysis. Statistical significance threshold was set at P = 0.05, and different levels were denoted as *, P < 0.05, **, P < 0.01, and ***, P < 0.001, respectively.
Supplementary information
Acknowledgements
The National Natural Science Foundation of China and the Samuel Waxman Cancer Research Foundation financially supported this study. Professor Li Xia, Proteomics Platform of Core Facility of Basic Medical Sciences, Shanghai Jiao Tong University School of Medicine (SJTU-SM), technically supported this study. This work was supported by the National Key Research and Development Program (2016YFC0902800 to R.R.), the Key Project of Natural Science Foundation of China (81530006 to R.R.), Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01 to R.R.), Natural Science Foundation of China (81870112 to R.R., 81770171 to B.J., and 81970134 to P.L.), Shanghai Science and Technology Development Funds (18ZR1423600 to P.L.), the Samuel Waxman Cancer Research Foundation (to R.R.), and Innovative Research Team of High-level Local Universities in Shanghai.
Author contributions
Z.X., B.J., and R.R. designed the study. Z.X. and B.J. developed key methodology of palmitoylation assays. Z.X. and X.Z. performed most of experiments and analyzed the data. P.L., R.Z., Z.S., D.L., M.W., N.N., X.X., and M.L. provided technical or material supports, such as reagents, animal experiments, and so on. Q.Z. and J.Z. helped on drug screen experiment. Z.X., B.J., and R.R. wrote and reviewed the manuscript. B.J. and R.R. supervised the study. All authors read and approved the final paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
All the animal experiments in this study were performed in accordance with the Declaration of Helsinki. All murine procedures were performed according to Shanghai Jiao Tong University Animal Care and Use Committee approved protocols.
Footnotes
Edited by M. Agostini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhizhou Xia, Xiuli Zhang
Contributor Information
Bo Jiao, Email: bjiao@sibs.ac.cn.
Ruibao Ren, Email: ren@brandeis.edu.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03311-1).
References
- 1.Kelly P, Casey PJ, Meigs TE. Biologic functions of the G12 subfamily of heterotrimeric G proteins: growth, migration, and metastasis. Biochemistry. 2007;46:6677–6687. doi: 10.1021/bi700235f. [DOI] [PubMed] [Google Scholar]
- 2.Muppidi JR, et al. Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma. Nature. 2014;516:254–258. doi: 10.1038/nature13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruppel KM, et al. Essential role for Gα13 in endothelial cells during embryonic development. Proc. Natl Acad. Sci. USA. 2005;102:8281–8286. doi: 10.1073/pnas.0503326102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivaraj KK, et al. G13 controls angiogenesis through regulation of VEGFR-2 expression. Dev. Cell. 2013;25:427–434. doi: 10.1016/j.devcel.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Zhang JJ, Rafii S, Huang XY. Suppression of tumor angiogenesis by Gα(13) haploinsufficiency. J. Biol. Chem. 2009;284:27409–27415. doi: 10.1074/jbc.M109.025460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu M, et al. Gα13 negatively controls osteoclastogenesis through inhibition of the Akt–GSK3β–NFATc1 signalling pathway. Nat. Commun. 2017;8:13700. doi: 10.1038/ncomms13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moers A, et al. G13 is an essential mediator of platelet activation in hemostasis and thrombosis. Nat. Med. 2003;9:1418–1422. doi: 10.1038/nm943. [DOI] [PubMed] [Google Scholar]
- 8.Shen B, et al. A directional switch of integrin signalling and a new anti-thrombotic strategy. Nature. 2013;503:131–135. doi: 10.1038/nature12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mu G, et al. Gastrin stimulates pancreatic cancer cell directional migration by activating the Gα12/13–RhoA–ROCK signaling pathway. Exp. Mol. Med. 2018;50:59. doi: 10.1038/s12276-018-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Casey PJ. MicroRNA-182 and microRNA-200a control G-protein subunit α-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. J. Biol. Chem. 2013;288:7986–7995. doi: 10.1074/jbc.M112.437749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi H, et al. GEP oncogene promotes cell proliferation through YAP activation in ovarian cancer. Oncogene. 2016;35:4471–4480. doi: 10.1038/onc.2015.505. [DOI] [PubMed] [Google Scholar]
- 12.Morin RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz R, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sha C, et al. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J. Clin. Oncol. 2019;37:202–212. doi: 10.1200/JCO.18.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao Y, Medeiros LJ, Li Y, Li J, Young KH. Genetic alterations and their clinical implications in DLBCL. Nat. Rev. Clin. Oncol. 2019;23:4117. doi: 10.1038/s41571-019-0225-1. [DOI] [PubMed] [Google Scholar]
- 16.Alizadeh AA, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 17.Swerdlow SH, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, et al. S1PR1 is an effective target to block STAT3 signaling in activated B cell-like diffuse large B-cell lymphoma. Blood. 2012;120:1458–1465. doi: 10.1182/blood-2011-12-399030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muppidi JR, Lu E, Cyster JG. The G protein-coupled receptor P2RY8 and follicular dendritic cells promote germinal center confinement of B cells, whereas S1PR3 can contribute to their dissemination. J. Exp. Med. 2015;212:2213–2222. doi: 10.1084/jem.20151250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldari CT. S1PR2 deficiency in DLBCL: a FOXy connection. Blood. 2016;127:1380–1381. doi: 10.1182/blood-2016-01-692129. [DOI] [PubMed] [Google Scholar]
- 21.O’Hayre M, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caeser R, et al. Genetic modification of primary human B cells to model high-grade lymphoma. Nat. Commun. 2019;10:4543–4516. doi: 10.1038/s41467-019-12494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattoretti G, et al. Targeted disruption of the S1P2 sphingosine 1-phosphate receptor gene leads to diffuse large B-cell lymphoma formation. Cancer Res. 2009;69:8686–8692. doi: 10.1158/0008-5472.CAN-09-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494.e415. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko PJ, Dixon SJ. Protein palmitoylation and cancer. EMBO Rep. 2018;19:e46666. doi: 10.15252/embr.201846666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linder ME, et al. Lipid modifications of G proteins: alpha subunits are palmitoylated. Proc. Natl Acad. Sci. USA. 1993;90:3675–3679. doi: 10.1073/pnas.90.8.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuiffo B, Ren R. Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood. 2010;115:3598–3605. doi: 10.1182/blood-2009-03-213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharyya R, Wedegaertner PB. Gα13 requires palmitoylation for plasma membrane localization, rho-dependent signaling, and promotion of p115-rhoGEF membrane binding. J. Biol. Chem. 2000;275:14992–14999. doi: 10.1074/jbc.M000415200. [DOI] [PubMed] [Google Scholar]
- 30.Ren J, et al. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng SL, Kao HJ, Huang CH, Lee TY. MDD-Palm: identification of protein S-palmitoylation sites with substrate motifs based on maximal dependence decomposition. PLoS ONE. 2017;12:e0179529. doi: 10.1371/journal.pone.0179529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshavsky A. The ubiquitin system, autophagy, and regulated protein degradation. Annu Rev. Biochem. 2017;86:123–128. doi: 10.1146/annurev-biochem-061516-044859. [DOI] [PubMed] [Google Scholar]
- 33.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/S0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 36.Julien O, Wells JA. Caspases and their substrates. Cell Death Differ. 2017;24:1380–1389. doi: 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016;48:4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mareschal S, et al. Whole exome sequencing of relapsed/refractory patients expands the repertoire of somatic mutations in diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2016;55:251–267. doi: 10.1002/gcc.22328. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, et al. Analysis of genomic alteration in primary central nervous system lymphoma and the expression of some related genes. Neoplasia. 2018;20:1059–1069. doi: 10.1016/j.neo.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonov AV, et al. PPISURV: a novel bioinformatics tool for uncovering the hidden role of specific genes in cancer survival outcome. Oncogene. 2014;33:1621–1628. doi: 10.1038/onc.2013.119. [DOI] [PubMed] [Google Scholar]
- 41.Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- 42.Bojarczuk K, et al. Targeted inhibition of PI3Kα/δ is synergistic with BCL-2 blockade in genetically defined subtypes of DLBCL. Blood. 2019;133:70–80. doi: 10.1182/blood-2018-08-872465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubois S, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin. Cancer Res. 2016;22:2919–2928. doi: 10.1158/1078-0432.CCR-15-2305. [DOI] [PubMed] [Google Scholar]
- 44.Wagener R, et al. The mutational landscape of Burkitt-like lymphoma with 11q aberration is distinct from that of Burkitt lymphoma. Blood. 2019;133:962–966. doi: 10.1182/blood-2018-07-864025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimono J, et al. Analysis of GNA13 protein in follicular lymphoma and its association with poor prognosis. Am. J. Surgical Pathol. 2018;42:1466–1471. doi: 10.1097/PAS.0000000000000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Hayre M, et al. Inactivating mutations in GNA13 and RHOA in Burkitt’s lymphoma and diffuse large B-cell lymphoma: a tumor suppressor function for the Gα13/RhoA axis in B cells. Oncogene. 2016;35:3771–3780. doi: 10.1038/onc.2015.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain N, et al. Ibrutinib and venetoclax for first-line treatment of CLL. N. Engl. J. Med. 2019;380:2095–2103. doi: 10.1056/NEJMoa1900574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delbridge ARD, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 49.Zhao X, et al. BCL2 amplicon loss and transcriptional remodeling drives ABT-199 resistance in B cell lymphoma models. Cancer Cell. 2019;35:752–766.e759. doi: 10.1016/j.ccell.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding H, et al. Systematic analysis of drug vulnerabilities conferred by tumor suppressor loss. Cell Rep. 2019;27:3331–3344 e3336. doi: 10.1016/j.celrep.2019.05.043. [DOI] [PubMed] [Google Scholar]
- 51.Greaves J, Chamberlain LH. DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochemical Sci. 2011;36:245–253. doi: 10.1016/j.tibs.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Ning N, et al. A novel microtubule inhibitor overcomes multidrug resistance in tumors. Cancer Res. 2018;78:5949–5957. doi: 10.1158/0008-5472.CAN-18-0455. [DOI] [PubMed] [Google Scholar]
- 53.Wu M, et al. N-arachidonoyl dopamine inhibits NRAS neoplastic transformation by suppressing its plasma membrane translocation. Mol. Cancer Ther. 2017;16:57–67. doi: 10.1158/1535-7163.MCT-16-0419. [DOI] [PubMed] [Google Scholar]
- 54.Zhuang L, Lin J, Lu ML, Solomon KR, Freeman MR. Cholesterol-rich lipid rafts mediate akt-regulated survival in prostate cancer cells. Cancer Res. 2002;62:2227–2231. [PubMed] [Google Scholar]
- 55.Adam RM, Yang W, Di Vizio D, Mukhopadhyay NK, Steen H. Rapid preparation of nuclei-depleted detergent-resistant membrane fractions suitable for proteomics analysis. BMC Cell Biol. 2008;9:30. doi: 10.1186/1471-2121-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.