Abstract
In the Gram-positive Firmicute bacterium Bacillus subtilis, amino acid starvation induces synthesis of the alarmone (p)ppGpp by the RelA/SpoT Homolog factor Rel. This bifunctional enzyme is capable of both synthesizing and hydrolysing (p)ppGpp. To detect amino acid deficiency, Rel monitors the aminoacylation status of the ribosomal A-site tRNA by directly inspecting the tRNA’s CCA end. Here we dissect the molecular mechanism of B. subtilis Rel. Off the ribosome, Rel predominantly assumes a ‘closed’ conformation with dominant (p)ppGpp hydrolysis activity. This state does not specifically select deacylated tRNA since the interaction is only moderately affected by tRNA aminoacylation. Once bound to the vacant ribosomal A-site, Rel assumes an ‘open’ conformation, which primes its TGS and Helical domains for specific recognition and stabilization of cognate deacylated tRNA on the ribosome. The tRNA locks Rel on the ribosome in a hyperactivated state that processively synthesises (p)ppGpp while the hydrolysis is suppressed. In stark contrast to non-specific tRNA interactions off the ribosome, tRNA-dependent Rel locking on the ribosome and activation of (p)ppGpp synthesis are highly specific and completely abrogated by tRNA aminoacylation. Binding pppGpp to a dedicated allosteric site located in the N-terminal catalytic domain region of the enzyme further enhances its synthetase activity.
INTRODUCTION
Alarmone nucleotides guanosine pentaphosphate (pppGpp) and tetraphosphate (ppGpp), collectively referred to as (p)ppGpp, regulate metabolism, virulence, stress responses and antibiotic tolerance in the vast majority of bacterial species (1–4). The cellular levels of (p)ppGpp are controlled by members of the RelA/SpoT Homolog (RSH) protein family (5). These enzymes both synthesize (p)ppGpp by transferring the pyrophosphate group of ATP onto the 3′ ribose position of either GDP or GTP, and degrade the alarmone by hydrolysing the nucleotide back to GDP or GTP, releasing inorganic pyrophosphate in the process.
RSH factors fall into two categories: ‘small’ single domain RSHs and ‘long’ multi-domain RSHs (5). In the majority of bacterial species, long RSHs are represented by a single ribosome-associated bifunctional enzyme called Rel that is capable of both synthesizing and degrading (p)ppGpp. In the lineage to Beta- and Gammaproteobacteria, duplication and functional divergence of the ancestral ribosome-associated Rel gave rise to a pair of specialized factors—the namesakes of the RSH protein family—RelA and SpoT (5,6). RelA is a synthesis-only enzyme incapable of hydrolysing (p)ppGpp (7), while SpoT and Rel are bifunctional (8,9). The (p)ppGpp synthesis activity of RelA and Rel is induced by so-called ‘starved’ ribosomal complexes (i.e. ribosomes accommodating deacylated tRNA in the A-site) (8,10). While deacylated tRNA dramatically stabilizes the association of Rel/RelA with the ribosome (11–13), RelA has significant affinity to the vacant ribosomal A-site as well. Indeed, Loveland et al., in addition to reporting structures of RelA associated with starved ribosomes, have also solved a structure of RelA bound to the ribosome without A-site tRNA (Structure 1, PDB 5KPS) (14). In the case of RelA, the pppGpp product is a potent inducer of the enzyme's synthesis activity (15). The mechanistic basis of this regulatory mechanism is currently unexplored, and it is unknown whether bifunctional RSH Rel is similarly regulated by pppGpp.
Long RSHs are comprised of an enzymatic N-terminal multi-domain region (NTD) and a regulatory C-terminal multi-domain region (CTD) (Figure 1). The NTD is subdivided into (p)ppGpp hydrolase (HD) and (p)ppGpp synthetase (SYNTH) catalytic domains, while the regulatory CTD is comprised of the TGS (ThrRS, GTPase and SpoT), Helical (equivalent to alpha-helical as per Loveland et al. (14)), ZFD (Zinc Finger Domain; equivalent to CC, Conserved Cysteine as per Atkinson et al. (5) and RIS, Ribosome-InterSubunit as per Loveland et al. (14)) and RRM (RNA recognition motif; equivalent to ACT, aspartokinase, chorismate mutase and TyrA as per Atkinson et al. (5)). Experiments with Rel from Streptococcus dysgalactiae subsp. equisimilis (16,17), Mycobacterium tuberculosis (18,19), Caulobacter crescentus (20), Staphylococcus aureus (21), Thermus thermophilus (22) and Bacillus subtilis (12) have established that (i) the synthetic and hydrolytic activities of the NTD are mutually exclusive, (ii) CTD suppresses the NTD synthetic activity in cis through autoinhibition, and (iii) the CTD is essential for Rel regulation by the ribosomal complexes.
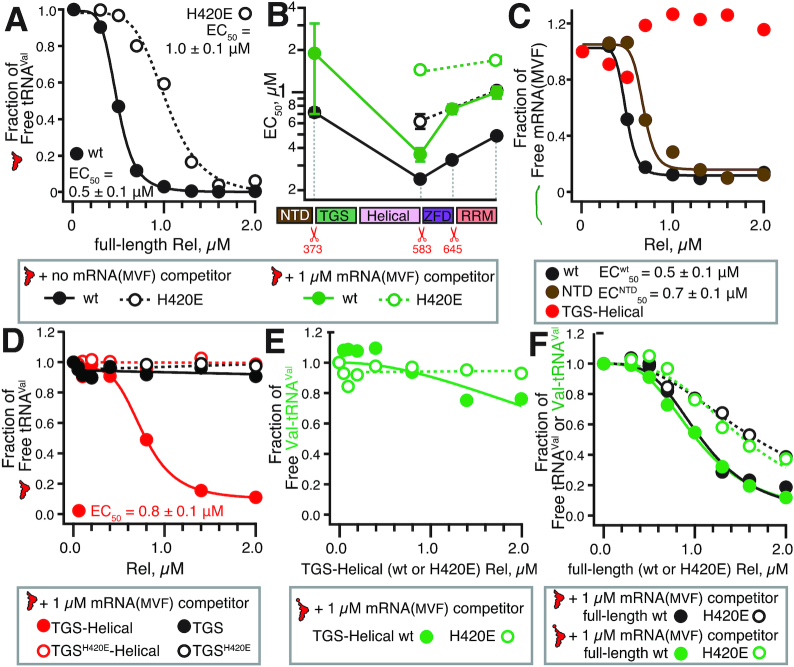
Figure 1.
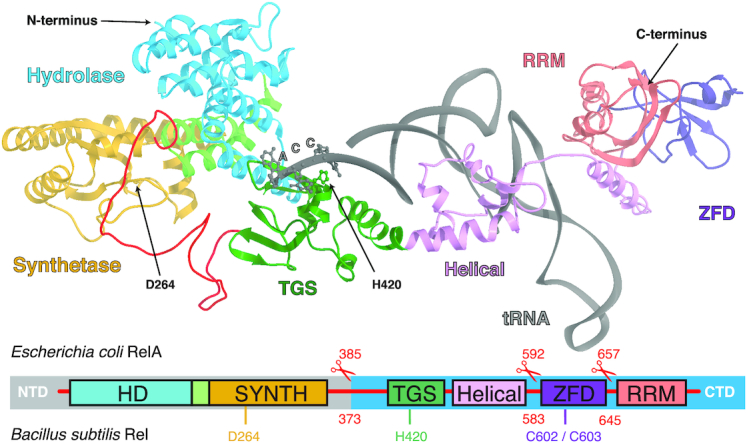
Molecular recognition of the deacylated A/R-tRNA by the stringent factor. The domain composition as well as the truncated versions of the enzymes (B. subtilis Rel: ΔRRM 1–645, ΔZFD-RRM 1–583 and NTD 1–373; E. coli RelA: ΔRRM 1-657, ΔZFD-RRM 1–592 and NTD 1–385) used for functional studies are highlighted, as well as functionally important residues D264 and H420 of B. subtilis Rel. The 3′ CCA end of the partially A-site accommodated and highly distorted deacylated tRNA (A/R-tRNA) is shown as spheres. The 3D model was constructed from using partial structure of B. subtilis Rel in complex with starved ribosomes (RDB accession number 6HTQ) and a swap dimeric state of C-terminally truncated B. subtilis Rel (6YXA) (32).
Our understanding of the relationship between the enzymatic activity of RelA/Rel and the factors’ interactions with starved ribosomal complexes is largely based on experiments with E. coli RelA. A family of related ‘hopping’ models was put forward (23–25). According to the original ‘hopping’ model, each act of (p)ppGpp synthesis fuels the displacement of RelA from the ribosome, upon which the factor ‘hops’ to the next starved complex (25). Single molecule tracking experiments using RelA C-terminal fusions with photoswitchable fluorescent proteins led to the formulation of the ‘extended hopping’ (23) and the ‘short hopping time’ (24) models. The former model suggests that upon activation by starved ribosomes RelA processively synthesizes (p)ppGpp off the ribosome and the latter states that upon activation, RelA spends prolonged periods of time catalytically active in association with starved ribosomal complexes. Finally, there are two ‘flavours’ of the ‘short hopping time’ model: RelA could first bind the vacant A-site and then recruit the deacylated tRNA (14,15) or form the RelA:tRNA complex off the ribosome and deliver deacylated tRNA to the A-site, similarly to how elongation factor EF-Tu delivers aminoacylated tRNA (26,27). Principally, one could also imagine a ‘tRNA-first’ mechanism, with deacylated tRNA binding first to the vacant A-site followed by the RelA association with the complex. However, this scenario was deemed unlikely given the structure of RelA locked on the starved ribosome (14).
The proposed delivery of deacylated tRNA to the ribosome by RelA/Rel is a controversial topic. In the case of M. tuberculosis Rel, binding to tRNA is readily detectible (8,18). While binding of tRNA to E. coli RelA is not detected by EMSA (15,27) unless the complex is crosslinked by formaldehyde (27), stable tRNA association was reported in the case of a truncated version of E. coli RelA containing just the TGS and part of the Helical domain (27). This suggests a possibility that in the full-length protein the tRNA interaction interface is not accessible and becomes accessible only upon ribosomal recruitment or truncation of the protein. Moreover, addition of A-site non-cognate deacylated tRNA in excess did not significantly inhibit RelA activation by starved complexes, as it would be expected if the factor would be sequestered in a complex with non-cognate tRNA off the ribosome (15). Importantly, the original report proposing tRNA delivery by E. coli RelA relied on the UV-induced crosslinking and analysis of cDNAs (CRAC) approach and the authors provided no biochemical or biophysical evidence of the RelA:tRNA complex formation (26). While CRAC is an extremely sensitive technique for detection of interactions, it is inherently unable to estimate the affinities or establish the sequence of binding events. The TGS and Helical domains of RelA form multiple contacts with deacylated A-site tRNA (14,28,29) while on the ribosome. However, it is unclear if these domains are accessible when RelA/Rel is off the ribosome.
Studies of the physiological effects of (p)ppGpp in the Gram-positive Firmicute bacterium B. subtilis have been fundamental to our understating of (p)ppGpp-mediated transcriptional regulation (30) and regulation of nucleotide metabolism (31). However, mechanistic understanding of B. subtilis Rel is limited (12,32). Using our reconstituted biochemical system (12), we have dissected the relationship between, on the one hand, Rel's association with deacylated tRNA and the ribosome and, on the other hand, (p)ppGpp synthesis and hydrolysis by the enzyme, and probed the roles of individual domains. We establish that the specific recognition of deacylated tRNA takes place after Rel associates with the vacant ribosomal A-site, and the strength of the interaction with deacylated tRNA fine-tunes the stability of the interaction of Rel/RelA enzymes with starved ribosomes. In the case of B. subtilis Rel, the factor forms a stable complex with starved ribosomes and this complex is not actively dissociated by processive synthesis of ppGpp. We demonstrate that pppGpp enhances synthetase activity of both B. subtilis Rel and E. coli RelA by binding to a dedicated allosteric site located in the enzymatic NTD region of the factor.
MATERIALS AND METHODS
Detailed description of experimental procedures is provided in Supplementary methods. All bacterial strains and plasmids used in this study are listed in Supplementary Tables 1 and 2. All proteins were expressed, purified and characterized using SUMO-tagging strategy described earlier for E. coli RelA (33) and B. subtilis Rel (12) (Supplementary Figure S1A-G). The monomeric nature of 50 nM B. subtilis Rel and 100 nM E. coli RelA was confirmed by mass photometry (34) using Refeyn OneMP instrument (Refeyn Ltd.) (Supplementary Figure S1H and I).
Enzymatic assays
3H-pppGpp/3H-ppGpp synthesis assays were performed at 37°C in HEPES:Polymix buffer (5 mM Mg2+ final concentration) as described earlier (12) using either E. coli RelA and B. subtilis Rel as well as (i) 70S initiation complexes assembled using either E. coli or B. subtilis and (ii) deacylated E. coli tRNAVal or tRNAPhe. Either 300 μM 3H-GTP or 300 μM 3H-GDP (both from PerkinElmer) as well as 1 mM ATP were used as substrates. 3H-pppGpp/3H-ppGpp hydrolysis assays were performed using either 300 μM 3H-pppGpp or 300 μM 3H-ppGpp as substrate as described earlier (12). Individual quenched timepoints were spotted PEI-TLC plates (Macherey-Nagel), nucleotides resolved in 1.5 M KH2PO4 pH 3.5 buffer, the TLC plates were dried and cut into sections as guided by UV-shadowing and 3H radioactivity was quantified by scintillation counting. Enzyme turnover was estimated by linear regression though individual kinetic points making sure that only the points on the linear kinetic regime are used (<50% substrate to product conversion). The error bars represent standard deviations of the turnover estimates using four data points; each experiment was performed at least two times.
Sucrose gradient fractionation and Western blotting
Sample preparation i: lysates: B. subtilis samples were prepared as described earlier (12) and supplemented with nucleotides GTP / GDP (0.5 mM) and ATP/AMPCPP (1 mM) as indicated on the figure legends and used for analytical ultracentrifugation and Western blotting.
Sample preparation ii: lysates: MG1655 relA::HTF E. coli strain (26) was grown in liquid LB media at 37°C. At OD600 of 0.2 the culture was treated 20 min with mupirocin added to final concentration of 70 μM, and the samples were processed as described for B. subtilis lysates.
Sample preparation iii: reconstituted system: 20 μl reaction mixtures containing 500 nM B. subtilis 70S IC(MVF), 140 nM Rel, E. coli tRNAVal (all in HEPES:Polymix buffer, 5 mM Mg2+ final concentration) were incubated at 37°C for 5 min and loaded onto sucrose gradients.
Sucrose gradient fractionation: clarified cell lysates (or reconstituted ribosomal complexes) were loaded onto 10–35% sucrose gradients in HEPES:Polymix buffer pH 7.5 (5 mM Mg2+ final concentration, supplemented with nucleotides GTP:Mg2+/GDP:Mg2+ (0.5 mM) and ATP:Mg2+/AMPCPP:Mg2+ (1 mM) as indicated on the figure legends), subjected to centrifugation (36 000 rpm for 3 h at 4°C, SW-41Ti Beckman Coulter rotor) and analysed using Biocomp Gradient Station (BioComp Instruments) with A260 and A280 as a readout.
Western blotting: experiments were performed as described earlier (12). Ribosomal protein L3 of the 50S ribosomal subunit was detected using anti-L3 primary antibodies (a gift from Fujio Kawamura (35)) combined with goat anti-rabbit IgG-HRP secondary antibodies.
Electrophoretic mobility shift assay (EMSA)
Before performing the experiment, stock mRNA(MVF) (5′-GGCAAGGAGGAGAUAAGAAUGGUUUUCUAAUA-3′) was incubated for 2 min at 60°C to denature possible secondary structures. Reaction mixtures (10 μl) in HEPES:Polymix buffer (36) with 5 mM Mg2+ were assembled by adding E. coli either tRNAVal/tRNAiMet (0.1 μM final concentration) or mRNA (0.1 μM final concentration) or both (0.1 μM tRNAVal/tRNAiMet and 1 μM mRNA(MVF) competitor), followed by the addition of Rel. After incubation for 5 min at 37°C, 4 μl of 50% sucrose was added per sample, and the samples were electrophoretically resolved on a 12% Tris:Borate:EDTA gel at 4°C (160–180V) for 1–1.5 h. Gels were stained with SYBR Gold nucleic acid stain (Life Technologies) for 30 min, followed by visualization using a Typhoon Trio Variable Mode Imager (Amersham Biosciences). Bands were quantified using ImageJ (37). The efficiency of complex formation (effective concentration, EC50 ± standart deviation) was calculated using the 4PL model (Hill equation) as per Sebaugh (38) using eight data points; each experiment was performed at least two times.
Isothermal titration calorimetry (ITC)
Dialyzed B. subtilis RelNTD/E. coli RelANTD was concentrated in Sartorius Ultrafiltration Centrifugal Concentrators (cut-off 30 kDa) at 3000 × g to a final concentration of 35 μM. 10 mM ppGpp and pppGpp (both from Jena Bioscience) were diluted to 450 μM in ITC buffer. Non-hydrolysable ATP analogue AMPCPP (ApCpp, Jena BioScience) and GDP were diluted with ITC buffer to 100 and 100 μM, respectively, and mixed with the protein prior to the experiment. 300 μM EDTA was incubated with B. subtilis RelNTD overnight at 4°C and supplemented with AMPCPP and GDP prior to the experiment. The samples were degassed and equilibrated at titration temperature and the ITC measurement were performed using Affinity ITC calorimeter (TA instruments) at 20°C. The stirring rate was set to 75 rpm and a constant injection volume of 2 μl of titrant (ppGpp or pppGpp) was injected into the cell (177 μl of B. subtilis RelNTD) with an injection interval time of 250 s. For the heat exchange measurements RelNTD protein was concentrated to 150 μM and 2.1 mM stock solutions of GDP and GTP were used. The stirring rate was set to 300 rpm and the measurements were performed at 20, 25 and 30°C. All data were processed and analysed using the NanoAnalyse and Origin software packages.
HPLC-based nucleotide quantification
HPLC-based nucleotide quantification was performed as per Varik et al. (39). Wild type MG1655 and MG1655 relA::HTF E. coli (26) strains were grown in MOPS minimal media at 37°C until OD600 0.5 and challenged with 150 μg/ml of mupirocin (3× the MIC).
RESULTS AND DISCUSSION
Off the ribosome the NTD region of B. subtilis Rel non-specifically binds RNA
When Rel/RelA is associated with the starved ribosomal complex, the TGS and Helical domains form specific contacts with the ribosome and tRNA (14,28,29,32) (Figure 1). To probe the accessibility of Rel's TGS and Helical domains for interaction with deacylated tRNA off the ribosome, we purified native, full-length RNA-free untagged B. subtilis Rel as well as a set of C-terminally truncated variants: lacking RRM (RelΔRRM, amino acids 1–645), lacking both RRM and ZFD (RelΔZFD-RRM, 1–583) and lacking all of the regulatory CTD domains (RelNTD, 1–373). For the sake of simplicity, throughout the text B. subtilis Rel—the main focus of the current study—is referred to simply as ‘Rel’.
As a specificity control for tRNA binding studies we substituted a conserved histidine residue (H420E) in the TGS domain. The corresponding histidine residue in E. coli RelA (H432) forms a stacking interaction with the 3′ CCA end of the uncharged A-site tRNA (28). The H432E substitution abrogates RelA’s functionality by abolishing RelA activation by deacylated tRNA (12,26). We have shown a similar loss-of-function effect of the H420E substitution in B. subtilis Rel (12). Therefore, the H420E substitution is an excellent tool to probe the specificity of Rel's interaction with deacylated tRNA.
We used electrophoretic mobility shift assays (EMSA) to study complex formation between native deacylated E. coli tRNAVal and B. subtilis Rel: full-length and C-terminally truncated, both wild-type and H420E variants. While analogous EMSAs failed to detect a stable complex between RelA and deacylated tRNA (15,27), a Rel:tRNA complex is readily observable (Figure 2A and Supplementary Figure S2). In the absence of a non-specific RNA competitor, full-length Rel forms a complex with EC50 of 0.5 μM. Surprisingly, the H420E substitution decreases the affinity only twice (from EC50 0.5 to 1.0 μM), suggesting a lack of specificity (Figure 2A and Supplementary Figure S2AB).
Figure 2.
The isolated TGS-Helical region of B. subtilis Rel, but not the full-length protein, can specifically recognize the tRNA 3′ CCA end. Complex formation between increasing concentrations of B. subtilis Rel (wild-type or mutants, H420E mutants shown as empty circles) and either 0.1 μM E. coli tRNAVal or synthetic mRNA(MVF) was monitored by EMSA. Representative full EMSA gels as well as EC50 quantifications are provided as Supplementary Figures S2–S5. (A) Complex formation between E. coli tRNAVal and either wild-type or the H420E Rel mutant in the absence of a non-specific RNA competitor. (B) Complex formation between C-terminally truncated Rel variants and E. coli tRNAVal. (C) Complex formation between mRNA(MVF) and full-length Rel, Rel NTD fragment or Rel TGS-Helical fragment. (D) Complex formation between E. coli tRNAVal and either the Rel TGS domain or the Rel TGS-Helical fragment. Complex formation between acetylated and deacylated E. coli tRNAVal and either isolated TGS-Helical fragment (E) or full-length (F) Rel. Analogous experiments using E. coli tRNAiMet are presented on Supplementary Figure S6.
The sequential deletion of RRM and ZFD domains increases the affinity to tRNAVal (EC50 of 0.3 and 0.2 μM), and even upon deletion of the entire CTD, RelNTD affinity for tRNAVal remains virtually unaffected (EC50 0.7 μM) (Figure 2B, black filled trace, and Supplementary Figure S2D–F). Consistent with the isolated NTD binding tRNAVal, while the H420E substitution in the full-length and ΔZFD-RRM backgrounds does decrease the factor's affinity to tRNA, it does not abrogate the interaction completely (Figure 2B, compare empty and filled traces). To further characterize the non-specific component of the Rel:tRNA interaction, we used single-stranded mRNA(MVF) as a competitor. In the presence of a 10-fold excess of mRNA over tRNA, the affinity for tRNA drops about two-fold (Figure 2B, green traces, and Supplementary Figure S3). At the same time, Rel binds both RNA species with a similar affinity, EC50 of 0.5 μM (Figure 2B and C). Using the set of truncated Rel variants described above, we localized the source of this non-specific affinity for mRNA to Rel's NTD region (Figure 2C and Supplementary Figure S4).
Taken together, these results suggest that when tested in the absence of ribosomes, complex formation between Rel and tRNA has a clear non-specific component, which is mediated by the protein's NTD region. This result is clearly at odds with the highly specific recognition of tRNA by the TGS domain in the cellular context as well as in the presence of starved ribosomal complexes, both of which are abrogated by the H420E substitution (12). The ability of E. coli RelA compromised in ribosomal binding by deletion of both RRM and ZFD domains to crosslink to tRNA in CRAC experiments was used as an argument for tRNA delivery by RelA to the ribosome (26). However, as we show here, these interactions lack specificity and therefore are unlikely to represent a productive on-path intermediate in the mechanism of stringent factor activation. To deconvolute RNA binding by the NTD from that mediated by the CTD, we next characterized the isolated Rel TGS-Helical region (amino acid positions 374–583) as well as the TGS domain alone (amino acid positions 374–469).
The isolated TGS-Helical region of Rel specifically recognizes the tRNA 3′ CCA end
While we do not detect formation of a stable complex in the case of TGS alone, the TGS-Helical fragment of Rel binds tRNAVal with EC50 of 0.8 μM (Figure 2D and Supplementary Figure S5AB). In stark contrast to the full-length protein, this interaction is completely abrogated upon introduction of the H420E substitution, suggesting that for these isolated regions (unlike the full-length protein), complex formation is driven by a specific recognition of the 3′ CCA tRNA end. Importantly, unlike the full-length enzyme and the NTD, the TGS-Helical fragment does not bind mRNA(MVF), further reinforcing the specificity of this interaction (Figure 2C and Supplementary Figure S4C). Finally, aminoacylation of tRNAVal completely abrogates the interaction with the isolated TGS-Helical domains (Figure 2E and Supplementary Figure S5EF), while the affinity of the full-length Rel remains largely unaffected (Figure 2F and Supplementary Figure S3GH). To rule out tRNA-specific effects, we performed an analogous set of binding assays with full-length Rel and the isolated TGS-Helical domains using both charged and deacylated E. coli initiator tRNAiMet (Supplementary Figure S6). The results are in excellent agreement with our experiments with tRNAVal: while tRNAiMet binding to full-length Rel is largely insensitive to the H420E substitution and tRNA aminoacylation, complex formation with isolated TGS-Helical domains is strictly dependent on deacylated tRNAiMet and is abrogated by the H420E substitution.
Our results demonstrate that, unlike the full-length protein, the isolated Rel TGS-Helical region is highly specific in its recognition of the tRNA 3′ CCA end. Taking into account cryo-EM studies dissecting RelA recruitment to the ribosome (14,28,29) and previous mechanistic studies (16), we hypothesize that it is ribosome binding that drives the transition of Rel from the ‘closed’ conformation, unable to specifically sense and bind tRNA, to an ‘open’ conformation in which the TGS-Helical region is primed to specifically recognize the deacylated 3′ CCA end. Therefore, we next investigated the effects of deacylated tRNA on the interaction of Rel with the ribosome.
Full-length Rel specifically recognizes the 3′ CCA of deacylated tRNA on the ribosome
To probe the specificity of tRNA-dependent recruitment of Rel to the ribosome, we performed 3H-pppGpp synthesis assays in the presence of 70S initiation complexes as well as increasing concentrations of either A-site cognate deacylated tRNAVal or non-cognate tRNAiMet (Figure 3A). The activation of Rel's synthetic activity by tRNAVal is potent (EC50 of 41 ± 7 nM) and specific: we detect no activation in the presence of tRNAiMet. Since non-cognate tRNA does not activate Rel in the presence of initiation complexes, formation of the stable non-cognate tRNA:Rel complex off the ribosome is expected to compromise the ribosome-dependent activation since the latter is strictly dependent on tRNA being cognate. However, even when the non-cognate tRNAPhe is added in a 50-fold excess of over cognate tRNAVal, we observe only a moderate inhibitory effect (30 %), demonstrating efficient selection of the cognate tRNA on the ribosome (Figure 3B). We have earlier performed analogous experiments with E. coli RelA with similar results (15).
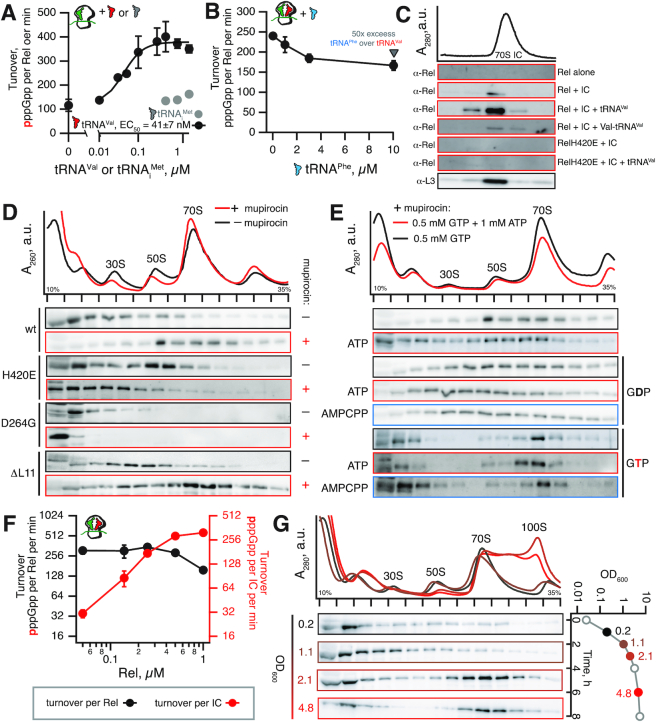
Figure 3.
Rel is stably recruited to the starved ribosomal complex to drive processive (p)ppGpp production. SYNTH activity of Rel assayed in the presence of (A) 70S IC(MVF) as well as increasing concentrations of cognate tRNAVal or non-cognate tRNAiMet or (B) starved ribosomal complexes (70S IC and sub-saturating concentration of cognate tRNAVal, 0.2 μM) as well increasing concentrations of non-cognate tRNAPhe. (C) Sucrose gradient centrifugation and immunobloting analyses probing against either Rel or ribosomal protein L3 of reconstituted Rel:ribosome complexes. Reaction mixtures containing 0.5 μM IC (MVF) and 140 nM of wild-type or H420E Rel protein were supplemented with either 2 μM tRNAVal or 2 μM Val-tRNAVal, incubated at 37°C for 5 min and resolved on 10 to 35% sucrose gradients in HEPES:Polymix buffer (pH 7.5, 5 mM Mg2+). (D) Polysome profile and Rel immunoblot analyses probing Rel's interaction with the ribosomes with or without the induction of acute isoleucine starvation by antibiotic mupirocin. Wild-type Rel was expressed in either wild-type 168 B. subtilis or in ΔL11 strain (VHB47). Rel H420E variant defective in CCA recognition was expressed from the native chromosomal locus in wild-type B. subtilis (VHB68). Synthesis-inactive D264G was ectopically expressed in Δrel B. subtilis under the control of Phy-spank promotor (VHB156), and expression was induced by 1 mM IPTG. (E) Effects of nucleotide substrates on Rel's association with starved ribosomes generated by B. subtilis by treatment with mupirocin. (F) Synthase activity of Rel activated by starved ribosomal complexes (0.5 μM 70S IC(MVF) and 2 μM tRNAVal) as a function increasing concentrations of Rel. Error bars represent standard deviations of the turnover estimates by linear regression and each experiment was performed at least three times. (G) Ribosomal association of Rel as a function of B. subtilis growth phase. All experiments were performed with B. subtilis grown in liquid LB media at 37°C.
To directly probe tRNA recruitment to the ribosome, we next resolved on sucrose gradients either purified stringent factor combined with reconstituted B. subtilis 70S initiation complexes (Figure 3C) or lysates of B. subtilis cells treated with the antibiotic mupirocin that induces acute isoleucine starvation (Figure 3D). We detected native, untagged Rel by immunoblotting with polyclonal anti-Rel antiserum (12). Deacylated tRNA promotes efficient and specific recruitment of Rel to the ribosome in both experimental systems (Figure 3C and D). In stark contrast with the non-specific Rel:tRNA interaction off the ribosome, tRNA-mediated locking of Rel on the ribosome is highly specific and is efficiently abrogated both by tRNA aminoacylation (Figure 3C; reconstituted system) and the H420E substitution (Figure 3C and D; both systems). The latter result is in agreement with the CRAC experiments detecting no RelA:rRNA crosslinks upon amino acid starvation of E. coli expressing the H432E RelA variant (26).
Next, we tested Rel recruitment upon mupirocin treatment in a B. subtilis strain lacking the ribosomal protein L11 (ΔrplK), which is crucial for activation of E. coli RelA by starved ribosomal complexes (7,25,40), as well as the functionality of C. crescentus (41) and B. subtilis (12) Rel. In good agreement with earlier results (25,41), the lack of L11 does not perturb Rel's recruitment to the ribosome (Figure 3D). This demonstrates that while association of Rel with starved complexes is driven by deacylated tRNA, recruitment to the ribosome is not sufficient for activation of Rel's synthetic activity.
Finally, we tested a synthesis-compromised D264G Rel variant (42). Surprisingly, D264G Rel is not stably recruited to the ribosome when acute amino acid starvation is induced by mupirocin (Figure 3D). Note that the centrifugation approach can only reliably detect stable complexes; for instance, while Rel lacking the C-terminal RRM domain is efficiently activated by starved ribosomes, stable ribosomal recruitment was not observed upon the mupirocin challenge (12). A possible explanation is that by driving the protein into the SYNTHOFF HDON state, this substitution compromises the binding of the stringent factor to the ribosome.
Deacylated tRNA locks Rel on the ribosome in stationary phase in B. subtilis
As bacteria enter the stationary phase and nutrients become limiting, the ribosome-associated RSHs RelA/Rel are activated and the intracellular concentration of (p)ppGpp increases (39). We characterized Rel association with ribosomes in B. subtilis throughout the growth curve, collecting the samples at OD600 of 0.2, 1.1, 2.1 and 4.8 (Figure 3G). In the last two samples, the bulk of Rel is recruited to ribosomes and the 100S ribosome dimer peak is prominent. The latter is a sign of high (p)ppGpp levels: expression of the ribosome dimerization factor HPF is under positive stringent control (43) resulting in formation of 100S ribosome dimers upon entry to the stationary phase (44).
Processive (p)ppGpp synthesis by ribosome-associated Rel does not efficiently dislodge Rel from the ribosome
Next, we reassessed the ‘hopping’ models by testing the effects of nucleotide substrates for (p)ppGpp synthesis on the interaction of wild-type Rel with starved ribosomes in cell lysates (Figure 3E). We supplemented both the lysates and the corresponding sucrose gradients with guanosine substrates (0.5 mM GTP or GDP), both added alone or together with either ATP or its non-hydrolysable analogue, α,β-methyleneadenosine 5′-triphosphate (AMPCPP, 1 mM). Importantly, since disrupted cells constitute <1% of the final gradient volume, the carryover of the intracellular nucleotides is negligible. This set of conditions was used to discriminate between the effects of active (p)ppGpp synthesis (GTP/GDP supplemented with ATP) and substrate binding per se (all the other conditions), since the former was suggested to actively fuel the dissociation of E. coli stringent factor RelA from the ribosome (25). In the presence of the GDP substrate, the Western blot signal of Rel is spread out in the ribosomal fractions, with further addition of ATP or AMPCPP having only a minor effect. Notably we see no difference between ATP and AMPCPP. In the case of GTP, Rel is detected both in the light fractions (free protein) and in the 70S ribosomal peak. Just as in the case of GDP, further addition of either ATP or AMPCPP does not have a significant effect. Taken together, these results suggest that the catalytic activity – or the lack of it (ATP or its substitution ATP for AMPCPP) – does not play a significant role in Rel association with starved ribosomes, as one would expect if acts of (p)ppGpp synthesis would drive the departure of Rel from the ribosome. There is, however, a clear difference between the effect of GDP and GTP on Rel association with ribosomes. This effect could arise from the GTP-dependent action of other A-site binding factors present in lysates, e.g. translational GTPases or different conformations induced by each nucleotide.
A potential drawback of sucrose gradient fractionation experiments is that the SYNTH-competence of the ribosome-associated Rel is not established simultaneously with immunodetection. Therefore, we have additionally probed the connection between ribosomal association and (p)ppGpp synthesis through enzymatic assays. We first performed a competition experiment. While keeping the concentration of wild-type Rel constant, we titrated either SYNTH-inactive D264G Rel (42) or the SYNTH-competent H420E variant compromised in recognition of the tRNA CCA (12) and assayed 3H-pppGpp production in the presence of starved ribosomes (Supplementary Figure S7). Both D264G and H420E Rel are compromised in their interaction with starved ribosomes (Figure 3D), and, when added 1:1 with wild-type Rel, do not inhibit (p)ppGpp synthesis. However, 10-fold excess of the catalytically-inactive D264G variant strongly inhibits production 3H-pppGpp, indicative of D264G Rel outcompeting wild-type Rel on starved ribosomes. Conversely, 10-fold excess of H420G Rel decreases the synthesis rate by merely 3.5-fold. This is probably due to, first, a competitive advantage of the wild-type protein which can be stabilized by tRNA, and, second, that the H420E variant still can be activated by the 70S, but not by deacylated tRNA (12).
To further discriminate between ‘hopping’ and processive synthesis on the ribosome, we titrated wild-type Rel in our reconstituted biochemical system. When the reaction turnover is calculated per starved ribosomal complex (Figure 3F, red trace), the enzymatic activity reaches a plateau when the concentration of Rel is equal to that of the ribosomes (0.5 μM). At higher Rel concentrations the efficiency of 3H-pppGpp production in the reconstituted system does not increase. Importantly, when turnover is calculated per Rel molecule, it decreases, consistent with only (stably) ribosome-bound Rel molecules being activated (Figure 3F, black trace). This behaviour is consistent with Rel processively synthesizing (p)ppGpp while associated with starved complexes rather than the enzyme spending prolonged periods off the ribosome in a catalytically active state upon departure from the ribosome. In the latter case one could expect that, acting catalytically, one starved ribosomal complex would fully activate several Rel molecules; this does not seem to be the case.
The hydrolysis substrate pppGpp does not promote Rel dissociation from starved ribosomal complexes
Given the well-documented antagonistic allosteric coupling between the SYNTH and HD catalytic domains of Rel enzymes (17,22), we tested the effect of the hydrolysis substrate pppGpp on Rel's interaction with ribosomes. However, due to the significant volume of sucrose gradients (12 mL) it is not feasible to perform the experiment in a way that makes the substrate available for the enzyme as it migrates though the gradient. Therefore, we resorted to supplementing only the lysate with 1 mM pppGpp. We see no effect on Rel's association with the ribosome (Supplementary Figure S8). There are at least two possible explanations for this result. First, pppGpp could be diluted in the test tube and thus lose its effect. Second, the association of Rel with the starved complexes could, by promoting its synthesis activity, inhibit pppGpp binding to the HD domain, thus counteracting the potential effect of this substrate.
The synthetase activity of Rel is activated by pppGpp binding to an allosteric site in the NTD region
Next, we characterized the effects of substrates (GDP or GTP) as well as regulators (ribosomal complexes, tRNA, ppGpp and pppGpp) on the synthesis activity of Rel. Since Mn2+ is universally essential for hydrolysis activity of long RSHs such as M. tuberculosis Rel (8), S. equisimilis Rel (16) T. thermophilus Rel (45) and E. coli SpoT (46), we characterized the synthesis activity of the full-length Rel in the absence of divalent manganese ions in order to avoid possible underestimation of the synthesis efficiency due to concomitant (p)ppGpp hydrolysis.
While the ribosome stimulates Rel's synthesis activity 5- to 10-fold, and the ultimate activator – the starved complex – has a significantly stronger effect, ∼50-fold (Figure 4A and B). In good agreement with our earlier results with E. coli RelA (15), deacylated tRNA by itself has no significant effect. As with Rel enzymes from S. equisimilis (16) and M. tuberculosis (19,47), B. subtilis Rel is moderately more efficient in converting GTP to pppGpp than converting GDP to ppGpp. Conversely, E. coli RelA prefers the GDP substrate (15,47). The preference for GTP likely plays a regulatory role since it is GTP consumption, rather than direct (p)ppGpp binding to RNA polymerase that effectuates the transcriptional regulation upon the stringent response in B. subtilis (30,48). Finally, similarly to E. coli RelA (15), while both ppGpp and pppGpp stimulate B. subtilis Rel synthetic activity, ppGpp has a weaker effect.
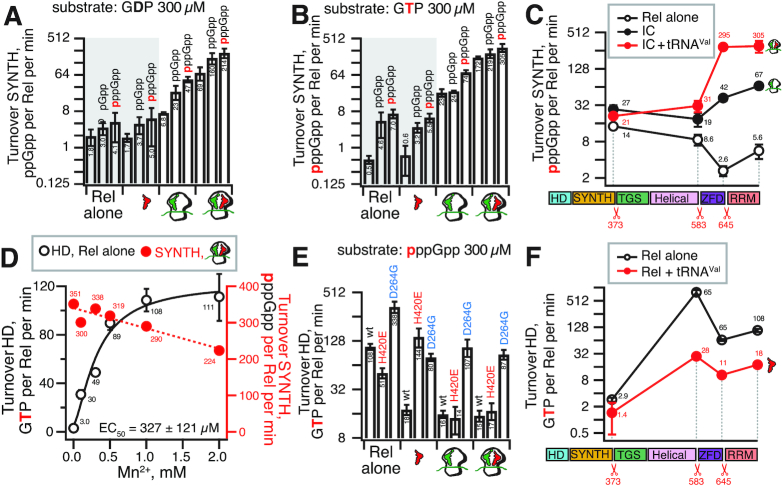
Figure 4.
Role of individual domains in the regulation of Rel's (p)ppGpp synthesis and hydrolysis activities by tRNA, ribosomes, starved ribosomal complexes and pppGpp. Synthase activity of 140 nM wild-type and C-terminally truncated Rel assayed in the presence of 1 mM ATP (A–C) and 0.3 mM of either 3H-labeled GDP (A) or GTP (B and C). Hydrolase activity of 140 nM wt and C-terminally truncated Rel variants assayed in the presence of 0.3 mM of 3H-labeled pppGpp (D–F). As indicated on the figure, the reaction mixtures were supplemented with combinations of 0.5 μM IC (MVF) and tRNAVal (2 μM: A-site) as well as 100 μM ppGpp (A, B and D) or pppGpp (A–C). All experiments were performed in HEPES:Polymix buffer, pH 7.5 at 37°C in the presence of either 5 mM Mg2+ (A–C), increasing concentrations of Mn2+ (D) or 5 mM Mg2+ supplemented with 1 mM Mn2+ (E and F). Error bars represent standard deviations of the turnover estimates by linear regression and each experiment was performed at least three times.
Stimulation of the synthesis activity of B. subtilis Rel by pppGpp implies complex formation between the alarmone and the protein. We used isothermal titration calorimetry (ITC) to study the binding of pppGpp to the NTD-only variant of Rel which is dramatically more soluble than the full-length protein and, just like the full-length protein, is activated by pppGpp (Supplementary Figure S9A). The Rel NTD region binds pppGpp with an affinity of 10.6 ± 0.9 μM (Table 1 and Supplementary Figure S9B). Importantly, the interaction has a stoichiometry close to unity, i.e. one Rel molecule binds one pppGpp molecule. To test the possibility of the heat signal being reflective of the alarmone binding to one of the two active sites, we titrated pppGpp into RelNTD in the presence of saturating concentrations of GDP and a non-hydrolysable ATP analogue AMPCPP (to prevent (p)ppGpp biding in the catalytic pocket of the SYNTH domain), or GDP and AMPCPP combined with EDTA (to remove the Mn2+ ion from the catalytic site of the HD domain and prevent the potential binding of (p)ppGpp by this site). In both cases, both the affinity (KD) or the stoichiometry (n) of pppGpp binding remained largely unperturbed. Taken together, our results suggest the existence of a dedicated pppGpp-binding allosteric site located in the NTD region of Rel.
Table 1.
Thermodynamic parameters of pppGpp binding to NTD domain region fragments of B. subtilis Rel and E. coli RelA as determined by ITC
| Titration | K D, μM | ΔG, kcal mol−1 | ΔH, kcal mol−1 | –TΔS, kcal mol−1 | n |
|---|---|---|---|---|---|
| pppGpp into RelNTD | 10.6 ± 0.9 | −6.67 ± 0.01 | −2.2 ± 0.8 | −4.5 ± 0.4 | 1.1 ± 0.1 |
| pppGpp into RelNTD + AMPCPP + GDP | 9 ± 1 | −6.74 ± 0.07 | −1.8 ± 0.5 | −4.9 ± 1 | 0.9 ± 0.2 |
| pppGpp into RelNTD + AMPCPP + GDP + EDTA | 10 ± 2 | −6.73 ± 0.01 | −2.4 ± 0.8 | −4.3 ± 0.5 | 1.0 ± 0.1 |
| pppGpp into RelANTD | 6.9 ± 0.9 | −6.92 ± 0.02 | −2.8 ± 0.5 | −4.1 ± 0.5 | 0.9 ± 0.5 |
| pppGpp into RelANTD + AMPCPP + GDP + EDTA | 11 ± 1 | −6.76 ± 0.01 | −14 ± 1 | 7.6 ± 0.5 | 0.9 ± 0.2 |
The binding affinities were determined from fitting a single interaction model to the experimental ITC data. Data represent mean values ± standard deviations.
Starved ribosomal complexes actively induce Rel's synthesis activity rather than merely release the auto-inhibition by CTD
Next, we used enzymatic assays in a reconstituted biochemical system to probe the roles of the individual domains in sensing the ribosome and A-site deacylated tRNA. Deletion of the RRM domain had modest effect on activation by starved complexes and pppGpp (Figure 4C). Progressive removal of RRM and ZFD domains abrogates activation by A-site tRNA, although activation by the initiation complex itself remains detectable. In good agreement with earlier results for M. tuberculosis Rel (18,19), when tested in the absence of the ribosomes, B. subtilis Rel NTD has a synthesis activity that is only about two-fold higher than the full-length protein. Thus the data do not support the autoinhibition model sensu stricto, where NTD synthesis activity is suppressed by the CTD, and release of this inhibition upon interaction with the starved ribosome explains the mechanism of Rel/RelA activation (16,49,50). Since the release of the autoinhibition of the NTD by the CTD does not account for the full activity of Rel observed in the presence of starved ribosomes, our results point toward ribosomal complexes playing an active role in induction of synthesis in addition to the release of auto-inhibition.
The ribosome and tRNA inhibit (p)ppGpp hydrolysis by Rel
The synthetic and hydrolytic activities of Rel are mutually exclusive (16,17,22). This motivated us to directly test the effects of the ligands that induce the synthetic activity (ribosomes, tRNA) on Rel's hydrolysis activity. The hydrolase activity of the enzyme tested alone is strictly Mn2+-dependent and peaks at 1 mM of Mn2+ (Figure 4D, empty black circles). Notably, when we tested the effects of increasing Mn2+ concentration on 3H-pppGpp production by Rel activated by starved complexes, only a modest decrease in net SYNTH activity was detected (Figure 4D, filled red circles). This result is in good agreement with suppression of Rel's HD activity by recruitment to the ribosome (see below).
We tested the effects of tRNA, ribosomes and starved complexes (Figure 4E). In good agreement with earlier results with M. tuberculosis Rel (8), tRNA inhibits the hydrolysis activity of B. subtilis Rel, and the inhibition is abrogated when the interaction with the tRNA CCA end is disrupted by the H420E substitution. Both initiation and starved complexes inhibit hydrolysis, and the H420E substitution does not overcome this effect, suggesting that the H420E Rel does bind to the ribosome, although can not be stabilized by the deacylated tRNA. Therefore, we conclude that it is the ribosome itself that inhibits hydrolysis. Synthesis-inactive D264G Rel has elevated hydrolysis activity, consistent with the antagonistic allosteric coupling between SYNTH and HD domains (17,22). This activity is inhibited by ribosomes, albeit, in agreement compromised ribosomal association of the D264G-subsituted protein, less efficiently than in the case of wild-type Rel.
Next, we tested the HD activity of our C-terminally truncated Rel variants, both in the presence and absence of tRNAVal (Figure 4F). Progressive deletion of both RRM and ZFD leads to induction of the hydrolysis activity (Figure 4F), while the synthesis activity is compromised (Figure 4C). Reduction of Rel to an NTD fragment lacking the regulatory CTD near-completely abrogates the hydrolysis activity, in good agreement with our microbiological (12) and ITC experiments (Supplementary Figure S9), as well as biochemical studies of S. equisimilis (16) and C. crescentus (20) Rel enzymes. The inhibitory effect of tRNAVal is lost only when Rel is reduced to its hydrolytically near-inactive NTD.
Taken together, our biochemical results demonstrate that the CTD-mediated transduction of the regulatory stimuli that induce (p)ppGpp synthesis inhibits the hydrolysis activity – and vice versa. This regulatory strategy in the full-length protein utilizes the antagonistic allosteric coupling within the NTD enzymatic core of the protein and efficiently prevents futile enzymatic activity.
E. coli RelA does not stably associate with starved ribosomal complexes due to low affinity to deacylated tRNA
Since E. coli RelA is, arguably, the most well-studied long RSH, we complemented our investigations of B. subtilis Rel with a set of experiments with the E. coli factor. Despite multiple contacts between tRNA and RelA on starved ribosomal complexes (14,28,29), our earlier EMSAs using full-length E. coli RelA failed to detect a stable complex with deacylated tRNA off the ribosome (15). At the same time, Kushwaha and colleagues have recently reported nM-range tRNA affinity of the E. coli RelA fragment containing the TGS domain and part of the Helical domain (27). Therefore, we tested tRNA binding to a set of C-terminal truncations of E. coli RelA (Supplementary Figure S10A), as well as the isolated TGS-Helical fragment (Supplementary Figure S10B). However, we did not detect stable complex formation in either of the cases. While the reason for this discrepancy is unclear, our EMSAs strongly suggest that E. coli RelA is a considerably weaker tRNA binder than B. subtilis Rel.
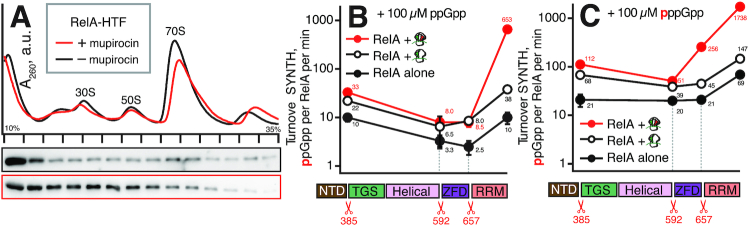
Since deacylated tRNA is the driving force promoting association of Rel and RelA with the ribosome, this implies that E. coli RelA association with starved ribosomes is also less stable. To detect RelA by Western blotting we used an E. coli strain encoding RelA C-terminally tagged with His6-TEV-FLAG3 (HTF) on the chromosome. This tagged RelA displays wild-type-like functionality in live cells (26) (Supplementary Figure S11). While the bulk of RelA is shifted to denser fractions upon amino acid starvation, the protein does not form a stable complex (Figure 5A), demonstrating that indeed E. coli RelA is a significantly weaker binder of starved complexes compared to B. subtilis Rel.
Figure 5.
E. coli RelA NTD region contains the allosteric regulatory site that binds pppGpp. (A) MG1655 E. coli (relA::HTF) expressing C-teminally HTF-tagged RelA was grown in LB liquid medium at 37°C. To induce amino acid starvation bacterial cultures exponentially growing at OD600 0.2 were treated for 20 min with mupirocin added to final concentration of 70 μM that completely abolishes bacterial growth. (B and C) Synthesis activity of wild-type E. coli RelA as well as RelAΔRRM (1–657), RelAΔZFD (1–592), and RelANTD (1–385) mutants by assayed in absence of ribosomes, in presence of 70S initiation complexes and 70S initiation complexes supplemented with cognate deacylated tRNAVal. Experiments were performed in the presence of either 100 μM ppGpp (B) or pppGpp (C). Error bars represent standard deviations of the turnover estimates by linear regression and each experiment was performed at least three times.
The pppGpp-binding site is conserved between B. subtilis Rel and E. coli RelA
To test if, similarly to B. subtilis RelNTD, the allosteric pppGpp binding site of E. coli RelA is also located in the NTD region, we performed a set of enzymatic assays with C-terminally truncated E. coli RelA variants in the presence of either ppGpp (the weaker activator allosteric activator of RelA (15)) or pppGpp (the stronger activator allosteric activator of RelA (15)) (Figure 5B and C). Since the ppGpp alarmone is produced in situ from 3H-GDP, we compared the effects of the two alarmones rather than omitting the alarmone altogether. While B. subtilis Rel lacking the RRM domain behaves very similarly to the wild-type protein (Figure 4C), deletion of the RRM domain strongly compromises activation of E. coli RelA by the A-site tRNA of the starved complex (Figure 5B). A likely explanation is that the low affinity of RelA to starved complexes renders it more sensitive to destabilization of the complex by deletion of the RRM domain. Notably, we do not observe an increase in its synthetic activity for RelANTD lacking the autoinhibitory CTD region as compared to the full-length protein.
All of the truncated variants of E. coli RelA are consistently more SYNTH-active in the presence of pppGpp and, similarly to Rel, this holds even for the NTD-only version. The strength of the effect varies from 2- to 32-fold depending on the construct and whether the protein is tested alone or in the presence of 70S initiation or starved complexes. Direct measurements by ITC confirmed that pppGpp binds to RelANTD with an affinity of 6.9 ± 0.9 μM, and this interaction is insensitive to the addition of GDP, AMPCPP and EDTA, (Table 1 and Supplementary Figure S9EF). This strongly suggests that the interaction is not mediated by the active site, consistent with the alarmone stimulating, rather than inhibiting RelA’s enzymatic activity as it would be expected in the case of direct competition with substrates. Taken together, these results demonstrate that as with B. subtilis Rel, the pppGpp-binding allosteric regulatory site of E. coli RelA is also located in the NTD region of the enzyme.
Model of Rel regulation by starved ribosomal complexes
The results presented in this study are compatible with a recent CRAC study of E. coli RelA (26). While the cryo-EM analysis of RelA bound to starved ribosomal complex suggested that delivery of deacylated tRNA to the ribosomal A-site in a complex with RelA is structurally unlikely (14), crosslinking of tRNA to RelA lacking RRM and ZDF domains was used as an argument for tRNA:RelA complex formation off the ribosome (26). While these truncations, indeed, compromise the association with and activation of Rel/RelA by starved ribosomal complexes (Figures 4C and 5BC), they do not abrogate the interaction with the ribosome, as evident from the increased crosslinking to the Sarcin-Ricin Loop, SRL, upon starvation (26). Simultaneously, removal of RRM and ZDF increases Rel's affinity to tRNA (Figure 2D), likely by rendering the TGS domain more accessible and promoting crosslinking to tRNA. Likewise, abrogation of both tRNA and rRNA crosslinks by the H432E (E. coli RelA numbering) substitution compromising interaction with the CCA end of uncharged A-site tRNA does not necessitate formation of the specific tRNA:RelA complex either: the lack of tRNA and rRNA crosslinks is readily explained by the instability of the mutant RelA:tRNA:ribosome complex (Figure 3C and D) which is strongly stabilized by the A-site tRNA (11–13).
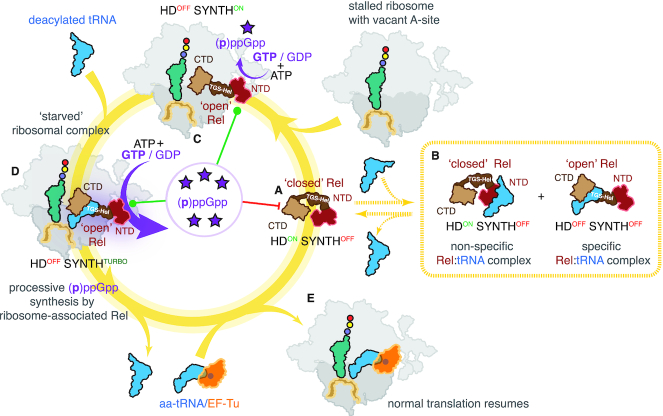
We propose the following model that rationalizes previously published studies (12,14,16,17,21,22,25–29) and our current results (Figure 6). In the absence of amino acid starvation Rel is not ribosome-associated (Figure 3D). Off the ribosome, Rel adopts a ‘closed’ conformation in which tRNA-binding TGS and Helical domains are sequestered (Figure 6A). In this ‘default’ state the protein has net hydrolase activity, i.e. HDON SYNTHOFF (Figure 4AB and E). Upon amino acid starvation ‘closed’ Rel could form a relatively weak (EC50 0.5 μM) complex with deacylated tRNA off the ribosome (Figure 6B) which suppresses the HD activity (HDOFF SYNTHOFF) in H420-dependent manner (Figure 4E). However – at least in the test tube – Rel:tRNA interaction has a strong non-specific component which is mediated by RelNTD region (Figure 2B) and, therefore, its physiological relevance is unclear. Amino acid starvation depletes the pool of ternary complexes formed by aminoacylated tRNA associated with elongation factor EF-Tu in a GTP-bound state, and the ribosomes stall with the A-site vacant. Rel samples the vacant A-site, and upon binding it assumes an ‘open’ conformation (Figure 6C). In this state, the protein presents its TGS and Helical domains which are responsible for specific recognition of tRNA CCA end (Figure 2D). (p)ppGpp hydrolysis is supressed in this conformation, and (p)ppGpp synthesis is moderately activated (HDOFF SYNTHON; Figure 4A, B and E). Subsequent binding of the cognate tRNA stabilizes Rel on the ribosome and serves as the ultimate inducer of (p)ppGpp synthesis (HDOFF SYNTHTURBO) (Figure 6D; Figures 3AC and 4AB). The product of the reaction, pppGpp, binds to a dedicated allosteric site located in the NTD region of both Rel (Figure 4C and F) and RelA (Figure 5B and C), and further stimulates the (p)ppGpp synthesis activity of the HDOFF ribosome-bound factor. Individual acts of catalysis are not incompatible with prolonged association with the ribosome (Figure 3E and F), and multiple turnovers are performed procesessively by the ribosome-associated Rel. Rel dissociates from the ribosome either as a complex with tRNA or by itself after departure of deacylated tRNA; currently available data do not lend decisive support to either of the two scenarios. As the cell exits stringency and the intracellular concentration of starved ribosomes decreases, the fraction of free, HDON SYNTHOFF, factor increases. Binding of the (p)ppGpp substrate to hydrolase-active Rel supresses its synthesis activity via intra-NTD allosteric coupling of the enzymatic domains, driving the efficient cellular switch from (p)ppGpp accumulation to (p)ppGpp degradation.
Figure 6.
Model of Rel regulation by starved ribosomal complexes. (A) Off the ribosome Rel assumes a ‘closed’ conformation concealing the tRNA-binding TGS and Helical domains. The factor is in hydrolytically active (HDON SYNTHOFF). (B) ‘Closed’ Rel can bind deacylated tRNA off the ribosome which suppresses the HD activity (HDOFF SYNTHOFF) as well as non-specifically bind tRNA through the NTD region. Dashed lines signify that while the interactions are observed in biochemical assays, their physiological relevance is unclear. (C) Upon binding to the vacant A-site of a starved ribosome, Rel switches to an ‘open’ conformation which stimulates the (p)ppGpp synthesis and inhibits (p)ppGpp hydrolysis (HDOFF SYNTHON). (D) Opening up primes Rel for specific recognition of deacylated tRNA by its TGS and Helical domains. Binding of deacylated tRNA stabilizes Rel on the ribosome and leads to full activation of the processive (p)ppGpp synthesis activity (HDOFF SYNTHTURBO). (E) Translation resumes once the aminoacylation levels are restored. Accumulating pppGpp acts as an allosteric regulator of Rel. In the case of the HDOFF factor bound to either vacant ribosomal A-site associated with starved complexes, the alarmone binds to the allosteric site in the NTD domain region to stimulate (p)ppGpp synthesis activity. In the case of the HDON free factor, the alarmone acts as an HD substrate, thus suppressing the synthesis activity via inter-NTD regulation.
CONCLUSIONS
Our study further nuances the understanding of how the enzymatic activities of the NTD region are regulated by the CTD region. It is well-established that the regulatory CTD region of the protein transduces signals from the ribosome and the tRNA, whereas the catalytic NTD ‘core’ of the protein is controlled via internal antagonistic allosteric control connecting the SYNTH and HD domains (17,22). While the two enzymatic activities of the NTD are antagonistic, the inactive state of one of the domains does not automatically necessitate activation of the other. For instance, the inhibition of hydrolysis by tRNA does not induce the activity of the SYNTH domain. At the same time, induction of one enzymatic domain efficiently represses the other, thus preventing non-productive idle cycles of synthesis and destruction of the alarmone. The main manifestation of this mechanism is suppression of hydrolysis activity upon association with a vacant ribosomal A-site or a starved ribosomal complex. Finally, our comparative biochemical investigation demonstrates that while the CTD-mediated interaction with the starved ribosome is essential for full activation of the Rel SYNTH domain, the connection between the activity and ribosomal binding is somewhat ‘loose’, with a broad range of acceptable ribosomal affinities for long RSHs. While E. coli RelA is a weak tRNA binder with a correspondingly low affinity to starved ribosomes, B. subtilis Rel has a significantly higher tRNA affinity and is stably recruited to the ribosome upon amino acid starvation. We hypothesize that the stable high-affinity interaction of Rel with starved ribosomal complexes is essential for efficient suppression of the hydrolysis activity upon amino acid starvation. In the case of the synthesis-only RelA this fail-safe mechanism is not needed, and the protein is evolutionary optimized for efficient sampling of the ribosomal population though dissociation and re-binding.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Protein Expertise Platform (PEP) at Umeå University and Mikael Lindberg for constructing plasmids, Gemma C. Atkinson for insightful comments on the manuscript, Tomás de Garay and Refeyn Ltd. for assistance in collecting mass photometry data, Kenn Gerdes and Kristoffer Skovbo Winther for sharing MG1655 relA::HTF E. coli strain (26).
Notes
Present address: Pavel Kudrin, Department of Molecular Biology and Genetics, Aarhus University, 8000 Aarhus C, Denmark.
Contributor Information
Hiraku Takada, Department of Molecular Biology, Umeå University, SE-901 87 Umeå, Sweden; Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, SE-901 87 Umeå, Sweden.
Mohammad Roghanian, Department of Molecular Biology, Umeå University, SE-901 87 Umeå, Sweden; Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, SE-901 87 Umeå, Sweden.
Julien Caballero-Montes, Cellular and Molecular Microbiology, Faculté des Sciences, Université Libre de Bruxelles (ULB), Building BC, Room 1C4 203, Boulevard du Triomphe, 1050 Brussels, Belgium.
Katleen Van Nerom, Cellular and Molecular Microbiology, Faculté des Sciences, Université Libre de Bruxelles (ULB), Building BC, Room 1C4 203, Boulevard du Triomphe, 1050 Brussels, Belgium.
Steffi Jimmy, Department of Molecular Biology, Umeå University, SE-901 87 Umeå, Sweden; Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, SE-901 87 Umeå, Sweden.
Pavel Kudrin, University of Tartu, Institute of Technology, 50411 Tartu, Estonia.
Fabio Trebini, Department of Molecular Biology, Umeå University, SE-901 87 Umeå, Sweden.
Rikinori Murayama, Akita Prefectural Research Center for Public Health and Environment, 6-6 Senshu-Kubotamachi, Akita, 010-0874, Japan.
Genki Akanuma, Department of Life Science, Graduate School of Science, Gakushuin University, Tokyo, Japan.
Abel Garcia-Pino, Cellular and Molecular Microbiology, Faculté des Sciences, Université Libre de Bruxelles (ULB), Building BC, Room 1C4 203, Boulevard du Triomphe, 1050 Brussels, Belgium; WELBIO, Avenue Hippocrate 75, 1200 Brussels, Belgium.
Vasili Hauryliuk, Department of Molecular Biology, Umeå University, SE-901 87 Umeå, Sweden; Laboratory for Molecular Infection Medicine Sweden (MIMS), Umeå University, SE-901 87 Umeå, Sweden; University of Tartu, Institute of Technology, 50411 Tartu, Estonia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Regional Development Fund through the Centre of Excellence for Molecular Cell Technology (to V.H.); Molecular Infection Medicine Sweden (MIMS) (to V.H.); Swedish Research council [2017-03783 to V.H.]; Ragnar Söderberg foundation (to V.H.); Umeå Centre for Microbial Research (UCMR) [postdoctoral grant 2017 to H.T.]; MIMS Excellence by Choice Postdoctoral Fellowship Programme [postdoctoral grant 2018 to M.R.]; Fonds National de Recherche Scientifique [FRFS-WELBIO CR-2017S-03, FNRS CDR J.0068.19, FNRS-PDR T.0066.18]; Joint Programming Initiative on Antimicrobial Resistance [JPI-EC-AMR -R.8004.18]; Program ‘Actions de Recherche Concertée’ 2016–2021 and Fonds d’Encouragement à la Recherche (FER) of ULB; Fonds Jean Brachet and the Fondation Van Buuren (to A.G.P.); the Fund for Research in Industry and Agronomy (FRIA) from the Fonds de la Recherche Scientifique of Belgium (FNRS) (to J.C.M.). Funding for open access charge: Swedish Research council [2017-03783].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hauryliuk V., Atkinson G.C., Murakami K.S., Tenson T., Gerdes K.. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015; 13:298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaca A.O., Colomer-Winter C., Lemos J.A.. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 2015; 197:1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu K., Bittner A.N., Wang J.D.. Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 2015; 24:72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ronneau S., Hallez R.. Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol. Rev. 2019; 43:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atkinson G.C., Tenson T., Hauryliuk V.. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011; 6:e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mittenhuber G. Comparative genomics and evolution of genes encoding bacterial (p)ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 2001; 3:585–600. [PubMed] [Google Scholar]
- 7. Shyp V., Tankov S., Ermakov A., Kudrin P., English B.P., Ehrenberg M., Tenson T., Elf J., Hauryliuk V.. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 2012; 13:835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avarbock D., Avarbock A., Rubin H.. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000; 39:11640–11648. [DOI] [PubMed] [Google Scholar]
- 9. Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M.. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 1991; 266:5980–5990. [PubMed] [Google Scholar]
- 10. Haseltine W.A., Block R.. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1973; 70:1564–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Agirrezabala X., Fernandez I.S., Kelley A.C., Carton D.G., Ramakrishnan V., Valle M.. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013; 14:811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Takada H., Roghanian M., Murina V., Dzhygyr I., Murayama R., Akanuma G., Atkinson G.C., Garcia-Pino A., Hauryliuk V.. The C-terminal RRM/ACT domain is crucial for fine-tuning the activation of ‘long’ RelA-SpoT Homolog enzymes by ribosomal complexes. Front. Microbiol. 2020; 11:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beljantseva J., Kudrin P., Jimmy S., Ehn M., Pohl R., Varik V., Tozawa Y., Shingler V., Tenson T., Rejman D. et al.. Molecular mutagenesis of ppGpp: turning a RelA activator into an inhibitor. Sci. Rep. 2017; 7:41839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loveland A.B., Bah E., Madireddy R., Zhang Y., Brilot A.F., Grigorieff N., Korostelev A.A.. Ribosome*RelA structures reveal the mechanism of stringent response activation. Elife. 2016; 5:e17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kudrin P., Dzhygyr I., Ishiguro K., Beljantseva J., Maksimova E., Oliveira S.R.A., Varik V., Payoe R., Konevega A.L., Tenson T. et al.. The ribosomal A-site finger is crucial for binding and activation of the stringent factor RelA. Nucleic Acids Res. 2018; 46:1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mechold U., Murphy H., Brown L., Cashel M.. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 2002; 184:2878–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R.. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell. 2004; 117:57–68. [DOI] [PubMed] [Google Scholar]
- 18. Jain V., Saleem-Batcha R., China A., Chatterji D.. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006; 15:1449–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Avarbock A., Avarbock D., Teh J.S., Buckstein M., Wang Z.M., Rubin H.. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry. 2005; 44:9913–9923. [DOI] [PubMed] [Google Scholar]
- 20. Ronneau S., Caballero-Montes J., Coppine J., Mayard A., Garcia-Pino A., Hallez R.. Regulation of (p)ppGpp hydrolysis by a conserved archetypal regulatory domain. Nucleic Acids Res. 2018; 47:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gratani F.L., Horvatek P., Geiger T., Borisova M., Mayer C., Grin I., Wagner S., Steinchen W., Bange G., Velic A. et al.. Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 2018; 14:e1007514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamman H., Van Nerom K., Takada H., Vandenberk N., Scholl D., Polikanov Y., Hofkens J., Talavera A., Hauryliuk V., Hendrix J. et al.. A nucleotide-switch mechanism mediates opposing catalytic activities of Rel enzymes. Nat. Chem. Biol. 2020; 16:834–840. [DOI] [PubMed] [Google Scholar]
- 23. English B.P., Hauryliuk V., Sanamrad A., Tankov S., Dekker N.H., Elf J.. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E365–E373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li W., Bouveret E., Zhang Y., Liu K., Wang J.D., Weisshaar J.C.. Effects of amino acid starvation on RelA diffusive behavior in live Escherichia coli. Mol. Microbiol. 2016; 99:571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wendrich T.M., Blaha G., Wilson D.N., Marahiel M.A., Nierhaus K.H.. Dissection of the mechanism for the stringent factor RelA. Mol. Cell. 2002; 10:779–788. [DOI] [PubMed] [Google Scholar]
- 26. Winther K.S., Roghanian M., Gerdes K.. Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-Site. Mol. Cell. 2018; 70:95–105. [DOI] [PubMed] [Google Scholar]
- 27. Kushwaha G.S., Bange G., Bhavesh N.S.. Interaction studies on bacterial stringent response protein RelA with uncharged tRNA provide evidence for its prerequisite complex for ribosome binding. Curr. Genet. 2019; 65:1173–1184. [DOI] [PubMed] [Google Scholar]
- 28. Brown A., Fernandez I.S., Gordiyenko Y., Ramakrishnan V.. Ribosome-dependent activation of stringent control. Nature. 2016; 534:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arenz S., Abdelshahid M., Sohmen D., Payoe R., Starosta A.L., Berninghausen O., Hauryliuk V., Beckmann R., Wilson D.N.. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 2016; 44:6471–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krasny L., Gourse R.L.. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004; 23:4473–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kriel A., Bittner A.N., Kim S.H., Liu K., Tehranchi A.K., Zou W.Y., Rendon S., Chen R., Tu B.P., Wang J.D.. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell. 2012; 48:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pausch P., Abdelshahid M., Steinchen W., Schafer H., Gratani F.L., Freibert S.A., Wolz C., Turgay K., Wilson D.N., Bange G.. Structural basis for regulation of the Opposing (p)ppGpp synthetase and hydrolase within the stringent response orchestrator rel. Cell Rep. 2020; 32:108157. [DOI] [PubMed] [Google Scholar]
- 33. Turnbull K.J., Dzhygyr I., Lindemose S., Hauryliuk V., Roghanian M.. Intramolecular interactions dominate the autoregulation of Escherichia coli stringent factor RelA. Front. Microbiol. 2019; 10:1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamara S., Franc V., Heck A.J.R.. A wealth of genotype-specific proteoforms fine-tunes hemoglobin scavenging by haptoglobin. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:15554–15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murina V., Kasari M., Takada H., Hinnu M., Saha C.K., Grimshaw J.W., Seki T., Reith M., Putrins M., Tenson T. et al.. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J. Mol. Biol. 2019; 431:3568–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Antoun A., Pavlov M.Y., Tenson T., Ehrenberg M.M.. Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol. Proced. Online. 2004; 6:35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sebaugh J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011; 10:128–134. [DOI] [PubMed] [Google Scholar]
- 39. Varik V., Oliveira S.R.A., Hauryliuk V., Tenson T.. HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 2017; 7:11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parker J., Watson R.J., Friesen J.D.. A relaxed mutant with an altered ribosomal protein L11. Mol. Gen. Genet. 1976; 144:111–114. [DOI] [PubMed] [Google Scholar]
- 41. Boutte C.C., Crosson S.. The complex logic of stringent response regulation in Caulobacter crescentus: starvation signalling in an oligotrophic environment. Mol. Microbiol. 2011; 80:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nanamiya H., Kasai K., Nozawa A., Yun C.S., Narisawa T., Murakami K., Natori Y., Kawamura F., Tozawa Y.. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 2008; 67:291–304. [DOI] [PubMed] [Google Scholar]
- 43. Tagami K., Nanamiya H., Kazo Y., Maehashi M., Suzuki S., Namba E., Hoshiya M., Hanai R., Tozawa Y., Morimoto T. et al.. Expression of a small (p)ppGpp synthetase, YwaC, in the (p)ppGpp(0) mutant of Bacillus subtilis triggers YvyD-dependent dimerization of ribosome. Microbiologyopen. 2012; 1:115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Akanuma G., Kazo Y., Tagami K., Hiraoka H., Yano K., Suzuki S., Hanai R., Nanamiya H., Kato-Yamada Y., Kawamura F.. Ribosome dimerization is essential for the efficient regrowth of Bacillus subtilis. Microbiology. 2016; 162:448–458. [DOI] [PubMed] [Google Scholar]
- 45. Van Nerom K., Tamman H., Takada H., Hauryliuk V., Garcia-Pino A.. The Rel stringent factor from Thermus thermophilus: crystallization and X-ray analysis. Acta Crystallogr. F Struct. Biol. Commun. 2019; 75:561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heinemeyer E.A., Geis M., Richter D.. Degradation of guanosine 3′-diphosphate 5′-diphosphate in vitro by the spoT gene product of Escherichia coli. Eur. J. Biochem. 1978; 89:125–131. [DOI] [PubMed] [Google Scholar]
- 47. Sajish M., Kalayil S., Verma S.K., Nandicoori V.K., Prakash B.. The significance of EXDD and RXKD motif conservation in Rel proteins. J. Biol. Chem. 2009; 284:9115–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ratnayake-Lecamwasam M., Serror P., Wong K.W., Sonenshein A.L.. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001; 15:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gropp M., Strausz Y., Gross M., Glaser G.. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J. Bacteriol. 2001; 183:570–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schreiber G., Metzger S., Aizenman E., Roza S., Cashel M., Glaser G.. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 1991; 266:3760–3767. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.