Abstract
The human genome contains hundreds of large, structurally diverse blocks that are insufficiently represented in the reference genome and are thus not amenable to genomic analyses. Structural diversity in the human population suggests that these blocks are unstable in the germline; however, whether or not these blocks are also unstable in the cancer genome remains elusive. Here we report that the 500 kb block called KRTAP_region_1 (KRTAP-1) on 17q12–21 recurrently demarcates the amplicon of the ERBB2 (HER2) oncogene in breast tumors. KRTAP-1 carries numerous tandemly-duplicated segments that exhibit diversity within the human population. We evaluated the fragility of the block by cytogenetically measuring the distances between the flanking regions and found that spontaneous distance outliers (i.e DNA breaks) appear more frequently at KRTAP-1 than at the representative common fragile site (CFS) FRA16D. Unlike CFSs, KRTAP-1 is not sensitive to aphidicolin. The exonuclease activity of DNA repair protein Mre11 protects KRTAP-1 from breaks, whereas CtIP does not. Breaks at KRTAP-1 lead to the palindromic duplication of the ERBB2 locus and trigger Breakage-Fusion-Bridge cycles. Our results indicate that an insufficiently investigated area of the human genome is fragile and could play a crucial role in cancer genome evolution.
INTRODUCTION
Genomic amplification pinpoints tumor-driving genes (oncogenes) in cancer genomes and provides information on diagnosis and therapeutic targets. Cancer genomics studies have mapped recurrently-amplified segments throughout the genome (1,2). Considering that genes with similar functions are elsewhere in the genome, why certain genes are recurrently amplified remains elusive. A potential mechanism underlying recurrence is the susceptibility of the locus to spontaneous DNA breaks, as breaks promote the amplification of nearby genes (3,4). This hypothesis would be validated if nearby genomic regions that break spontaneously and frequently (fragile sites) were identified. With the mild inhibition of DNA replication by aphidicolin and hydroxyurea, fragile sites have been mapped throughout the genome in human and mouse cells (5,6). It remains unclear whether these approaches can uncover spontaneous fragile sites comprehensively throughout the genome. Limitations of such methods include that (1) these sites may not represent spontaneous fragility, given the use of replication inhibitors, and (2) genomic approaches typically cover the portions of the reference genomes where short sequencing reads can be mapped uniquely and confidently but do not cover regions of duplicated sequences (segmental duplications, also called low copy repeats) (7,8), where mappability of short reads are compromised. The link between genomic amplification and known fragile sites is rare in tumors (9).
Segmental duplications in the human genome arise during primate evolution and retain high sequence identities (>90%) between duplicated pairs (10,11). Segmental duplications account for ∼5% of the human genome and are more than 1 kb in size. Duplicated sequences tend to cluster in ∼400 distinct regions of the genome (duplication blocks) where duplications are juxtaposed to each other. Such duplication architectures could result in the formation of secondary structures via base pairing within strands, which are obstacles for DNA replication. Indeed, stalled forks have been attributed as a cause of complex genome rearrangements within duplication blocks in patients with neurological disorders (8,12). Furthermore, duplication blocks are significantly associated with copy number variations in humans (7,13). Therefore, duplication blocks are structurally diverse and are likely unstable in the germline. With such diversity in the germline, representing duplication blocks in a single reference genome is not possible. Even for some regions that appear to be complete, multiple haplotypes (alternate haplotypes) exist (14). In addition, there are 819 assembly gaps in the most recent version (hg38) that account for >5% of the genome, and duplicated sequences flank the majority of these assembly gaps (15). It remains unknown whether or not these blocks are unstable in somatic cells and play any role in disease etiology such as cancer development.
With complex duplication blocks in the genome, DNA replication needs to be supported by repair mechanisms that act quickly when adverse lesions arise. The DNA repair protein Mre11 could play such a role (16,17). Mre11 and the binding partner RAD50 form a highly conserved protein complex in the three domains of life. This Mre11 complex is associated with replication forks, and the association is enhanced when replication forks stall (18,19). The depletion of Mre11 results in spontaneous DNA breaks during replication in metazoans (20,21). Spontaneous replication-associated breaks halt cell cycle progression and render Mre11-null vertebrate cells proliferation-deficient (22,23). Simple organisms such as Escherichia coli and Saccharomyces cerevisiae can tolerate (SbcCD and) Mre11 deficiency (24,25), although the retarded growth is evident. Thus, the essentiality of Mre11 seems to be proportional to genome complexity. Mre11 possesses robust 3′ to 5′ exonuclease activity and single-strand endonuclease activity (26,27). Recent studies revealed a concerted action of these two nuclease activities for the removal of protein-bound DNA ends that arise during meiotic recombination and DNA double strand break (DSB) repair (28,29). This process is initiated by the endo-nucleolytic incision of DNA that is internal to a protein-bound end followed by the exo-nucleolytic degradation of DNA towards the end (endo-then-exo model). Two other proteins regulate Mre11 nuclease activity. Nbs1, the third component complexed with Mre11–Rad50, promotes the endo-nuclease activity and restrains the exo-nuclease activity (29). Another protein CtIP is also stimulatory to the Mre11 endonuclease activity (30,31). The endo-nuclease activity alone has an independent role in processing DNA secondary structures such as hairpin loops arising from the intra-strand annealing between the arms of inverted repeats (32,33).
We sought to understand how fragile a duplication block is, how it is protected, and whether or not the fragility of a block is linked to genomic amplification. To address these questions, we developed a fluorescence in situ hybridization (FISH)-based approach and investigated the fragility of a duplication block KRTAP_Region_1 (KRTAP-1). KRTAP-1, consisting of the gene cluster of keratin-associated protein genes, is located 1.12 Mb telomeric to the oncogene ERBB2 (encoding the Human Epidermal Growth Factor Receptor 2, HER2). Among the four EGFR receptor family members, ERBB2 is the most frequently amplified gene. ERBB2 is amplified in 10–30% of breast tumors (HER2-positive breast tumors) as well as in stomach, bladder, and esophageal cancers (34,35), although the underlying mechanism of the recurrence remains elusive. We report here that the KRTAP-1 duplication block is indeed fragile. The fragility is not exacerbated by aphidicolin suggesting that the mechanism of fragility is distinct from the aphidicolin-sensitive common fragile sites (CFS). Mre11, in particular its exo-nuclease activity, is crucial in protecting KRTAP-1. Breaks at KRTAP-1 initiate the formation of inverted duplications and copy number gains of the ERBB2 oncogene. Our study reveals the fragility of a complex duplication block in the human genome and its role in recurrent genomic amplification.
MATERIALS AND METHODS
Cells
Primary human mammary epithelial cells were established from five normal breast tissues from prophylactic mastectomy by following a previously-reported method (36): NBC 16–131, NBC 16–172, NBC 16–634, NBC 14–668 and NBC 16–805.
Generation of the TK6 cell lines with conditionally disrupted Mre11 and CtIP genes were reported in detail previously (37). To generate the MRE11−/− and MRE11−/H129N cells from MRE11loxP/loxP and MRE11loxP/H129N, respectively, 4-hydroxytamoxifen (Sigma-Aldrich, H7904) was added to the culture medium at a final concentration of 200 nM. For the conditional disruption of the CtIP gene using the auxin-inducible degron (AID) system, 3-Indoleacetic acid (Sigma-Aldrich, I2886) was added to the culture medium at a final concentration of 200 μM. RKO cells with IPTG-inducible Mre11 knockdown system were established using shRNA-expressing vector from Sigma Mission TRC1 lentiviral shRNA Library (Sigma-Aldrich).
Flow cytometry
The BD FACSAria III sorter (BD Biosciences) equipped with five excitation lasers (375, 405, 488, 561 and 633 nm) was used for fluorescence-activated cell sorting (FACS). For the fluctuation tests, single TK6 cells were sorted on 96 well plates to establish single-cell derived clones. For the selection of Cas9-expressing cells for the subsequent detection of CRISPR/Cas9-induced DSBs, the Enhanced Green Fluorescent Protein (EGFP) fused with Cas9 (in pX458 from Addgene #48138) was excited with 488 nm irradiation for sorting. For the ImmunoFISH experiments, U-2 OS cells were stained with Hoechst 33342 (Thermo Fisher Scientific, 62249) and were excited with 405 nm irradiation to quantify DNA content. For cell cycle analysis of cells treated with Mre11 inhibitors, the Muse Cell Analyzer (Millipore) and Muse Cell Cycle Kit (Millipore Sigma, MCH100106) were used according to the manufacturer's instructions. Cell cycle profiles were analyzed with the FlowJo software.
Dual-color FISH
The fixation, denaturation and hybridization of samples were conducted as described previously (38). Briefly, for interphase nuclei, a semi-confluent cell population was harvested and resuspended in phosphate-buffered saline (PBS). The cells were fixed, suspended by Carnoy's fixative, and dropped onto glass slides. For the preparation of metaphase spreads, mitotic IMR-90 cells were collected from 175 cm2 flasks by shake-off after 12 h of treatment with 100 ng/ml colcemid (Sigma-Aldrich). The Chromosome Resolution Additive (Genial Genetic Solutions, Chester, UK) was added 1.5 h before cell harvest. The cells were subjected to the hypotonic treatment with Pre-hypotonic and Optimal hypotonic (Genial Genetic Solutions). After hypotonization, the cells were fixed and suspended with Carnoy's fixative. The cells were then dropped onto glass slides and spread.
FISH probes were prepared from the BAC clones (Supplementary Table S1). The DNA was labeled with Biotin- or DIG-Nick Translation Mix (Roche) and hybridized onto the interphase or metaphase samples at 37°C for 2 days. The biotin- and digoxigenin-labeled DNA probe were visualized by Alexa488-conjugated streptavidin (Thermo Fisher Scientific, S32354) and by anti-digoxigenin-DyLight 594 (Vector Laboratories, DI-7594), respectively. Cells were mounted in ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific). Images were captured using a fluorescence microscope BZ-X 700 (Keyence).
Scoring chromosome breaks (break-apart FISH)
Only diploid cells (which have two red and two green signals) were considered for scoring breaks in order to minimize the influence of spurious or missed spots. The minimum distances between two probes (red and green) in captured images were measured using the BZ-X Analyzer (Keyence). To identify breaks, we employed the statistical approach of quartile analysis. The widely accepted threshold for the detection of statistical outliers, which is the sum of the third quartile and 1.5 times the interquartile range, was determined. Based on the thresholds, statistical outliers were scored as proxies for spontaneous breaks. For the experiments with interventions, the thresholds of control groups (without interventions) were applied to experimental groups to score outliers. This refinement is expected to mitigate underestimation or overestimation that is attributable to the artificial shift of thresholds due to the shift of the overall distribution.
We employed CRISPR/Cas9-induced breaks to verify the feasibility of our method for scoring breaks. Single guide RNAs (sgRNAs) were designed to target the exons of the WWOX gene within the common fragile site FRA16D on chromosome 16: sgRNA1 targets 5′-GTTTTTTAACAGTCACACCG-3′ in exon 2 and sgRNA2 targets 5′-ACCACCCGGCAAAGATACGA-3′ in exon 4. The sgRNA1 or the sgRNA2 were cloned into pSpCas9(BB)-2A-GFP (pX458) from Addgene (#48138). HeLa S3 and TK6 cells were transfected with either of the plasmids using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer's instructions. Cas9-expressing cells were sorted with FACS 48 hours after transfection by using the fused EGFP as the marker (Figure 2B, left) and were subject to FISH analysis.
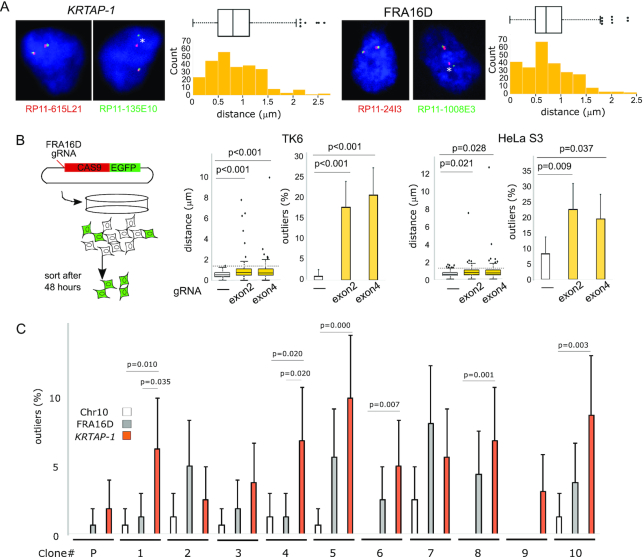
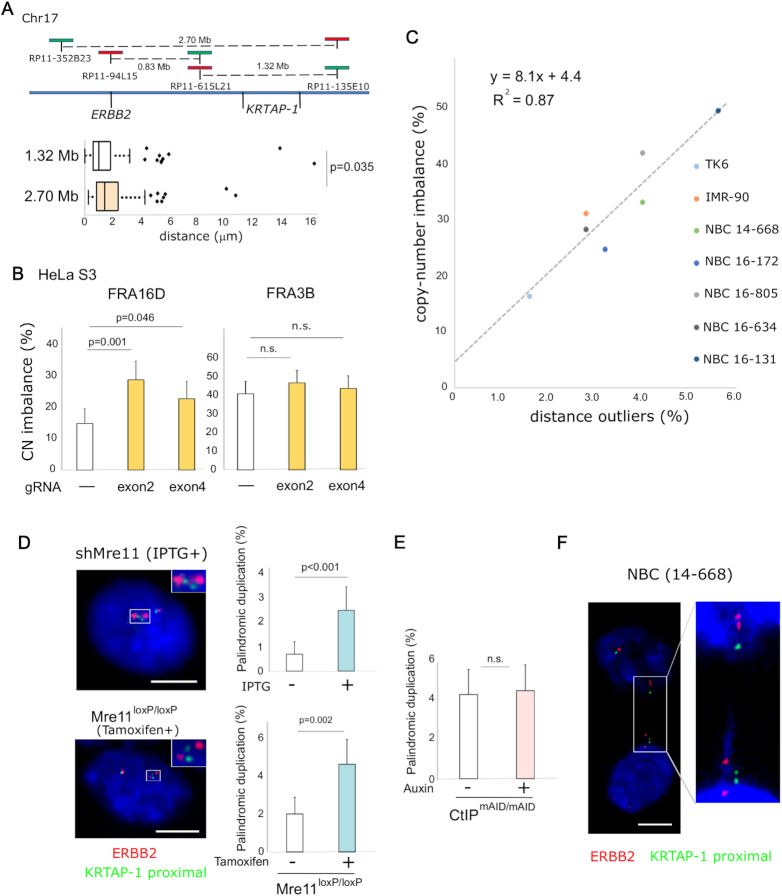
Figure 2.
Spontaneous breaks at KRTAP-1. (A) Measurements of the distance between both ends of KRTAP-1 and FRA16D. Representative photomicrographs of FISH of the nuclei of IMR-90 are shown. The asterisks (*) show that the distance between the probe pair is a statistical outlier. The distributions of the distances (n = 250) are depicted by box plots and histograms. Outliers are plotted at their exact lengths. (B) The distribution of distances between probes before and after CRISPR/Cas9-targeted breaks at FRA16D in TK6 and HeLa S3 cells. (left) A schematic drawing of the experimental procedure. (right) The box plots depict distributions of the distances (n = 100), with outliers plotted at their exact distances. P-values were determined by one-tailed Mann-Whitney U test. The horizontal dotted lines indicate thresholds for outliers determined by the cells without the CRISPR/Cas9-targeted breaks. The histograms depict the frequencies of outliers. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. (C) Frequencies of breaks in 10 single cell-derived TK6 clones. The histogram depicts the frequency of outliers. (P): parental TK6 cell population. Error bars represent 95% confidence intervals. P-values determined by Fisher's exact test are shown for the clones with a statistically significant difference between KRTAP-1 and FRA16D or Chr10 (the region centromeric to a rare fragile site FRA10A).
Fluctuation test
Single-cell derived clones were isolated from the unstained TK6 cell population. FACS sorted single cells were plated into each well of 96-well plates. Clones were re-plated into 24-well plates and expanded until the population size reached 1 million cells (±20%). The distances between two flanking probes were measured for KRTAP-1, FRA16D, and the region centromeric to a rare fragile site FRA10A at 10q23.23 to study the frequency of breakage. The breakage rate per cell per division was calculated with bz-rates (http://www.lcqb.upmc.fr/bzrates), a web-based program for estimation of mutation rates from fluctuation analysis (39).
Western blotting
To monitor the depletion of Mre11 and CtIP proteins in the inducible systems, cells were fixed with 10% trichloroacetic acid for 30 min on ice, and the total cell lysates were prepared with LDS Sample Buffer (Thermo Fisher Scientific). Western blotting was performed with NuPAGE protein electrophoresis system (Thermo Fisher Scientific). Rabbit anti-Mre11 (1:200, Novus Biologicals, NB 100-1420), rabbit anti-CtIP (1:1000, Bethyl, A300-488A), and rabbit anti-Rad51 (1:500, Santa Cruz, sc-8349) were used as primary antibodies. Bands were visualized with Immun-Star AP Chemiluminescence Kits (Bio-Rad), and were detected with ChemiDoc Touch Imaging System (Bio-Rad).
Drug treatments
For low dose aphidicolin (Sigma-Aldrich) treatment, we employed the common doses and duration: 0.2 μM for IMR-90 and 0.1 μM for TK6 for 24 h (40). For Mre11 inhibitors, Mirin, PFM01, and PFM39 were obtained from Sigma-Aldrich, and PFM03 was a gift from Dr John Tainer (The University of Texas MD Anderson Cancer Center). We carefully monitored cells at various doses of drugs for 24 h and chose the highest dose that did not disturb cell viability and cell cycle profiles. The concentrations for the Mre11 inhibitors were as follows; 25 μM Mirin for IMR-90, 5 μM Mirin for TK6, 25 μM PFM01 for IMR-90, 5 μM PFM01 for TK6, 25 μM PFM39 for TK6, and 5 μM PFM03 for TK6 cells.
ImmunoFISH
U-2 OS cells in S-phase or G1-phase were sorted by FACS with DNA content as an indicator. Cells were plated on coverslips coated with poly-L-lysine and were subjected to immunostaining with mouse anti-Mre11 antibody (1:500, GeneTex, GTX70212) or mouse anti-phospho-Histone H2A.X (Ser139) antibody (1:500, Sigma-Aldrich, 05–636). Following refixation and ethanol dehydration, samples were denatured along with DIG-labeled probes flanking KRTAP-1 (Supplementary Table S1; RP11–615L21 and RP11–135E10). Slides were then incubated in a humidified chamber at 70°C for 30 min followed by overnight incubation at 37°C. The DIG-labeled FISH probes were detected using anti-digoxigenin-DyLight 594 (Vector Laboratories, DI-7594). Images were captured using the fluorescence microscope BZ-X 700 (Keyence). The minimum red-green distances were measured (n = 200) using the BZ-X Analyzer (Keyence). The experiments were repeated three times.
Whole-genome sequencing (WGS) analyses
Breast tumor WGS data were obtained from the European Genome Phenome Archive (accession number EGAD00001001334) (41). The reads were trimmed to remove adapters using Trim Galore (v.0.6.1) and Cut Adapt (v2.3) and aligned to hg38 using Bowtie2 (v2.3.5). Reads were converted to BAM format, reordered, and sorted using Samtools (v1.9). Duplicate reads and PCR artifacts were removed using the Genome Analysis Toolkit (GATK) (v4.1.2.0). The hg38 reference genome was divided into 1 kb non-overlapping bins using Bedtools (v2.28.0) and reads were counted in each bin. Browser extensible data format (BED) files were generated using Bedtools (v2.28.0) and visualized using the Interactive Genome Viewer (IGV) (v2.5.0). For isolating reads mapping uniquely to the genome, only reads with mapping quality score >40 (MAPQ>40) were considered.
For presenting the read depth of the ERBB2 locus (Figure 1C), the number of sequencing reads was counted for each 1 kb bin throughout the genome. The number of reads for each 1 kb bin was then normalized across the tumor and normal samples by a per million scaling factor for each WGS dataset (i.e. if the WGS dataset has 100 million reads, the per million scaling factor is 100).
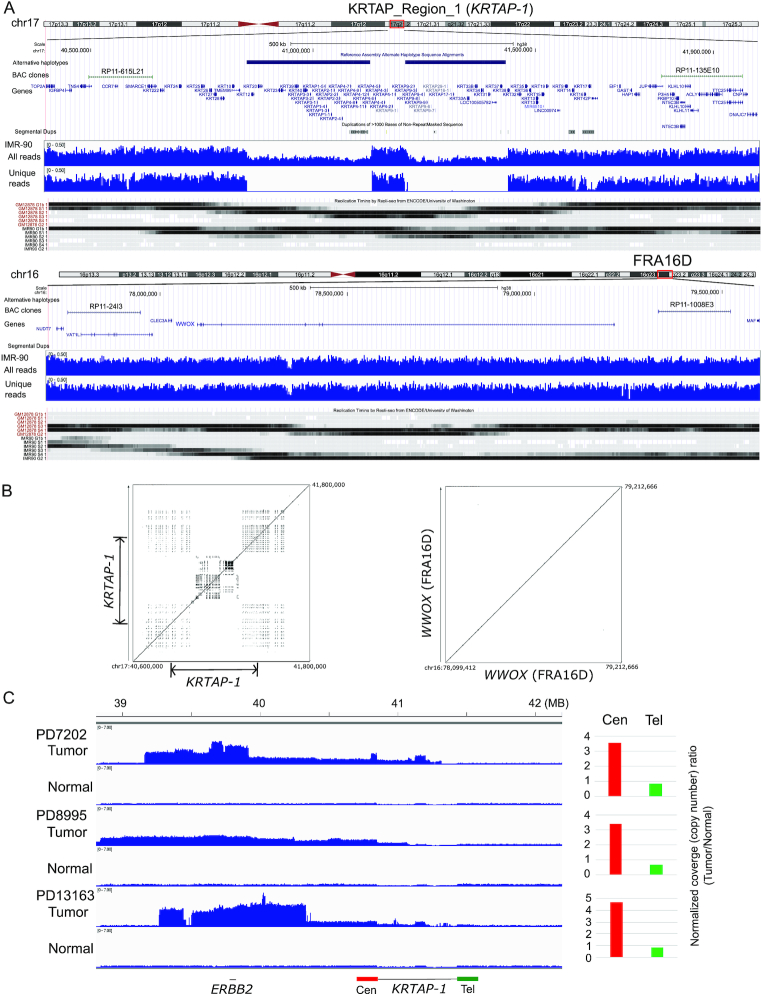
Figure 1.
KRTAP_Region_1 is a 500 kb block on chromosome 17q21.2. (A) KRTAP-1, FRA16D, and their flanking regions in hg38. (top) The locations of alternative haplotypes, BAC clones for Break-apart FISH probes, and genes within the regions are shown. (middle) Read depth of IMR-90 whole genome sequencing for total reads and for reads that are uniquely mapped (unique reads, MAPQ > 40) to the regions are shown. (bottom) The replication timings of GM12878 and IMR-90 cells from Repli-seq data (ENCODE), with the regions replicating in late G1 (top), S (four phases) and early G2 (bottom) phases are shown by shades. (B) Advanced PipMaker dot plot of KRTAP-1 and WWOX gene (FRA16D). The coordinates in the hg38 are shown. Dots represent sequences (≥100 bp without a gap) with ≥70% nucleotide identity. (C) Read coverage of the 17q12–21 region for the whole genome sequencing (WGS) data of breast tumors (European Genome-Phenome Archive data, EGAD00001001334). Normalized coverage (# of reads/kb/million reads) from three tumor/normal pairs are shown. Note that in all three tumors, the side centromeric to KRTAP-1 has greater read depth than the region telomeric to KRTAP-1. Histograms show the normalized coverage ratio between tumor DNA and normal DNA for the region centromeric to KRTAP-1 (200 kb region, Cen, in red) and telomeric to KRTAP-1 (Tel, in green).
RESULTS
Copy number breakpoints within the complex duplication block KRTAP-1 in cancer genomes
A whole genome sequencing (WGS) study of HER2-positive breast tumors reports that Breakage-Fusion-Bridge (BFB) cycles could underlie the amplification of ERBB2 (41). BFB cycles can be initiated by a spontaneous break (Supplementary Figure S1A). Following DNA replication, two sister chromatids can then fuse at the broken ends to become an invertedly-duplicated (palindromic) giant chromosome with two centromeres. The two centromeres move to opposite poles during cell division and subsequently cause another break somewhere between the two centromeres resulting in the uneven distribution of chromosomal DNA into the daughter cells. Repeating this cycle establishes chromosomal regions with genomic amplification. If the region harbors an oncogene such as ERBB2, the clone with genomic amplification becomes dominant in a tumor cell population. Thus, according to this model, the initial break is crucial because the following copy number gains are streamlined by the cycles.
The location of an initial break can be identified as a transition point between a region of normal copy number and a region of increased copy number (Supplementary Figure S1B). Using European Genome-Phenome Archive WGS data of HER2-positive breast tumors (41), we found that copy number transition points occur in KRTAP-1 in multiple tumors (Figure 1C). Consistent with the model (Supplementary Figure S1B), tumors gain extra copies on the centromeric side (Cen) of KRTAP-1 and lose copy number on the telomeric side (Tel). Within KRTAP-1, there are two large (250 kb) segments with alternative haplotypes. Alternative haplotypes have been recently introduced into the reference human genome (hg38) for genomic regions that are too diverse and complex to be represented by a single tiling path (14). The diversity is associated with abundant tandemly-duplicated sequences (duplication block) (Figure 1B) which could suggest an unstable nature (42,43). When closely juxtaposed to each other, duplicated sequences can form secondary structures by intrastrand base pairing and become obstacles to DNA replication (44), leading to spontaneous breaks and promoting recurrent genomic amplification.
Spontaneous breaks within KRTAP-1
To test the instability, we set to define the relative fragility of KRTAP-1 to representative fragile sites in human chromosomes, the Common Fragile Sites (CFSs) FRA16D and FRA3B (Figure 1A and Supplementary Figure S1C). In contrast to KRTAP-1, both CFSs carry uniquely mappable sequences and lack duplications (Figure 1A, B and Supplementary Figure S1C). To assess the fragility, we took a cytogenetic approach. A pair of probes was prepared for each locus from BAC clones (Supplementary Table S1), with the physical distances between the two probes of 1.32 Mb for KRTAP-1, 1.58 Mb for FRA16D and 1.54 Mb for FRA3B (Figure 1A, Supplementary Figure S1C, and S1D). Probe pairs were differentially labeled and hybridized to fixed IMR-90 normal primary fibroblasts, and the distances between the two probes in interphase nuclei were measured (Figure 2A and Supplementary Figure S2A). In the majority of the nuclei, two signals (green and red) were very close to each other, indicating that these loci were intact. However, there were outliers (> Q3 + 1.5 × IQR, in which Q3 is the 75 percentile and IQR is the interquartile range Q3–Q1, 25 percentile) with increased distances that indicate (the history of) breaks between the probes. To further establish the causal relationship between breaks and statistical outliers, we employed CRISPR/Cas9-induced breaks and quantified the outliers (Figure 2B and Supplementary Figure S2B). By targeting FRA16D, we observed an increase in outliers in FRA16D, but not in the non-targeted FRA3B. The median distances between probes at FRA16D also increased by targeting FRA16D: 0.56–0.75 μm (exon 2) and 0.56–0.73 μm (exon 4) in TK6, and 0.56–0.75 μm (exon 2) and 0.56–0.73 μm (exon 4) in HeLa S3. The increases of the median distances were associated with the increases of the third quartile distances (Q3) in cells with CRISPR/CAS9-induced breaks. The increases of Q3 indicate that induced breaks increased the distances between the probes in a significant fraction of cells. However, the increase would also lead to higher thresholds for calling outliers than the control, uninduced cells. Therefore, scoring breaks independently in each dataset based on the outliers could underestimate the breaks in cells with CRISPR/CAS9-induced breaks because the higher threshold would disregard some of the distances that would be considered outliers in uninduced cells but are not in cells with induced breaks. To avoid this confounding effect, we applied outlier thresholds from the un-induced control group to the experimental groups. Outliers at FRA16D increased from 1.0% to 18.0% (exon 2) and 21.0% (exon 4) in TK6 cells, and 8.0% to 22.0% (exon 2) and 19.0% (exon 4) in HeLa S3 cells.
We first sought to address how frequently KRTAP-1 breaks spontaneously using an approach that is similar to the fluctuation test (45). We isolated single-cell clones from TK6 cells and expanded the clones for 20 generations until the population reached 1 million cells (Figure 2C and Supplementary Figure S2D). With less than 2% of outliers in the parental cell population, a break at KRTAP-1 in priori in each single-cell clone is unlikely, and the distance outliers detected after 20 generations most likely arise during the propagation of each clone. In eight of the ten clones, the distance outliers of KRTAP-1 were more frequent than outliers of FRA16D, with two of them showing statistical significance (Figure 2C, right, P < 0.05). With the ten independent clones, the spontaneous break rate per cell division, estimated by bz-rates (39), is 0.436% for KRTAP-1 and 0.030% for FRA16D. These rates would likely be underestimated, because not all the breaks result in generating cytologically separate loci. We further examined breaks at the region centromeric to a rare fragile site FRA10A at 10q23.23 (Supplementary Figure S2C), which expresses fragility by folate deprivation for only one in 500 individuals (46). Given the rarity, the centromeric flanking site would represent a potential ‘stable’ site of the genome. Indeed, outliers were extremely rare, with the estimated break rate of 0.00012%.
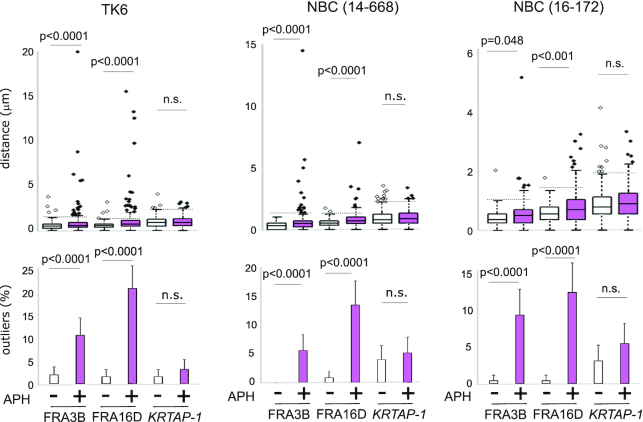
A common characteristic of CFS is late replication (6,47). FRA16D and FRA3B are replicated in late-S and G2 phases in both IMR-90 and GM12878 cells (Figure 1A and Supplementary Figure S1C). In contrast, KRTAP-1 replicates in mid-S phase in both cells, which led us to hypothesize that breaks at KRTAP-1 are mediated by a different mechanism from CFSs. We tested the hypothesis by treating seven different cell types with aphidicolin for 24 h; TK6, IMR-90, and five primary human mammary epithelial cells (NBCs) (Figure 3 and Supplementary Figure S3). For KRTAP-1, average distances between the probes did not significantly increase except in one of the NBCs (16–131). For CFSs, the distances increased significantly in four (FRA16D) and three (FRA3B) cell types (Figure 3 and Supplementary Figure S3). Consistently, outliers were not significantly increased at KRTAP-1 in any of the cell types, whereas outliers were found to be induced significantly by aphidicolin in four (FRA16D) and three (FRA3B) cell types for CFS. Therefore, KRTAP-1 is unlikely to be sensitive to aphidicolin. The insensitivity to aphidicolin sensitivity could likely be due to the early replicating nature of KRTAP-1 (Figure 1A), as, unlike the late replicating CFSs, KRTAP-1 has sufficient time to complete replication before the end of S-phase and entering into mitosis.
Figure 3.
Aphidicolin-induced breaks at KRTAP-1. The box plots (top) showing the distribution of distances between the flanking probes for KRTAP-1, FRA16D, and FRA3B in TK6 and a primary human mammary epithelial cell (NBC 14–668 and 16–172) with or without aphidicolin treatment (24 h). For each locus, the distances between the pair of probes were measured for 250 pairs. P-values were determined by Mann–Whitney U test. The horizontal dotted lines indicate thresholds for the outliers for cells without aphidicolin treatments. The histograms (bottom) depict the frequencies of outliers. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. n.s.: not significant.
Mre11 protects KRTAP-1 from breaks through its exonuclease activity
In addition to KRTAP-1, there are 177 regions with alternative haplotypes in hg38 (14). Considering the prevalence of large duplication blocks in the human genome (48), there should be a mechanism that protects these duplication blocks from breaks. The Mre11/Rad50/Nbs1 (MRN) complex is a good candidate because of (I) the essentiality of Mre11 in the viability of organisms is proportional to genome complexity (22–25) and (II) the depletion of Mre11 in metazoans results in spontaneous breaks during replication (20,21). We employed inducible systems to monitor distance outliers before and after Mre11 modulation. In the RKO colorectal cancer cell line with an IPTG-inducible expression of Mre11 shRNA (Supplementary Figure S4A), outliers significantly increased for KRTAP-1 in two independent clones (3.6–11.2% in clone 1 and 4.6–11.6% in clone 2). In the TK6 cells expressing the Cre recombinase fused to estrogen receptor binding domain (ER-Cre), loxP-flanked Mre11 can be deleted by the addition of tamoxifen (Figure 4A) (37). Consistent with the shRNA-inducible knockdown of Mre11 in RKO cells, outliers increased significantly from 0.4% to 3.2%.
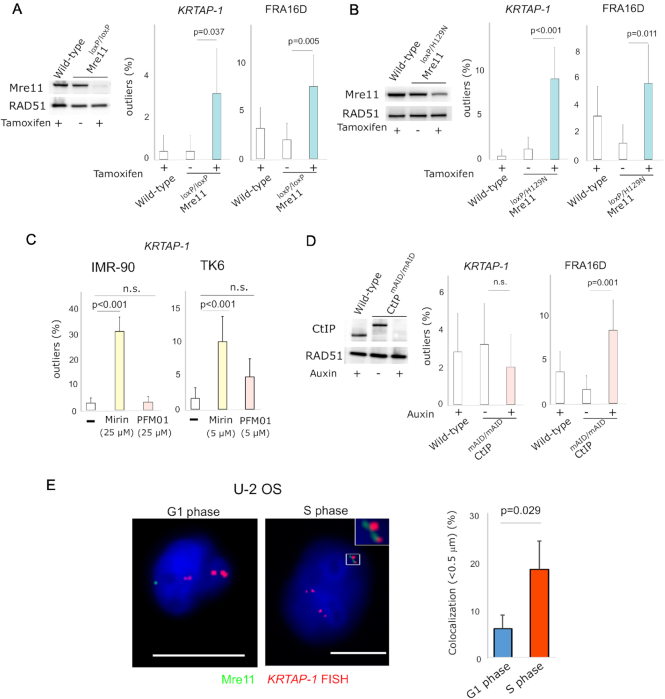
Figure 4.
Depletion of Mre11 induces breakage in KRTAP-1. (A) TK6 cells with tamoxifen-inducible knockout of Mre11. Western blot shows Mre11 protein level before and after 5 days of tamoxifen treatment. RAD51 protein level is shown as a control. The histograms depict the frequencies of outliers. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. (B) TK6 cells with tamoxifen-inducible knockout of wild-type Mre11 for the exclusive expression of the nuclease-deficient Mre11 (H129N). Western blot shows Mre11 protein level before and after 5 days of tamoxifen treatment. RAD51 protein level is shown as a control. The histograms depict the frequencies of outliers. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. (C) Distinct effects of an exonuclease (Mirin) and an endonuclease (PFM01) inhibitor of Mre11 on breaks in KRTAP-1 in IMR-90 and TK6 cells. The histograms depict frequencies of outliers at KRTAP-1 and FRA16D. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. n.s.: not significant. (D) Auxin-inducible depletion of CtIP in TK6 cells expressing CtIP fused with mAID. Western blot shows the CtIP protein level before and after 3 days of auxin treatment. RAD51 protein level is shown as a control. The histograms depict the frequencies of outliers. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. n.s.: not significant. (E) Representative photomicrographs of the nuclei of U-2 OS cells in G1 phase and S phase after staining with anti-Mre11 antibodies (green) and hybridization of FISH probes for KRTAP-1 (red). Scale bars = 10 μm. The histogram depicts the fraction of nuclei with the co-localization of Mre11 foci and KRTAP-1. The averages of three experiments are shown. P-value was calculated with two-tailed t-test.
Mre11 possesses very robust 3′ to 5′ exonuclease activity, as well as single-strand endonuclease activity (17,26,27). We tested the role of Mre11 nuclease activity using a TK6 cell line that exclusively expresses a nuclease-deficient form of Mre11 (H129N) after ER-Cre-dependent removal of the wild-type Mre11 allele (Figure 4B) (37). The number of outliers significantly increased for KRTAP-1 upon the deletion of the wild-type allele (1.2% to 9.2%), indicating that the nuclease activity of Mre11 is crucial in protecting KRTAP-1. We next tested Mre11 exo- (Mirin and PFM39) and endo-nuclease specific inhibitors (PFM01 and PFM03) (49) to examine the roles of each type of nuclease activity in protecting KRTAP-1. A serial dose of the inhibitors was tested for TK6 and IMR-90, and the optimal doses were selected, which were the highest doses without signs of cell cycle arrest and abnormality in morphology after 24 h (Supplementary Figure S5A and S5B). With Mirin or PFM39, outliers significantly increased for KRTAP-1 in both TK6 and IMR-90 cells (Figure 4C and Supplementary Figure S5C and S5D). In contrast, neither PFM01 nor PFM03 promoted outliers significantly, whereas both did so for FRA16D (Supplementary Figure S5E). To further evaluate the endonuclease activity, we used the TK6 cell line with the inducible degradation of CtIP with mini-auxin-inducible degron (Figure 4D) (37). Because CtIP stimulates the endonuclease activity of Mre11 (31), degrading CtIP could mimic endonuclease-deficient Mre11. Consistent with the experiments with inhibitors, outliers increased at FRA16D (1.6–8.4%) (33), but not in KRTAP-1 (3.2–2.0%). Collectively, these results indicate that Mre11 protects KRTAP-1 from breaks, primarily through its exonuclease activity.
Mre11 foci colocalize with PCNA, a homotrimeric sliding clamp of replication forks (18,19). Mre11 could protect KRTAP-1 when replication machinery goes through challenging DNA secondary structures. In this scenario, Mre11 foci should be spatially associated with KRTAP-1 at points during S phase. To investigate this association, we visualized Mre11 proteins and the KRTAP-1 locus simultaneously (immunoFISH) in the U-2 OS cell line, in which Mre11 foci have been extensively investigated (50). We isolated S-phase and G1-phase populations and, in each population, measured the minimal distance between Mre11 foci (green) and KRTAP-1 loci (red). We assume co-localization when Mre11 and KRTAP-1 are very close to each other, which we defined as <0.5 μm. The fraction of nuclei exhibiting co-localization was higher in S-phase cells than G1 cells (18.8% versus 6.2%, P <0.05) (Figure 4E and Supplementary Figure S4F). Phosphorylated histone H2AX at serine 139 (γ-H2AX) foci also co-localized with KRTAP-1 more frequently in S-phase cells than in G1 cells (18.2% versus 5.3%, P < 0.01) (Supplementary Figure S4G), suggesting that DNA damage at KRTAP-1 is more common in cells undergoing replication than in cells before replication.
Breaks within KRTAP-1 initiate palindromic duplication and BFB cycles
Frequent copy number transitions within KRTAP-1 in breast tumors (Figure 1C) imply that KRTAP-1 is a primary fragile site within the ERBB2 locus. To further corroborate this idea, we designed an additional probe on the centromeric side of KRTAP-1 that was twice as far apart (2.70 Mb) from the telomeric probe (Figure 5A). Probing a genomic region that was twice the size did increase the median distance between the two probes (0.96 versus 1.36 μm, P = 0.035). However, extreme outliers (>5 μm) did not increase (seven outliers), suggesting that within the ERBB2 locus, a majority of outliers came from KRTAP-1.
Figure 5.
Breaks at KRTAP-1 trigger palindromic duplication and BFB cycles for the amplification of ERBB2. (A) KRTAP-1 is the primary fragile site in the region surrounding ERBB2. A schematic drawing of the locations of probes (top). The box plot (bottom) depicts the distributions of distances between probes in interphase nuclei of NBC (16–131; n = 170). Outliers are plotted at their exact distances. A P-value determined by one-tailed Mann-Whitney U test is shown. (B) Frequencies of copy-number imbalances between two flanking probes for FRA16D and FRA3B in HeLa S3 cells 7 days after the introduction of breaks at FRA16D with CRISPR/Cas9. The fraction of nuclei exhibiting copy number imbalances (n = 200) is shown in the histogram. Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. n.s.: not significant. (C) A scatter plot showing a positive correlation (R2 = 0.87) between frequencies of outliers and frequencies of copy-number imbalances at KRTAP-1. Data were collected from untreated TK6, IMR-90, and five NBCs (14–668, 16–172, 16–805, 16–634 and 16–131; n = 250). (D) Frequency of palindromic duplication before and after depletion of Mre11 in the IPTG-inducible Mre11 knockdown in RKO cells (top) and the Cre/loxP-inducible Mre11 knockout in TK6 cells (bottom). Histograms show the frequency of cells with two ERBB2 (red; RP11–94L15 in Figure 5A top) signals flanking one or two green (KRTAP-1 proximal; RP11–615L21 in A, top) signals (as shown by an asterisk in Supplementary Figure S1A). Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. (E) Frequency of palindromic duplication before and after depletion of CtIP in the mAID-inducible depletion of CtIP in TK6 cells. Histograms show the frequency of nuclei with palindromic duplication: two ERBB2 (red; RP11–94L15 in Figure 5A top) signals flanking one or two green (KRTAP-1 proximal; RP11–615L21 in Figure 5A top) signals (as shown by an asterisk in Supplementary Figure S1A). Error bars represent 95% confidence intervals. P-values were determined by Fisher's exact test. n.s.: not significant. (F) Photomicrographs showing dividing nuclei with a chromatin bridge carrying multiple ERBB2 and KRTAP-1 loci in NBC (14–668). FISH visualizes ERBB2 (red; RP11–94L15 in A, top) and the centromere-proximal region of KRTAP-1 (KRTAP-1 proximal; green; RP11–615L21 in A, top). Scale bars = 10 μm.
A possible outcome of chromosome breaks is a copy number imbalance between each side of the break. Copy number imbalance was demonstrated by the CRISPR/Cas9-induced breaks; the frequency of copy-number imbalance between the two sides of FRA16D increases from 16.0% (empty vector) to 31.0% (exon 2 sgRNA) and 24.5% (exon 4 sgRNA) in HeLa S3 cells (Figure 5B). We also observed a strong positive correlation between the frequency of spontaneous breaks (outliers) and copy-number imbalance at KRTAP-1 in the seven cell types (R2 = 0.87, Figure 5C). BFB cycles could contribute to copy number imbalance by the gain (via palindromic duplication) of centromere-proximal regions of the breaks and the loss of centromere-distal regions (Supplementary Figure S1A and S1B). To test whether breaks at KRTAP-1 initiate palindromic duplication, dual-color FISH was performed using a probe covering the ERBB2 gene (red; RP11–94L15 in Figure 5A) and a probe covering the centromere-proximal region of KRTAP-1 (KRTAP-1 proximal, green; RP11–615L21 in Figure 5A). Palindromic ERBB2 duplication resulting from the breaks at KRTAP-1 would constitute one or two KRTAP-1 proximal signals flanked by two ERBB2 (red) signals (* in Supplementary Figure S1A). The depletion of Mre11, which increases the frequency of breaks at KRTAP-1 (Figure 4A and Supplementary Figure S4A), significantly increased palindromic duplication (from 0.7% to 2.5%, in IPTG-inducible RKO, P <0.001; from 2.0% to 4.6% in TK6, P = 0.002) (Figure 5D). Without the increase of breaks (in CtIP-depleted TK6 cells, Figure 4D), palindromic duplication did not increase (Figure 5E). These results demonstrate that breaks at KRTAP-1 lead to copy number imbalance, at least in part through palindromic duplication. We also found a pair of ERBB2 signals associated with one or two KRTAP-1 proximal (green) on each edge of a chromatin bridge between dividing nuclei in a normal breast cell culture (Figure 5F). Since the normal breast culture is free from experimental interventions, this example indicates a dicentric chromosome spontaneously arising by a break at KRTAP-1. Finally, Mre11 depletion did not cause breaks between the centromere proximal region of KRTAP-1 and ERBB2 (Supplementary Figure S6), further supporting that KRTAP-1 is the primary fragile site in the ERBB2 locus.
DISCUSSION
In this study, we evaluated the fragility of a large duplication block and its role in promoting recurrent genomic amplification. We show that the fragility of KRTAP-1 can lead to copy number imbalance and palindromic duplication of the genomic region harboring ERBB2. These results provide a seminal example of the critical role of large duplication blocks in shaping cancer genomes. Whether other duplication blocks play similarly crucial roles needs to be tested in future studies. KRTAP-1 is one of the 26 regions in the entire genome that contain two or more alternate loci (14). Most of the 26 regions are located very close to the ends of chromosomes. However, chromosome 17 has three regions in the middle of the long arm, including KRTAP-1, that flank cancer-related genes such as ERBB2 and BRCA1. Another region with an alternative haplotype (Region 238) is a part of a massive duplication block at 17p11 that is known to mediate the formation of isochromosome 17q (an inverted duplication of the long arm of chr17), a common abnormality in myeloid malignancies (51). Therefore, duplication blocks could play more prominent roles in cancer genome rearrangements than we currently recognize. Rearrangements at these large complex areas are impossible to detect by reference genome- and short sequencing reads-based approaches (13). Indeed, long homology-based rearrangements have been excluded from the structural variant analysis in Pan-Cancer studies (52).
Emerging technologies, such as single-molecule sequencing, produce long reads and could provide better resolution of sequence contexts in duplication blocks (7). Such information is essential to understand how Mre11, in particular its exonuclease activity, protects KRTAP-1. The frequency of breaks at KRTAP-1 increased significantly in cells expressing nuclease-deficient Mre11 (H129N) and in cells treated with Mre11 exonuclease inhibitors Mirin or PFM39 (Figure 4B and C). However, breaks did not increase when CtIP is depleted (Figure 4D) and when cells were treated with endonuclease inhibitors PFM01 or PFM03 (Figure 4C and Supplementary Figure S5C). This is in contrast to FRA16D, for which breaks increased when either endo- or exo-nuclease function is blocked (Supplementary Figure S5E). A previous study also reported that both CtIP and Mre11 is essential in protecting FRA16D, possibly at the small, hairpin forming AT-rich repeat. Therefore, the mechanisms causing breaks at KRTAP-1 and FRA16D appear to be different. The exonuclease activity has been implicated in the uncontrolled degradation of reversed replication forks in cells deficient in proteins engaged in homology-directed repair (53). Considering the essentiality of Mre11 in the proliferation of vertebrate cells, Mre11 must have a beneficial function in repair-proficient cells. In vitro, Mre11 exonuclease activity is robust and 50 times more efficient than the hairpin opening activity by endonuclease activity (26). Very recently, Mirin-treated vertebrate cells were shown to be incapable of removing synthetic chain-terminating nucleotide analogs from newly synthesized strands, resulting in replication fork collapse (54). Whether naturally occurring chain-terminators are a significant obstacle to replication forks at duplication blocks remains to be determined, although such evidence points to yet undefined roles of Mre11 exonuclease activity during DNA replication.
Naturally-occurring breaks are a source of genome instability. Despite extensive investigations, our knowledge of the hotspots for spontaneous chromosome breaks is very limited. We focused on duplication blocks, given the diversity within humans. Because genomic approaches have limitations in duplication blocks, we employed a cytogenetic approach. To quantitatively evaluate spontaneous breaks occurring during cell proliferation, but not inherited ones in the cell population, single cell-derived clones were investigated (Figure 2C). We found that the rate of breaks seems higher in KRTAP-1 than FRA16D, raising the possibility that genetically-diverse duplication blocks represent a new class of fragile sites. However, to reach a definitive conclusion, further examination is needed for two reasons. First, the potential difference of adverse effects of breaks on cell viability between these two loci may affect the outcomes. Second, although a large number of measurements are taken for each clone, the number of independent clones (10 clones) may not be sufficient to gain statistical power. Thus, further development of an optimal approach for measuring spontaneous breaks is crucial for a full understanding of fragility in human chromosomes.
A remarkable association between duplication contents, spontaneous breaks, copy number imbalance, and the propensity for palindromic duplication at KRTAP-1 could support a model of homology-driven formation of dicentric chromosomes (4,55), in which broken DNA initiates the intra-strand annealing within long inverted repeats. DNA synthesis from an annealed broken end and following replication would complete the formation of a palindromic, giant chromosome. Alternatively, duplication architectures within KRTAP-1 could simply trigger stalled forks and breaks (32), as intra-strand annealing can occur without extensive homologies (4,56). Distinguishing these possibilities requires a full understanding of the repeat contents within the blocks and rearrangements at a nucleotide-level.
DATA AVAILABILITY
All the reagents are available upon request.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr John Tainer at The University of Texas MD Anderson Cancer Center for the generous gift of PFM03. We also thank the Flow Cytometry Core at Cedars-Sinai Medical Center for technical support.
Notes
Present address: Takaaki Watanabe, Department of Molecular Life Science, Division of Basic Molecular Science and Molecular Medicine, School of Medicine, Tokai University, Isehara, Kanagawa 259-1193, Japan.
Contributor Information
Ryusuke Suzuki, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Michael M Murata, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Nicholas Manguso, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Takaaki Watanabe, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Lila Mouakkad-Montoya, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Fumie Igari, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Md Maminur Rahman, Department of Radiation Genetics, Graduate School of Medicine, Kyoto University, Kyoto 606-8501, Japan.
Ying Qu, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Xiaojiang Cui, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA; Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Armando E Giuliano, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA; Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA; Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Shunichi Takeda, Department of Radiation Genetics, Graduate School of Medicine, Kyoto University, Kyoto 606-8501, Japan.
Hisashi Tanaka, Department of Surgery, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA; Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA; Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [2 R01 CA149385 to H.T.]; Department of Defense [W81XWH-18-1-0058 to H.T.]; Cedars-Sinai Medical Center [to H.T.]; Japan Society for the Promotion of Science (JSPS) Core-to-Core Program, A. Advanced Research Networks (to S.T.); Margie and Robert E. Petersen Foundation [to A.E.G.]; Fashion Footwear Charitable Foundation of New York, Inc. [to A.E.G.]; Avon Foundation [to A.E.G.]; Associates for Breast and Prostate Cancer Studies [to A.E.G.]. Funding for open access charge: Cedars-Sinai Medical Center.
Conflict of interest statement. None declared.
REFERENCES
- 1. Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M. et al.. The landscape of somatic copy-number alteration across human cancers. Nature. 2010; 463:899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanaka H., Watanabe T.. Mechanisms underlying recurrent genomic amplification in human cancers. Trends Cancer. 2020; 6:462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tanaka H., Tapscott S.J., Trask B.J., Yao M.C.. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:8772–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tanaka H., Cao Y., Bergstrom D.A., Kooperberg C., Tapscott S.J., Yao M.C.. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol. Cell. Biol. 2007; 27:1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barlow J.H., Faryabi R.B., Callen E., Wong N., Malhowski A., Chen H.T., Gutierrez-Cruz G., Sun H.W., McKinnon P., Wright G. et al.. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013; 152:620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glover T.W., Wilson T.E., Arlt M.F.. Fragile sites in cancer: more than meets the eye. Nat. Rev. Cancer. 2017; 17:489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Audano P.A., Sulovari A., Graves-Lindsay T.A., Cantsilieris S., Sorensen M., Welch A.E., Dougherty M.L., Nelson B.J., Shah A., Dutcher S.K. et al.. Characterizing the major structural variant alleles of the human genome. Cell. 2019; 176:663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carvalho C.M., Lupski J.R.. Mechanisms underlying structural variant formation in genomic disorders. Nat. Rev. Genet. 2016; 17:224–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hellman A., Zlotorynski E., Scherer S.W., Cheung J., Vincent J.B., Smith D.I., Trakhtenbrot L., Kerem B.. A role for common fragile site induction in amplification of human oncogenes. Cancer Cell. 2002; 1:89–97. [DOI] [PubMed] [Google Scholar]
- 10. Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E.. Recent segmental duplications in the human genome. Science. 2002; 297:1003–1007. [DOI] [PubMed] [Google Scholar]
- 11. Eichler E.E. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001; 17:661–669. [DOI] [PubMed] [Google Scholar]
- 12. Harel T., Lupski J.R.. Genomic disorders 20 years on-mechanisms for clinical manifestations. Clin. Genet. 2018; 93:439–449. [DOI] [PubMed] [Google Scholar]
- 13. Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R. et al.. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005; 77:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Church D.M., Schneider V.A., Graves T., Auger K., Cunningham F., Bouk N., Chen H.-C., Agarwala R., McLaren W.M., Ritchie G.R.S. et al.. Modernizing reference genome assemblies. PLoS Biol. 2011; 9:e1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eichler E.E., Clark R.A., She X.. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 2004; 5:345–354. [DOI] [PubMed] [Google Scholar]
- 16. Stracker T.H., Petrini J.H.J.. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011; 12:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paull T.T. 20 Years of Mre11 biology: no end in sight. Mol. Cell. 2018; 71:419–427. [DOI] [PubMed] [Google Scholar]
- 18. Maser R.S., Mirzoeva O.K., Wells J., Olivares H., Williams B.R., Zinkel R.A., Farnham P.J., Petrini J.H.. Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 2001; 21:6006–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dungrawala H., Rose K.L., Bhat K.P., Mohni K.N., Glick G.G., Couch F.B., Cortez D.. The replication checkpoint prevents two types of fork collapse without regulating eplisome stability. Mol. Cell. 2015; 59:998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costanzo V., Robertson K., Bibikova M., Kim E., Grieco D., Gottesman M., Carroll D., Gautier J.. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 2001; 8:137–147. [DOI] [PubMed] [Google Scholar]
- 21. Kondratova A., Watanabe T., Marotta M., Cannon M., Segall A.M., Serre D., Tanaka H.. Replication fork integrity and intra-S phase checkpoint suppress gene amplification. Nucleic Acids Res. 2015; 43:2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamaguchi-Iwai Y., Sonoda E., Sasaki M.S., Morrison C., Haraguchi T., Hiraoka Y., Yamashita Y.M., Yagi T., Takata M., Price C. et al.. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999; 18:6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xiao Y., Weaver D.T.. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997; 25:2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moreau S., Ferguson J.R., Symington L.S.. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999; 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gibson F.P., Leach D.R., Lloyd R.G.. Identification of sbcD mutations as cosuppressors of recBC that allow propagation of DNA palindromes in Escherichia coli K-12. J. Bacteriol. 1992; 174:1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paull T.T., Gellert M.. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998; 1:969–979. [DOI] [PubMed] [Google Scholar]
- 27. Trujillo K.M., Sung P.. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50/Mre11 complex. J. Biol. Chem. 2001; 276:35458–35464. [DOI] [PubMed] [Google Scholar]
- 28. Connelly J.C., de Leau E.S., Leach D.R.F.. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst.). 2003; 2:795–807. [DOI] [PubMed] [Google Scholar]
- 29. Deshpande RA., Lee J.-H., Arora S., Paull TT.. Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Mol. Cell. 2016; 64:593–606. [DOI] [PubMed] [Google Scholar]
- 30. Cannavo E., Cejka P.. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature. 2014; 514:122–125. [DOI] [PubMed] [Google Scholar]
- 31. Anand R., Ranjha L., Cannavo E., Cejka P.. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell. 2016; 64:940–950. [DOI] [PubMed] [Google Scholar]
- 32. Lobachev K.S., Gordenin D.A., Resnick M.A.. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002; 108:183–193. [DOI] [PubMed] [Google Scholar]
- 33. Wang H., Li Y., Truong LN., Shi LZ., Hwang PY.-H., He J., Do J., Cho MJ., Li H., Negrete A. et al.. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014; 54:1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan M., Schwaederle M., Arguello D., Millis S.Z., Gatalica Z., Kurzrock R.. HER2 expression status in diverse cancers: review of results from 37,992 patients. Cancer Metastasis Rev. 2015; 34:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L.. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987; 235:177–182. [DOI] [PubMed] [Google Scholar]
- 36. Jin L., Qu Y., Gomez L.J., Chung S., Han B., Gao B., Yue Y., Gong Y., Liu X., Amersi F. et al.. Characterization of primary human mammary epithelial cells isolated and propagated by conditional reprogrammed cell culture. Oncotarget. 2018; 9:11503–11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoa N.N., Akagawa R., Yamasaki T., Hirota K., Sasa K., Natsume T., Kobayashi J., Sakuma T., Yamamoto T., Komatsu K. et al.. Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells. 2015; 20:1059–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watanabe T., Tanabe H., Horiuchi T.. Gene amplification system based on double rolling-circle replication as a model for oncogene-type amplification. Nucleic. Acids. Res. 2011; 39:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gillet-Markowska A., Louvel G., Fischer G.. bz-rates: a web tool to estimate mutation rates from fluctuation analysis. G3 (Bethesda). 2015; 5:2323–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okamoto Y., Iwasaki W.M., Kugou K., Takahashi K.K., Oda A., Sato K., Kobayashi W., Kawai H., Sakasai R., Takaori-Kondo A. et al.. Replication stress induces accumulation of FANCD2 at central region of large fragile genes. Nucleic. Acids. Res. 2018; 46:2932–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrari A., Vincent-Salomon A., Pivot X., Sertier A.S., Thomas E., Tonon L., Boyault S., Mulugeta E., Treilleux I., MacGrogan G. et al.. A whole-genome sequence and transcriptome perspective on HER2-positive breast cancers. Nat. Commun. 2016; 7:12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Antonacci F., Dennis M.Y., Huddleston J., Sudmant P.H., Steinberg K.M., Rosenfeld J.A., Miroballo M., Graves T.A., Vives L., Malig M. et al.. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 2014; 46:1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nuttle X., Giannuzzi G., Duyzend M.H., Schraiber J.G., Narvaiza I., Sudmant P.H., Penn O., Chiatante G., Malig M., Huddleston J. et al.. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 2016; 536:205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mirkin E.V., Mirkin S.M.. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 2007; 71:13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luria S.E., Delbruck M.. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943; 28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petit P., Fryns J.P., Berghe H.v.d, Hecht F.. Population cytogenetics of autosomal fragile sites. Clin. Genet. 1986; 29:96–100. [DOI] [PubMed] [Google Scholar]
- 47. Letessier A., Millot G.A., Koundrioukoff S., Lachages A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M.. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011; 470:120–123. [DOI] [PubMed] [Google Scholar]
- 48. Jiang Z., Tang H., Ventura M., Cardone M.F., Marques-Bonet T., She X., Pevzner P.A., Eichler E.E.. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007; 39:1361–1368. [DOI] [PubMed] [Google Scholar]
- 49. Shibata A., Moiani D., Arvai A.S., Perry J., Harding S.M., Genois M.M., Maity R., van Rossum-Fikkert S., Kertokalio A., Romoli F. et al.. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell. 2014; 53:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lovejoy C.A., Lock K., Yenamandra A., Cortez D.. DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 2006; 26:7977–7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barbouti A., Stankiewicz P., Nusbaum C., Cuomo C., Cook A., Hoglund M., Johansson B., Hagemeijer A., Park S.S., Mitelman F. et al.. The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am. J. Hum. Genet. 2004; 74:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li Y., Roberts N.D., Wala J.A., Shapira O., Schumacher S.E., Kumar K., Khurana E., Waszak S., Korbel J.O., Haber J.E. et al.. Patterns of somatic structural variation in human cancer genomes. Nature. 2020; 578:112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L. et al.. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 2017; 67:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boeckemeier L., Kraehenbuehl R., Keszthelyi A., Gasasira M.U., Vernon E.G., Beardmore R., Vågbø C.B., Chaplin D., Gollins S., Krokan H.E. et al.. Mre11 exonuclease activity removes the chain-terminating nucleoside analog gemcitabine from the nascent strand during DNA replication. Sci. Adv. 2020; 6:eaaz4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mizuno K., Miyabe I., Schalbetter S.A., Carr A.M., Murray J.M.. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature. 2013; 493:246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deng S.K., Yin Y., Petes T.D., Symington L.S.. Mre11-Sae2 and RPA collaborate to prevent palindromic gene amplification. Mol. Cell. 2015; 60:500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the reagents are available upon request.