Abstract
The increasing availability of detailed structural information on many biological systems provides an avenue for manipulation of these structures, either for probing mechanism or for developing novel therapeutic agents for treating disease. This has been accompanied by the advent of several powerful new methods, such as the ability to incorporate non-natural amino acids or perform fragment screening, increasing the capacity to leverage this new structural information to aid in these pursuits. The abundance of structural information also provides new opportunities for protein engineering, which may become more and more relevant as treatment of diseases using gene therapy approaches become increasingly common. This is illustrated by example with the TGF-β family of proteins, for which there is ample structural information, yet no approved inhibitors for treating diseases, such as cancer and fibrosis that are promoted by excessive TGF-β signaling. The results presented demonstrate that through several relatively simple modifications, primarily involving the removal of an α-helix and replacement of it with a flexible loop, it is possible to alter TGF-βs from being potent signaling proteins into inhibitors of TGF-β signaling. The engineered TGF-βs have improved specificity relative to kinase inhibitors and a much smaller size compared to monoclonal antibodies, and thus may prove successful as either as an injected therapeutic or as a gene therapy-based therapeutic, where other classes of inhibitors have failed.
Keywords: TGF-beta, protein engineering, cell signaling, inhibitor
1. Introduction
There has been an exponential increase in the amount of detailed structural information about all classes of proteins, including membrane proteins, that has occurred over the past 20 years 1. This increase has largely been driven by improvements in the capacities for structure determination using X-ray crystallography 2, 3, and while there were some indications this trend was beginning to ebb, it is likely only to be momentary, as recent advances in single particle cryo-electron microscopy 4 and cryo-electron tomography 5 are likely to push boundaries to enable the determination of larger and even more complex macromolecular structures at atomic resolution.
The increasing availability of detailed structural information on many biological systems provides an avenue for manipulation of these structures, either for probing mechanism or for developing novel therapeutic agents for treating disease. Traditionally, to probe mechanism, prominent residues in the active site or regulatory regions are substituted and evaluated in functional assays. To develop novel therapeutic agents, compounds are identified by screening chemical libraries and in turn the activity of the hit compounds are optimized by establishing structure activity relationships (SAR) and by altering the physicochemical properties for drug-likeness. Though these still represent important avenues, innovative new approaches, such as incorporation of non-natural amino acids for probing mechanism 6, 7, or fragment screening for discovery of novel therapeutic agents 8, have emerged and are now becoming increasingly used as alternatives to conventional approaches.
The objective of this article is to present, by way of example with the TGF-β family of secreted signaling proteins, how protein engineering approaches can be used for probing mechanism or for developing novel therapeutic agents. The idea of protein engineering is by no means new, and indeed some of the most important therapeutic agents used today, such as recombinant insulin for treating diabetes 9 or vaccines protecting us from deadly viruses 10, are a product of protein engineering. There is nonetheless a renewed interest in protein engineering for development of novel therapeutic agents, based on the recent success in reprogramming patient-derived T-cells with engineered chimeric antigen receptors 11 for treatment of hematologic malignancies, such as relapsed acute lymphoblastic leukemia 12 and refractory large B-cell lymphoma 13. This opens the door to an entirely new era in medicine, in which drugs are not manufactured and administered, but instead patient’s own cells are genetically modified with coding sequences for engineered proteins that confer a therapeutic benefit 14. To illustrate how protein engineering has been used with the TGF-β family of growth factors, which are studied in my lab, I will first present an overview of the TGF-β pathway and how TGF-βs bind their receptors to initiate a signaling response. I will then describe the engineered TGF-β monomer my lab has developed, which inhibits TGF-β signaling and has potential use as a therapeutic agent for treating cancer or fibrosis, or for probing mechanism.
2. TGF-β family signaling
Transforming growth factor beta isoforms, TGF-β1, −β2, and −β3, are 25 kDa secreted homodimeric signaling proteins. They are present only in vertebrates and perform functions essential for the long-term survival of humans and other higher vertebrates, including regulation of the adaptive immune system 15 and coordination of wound healing 16. The TGF-βs belong to a highly diversified family of signaling proteins, with 33 members in humans, known as the TGF-β family 17. The other members of the family include the bone morphogenetic proteins (BMPs) which regulate embryonic patterning, the closely related growth and differentiation factors (GDFs) which regulate cartilage and skeletal development, the activins (Acts) and inhibins (Inhs) which regulate the release of pituitary hormones, and others, such as Müllerian inhibiting substance (MIS) that regulate sex determination during embryonic development.
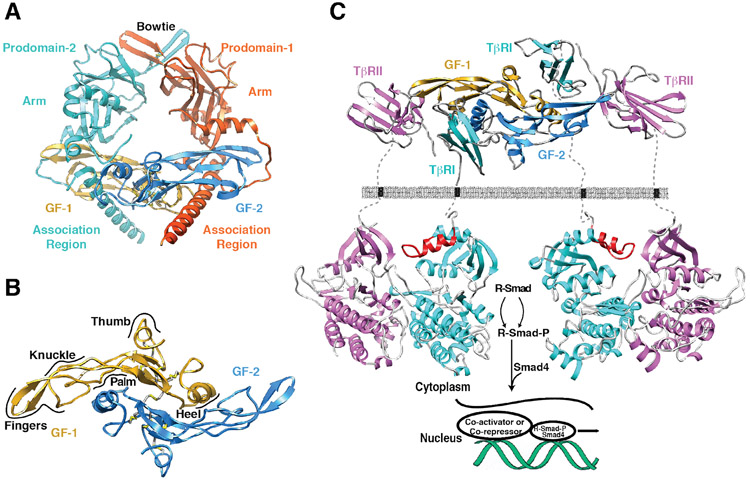
The TGF-βs, and other proteins of the family, are synthesized are pre-proproteins, and after maturation and secretion, are found as pro-complexes in which the mature growth factor homodimer is non-covalently bound by a homodimer of its pro-domain (Fig. 1A) 18-21. The mature growth factor homodimers, which consist of two cystine-knotted monomers held together in most, but not all cases by a single inter-chain disulfide bond (Fig. 1B) 22-34, signal by binding and bringing together two serine-threonine kinase receptors, known as receptor types I and II (Fig. 1C) 35. The assembly of receptor types I and II into heterotetrameric receptor complexes 36 leads to the activation of the type I receptor kinase, which in turn activates cytoplasmic effectors, known as receptor-regulated Smads or R-Smads 37 (Fig. 1C). R-Smads, together with the co-mediator Smad, Smad4, are dependent on other co-activators and co-repressors to effect transcriptional responses 38. This dependence on co-activators and co-repressors, coupled with the variation of such factors from cell to cell, is thought to underlie the pronounced cell- and context-dependent activities characteristic of GFs of the family 39.
Figure 1.
Structure of a representative TGF-β family member, its type I and type II receptors, and a schematic of it signaling mechanism. A. Structure of proTGF-β1 (PDB 3RJR) 20, in which the disulfide-linked growth factor homodimer, is surrounded and non-covalently bound by a homodimer of its prodomain. Growth factor (GF) monomers (GF-1 and GF-2) are depicted in dark blue and gold, while the prodomain monomers are depicted in cyan and orange. B. Structure of the disulfide-linked TGF-β2 homodimer (PDB 2TGI) 24. GF-1 and GF-2 are shaded as in panel A and the four intramolecular disulfides, and one inter-molecular disulfide, are depicted in yellow. GF monomers are described as a curled left hand, in which the heel of one hand packs into the palm of the other. Other features of the of GF fold, including the fingers, the knuckles, and the thumb, are labeled. C. Structure of the heterotetrameric signaling complex formed by a TGF-β3 homodimer, and two molecules each of its cognate type I and type II receptor, TβRI and TβRII. Structure of the TGF-β3 homodimer bound to the TβRI and TβRII ectodomains was determined experimentally. Structure of the TβRI kinase domain was determined experimentally (PDB 1IAS), while that of TβRII or the TβRI:TβRII kinase domain complex has not been reported (structure shown for the TβRII kinase domain is that of ActRIIB, PDB 2QLU). GF-1 and GF-2 are shaded as in panel A and TβRI and TβRII are depicted in cyan and magenta, respectively. GS domain of TβRI, which regulates its activity, is depicted in red.
There are seven type I and five type II receptors in mammals 17. Thus, there are many more GFs than receptors, and by necessity, there is significant growth factor-receptor promiscuity. The type I receptors of the family additionally couple to and activate only two classes of R-Smads: the more recently evolved members of the family, such as TGF-βs, activins, nodal and some of the GDFs and BMPs, such as GDF-9, −11, and −15 and BMP-15, bind and signal through type I receptors that activate R-Smads 2, 3, while the more distantly-related GDFs and BMPs, such as GDF-1, −3, −5, −7 and BMP-2, −3, −4, −5, −6, and −7, bind and signal through type I receptors that couple to and activate R-Smads 1, 5, and 8 17. The functional diversity that can be attained through intrinsic differences in signaling is therefore limited as there only two types of intracellular Smad signals induced by the 33 GFs of the family. The interactions of the GFs with the signaling receptors therefore represents only a portion of the ‘molecular recognition’ that results in the distinctive activities of the proteins of the family. The multitude of accessory binding proteins, which regulate access of the GFs to the signaling receptors and thereby determine which cells and groups of cells are targeted by TGF-β family GFs, instead provide much of the molecular recognition that results in the distinctive activities of GFs of the family.
3. TGF-βs bind and assemble their receptors in a distinct manner
TGF-β1, −β2, and −β3 are unique among the proteins of the family in that they are the only known GFs that bind and signal through the TGF-β type II receptor, TβRII17, 40, 41. Through purified component binding studies, it also known that TGF-β1 and TGF-β3, the two TGF-βs that have high intrinsic affinity for TβRII, bind and assemble TβRII, and their cognate type I receptor TβRI, in an ordered manner, first by binding TβRII and then by recruiting TβRI 42, 43. The BMPs and GDFs, which exhibit promiscuous binding to both type I and type II receptors, have in contrast been shown through purified component binding studies to bind their type I and type II receptors in manner that is largely independent of one another 25, 44.
These findings, together with differences in the promiscuity of receptor binding, hinted that the TGF-βs/activins and BMPs/GDFs might differ in the manner by which they bind and assemble their receptors into signaling complexes.
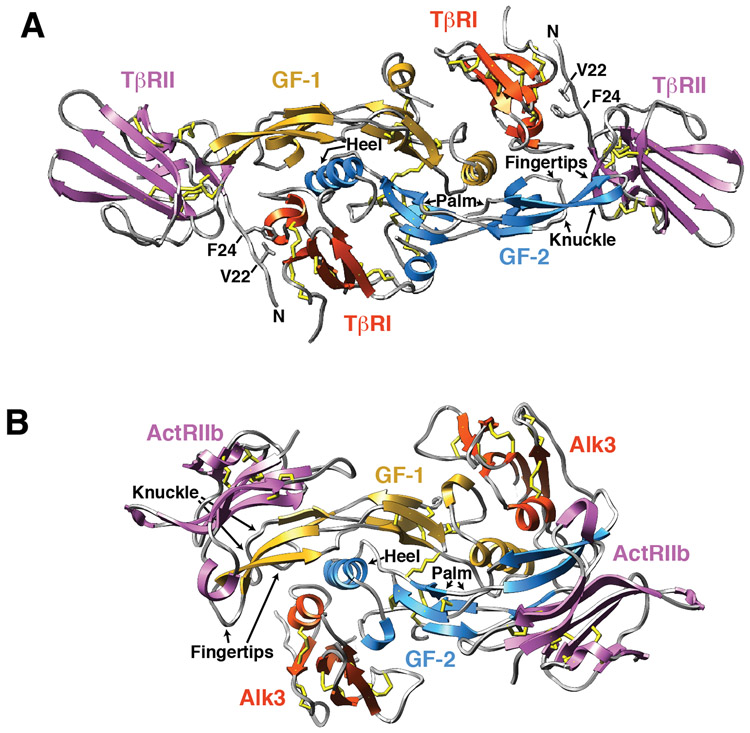
These differences in receptor binding have been borne out by the structures of the TGF-β and BMP type I type II receptor ternary complexes determined over the past several years. The structures of the ternary complexes show that although the signaling proteins and receptors of the BMP and TGF-β subfamilies share the same overall fold (Fig. 2 and Fig. 3, respectively), they nevertheless bind their receptors in a distinct manner (Fig. 4A-B) 25, 44-50. The BMP type I and type II receptors bind to the underside of “fingers” and “knuckle”, respectively, and do not contact one another (Fig. 4b), while the TGF-β type I and type II receptors bind to the underside of the “fingers” and to the “fingertips”, respectively, and have extensive contact (Fig. 4A). The direct contact between the type I and type II receptors in the TGF-β complex, but not the BMP, has been shown to be responsible for the pronounced stepwise manner by which TGF-βs bind and assemble their type I and type II receptors into a signaling complex 46, 51, thus distinguishing them both structurally and functionally from all other proteins of the family.
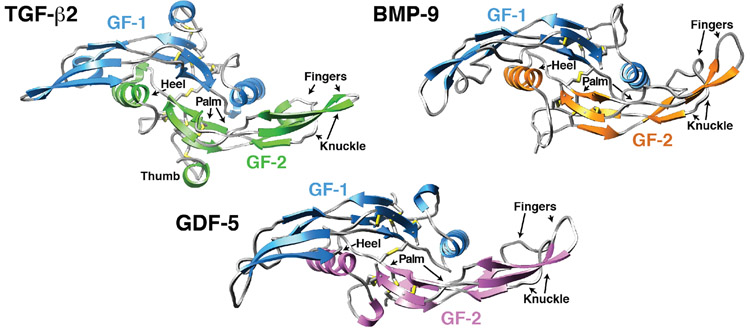
Figure 2.
Structures of three representative TGF-β family GFs. Structures are disulfide-bonded homodimers, with a single inter-chain disulfide connecting each of the monomers. Structures are color coded, with one of the monomers in blue and the other monomer in another shade. Sulfur atoms of the intramolecular and intermolecular disulfides are depicted in yellow. Structures shown correspond to the following PDB entries: TGF-β2 – 2TGI 24, BMP-9 – IZKZ 23, and GDF-5 – 1WAQ 31.
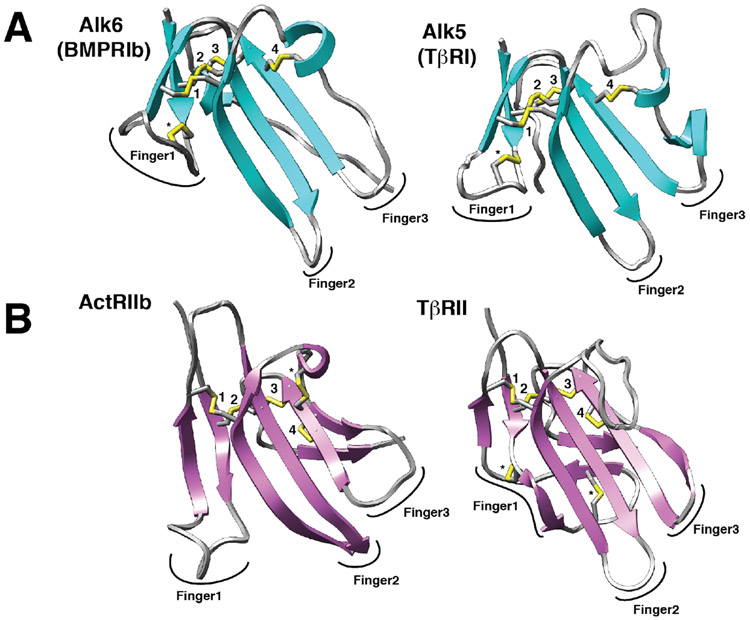
Figure 3.
Structures of representative type I and type II receptors of the TGF-β family. A, B. Structures of type I and type II receptors (A and B, respectively) shown as ribbon diagrams. Disulfide bonds are depicted as sticks with yellow sulfur atoms. Four disulfide bonds that are positionally conserved in all type I and type II receptors of the TGF-β family are labeled 1 – 4. Disulfide bonds that are unique are labeled with an asterisk (*). The three fingers of the type I and type II receptors that define them as having a three-finger toxin fold are indicated. Structures shown correspond to the following PDB entries: Alk6 – 3EVS 103, Alk5 – 2L5S 104, ActRIIb – 1NYU 105, and TβRII – 1M9Z 106.
Figure 4.
Structures of two representative GF:type I receptor:type II receptor complexes. A. Structure of the TGF-β receptor complex shown as a ribbon diagram with the two growth factor monomers shown in blue and gold, the two bound TβRII ectodomains in magenta, and the two bound TβRI ectodomains in orange. Disulfides are shown as sticks, with the sulfur atoms depicted in yellow. Structure shown is from PDB 2PJY 46. Structure of the BMP receptor complex shown as a ribbon diagram with the same shading scheme as that used in panel A. Structure shown is from PDB 2H62 44.
4. TGF-βs in human disease and the need for novel inhibitors
TGF-β1, −β2-, and −β3 play essential roles in tumor suppression 52, maintain the balance between the tolergenic and immunogenic arms of the adaptive immune system 15, maintain the extracellular matrix 53, and orchestrate wound healing 16. The importance of the TGF-βs in these processes is demonstrated through inherited human diseases, such as hereditary nonpolyposis colorectal cancer (HNPCC) which is caused by mutations in TβRI 54 and the connective tissue disorders, Marfan’s and Loey’s-Dietz syndromes, which are caused by mutations in the proTGF-β binding matrix protein fibrillin-1, or TβRI, that lead to too much or too little TGF-β signaling, respectively 55, 56. TGF-β signaling also plays a significant role in soft tissue cancers and fibrotic disorders 52, 53, 57, where the TGF-β pathway remains intact, but excessive TGF-β signaling drives disease progression. Though a role for excessive TGF-β signaling in promoting cancer progression may seem at odds with its tumor suppressive growth inhibitory activity, many cancer cells dysregulate their cell cycle, which antagonizes TGF-β’s ability to inhibit cell growth 58. The many tumor promoting activities of the TGF-β’s, including their ability to potently suppress the immune system, induce EMT, promote cell migration and invasion, and increase the population of cancer stem cells, nonetheless remain intact 59. This, coupled with overexpression of TGF-βs at levels 10- to 100-fold higher than in non-transformed cells, is thought to underlie TGF-β’s role in the growth and invasiveness of many cancers, including those of the breast, brain, bone, prostate, pancreas, colon, lung, and bladder 60-66. The fibrotic disorders caused by dysregulated TGF-β signaling, which include idiopathic pulmonary fibrosis (IPF), renal fibrosis, cardiac fibrosis, and coronary restenosis, are a result of hyperactive TGF-β signaling following tissue injury or disease progression that leads to the accumulation of extracellular matrix proteins 53.
The disease promoting activities of the TGF-βs have prompted vigorous TGF-β inhibitor development and testing in clinical trials 67, 68. TGF-β inhibitors include small molecule TGF-β receptor kinase inhibitors, antisense oligonucleotides, and synthetic peptides, or protein-based biologics, such as TGF-β or TGF-β receptor neutralizing antibodies, TGF-β receptor traps, or antibodies that interfere with TGF-β maturation. Though pre-clinical and clinical studies have shown that TGF-β inhibitors offer significant promise for the treatment of cancer and fibrosis 59, 69-81, to date none have been approved for use in humans 67. Small molecule TGF-β receptor kinase inhibitors have progressed slowly in clinical settings, primarily because they lack specificity for TβRI 82, resulting in a narrow therapeutic window 83. Biologics offer promise as alternatives to SMRKIs due to their inherent high specificity, but they also have the limitation that they tend to occupy the vascular space and penetrate poorly into dense tissues such as tumors 84, 85.
5. TGF-βs high specificity for TβRII provides an avenue for inhibitor design
To address the need for novel TGF-β inhibitors, it occurred to us that it might be possible to specifically inhibit TGF-β signaling by developing a modified form of TGF-β1 or −β3 that retained its high inherent affinity for TβRII, but was fully disrupted in terms of its ability to bind and recruit TβRI. This approach, if successful, would have several advantages over existing inhibitors, for example it would be expected to have high target selectivity, like antibodies and unlike small molecule TGF-β receptor kinase inhibitors, but at the same time to have a greater propensity to penetrate dense tissues, like small molecule TGF-β receptor kinase inhibitors, but unlike antibodies.
Though one might consider designing such an inhibitor by introducing residue substitutions that selectively disrupt TβRI binding, this approach was considered unlikely to be viable since our previous experience showed us that the folding of the TGF-βs to form the intricate cystine knot within each monomer and inter-chain disulfide was very sensitive to residue substitutions 86, with single substitutions sometimes leading to the formation of little to no native disulfide-liked dimer. The other reason is that TGF-β homodimers are notoriously insoluble, thus posing a significant challenge for delivery, even if variants unable to bind TβRI could be generated.
To overcome these challenges, we considered covalently monomeric TGF-β1 or TGF-β3, formed by substituting the cysteine residue that normally forms the inter-chain disulfide bond, Cys77, with serine. TGF-β1 C77S and TGF-β3 C77S had been previously reported 51, 87 and importantly these monomers had at least one of the essential characteristics for the development of a dominant negative inhibitor, specifically the retention of high affinity TβRII binding. This characteristic was predicted from the structure of the TGF-β receptor complex, which showed that TβRII binds to residues from a single TGF-β monomer 41, 46, 49. Though possessing this one very desirable characteristic, they nonetheless possessed one very undesirable characteristic, namely sinaling activity that was only slightly (ca. 10-fold) diminished relative to the corresponding disulfide-linked homodimer 51, 87. This was not predicted from the structure of the TGF-β receptor complex since TβRI was shown to bind to a composite interface formed by both monomers of TGF-β, as well as TβRII.
To reconcile this, we hypothesized that that the signaling capacity of TGF-β1 C77S or TGF-β3 C77S arise from their ability to non-covalently dimerize and in turn assemble the same heterotetrameric receptor complex as native disulfide-linked TGF-β homodimers 51. To prevent the formation of such complexes, we reasoned this might be possible by replacing the heel helix, which connect β-strands 4 and 5 and forms a substantial component of both the hydrophobic dimer interface and TβRI binding site, with a flexible loop. The elimination of α-helix 3 should interfere with self-association of the monomers, and binding and recruitment of TβRI, but should not impair TαRII binding. Though uncertain, it was hypothesized that removal of the heel helix, which includes a large number of hydrophobic residues that interdigitate and pack against hydrophobic residues in the palm of the opposing monomer and replacement of this with a flexible loop bearing several charged residues, might decrease the hydrophobic character enough so that solubility of the engineered monomer is improved relative to that of either TGF-β homodimers or full TGF-β monomers (TGF-β1 or −β3 C77S), which are also poorly soluble.
6. TGF-β engineered monomer
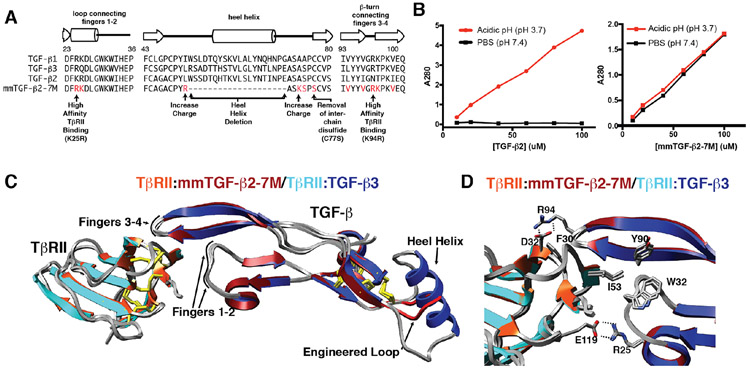
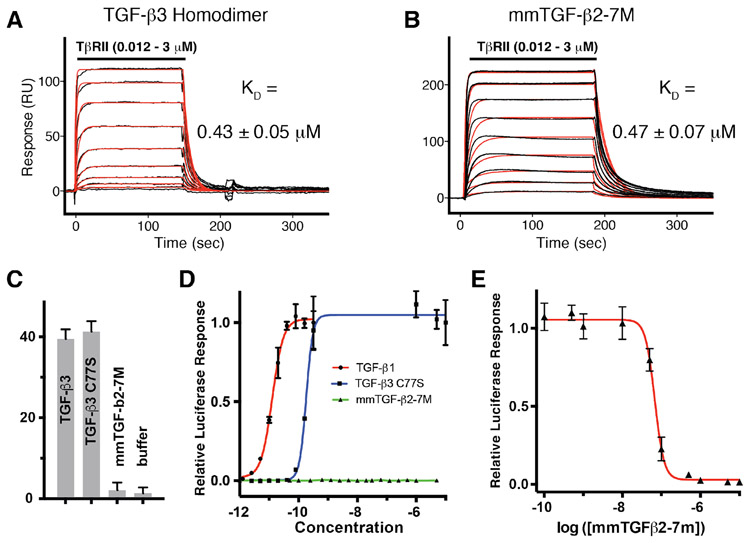
The simple design described above, in which Cys77 was substituted with serine and the heel helix of the TGF-βs was removed and replaced with a short flexible loop to form a so-called TGF-β mini monomer (Fig. 5A), was investigated and remarkably had all the desired design characteristics, including retention of the same growth factor fold and manner of TβRII binding relative to that of TGF-β1 and −β3 dimers (Fig. 5C-D), the same high affinity TβRII binding as TGF-β1 and TGF-β3 dimers (Fig. 6A-B), an inability to bind TβRI (Fig. 6C) and signal (Fig. 6D), and inhibitory activity against TGF-β1, −β2, and −β3, with potencies (IC50s) in the range of 20 – 60 nM (Fig. 6E) 88 Though only a minor component of the design, an additional property was greatly improved solubility relative to TGF-β2 dimers (Fig. 5B) 88.
Figure 5.
Engineered TGF-β monomer and characterization of its structure and solubility. A. Design of the engineered TGF-β monomer, known as mmTGF-β2-7M, which is based on the backbone of TGF-β2. mmTGF-β2-7M, in addition to lacking the heel helix and bearing a substitution of Cys77 with Ser, also has two substitutions to increase the charge in the loop that serves to replace the heel helix and seven substitutions in the loops connecting fingers 1-2 and 3-4 that contact TβRII. B. Solubility of TGF-β2 dimers (left panel) and mmTGF-β2-7M (right panel). Solubility was assessed by measuring the absorbance at 280 nm of the supernatant after TGF-β2 or mmTGF-β2-7M are diluted from an acidic stock where they are highly soluble into either acidic solution (pH 3.7) or phosphate buffered saline (PBS) at neutral pH (pH 7.4) and centrifuged. C. Overlay of the 1.8 Å crystal structure of mmTGF-β2-7M:TβRII complex (dark red and orange ribbons, respectively) with one of the TGF-β3 monomers and its bound TβRII from the 3.0 Å crystal structure of the TGF-β3:TβRII:TβRI complex (PDB 2PJY, TGF-β3 monomer and TβRII shown in dark blue and cyan ribbon, respectively; TβRI not shown for clarity). Newly created loop in mmTGF-β2 (red) which takes the place of the heel (α3) helix in TGF-β2 is depicted in red. D. Overlay as in panel C, but expanded to show the near identity of critical hydrophobic and hydrogen-bonding/electrostatic interactions shown previously to be essential for high affinity TGF-β3:TβRII binding 40, 42. Figure is adapted and reproduced with permission from Kim, et. al, J. Biol. Chem., 292, 7173-7188 (2017).
Figure 6.
Receptor binding properties and cell-based inhibitory activity of the engineered monomer. A-B. SPR sensorgrams for injection of a two-fold dilution series from 3 – 0.012 μM of TβRII ectodomain monomer over immobilized avi-TGF-β3 or avi-mmTGF-β2-7M. Sensorgrams were fitted to a 1:1 binding model – raw data is shown in black and the fitted curve is shown in red. C. Time resolved FRET assay for ligand-mediated assembly of TβRI:TβRII complexes. Preassembled TGF-β3:TβRII-His (1:2), TGF-β3 C77S:TβRII-His (1:1), and mmTGF-β2-7M:TβRII-His (1:1) complexes at a concentration of 250 nM were incubated with 50 nM biotinylated TβRI-Avi and 2 nM Tb-anti-His and 30nM SA-665 for 2 hours at room temperature as previously described. Buffer control (orange bars) contained only 2 nM Tb-anti-His and 30nM SA-665. D. TGF-β luciferase reporter activity for TGF-β1, TGF-β3 C77S, and mmTGF-β2-7M. The solid lines, colored red and blue, correspond to the fitted curves to derive the EC50 (green line for mmTGF-β2-7M was not fit due to the lack of signaling activity for this variant). E. TGF-β luciferase reporter activity for cells treated with a sub-saturating concentration of TGF-β1 (8 pM) with increasing concentration of mmTGF-β2-7M added. The solid red line corresponds to the fitted curve for mTGF-β2-7M to derive the IC50. Figure is adapted and reproduced with permission from Kim, et. al, J. Biol. Chem., 292, 7173-7188 (2017).
The combination of properties that make the TGF-β mini monomer a favorable inhibitor may seem like a stroke of luck, but in fact several of these, including the ability to fold and bind TβRII with high affinity, were anticipated based on previous studies. The ability to fold was anticipated based on the previous structure of an “open” TGF-β3 homodimer previously determined in our laboratory 41, in which the core of monomer, including the TβRII binding fingertips, retained its structure in the absence of any contacts with the other monomer. To increase the likelihood of isolating natively folded protein, the TGF-β mini monomer was based on the backbone of TGF-β2 88, which from our previous studies was known to fold the most efficiently of all three TGF-βs 86, but at the same time to bind TβRII with low affinity 42. To engender the TGF-β2 mini monomer with high affinity TβRII binding, Lys25 and Lys94, along with five other residues, were substituted with the corresponding residues from TGF-β1 and TGF-β3, which we knew from our earlier studies could confer TGF-β2 with ability to bind TβRII with high affinity 40, 42.
The properties of design of which we were less certain were the inability to bind and recruit TβRI and the improved solubility. These properties were anticipated based on the structure, but we had no a priori information to indicate this would be successful or not. To investigate TβRI binding, we developed a highly sensitive time-resolved fluorescence energy resonance energy transfer (TR-FRET) assay to monitor assembly of TβRI and TβRII into a complex by TGF-βs and showed that the TGF-β3 homodimer, but as well the full TGF-β3 monomer, TGF-β3 C77S, led to a TR-FRET signal 38 - 42-fold above background (Fig. 6C) 88. The engineered monomer, which we designated as the TGF-β2 mini monomer with 7 substitutions, or mmTGF-β2-7M, in contrast, led to a TR-FRET signal indistinguishable from background (Fig. 6B) 88. The fact that TGF-β3 C77S could assemble a complex under these assay conditions, but mmTGF-β2-7M could not, indicated that mmTGF-β2-7M was likely devoid of any capacity to bind and recruit TβRI. To investigate solubility, we diluted TGF-β dimers, as well as the full TGF-β monomer and mmTGF-β2-7M from acidic stocks where they were highly soluble into phosphate buffer at neutral pH and quantitated how much protein precipitated versus remained in solution (Fig. 5B) 88 Though an improvement in solubility was expected, based on the removal of the heel helix, which includes a large number of hydrophobic residues, the degree of this improvement was nonetheless striking. The reason for this has is not known, but may be that elimination of the heel helix reduced the longest possible stretch of hydrophobic amino acids to below a critical value, a property well known to be important increasing protein solubility 89.
Though we describe this protein as an engineered monomer, it is in fact only covalently monomeric, not monomeric in solution. This was shown by sedimentation velocity analytical ultracentrifugation experiments in which the sedimentation profiles could only be fit assuming mmTGF-β2-7M was undergoing a monomer/dimer equilibrium in solution 88. The fact that mmTGF-β2-7M has some propensity to non-covalently dimerize is not surprising given that some remnants (i.e. hydrophobic residues) of the dimer interface remain. The fact that mmTGF-β2-7M has some propensity to dimerize, does however, show that perhaps the most important element of its design is elimination of a large portion of the TβRI binding site – in other words, the design of mmTGF-β2-7M does not require that the engineered protein behave as a perfect monomer in solution, but instead that enough of its TβRI binding site be removed so that if the molecule does dimerize, it is still unable to bind and recruit TβRI.
7. Perspective
The results presented here have demonstrated how a simple a modification, including substitution of a single cysteine residue with serine and replacement of a helix with a flexible loop, can change a potent signaling protein into a potent specific inhibitor of the same signaling protein. Though further studies are required, this relatively small and highly specific TGF-β inhibitor has several possible uses, such as an injected therapeutic, possibility conjugated with the Fc domain of an antibody or with albumin to diminish renal filtration, or as a secreted protein, either alone or conjugated to an Fc domain or albumin, for gene therapy applications. Though not demonstrated or described, this engineered form of TGF-β also has several potential uses for further mechanistic studies β for example, this form of TGF-β could be used to probe and possibly localize the binding site for other proteins known to bind and alter the functions of TGF-βs, such as the non-signaling type III receptor 90, 91, or soluble TGF-β binding proteins, such as decorin or biglycan 92, 93.
Though the changes required to completely alter the function of this protein are simple, it in fact required significant knowledge of the structure, and accompanying binding properties of the receptors, to enable the design. This type of detailed structural and mechanistic knowledge is being increasingly employed to develop other types of engineered growth factors, such as vascular endothelial growth factor 94-97, fibroblast growth factor 98-100, and Wnts 101, 102, with altered functions for use as therapeutic agents. Through advances in capabilities for structure determination by NMR, X-ray, and EM continue, it is expected that the type of detailed structural and biophysical knowledge that enabled the design of the engineered TGF-β monomer, as well as other growth factors, will become more commonplace, thus expanding a potentially important avenue for the altering the function of many different classes of growth factors for therapeutic gain.
8. Acknowledgments
APH is supported by NIH R01GM58670, NIH CA172886, DoD W81XWH-17-1-0429, and by the University of Pittsburgh Vascular Medicine Institute.
Abbreviations:
- TGF-β
transforming growth factor-beta
- GDF
growth and differentiation factor
- BMP
bone morphogenetic protein
- SPR
surface plasmon resonance
- EM
electron microscopy
- NMR
nuclear magnetic resonance
- EMT
epithelial-to-mesenchymal transition
- TβRI
TGF-β type I receptor
- TβRII
TGF-β type II receptor
- SA
streptavidin
References
- [1].Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, and Velankar S (2017) Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive, Methods Mol Biol 1607, 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Martin-Garcia JM, Conrad CE, Coe J, Roy-Chowdhury S, and Fromme P (2016) Serial femtosecond crystallography: A revolution in structural biology, Arch Biochem Biophys 602, 32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Owen RL, Juanhuix J, and Fuchs M (2016) Current advances in synchrotron radiation instrumentation for MX experiments, Arch Biochem Biophys 602, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Frank J (2017) Advances in the field of single-particle cryo-electron microscopy over the last decade, Nat Protoc 12, 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Baker LA, Grange M, and Grunewald K (2017) Electron cryo-tomography captures macromolecular complexes in native environments, Curr Opin Struct Biol 46, 149–156. [DOI] [PubMed] [Google Scholar]
- [6].Liu W, Brock A, Chen S, Chen S, and Schultz PG (2007) Genetic incorporation of unnatural amino acids into proteins in mammalian cells, Nat Methods 4, 239–244. [DOI] [PubMed] [Google Scholar]
- [7].Young TS, Ahmad I, Yin JA, and Schultz PG (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli, J Mol Biol 395, 361–374. [DOI] [PubMed] [Google Scholar]
- [8].Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, and Jhoti H (2016) Twenty years on: the impact of fragments on drug discovery, Nat Rev Drug Discov 15, 605–619. [DOI] [PubMed] [Google Scholar]
- [9].Zaykov AN, Mayer JP, and DiMarchi RD (2016) Pursuit of a perfect insulin, Nat Rev Drug Discov 15, 425–439. [DOI] [PubMed] [Google Scholar]
- [10].Kulp DW, and Schief WR (2013) Advances in structure-based vaccine design, Curr Opin Virol 3, 322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ye B, Stary CM, Li X, Gao Q, Kang C, and Xiong X (2018) Engineering chimeric antigen receptor-T cells for cancer treatment, Mol Cancer 17, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ronson A, Tvito A, and Rowe JM (2016) Treatment of Relapsed/Refractory Acute Lymphoblastic Leukemia in Adults, Curr Oncol Rep 18, 39. [DOI] [PubMed] [Google Scholar]
- [13].Brudno JN, and Kochenderfer JN (2018) Chimeric antigen receptor T-cell therapies for lymphoma, Nat Rev Clin Oncol 15, 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, and Sadelain M (2018) Gene therapy comes of age, Science 359. [DOI] [PubMed] [Google Scholar]
- [15].Sanjabi S, Oh SA, and Li MO (2017) Regulation of the Immune Response by TGF-beta: From Conception to Autoimmunity and Infection, Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Penn JW, Grobbelaar AO, and Rolfe KJ (2012) The role of the TGF-beta family in wound healing, burns and scarring: a review, Int J Burns Trauma 2, 18–28. [PMC free article] [PubMed] [Google Scholar]
- [17].Hinck AP, Mueller TD, and Springer TA (2016) Structural Biology and Evolution of the TGF-beta Family, Cold Spring Harb Perspect Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cotton TR, Fischer G, Wang X, McCoy JC, Czepnik M, Thompson TB, and Hyvonen M (2018) Structure of the human myostatin precursor and determinants of growth factor latency, EMBO J 37, 367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mi LZ, Brown CT, Gao Y, Tian Y, Le VQ, Walz T, and Springer TA (2015) Structure of bone morphogenetic protein 9 procomplex, Proc Natl Acad Sci U S A 112, 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, and Springer TA (2011) Latent TGF-beta structure and activation, Nature 474, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang X, Fischer G, and Hyvonen M (2016) Structure and activation of pro-activin A, Nat Commun 7, 12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Allendorph GP, Isaacs MJ, Kawakami Y, Izpisua Belmonte JC, and Choe S (2007) BMP-3 and BMP-6 structures illuminate the nature of binding specificity with receptors, Biochemistry 46, 12238–12247. [DOI] [PubMed] [Google Scholar]
- [23].Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, Roschke V, Sanyal I, and Choe S (2005) Crystal structure of BMP-9 and functional interactions with pro-region and receptors, The Journal of biological chemistry 280, 25111–25118. [DOI] [PubMed] [Google Scholar]
- [24].Daopin S, Piez KA, Ogawa Y, and Davies DR (1992) Crystal structure of transforming growth factor-beta 2: an unusual fold for the superfamily, Science 257, 369–373. [DOI] [PubMed] [Google Scholar]
- [25].Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, and Choe S (2003) The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly, Mol Cell 11, 605–617. [DOI] [PubMed] [Google Scholar]
- [26].Griffith DL, Keck PC, Sampath TK, Rueger DC, and Carlson WD (1996) Three-dimensional structure of recombinant human osteogenic protein 1: structural paradigm for the transforming growth factor beta superfamily, Proceedings of the National Academy of Sciences of the United States of America 93, 878–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S, and Hyvonen M (2006) Structural basis for the inhibition of activin signalling by follistatin, The EMBO journal 25, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, Tsang ML, Lucas R, Zhang BL, Wenker J, and Torchia DA (1996) Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2, Biochemistry 35, 8517–8534. [DOI] [PubMed] [Google Scholar]
- [29].Keller S, Nickel J, Zhang JL, Sebald W, and Mueller TD (2004) Molecular recognition of BMP-2 and BMP receptor IA, Nat Struct Mol Biol 11, 481–488. [DOI] [PubMed] [Google Scholar]
- [30].Mittl PR, Priestle JP, Cox DA, McMaster G, Cerletti N, and Grutter MG (1996) The crystal structure of TGF-beta 3 and comparison to TGF-beta 2: implications for receptor binding, Protein Sci 5, 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nickel J, Kotzsch A, Sebald W, and Mueller TD (2005) A single residue of GDF-5 defines binding specificity to BMP receptor IB, Journal of molecular biology 349, 933–947. [DOI] [PubMed] [Google Scholar]
- [32].Scheufler C, Sebald W, and Hulsmeyer M (1999) Crystal structure of human bone morphogenetic protein-2 at 2.7 A resolution, Journal of molecular biology 287, 103–115. [DOI] [PubMed] [Google Scholar]
- [33].Schlunegger MP, and Grutter MG (1992) An unusual feature revealed by the crystal structure at 2.2 A resolution of human transforming growth factor-beta 2, Nature 358, 430–434. [DOI] [PubMed] [Google Scholar]
- [34].Schreuder H, Liesum A, Pohl J, Kruse M, and Koyama M (2005) Crystal structure of recombinant human growth and differentiation factor 5: evidence for interaction of the type I and type II receptor-binding sites, Biochem Biophys Res Commun 329, 1076–1086. [DOI] [PubMed] [Google Scholar]
- [35].Laiho M, Weis FM, Boyd FT, Ignotz RA, and Massague J (1991) Responsiveness to transforming growth factor-beta (TGF-beta) restored by genetic complementation between cells defective in TGF-beta receptors I and II, J Biol Chem 266, 9108–9112. [PubMed] [Google Scholar]
- [36].Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, and Massague J (1992) TGF beta signals through a heteromeric protein kinase receptor complex, Cell 71, 1003–1014. [DOI] [PubMed] [Google Scholar]
- [37].Wrana JL, Attisano L, Wieser R, Ventura F, and Massague J (1994) Mechanism of activation of the TGF-beta receptor, Nature 370, 341–347. [DOI] [PubMed] [Google Scholar]
- [38].Hill CS (2016) Transcriptional Control by the SMADs, Cold Spring Harb Perspect Biol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Massague J, and Wotton D (2000) Transcriptional control by the TGF-beta/Smad signaling system, EMBO J 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Baardsnes J, Hinck CS, Hinck AP, and O'Connor-McCourt MD (2009) TbetaR-II discriminates the high- and low-affinity TGF-beta isoforms via two hydrogen-bonded ion pairs, Biochemistry 48, 2146–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, and Hinck AP (2002) Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex, Nat Struct Biol 9, 203–208. [DOI] [PubMed] [Google Scholar]
- [42].De Crescenzo G, Hinck CS, Shu Z, Zuniga J, Yang J, Tang Y, Baardsnes J, Mendoza V, Sun L, López-Casillas F, O'Connor-McCourt M, and Hinck AP (2006) Three key residues underlie the differential affinity of the TGFbeta isoforms for the TGFbeta type II receptor, J Mol Biol 355, 47–62. [DOI] [PubMed] [Google Scholar]
- [43].Huang T, David L, Mendoza V, Yang Y, Villarreal M, De K, Sun L, Fang X, López-Casillas F, Wrana J, and Hinck AP (2011) TGF-β signaling is mediated by two autonomously functioning TβRI:TβRII pairs, EMBO J in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, Sebald W, and Mueller TD (2007) A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor, BMC Struct Biol 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Allendorph GP, Vale WW, and Choe S (2006) Structure of the ternary signaling complex of a TGF-beta superfamily member, Proc Natl Acad Sci U S A 103, 7643–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, and Hinck AP (2008) Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding, Mol Cell 29, 157–168. [DOI] [PubMed] [Google Scholar]
- [47].Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, and Hinck AP (2002) Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex, Nature structural biology 9, 203–208. [DOI] [PubMed] [Google Scholar]
- [48].Kirsch T, Sebald W, and Dreyer MK (2000) Crystal structure of the BMP-2-BRIA ectodomain complex, Nature structural biology 7, 492–496. [DOI] [PubMed] [Google Scholar]
- [49].Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, and Sun PD (2010) Ternary complex of transforming growth factor-β1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily, J Biol Chem 285, 14806–14814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Thompson TB, Woodruff TK, and Jardetzky TS (2003) Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand:receptor interactions, The EMBO journal 22, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zuniga JE, Groppe JC, Cui Y, Hinck CS, Contreras-Shannon V, Pakhomova ON, Yang J, Tang Y, Mendoza V, López-Casillas F, Sun L, and Hinck AP (2005) Assembly of TbetaRI:TbetaRII:TGFbeta ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent, J Mol Biol 354, 1052–1068. [DOI] [PubMed] [Google Scholar]
- [52].Seoane J, and Gomis RR (2017) TGF-beta Family Signaling in Tumor Suppression and Cancer Progression, Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kim KK, Sheppard D, and Chapman HA (2017) TGF-beta1 Signaling and Tissue Fibrosis, Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Markowitz SD, and Roberts AB (1996) Tumor suppressor activity of the TGF-beta pathway in human cancers, Cytokine Growth Factor Rev 7, 93–102. [DOI] [PubMed] [Google Scholar]
- [55].Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, and et al. (1991) Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene, Nature 352, 337–339. [DOI] [PubMed] [Google Scholar]
- [56].Loeys BL, Schwarze U, Holm T, Callewaert BL, Thomas GH, Pannu H, De Backer JF, Oswald GL, Symoens S, Manouvrier S, Roberts AE, Faravelli F, Greco MA, Pyeritz RE, Milewicz DM, Coucke PJ, Cameron DE, Braverman AC, Byers PH, De Paepe AM, and Dietz HC (2006) Aneurysm syndromes caused by mutations in the TGF-beta receptor, N Engl J Med 355, 788–798. [DOI] [PubMed] [Google Scholar]
- [57].Massagué J (2008) TGFbeta in Cancer, Cell 134, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chen CR, Kang Y, and Massague J (2001) Defective repression of c-myc in breast cancer cells: A loss at the core of the transforming growth factor beta growth arrest program, Proc Natl Acad Sci U S A 98, 992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang M, Kleber S, Rohrich M, Timke C, Han N, Tuettenberg J, Martin-Villalba A, Debus J, Peschke P, Wirkner U, Lahn M, and Huber PE (2011) Blockade of TGF-beta signaling by the TGFbetaR-I kinase inhibitor LY2109761 enhances radiation response and prolongs survival in glioblastoma, Cancer Res 71, 7155–7167. [DOI] [PubMed] [Google Scholar]
- [60].Adler HL, McCurdy MA, Kattan MW, Timme TL, Scardino PT, and Thompson TC (1999) Elevated levels of circulating interleukin-6 and transforming growth factor-beta1 in patients with metastatic prostatic carcinoma, J Urol 161, 182–187. [PubMed] [Google Scholar]
- [61].Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, Aura C, Barba I, Peg V, Prat A, Cuartas I, Jimenez J, Garcia-Dorado D, Sahuquillo J, Bernards R, Baselga J, and Seoane J (2012) USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma, Nat Med 18, 429–435. [DOI] [PubMed] [Google Scholar]
- [62].Lamora A, Talbot J, Mullard M, Brounais-Le Royer B, Redini F, and Verrecchia F (2016) TGF-beta Signaling in Bone Remodeling and Osteosarcoma Progression, J Clin Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, Garcia-Dorado D, Poca MA, Sahuquillo J, Baselga J, and Seoane J (2009) TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma, Cancer Cell 15, 315–327. [DOI] [PubMed] [Google Scholar]
- [64].Sato M, Matsubara T, Adachi J, Hashimoto Y, Fukamizu K, Kishida M, Yang YA, Wakefield LM, and Tomonaga T (2015) Differential Proteome Analysis Identifies TGF-beta-Related Pro-Metastatic Proteins in a 4T1 Murine Breast Cancer Model, PLoS One 10, e0126483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Stuelten CH, Busch JI, Tang B, Flanders KC, Oshima A, Sutton E, Karpova TS, Roberts AB, Wakefield LM, and Niederhuber JE (2010) Transient tumor-fibroblast interactions increase tumor cell malignancy by a TGF-Beta mediated mechanism in a mouse xenograft model of breast cancer, PLoS One 5, e9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wu Q, Tian Y, Zhang J, Zhang H, Gu F, Lu Y, Zou S, Chen Y, Sun P, Xu M, Sun X, Xia C, Chi H, Ying Zhu A, Tang D, and Wang D (2017) Functions of pancreatic stellate cell-derived soluble factors in the microenvironment of pancreatic ductal carcinoma, Oncotarget 8, 102721–102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Akhurst RJ (2017) Targeting TGF-beta Signaling for Therapeutic Gain, Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Akhurst RJ, and Hata A (2012) Targeting the TGFbeta signalling pathway in disease, Nat Rev Drug Discov 11, 790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, Cuartas I, Raventos C, Martinez-Ricarte F, Poca MA, Garcia-Dorado D, Lahn MM, Yingling JM, Rodon J, Sahuquillo J, Baselga J, and Seoane J (2010) TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma, Cancer Cell 18, 655–668. [DOI] [PubMed] [Google Scholar]
- [70].Anscher MS, Thrasher B, Zgonjanin L, Rabbani ZN, Corbley MJ, Fu K, Sun L, Lee WC, Ling LE, and Vujaskovic Z (2008) Small molecular inhibitor of transforming growth factor-beta protects against development of radiation-induced lung injury, Int J Radiat Oncol Biol Phys 71, 829–837. [DOI] [PubMed] [Google Scholar]
- [71].Bandyopadhyay A, Agyin JK, Wang L, Tang Y, Lei X, Story BM, Cornell JE, Pollock BH, Mundy GR, and Sun LZ (2006) Inhibition of pulmonary and skeletal metastasis by a transforming growth factor-beta type I receptor kinase inhibitor, Cancer Res 66, 6714–6721. [DOI] [PubMed] [Google Scholar]
- [72].Bandyopadhyay A, Wang L, Agyin J, Tang Y, Lin S, Yeh IT, De K, and Sun LZ (2010) Doxorubicin in combination with a small TGFbeta inhibitor: a potential novel therapy for metastatic breast cancer in mouse models, PLoS One 5, e10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bouquet F, Pal A, Pilones KA, Demaria S, Hann B, Akhurst RJ, Babb JS, Lonning SM, DeWyngaert JK, Formenti SC, and Barcellos-Hoff MH (2011) TGFbeta1 inhibition increases the radiosensitivity of breast cancer cells in vitro and promotes tumor control by radiation in vivo, Clin Cancer Res 17, 6754–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Flechsig P, Dadrich M, Bickelhaupt S, Jenne J, Hauser K, Timke C, Peschke P, Hahn EW, Grone HJ, Yingling J, Lahn M, Wirkner U, and Huber PE (2012) LY2109761 attenuates radiation-induced pulmonary murine fibrosis via reversal of TGF-beta and BMP-associated proinflammatory and proangiogenic signals, Clin Cancer Res 18, 3616–3627. [DOI] [PubMed] [Google Scholar]
- [75].Ganapathy V, Ge R, Grazioli A, Xie W, Banach-Petrosky W, Kang Y, Lonning S, McPherson J, Yingling JM, Biswas S, Mundy GR, and Reiss M (2010) Targeting the Transforming Growth Factor-beta pathway inhibits human basal-like breast cancer metastasis, Mol Cancer 9, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Park J, Wrzesinski SH, Stern E, Look M, Criscione J, Ragheb R, Jay SM, Demento SL, Agawu A, Licona Limon P, Ferrandino AF, Gonzalez D, Habermann A, Flavell RA, and Fahmy TM (2012) Combination delivery of TGF-beta inhibitor and IL-2 by nanoscale liposomal polymeric gels enhances tumour immunotherapy, Nat Mater 11, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Shinto O, Yashiro M, Kawajiri H, Shimizu K, Shimizu T, Miwa A, and Hirakawa K (2010) Combination effect of a TGF-beta receptor kinase inhibitor with 5-FU analog S1 on lymph node metastasis of scirrhous gastric cancer in mice, Cancer Sci 101, 1846–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Takeuchi K, Abe M, Hiasa M, Oda A, Amou H, Kido S, Harada T, Tanaka O, Miki H, Nakamura S, Nakano A, Kagawa K, Yata K, Ozaki S, and Matsumoto T (2010) Tgf-Beta inhibition restores terminal osteoblast differentiation to suppress myeloma growth, PLoS One 5, e9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Triplett TA, Tucker CG, Triplett KC, Alderman Z, Sun L, Ling LE, Akporiaye ET, and Weinberg AD (2015) STAT3 Signaling Is Required for Optimal Regression of Large Established Tumors in Mice Treated with Anti-OX40 and TGFbeta Receptor Blockade, Cancer Immunol Res 3, 526–535. [DOI] [PubMed] [Google Scholar]
- [80].Van der Jeught K, Joe PT, Bialkowski L, Heirman C, Daszkiewicz L, Liechtenstein T, Escors D, Thielemans K, and Breckpot K (2014) Intratumoral administration of mRNA encoding a fusokine consisting of IFN-beta and the ectodomain of the TGF-beta receptor II potentiates antitumor immunity, Oncotarget 5, 10100–10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wan X, Li ZG, Yingling JM, Yang J, Starbuck MW, Ravoori MK, Kundra V, Vazquez E, and Navone NM (2012) Effect of transforming growth factor beta (TGF-beta) receptor I kinase inhibitor on prostate cancer bone growth, Bone 50, 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yingling JM, Blanchard KL, and Sawyer JS (2004) Development of TGF-beta signaling inhibitors for cancer therapy, Nat Rev Drug Discov 3, 1011–1022. [DOI] [PubMed] [Google Scholar]
- [83].Gueorguieva I, Cleverly AL, Stauber A, Sada Pillay N, Rodon JA, Miles CP, Yingling JM, and Lahn MM (2014) Defining a therapeutic window for the novel TGF-beta inhibitor LY2157299 monohydrate based on a pharmacokinetic/pharmacodynamic model, Br J Clin Pharmacol 77, 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Meibohm B (2012) Pharmacokinetics and Half-Life of Protein Therapeutics, In Therapeutic Proteins: Strategies to Modulate Their Plasma Half-Lives (Kontermann R, Ed.), pp 23–38, Wiley-Blackwell, Weinheim, Germany. [Google Scholar]
- [85].Meibohm B, and Braeckman RA (2007) Pharmacokinetics and Pharmacodynamics of Peptides and Proteins, In Pharmaceutical Biotechnology: Concepts and Applications (Crommelin DJA, Sindelar RD, and Meibohm B, Eds.), pp 95–123. [Google Scholar]
- [86].Huang T, and Hinck AP (2016) Production, Isolation, and Structural Analysis of Ligands and Receptors of the TGF-beta Superfamily, Methods Mol Biol 1344, 63–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Amatayakul-Chantler S, Qian SW, Gakenheimer K, Bottinger EP, Roberts AB, and Sporn MB (1994) [Ser77]transforming growth factor-beta 1. Selective biological activity and receptor binding in mink lung epithelial cells, J Biol Chem 269, 27687–27691. [PubMed] [Google Scholar]
- [88].Kim SK, Barron L, Hinck CS, Petrunak EM, Cano KE, Thangirala A, Iskra B, Brothers M, Vonberg M, Leal B, Richter B, Kodali R, Taylor AB, Du S, Barnes CO, Sulea T, Calero G, Hart PJ, Hart MJ, Demeler B, and Hinck AP (2017) An engineered transforming growth factor beta (TGF-beta) monomer that functions as a dominant negative to block TGF-beta signaling, J Biol Chem 292, 7173–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Smialowski P, Doose G, Torkler P, Kaufmann S, and Frishman D (2012) PROSO II--a new method for protein solubility prediction, FEBS J 279, 2192–2200. [DOI] [PubMed] [Google Scholar]
- [90].López-Casillas F, Payne HM, Andres JL, and Massague J (1994) Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: mapping of ligand binding and GAG attachment sites, J Cell Biol 124, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Villarreal MM, Kim SK, Barron L, Kodali R, Baardsnes J, Hinck CS, Krzysiak TC, Henen MA, Pakhomova O, Mendoza V, O'Connor-McCourt MD, Lafer EM, López-Casillas F, and Hinck AP (2016) Binding Properties of the Transforming Growth Factor-beta Coreceptor Betaglycan: Proposed Mechanism for Potentiation of Receptor Complex Assembly and Signaling, Biochemistry 55, 6880–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Hildebrand A, Romaris M, Rasmussen LM, Heinegard D, Twardzik DR, Border WA, and Ruoslahti E (1994) Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta, Biochem J 302 ( Pt 2), 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Takeuchi Y, Kodama Y, and Matsumoto T (1994) Bone matrix decorin binds transforming growth factor-beta and enhances its bioactivity, J Biol Chem 269, 32634–32638. [PubMed] [Google Scholar]
- [94].Guc E, Briquez PS, Foretay D, Fankhauser MA, Hubbell JA, Kilarski WW, and Swartz MA (2017) Local induction of lymphangiogenesis with engineered fibrin-binding VEGF-C promotes wound healing by increasing immune cell trafficking and matrix remodeling, Biomaterials 131, 160–175. [DOI] [PubMed] [Google Scholar]
- [95].Kapur S, Silverman AP, Ye AZ, Papo N, Jindal D, Blumenkranz MS, and Cochran JR (2017) Engineered ligand-based VEGFR antagonists with increased receptor binding affinity more effectively inhibit angiogenesis, Bioeng Transl Med 2, 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Papo N, Silverman AP, Lahti JL, and Cochran JR (2011) Antagonistic VEGF variants engineered to simultaneously bind to and inhibit VEGFR2 and alphavbeta3 integrin, Proc Natl Acad Sci U S A 108, 14067–14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang C, Poon S, Murali S, Koo CY, Bell TJ, Hinkley SF, Yeong H, Bhakoo K, Nurcombe V, and Cool SM (2014) Engineering a vascular endothelial growth factor 165-binding heparan sulfate for vascular therapy, Biomaterials 35, 6776–6786. [DOI] [PubMed] [Google Scholar]
- [98].Andrades JA, Wu LT, Hall FL, Nimni ME, and Becerra J (2001) Engineering, expression, and renaturation of a collagen-targeted human bFGF fusion protein, Growth Factors 18, 261–275. [DOI] [PubMed] [Google Scholar]
- [99].Huang C, Liu Y, Beenken A, Jiang L, Gao X, Huang Z, Hsu A, Gross GJ, Wang YG, Mohammadi M, and Schultz JEJ (2017) A novel fibroblast growth factor-1 ligand with reduced heparin binding protects the heart against ischemia-reperfusion injury in the presence of heparin co-administration, Cardiovasc Res 113, 1585–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Yoneda A, Asada M, Oda Y, Suzuki M, and Imamura T (2000) Engineering of an FGF-proteoglycan fusion protein with heparin-independent, mitogenic activity, Nat Biotechnol 18, 641–644. [DOI] [PubMed] [Google Scholar]
- [101].Sopko R, Mugford JW, Lehmann A, Shapiro RI, Rushe M, Kulkarni A, Worrall J, Amatucci J, Wen D, Pederson NE, Minesinger BK, Arndt JW, and Pepinsky B (2017) Engineering potent long-acting variants of the Wnt inhibitor DKK2, Protein Eng Des Sel 30, 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Warner ML, Bell T, and Pioszak AA (2015) Engineering high-potency R-spondin adult stem cell growth factors, Mol Pharmacol 87, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kotzsch A, Nickel J, Sebald W, and Mueller TD (2009) Purification, crystallization and preliminary data analysis of ligand-receptor complexes of growth and differentiation factor 5 (GDF5) and BMP receptor IB (BRIB), Acta Crystallogr Sect F Struct Biol Cryst Commun 65, 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Zuniga JE, Ilangovan U, Mahlawat P, Hinck CS, Huang T, Groppe JC, McEwen DG, and Hinck AP (2011) The TbetaR-I pre-helix extension is structurally ordered in the unbound form and its flanking prolines are essential for binding, J Mol Biol 412, 601–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Thompson TB, Woodruff TK, and Jardetzky TS (2003) Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand:receptor interactions, EMBO J 22, 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Boesen CC, Radaev S, Motyka SA, Patamawenu A, and Sun PD (2002) The 1.1 A crystal structure of human TGF-beta type II receptor ligand binding domain, Structure 10, 913–919. [DOI] [PubMed] [Google Scholar]