Abstract
Histone modifications and their functional readout serve as an important mechanism for gene regulation. Lysine benzoylation (Kbz) on histones is a recently identified acylation mark associated with active transcription. However, it remains to be explored whether putative readers exist to recognize this epigenetic mark. Here, our systematic binding studies demonstrated that the DPF and YEATS, but not the Bromodomain family members, are readers for histone Kbz. Co-crystal structural analyses revealed a ‘hydrophobic encapsulation’ and a ‘tip-sensor’ mechanism for Kbz readout by DPF and YEATS, respectively. Moreover, the DPF and YEATS family members display subtle yet unique features to create somewhat flexible engagements of different acylation marks. For instance, YEATS2 but not the other YEATS proteins exhibits best preference for Kbz than lysine acetylation and crotonylation due to its wider ‘tip-sensor’ pocket. The levels of histone benzoylation in cultured cells or in mice are upregulated upon sodium benzoate treatment, highlighting its dynamic regulation. In summary, our work identifies the first readers for histone Kbz and reveals the molecular basis underlying Kbz recognition, thus paving the way for further functional dissections of histone benzoylation.
INTRODUCTION
Covalent histone and DNA modifications index the eukaryotic genome and carry key epigenetic messages to regulate the decoding of the genetic information stored in DNA sequence. In this process, modification type- and site-specific readout by effector modules, so-called readers, often serves as an important mechanism to mediate downstream events in cells (1,2).
Histone acetylation (Kac) was the first histone modification identified in 1963 (3). Up to date, over ten types of histone acylation have been identified, including formylation (Kfo) (4), propionylation (Kpr) and butyrylation (Kbu) (5), crotonylation (Kcr) (6), malonylation (Kma) and succinylation (Ksucc) (7), glutarylation (Kglu) (8), 2-hydroxyisobutyrylation (Khib) (9), β-hydroxybutyrylation (Kbhb) (10), benzoylation (Kbz) (11) and lactylation (Kla) (12) (Figure 1A). Sodium benzoate (NaBz) can stimulate histone Kbz by generating benzoyl-CoA (11). NaBz, which can combine with glycine to form hippuric acid, is an FDA-approved drug for hyperammonemia treatment, and a widely used food preservative. There have been a few studies that focus on NaBz effects and safety. Some studies affirm its neuroprotection function, anti-inflammatory activity, therapeutic importance in multiple sclerosis and induction of cancer cell apoptosis (13–16). However, other studies have found that NaBz induced developmental defects and behavior abnormalities in zebrafish larva, significantly impaired memory and motor coordination in mice, and increased attention-deficit hyperactivity disorder symptoms in humans (17–19). The discovery of histone Kbz opens up a new direction for investigating the molecular mechanisms underlying the cellular and physiological impact of NaBz treatment.
Figure 1.
Identification of DPF and YEATS domains as readers for histone benzoylation. (A) Chemical formula of twelve histone lysine acylations. The acylation group is color shaded and benzoylation is highlighted in bold. (B) ITC fitting curves of indicated histone peptides with human DPF domains. Mean KD and standard deviation are shown (N ≥ 3). Un, unmodified peptide. (C) Affinity comparison of Kbz with Kcr or Kac among DPF family members. Red dashes, reference line of equal affinity; Blue dashes, reference line of 1:4 preference. (D) ITC fitting curves of indicated histone peptides with human YEATS domains. Mean KD and standard deviation are shown (N ≥ 3). N.D., not detectable. (E) Affinity comparison of Kbz with Kcr or Kac among YEATS family members. Red dashes, reference line of equal affinity.
The benzoylation group—larger in size, aromatic and hydrophobic—is structurally distinct from acetylation and crotonylation. In vitro studies showed that Kbz can be erased by SIRT2, but not by other sirtuins or histone deacetylases (HDACs); moreover, unlike Kac and Kcr, the Kbz modification mostly occurs to the N-terminal tails of histones, indicating a unique regulatory pathway of Kbz (11). To better explore the biological function of Kbz, there is an urgent need to determine functional reader proteins of histone Kbz.
In humans, there are three major acylation reader families: Bromodomain (BRD), double PHD finger (DPF) domain, and YEATS (Yaf9, ENL, AF9, Taf14 and Sas5) domain proteins. Recently, the ZZ domain of ZZZ3 and the PHD6 domain of MLL4 were also reported to have histone Kac reader activity (20,21). BRD was the first identified histone acetylation reader, which binds Kac through an Asn-lined ‘side-open’ hydrophobic pocket (22,23). In 2008, the DPF domain was reported as a novel family of Kac reader, and structural evidence revealed a ‘dead-end’ hydrophobic pocket for acetyl recognition (22,24–26). In 2014, we found that the YEATS domain can bind Kac through an ‘open-end’ aromatic sandwich pocket (22,27). Subsequent studies from our and other groups further showed that the pockets of YEATS and DPF domains favor the bulkier Kcr over Kac, in contrast with most BRDs (28–33). Therefore, we propose that YEATS and DPF domains have greater potential to accommodate Kbz.
Here we conducted a systematic analysis to explore the reader activities of BRD, DPF and YEATS domains for Kbz. Quantitative binding studies established that DPF and YEATS but not BRD proteins are Kbz readers. The complex structure of MOZDPF bound to H3K14bz revealed a ‘hydrophobic encapsulation’ mechanism for Kbz readout. Complex structures of AF9YEATS-H3K9bz and YEATS2YEATS-H3K27bz revealed a ‘tip-sensor’ mechanism, in which a wider ‘tip-sensor’ pocket contributes to the observed Kbz preference of YEATS2YEATS. Furthermore, we revealed inducible generation of histone benzoylation in cells or in mice upon NaBz treatment, suggesting its dynamic regulation. Collectively, our work identified downstream readers of histone Kbz and provided new clues to the cellular function of histone benzoylation.
MATERIALS AND METHODS
Materials
All histone peptides bearing different modifications (>95% purity) were synthesized at SciLight Biotechnology and are summarized in Supplementary Table S1. Anti-histone antibodies including anti-H4 (PTM-1009, WB 1:2000), anti-pan Kac (PTM101, WB 1:2000) and anti-pan Kbz (PTM762, WB 1:1000, IF 1:500) were obtained from PTM Biolabs. Alexa Fluor 594 (A-11032, 1:1000) antibody was purchased from Invitrogen.
Preparation of expression constructs
The YEATS domains of AF9 (2–138), ENL (1–148) and GAS41 (14–159) were cloned into pET28b vector, and the DPF domains of MOZ (194–323), MORF (211–322), PHF10 (289–410), DPF1 (331–373), DPF2 (270–391) and DPF3 (254–368) as well as the YEATS domain of YEATS2 (201–332) were cloned into a pSUMOH10 vector (modified from pET28b) containing an N-terminal 10 × His-SUMO tag. The bromodomain of BRD9 (131–250) and BRD3 (306–416) were cloned into pGEX-6P-1 vector (Novagen). All mutations were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
Protein expression and purification
All proteins were expressed in Escherichia coli BL21 (Novagen) and induced overnight by 0.2 mM isopropyl β-D-thiogalactoside at 16°C in LB or TB medium. For recombinant YEATS domain and mutant proteins, collected cells were resuspended in 20 mM Tris (pH 7.5), 500 mM NaCl and 5% glycerol and purified as previously described (27). For DPF domain proteins, collected cells were resuspended in 20 mM Tris (pH 7.5), 100 mM NaCl and 5% glycerol and purified as previously described (28). For BRD proteins, collected cells were resuspended in 50 mM HEPES (pH 7.5), 500 mM NaCl and 5% glycerol and purified as previously described (34).
Briefly, harvested cells were disrupted by an EmulsiFlex-C3 homogenizer (Avestin) in their respective resuspension buffers containing 1 mM fresh PMSF. The lysate was further cleared by centrifugation, and the supernatant was applied to a HisTrap column (GE Healthcare) or GST column (GE Healthcare) depending on the protein tag. Next, the column was subjected to extensive washing with high salt (750 and 1000 mM NaCl) resuspension buffer. GST-tagged proteins and His-SUMO-tagged proteins were digested overnight by PreScission protease and ULP1 SUMO protease, respectively. His-tagged proteins were eluted with eluent buffer: 20 mM Tris (pH 7.5), 500 mM NaCl, 5% glycerol and 300 mM imidazole, and then the His tag was cleaved overnight by thrombin (Sigma). All digested proteins were further polished by size exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare) using the AKTA Purifier 10 system. Purified peak fractions were pooled, concentrated, aliquoted and stored at −80°C for future use.
Isothermal titration calorimetry (ITC)
ITC measurement was performed as previously described (27). Briefly, synthetic histone H3 peptides (modified at H32–16K9, H31–25K14, H31–25K18, and H315–39K27) and recombinant proteins were extensively dialyzed against buffer. ITC buffers were the following: 25 mM Tris (pH 7.5), 100 mM NaCl, 5% glycerol for DPF proteins; 25 mM HEPES (pH 7.5), 500 mM NaCl, 5% glycerol for BRD proteins; 25 mM Tris (pH 7.5), 500 mM NaCl, 5% glycerol, 2 mM β-ME for AF9, ENL, GAS41 and related mutant proteins; 25 mM Tris (pH 7.5), 500 mM sodium citrate, 5% glycerol for YEATS2 and its mutant proteins. Protein concentration was measured by A280 nm. Peptide concentrations were measured by weighing of large quantities. The titration was performed using the MicroCal iTC200 system (GE Healthcare) at 15°C for YEATS proteins and 25°C for DPF and BRD proteins. Each ITC titration consisted of 17 successive injections, with 0.4 μl used for the first and 2.41 μl used for the rest. Usually, H3 peptides at 1.0–1.2 mM were titrated into proteins at 0.07–0.1 mM. The resultant ITC curves were processed with Origin 7.0 software (OriginLab) using the ‘One Set of Sites’ fitting model. Detailed thermodynamic parameters of each titration are summarized in Supplementary Table S2.
Crystallization, data collection, and structure determination
Crystallization was performed via the sitting or hanging drop vapor diffusion method at 18°C by mixing equal volumes (0.2–1.0 μl) of AF9YEATS-H32–16K9bz (1:3 molar ratio, 8 mg/ml) or YEATS2YEATS-H324–31K27bz (1:10 molar ratio, 7 mg/ml) or MOZDPF-H31–25K14bz (1:3 molar ratio, 10 mg/ml) with reservoir solution. Reservoir solutions were the following: 0.1 M Bis-Tris (pH 5.5), 200 mM lithium sulphate, 25% w/v PEG 3350 for AF9YEATS-H32–16K9bz complex; 0.1 M Bicine (pH 9.0), 2 M ammonium sulphate for YEATS2YEATS-H324–31K27bz complex; 0.1 M HEPES (pH 7.5), 1.4 M sodium citrate tribasic dihydrate for MOZDPF-H31–25K14bz complex. The co-crystals were briefly soaked in cryoprotectant, comprised of reservoir solution supplemented with 10% glycerol (AF9YEATS-H32–16K9bz complex) or reservoir solution supplemented with 30% glycerol (YEATS2YEATS-H324–31K27bz and MOZDPF-H31–25K14bz complexes), and then flash frozen in liquid nitrogen for data collection.
The diffraction data set was collected at beamline BL17U of the Shanghai Synchrotron Radiation Facility at 0.9791 Å. Selected diffraction images were indexed, integrated, and merged using HKL2000 (35). The structures were determined by molecular replacement using MOLREP (36) with AF9-H3K9ac (PDB ID: 4TMP), YEATS2-H3K27cr (PDB ID: 5IQL), and MOZ-H3K14cr (PDB ID: 5B76) as the search model for AF9YEATS-H32–16K9bz, YEATS2YEATS-H324–31K27bz, and MOZDPF-H31–25K14bz, respectively. Structural refinement was carried out using PHENIX (37), and iterative model building was performed with COOT (38). Detailed data collection and refinement statistics are summarized in Table 1. Structural figures were created using Chimera, LigPlus or PYMOL (http://www.pymol.org/).
Table 1.
Data collection, processing and refinement statistics
| MOZDPF-H3K14bz | AF9YEATS-H3K9bz | YEATS2YEATS-H3K27bz | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 |
| Space group | P3221 | C2 | P422 |
| Cell dimensions | |||
| a, b, c (Å) | 48.2, 48.2, 116.8 | 90.3, 42.2, 90.3 | 89.7, 89.7, 86.9 |
| α, β, γ (°) | 90, 90, 120 | 90, 96.6, 90 | 90, 90, 90 |
| Resolution (Å) | 50–2.0 (2.03–2.00)* | 50.0–2.2 (2.24–2.20) | 50–2.05 (2.09–2.05) |
| No. of reflections | 10 923 | 16 939 | 22,927 |
| Rmerge (%) | 9.1 (41.2) | 13.9 (42.2) | 7.8 (78.7) |
| I/σ (I) | 31.5 (5.3) | 13.3 (2.1) | 26.3 (2.1) |
| Completeness (%) | 97.7 (98.7) | 96.6 (94.0) | 100.0 (100.0) |
| Redundancy | 4.9 (5.5) | 4.0 (3.3) | 9.6 (9.1) |
| Refinement (F > 0) | |||
| Resolution (Å) | 39.3–2.00 | 44.8–2.20 | 44.8–2.05 |
| No. of reflections | 10 882 | 16 874 | 22 922 |
| Rwork/Rfree (%) | 15.8/20.5 | 22.7/27.2 | 19.1/23.4 |
| No. of atoms | |||
| Protein | 943 | 2306 | 2234 |
| Peptide | 193 | 140 | 130 |
| H2O | 89 | 31 | 168 |
| Other ligands | 4 | 51 | |
| B-factors (Å2) | |||
| Protein | 36.9 | 55.7 | 39.7 |
| Peptide | 50.0 | 55.7 | 33.2 |
| H2O | 41.5 | 46.2 | 43.4 |
| Other ligands | 28.8 | 60.3 | |
| RMSD bonds (Å) | 0.006 | 0.003 | 0.008 |
| RMSD angles (°) | 0.773 | 0.65 | 0.899 |
| Ramachandran quality | |||
| Favored (%) | 89.4 | 88.8 | 92.2 |
| Allowed (%) | 10.6 | 11.2 | 7.8 |
| Outliers (%) | 0 | 0 | 0 |
*Values in parentheses are for the highest-resolution shell.
Cell culture, NaBz treatment and histone extraction
All cell lines were tested for mycoplasma contamination according to manufacturer's guide (MycoBlue Mycoplasma Detector, D101–01). Human HEK 293 T, HeLa, HepG2, and colorectal cancer cell line HCT116 were purchased from ATCC (www.atcc.org). Human HEK 293 T and HeLa cells were maintained in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco). HepG2 and HCT116 cells were cultured in ATCC 30–2003 medium and McCoy's 5A Medium (Gibco) respectively, both supplemented with 10% fetal bovine serum. Cells were passaged and cultured for 24 h in indicated medium. Then NaBz was added to the medium and after a 24 h treatment, cells were collected and histone proteins were extracted according to manufacturer's protocols using the epiQuik total histone extraction kit (Epigentek, OP-0006). Histone protein concentration was measured by the BCA method.
Mice experiments
C57BL/6N mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. and housed under SPF conditions. All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee. NaBz was delivered via drinking water for 10 days. Mice were killed by cervical dislocation, and the small intestine was dissected. After removal of mesenteric materials and fats, the small intestine was cut into 1 cm pieces and transferred into a conical tube in 15 ml pre-warmed phosphate buffered saline (PBS) following vigorous shaking to remove the remaining feces and mucus. Next, the sample was applied onto a kitchen strainer, and the tissues were transferred to a new conical tube in 15 ml pre-warmed washing buffer (1 × Hank's Balanced Salt Solution, without Ca2+ and Mg2+, 10 mM HEPES buffer, 5 mM EDTA, 1 mM DTT, 3% FBS). To isolate intestinal epithelial cells, the sample was placed on a platform shaker at 200 rpm, 37°C for 20 min. Then the sample was further applied onto a new kitchen strainer and the flown through cell suspension containing intestinal epithelial cells was centrifuged by 1500 rpm at 4°C for 5 min. Histone proteins’ extraction and quantification were performed as above mentioned.
Immunofluorescence staining
HEK 293 T and HCT116 cells were seeded on coverslips before experiment. After 10 mM NaBz treatment (24 h) or not, cells were washed twice with PBS. Then cells were fixed in 4% formaldehyde for 20 min at room temperature, washed, and permeabilized in 0.5% PBS-T (Triton X-100) on ice for 10 min. Next, cells were washed, and blocked in 3% PBS-B (BSA) for 1 h. After incubation with anti-pan Kbz antibody (1:500) in 3% PBS-B overnight at 4°C, cells were washed and probed with goat anti-mouse, Alexa Fluor 594 (1:1000) in 3% PBS-B (BSA) for 1 h at room temperature. Cells on the coverslips were then washed, covered with mounting medium (with DAPI), and observed under Olympus confocal microscope FluoView FV1000.
Western blot analysis
Protein extracts were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane using a transfer apparatus. After 1 h blocking with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Tween 20) at room temperature, the membrane was incubated with the indicated primary antibodies at 4°C overnight. Then, the membrane was washed three times (10 min for each) with TBST and incubated with a 1: 5,000 dilution of horseradish peroxidase conjugated anti-mouse or anti-rabbit antibody at room temperature for 1 h. Next, the membrane was washed three times (10 min for each) with TBST. Finally, the membrane was developed with an enhanced chemiluminescence detection system. Uncropped western blots are shown in Supplementary Figure S8.
RESULTS
DPF and YEATS domains are readers of histone benzoylation
To screen for histone benzoylation readers, we expressed and purified twelve reader domains of all human DPF and YEATS proteins as well as representative BRD family members (Supplementary Figure S1A), and carried out binding assay using (ITC). Histone peptides with various acylation sites were selected based on the sequence preference of each reader as previously reported (28,31,34,39,40). We found that DPF domains can recognize Kbz (Figure 1B; Supplementary Table S3; Figure S1B). Compared with unmodified peptides, ITC experiments showed that Kbz modification leads to 5- to 10-fold binding enhancement, with KD values of 10.25, 8.43 μM for the MOZ/MORF DPFs and 0.76, 0.64, 0.37, 0.45 μM for the PHF10/DPF1/2/3 (a.k.a. BAF45a/b/d/c) DPFs, respectively (Figure 1B). The binding affinities of DPF domains for Kbz were generally comparable to those for Kac, but weaker than those for Kcr (Figure 1B and C). In particular, the DPF1/2/3 DPFs displayed a clear preference for Kcr than Kbz, especially in regard to enthalpy (Figure 1B).
The YEATS domain also binds benzoylation-modified histones (Figure 1D; Supplementary Table S3; Figure S1C). The binding affinities for Kbz varied substantially among the four human YEATS domain-containing proteins. AF9YEATS showed the strongest binding to H3K9bz (5.97 μM) than that of ENLYEATS (64.68 μM), YEATS2YEATS (21.57 μM) and GAS41YEATS (62.96 μM) to Kbz (Figure 1D). The bindings of AF9YEATS and GAS41YEATS to Kbz were 1.4- to 5.5-fold weaker than their bindings to Kcr or Kac (Kcr > Kac > Kbz), whereas ENLYEATS exhibited stronger binding to Kcr than Kbz or Kac (Kcr > Kbz > Kac). Interestingly, YEATS2YEATS showed the strongest preference for Kbz, with nearly 6- and 2-fold stronger than Kac and Kcr binding, respectively (Kbz > Kcr > Kac) (Figure 1D and E).
BRDs possess a ‘side-open’ pocket (22), and we speculated that they cannot accommodate the bulkier Kbz modification. Meanwhile, Flynn et al. has profiled nearly all human BRDs in order to reveal their binding capacity for larger histone acyl modifications (29). Although the histone Kbz modification had not been reported at that time, they investigated the binding of BRDs to Kbz. Peptide array results showed no appreciable recognition of Kbz by BRDs, albeit BRD9, CECR2 and TAF1 BRDs could bind a larger butyryllysine. On the basis of our structural predictions and the results of others, we selected BRD9 and BRD3 proteins as representatives to study BRD recognition of Kbz. As expected, neither BRD9BRD nor the second bromodomain of BRD3, hereafter BRD3BRD2, could bind benzoylation (Supplementary Figure S1D).
Taken together, we demonstrated for the first time that YEATS and DPF, but not BRD domains, act as readers of histone benzoylation, and that YEATS2YEATS displays the strongest binding preference for benzoyllysine over other acyl modifications.
Molecular recognition of histone H3K14bz by MOZDPF
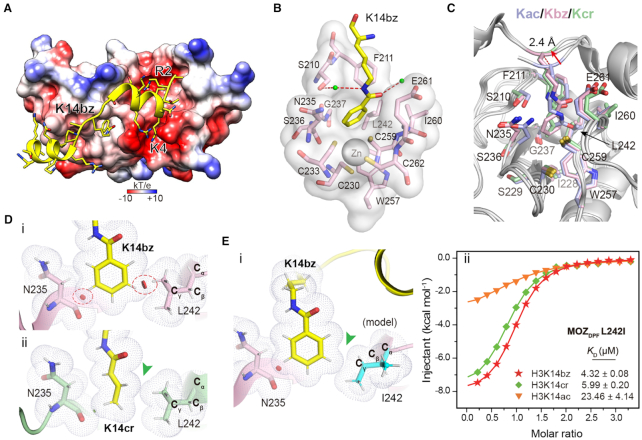
To elucidate the molecular basis for histone benzoylation recognition by DPF domains, we solved the co-crystal structure of MOZDPF with H31–25K14bz peptide at 2.0 Å resolution (Table 1). Upon complex formation, the flexible histone H3 tail forms two induced α-helical structure with the K14bz mark inserted into a central pocket of MOZDPF (Figure 2A and Supplementary Figure S2A). Positively charged histone H3 R2 and K4 residues are inserted into two acidic pockets as described previously (Figure 2A) (24,28,41). The K14bz mark is encapsulated by a hydrophobic pocket constituted by N235–G237, I228–C230, C259–E261 and S210, F211, L242, W257 (Figure 2B and C; Supplementary Figure S2B). In addition to hydrophobic interactions, K14bz is also stabilized by a water-mediated hydrogen bonding interaction with S210 (Figure 2B). Compared with the H3K14ac complex, we observed a 2.4 Å up-lift of the H3K14bz backbone and an expansion of the reader pocket involving residues N235, E261 and S210 (Figure 2C). The overall engagement mode of H3K14bz with MOZDPF is similar to that of H3K14cr. However, the insertion of a bulkier benzoylation group leads to unfavorable close contacts between the benzene ring and residues L242 (side chain) and N235 (main chain) (Figure 2Di). Such high-energy close contacts do not occur to Kcr complex (Figure 2Dii), which may account for the observed Kcr > Kbz preference of MOZDPF.
Figure 2.
Molecular details for histone H3K14bz recognition by MOZDPF. (A) Overall structure of MOZDPF in complex with H31–25K14bz. MOZDPF is shown as electrostatic potential surface ranging from −10 (red) to +10 (blue) kT/e. Histone H3 is shown as yellow cartoon with residues depicted as sticks. (B) Recognition of K14bz mark by the hydrophobic reader pocket of MOZDPF. Small green balls, water molecules; Red dashes, hydrogen bonds; Gray sphere, zinc ion. (C) Superimposition of K14bz-bound (pink), K14ac-bound (light blue, PDB: 4LLB) and K14cr-bound (pale green, PDB ID: 5B76) MOZDPF structures. Key pocket residues are depicted as sticks; Red arrow highlights the up-lift of the H3K14bz backbone. (D) Close contact analyses of MOZDPF structure bound to (i) H3K14bz and (ii) H3K14cr (PDB ID: 5B76). Gray dots denote van der Waals surface of the indicated residues. Large red disk, severe van der Waals overlap; Small green disk, slight van der Waals overlap. Red circles and green arrowhead highlight steric clashes in H3K14bz imbedded structure and spatial compatibility in H3K14cr imbedded structure, respectively. Hydrogens (white sticks) were added for analysis. (E) i, structural modeling of L242I mutation of MOZDPF bound to H3K14bz. Note the loss of ‘close contact’ in L242I mutant (green arrowhead); ii, ITC fitting curves of the indicated histone peptides with L242I mutant. Mean KD and standard deviation are shown (N = 2).
Residues L242 and N235 are located at the bottom of the hydrophobic pocket and they are relatively buried upon Kbz insertion (Figure 2C). We reasoned that the limited conformational freedom of L242 and N235 likely led to engagement tension as reflected by the local close contact detected in the complex structure (Figure 2D). As structural isomers, isoleucine and leucine are different in that a Cγ methyl group in Leu is moved to the Cβ position in Ile. Our modeling analysis suggested that L242I mutation could likely release the close contact between the Cγ methyl of L242 and the benzene ring of Kbz (Figure 2Ei). Binding studies validated such an idea of molecular engineering. Compared with wild-type (WT) MOZDPF, the L242I mutant displayed best preference for histone H3K14bz mark (Kbz > Kcr > Kac) with 2.4-fold binding KD decrease from 10.25 to 4.32 μM and 1.7-fold gain of binding ΔH from −5.02 to −8.33 kcal/mol (Figures 1B and 2Eii; Supplementary Table S2; Figure S2C). We also generated N235S mutant and performed ITC titrations. However, the binding affinity and preference were essentially unaffected (Supplementary Figure S2C and D), suggesting a less dominant role of N235 in determining Kbz preference of MOZDPF.
Molecular recognition of histone H3K9bz by AF9YEATS
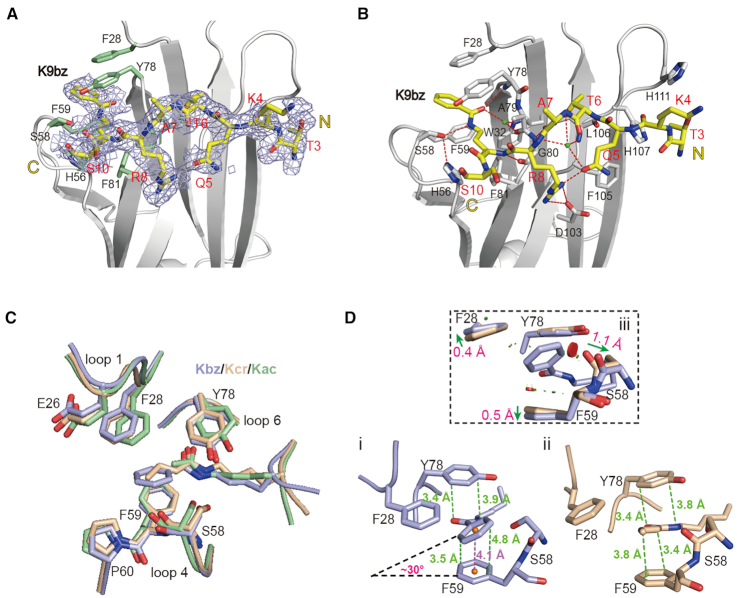
Calorimetric titration revealed that AF9YEATS recognizes histone H3K9bz at 5.97 μM, although the affinity is about 1.4- and 2.6-fold weaker than that of H3K9ac and H3K9cr, respectively (Figure 1E). To explore the underlying structural basis, we crystalized AF9YEATS bound to H32–16K9bz peptide and solved the co-crystal structure at 2.2 Å resolution (Table 1). Based on electron densities, residues 3–10 of histone H3 can be modeled with K9bz sandwiched by aromatic residues H56, F59, F81, F28 and Y78 (Figure 3A and Supplementary Figure S3A). The overall recognition mode of AF9YEATS with H3K9bz is quite similar to that of H3K9ac and H3K9cr (27,32). These include hydrogen bond-stabilized sequence motif recognition involving R8, Q5 and T6 of histone H3 as well as relayed hydrogen bonding interactions with the amide group of acylations by S58 and Y78 of AF9 (Figure 3B and Supplementary Figure S3B). Structural alignment of H3K9bz, H3K9cr and H3K9ac complexes revealed local conformational adjustments of the Kbz insertion pocket (Figure 3C). The benzene ring of Kbz adopts a tilt rotamer conformation that results in an imperfect π–π stacking with F59 as reflected by a dihedral angle of ∼30° (Figure 3Di), which is in sharp contrast with a parallel π–π stacking between the crotonylamide group and F59 (Figure 3Dii). Comparing the Kbz and Kcr complex structures, we detected a 1.1 Å displacement of the main chain carbonyl oxygen of S58, and 0.4–0.5 Å shift of the F28 and F59 benzene rings so as to avoid steric clashes (Figure 3Diii). Our structural comparison suggests the existence of steric tension between the bulkier benzoyl group and pocket residues F28, S58 and F59 (Figure 3D), which together with the imperfect π–π stacking of Kbz with F59 explain the observed Kcr > Kbz preference of AF9YEATS.
Figure 3.
Molecular recognition of histone benzoylation by AF9YEATS. (A) Fo-Fc omit map of H3K9bz peptide contoured at 2.0 σ level. Histone peptide is shown as yellow sticks. Key residues of AF9YEATS are depicted as pale green sticks. Light blue meshes, Fo-Fc omit map. (B) Hydrogen bonding networks between H3K9bz peptide and AF9YEATS. (C) Structure alignment of K9bz-bound (light blue), K9ac-bound (pale green, PDB ID: 4TMP) and K9cr-bound (wheat, PDB ID: 5HJB) structures. (D) π–π stacking analyses of (i) K9bz and (ii) K9cr in AF9YEATS reader pocket and its local conformational adjustments (iii). Distances measured between atoms or centroids are color-coded in green or magenta, respectively. Symbols of close contacts are as described for Figure 2D. Green arrows, conformational displacement comparing Kbz and Kcr readout.
Structural basis for preferential H3K27bz readout by YEATS2YEATS
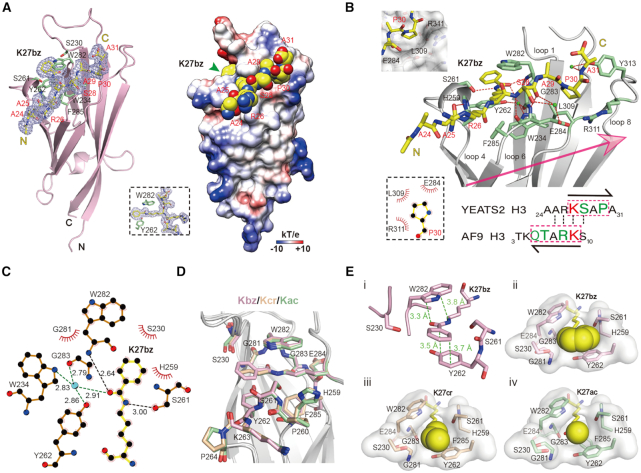
Unlike other human YEATS family members, YEATS2YEATS displayed a unique preference for Kbz mark in our quantitative binding assays (Figure 1E). We also solved the complex structure of YEATS2YEATS bound to H324–31K27bz peptide at 2.05 Å resolution (Table 1). Residues ‘A24-A25-R26-K27bz-S28-A29-P30-A31’ of H3 can be traced according to electron densities; H3K27bz is stapled into a deep aromatic pocket and sandwiched by residues W282 and Y262 (Figure 4A). Unlike AF9 and ENL that recognize acylations of both H3K9 and H3K27 that share a common ‘ARKS’ motif, YEATS2 is specific for H3K27 but not H3K9 (31,32,40). Our complex structure revealed that the H3K27bz peptide binds to YEATS2 in an opposite peptide orientation as compared with the AF9 complex (Figures 4A and 3A). As such, the sequence motif of ‘27KSAP30’ determines the site-specificity of H3K27bz, in which S28 at +1 position is registered by hydrogen bonding interactions and P30 at +3 position is stabilized by hydrophobic contacts with sound shape complementarity (Figure 4B). The ‘KSAP’ motif does not occur to H3K9, thereby YEATS2 is not an effective H3K9 acylation reader (31). In the aromatic sandwich pocket of YEATS2YEATS, K27bz is recognized by relayed hydrogen bonding interactions with S261 and W282, hydrophobic contacts involving S230, G281 and H259, as well as π–π stacking with Y262 and W282 (Figure 4B, C and Ei). The mutant binding assay further confirms the importance of sandwich pocket in Kbz recognition (Supplementary Figure S4A and B). Structural alignment of K27bz, K27cr and K27ac bound complexes of YEATS2YEATS revealed no obvious conformational changes (Figure 4D). Pocket dimension analysis showed that YEATS2 has a wider Kbz binding pocket. Unlike the case of the AF9-K9bz complex, the bulkier benzene ring of K27bz recognized by YEATS2 forms a near perfect π-π stacking with Y262 without apparent tilt (Figure 4Ei). The benzoyl group of K27bz snugly occupies the wide reader pocket of YEAST2YEATS (Figure 4Eii). By contrast, K27cr and K27ac bind loosely within the reader pocket (Figure 4Eiii and iv). Collectively, the observed intimate encapsulation and ideal aromatic stacking of Kbz establish the molecular basis for preferential H3K27bz readout by YEATS2YEATS.
Figure 4.
Structural basis for Kbz recognition by YEATS2YEATS. (A) Overall structure of YEATS2YEATS bound to H3K27bz peptide. Left, ribbon view; Right, electrostatic potential surface view. H3K27bz peptide was covered by Fo-Fc omit map contoured at 2.0 σ level. (B) Binding details between YEATS2YEATS and H3K27bz peptide. A sequence comparison of H3K27 and H3K9 is shown below. Note the positioning of P30 in a hydrophobic pocket of YEATS2YEATS. (C) LigPlot diagram listing critical contacts between K27bz and YEATS2YEATS. The schematic symbols are described in Supplementary Figure S2B. (D) Alignment of K27bz-bound (light pink), K27ac-bound (pale green, PDB ID: 5XNV) and K27cr-bound (wheat, PDB ID: 5IQL) structures. (E) i, π–π stacking analysis of K27bz bound to YEATS2YEATS; Positioning of (ii) Kbz, (iii) Kcr and (iv) Kac into the YEATS2YEATS reader pocket. Green dashes, π–π distances; Yellow spheres, different acyl groups.
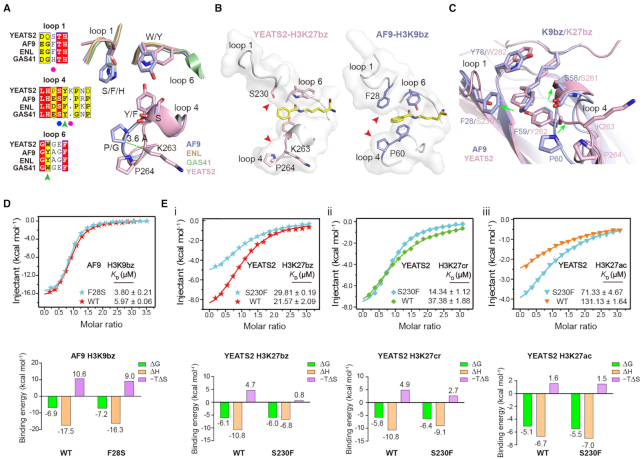
A ‘tip-sensor’ mechanism determines the acylation type preference of YEATS proteins
Despite the conservation of an aromatic sandwich pocket, YEATS2YEATS is quite unique compared with other YEATS family members in term of residue composition of the reader pocket (Figure 5A). The reader pocket of YEATS2YEATS is characteristic of a smaller residue S230 (F28 in AF9/ENL and H43 in GAS41) in loop1 and an insertion of K263 in loop4 (Figure 5A), which leads to a wider end opening of the pocket (Figure 5B). In the AF9-H3K9bz structure, the tip of Kbz is confined by residues F28 from left and ‘S58-F59-P60’ from right (Figure 5C). The tilting of Kbz benzene ring suggests engagement tension between the benzoylation mark and AF9 reader pocket, which explains why the less bulkier and hydrophobic crotonylation mark is most favored by AF9 (Figure 1D). By contrast, the small side chain of S230 and outer shift of the ‘Y262-K263-P264’ segment following K263 insertion create a wider pocket of YEATS2YEATS for optimal accommodation of the H3K27bz mark (Figures 4E and 5C), which explains the observed Kbz > Kcr > Kac mark preference of YEATS2 (Figure 1D).
Figure 5.
A ‘tip-sensor’ mechanism determines acylation type selectivity of YEATS domains. (A) Sequence and structural alignments of reader pocket loops among four human YEATS domains. Magenta circles, ‘tip-sensor’ residues; Blue hexagon, conserved Ser residue for acyllysine amide recognition; Green arrowhead, aromatic sandwiching residues; Light blue, AF9YEATS; Wheat, ENLYEATS (PDB ID: 5J9S); Pale green, GAS41YEATS (PDB: 5XTZ); Light pink, YEATS2YEATS. (B) Comparison of reader pocket between YEATS2YEATS and AF9YEATS. Red arrowheads indicate a wide and narrow end-opening of YEATS2 and AF9, respectively. (C) Structure alignment of YEATS2YEATS-H3K27bz with AF9YEATS-H3K9bz centred on the recognition pocket. Green arrows indicate conformational variations between the two proteins. (D) ITC assays of Kbz binding by WT AF9YEATS and its F28S mutant. Mean KD and standard deviation are shown (N = 4). Thermodynamic parameters of each titration are illustrated below. (E) ITC assays of (i) Kbz, (ii) Kcr and (iii) Kac binding by WT YEATS2YEATS and its S230F mutant. Mean KD and standard deviation are shown (N ≥ 2). Thermodynamic parameters of each titration are illustrated below.
We next performed mutagenesis studies by introducing F28S of AF9YEATS and S230F of YEATS2YEATS. As expected, AF9YEATS F28S displayed ∼1.6-fold binding enhancement for H3K9bz peptide in ITC assay, suggesting relief of engagement tension of the Kbz mark with reduced size of residue 28 (Figure 5D; Supplementary Figure S5A and B). Consistently, YEATS2YEATS S230F displayed ∼1.38-fold binding reduction and ∼37% loss of enthalpy change (ΔH = −10.8 versus −6.8 kcal/mol) when compared with WT YEATS2YEATS (Figure 5Ei), which suggests existence of steric repulsion against Kbz due to the increased bulkiness of S230F. Despite this, an effect of enthalpy–entropy compensation was observed given a lower entropy cost of the S230F mutant (−TΔS = 0.8 versus 4.7 kcal/mol), indicative of favorable hydrophobic contacts between F230 and Kbz after some conformational adjustments (Figure 5Ei). Interestingly, S230F mutant YEATS2YEATS displayed 1.8- and 2.6-fold binding enhancement toward H3K27ac and H3K27cr, respectively, as well as a rearranged mark preference of Kcr > Kbz > Kac (Figure 5Eii and iii; Supplementary Figure S5B and C), which suggests less engagement tension and favorable hydrophobic contacts of S230F with the smaller yet hydrophobic Kcr and Kac marks.
Collectively, our structural comparison and mutagenesis studies revealed a ‘tip-sensor’ mechanism of the YEATS proteins, in which the acylation type preference is achieved by a set of loop residues that can sense the molecular and chemical properties of the tip group of a mark for recognition.
Histone Kbz is an inducible mark in cells and mice
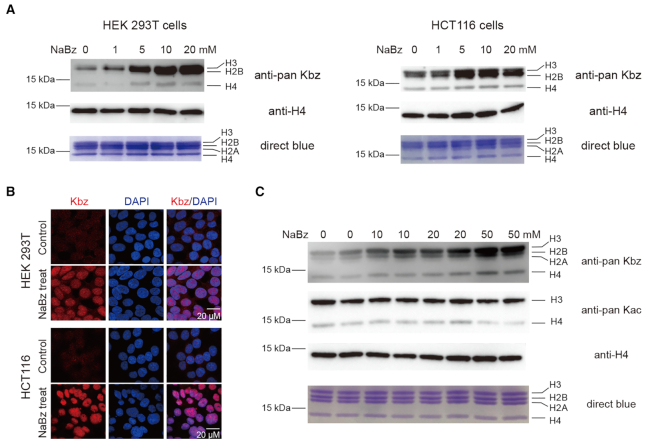
Benzoylation is coupled with anabolic pathways of aromatic compounds. It is distinct from other aliphatic acylations that are mostly coupled with energy metabolic pathways (6,10,12,42–44). We next performed immunoblotting using pan Kbz antibody in mammalian cells and explored the distributions and levels of histone Kbz with or without NaBz treatment. Histone Kbz signal could be detected in multiple mammalian cell lines such as HEK 293T, HeLa, HCT116 and HepG2 (Figure 6A and Supplementary Figure S6). Following 24-h treatment with NaBz at various concentrations, the signals of histone Kbz were upregulated in a dose-dependent manner. The inducible histone Kbz signal is most obvious on H3 and H2B, suggesting an active regulation centred on H3 and H2B among all four core histones. Immunofluorescence staining studies in HEK 293T and HCT116 cells revealed that the nuclear Kbz signals were lightened up by 10 mM NaBz treatment, further confirming the inducible nature of histone Kbz in cells (Figure 6B).
Figure 6.
Histone benzoylation is an inducible epigenetic mark in cells and mice. (A) Western blot analysis of core histone Kbz levels in response to the indicated concentrations of NaBz treatment in HEK 293T and HCT116 cells. Unmodified histone H4 was used as a loading control. (B) Immunofluorescence analysis of Kbz levels in HEK 293T and HCT116 cells. DNA was stained with DAPI. Control, cells without NaBz treatment. (C) Western blot analysis of core histone Kbz and Kac levels in mouse intestinal epithelial cells after the indicated concentrations of NaBz water feeding. Unmodified histone H4 was used as a loading control. Assays were performed in duplicates by using two mice for each concentration.
NaBz has been widely used as a food additive (usually at a concentration of around 0.1% w.t.) (45), which raises an interesting question regarding the occurrence and inducible nature of histone Kbz in animal. To explore this, we performed NaBz water feeding studies in mice. As shown in Figure 6C, without NaBz water feeding, weak histone Kbz signal could be detected in mouse intestinal epithelial cells. Remarkably, after ten days of NaBz water feeding at concentrations of 10, 20 and 50 mM, the histone Kbz signal is clearly boosted in mouse intestinal epithelial cells. The upregulated histone Kbz signal is most abundant in H2B, H3 and to a lesser extent in H4. By contrast, the pan Kac level of histone was not stimulated by NaBz treatment and even displayed slight downregulation in mouse intestinal epithelial cells (Figure 6C). Collectively, the above results support inducible and dynamic regulation of histone Kbz in cells and animal; and the reader-mediated Kbz recognition events reported in this study likely play a functional role in the yet to be established Kbz regulatory pathways.
DISCUSSION
Over the past two decades, a diverse array of histone modifications have been characterized in distinct chemical types and modification sites, among which histone benzoylation, serotonylation and lactylation are recently discovered ones (11,12,46). A major working mechanism centred on histone modification and its regulation is that so-called ‘writer’, ‘eraser’ and ‘reader’ exist in cells to generate, eliminate and interpret such an ‘epigenetic code’ for genetic message decoding at the chromosomal level. More than twenty different histone Kbz marks have been identified in human and mouse cell lines, and they are enriched at promoter regions of actively transcribed genes. Based on mass spectrum counting, global Kbz level at sites like H3K9 and H3K14 could be upregulated to a level comparable to Kac upon NaBz treatment (11). Thus, the abundance and the inducible feature of histone Kbz call attention to an important topic regarding the functional readout of histone Kbz. In this study, we performed binding profiling and structural studies to search for histone Kbz readers through a candidate approach. We demonstrated here that human DPF and YEATS but not BRD domains are the readers for histone benzoylation. Remarkably, YEATS2 showed best preference for Kbz (Figure 1E), suggesting an important role of YEATS2-Kbz interaction in gene regulation.
Our structural analyses revealed the molecular basis underlying Kbz readout. Firstly, the BRD domains has a ‘side-open’ pocket with limited dimension, thus being good for Kac but not for bulkier Kcr/Kbz readout. Secondly, the DPF domains contain a ‘dead-end’ hydrophobic pocket that best fits an aliphatic Kcr mark of 4-carbon in length. The aromatic Kbz group can also be accommodated by the reader pocket of DPFs through hydrophobic encapsulation. Yet, an optimal binding with Kbz is compromised by a few repulsive close contacts involving residues N235 and L242 in the case of MOZDPF (Figure 2E), which may explain the observed binding preference of Kbz < Kcr by DPFs (Figure 1B). Based on structural analyses, we successfully designed a MOZDPF mutant, L242I, which displayed a switched preference and enhanced binding for Kbz likely due to released engagement tension (Figure 2E).
Most residues constituting the acylation reader pocket among the DPF family members are conserved, except for residues at positions S210 and N235 of MOZDPF (Supplementary Figure S7A). S210 is replaced by a Gly residue in PHF10 and an Asp residue in DPF1/2/3, while N235 is replaced by a Ser residue in MORF and an Arg residue in DPF1/2/3. Structurally, the absolutely conserved residue L242 constitutes one contact surface for Kbz and the more divergent ‘S210-N235’ residue pair constitutes another (Supplementary Figure S7B). The subtle yet characteristic pocket residue difference may cooperatively modulate acylation mark preference among different DPF family members (Figure 1C).
The YEATS domain is unique as its ‘open-end’ reader pocket allows a ‘tip-sensor’ mechanism for Kbz recognition. The ‘tip-sensor’ loops 1 and 4 of YEATS2 are characteristic of a small residue S230 and insertion of K263 such that a wider pocket is created for Kbz-favorable readout (Figure 5A and B). Although benzoylation promoted histone binding by AF9/ENL/GAS41 YEATS domains, these YEATS family members are better ‘designed’ as Kcr rather than Kbz readers given the observed 2.0- to 5.5-fold binding reduction (Figure 1E). In molecular detail, the ‘tip-sensor’ loops of AF9/ENL/GAS41 have a bulkier residue (Phe or His) in place of S230 and do not contain an inserted K263, which leads to engagement tension upon Kbz but not Kcr insertion (Figure 3C and D). The occurrence of epigenetic reader module is under evolution pressure for the benefit of recognition and regulation. The YEATS domain adopts an immunoglobin fold and we have proposed the antibody-like ‘loop evolution’ hypothesis regarding the reader pocket design of YEATS (22). The observed Kbz > Kcr > Kac preference of YEATS2, though being mildly selective, has an exquisite and conserved molecular set-up of the ‘evolved’ pocket residues, suggesting a functional adaption of YEATS2 to the prevalence of Kbz/Kcr-regulated pathways. Such a fact is reminiscent of the molecular ‘design’ of the PWWP domain of the de novo DNA methyltransferase DNMT3A that displayed a mild yet conserved preference for H3K36me2 over H3K36me3, reflecting the prevalence of NSD1/2, a histone H3K36me2 methyltransferase and DNMT3A co-regulated pathways in shaping the intergenic DNA methylation landscape (47).
Metabolism and gene expression reciprocally regulate each other, and epigenetic modifications often serve as a molecular link of the two fundamental processes (44,48,49). The recently characterized non-acetyl histone acylations underscore a role of metabolites as a message carrier once they are deposited into chromatin as epigenetic marks. The level of histone acylation usually depends on the availability of its cognate metabolic precursor and is regulated in a dynamic manner. For example, a recent study showed that histone crotonylation and acetylation oscillate inversely during the yeast metabolic cycle, and elevated histone crotonylation upon nutrient limitation is coincident with periodical upregulation of fatty acid β-oxidation pathway and suppression of pro-growth genes; remarkably, a normal crotonyl-CoA metabolism as well as Kcr readout by the YEATS domain of Taf14 are necessary to maintain transcription oscillations during this process (50). Here, we confirmed inducible upregulation of histone benzoylation in cell and animal in response to NaBz treatment, and characterized DPF and YEATS family proteins, especially YEATS2, as histone benzoylation readers. Though future work is warranted, these findings offer a new avenue to pinpoint the role of benzoyl-CoA metabolism in gene regulation and beyond. Moreover, our structural studies provide key insights into molecular engineering of preference-switched acylation readers, an intriguing strategy that has been adopted to explore the functional importance of histone modification and its readout (51,52).
DATA AVAILABILITY
The atomic coordinates and structure factors of MOZDPF-H3K14bz, AF9YEATS-H3K9bz and YEATS2YEATS-H3K27bz have been deposited in the Protein Data Bank under accession codes 6LSB, 6LS6 and 6LSD, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff members at beamline BL17U of the Shanghai Synchrotron Radiation Facility and Dr S.F. at the Tsinghua Center for Structural Biology for their data collection assistance and the China National Center for Protein Sciences in Beijing for facility support.
Author Contributions: H.L. conceived the project; X.R. designed and performed the experiments under the guidance of H.L.. Y.Z., Z.X. and N.H. helped with the experiments. Y.L., X.G., D.W. and X.S. offered expert comments. H.L. and X.R. wrote the manuscript with input from X.S.
Contributor Information
Xiangle Ren, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Yang Zhou, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Zhaoyu Xue, Center for Epigenetics, Van Andel Institute, Grand Rapids, MI 49503, USA.
Ning Hao, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Yuanyuan Li, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Xiaohuan Guo, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Daliang Wang, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China.
Xiaobing Shi, Center for Epigenetics, Van Andel Institute, Grand Rapids, MI 49503, USA.
Haitao Li, MOE Key Laboratory of Protein Sciences, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing 100084, China; Tsinghua-Peking Center for Life Sciences, Beijing 100084, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [91753203 and 31725014 to H.L., 31871283 and 31922016 to Y.L.]; National Key Research Development Program of China [2020YFA0803300 and 2016YFA0500700 to H.L.]; Beijing Natural Science Foundation [5182014 to Y.L.]; Beijing Municipal Science & Technology Commission for Beijing Nova Program [Z181100006218068 to Y.L.]; Young Elite Scientists Sponsorship Program by China Association for Science and Technology [YESS20170075 to Y.L.]. Funding for open access charge: National Natural Science Foundation of China [31725014].
Conflict of interest statement. None declared.
REFERENCES
- 1. Strahl B.D., Allis C.D.. The language of covalent histone modifications. Nature. 2000; 403:41–45. [DOI] [PubMed] [Google Scholar]
- 2. Waddington C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012; 41:10–13. [DOI] [PubMed] [Google Scholar]
- 3. Phillips D.M. The presence of acetyl groups of histones. Biochem. J. 1963; 87:258–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiang T., Zhou X., Taghizadeh K., Dong M., Dedon P.C.. N-formylation of lysine in histone proteins as a secondary modification arising from oxidative DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C., Falck J.R., Peng J., Gu W., Zhao Y.. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics. 2007; 6:812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N. et al.. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011; 146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J.D., Zhao Y.. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics. 2012; 11:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L., Park J., Chen Y., Huang H., Zhang Y. et al.. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014; 19:605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dai L., Peng C., Montellier E., Lu Z., Chen Y., Ishii H., Debernardi A., Buchou T., Rousseaux S., Jin F. et al.. Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 2014; 10:365–370. [DOI] [PubMed] [Google Scholar]
- 10. Xie Z., Zhang D., Chung D., Tang Z., Huang H., Dai L., Qi S., Li J., Colak G., Chen Y. et al.. Metabolic regulation of gene expression by histone lysine beta-Hydroxybutyrylation. Mol. Cell. 2016; 62:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang H., Zhang D., Wang Y., Perez-Neut M., Han Z., Zheng Y.G., Hao Q., Zhao Y.. Lysine benzoylation is a histone mark regulated by SIRT2. Nat. Commun. 2018; 9:3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang D., Tang Z., Huang H., Zhou G., Cui C., Weng Y., Liu W., Kim S., Lee S., Perez-Neut M. et al.. Metabolic regulation of gene expression by histone lactylation. Nature. 2019; 574:575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brahmachari S., Jana A., Pahan K.. Sodium benzoate, a metabolite of cinnamon and a food additive, reduces microglial and astroglial inflammatory responses. J. Immunol. 2009; 183:5917–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khasnavis S., Pahan K.. Sodium benzoate, a metabolite of cinnamon and a food additive, upregulates neuroprotective Parkinson disease protein DJ-1 in astrocytes and neurons. J. Neuroimmun. Pharmacol. 2012; 7:424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Modi K.K., Jana M., Mondal S., Pahan K.. Sodium benzoate, a metabolite of cinnamon and a food additive, upregulates ciliary neurotrophic factor in astrocytes and oligodendrocytes. Neurochem. Res. 2015; 40:2333–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yilmaz B., Karabay A.Z.. Food Additive Sodium Benzoate (NaB) activates NFkappaB and induces apoptosis in HCT116 Cells. Molecules. 2018; 23:723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khoshnoud M.J., Siavashpour A., Bakhshizadeh M., Rashedinia M.. Effects of sodium benzoate, a commonly used food preservative, on learning, memory, and oxidative stress in brain of mice. J. Biochem. Mol. Toxic. 2018; 32:e22022. [DOI] [PubMed] [Google Scholar]
- 18. Gaur H., Purushothaman S., Pullaguri N., Bhargava Y., Bhargava A.. Sodium benzoate induced developmental defects, oxidative stress and anxiety-like behaviour in zebrafish larva. Biochem. Biophys. Res. Commun. 2018; 502:364–369. [DOI] [PubMed] [Google Scholar]
- 19. Beezhold B.L., Johnston C.S., Nochta K.A.. Sodium benzoate-rich beverage consumption is associated with increased reporting of ADHD symptoms in college students a pilot investigation. J. Atten. Disord. 2014; 18:236–241. [DOI] [PubMed] [Google Scholar]
- 20. Mi W., Zhang Y., Lyu J., Wang X., Tong Q., Peng D., Xue Y., Tencer A.H., Wen H., Li W. et al.. The ZZ-type zinc finger of ZZZ3 modulates the ATAC complex-mediated histone acetylation and gene activation. Nat. Commun. 2018; 9:3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Jang Y., Lee J.E., Ahn J., Xu L., Holden M.R., Cornett E.M., Krajewski K., Klein B.J., Wang S.P. et al.. Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF. Nat. Commun. 2019; 10:2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao D., Li Y., Xiong X., Chen Z., Li H.. YEATS domain-a histone acylation reader in health and disease. J. Mol. Biol. 2017; 429:1994–2002. [DOI] [PubMed] [Google Scholar]
- 23. Owen D.J., Ornaghi P., Yang J.C., Lowe N., Evans P.R., Ballario P., Neuhaus D., Filetici P., Travers A.A.. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 2000; 19:6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dreveny I., Deeves S.E., Fulton J., Yue B., Messmer M., Bhattacharya A., Collins H.M., Heery D.M.. The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification. Nucleic Acids Res. 2014; 42:822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lange M., Kaynak B., Forster U.B., Tonjes M., Fischer J.J., Grimm C., Schlesinger J., Just S., Dunkel I., Krueger T. et al.. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008; 22:2370–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeng L., Zhang Q., Li S., Plotnikov A.N., Walsh M.J., Zhou M.M.. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010; 466:258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y., Wen H., Xi Y., Tanaka K., Wang H., Peng D., Ren Y., Jin Q., Dent S.Y., Li W. et al.. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014; 159:558–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiong X., Panchenko T., Yang S., Zhao S., Yan P., Zhang W., Xie W., Li Y., Zhao Y., Allis C.D. et al.. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016; 12:1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flynn E.M., Huang O.W., Poy F., Oppikofer M., Bellon S.F., Tang Y., Cochran A.G.. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure. 2015; 23:1801–1814. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Q., Zeng L., Zhao C.C., Ju Y., Konuma T., Zhou M.M.. Structural insights into histone crotonyl-lysine recognition by the AF9 YEATS domain. Structure. 2016; 24:1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao D., Guan H., Zhao S., Mi W., Wen H., Li Y., Zhao Y., Allis C.D., Shi X., Li H.. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016; 26:629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y., Sabari B.R., Panchenko T., Wen H., Zhao D., Guan H., Wan L., Huang H., Tang Z., Zhao Y. et al.. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell. 2016; 62:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andrews F.H., Shinsky S.A., Shanle E.K., Bridgers J.B., Gest A., Tsun I.K., Krajewski K., Shi X.B., Strahl B.D., Kutateladze T.G.. The Taf14 YEATS domain is a reader of histone crotonylation. Nat. Chem. Biol. 2016; 12:396–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Muller S., Pawson T. et al.. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012; 149:214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 36. Vagin A., Teplyakov A.. Molecular replacement with MOLREP. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:22–25. [DOI] [PubMed] [Google Scholar]
- 37. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 39. Hsu C.C., Shi J., Yuan C., Zhao D., Jiang S., Lyu J., Wang X., Li H., Wen H., Li W. et al.. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev. 2018; 32:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wan L., Wen H., Li Y., Lyu J., Xi Y., Hoshii T., Joseph J.K., Wang X., Loh Y.E., Erb M.A. et al.. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature. 2017; 543:265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qiu Y., Liu L., Zhao C., Han C., Li F., Zhang J., Wang Y., Li G., Mei Y., Wu M. et al.. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev. 2012; 26:1376–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Porter A.W., Young L.Y.. Benzoyl-CoA, a universal biomarker for anaerobic degradation of aromatic compounds. Adv. Appl. Microbiol. 2014; 88:167–203. [DOI] [PubMed] [Google Scholar]
- 43. Hirschey M.D., Zhao Y.. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteomics. 2015; 14:2308–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao T., Diaz-Hirashi Z., Verdeguer F.. Metabolic signaling into chromatin modifications in the regulation of gene expression. Int. J. Mol. Sci. 2018; 19:4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Del Olmo A., Calzada J., Nunez M.. Benzoic acid and its derivatives as naturally occurring compounds in foods and as additives: Uses, exposure, and controversy. Crit. Rev. Food Sci. Nutr. 2017; 57:3084–3103. [DOI] [PubMed] [Google Scholar]
- 46. Farrelly L.A., Thompson R.E., Zhao S., Lepack A.E., Lyu Y., Bhanu N.V., Zhang B., Loh Y.E., Ramakrishnan A., Vadodaria K.C. et al.. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature. 2019; 567:535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weinberg D.N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K.N., Horth C., McGuire J.T., Xu X., Nikbakht H. et al.. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019; 573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li X., Egervari G., Wang Y., Berger S.L., Lu Z.. Regulation of chromatin and gene expression by metabolic enzymes and metabolites. Nat. Rev. Mol. Cell Biol. 2018; 19:563–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ye C.Q., Tu B.P.. Sink into the epigenome: histones as repositories that influence cellular metabolism. Trends Endocrin. Met. 2018; 29:626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gowans G.J., Bridgers J.B., Zhang J., Dronamraju R., Burnetti A., King D.A., Thiengmany A.V., Shinsky S.A., Bhanu N.V., Garcia B.A. et al.. Recognition of histone crotonylation by Taf14 links metabolic state to gene expression. Mol. Cell. 2019; 76:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Noh K.M., Wang H., Kim H.R., Wenderski W., Fang F., Li C.H., Dewell S., Hughes S.H., Melnick A.M., Patel D.J. et al.. Engineering of a histone-recognition domain in Dnmt3a alters the epigenetic landscape and phenotypic features of mouse ESCs. Mol. Cell. 2015; 59:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klein B.J., Vann K.R., Andrews F.H., Wang W.W., Zhang J., Zhang Y., Beloglazkina A.A., Mi W., Li Y., Li H. et al.. Structural insights into the pi-pi-pi stacking mechanism and DNA-binding activity of the YEATS domain. Nat. Commun. 2018; 9:4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of MOZDPF-H3K14bz, AF9YEATS-H3K9bz and YEATS2YEATS-H3K27bz have been deposited in the Protein Data Bank under accession codes 6LSB, 6LS6 and 6LSD, respectively.