Abstract
Chromatin properties are regulated by complex networks of epigenome modifications. Currently, it is unclear how these modifications interact and if they control downstream effects such as gene expression. We employed promiscuous chromatin binding of a zinc finger fused catalytic domain of DNMT3A to introduce DNA methylation in HEK293 cells at many CpG islands (CGIs) and systematically investigated the dynamics of the introduced DNA methylation and the consequent changes of the epigenome network. We observed efficient methylation at thousands of CGIs, but it was unstable at about 90% of them, highlighting the power of genome-wide molecular processes that protect CGIs against DNA methylation. Partially stable methylation was observed at about 1000 CGIs, which showed enrichment in H3K27me3. Globally, the introduced DNA methylation strongly correlated with a decrease in gene expression indicating a direct effect. Similarly, global but transient reductions in H3K4me3 and H3K27ac were observed after DNA methylation but no changes were found for H3K9me3 and H3K36me3. Our data provide a global and time-resolved view on the network of epigenome modifications, their connections with DNA methylation and the responses triggered by artificial DNA methylation revealing a direct repressive effect of DNA methylation in CGIs on H3K4me3, histone acetylation, and gene expression.
INTRODUCTION
Chromatin functions are regulated by a large number of reversible chemical modifications of the DNA and the histone proteins, which altogether constitute the epigenome (1,2). DNA modifications include methylation and hydroxymethylation of cytosine residues mainly within CpG sites. DNA is methylated to ∼70–80% at all CpG sites (3), but the so-called CpG islands (CGIs) are usually unmethylated (4). Local methylation levels of the genome are determined by a dynamic equilibrium of methylation and demethylation activities at the target sites (5,6). In many cancer cells and cell lines, a CGI methylator phenotype associated with the aberrant methylation of thousands of CGIs has been described (7). Histone protein modifications include methylation of lysine and arginine, acetylation and ubiquitination of lysine and phosphorylation of serine and threonine at various sites (8). The different chromatin marks are connected with each other forming a complex epigenome network, which regulates essentially all chromatin-based processes during normal development and disease (9–12).
Epigenome editing refers to the rewriting of epigenome modifications with artificial EpiEditors comprising a DNA binding part that directs the fusion construct to its genomic target sites and an effector domain that recruits other cellular factors or directly alters a chromatin modification (13–17). In the past, different DNA binding domains, viz. zinc finger proteins (ZnF), TAL effectors and deactivated Cas9 in complex with single guide RNAs have been employed for epigenome editing and combined with several different effector domains to introduce or remove DNA methylation, histone methylation or histone acetylation (13–17). While the general approach of epigenome editing has been proven successful and able to achieve gene regulation and chromatin state modulation in many examples, critical questions have remained unsolved, including the specificity of epigenome editing and the stability of the introduced epigenome changes. Seminal studies have demonstrated locus specific maintenance of H3K9 methylation and its causative role in silencing of gene expression in fission yeast (18,19). However, the mutual connections of different chromatin modifications within the human epigenome network are not well established and the mechanisms leading to stable switching of chromatin states are not clear.
For the application of epigenome editing in basic science and in the clinic, the underlying mechanisms leading to stable reprogramming of gene expression are of particular importance. In this respect, partially contradictory observations have been made in the past. For instance, DNA methylation introduced at different loci was found to be unstable in HEK293 cells even if high initial methylation levels were reached (20,21). Similarly, editing of H3K9me3 was found not to be durable (20,22), while the targeted introduction of H3K4me3 was found to cause stable effects (23). In primary breast cell lines, methylation of the promoters of cancer-associated genes was found to be stable (24). Dynamics of epigenome regulation were studied at the single-cell level in CHO-K1 cells with a stably integrated reporter gene showing different kinetics of reversible and irreversible silencing (25). Silencing with DNMT3B was slow, but it led to the highest levels of irreversible effects, while silencing with the Krüppel-associated box (KRAB) was fast but reversible. In K-562 cells expressing a reporter gene, transient gene silencing was observed with DNMT3A or the KRAB domain (leading to the deposition of H3K9me3) alone, but the combined introduction of DNA methylation and H3K9me3 led to stable silencing (26). In contrast, the combination of DNMT3A and KRAB targeting was not reported to cause stable repression of the endogenous HER2 locus in HCT116 cells despite strong transient silencing (27). It is not known at present to what extent these discrepancies were caused by the variation in the target cells, target genes, targeting strategies (such as choice of the DNA binding domain, direct targeting of enzyme activity versus targeting of a recruiting factor) or delivery systems of the EpiEditors (regulated expression vs. viral delivery vs. transient transfection).
In this work, we employed promiscuous DNA binding of a fusion protein comprising a ZnF and the catalytic domain of DNMT3A (ZnF-3AC) to introduce DNA methylation at several thousand CGIs in one experiment, in order to investigate the influence of the target site on the efficiency and stability of the introduced DNA methylation in a systematic manner. Expression of the ZnF-3AC protein was under the control of doxycycline (dox) allowing us to deliver DNA methylation to target CGIs in a time-controlled manner. After the removal of dox, we determined the stability of the introduced DNA methylation at the target CGIs and studied the state of other chromatin modifications at the target sites before DNA methylation editing as well as their dynamic changes. Similarly, the dynamics of the expression of genes associated with the target CGIs were investigated as well. Our data show efficient methylation at thousands of CGIs, followed by rapid loss of the methylation at most (90%) of the target sites. Partially stable methylation was observed at ∼1000 CGIs, which were enriched in H3K27me3. Overall in the targeted CGIs, the introduction of DNA methylation strongly correlated with a decrease in gene expression, as well as reductions in H3K4me3 and H3K27ac. No changes were observed in the levels of H3K9me3 and H3K36me3. Our findings contribute to the understanding of the network properties of epigenome regulation and the mutual connections of epigenome modifications. Moreover, they demonstrate a direct antagonistic effect of DNA methylation in CGIs on H3K4me3, histone acetylation and gene expression.
MATERIALS AND METHODS
Cloning
In this work, the gene for the catalytic domain of DNMT3A (3AC) from Mus musculus was used, which encodes an identical protein sequence as its human homolog. ZnF-3AC wild type and the catalytically inactive E756A mutant were amplified from pAdTrackCMV-ZnF-3AC vectors (20) and cloned into a pSIN-TRE3G-IRES-GFP-PGK-Neo based vector via Gibson Assembly. 3AC without ZnF was amplified from the previously obtained vector and introduced into the same target vector as before via Gibson Assembly. The final plasmids with variants of 3AC (with and without ZnF) are hereinafter referred to as pSIN-TRE3G-(ZnF-)3AC-IRES-GFP-PGK-Neo. pAdTrack-ZnF-3xHA was cloned by amplifying ZnF from the pAdTrackCMV-ZnF-3AC vector as well as a short backbone-fragment from the same vector. The empty vector pAdTrackCMV was linearized and the amplicons were introduced via Gibson assembly. The sequences of all constructs were confirmed by Sanger sequencing (Microsynth AG) and the nucleotide and protein sequences of ZnF-3AC are shown in Supplementary Text S1.
Cell maintenance and generation of stable cell lines
Human Embryonic Kidney cells 293 (HEK293) were obtained from DSMZ (Braunschweig, Germany). Platinum-E (PlatE) cells were from Cell Biolabs. Both cell lines were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma-Aldrich), 4 mM l-glutamine (Sigma-Aldrich) and 10 ml·l−1 penicillin/streptomycin (Sigma-Aldrich), hereinafter referred to as DMEM+. The cells were grown at 95% relative humidity, 5% CO2 and 37°C and split every 2–3 days to keep confluency <90%. Therefore, the cells were washed with Dulbecco's Phosphate Buffered Saline without MgCl2 and CaCl2 (Sigma-Aldrich), immersed with Trypsin-EDTA solution (Sigma Aldrich) and incubated at 37°C for 5 min. Trypsinization was stopped by resuspension in DMEM+ and cells were usually split in a 1:4–1:8 ratio.
Stable cell lines were generated as described (28). In brief, HEK293 cells were lentivirally transduced with pWPXLd-RIEP to express the ecotropic receptor (EcoR) (29) and the rtTA3 transcriptional activator for dox induced gene expression (30). Cells expressing the receptor (HEK293R) were selected with 1 μg/ml puromycin (Sigma-Aldrich) and frozen stocks were prepared by resuspending 1 × 106 cells in 1 ml freezing medium (50% DMEM, 40% FBS, 10% DMSO). 20 μg of the previously cloned pSIN-TRE3G-(ZnF-)3AC-IRES-GFP-PGK-Neo plasmids and 10 μg of helper DNA (pCMV-Gag-Pol, Cell Biolabs) were transfected into PlatE cells using calcium phosphate transfection for retroviral packaging. Forty hours after transfection, HEK293R cells were infected with the supernatant from the transfected PlatE cells, containing the retroviral particles. The selection of transduced cells was started 48 post-transduction using 0.5 mg/ml G-418 (Roche). Cells were expanded post-selection and frozen stocks were prepared as described above.
Cell culture experiments
Fresh stocks of HEK293R (pSIN-TRE3G-(ZnF-)3AC-IRES-GFP-PGK-Neo) cells were cultured as described in the previous chapter. To obtain cells for the time point ‘no Dox’, cells were harvested before induction by washing twice with PBS without MgCl2 and CaCl2, trypsinization for 5 min at 37°C and resuspending in DMEM+. The cell suspension was centrifuged for 5 min at 300 g, washed again and distributed into 1.5 ml reaction tubes to yield 0.5–2 × 106 cells per tube. For experiments aiming to analyze acetylation signals, PBS without MgCl2 and CaCl2 was supplemented with 100 nM Trichostatin-A (Sigma-Aldrich). Cell pellets were snap-frozen in liquid nitrogen and stored at −80°C.
For induction of ZnF-3AC variants or 3AC, 6 × 106 cells were seeded into a T175 flask in 20 ml DMEM+ supplemented with 1 μg/ml doxycycline (dox) (Sigma-Aldrich). DMEM+ and dox were refreshed after 48 h. After 3 days, cells were harvested and filtered through a 70 μm pre-separation filter (Miltenyi Biotec). Cell sorting was performed using either Cell Sorter SF800S (Sony Biotechnology) or the BD FACSAria III (BD Biosciences). FACS settings were chosen to collect only single, GFP-positive cells. After sorting, 7.5 × 105 cells were seeded back into a well of a six-well cell culture plate (Greiner Bio-One) and grown in DMEM+ without dox for analysis of later time points. The other cells designated as ‘3d dox’ were spun down and frozen as described for ‘no dox’. Five days after sorting, 80% of the cells were harvested (‘5d off’) and 20% were seeded back. This procedure was repeated eight days after sorting (‘8d off’) and eleven days after sorting (‘11d off’), the experiment was stopped after harvesting.
Flow cytometry experiment (GFP dynamics)
HEK293R (pSIN-TRE3G-(ZnF-)3AC-IRES-GFP-PGK-Neo) cells were induced with dox as described above to either express ZnF-3ACWT or ZnF-3ACE756A. After 3 days of induction, cells were sorted as described in the previous chapter and 7.5 × 105 cells were seeded back into 6-Well plates. A fraction of the sorted cells, as well as uninduced cells, were set aside for flow cytometry analysis using the MACSQuant® VYB Flow Cytometer (Miltenyi Biotec) using FACS settings to measure the GFP-signal only from single, live cells. At the time points ‘1d off’, ‘2d off’, ‘3d off’ and ‘4d off’, these measurements were repeated with the same cytometer settings. Medians of the GFP-signal were determined using the Flowing Software V2.5.1 (http://flowingsoftware.btk.fi) and normalized to ‘3d dox’.
Transient transfection and crosslink-ChIP (XChIP) of ZnF-3xHA
HEK293 cells were transiently transfected with the pAdTrack-CMV-ZnF-3xHA plasmid. Therefore, cells were seeded into a T75 flask in 14 ml total volume DMEM+ to reach a confluency of 70–80% 24 h after seeding. 17.5 μg plasmid DNA was mixed with serum-free DMEM in a total volume of 1.75 ml. In parallel, 52.5 μg of Polyethylenimine (PEI) MAX 40k (Polysciences) was mixed thoroughly with serum-free DMEM and incubated for 5 min. The DNA mixture was added dropwise to the PEI mixture and incubated for 20 min at room temperature. Then, the DNA-PEI-mixture was pipetted dropwise to the cells. 16 h post-transfection, the medium was exchanged by fresh DMEM+. Three days post-transfection, the transfection efficiency was checked by microscopy and the cells were harvested for XChIP.
Crosslinking was performed by adding formaldehyde (Sigma-Aldrich, Cat. No. 25,254-9) to a final concentration of 1% (w/v). After incubation with rotation for 10 min, the reaction was quenched by adding glycine to a final concentration of 125 mM followed by another incubation step with rotation for 10 min. Cells were washed and lysis was performed in Cell Lysis Buffer (10 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.2% IGEPAL® CA-630, EDTA-free Protease Inhibitor Cocktail tablet (Roche)). Nuclei were resuspended in Nuclei Lysis Buffer (Tris–HCl pH 8.0, 10 mM EDTA, 1% SDS, EDTA-free Protease Inhibitor Cocktail tablet). To obtain DNA fragments in a size range of 100–500 bp, the crosslinked chromatin was sheared using the EpiShear™ probe sonicator (Active Motif). The sonication efficiency was checked on an 1% agarose gel. Subsequently, the chromatin was separated from nuclei debris by centrifugation. For pre-clearing, IP Dilution Buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.01% SDS, EDTA-free Protease Inhibitor Cocktail tablet), Protein A/G magnetic beads (Thermo Fisher Scientific) and 5 μg normal rabbit IgG (Santa Cruz Biotechnology, sc-2027) were added to the sample and incubated for 2 h at 4°C. Afterward, 5% of the pre-cleared lysate was set aside as ‘input’ and stored at -20°C. 5 μg anti-HA tag antibody (Abcam, ab9110) was added to the IP and incubated overnight at 4°C. The next day, Protein A/G magnetic beads were added and incubated for 2 h at 4°C. Next, the beads were washed once with IP Wash 1 Buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 50 mM NaCl, 1% Triton X-100, 0.1% SDS), twice with High Salt Buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.01% SDS), once with IP Wash 2 Buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 1% IGEPAL® CA-630, 1% deoxycholic acid) and twice with TE-Buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA). Two elution steps were performed by adding Elution Buffer (1% SDS, 100 mM NaHCO3) to the beads and incubating for 30 min at room temperature. After pooling the elution fractions, 20 μg RNase A (Machery-Nagel) and NaCl to a final concentration of 300 mM were added to the IP and the input sample and both were incubated overnight at 65°C. The next day, 2.4 U Proteinase K (NEB) was added and samples were incubated for 2 h at 45°C. Subsequently, the DNA was extracted using the ChIP DNA Purification Kit (Active Motif) according to the manufacturer's protocol and eluted in 50 μl elution buffer provided in the kit.
MBD2-pulldown and quantitative PCR (qPCR)
Frozen cell pellets (0.5–1 × 106 cells) were thawed on ice and genomic DNA (gDNA) was extracted using the QIAmp DNA Mini Kit (Qiagen) according to the manufacturer's protocol. To obtain DNA fragment sizes between 100 and 1000 bp, 200 μl of gDNA was sonicated in a 1.5 ml reaction tube utilizing the EpiShear™ probe sonicator (Active Motif) and using the 2 mm tip (settings: 25% amplitude, 20 × 20-s pulse/30-s pause). Subsequently, the gDNA was concentrated using the NucleoSpin® Gel and PCR Clean-up Kit (Macherey-Nagel) and eluted in 50 μl pre-warmed ddH2O. The level of fragmentation was checked on a 1% agarose gel or via LabChip® GXII Touch™ HT system (Perkin Elmer). 1 μg of gDNA per sample was used per pulldown and adjusted to a final volume of 250 μl with pre-cooled PB150 buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5% IGEPAL® CA-630, 2 mM DTT). After the addition of 8.75 μg GST-MBD2, samples were rotated overnight at 4°C. 50 μl of Glutathione Agarose beads (Macherey-Nagel) per sample were prepared the next morning by washing four times with 200 μl pre-cooled PB150 buffer and spinning down for 1 min at 2000 g. After the final wash, the supernatant was discarded and the GST-MBD2/gDNA mix was transferred to the beads. Samples were incubated for 2 h at 4°C under constant rotation. Subsequently, the samples were spun down in a pre-cooled centrifuge for 2 min at 2000 g and the supernatant was discarded. 200 μl of ice-cold PB500 buffer (50 mM Tris–HCl pH 8.0, 500 mM NaCl, 1 mM EDTA, 0.5% IGEPAL® CA-630, 2 mM DTT) was added and the samples were rotated for 5 min at 4°C. Washing was repeated twice and the supernatant was removed after the final wash. 150 μl of unchilled PB2000 elution buffer (10 mM Tris–HCl pH 8.0, 2000 mM NaCl, 1 mM EDTA) was added to the beads. After 15 min of constant rotation at room temperature, the samples were spun down for 2 min at 2000 g and the supernatant was transferred into a fresh 2 ml reaction tube. A second elution was performed with 150 μl PB2000 and after centrifugation, both elution fractions were pooled. The precipitated DNA was purified using the ChIP DNA Purification Kit (Active Motif) and eluted in 50 μl elution buffer provided in the kit. Samples were stored at 4°C short-term or at -20°C long-term.
For qPCR, a 1:5 dilution series of input gDNA was prepared for normalization of pulldown DNA and evaluation of PCR efficiency. Per region of interest, a master mix with 7.5 μl 2× ORA™ See qPCR Probe Mix (highQu), 0.4 μl forward primer, 0.4 μl reverse primer and 5.7 μl ddH2O per reaction was prepared. All samples were pipetted in triplicates and 1 μl of pulldown sample was added per reaction. For each primer set (Supplementary Table S1), non-template-controls were included and the PCR was run with the following program: 95°C for 3 min, 40 cycles of 95°C for 3 s, 58°C for 20 s, 72°C for 4 s and finally a 65–95°C ramp (0.5°C steps every 5 s). After normalization to input, the signal of the VEGFA locus was internally re-normalized on the SLC6A3 locus which shows almost complete methylation (Supplementary Figure S1C, Table S6).
Native chromatin immunoprecipitation (NChIP)
1 × 106 cells were resuspended in 50 μl Lysis Buffer (10 mM Tris–HCl pH 7.4, 2 mM MgCl2, 0.6% IGEPAL® CA-630, 0.5 mM PMSF, 1 mM DTT, 100 μl/ml EDTA-free Protease Inhibitor Cocktail tablet (Roche) dissolved in 1 ml H2O) and incubated on ice. A 10× MNase Dilution Buffer (1× Micrococcal Nuclease Reaction Buffer (NEB), 200 μg/ml BSA (NEB), 5 U/μl Micrococcal Nuclease (NEB)) was prepared. 10 μl of MNase Master Mix (6 μl Micrococcal Nuclease Reaction Buffer (NEB), 1.2 μl MNase Dilution Buffer, 2.8 μl H2O) was added to the sample and put at room temperature. MNase digestion was performed at 37°C for 10 min and the reaction was quenched by adding 6.6 μl of 100 μM EDTA. 6.6 μl of 1% Triton X-100/1% deoxycholate solution was added and the sample was kept on ice for 15 min. After vortexing thoroughly, 250 μl Complete Immunoprecipitation (IP) Buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 0.1% Triton X-100. 1 mM PMSF, 100 μl/ml EDTA-free Protease Inhibitor Cocktail tablet dissolved in 1 ml H2O) was added. The sample was incubated for 1 h at 4°C under constant rotation. Meanwhile, antibody-bead-complexes were prepared by washing 20 μl Protein A/G magnetic beads (Thermo Fisher Scientific) three times with Complete IP Buffer and subsequently resuspending in 200 μl Complete IP Buffer. Suitable amounts of an antibody following the manufacturer's recommendations were added to the beads suspension and rotated for 3 h at 4°C. The following antibodies were used in NChIP experiments: H3K4me3, ab8580 (Abcam), 2.5 μg per IP; H3K27me3, #39155 (Active Motif), 5 μg per IP; H3K9me3, ab8898 (Abcam), 2.5 μg per IP; H3K36me3, ab9050 (Abcam), 3.5 μg per IP; H3K27ac, ab4729 (Abcam), 3 μg per IP.
The cell lysate was vortexed and centrifuged for 10 min at 14 000 g and 4°C. From supernatant, the DNA concentration was measured using NanoDrop 1000 (Thermo Fisher Scientific). 20–25 μg chromatin was used per IP and the volume was adjusted to 200 μl with Complete IP Buffer. For pre-clearing of chromatin, 20 μl Protein A/G magnetic beads were washed three times with 200 μl Complete IP Buffer. The chromatin lysate was transferred to the beads and 2.5 μg Normal Rabbit IgG (R&D Systems, #AB-105-C) was supplemented and rotated at 4°C for 2 h. Afterward, the supernatant was removed from antibody-beads complexes. Ten percent of the volume from the pre-cleared lysate (‘Input’) was set aside (stored at –20°C), the chromatin was transferred to the antibody-beads complexes and then incubated overnight at 4°C under constant rotation. The next day, the beads were washed twice with 400 μl Low Salt Buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.1% SDS) and twice with High Salt Buffer (20 mM Tris–HCl pH 8.0, 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS) for 10 min at 4°C. For elution, the supernatant was removed after the final washing step and 200 μl ChIP Elution Buffer (1% SDS, 100 mM NaHCO3) as well as 2.4 U Proteinase K (NEB) were added. The input was also included by adjusting the volume to 200 μl with ChIP Elution Buffer and by adding 2.4 U Proteinase K. After 2 h incubation at 65°C, the IP sample was separated from the beads and transferred into a fresh reaction tube. DNA from IP and Input was purified using the ChIP DNA Purification Kit (Active Motif).
SDS-PAGE and western blot
Preparation of whole cell lysates from 1.5 × 106 cells per time point was performed by resuspending the pelleted cells in 50 μl of 2× Laemmli buffer (4% SDS, 10% β-mercaptoethanol, 20% glycerol, 0.01% bromophenol blue, 1.25 M Tris–HCl pH 6.8) and incubation at 95°C for 10 min. The cell lysates were spun down for 1 min at 13 000 g and the supernatants were transferred into a fresh vial. The proteins were separated by SDS-PAGE by loading 12 μl of lysate per sample on a 12% SDS gel. Afterwards, the proteins were blotted on a nitrocellulose membrane (Amersham Biosciences). The membrane was cut at the 55 kDa marker band and blocked with 5% (w/v) milk powder in PBS-T (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.1% (v/v) Tween 20). After three washes with PBS-T, the membranes were incubated with either anti-DNMT3A-antibody (1:750, NB120-13888, Novus Biologicals) or anti-Lamin B1 (1:500, sc-6217, Santa Cruz Biotechnology Inc.) in PBS-T with 1% (w/v) milk powder and incubated overnight at 4°C. The next day, the membranes were washed three times with PBS-T and incubated for two hours with anti-mouse IgG (1:5000, NXA931V, GE Healthcare) or anti-Goat IgG (1:10 000, A4174, Sigma-Aldrich) secondary antibodies both conjugated with horseradish peroxidase. After three final washes with PBS-T, the membrane was developed using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) in 1:10 dilution and the signal was captured with the FUSION advance solo 4 (Peqlab).
RNA isolation, cDNA synthesis, RT-qPCR
Frozen cell pellets containing 0.5–1 × 106 cells were thawed on ice and RNA was purified using the RNeasy® Plus Mini Kit (Qiagen) according to the manufacturer's protocol. The RNA was eluted in 50 μl RNase-free water provided in the kit and the concentration was determined using NanoDrop 1000 (Thermo Fisher Scientific). For complementary DNA (cDNA) synthesis, 1 μg RNA was adjusted to 10 μl final volume with RNase-free water and combined with 10 μl of a 2× reaction mix (2× PCR Buffer II (Thermo Fisher Scientific), 3.2 mM dNTPs RNase-free (Genaxxon), 0.5 A260 units Oligo d(T)18 mRNA Primer (NEB), 10 mM MgCl2 Solution (Thermo Fisher Scientific), 4 U RNasin® Plus RNase Inhibitor (NEB), 50 U MultiScribe™ Reverse Transcriptase (RT) (Thermo Fisher Scientific)). For all samples, a non-RT control was implemented. The cDNA was synthesized with the following program: 25°C for 10 min, 37°C for 120 min, 85°C for 5 min. Subsequently, the samples were diluted threefold with ddH2O.
For RT-qPCR, a master mix with 7.5 μl 2× ORA™ See qPCR Probe Mix (highQu), 0.4 μl forward primer, 0.4 μl reverse primer and 5.7 μl ddH2O per reaction was prepared. For analysis of ZnF-3AC expression, primers were designed to bind in the zinc finger and the methyltransferase part of the construct to discriminate between potentially endogenously expressed DNMT3A and the induced construct. As a reference gene, RAB13 was selected due to its lack of a CGI in the promoter (primers are listed in Supplementary Table S2). All samples were pipetted in technical triplicates and non-RT controls, as well as non-template controls, were included. To determine the PCR efficiency for each primer set, a 1:5 dilution series of pooled cDNA was pipetted. The PCR was performed using the CFX96 Real-Time PCR detection system (Bio-Rad Laboratories) with the following cycling condition: 95°C for 3 min, 40 cycles of 95°C for 3 s, 60°C for 20 s, 72°C for 4 s and finally a 65–95°C ramp (0.5°C steps every 5 s). Relative expression ratios were calculated using the Pfaffl method for qPCR (31).
Library preparation for RNA-seq, downstream analysis
For the generation of RNA-seq libraries, 100 ng RNA per sample (two biological replicates per time point) were processed using the NEBNext® Single Cell/Low Input RNA Library Prep Kit for Illumina® (NEB) according to the manufacturer's protocol. The quality and quantity of the samples were checked during processing using a LabChip® GXII Touch™ HT system (Perkin Elmer). Samples were indexed and barcoded using the NEBNext® Multiplex Oligos for Illumina® (Index Primers Set 1) (NEB). Finally, 250 ng per barcoded sample (n = 6) were pooled and sequenced at the Max Planck-Genome-Centre Cologne using the Illumina HiSeq3000 platform with a depth of 120 million reads and in 1 × 150 bp mode. Sequencing data were received in FASTQ format and all data processing steps were performed on the European Galaxy web platform (32). The quality of the reads was assessed using the FastQC tool and the reads were mapped on Hg19 using the gapped-read mapper TopHat (33) under default settings. Then, the transcripts were assembled with the Cufflinks tool (34) in default settings using the previously obtained BAM files and the reference annotation for genes from UCSC in gtf format. Utilizing the Cuffmerge tool, the transcripts were merged afterward. Finally, the expression levels of genes were obtained in fragments per kilobase of exon model per million reads mapped (FPKM) using the Cuffdiff tool under default settings. Gene ontology (GO) analysis of selected gene sets was performed using the GO website (35) (http://geneontology.org/).
Library preparation for MBD2-pulldown, XChIP and NChIP
Libraries were generated in either triplicates (ZnF-3AC) or duplicates (3AC) for MBD2-seq (Supplementary Figure S2A). In case of NChIP to analyze global distribution of histone modifications, one repeat was conducted for several time points. However, ChIP-qPCR was performed in two repeats for H3K27me3, H3K27ac and H3K36me3 and one repeat for H3K4me3 and H3K9me3 to validate the quality of the ChIP-seq experiments (Supplementary Figure S2B). The same accounts for XChIP of the zinc finger, where ChIP-qPCR was conducted prior to Next-Generation-Sequencing.
For all MBD2-pulldown experiments comprising ZnF-3AC, as well as ZnF-ChIP and NChIP for H3K4me3, H3K27me3, H3K9me3 and H3K36me3, libraries were prepared by the Max Planck-Genome-centre Cologne. In the case of MBD2-pulldowns of 3AC and NChIP of H3K27ac, libraries were prepared using the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (NEB) according to the manufacturer's protocol. Indices and barcodes were introduced using the NEBNext® Multiplex Oligos for Illumina® (Index Primers Set 1) (NEB). The quality of the libraries was finally checked using the LabChip® GXII Touch™ HT system (Perkin Elmer) and 200 ng (H3K27ac-ChIP libraries) or 1000 ng (MBD2-pulldown libraries) per sample were pooled and sequenced at the Max Planck-Genome-Centre Cologne using the Illumina HiSeq3000 platform in 1 × 150 bp (MBD2-seq, ZnF-ChIP) or 2 × 150 bp (histone modifications) mode.
Next-Generation-Sequencing of MBD2-pulldown, XChIP and NChIP samples
Samples from MBD2-pulldowns or XChIP were sequenced in 1 × 150 bp mode, samples from NChIP usually in 2 × 150 bp mode using either the Illumina HiSeq2500 or Illumina HiSeq3000 platform. The sequencing depth was generally around 10 million reads per sample and data files were obtained in FASTQ format. All data were processed on the European Galaxy web platform (32). The quality of the reads was checked using the FastQC tool. Reads were mapped on the human reference genome Hg19 using the Bowtie2 alignment tool (36) with default settings. The genomic coverage of the reads was calculated using the bamCoverage tool from the DeepTools package (37) in 25 bp bins and normalized by reads per kilobase per million mapped reads (RPKM). For each epigenome modification, an internal normalization was applied by selecting regions with presumably stable signals over the time course of the experiment (Supplementary Figure S3). Therefore, the signals of the previously obtained bigWig files were determined in these regions using the multiBigwigSummary tool. Normalization factors were calculated in Excel and bamCoverage was repeated with the same settings as before (25 bp bins, RPKM), however, with an additional scaling factor included.
Heatmaps were created by running first the computeMatrix tool in regions of interest in scale-regions mode (regions fit in 1 kb, 5 kb flanks, 50 bp bins) and subsequently using the plotHeatmap tool with individually adjusted settings, but always without sorting of regions. Peaks for ZnF-3AC-XChIP were called using the Model-based Analysis of ChIP-Seq 2 (MACS2) (38) tool with a q-value of 0.1 and mfold bounds of 3 and 1000 respectively. Enrichment of motifs in ZnF-3xHA peaks was calculated with the Discriminative Regular Expression Motif Elicitation (DREME) tool (http://meme-suite.org/tools/dreme) (39). Statistical significance was assessed using the E-value, which is the enrichment P-value multiplied with the number of candidate motifs tested. Genomic distribution of ZnF-3xHA peaks was analyzed using the CEAS tool available at the Cistrome Project web platform (http://cistrome.org/) (40). Clusters of differential DNA methylation stability were obtained by K-means clustering using the Chromatin Analysis & Exploration (ChAsE) tool (http://chase.cs.univie.ac.at/overview) (41). Browser views of NGS data were generated utilizing the Integrative Genomics Viewer (IGV) (http://software.broadinstitute.org/software/igv/home) (42). For the generation of boxplots and correlation matrices, the average of ChIP or pulldown signals in target regions were determined via the multiBigwigSummary tool.
The quality of the NChIP data was further assessed by comparison with publicly available datasets (Supplementary Figure S4B). Clustering of these datasets side by side showed a good comparability of their local enrichment (data not shown). In many cases, the ChIP-seq data of different time points after dox removal were very similar to each other also supporting their quality.
Bisulfite sequencing
Bisulfite sequencing experiments were performed in triplicates for most of the analyzed regions. As described in the MBD2-pulldown section, gDNA was isolated from frozen cell pellets and sonicated to obtain fragment sizes in the range of 100–1000 bp. Bisulfite conversion was carried out with 500 ng sheared DNA using the EZ DNA Methylation-Lightning Kit (Zymo Research) as described in the manufacturer's protocol. The converted DNA was eluted in 20 μl elution buffer provided in the kit and served as a template for the following PCR mixture: 1 μl converted DNA, 0.4 μM fwd. primer, 0.4 μM rev. primer, 1× Q-Solution, 1× PCR Buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 2.5 U HotStartTaq DNA Polymerase (Qiagen). The reverse primers were designed to carry a 6 bp barcode (Supplementary Table S3). For amplification, a Touchdown PCR program was chosen with the following conditions: 95°C for 15 min, 5 cycles of 30 s at 94°C, 45 s at 60°C, 30–45 s at 72°C, then 5 cycles of 30 s at 94°C, 45 s at 55°C, 30–45 s at 72°C, subsequently 30 cycles of 30 s at 94°C, 45 s at 50°C, 30–45 s at 72°C and finally 5 min at 72°C. PCR products were loaded on a 1% agarose gel, bands were cut from the gel and the DNA was extracted using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel). Concentrations were measured with NanoDrop 1000, then 20–40 ng per sample were pooled in equimolar amounts. Libraries were prepared by the Max Planck-Genome-Centre Cologne and sequencing was performed using the Illumina HiSeq2500 platform with 2 × 250 bp and sequencing depth of 4.7 × 106 reads. Sequencing results were submitted in FASTQ format and all data were processed on the European Galaxy web platform (32). The quality of the reads was checked using the FastQC tool and merged using PEAR (43) with a quality score of at least 20 and a minimum overlap size of 10 bp. The barcoded reads were filtered out using the Barcode Splitter tool not allowing any mismatches in the barcode sequence. Reads were mapped on the corresponding amplicon sequences uploaded in FASTA format using the bwameth tool (arXiv: 1401.1129v2). Finally, methylation levels of CpGs were extracted using the MethylDackel tool (https://github.com/dpryan79/MethylDackel).
RESULTS
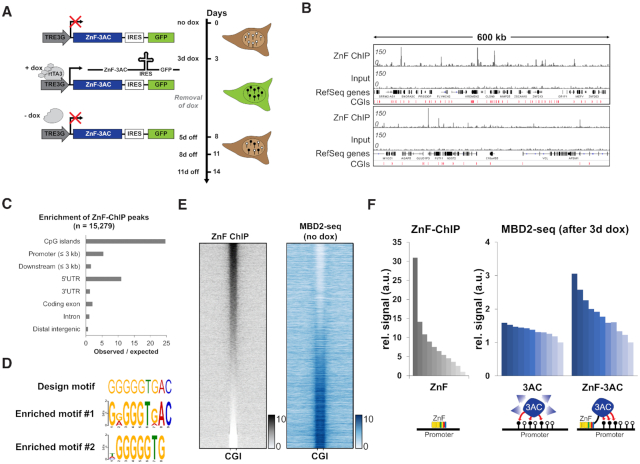
In this work, promiscuous chromatin binding of a fusion of a ZnF and the catalytic domain of DNMT3A (3AC) was used to introduce DNA methylation at several genomic target sites. The ZnF had been originally designed to bind a 9-bp motif in the VEGFA promoter (44) and it was previously used to deliver DNA and H3K9 methylation to this site (20,45,46). Stable HEK293 cells with regulated expression of the ZnF-3AC construct were generated by viral transduction. After induction of ZnF-3AC expression for 3 days by addition of doxycycline (dox), expressing cells were enriched by cell sorting, and samples before and after induction were retrieved for DNA methylation analysis, ChIP and expression analysis. Afterward, the cells were cultured for up to 11 days without dox, and samples were collected on day 5, day 8 and day 11 after dox removal (Figure 1A). DNA methylation was measured by MBD2-pulldown coupled to deep sequencing (MBD2-seq), which is a reliable and cost-efficient method for genome-wide DNA methylation analysis at medium resolution (47,48). In addition, cells expressing the DNMT3A catalytic domain (3AC) not fused to a ZnF were prepared and DNA methylation was determined after expression of this construct as well. Two or three repetitions of the MBD2 pulldown experiments were performed per construct and the corresponding MBD2-seq data showed a high correlation with each other and low correlation with input (Supplementary Figure S2A). The reliability of the MBD2-seq data was confirmed by bisulfite analysis of selected regions (Supplementary Figure S1A and B).
Figure 1.
Zinc finger targeted de novo DNA methylation by the catalytic domain of DNMT3A in HEK293 cells. (A) Stable HEK293 cell line containing a zinc finger (ZnF) fused to the catalytic domain of DNMT3A (3AC) under the control of a doxycycline (dox) inducible promoter. By the addition of dox, the reverse tetracycline-controlled transactivator 3 (rtTA3) can bind the TRE3G promoter and induce the expression of ZnF-3AC. Green fluorescent protein (GFP) is co-expressed with ZnF-3AC using an internal ribosomal entry site (IRES). After 3d of dox treatment, cells are sorted by FACS and grown after removal of dox for another 5–11 days (5–11 days off). Black lollipops indicate methylated DNA, white lollipops unmethylated DNA. (B) Representative browser views of crosslinked-ChIP of HA-tagged ZnF in transiently transfected HEK293 cells with the corresponding input. Two genomic regions are shown, chr16:2761829–3371040 (upper panel) and chr10:75371741–75993358 (lower panel, both in hg19). CGIs, CpG islands. (C) ZnF-ChIP peaks were called by MACS2 and their observed over expected occurrence in genomic elements was analyzed. (D) Originally designed target motif of the ZnF compared to the motifs enriched in ZnF-ChIP peaks as determined by DREME. (E) Heatmap of ZnF-ChIP signals in CGIs with 5 kb flanks sorted by intensity. MBD2-seq data in ‘no dox’ state are displayed in the same order. (F) Signal of ZnF-ChIP in unmethylated CGIs (n = 10 910) sorted and binned into groups of 1000 (left panel). Means of the groups are depicted in the bar diagram. Mean signals of MBD2-seq data of 3AC without ZnF (middle panel) and 3AC with ZnF fusion (right panel) are displayed in the same order. The schemes below the bar diagrams illustrate the corresponding experimental setting.
We initially analyzed the methylation levels of CGIs in cells before dox treatment. The ∼28 000 CGIs in the human genome were subdivided into two groups on the basis of the MBD2-seq data using K-means clustering showing that ∼17 000 of them were already methylated in HEK293 cells and 11 000 were in an unmethylated state (Figure 1E and Supplementary Figure S4A). This result indicates that the HEK293 cells have a CpG island methylator phenotype with a large fraction of CGIs being methylated, which is in agreement with an earlier bisulfite-based analysis of CGI methylation on Chromosome 21 in this cell line (49). For internal calibration of the MBD2-seq data, regions with high methylation were used, which represent fully methylated DNA where no further gain in methylation is possible. To this end, the 1000 CGIs with the highest MBD2-seq signals before dox addition were extracted and the corresponding signals used for internal normalization of the different data sets (Supplementary Figure S3A). Bisulfite-seq analysis of a representative calibration region confirmed its high methylation (∼95%, Supplementary Figure S1C).
Analysis of ZnF binding and targeted DNA methylation
We studied the genome-wide binding of the ZnF by ChIP-seq using an antibody against its HA-tag (Figure 1B). Peak calling revealed 15,279 peaks, which were highly enriched in CGIs and promoters (Figure 1C). Based on the preferred binding of the ZnF at CGIs and their important biological role in the promoters of many genes, we focused the following analyses on targeted DNA methylation in CGIs. These genmic elements represent highly suitable targets for de novo DNA methylation, because they are rich in unmethylated CpG sites. Analysis of the ZnF peaks with DREME identified that two motifs were highly enriched. Motif 1 (E-value = 6.8 × 10−902) matches the original design sequence of the ZnF and motif 2 (E-value = 2.5 × 10−241) represents the core element of the design sequence (Figure 1D), indicating that chromatin binding is reduced in specificity as compared to the 9 bp motif used in the ZnF design. DNA methylation data in the initial state before dox addition showed that strong ZnF binding mainly occurred at unmethylated CGIs (Figure 1E). As a direct effect of DNA methylation on ZnF binding is unlikely, because the ZnF does not contain a CpG site in its recognition sequence, this trend probably reflects the better accessibility of the unmethylated CGIs.
We next compared DNA methylation levels before dox induction and after 3 days of dox treatment and strikingly observed methylation of about 11,000 CGIs, 7000 of them at relatively high levels (see below). The high methylation levels introduced at exemplary CGIs were confirmed by amplicon-based bisulfite sequencing showing close to 100% methylation for several of the covered CpG sites (Supplementary Figure S1A). Control reactions with a catalytically inactive ZnF-3AC E756A mutant showed that the introduced DNA methylation is fully dependent on the catalytic activity of the DNMT3AC part (Supplementary Figure S7A). The strong and almost ubiquitous methylation of CGIs after expression of the ZnF-3AC protein is in agreement with results of a previous study where epigenome modulators were fused to a ZnF targeting Sox2 in a similar dox-regulated expression system as used here, which also identified thousands of ZnF binding sites mostly in promoter regions (50).
To investigate if ZnF targeting contributed to the delivery of methylation, we compared the ZnF-ChIP data with DNA methylation data determined after 3 days of dox induction of either ZnF-3AC or the DNMT3A catalytic domain alone without fusion to ZnF (3AC) (Figure 1F). After expression of 3AC, around 9300 new methylation peaks were observed compared to uninduced cells, among them 78% were located in CGIs and 18% at transcriptional start sites (TSS). This strong, unspecific activity of untargeted 3AC is in good agreement with similar data showing massive and global DNA methylation after ectopic expression of untargeted DNMT3B in MEF cells (51) and massive off-target methylation in dCas9 fused DNMT3A (52). However, regarding the methylation introduced by 3AC and ZnF-3AC into the CGI of the VEGFA gene (the primary target of the ZnF), a 36% higher signal was observed for ZnF-3AC close to the ZnF binding motif (Supplementary Figure S7A). These constructs also expressed GFP together with ZnF-3AC or 3AC using an IRES sequence, which was used to monitor ZnF-3AC and 3AC expression by cytometry after 3 days of dox treatment (Supplementary Figure S7B). The GFP signal levels showed that the higher methylation at the VEGFA promoter after expression of ZnF-3AC was observed despite a reduced expression of ZnF-3AC when compared to 3AC. On a more global view, the DNA methylation introduced by ZnF-3AC showed a moderate Spearman's correlation factor (ρ = 0.43) with the ZnF-ChIP signal (Figure 1F), suggesting a targeting effect of the ZnF module. This effect was most prominent for the 10% strongest ZnF binding sites which showed particularly high DNA methylation after expression of ZnF-3AC. In contrast, the methylation introduced by the untargeted 3AC showed a weaker correlation with the ZnF-ChIP signal (ρ = 0.24), presumably reflecting the preference of both proteins for binding to highly accessible genomic sites.
Efficiency and stability of targeted DNA methylation
Next, we were interested in determining the stability of the DNA methylation introduced at the different CGIs. To this end, the cells were cultivated in the absence of dox after the 3 days expression of ZnF-3AC and DNA methylation was investigated 5, 8 and 11 days after dox removal. RT-qPCR RNA data showed loss of the mRNA for ZnF-3AC after 5 days (Supplementary Figure S8A), which was in agreement with Western Blots showing complete loss of the ZnF-3AC protein 5 days after dox removal (Supplementary Figure S8B). In agreement with these findings, flow cytometry analysis confirmed an almost complete loss of GFP expression already 4 days after dox removal indicative of an equivalent reduction in ZnF-3AC expression (Supplementary Figure S8C). GFP expression data were also determined for the construct containing the ZnF fused to the catalytically inactive E756A mutant, used as a negative control as mentioned above. Importantly, the kinetics of the loss of GFP fluorescence were identical for both constructs no matter if they contained the active or inactive DNMT3A protein (Supplementary Figure S8C). As the inactive mutant did not introduce DNA methylation, this result indicates that there was no detectable counter-selection against cells with high methylation in our experiment. This is an important control, because counter-selection against cells with high methylation would cause a shift in cell populations, which then would lead to a bias in our data.
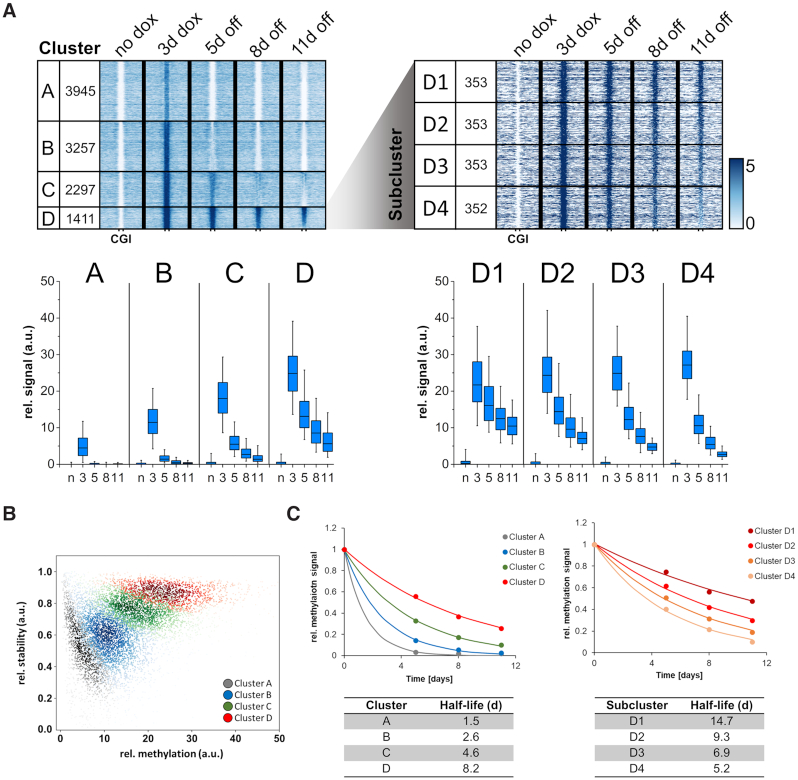
Strikingly, our data show that the introduced DNA methylation was rapidly lost at most of the CGIs over 11 days (Figure 2A) and additional studies at day 14 showed a continuation of the decline at later time points (data not shown). K-means clustering of the CGIs into four groups was performed on the basis of the gain in methylation after 3 days of dox treatment and stability of methylation at days 5, 8 and 11 after dox removal (Figure 2A). Successful clustering was confirmed by a scatter plot showing the CGIs from different clusters in clearly defined areas of the plot (Figure 2B). Cluster A contains CGIs with low methylation that is rapidly lost. Cluster B contains CGIs in which DNA methylation was strong but still rapidly lost. CGIs in cluster C show even stronger methylation and some residual stability. Finally, cluster D comprises 1411 CGIs showing the strongest overall methylation and at least partial stability of the introduced methylation mark up to day 11 after dox removal. It is interesting to note, that the about 7000 CGIs in clusters B–D acquired methylation at levels corresponding to the range of methylation levels observed in CGIs which are already methylated in untreated cells (Supplementary Figure S5).
Figure 2.
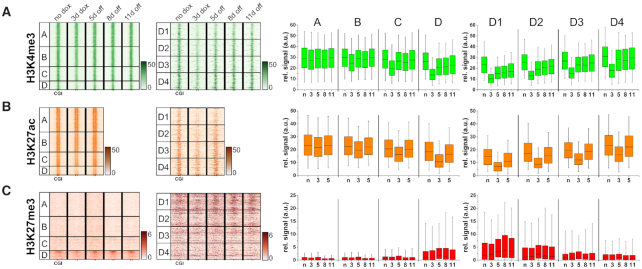
Clustering of CGIs showing differential stability after de novo DNA methylation by ZnF-3AC. (A) Heatmaps of MBD2-seq data are centered around CGIs with 2.5 kb flanks on either side. All CGIs selected for this study were originally unmethylated in ‘no dox’ state and gain methylation after 3 days dox induction of ZnF-3AC. Clusters A–D show differential stability of DNA methylation 5, 8 and 11 days after dox removal. The most stable cluster D was sorted by relative methylation stability at the time point ‘11d off’ to ‘3d dox’ and subclustered into four groups, D1 (most stable) to D4 (least stable). Cluster names and the numbers of CGIs in the clusters are displayed in the first two columns of each heatmap. Boxplots show relative methylation signals of all clusters at different time points. The line in the box indicates the median, the box indicates the 25th and 75th percentile, the whiskers the 5th and 95th percentile. N, no dox; 3, 3d dox; 5/8/11, 5d/8d/11d off. Confidence intervals of average signals and significance of pairwise MBD2-seq signal differences are shown in Supplementary Figure S6A. (B) Scatter plot of the CGIs in the different clusters showing their methylation levels and stability of the methylation between ‘3d dox’ and ‘5d off’. (C) Quantitative analysis of the rates of methylation loss in the different clusters. The average methylation levels of cluster A–D and D1–D4 were fitted to a single exponential decay curve to determine the half-lives of the introduced methylation. The figure shows the average methylation levels as data points and fits as lines. The corresponding half-lives of DNA methylation are indicated below.
Fitting of the average methylation levels of each cluster to an exponential decay curve revealed striking differences in the average half-lives of the methylation signals in the different clusters ranging in between 1.5 days in the case of cluster A to 8.2 days in the case of cluster D (Figure 2C). DNA sequence analysis revealed highly overlapping ranges of CpG density and GC content among all clusters (Supplementary Figure S9). However, the GC and CpG content of cluster A was slightly reduced suggesting that the weaker methylation of this cluster might be caused by the lower binding of the ZnF with its very G-rich recognition sequence. We, therefore, analyzed ZnF binding to the CGIs in the four clusters revealing that overall methylation levels at day 3 (increasing from cluster A to D) are indeed correlated with ZnF binding (Supplementary Figure S10A). Further subdividing of cluster D into four equal bins based on the stability of the methylation, revealed stable DNA methylation over 11 days at about 1000 CGIs in the subclusters D1 to D3 (Figure 2A). Quantitative data analysis revealed a half-life of the methylation signal of 14.7 days for cluster D1 which contains the CGIs with highest stability of introduced DNA methylation (Figure 2C).
Grouping of the CGIs by their methylation signal after 3 days of dox induction followed by splitting of each group into three subgroups by the stability of the DNA methylation at 11 days without dox clearly indicates that regions with similar levels of introduced methylation can show drastically different kinetics of methylation loss. This observation indicates that the initial gain in methylation alone is not sufficient for the prediction of the stability of targeted methylation (Supplementary Figure S11A). This observation is also supported by the finding that the gains in methylation are slightly increasing from subcluster D1 to D4, but stability of the introduced DNA methylation has the opposite trend being most stable in subcluster D1 (14.7 days half-life) and least stable in D4 (5.2 days half-life) (Figure 2A).
To further explore the correlation of ZnF binding and stability of DNA methylation, all CGIs were subdivided into three groups based on the ZnF ChIP-seq signal. The distribution of these groups in the clusters A-D (Supplementary Figure S11B) showed that high ZnF binding supports stable methylation (as evident from the enrichment of high ZnF binding CGIs in cluster D). This effect was expected, because high ZnF binding was correlated with strong methylation gain (Figure 1F). However, CGIs with high ZnF ChIP signals were also present in clusters A–C indicating that strong ZnF binding alone is not a sufficient condition for stable DNA methylation. Moreover, ZnF binding was almost equal in all subclusters D1 to D4 (Supplementary Figure S10B) indicating that in this case, the stability of DNA methylation is not correlated with the ZnF binding strength, but it must depend on other properties of the target sites (see below).
Chromatin state analysis of cluster A–D CGIs
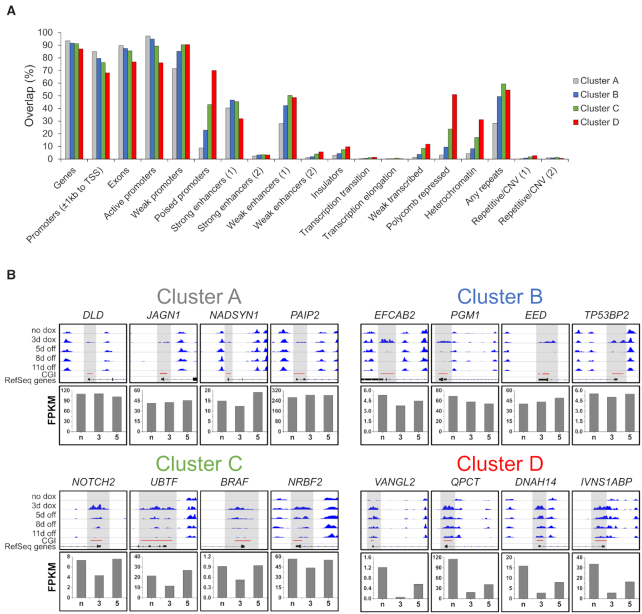
After the definition of four main clusters, which display differential dynamics in DNA methylation stability, we attempted to figure out the specific features of these regions. We therefore performed a chromatin state segmentation analysis of these clusters using EpiExplorer (53) and observed a trend that CGIs with increased stability of methylation (cluster A to D) were less associated with genes with high expression or strong enhancers (Figure 3A). Interestingly, the most stable cluster D showed strong enrichment in chromatin states designated as ‘poised promoters’ or ‘Polycomb repressed’, which are characterized by the presence of H3K27me3 and usually connected to cell development or differentiation. This observation was further supported by gene ontology analysis where the associated genes from cluster D were highly enriched in biological processes involved in development or neuronal differentiation (Supplementary Figure S12). In contrast, the low methylation stability clusters A and B were linked to genes that are mainly involved in processes which can be summarized as housekeeping. These observations can already serve as a hint for the highly diverse epigenetic mechanisms that regulate the levels of DNA methylation within CGIs and the sensitivity of gene promoters for this modification.
Figure 3.
Enrichment of the differentially methylated clusters A–D in various chromatin states and examples for the dynamics in gene expression. (A) Chromatin state segmentation for clusters A–D prepared using EpiExplorer (https://epiexplorer.mpi-inf.mpg.de/) (53). (B) Representative browser views of MBD2-seq data in different clusters and expression data of corresponding genes. The upper boxes show browser views of MBD2-seq data (data range 0–80) generated using the Integrative Genomics Viewer (IGV). Regions showing de novo methylation are highlighted in gray. Clusters A–D exhibit differential DNA methylation dynamics over the time course of the experiment. The lower boxes show the RNA-seq FPKM values of genes associated with the CGIs displayed as bar diagrams. The expression of the genes shows a methylation-dependent dynamic. N, no dox; 3, 3d dox; 5, 5d off. The corresponding genomic regions are listed in Supplementary Table S4.
Dynamic changes of gene expression after CGI methylation
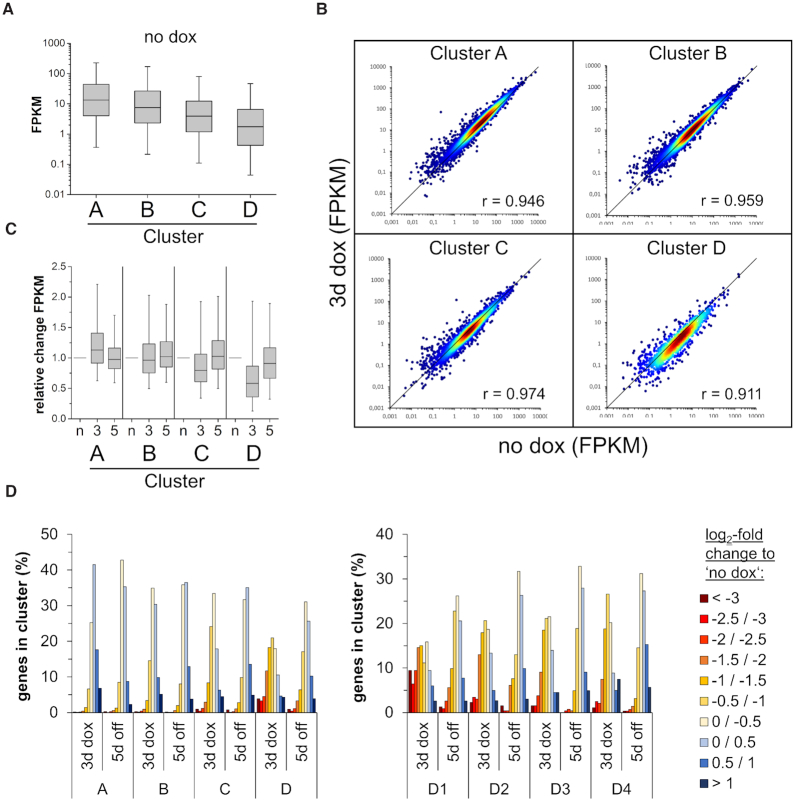
We determined the expression of genes associated with the CGIs studied here before dox addition, after 3 days of dox treatment and 5 days after dox removal (Figures 3B and 4). Similarly as observed in the chromatin segmentation analysis (Figure 3A), the gene expression studies revealed that the efficiency of DNA methylation as determined by the DNA methylation level after 3 days of dox treatment anticorrelated with gene expression in the initial state (before dox), suggesting that lowly expressed genes are more amenable for DNA methylation (Figure 4A). At later time points, the dynamic changes of gene expression were highly correlated with the level and stability of the DNA methylation gain. In cluster A and B, where only moderate and transient CGI methylation was introduced, the effect of the methylation on gene expression was marginal (Figure 4B and C). However, in the case of cluster C and D, where strong and (in cluster D) partially stable CGI methylation was introduced, a global trend towards gene silencing was observed. This effect was especially pronounced in the subclusters D1 and D2 where a fraction of genes showed a >8-fold reduction in expression (Figure 4D). Even after 5 days of dox removal, the majority of genes did not recover to original expression levels and ∼20% of them were still reduced by at least 2-fold. These results reveal that the extent and stability of CGI methylation and gene repression are highly target-gene dependent. However, the level and stability of DNA methylation gain showed a striking correlation with the level and stability of gene repression at the genome-wide level. These findings suggest that the delivery of DNA methylation at CGIs directly leads to the reduction of gene expression, as it is not plausible to assume indirect effects acting in the same direction at so many different loci.
Figure 4.
Genome-wide introduction of DNA methylation in CGIs affects the expression of hundreds of genes. (A) Boxplot showing the FPKM values of genes associated with CGIs of the clusters A–D at the time point ‘no dox’. The line in the box indicates the median, the box indicates the 25th and 75th percentile, the whiskers the 5th and 95th percentile. (B) Scatter plots of FPKMs at time points ‘no dox’ versus ‘3d dox’ in clusters A–D. Pearson correlation coefficients are displayed in each cluster. (C) Relative expression of genes (FPKM ≥ 0.1 at ‘no dox’) normalized to ‘no dox’ in clusters A to D. N, no dox; 3, 3d dox; 5, 5d off. The line in the box indicates the median, the box indicates the 25th and 75th percentile, the whiskers the 5th and 95th percentile. (D) Histogram showing the fraction of genes with their relative expression change at time point ‘3d dox’ and ‘5d off’ compared to ‘no dox’ in each cluster and subcluster.
Dynamic changes of other chromatin modifications after CGI methylation
As a next step, the genome-wide distribution of several chromatin marks (H3K4me3, H3K9me3, H3K27me3, H3K27ac and H3K36me3) was determined by ChIP-seq. After quality control and normalization (Supplementary Figure S3B–F), the data were analyzed using the clusters defined previously based on the level and stability of DNA methylation. Strikingly, both H3K4me3 and H3K27ac showed a genome-wide response that was strongly anti-correlated with the level of DNA methylation (Figure 5A and B). Both modifications showed strong reductions after 3 days of dox treatment in clusters B to D, but rapidly returned to normal levels in cluster B and C. A relatively stable reduction of H3K4me3 was observed in cluster D, mainly in the subclusters D1 and D2, while in case of H3K27ac, stability was low in all clusters. These findings suggest that the delivery of DNA methylation at CGIs directly leads to the reduction of H3K4me3 and H3K27ac.
Figure 5.
DNA methylation is most stable in CGIs containing H3K27me3 and it leads to a differential reduction of activating marks H3K4me3 and H3K27ac. The same clusters were used for the analysis of histone modifications as previously in MBD2-seq. Heatmaps are centered around CGIs with 5 kb flanks. Boxplots show the signals of the histone modifications in CGIs. The line in the box indicates the median, the box indicates the 25th and 75th percentile, the whiskers the 5th and 95th percentile. N, no dox; 3, 3d dox; 5/8/11, 5d/8d/11d off. ChIP-seq signals are shown for: (A) H3K4me3, (B) H3K27ac and (C) H3K27me3. Confidence intervals of average signals and significance of pairwise ChIP-seq signal differences are shown in Supplementary Figure S6B–D.
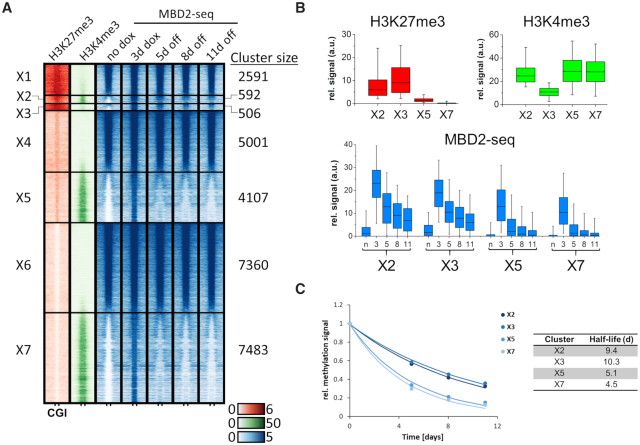
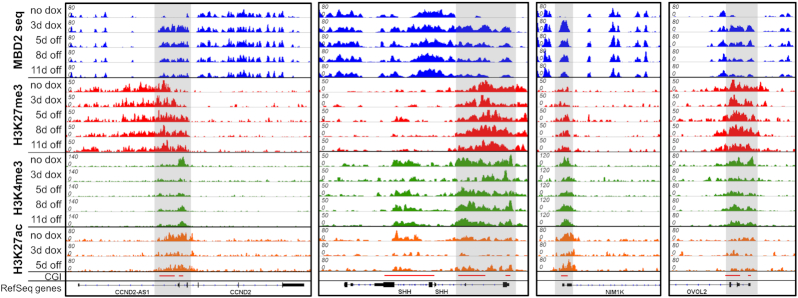
Analysis of the density of H3K27me3 revealed a clear enrichment of this modification in cluster D (Figure 5C) with the highest enrichment in subcluster D1 and D2, which contain the most stable DNA methylation after its installment. This finding confirms the conclusions from the initial chromatin segmentation analysis (Figure 3A). As some of these regions also contain H3K4me3, we prepared an additional clustering on the basis of initial H3K27me3, H3K4me3 and DNA methylation signals (no dox conditions) resulting in 7 groups (X1 to X7) (Figure 6A–C). Among them, cluster X1, X4 and X6 show strong, medium and weak H3K27me3, no H3K4me3 and strong DNA methylation already in the untreated cells. Cluster X5 and X7 contain CGIs with strong H3K4me3, medium or weak H3K27me3, and no methylation in untreated cells. These regions showed strong, but unstable introduction of DNA methylation. Cluster X2 contains bivalent CGIs with strong H3K27me3 and strong H3K4me3. Cluster X3 contains CGIs with strong H3K27me3 but no or weak H3K4me3. Both clusters were unmethylated in the untreated cells and showed strong and partially stable methylation. Interestingly, the introduced methylation was most stable in cluster X3, indicating that H3K27me3 was relevant for stability, while H3K4me3 was slightly reducing stability. Inspection of representative genomic loci illustrates these global observations revealing presence of H3K27me3 in regions with stable gain in DNA methylation (Figure 7). At the same time, a strong depletion of H3K4me3 and H3K27ac can be observed with a gradual recovery of the signal over time.
Figure 6.
Alternative CGI clustering based on initial H3K27me3, H3K4me3 and DNA methylation (no dox) revealed an influence of H3K27me3 on DNA methylation stability. (A) Heatmaps are centered around CGIs with 2.5 kb flanks on either side showing H3K27me3 and H3K4me3 ChIP-seq signals at ‘no dox’ and MBD2-seq signals at the different time points of ZnF-3AC induction. K-means clustering was performed first for H3K27me3, then based on H3K4me3 and MBD2-seq. Clusters are labeled with ‘X1–7’. (B) Boxplots showing the H3K27me3, H3K4me3 and MBD2-seq signals in the selected clusters X2, X3, X5 and X7. The line in the box indicates the median, the box indicates the 25th and 75th percentile, the whiskers the 5th and 95th percentile. n, no dox; 3, 3d dox; 5/8/11, 5d/8d/11d off. (C) The average methylation levels of clusters X2, X3, X5 and X7 were fitted to a single exponential decay curve to determine the half-lives of the methylation. The figure shows the average methylation levels as data points and fits as lines.
Figure 7.
Example of the epigenome landscape in regions showing stable DNA methylation. Browser views of MBD2-seq, H3K27me3-ChIP, H3K4me3-ChIP and H3K27ac-ChIP at representative loci generated with IGV. Regions showing strong de novo DNA methylation are highlighted in gray. Shown are example regions illustrating the stable introduction of DNA methylation in CGIs with high H3K27me3 signal. Strong, but transient reductions of H3K4me3 and H3K27ac were observed at these loci. The corresponding genomic regions are listed in Supplementary Table S5.
In the case of H3K9me3, we did not observe strong signals in the tested CGIs and DNA methylation did not lead to a detectable deposition of this mark (Supplementary Figure S13A). However, some of the already methylated CGIs showed co-occurrence of H3K9me3 (Supplementary Figure S4A). Similar to H3K9me3, no strong H3K36me3 signals and no deposition of this chromatin mark was observed in the CGIs investigated in our study (Supplementary Figure S13B). However, we did observe a general trend in the initial state of the cells, where decreasing H3K36me3 signal in the regions downstream of the CGI from cluster A to D was correlated with the expression levels of the associated genes. Moreover, the H3K36me3 signals in the regions downstream of the CGI were higher for CGIs in unmethylated state than for those in methylated state (Supplementary Figure S4A). This result is in agreement with the general observation that H3K36me3 accumulates in the bodies of expressed genes where it is introduced by the SETD2 protein lysine methyltransferase after recruitment by the active RNAPII (54,55).
DISCUSSION
Chromatin properties are regulated by numerous epigenome modifications that occur in various combinations and establish a complex network. Currently, it is unclear how these modifications interact with each other and to which extent each of them is causative for downstream effects such as changes in gene expression. While in many cases clear correlations between different modifications and properties such as gene expression were documented, it is difficult to find out if these reflect causative effects or if the affected modifications are indirectly connected through secondary effects. Another important open question is related to the switching of biological states encoded in epigenome networks. Previous studies have documented that DNA methylation introduced at promoters was transient in some cases while stable in others. It remained unclear if these inconsistencies are due to the editing of other modifications together with the DNA methylation, other technical peculiarities like different cell lines, different epigenome editing strategies and delivery methods, if they are related to the specific target loci, or if any combination of these factors caused the variable outcome. Yet, this question is highly relevant, because clinical applications of epigenome editing typically aim for stable reprogramming, meaning that the conditions leading to stable changes of the epigenome in a predictable manner need to be elucidated.
Epigenome editing is a powerful approach to study the mutual interaction of chromatin modifications and their direct biological effects, because it allows introducing a defined perturbation into the epigenome network at a specific genomic locus. In this work, the catalytic domain of the DNMT3A methyltransferase was selected as an epigenome effector, because it is able to introduce DNA methylation after genome targeting (20,56). On the other hand, it does not interact with other endogenous chromatin factors, thereby restricting the epigenome modification to the introduced DNA methylation, and thus avoiding the undesired introduction of additional modifications by endogenous enzymes bound to the DNMT3A catalytic domain. In addition, the catalytic domain of DNMT3A does not contain two additional domains of the DNMT3A full-length protein (the PWWP domain and the ADD domain) which are known to interact with chromatin and read histone tail modifications, thus avoiding a direct crosstalk of the existing epigenome modifications at the target sites with the recruitment or activity of the EpiEditor (57,58).
For targeting, we exploited the binding of a ZnF with promiscuous specificity allowing us to introduce DNA methylation at several thousand CGIs in one experiment. Using this approach, we investigated the influence of the target sites and the epigenome state on the dynamic outcome of targeted DNA methylation in a systematic manner. Our work revealed that DNA methylation can be efficiently introduced at thousands of CGIs, but the introduced DNA methylation is rapidly lost at most of them, with partial stability observed at a fraction of about 10% of CGIs which are enriched in H3K27me3. Strikingly, this rapid demethylation of CGIs occurs in cultivated cell lines with stable DNA methylation, indicating that overall maintenance methylation is intact, but it fails at artificially methylated CGIs. Comparison of the stability of methylation at the different CGIs indicates that regions with similar levels of the introduced methylation still show very different kinetics of methylation loss indicating that the demethylation of CGIs is not a global event, but its kinetics depend on the local sequence and epigenome context of each individual region (Supplementary Figure S11A). Our data agree with previous results showing that CGIs are refractory to DNA methylation due to their DNA sequence (59). The local CGI specific demethylation observed here could be caused by two effects, the prevention of the recruitment or activity of DNMT1 leading to a failure in maintenance methylation or the presence of TET enzymes at the CGI triggering active demethylation (60,61). It will be an interesting topic for future work to distinguish between these alternative but not mutually exclusive mechanisms.
Regarding the crosstalk of DNA methylation and H3K27me3, these two marks are non-overlapping in ES cells, where H3K27me3 occurs in discrete, punctate regions mostly overlapping with CpG islands, which are unmethylated in these cells (62). However, in somatic cell types and cancer cell lines, extensive overlap between DNA methylation and H3K27me3 methylation has been observed indicating that both modifications are no longer mutually exclusive (62–64). Coexistence of H3K27me3 and DNA methylation in untreated cells is also visible in our data (cluster X1 in Figure 6A). Importantly, our data show that the presence of H3K27me3 predisposes CGIs for stable introduction of DNA methylation and reprogramming (cluster X2 and X3 in Figure 6). This finding is in very good agreement with several studies documenting that H3K27me3 marked CGIs tend to acquire aberrant methylation in tumors and cell lines (49,65,66). Similarly, expression of different DNMT3A variants with mutations in the PWWP domain, which abrogate chromatin targeting caused preferential methylation at H3K27me3 containing regions, suggesting that they are most prone to de novo methylation (67,68). Recently, the positive correlation of DNA methylation and H3K27me3 has also been reported in a genetic study in mice expressing a catalytically inactive DNMT3B enzyme (69). A similar effect was also observed after ectopic expression of untargeted DNMT3B (51). In agreement with our data, a recent study showed that the combined editing of DNA and H3K27me3 leads to more stable gene silencing (70). Regarding the molecular mechanisms connecting H3K27me3 and DNA methylation, it had been reported that PRC2, the complex responsible for setting H3K27me3, can recruit DNMTs (71) and DNMT3A interacts with PRC2 components (72,73), which could explain enhanced DNA methylation at H3K27me3 regions. Strikingly, the opposite connection has recently been established as well, because the AEBP2 Zinc finger protein that is associated with PRC2 binds CpG methylated DNA (74) suggesting the existence of a synergistic mutual enhancement loop of DNA methylation and H3K27me3.
Interestingly, we could not detect any crosstalk of DNA methylation at CGIs to H3K9me3, another prominent silencing chromatin modification. This genome-wide lack of effect was observed although DNA methylation and H3K9me3 co-occur in heterochromatic repeats and H3K9 methylation has been observed to target DNA methylation to these sites (75). Different pathways have been described that could connect H3K9me3 to DNA methylation, including the recruitment of the SUV39H1-HP1 complex responsible for setting H3K9me3 by the methyl CpG binding proteins MBD1 (76) and MeCP2 (77,78) and the interaction of HP1 (reading H3K9me3) with DNMT3B (75). Our data suggest that the crosstalk of DNA methylation and H3K9me3 is not efficient at CGIs in HEK293 cells, but perhaps restricted to heterochromatic regions. Nevertheless, co-editing of DNA and H3K9 methylation at CGI was found to enhance and stabilize gene repression, suggesting that an artificial deposition of both marks at CGIs is possible and it can be effective (26).
We show that targeted DNA methylation at thousands of CGIs directly led to the fast but transient decline of H3K4me3 and H3K27ac modifications. The parallel response at multiple sites suggests that a direct molecular link exists between the introduced DNA methylation and the decline in these two marks, because indirect or promoter-specific effects should not result in such a concerted global response. Several molecular pathways have been identified that could directly contribute to this effect. With respect to H3K4me3 reduction, the CXXC domains of several H3K4 specific protein lysine methyltransferases (KMT2A, KMT2B) as well as CFP1 (a member of KMT2F and KMT2G complexes) were shown to specifically interact with unmethylated DNA (79–82). This finding can explain the loss of the KMT2 enzymes after CGI methylation leading to the subsequent loss of H3K4me3. Regarding the loss of histone acetylation, DNA methylation has been shown to recruit HDACs through the methyl binding proteins MeCP2 (83) and MBD2 (84), which can explain the temporal coupling of histone deacetylation following DNA methylation. Strikingly, our data suggest that these changes are part of a ‘default’ response of the epigenome network to CGI methylation, although their intensity is modulated by CGI-specific additional factors.
Finally, our data allow us answering the long-standing question of whether DNA methylation at CGIs can cause gene repression or if it is a consequence of a reduction in gene expression triggered by other processes. We observed a decline in gene expression that occurred genome-wide at hundreds of promoters after methylation of their associated CGIs. The kinetics and stability of this effect were tightly correlated with the dynamics of the DNA methylation deposition and its stability implicating that our results cannot be explained by promoter-specific local effects, but they indicate that introduced CGI methylation has the capacity to directly silence gene expression. The possible molecular pathways leading to the decline in gene expression are the recruitment of diverse repressive MBD protein-containing complexes, which induce chromatin remodeling and also lead to the decline in H3K4me3 and histone acetylation as described above.
CONCLUSIONS
Chromatin properties are regulated by numerous epigenome modifications, which occur in various combinations creating a complex network. Currently, it is unclear how these modifications interact with each other and to which extent each of them is causative for downstream effects like changes in gene expression. In this work, we employed the promiscuous DNA binding of a fusion protein comprising a ZnF and the catalytic domain of DNMT3A to introduce DNA methylation at several thousand CGIs in one experiment. This allowed us to investigate in a systematic manner the influence of the epigenome environment on the efficiency and stability of targeted DNA methylation and the subsequent changes of the epigenome network triggered by the introduced DNA methylation. Our data show efficient methylation at many thousand CGIs, but the rapid loss of the methylation at most of the target sites. This finding illustrates the power of the molecular processes that protect CGIs against aberrant DNA methylation. However, partially stable DNA methylation was observed at ∼1000 target sites and CGIs with stable DNA methylation showed an enrichment in H3K27me3. Globally, the introduction of DNA methylation strongly correlated with a decrease in gene expression, indicating a direct role of DNA methylation in gene expression control. Similarly, global reductions in H3K4me3 and H3K27ac were observed after DNA methylation at CGIs, which can be explained by the repulsion of HATs and KMT2 enzymes and recruitment of HDACs. In contrast, no changes were observed for H3K9me3 and H3K36me3 despite the existence of potential molecular pathways connecting these modifications. All in all, our data provide a global and time-resolved view on the network of epigenome modifications, their interconnection and the concerted response to the artificial DNA methylation of thousands of CGIs and they document a direct connection of DNA methylation at CGIs to reduced H3K4me3, histone acetylation and gene expression.
DATA AVAILABILITY
Raw data are deposited at the NCBI GEO database under the following accession numbers:
SuperSeries GSE144904: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing.
This Series is composed of the following SubSeries:
GSE144903: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing [RNA-seq]
GSE144928: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing [ChIP-Seq]
GSE144331: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing [MBD-seq]
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge support by the Max Planck-Genome-Centre Cologne in NGS sequencing. Thanks are due to Dr Stephan Eisler (SRCSB, Stuttgart University) for support in Cell sorting at the beginning of the project.
Author contributions: J.B. conducted all experiments with support from G.K., P.B. and P.R. A.J. and G.K. conceived and devised the study. A.J., G.K., P.R. and P.B. supervised the work. J.B. and A.J. prepared the manuscript draft and figures. All authors contributed to data interpretation and editing of the manuscript. The final manuscript was approved by all authors.
Notes
Present address: Dr Goran Kungulovski, Bio Engineering LLC, Skopje, Macedonia.
Contributor Information
Julian Broche, Department of Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, 70569 Stuttgart, Germany.
Goran Kungulovski, Department of Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, 70569 Stuttgart, Germany.
Pavel Bashtrykov, Department of Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, 70569 Stuttgart, Germany.
Philipp Rathert, Department of Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, 70569 Stuttgart, Germany.
Albert Jeltsch, Department of Biochemistry, Institute of Biochemistry and Technical Biochemistry, University of Stuttgart, 70569 Stuttgart, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
BW Foundation [BWST-ncRNA-007]. The funder had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. Funding for open access charge: University of Stuttgart.
Conflict of interest statement. None declared.
REFERENCES
- 1. Allis C.D., Jenuwein T.. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016; 17:487–500. [DOI] [PubMed] [Google Scholar]
- 2. Huang H., Lin S., Garcia B.A., Zhao Y.. Quantitative proteomic analysis of histone modifications. Chem. Rev. 2015; 115:2376–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M. et al.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009; 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeltsch A., Broche J., Bashtrykov P.. Molecular processes connecting DNA methylation patterns with DNA methyltransferases and histone modifications in mammalian genomes. Genes. 2018; 9:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jeltsch A., Jurkowska R.Z.. New concepts in DNA methylation. Trends Biochem. Sci. 2014; 39:310–318. [DOI] [PubMed] [Google Scholar]
- 6. Ginno P.A., Gaidatzis D., Feldmann A., Hoerner L., Imanci D., Burger L., Zilbermann F., Peters A., Edenhofer F., Smallwood S.A. et al.. A genome-scale map of DNA methylation turnover identifies site-specific dependencies of DNMT and TET activity. Nat. Commun. 2020; 11:2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baylin S.B., Jones P.A.. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011; 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao Y., Garcia B.A.. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb. Perspect. Biol. 2015; 7:a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev.. Genet. 2012; 13:484–492. [DOI] [PubMed] [Google Scholar]
- 10. Bergman Y., Cedar H.. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013; 20:274–281. [DOI] [PubMed] [Google Scholar]
- 11. Suva M.L., Riggi N., Bernstein B.E.. Epigenetic reprogramming in cancer. Science. 2013; 339:1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schubeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. [DOI] [PubMed] [Google Scholar]
- 13. Kungulovski G., Jeltsch A.. Epigenome Editing: State of the art, concepts, and perspectives. Trends Genet. 2016; 32:101–113. [DOI] [PubMed] [Google Scholar]
- 14. Pulecio J., Verma N., Mejia-Ramirez E., Huangfu D., Raya A.. CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell. 2017; 21:431–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Holtzman L., Gersbach C.A.. Editing the epigenome: reshaping the genomic landscape. Annu. Rev. Genomics Hum. Genet. 2018; 19:43–71. [DOI] [PubMed] [Google Scholar]
- 16. Sen D., Keung A.J.. Designing epigenome editors: considerations of biochemical and locus specificities. Methods Mol. Biol. 2018; 1767:65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tadic V., Josipovic G., Zoldos V., Vojta A.. CRISPR/Cas9-based epigenome editing: An overview of dCas9-based tools with special emphasis on off-target activity. Methods. 2019; 164–165:109–119. [DOI] [PubMed] [Google Scholar]
- 18. Ragunathan K., Jih G., Moazed D.. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science. 2015; 348:1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Audergon P.N., Catania S., Kagansky A., Tong P., Shukla M., Pidoux A.L., Allshire R.C.. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science. 2015; 348:132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kungulovski G., Nunna S., Thomas M., Zanger U.M., Reinhardt R., Jeltsch A.. Targeted epigenome editing of an endogenous locus with chromatin modifiers is not stably maintained. Epigenet. Chromatin. 2015; 8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vojta A., Dobrinic P., Tadic V., Bockor L., Korac P., Julg B., Klasic M., Zoldos V.. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016; 44:5615–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braun S.M.G., Kirkland J.G., Chory E.J., Husmann D., Calarco J.P., Crabtree G.R.. Rapid and reversible epigenome editing by endogenous chromatin regulators. Nat. Commun. 2017; 8:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cano-Rodriguez D., Gjaltema R.A., Jilderda L.J., Jellema P., Dokter-Fokkens J., Ruiters M.H., Rots M.G.. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat. Commun. 2016; 7:12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saunderson E.A., Stepper P., Gomm J.J., Hoa L., Morgan A., Allen M.D., Jones J.L., Gribben J.G., Jurkowski T.P., Ficz G.. Hit-and-run epigenetic editing prevents senescence entry in primary breast cells from healthy donors. Nat. Commun. 2017; 8:1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bintu L., Yong J., Antebi Y.E., McCue K., Kazuki Y., Uno N., Oshimura M., Elowitz M.B.. Dynamics of epigenetic regulation at the single-cell level. Science. 2016; 351:720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amabile A., Migliara A., Capasso P., Biffi M., Cittaro D., Naldini L., Lombardo A.. Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell. 2016; 167:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O’Geen H., Ren C., Nicolet C.M., Perez A.A., Halmai J., Le V.M., Mackay J.P., Farnham P.J., Segal D.J.. dCas9-based epigenome editing suggests acquisition of histone methylation is not sufficient for target gene repression. Nucleic Acids Res. 2017; 45:9901–9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rathert P., Roth M., Neumann T., Muerdter F., Roe J.S., Muhar M., Deswal S., Cerny-Reiterer S., Peter B., Jude J. et al.. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015; 525:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qing K., Bachelot T., Mukherjee P., Wang X.S., Peng L., Yoder M.C., Leboulch P., Srivastava A.. Adeno-associated virus type 2-mediated transfer of ecotropic retrovirus receptor cDNA allows ecotropic retroviral transduction of established and primary human cells. J. Virol. 1997; 71:5663–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Markusic D., Oude-Elferink R., Das A.T., Berkhout B., Seppen J.. Comparison of single regulated lentiviral vectors with rtTA expression driven by an autoregulatory loop or a constitutive promoter. Nucleic Acids Res. 2005; 33:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Afgan E., Baker D., Batut B., van den Beek M., Bouvier D., Cech M., Chilton J., Clements D., Coraor N., Gruning B.A. et al.. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018; 46:W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y. et al.. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011; 12:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Younesy H., Nielsen C.B., Lorincz M.C., Jones S.J., Karimi M.M., Moller T.. ChAsE: chromatin analysis and exploration tool. Bioinformatics. 2016; 32:3324–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang J., Kobert K., Flouri T., Stamatakis A.. PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics. 2014; 30:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu P.Q., Rebar E.J., Zhang L., Liu Q., Jamieson A.C., Liang Y., Qi H., Li P.X., Chen B., Mendel M.C. et al.. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J. Biol. Chem. 2001; 276:11323–11334. [DOI] [PubMed] [Google Scholar]
- 45. Snowden A.W., Gregory P.D., Case C.C., Pabo C.O.. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 2002; 12:2159–2166. [DOI] [PubMed] [Google Scholar]
- 46. Siddique A.N., Nunna S., Rajavelu A., Zhang Y., Jurkowska R.Z., Reinhardt R., Rots M.G., Ragozin S., Jurkowski T.P., Jeltsch A.. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J. Mol. Biol. 2013; 425:479–491. [DOI] [PubMed] [Google Scholar]
- 47. Jeltsch A., Broche J., Lungu C., Bashtrykov P.. Biotechnological applications of MBD domain proteins for DNA methylation analysis. J. Mol. Biol. 2019; doi:10.1016/j.jmb.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 48. Aberg K.A., Chan R.F., van den Oord E.. MBD-seq - realities of a misunderstood method for high-quality methylome-wide association studies. Epigenetics. 2020; 15:431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y., Rohde C., Tierling S., Jurkowski T.P., Bock C., Santacruz D., Ragozin S., Reinhardt R., Groth M., Walter J. et al.. DNA methylation analysis of chromosome 21 gene promoters at single base pair and single allele resolution. PLos Genet. 2009; 5:e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grimmer M.R., Stolzenburg S., Ford E., Lister R., Blancafort P., Farnham P.J.. Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation. Nucleic Acids Res. 2014; 42:10856–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Charlton J., Karnik R., Beerman I., Smith Z.D., Gu H., Boyle P., Mi X., Clement K., Pop R. et al.. Targets and genomic constraints of ectopic Dnmt3b expression. eLife. 2018; 7:e40757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Galonska C., Charlton J., Mattei A.L., Donaghey J., Clement K., Gu H., Mohammad A.W., Stamenova E.K., Cacchiarelli D., Klages S. et al.. Genome-wide tracking of dCas9-methyltransferase footprints. Nat. Commun. 2018; 9:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Halachev K., Bast H., Albrecht F., Lengauer T., Bock C.. EpiExplorer: live exploration and global analysis of large epigenomic datasets. Genome Biol. 2012; 13:R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wagner E.J., Carpenter P.B.. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012; 13:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McDaniel S.L., Strahl B.D.. Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci.: CMLS. 2017; 74:3317–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nunna S., Reinhardt R., Ragozin S., Jeltsch A.. Targeted methylation of the epithelial cell adhesion molecule (EpCAM) promoter to silence its expression in ovarian cancer cells. PLoS One. 2014; 9:e87703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jeltsch A., Jurkowska R.Z.. Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm. Nucleic Acids Res. 2016; 44:8556–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gowher H., Jeltsch A.. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem. Soc. Trans. 2018; 46:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Long H.K., King H.W., Patient R.K., Odom D.T., Klose R.J.. Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res. 2016; 44:6693–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wu S.C., Zhang Y.. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010; 11:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rasmussen K.D., Helin K.. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016; 30:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rose N.R., Klose R.J.. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim. Biophys. Acta. 2014; 1839:1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Statham A.L., Robinson M.D., Song J.Z., Coolen M.W., Stirzaker C., Clark S.J.. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012; 22:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brinkman A.B., Gu H., Bartels S.J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A. et al.. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012; 22:1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Widschwendter M., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I. et al.. Epigenetic stem cell signature in cancer. Nat. Genet. 2007; 39:157–158. [DOI] [PubMed] [Google Scholar]
- 66. Schlesinger Y., Straussman R., Keshet I., Farkash S., Hecht M., Zimmerman J., Eden E., Yakhini Z., Ben-Shushan E., Reubinoff B.E. et al.. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007; 39:232–236. [DOI] [PubMed] [Google Scholar]
- 67. Heyn P., Logan C.V., Fluteau A., Challis R.C., Auchynnikava T., Martin C.A., Marsh J.A., Taglini F., Kilanowski F., Parry D.A. et al.. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 2019; 51:96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sendzikaite G., Hanna C.W., Stewart-Morgan K.R., Ivanova E., Kelsey G.. A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 2019; 10:1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nowialis P., Lopusna K., Opavska J., Haney S.L., Abraham A., Sheng P., Riva A., Natarajan A., Guryanova O., Simpson M. et al.. Catalytically inactive Dnmt3b rescues mouse embryonic development by accessory and repressive functions. Nat. Commun. 2019; 10:4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. O’Geen H., Bates S.L., Carter S.S., Nisson K.A., Halmai J., Fink K.D., Rhie S.K., Farnham P.J., Segal D.J.. Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenet. Chromatin. 2019; 12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M. et al.. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006; 439:871–874. [DOI] [PubMed] [Google Scholar]
- 72. Neri F., Krepelova A., Incarnato D., Maldotti M., Parlato C., Galvagni F., Matarese F., Stunnenberg H.G., Oliviero S.. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013; 155:121–134. [DOI] [PubMed] [Google Scholar]
- 73. Manzo M., Wirz J., Ambrosi C., Villasenor R., Roschitzki B., Baubec T.. Isoform-specific localization of DNMT3A regulates DNA methylation fidelity at bivalent CpG islands. EMBO J. 2017; 36:3421–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang X., Paucek R.D., Gooding A.R., Brown Z.Z., Ge E.J., Muir T.W., Cech T.R.. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 2017; 24:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lehnertz B., Ueda Y., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H.. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003; 13:1192–1200. [DOI] [PubMed] [Google Scholar]
- 76. Fujita N., Watanabe S., Ichimura T., Tsuruzoe S., Shinkai Y., Tachibana M., Chiba T., Nakao M.. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 2003; 278:24132–24138. [DOI] [PubMed] [Google Scholar]
- 77. Fuks F., Hurd P.J., Deplus R., Kouzarides T.. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003; 31:2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fuks F., Hurd P.J., Wolf D., Nan X., Bird A.P., Kouzarides T.. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003; 278:4035–4040. [DOI] [PubMed] [Google Scholar]
- 79. Allen M.D., Grummitt C.G., Hilcenko C., Min S.Y., Tonkin L.M., Johnson C.M., Freund S.M., Bycroft M., Warren A.J.. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006; 25:4503–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hashimoto H., Vertino P.M., Cheng X.. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010; 2:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Xu C., Bian C., Lam R., Dong A., Min J.. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat. Commun. 2011; 2:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu C., Liu K., Lei M., Yang A., Li Y., Hughes T.R., Min J.. DNA sequence recognition of human CXXC domains and their structural determinants. Structure. 2018; 26:85–95. [DOI] [PubMed] [Google Scholar]
- 83. Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P.. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998; 19:187–191. [DOI] [PubMed] [Google Scholar]
- 84. Ng H.H., Zhang Y., Hendrich B., Johnson C.A., Turner B.M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A.. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999; 23:58–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data are deposited at the NCBI GEO database under the following accession numbers:
SuperSeries GSE144904: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing.
This Series is composed of the following SubSeries:
GSE144903: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing [RNA-seq]
GSE144928: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing [ChIP-Seq]
GSE144331: Genome-wide investigation of the dynamic changes of epigenome modifications after DNA methylation editing [MBD-seq]