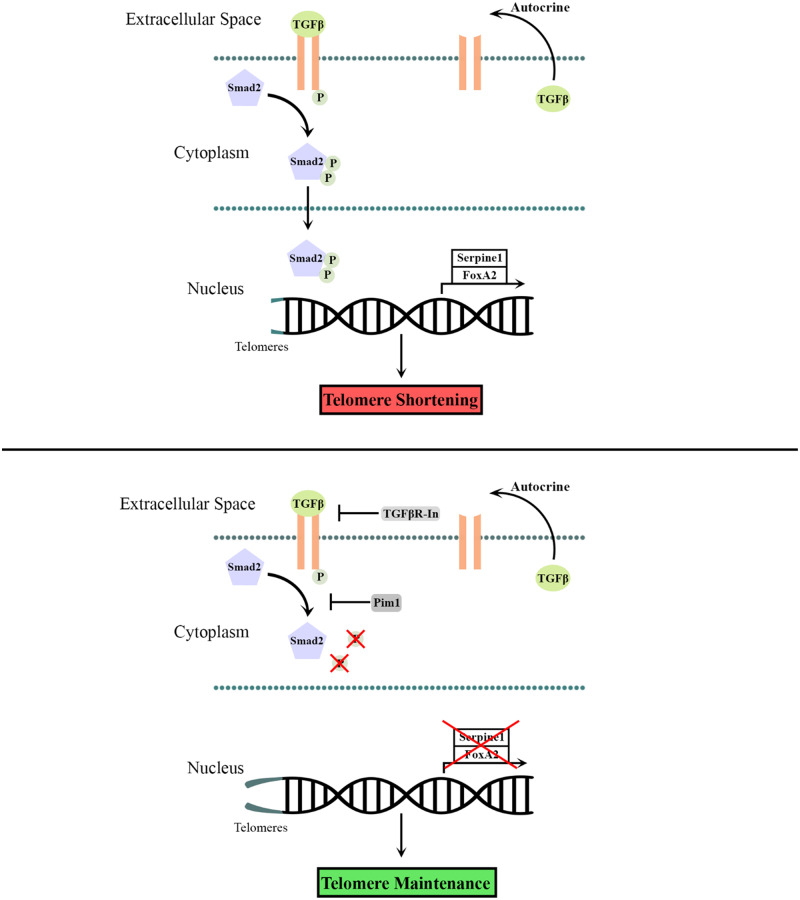
Abstract
Aims
Telomere attrition in cardiomyocytes is associated with decreased contractility, cellular senescence, and up-regulation of proapoptotic transcription factors. Pim1 is a cardioprotective kinase that antagonizes the aging phenotype of cardiomyocytes and delays cellular senescence by maintaining telomere length, but the mechanism remains unknown. Another pathway responsible for regulating telomere length is the transforming growth factor beta (TGFβ) signalling pathway where inhibiting TGFβ signalling maintains telomere length. The relationship between Pim1 and TGFβ has not been explored. This study delineates the mechanism of telomere length regulation by the interplay between Pim1 and components of TGFβ signalling pathways in proliferating A549 cells and post-mitotic cardiomyocytes.
Methods and results
Telomere length was maintained by lentiviral-mediated overexpression of PIM1 and inhibition of TGFβ signalling in A549 cells. Telomere length maintenance was further demonstrated in isolated cardiomyocytes from mice with cardiac-specific overexpression of PIM1 and by pharmacological inhibition of TGFβ signalling. Mechanistically, Pim1 inhibited phosphorylation of Smad2, preventing its translocation into the nucleus and repressing expression of TGFβ pathway genes.
Conclusion
Pim1 maintains telomere lengths in cardiomyocytes by inhibiting phosphorylation of the TGFβ pathway downstream effectors Smad2 and Smad3, which prevents repression of telomerase reverse transcriptase. Findings from this study demonstrate a novel mechanism of telomere length maintenance and provide a potential target for preserving cardiac function.
Keywords: Pim1, Telomere, TGFβ, Cardiomyocyte, Smad2
Graphical Abstract
1. Introduction
Critically short telomeres are associated with cellular senescence and cardiovascular disease.1–3 Cardiomyocyte-specific telomere attrition is a characteristic of heart failure in humans and has been associated with decreased contractile ability, hypertrophy, and senescence.4,5 In proliferative cells, multiple rounds of cell division promote telomeric shortening, activating DNA damage response that results in cellular senescence or apoptosis.6 Cardiomyocytes also exhibit telomere attrition as a result of biomechanical stress, oxidative stress, or inflammation.5 Unveiling molecular mechanisms responsible for preserving telomeres in cardiomyocytes could provide valuable insights into antagonizing cellular aging and preserving cardiac function.
Pim1 kinase plays a role in maintaining a youthful cellular phenotype as it regulates a myriad of physiological responses such as anti-apoptotic signalling,7 preservation of mitochondrial integrity, enhancing metabolic activity,8 and importantly, maintenance of telomere length.9,10 Pim1-mediated maintenance of telomeres was established in proliferating cardiac interstitial cells (CICs) derived from murine hearts9 or cardiac explants from heart failure patients.10 Additionally, telomere shortening was observed in response to knockout of Pim1 in mouse cardiomyocytes.11 Although the role of Pim1 in maintaining telomeres has been established, the underlying mechanisms of Pim1-induced telomere maintenance are largely unknown.
Another pathway responsible for regulating telomere length is the transforming growth factor beta (TGFβ) signalling pathway, where TGFβ activation inhibits telomerase activity.12,13 TGFβ is a cytokine that mediates a host of cellular responses through stimulation of TGFβ receptors and downstream phosphorylation and activation of receptor-associated Smad proteins including Smad2. Activated Smad2 translocates into the nucleus and regulates expression of TGFβ pathway target genes responsible for inducing hypertrophic effects on cardiomyocytes and promoting myocardial fibrosis in the failing heart.14,15 Although TGFβ-mediated regulation of telomeres has been elucidated in proliferating cells,16,17 the impact of TGFβ signalling inhibition upon telomere maintenance in post-mitotic cells is yet to be explored. Moreover, the intracellular molecular interactions between TGFβ signalling and other pathways involved in telomere regulation have not been fully unravelled.
Independent studies showing opposing effects of Pim1 kinase and TGFβ signalling upon telomere length point to a potential interaction between Pim1 and TGFβ signalling cascades. This study was designed to assess the role of signalling interplay between Pim1 kinase and TGFβ in regulating telomere length. Findings from this study demonstrate that both Pim1 overexpression and TGFβ signalling inhibition maintains telomere length. Mechanistically, Pim1 maintains telomere length by inhibiting TGFβ signalling as revealed by decreased phosphorylation of its downstream effector Smad2. TGFβ signalling inhibition by Pim1 was further demonstrated by decreased expression of TGFβ pathway target genes Serpine1 and FOXA2. Signalling events were first established in A549 human lung carcinoma cells and recapitulated in post-mitotic adult murine cardiomyocytes, indicating that the Pim1/TGFβ signalling axis extends to proliferating and non-proliferating. cells. Overall, findings from this article introduce a novel molecular link between Pim1 and TGFβ signalling cascades, suggesting a potential mechanism by which Pim1 maintains telomere length.
2. Methods
2.1 Cell culturing
About 200 000 A549 cells (ATCC; Manassas, VA, USA; CCL-185) were plated in 60 mm dishes and were cultured in high glucose Dulbecco’s Modified Eagle Medium (ThermoFisher; Waltham, MA, USA; 11965092) supplemented with 10% foetal bovine serum (Omega Scientific; Tarzana, CA, USA; FB-11) and 1% Penicillin–Streptomycin–Glutamine (ThermoFisher; Waltham, MA, USA; 10378016) in a 37°C, 5% CO2 humidified incubator.
2.2 Viral infections
Lentiviral plasmids were created as previously described.18 A549 cells were infected with lentivirus encoding enhanced green fluorescent protein (GFP) or human PIM1-GFP. GFP and PIM1 overexpression were confirmed via immunoblotting. Cardiomyocytes were infected with lentivirus harbouring sh-RNA against telomerase reverse transcriptase (TERT) (Lt-sh-TERT) (Dharmacon; Lafayette, CO) or a scramble (Lt-sh-Control).
2.3 Animal experiments
All animal protocols were approved by the Institutional Animal Care and Use Committee of San Diego State University and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Transgenic mice with cardiac-specific overexpression of human PIM1 (PIM1) and global deletion of Pim1 (Pim1-KO) have been described before.7 Non-transgenic (NTg) age- and gender-matched mice of the same strain (FVB) were used as controls. Male adult mice aged 3 months and older were used for the study.
2.4 Isolation and culture of adult mouse cardiomyocytes
Cardiomyocytes were isolated from 3-month-old FVB mice as previously described.19 Briefly, mice were anaesthetized with ketamine (100 mg/kg) and injected with Heparin (100 U/kg) (Sigma–Aldrich; Burlington, MA, USA; H3393) intraperitoneally to prevent blood clots. After mice displayed no pedal reflex in response to a firm toe pinch, hearts were cannulated via the aorta and digested on a Langendorff apparatus using Collagenase II (230 U/mL) (Worthington; Lakewood, NJ, USA; LS004147). Hearts were finely minced and triturated until no large tissue remained. Following calcium introduction, 100 000 cardiomyocytes were pre-plated on laminin (ThermoFisher; Waltham, MA, USA; 23017015) -coated 35 mm dishes. After 2 h, cardiomyocytes were cultured in serum-free media overnight at 37°C following which pharmacological treatments were conducted.
2.5 Pharmacologic treatments
About 50 000 A549 cells were plated in 35 mm dishes. The following day, cells were treated with 10 μM LY-364947 (TGFR-In) (Selleck Biochem; Houston, TX, USA; S2805) for 48 h or with 10 ng/mL TGF-β1 (R&D Systems; Minneapolis, MN, USA; 766-MB) for 1 h. About 100 000 cardiomyocytes were plated in 35 mm dishes and were treated with 10 ng/mL TGF-β1 10 μM TGFR-In, 3 mM N-acetyl-cysteine (NAC) (Sigma–Aldrich; Burlington, MA; A9165), 200 μM 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox) (Sigma–Aldrich; Burlington, MA; 238813), or 25 μM blebbistatin (Sigma–Aldrich; Burlington, MA; B0560) for 48 h.
2.6 Quantitative RT–PCR
RNA was isolated using Quick-RNA Miniprep Kit (Zymo Research; Irvine, CA, USA; R1054) following the manufacturer’s instructions. RNA concentrations were determined using a Nanodrop 2000 spectrophotometer (ThermoFisher; Waltham, MA; ND-2000) and cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad; Hercules, CA, USA; 1708890). Reactions were prepared in triplicate using 6.5 ng cDNA per reaction and iQ SYBER Green (Bio-Rad; Hercules, CA, USA; 1708880) on a CFX Real-Time PCR thermocycler (Bio-Rad; Hercules, CA, USA; 1855201). Samples were normalized to 18S and data were analysed by ΔΔCt method. Primers sequences are listed in Table 1.
2.7 Telomere measurement using quantitative RT–PCR
Telomere lengths were measured based on the monochrome multiplex qPCR method established by Cawthon et al.20 Genomic DNA was isolated from A549 cells, myocardial tissue, or isolated adult myocytes using the NucleoSpin Tissue kit (Machery-Nagel; Bethlehem, PA, USA; 740952) following the manufacturer’s instructions. Reactions were prepared in quintuplicate using 1.67 ng DNA and iQ SYBER Green (Bio-Rad; Hercules, CA, USA; 1708880) on a CFX Real-Time PCR thermocycler (Bio-Rad; Hercules, CA, USA; 1855201). Samples were normalized to albumin and data were analysed by ΔΔCt method. The telomere length was calculated as 2−ΔΔCt multiplied by the average telomere length for the mouse strain, determined from previous studies to be 75 000 bp for FVB mice.21 Primer sequences are listed in Table 1.
Table 2.
Antibody list
| Antibody | Catalogue number | Dilution | Application |
|---|---|---|---|
| Smad2 | Cell Signalling 5339 | 1:500 | IB |
| Phospho-Smad2 (Ser465/467) | Cell Signalling 3108 | 1:500 | IB |
| Smad2/3 | Cell Signalling 3102 | 1:500 | IB |
| Phospho-Smad3 (Ser423/425) | Abcam Ab52903 | 1:500 | IB |
| Pim-1 | ThermoFisher 39-4600 | 1:1000 | IB |
| GFP | Invitrogen G10362 | 1:1000 | IB |
| GFP | Rockland 600-101-215 | 1:100 | IHC |
| TGFβ | R&D Systems MAB240 | 1:500 | IB |
| TGFβ | Cell Signalling 3709 | 1:50 | IHC |
| TERT | ThermoFisher PA5-80105 | 1:500 | IB |
| Cardiac troponin T | ThermoFisher MA5-12960 | 1:100 | IHC |
| GAPDH | Millipore AB2302 | 1:5000 | IB |
| Myosin light chain 2 | Santa Cruz Sc-34490 | 1:20 | IHC |
2.8 Immunoblot
Cells were lysed in ice-cold radioimmunoprecipitation assay buffer (ThermoFisher; Waltham, MA, USA; 89900) containing protease and phosphatase inhibitor cocktails (Sigma–Aldrich; Burlington, MA, USA; P8340, P5726, and P2850). Protein concentrations were analysed and normalized by Bradford Assay and lysates were prepared by addition of NuPAGE LDS Sample Buffer (ThermoFisher; Waltham, MA, USA; NP0007). Samples were sonicated and boiled then loaded onto a 4–12% NuPAGE Bis–Tris gel (ThermoFisher; Waltham, MA, USA; NP0321BOX). Proteins were transferred onto an Immobilon-FL Polyvinylidene difluoride membrane (EMD Millipore; Burlington, MA, USA; IPFL0010) and the membrane was blocked with Odyssey Blocking Buffer (Li-Cor; Lincoln, NE, USA; 927-50000) for 1 h at room temperature and incubated with primary antibodies overnight at 4°C (Table 2). Secondary antibodies were applied for 2 h at room temperature. Fluorescent signal was detected using an Odyssey CLx imaging system (Li-Cor; Lincoln, NE, USA) and bands were quantified using Image Studio.
Table 1.
Primer list
| Target | Forward | Reverse |
|---|---|---|
| Human SERPINE1 | GCAACGTGGTTTTCTCACCC | GGCCATGCCCTTGTCATCAA |
| Human N-CADHERIN | ATTGATGCTGACGATCCCAATGCC | TCAAGTCCAGCTGCCACTGTGATCA |
| Human 18S | CGAGCCGCCTGGATACC | CATGGCCTCAGTTCCGAAAA |
| Mouse Serpine1 | TTCAGCCCTTGCTTGCCTC | ACACTTTTACTCCGAAGTCGGT |
| Mouse Foxa2 | CCCTACGCCAACATGAACTCG | GTTCTGCCGGTAGAAAGGGA |
| Mouse Pim1 | ATCCGCGTCGCCGACAACTT | TCGGGTGCCATTGGGCAGTT |
| Mouse 18S | CGAGCCGCCTGGATACC | CATGGCCTCAGTTCCGAAAA |
| Telomere | ACACTAAGGTTTGGGTTTGGGTTTGGGTTTGGGTTAGTGT | TGTTAGGTATCCCTATCCCTATCCCTATCCCTATCCCT AACA |
| Human ALB | CGGCGGCGGGCGGCGCGGGCTGGGCGGAAATGCTGCA CAGAATCCTTG | GCCCGGCCCGCCGCGCCCGTCCCGCCGGAAAAGCATGG TCGCCTGTT |
| Mouse Alb | CGGCGGCGGGCGGCGCGGGCTGGGCGGTTGCATGAA GTTGCCAGAAG | GCCCGGCCCGCCGCGCCCGTCCCGCCGGTCAGGCAGCT TTCCTTGTC |
2.9 Immunofluorescence
Immunohistochemistry was performed on paraffin heart sections as previously described.22,23 Briefly, slides were deparaffinized in three changes of xylene for 5 min, followed by three changes of 100% ethanol for 3 min, two changes of 95% ethanol for 3 min, 70% ethanol for 3 min, then rehydrated in three changes of de-ionized water for 3 min. The sections were boiled in 10 mmol/L citrate buffer (pH 6.0) for 15 min then were blocked with Tris–NaCl-Blocking buffer (TNB) (Perkin Elmer; Waltham, MA, USA; FP1020) for 30 min. Primary antibody (Table 2) was applied overnight at 4°C in TNB. The following day, slides were washed with three changes of Tris–NaCl (1XTN), stained with secondary antibody for 2 h at room temperature, washed in three changes of 1XTN for 5 min, and then stained with 1 µg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma–Aldrich; Burlington, MA, USA; D8417). Slides were mounted with Vectashield (Vector Laboratories; Burlingame, CA, USA; H-1000), following which images were acquired on an SP8 confocal microscope (Leica; Buffalo Grove, IL, USA).
2.10 Quantitative-fluorescence in situ hybridization
Telomere lengths were measured in heart samples by quantitative-fluorescence in situ hybridization (qFISH) using a modified version of a previously published protocol.24 Paraffin sections were subjected to deparaffinization and antigen retrieval as described previously.23 The hybridization probe was warmed to 37°C for 5 min and the hybridization buffer (20 mM Tris at pH 7.4, 60% formamide, 0.1 µg/mL salmon sperm DNA) (Sigma–Aldrich; Burlington, MA, USA; D1626) was heated to 85°C for 5 min. The telomere probe (PNA Bio; Newbury Park, CA, USA; F1002) was mixed with hybridization buffer at a ratio of 1:12.5 by volume and heated to 85°C for 10 min. Simultaneously, the dried slides were preheated at 85°C for 5 min. Following application of the mix containing the telomere probe and hybridization buffer, slides were heated at 85°C for 10 min and subsequently incubated overnight at 37°C in a covered, humidified chamber. The next day, the slides were washed once with 37°C 2× saline-sodium citrate (2XSSC) (Sigma–Aldrich; Burlington, MA, USA; S6639), twice with 37°C 2XSSC containing 50% formamide for 5 min each, and twice with 37°C 2XSSC for 5 min each. Slides were then blocked with TNB (Perkin Elmer; Waltham, MA, USA; FP1020) for 1 h at room temperature, and incubated with primary antibody overnight at 4°C. The next day, slides were washed, incubated with secondary antibody for 1 h at room temperature, washed, stained with 1 µg/mL DAPI (Sigma–Aldrich; Burlington, MA, USA; D8417), then mounted, and imaged.
To measure telomere length in vitro, slides of cultured adult cardiomyocytes were treated with fixative containing 3:1 ratio of ethanol: acetic acid for 30 min. Slides were treated with RNase solution (2XSSC containing 100 µg/mL ribonuclease A) and incubated at 37°C for 1 h then washed with 2XSSC and phosphate-buffered saline for 5 min each. Slides were then treated with 37°C 0.005% pepsin (Sigma–Aldrich; Burlington, MA, USA; P7000) for 3 min and washed with phosphate-buffered saline for 5 min. The slides were dehydrated using cold 70%, 90%, and 100% ethanol for 2 min each then air dried. The PNA probe was warmed to 37°C for 5 min and mixed with hybridization buffer at a ratio of 1:12.5 by volume and heated to 85°C for 10 min. Simultaneously, the dried slides were preheated at 85°C for 5 min. Following application of the mix containing the telomere probe and hybridization buffer, slides were heated at 85°C for 5–10 min and subsequently incubated overnight at 37°C in a covered, humidified chamber. The next day, the slides were washed once with 37°C 2XSSC for 5 min, twice with 37°C 2XSSC containing 50% formamide for 5 min each, and twice with warm 2XSSC for 5 min each. Following addition of 1 µg/mL DAPI, slides were mounted and cells were imaged.
2.11 Telomere length measurements using qFISH
qFISH images were acquired on the Leica SP8 confocal microscope. Mean values provided by the Leica software represent the pixel sum/sum processed pixel that accounts for the area of the region of interest, therefore, no additional normalizations to the nuclear area were done.
2.12 Statistical analyses
Statistical analyses were performed using paired or unpaired Student’s t-test or one-way ANOVA with Tukey’s multiple comparison test on GraphPad Prism v5.0. A P-value of <0.05 was considered statistically significant.
3. Results
3.1 Telomere length is elongated by PIM1 overexpression or TGFβ receptor inhibition
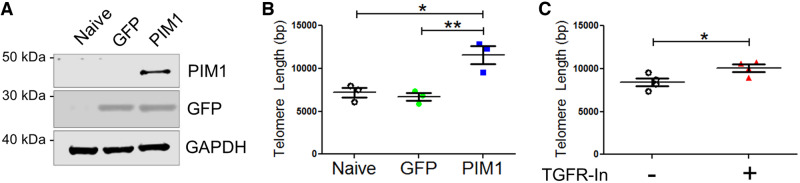
Previous studies demonstrate that telomere length is maintained by PIM1 overexpression in mouse cardiac non-myocyte cells,9 and that telomeric shortening is associated with TGFβ signalling.13,16,17,25 To recapitulate these results, A549 cells were used because these cells express low levels of endogenous PIM1. GFP or GFP-PIM1 (PIM1) was overexpressed via lentiviral infection in A549 cells and overexpression was confirmed via immunoblot (Figure 1A). PIM1 cells contained 1.55-fold longer telomeres than GFP cells after three passages as revealed by quantitative RT–PCR (Figure 1B). To determine basal level of telomere length, telomeres were measured in A549 cells before and after three passages in culture. There was no significant change in telomere length over three passages in culture suggesting that telomere length is already in equilibrium in A549 cells (see Supplementary material online, Figure S1A). Telomeres are dynamic in length as they are progressively eroded during rounds of cell division and elongated by telomerase.26 PIM1 influences the equilibrium of telomeres in A549 cells, shifting it towards elongation. The role of TGFβ signalling on telomere length was then evaluated by pharmacologic inhibition of TGFβ receptor-1 (TGFR1) using LY-364947 (TGFR-In), a selective, ATP-competitive inhibitor of TGFR1. Inhibition of TGFR1 resulted in 1.22-fold longer telomeres compared to the vehicle control (Figure 1C). Collectively, these results demonstrate that telomeres can be elongated by either PIM1 overexpression or TGFβ signalling inhibition in A549 cells.
Figure 1.
Telomere length is maintained by PIM1 overexpression or TGFβ receptor inhibition. (A) Representative western-blot bands of GFP and PIM1 in A549 cells transduced with GFP or PIM1-GFP. (B) qRT–PCR quantification of telomere lengths in transduced A549 cells. n = 3, error bars represent SEM, **P < 0.01 vs. GFP, *P < 0.05 vs. naive as measured by one-way ANOVA followed by Tukey’s multiple comparison test. (C) qRT–PCR quantification of telomere lengths in naive A549 cells treated with TGFR-In. n = 4, error bars represent SEM, *P < 0.05 vs. untreated as measured by Student’s t-test.
3.2 Tgfβ signalling is blunted by PIM1
The link between PIM1 and TGFβ signalling was established by examining the TGFβ pathway downstream targets, pSMAD2, SERPINE1, and N-CADHERIN expression following treatment of 10 ng/mL TGFβ1 on GFP and PIM1 cells. In the presence of TGFβ1 agonist, PIM1 exhibited a 41% reduction in pSMAD2 levels compared to GFP as evaluated by immunoblotting (Figure 2A and B). Inhibitory effects of PIM1 on TGFβ signalling were confirmed by expression of canonical downstream target SERPINE1 and non-canonical target N-CADHERIN as measured via qRT–PCR. PIM1 displayed a 64% and 42% reduction in mRNA expression of SERPINE1 and N-CADHERIN, respectively, compared to GFP (Figure 2C and D). These findings indicate that TGFβ signalling is inhibited by PIM1 overexpression, suggesting a potential mechanism by which telomere length is maintained by PIM1 in proliferating A549 cells.
Figure 2.
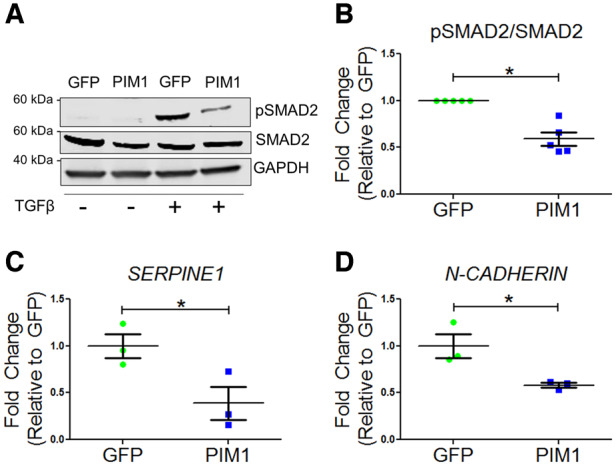
TGFβ signalling is blunted by PIM1. (A) Representative western blot of pSMAD2 and total SMAD2 in GFP and PIM1 cells with and without treatment of 4 ng/mL TGFβ. (B) Quantification of pSMAD2 over total SMAD2. n = 5, error bars represent SEM, *P < 0.01 vs. GFP as measured by Student’s t-test. (C) Quantification of SERPINE1 and (D) N-CADHERIN mRNA expression via qRT–PCR. n = 3, error bars represent SEM, *P < 0.05 vs. GFP as measured by Student’s t-test.
3.3 Telomere length is maintained by PIM1 in adult murine cardiomyocytes
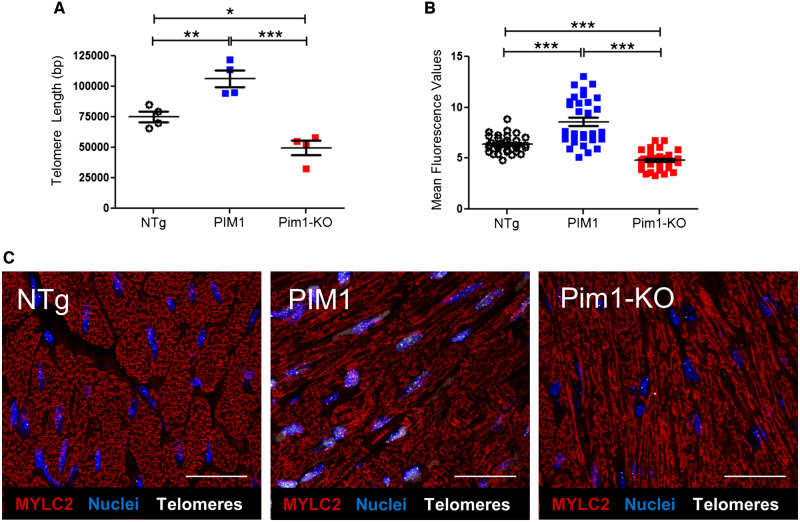
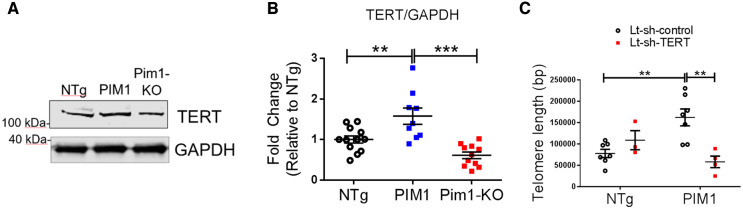
Pim1 exerts cardioprotective effects in mouse myocardium.7 To determine if telomere length is maintained by Pim1 in cardiomyocytes, telomeres were measured in hearts of 3-month-old non-transgenic mice (NTg), transgenic mice with cardiac-specific overexpression of human PIM1 (PIM1),27 and knockout mice with global deletion of Pim1 (Pim1-KO).7 Telomeres were 27% longer in PIM1 myocardium and 31% shorter in Pim1-KO hearts compared to NTg as revealed by qRT–PCR (Figure 3A). Telomere lengths were then analysed in cardiomyocytes from NTg, PIM1, and Pim1-KO heart sections via qFISH (Figure 3C). Consistent with findings from whole heart analysis, telomere fluorescence intensity was 28% greater in PIM1 cardiomyocytes and reduced by 30% in Pim1-KO cardiomyocytes compared to NTg controls (Figure 3B). Collectively, these results indicate that telomere length is maintained by PIM1 in adult mouse cardiomyocytes.
Figure 3.
Telomere length is maintained by PIM1 in adult murine cardiomyocytes. (A) Analysis of telomere lengths in isolated adult cardiomyocytes from PIM1 or Pim1-KO mice via qRT–PCR. n = 4, error bars represent SEM, *P < 0.05, **P < 0.01 vs. NTg, ***P < 0.001 vs. PIM1 as measured by one-way ANOVA followed by Tukey’s multiple comparison test. (B) Quantification of telomere fluorescence intensity indicated as mean fluorescence values from qFISH images. n = 32 nuclei per group, error bars represent SEM, ***P < 0.001 vs. NTg and vs. PIM1 as measured by one-way ANOVA followed by Tukey’s multiple comparison test. (C) Representative qFISH images of telomeres (white) from NTg, PIM1, and Pim1-KO mouse heart sections co-stained with myosin light chain 2 (MYLC2, red) and DAPI (blue). Scale bar represents 25 μm.
3.4 Telomere length is maintained by TGFβ inhibition in cardiomyocytes
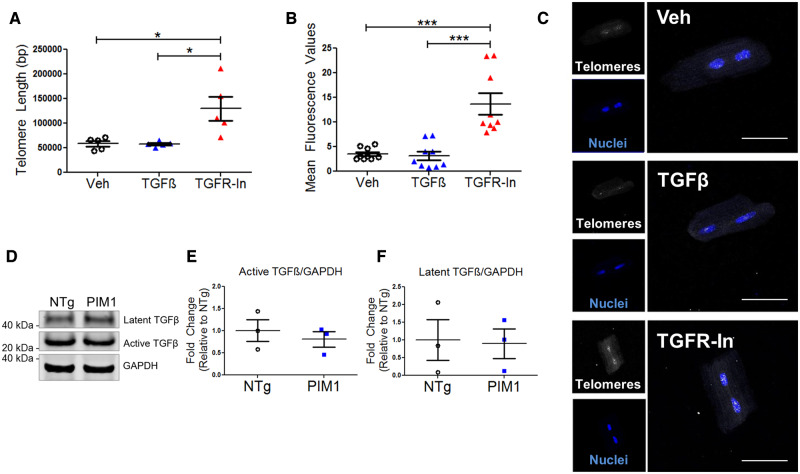
The link between TGFβ signalling and telomere length maintenance has not been established in cardiomyocytes. Therefore, telomere lengths were measured by qRT–PCR in isolated adult NTg mouse cardiomyocytes following treatment with TGFβ or TGFR-In for 48 h. Telomeres were 1.87-fold longer following treatment with TGFR-In, whereas no significant change to telomere length was observed with TGFβ treatment compared to the vehicle control (Figure 4A). Furthermore, treatment of Pim1-KO with TGFR-In rescued the telomere shortening observed (see Supplementary material online, Figure S2A). Addition of TGFR-In to PIM1 cardiomyocytes did not further maintain the telomeres, suggesting that PIM1 and TGFR-In act via similar or concurrent pathways to maintain the telomeres in cardiomyocytes (see Supplementary material online, Figure S2B). Telomeres rapidly eroded in isolated cardiomyocytes over 48 h in culture (see Supplementary material online, Figure S2A). Treatment with the antioxidant Trolox maintained telomere length in culture, whereas treatment with blebbistatin had no effect on telomere length, suggesting that telomere attrition in culture was due to oxidative stress and not biomechanical stress (see Supplementary material online, Figure S2C). To further confirm the effect of TGFβ signalling on telomere length, qFISH analysis of telomeres was performed on isolated myocytes following treatment with TGFβ or TGFR-In. Telomere fluorescence intensity was 3.84-fold greater following treatment with TGFR-In compared to the vehicle control (Figure 4B and C). Endogenous TGFβ expression was measured to account for the lack of change in telomere length in TGFβ-treated cells. Indeed, endogenous TGFβ expression was detected in cardiomyocytes as determined by immunoblot (Figure 4D). TGFβ is synthesized as a latent precursor protein that cannot interact with its receptor until it is cleaved to yield the active form.28 There was no significant difference in latent and active TGFβ expression between NTg and PIM1 cardiomyocytes (Figure 4E and F). Furthermore, TGFβ expression was evaluated in NTg and PIM1 hearts 7-day post-myocardial infarction. Conditions of stress up-regulated TGFβ expression in both NTg and PIM1, however, there was no significant difference in TGFβ expression between NTg and PIM1 indicating that PIM1 does not directly affect ligand expression (see Supplementary material online, Figure S3B and C). Likewise, treatment of NTg cardiomyocytes with TGFβ had no effect on Pim1 expression suggesting that Pim1 and TGFβ do not exhibit feedback relationships (see Supplementary material online, Figure S3C). Overall, these results demonstrate that telomere length is maintained by inhibition of the TGFβ signalling pathway in cardiomyocytes.
Figure 4.
Telomere length is maintained by TGFβ inhibition in cardiomyocytes. (A) Quantification of telomere lengths in isolated adult mouse cardiomyocytes treated with 4 ng/mL TGFβ or 10 μM TGFR-In for 48 h via qRT–PCR. n = 5, error bars represent SEM, *P < 0.05 vs. Veh and vs. TGFβ as measured by one-way ANOVA followed by Tukey’s multiple comparison test. (B) Quantification of telomere fluorescence intensity indicated as mean fluorescence values from qFISH in isolated cardiomyocytes. n = 3 biological×3 technical replicates per group, error bars represent SEM, ***P < 0.001 vs. Veh and vs. TGFβ as measured by one-way ANOVA followed by Tukey’s multiple comparison test. (C) Representative qFISH images of telomeres (white) in cardiomyocytes stained with DAPI (blue). Scale bar represents 75 μm. (D) Immunoblot of TGFβ expression in NTg and PIM1 cardiomyocytes with corresponding quantification of (E) latent and (F) active TGFβ expression. n = 3, no significance as measured by Student’s t-test.
3.5 Expression of TGFβ downstream targets is reduced by PIM1 in cardiomyocytes
To determine whether TGFβ signalling inhibition is mediated by PIM1 in cardiomyocytes as revealed in A549 cells, phosphorylation of the downstream effector Smad2 was evaluated in PIM1 cardiomyocytes by immunoblot analysis. Cardiac-specific overexpression of PIM1 resulted in a 39% reduction in pSmad2 levels compared to NTg (Figure 5A and B). Pim1-KO cardiomyocytes also displayed an up-regulation of pSmad2 compared to PIM1 (see Supplementary material online, Figure S4A and B). However, there was no significant difference in pSmad2/Smad2 between the NTg and Pim1-KO (see Supplementary material online, Figure S4A and B). Adult cardiomyocytes already have very low basal levels of Pim1 which could account for the lack of difference observed between the NTg and Pim1-KO. Immunoblot analysis of pSmad3 also revealed a reduction of TGFβ signalling in PIM1 compared to NTg and Pim1-KO (see Supplementary material online, Figure S4C and D). Additionally, PIM1 cardiomyocytes showed significant down-regulation of TGFβ pathway target genes Serpine1 and FoxA2 (Figure 5C and D). Taken together, these results demonstrate the role of PIM1 in maintaining telomeres in cardiomyocytes through inhibition of the canonical TGFβ signalling pathway.
Figure 5.
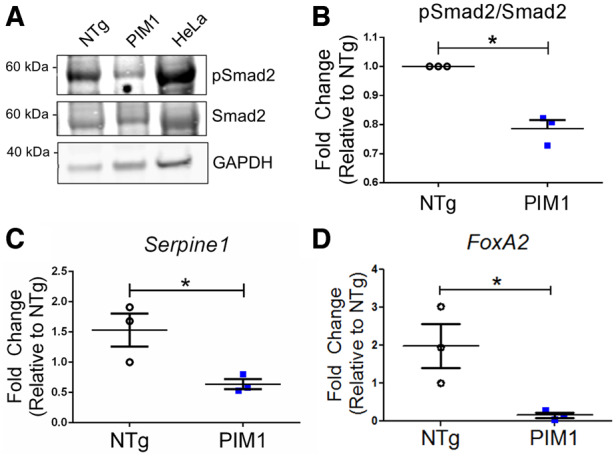
Expression of TGFβ downstream targets is reduced by PIM1 in cardiomyocytes. (A) Western-blot image of pSmad2 and total Smad2 in non-transgenic (NTg) and PIM1 cardiomyocytes. (B) Quantification of pSmad2 over total Smad2 protein expression in cardiomyocytes. n = 3, error bars represent SEM, *P < 0.05 vs. NTg as measured by Student’s t-test. (C) Serpine1 and (D) FoxA2 gene expression in NTg and PIM1 cardiomyocytes. n = 3, error bars represent SEM, *P < 0.05 vs. NTg as measured by Student’s t-test.
3.6 Pim1-mediated maintenance of telomere length is TERT dependent
Upon TGFβ stimulation, Smad2/3 is phosphorylated and translocated into the nucleus where it binds to the TERT gene promotor, repressing transcriptional activation of TERT.13 To determine if TERT is involved in PIM1-mediated telomere maintenance in cardiomyoctes, TERT protein expression was analysed in PIM1 and Pim1-KO cardiomyocytes. Expression of TERT was coincident with telomere length such that PIM1 mice had increased TERT levels compared to NTg (Figure 6A and B). Conversely Pim1-KO mice had significant reduction in TERT protein expression relative to Ntg (Figure 6A and B). The mechanism underlying PIM1-mediated telomere maintenance was further assessed by knocking down TERT expression in cardiomyocytes isolated from NTg and PIM1 mice. Knocking down TERT blunted PIM1-mediated increases in telomere length indicating that TERT is a critical intermediate in PIM1-mediated telomere maintenance in cardiomyocytes (Figure 6C).
Figure 6.
PIM1-mediated maintenance of telomere length is TERT dependent. (A) Western-blot image of TERT expression in NTg, PIM1, and Pim1-KO cardiomyocytes. (B) Quantification of TERT protein expression in cardiomyocytes. n = 5 biological×1–3 technical replicates per group, error bars represent SEM, **P < 0.01 and ***P < 0.001 as measured by one-way ANOVA followed by Tukey’s multiple comparison test. (C) qRT–PCR quantification of telomere lengths in transduced cardiomyocytes. n = 3–7 per group, error bars represent SEM, **P < 0.01 as measured by one-way ANOVA followed by Tukey’s multiple comparison test.
4. Discussion
Telomere-associated cellular senescence of cardiomyocytes remains a hallmark of heart failure that contributes to long-term deterioration of the heart.4 Telomere attrition in mouse cardiomyocytes resulted in increased cardiomyocyte apoptosis, cellular hypertrophy, up-regulation of the senescence marker, p16, and increased levels of the proapoptotic transcription factor, p53.5,29 Functionally, hearts with critically short telomeres in cardiomyocytes displayed cardiac dysfunction characterized by increased end diastolic left ventricular pressure and decreased contractility and relaxation of the left ventricle.30 Delineating mechanisms of telomere length maintenance are key to antagonizing telomere-associated cellular aging and preserving cardiac function. Herein, we establish a novel link between two major signalling molecules that regulate telomere length. PIM1 inhibits phosphorylation of Smad2, the downstream effector of TGFβ signalling, which in turn maintains telomere length in vitro. These findings provide novel insights into PIM1-mediated mechanisms in maintaining telomere length and antagonizing cardiac cellular aging.
Previous reports have demonstrated PIM1-mediated telomere maintenance in CICs depends upon TERT activity.9 CICs overexpressing PIM1 showed no increase in telomere length following treatment with a TERT inhibitor, suggesting that PIM1 maintains telomeres through a TERT mediated pathway.9 Telomeres are maintained by telomerase enzyme that comprises telomerase RNA template (TER), and the protein component, TERT.13 TGFβ signalling has been shown to negatively regulate telomerase activity by repressing expression of the TERT gene.13 In the presence of TGFβ, Smad2/3 is phosphorylated and translocated into the nucleus where it binds to the TERT gene promotor, repressing transcriptional activation of TERT.13 Inhibition of TGFβ signalling is likely influencing telomere length in a similar manner in cardiomyocytes. Findings from this article suggest PIM1-mediated telomere maintenance through inhibition of TGFβ signalling potentially occurs in a TERT-dependent pathway.
Collectively, findings from this study along with previously published reports emphasize the crucial role of Pim1 kinase in reversing aging phenotypes in different cardiac cell populations and antagonizing myocardial aging by preserving telomeres. Additionally, Pim1-mediated inhibition of TGβF signalling further preserves cardiac function. Endogenous TGFβ signalling in the heart contributes to the pathogenesis of cardiac fibrotic and hypertrophic re-modelling in the pressure-overloaded heart.15 TGFβ signalling drives pathological hypertrophy through suppression of antioxidant genes Mt1/2.31 PIM1-mediated inhibition of the TGFβ signalling pathway in cardiomyocytes (Figure 5) suggests a plausible mechanism by which Pim1 inhibits pathological re-modelling and preserves cardiac function with age and post-injury. On the other hand, a recent report documented the involvement of PIM1 in Smad2/3 activation in clear-cell renal-cell carcinoma,32 indicating differential regulation of Smad2/3 by Pim1 in different cell types. PIM1 has been demonstrated to play a dual role in cancers, acting as either an activator or an suppressor depending on the type of cancer.32 PIM1 is overexpressed in breast cancer, mesothelioma, glioblastoma, and clear-cell renal-cell carcinoma and contributes to increased Smad2 phosphorylation and EMT.32 Conversely, PIM1 is down-regulated in other cancers such as pancreatic ductal carcinoma and non-small cell lung cancer.32–34 Therefore, the interaction between PIM1 and Smad2 is highly dependent on the cell type and the associated signalling pathways. In this study A549 cells, a non-small cell lung cancer line, were utilized because these cells have very low basal expression of PIM1, similar to adult cardiomyocytes.
While the present study introduces a novel mechanistic link between Pim1 and TGFβ signalling pathways in telomere regulation, the mechanism in the signalling cascade remain to be elucidated. Pim1 is a serine/threonine kinase that phosphorylates a multitude of targets including Bad, p21, and Cdc25A.35–37 However, findings demonstrate that overexpression of Pim1 results in decreased levels of phosphorylated Smad2 (Figures 2 and 5). The antagonistic relationship between Pim1 and Smad phosphorylation supports previous reports demonstrating a repressive effect of Akt upon the TGFβ pathway. The mechanism of Pim1-mediated inhibition of SMAD2 phosphorylation has not been identified, but it hypothesized that Pim1 acts in a similar manner as Akt. Akt has been shown to inhibit TGFß signalling through direct interaction with Smad3.38,39 Co-immunoprecipitation assays show that Akt physically binds unphosphorylated Smad3 to sequester it outside the nucleus and to prevent its phosphorylation.38,39 This interaction is constitutive but is inhibited by treatment with TGFß.38,39 Furthermore, inhibition of Smad3 phosphorylation by Akt occurs in a kinase-independent manner. A catalytically inactive Akt mutant is effective at suppressing TGFß-mediated apoptosis.39 Pim1 operates downstream of Akt and mediates the cardioprotective signalling of Akt.7 Akt is a serine-threonine kinase involved in cell proliferation and survival.7 Nuclear accumulation of Akt induces Pim1 expression, mediating the cardioprotective effects of Akt.40 Ablation of Pim1 induces Akt expression and activation, but the increase in Akt activation does not mediate enhanced recovery or reduced apoptosis following infarction, indicating that the cardioprotective effects of Akt depend on Pim1.7 Future studies could assess whether Pim1-induced Smad inhibition is a consequence of physical interaction and sequestration of Smad proteins. Additionally, identifying downstream effectors of Pim1/TGFβ signalling in telomere regulation remains a topic for future investigations.
In summary, findings from this article demonstrate a novel mechanism by which PIM1-mediated inhibition of the canonical TGFβ signalling pathway contributes to telomere maintenance, consistent with cumulative evidence of Pim1-induced reversal of aging phenotypes, particularly in myocardial aging. Preservation of a youthful cellular phenotype is key to blunting damage in the injured myocardium and preserving cardiac function. Profound mechanistic understanding of telomere maintenace could provide valuable information towards developing novel therapeutic strategies for heart failure as well as a variety of age-related disorders.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
M.S., N.G., and M.G. designed and directed the project. D.E., F.K., K.M., M.M., C.E., V.S., N.H., K.K., M.M., J.E., C.C., P.Q., J.N., R.A., M.V., M.K., B.W., F.F., J.N., performed the experiments. D.E., F.K., K.B., M.M., V.S., N.H., K.K., M.M., J.E., C.C., P.Q., J.N., R.A., M.V., M.K., B.W., F.F., N.G., and M.S. analysed and interpreted the data. D.E. and F.K. wrote the paper with input from all the authors.
Supplementary Material
Acknowledgements
The authors thank all former and current members of the Sussman lab for their critical feedback and review of the article.
Conflict of interest: M.S. is Chief Scientific Officer and a founding member of CardioCreate, Inc. N.H. is an employee of Roche Molecular Systems, Inc., where her work is unrelated to the content of this article.
Funding
This work was supported by National Institutes of Health (4R37HL091102, 5R01HL105759, 5R01HL067245, 1R01HL113647, 1R01HL117163, and 2P01HL085577 to M.A.S.), American Heart Association Scientist Development Grant (16SDG30970046 to N.H.), and Foundation Leducq Transatlantic Network Consortium.
Time for primary review: 36 days
Translational perspective
Telomere maintenance is associated with preservation of cardiomyocyte functional competency and survival, with telomeric shortening linked to cardiomyopathic disease. Aging also contributes to telomere erosion that contributes to deterioration of myocardial performance. Pim1 kinase mediates beneficial effects to preserve cardiomyocyte survival and function and this report demonstrates a novel molecular mechanism of Pim1 action to mitigate telomeric shortening. Findings linking Pim1 to telomere biology introduce another facet of cardioprotection that can be developed as a molecular interventional approach to treat myocardial injury and heart failure.
References
- 1. Lansdorp PM. Telomeres and disease. EMBO J 2009;28:2532–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blasco MA. Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet 2005;6:611–622. [DOI] [PubMed] [Google Scholar]
- 3. Yeh J-K, Wang C-Y.. Telomeres and telomerase in cardiovascular diseases. Genes 2016;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharifi-Sanjani M, Oyster NM, Tichy ED, Bedi KC, Harel O, Margulies KB, Mourkioti F.. Cardiomyocyte-specific telomere shortening is a distinct signature of heart failure in humans. J Am Heart Assoc 2017;6:e005086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Booth SA, Charchar FJ.. Cardiac telomere length in heart development, function, and disease. Physiol Genomics 2017;49:368–384. [DOI] [PubMed] [Google Scholar]
- 6. Armanios M. Telomeres and age-related disease: how telomere biology informs clinical paradigms. J Clin Invest 2013;123:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muraski JA, Rota M, Misao Y, Fransioli J, Cottage C, Gude N, Esposito G, Delucchi F, Arcarese M, Alvarez R, Siddiqi S, Emmanuel GN, Wu W, Fischer K, Martindale JJ, Glembotski CC, Leri A, Kajstura J, Magnuson N, Berns A, Beretta RM, Houser SR, Schaefer EM, Anversa P, Sussman MA.. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nat Med 2007;13:1467–1475. [DOI] [PubMed] [Google Scholar]
- 8. Borillo GA, Mason M, Quijada P, VöLkers M, Cottage C, McGregor M, Din S, Fischer K, Gude N, Avitabile D, Barlow S, Alvarez R, Truffa S, Whittaker R, Glassy MS, Gustafsson AB, Miyamoto S, Glembotski CC, Gottlieb RA, Brown JH, Sussman MA.. Pim-1 kinase protects mitochondrial integrity in cardiomyocytes. Circ Res 2010;106:1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cottage CT, Neidig L, Sundararaman B, Din S, Joyo AY, Bailey B, Gude N, Hariharan N, Sussman MA.. Increased mitotic rate coincident with transient telomere lengthening resulting from Pim-1 overexpression in cardiac progenitor cells. Stem Cells 2012;30:2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohsin S, Khan M, Nguyen J, Alkatib M, Siddiqi S, Hariharan N, Wallach K, Monsanto M, Gude N, Dembitsky W, Sussman MA.. Rejuvenation of human cardiac progenitor cells with Pim-1 kinase. Circ Res 2013;113:1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Din S, Konstandin MH, Johnson B, Emathinger J, Völkers M, Toko H, Collins B, Ormachea L, Samse K, Kubli DA, La Torre AD, Kraft AS, Gustafsson AB, Kelly DP, Sussman MA.. Metabolic dysfunction consistent with premature aging results from deletion of Pim kinases. Circ Res 2014;115:376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang H, Kyo S, Takatura M, Sun L.. Autocrine transforming growth factor beta suppresses telomerase activity and transcription of human telomerase reverse transcriptase in human cancer cells. Cell Growth Differ 2001;12:119–127. [PubMed] [Google Scholar]
- 13. Li H, Xu D, Li J, Berndt MC, Liu JP.. Transforming growth factor β suppresses human telomerase reverse transcriptase (hTERT) by Smad3 interactions with c-Myc and the hTERT gene. J Biol Chem 2006;281:25588–25600. [DOI] [PubMed] [Google Scholar]
- 14. Shi Y, Massagué J.. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 2003;113:685–700. [DOI] [PubMed] [Google Scholar]
- 15. Dobaczewski M, Chen W, Frangogiannis NG.. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J Mol Cell Cardiol 2011;51:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cassar L, Nicholls C, Pinto AR, Chen R, Wang L, Li H, Liu JP.. TGF-beta receptor mediated telomerase inhibition, telomere shortening and breast cancer cell senescence. Protein Cell 2017;8:39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lacerte A, Korah J, Roy M, Yang XJ, Lemay S, Lebrun JJ.. Transforming growth factor-β inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal 2008;20:50–59. [DOI] [PubMed] [Google Scholar]
- 18. Fischer KM, Cottage CT, Wu W, Din S, Gude NA, Avitabile D, Quijada P, Collins BL, Fransioli J, Sussman MA.. Enhancement of myocardial regeneration through genetic engineering of cardiac progenitor cells expressing pim-1 kinase. Circulation 2009;120:2077–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubli DA, Cortez MQ, Moyzis AG, Najor RH, Lee Y, Gustafsson ÅB.. PINK1 is dispensable for mitochondrial recruitment of parkin and activation of mitophagy in cardiac myocytes. PLoS One 2015;10:e0130707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 2009;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hemann MT. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res 2000;28:4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hariharan N, Quijada P, Mohsin S, Joyo A, Samse K, Monsanto M, La Torre AD, Avitabile D, Ormachea L, McGregor MJ, Tsai EJ, Sussman MA.. Nucleostemin rejuvenates cardiac progenitor cells and antagonizes myocardial aging. J Am Coll Cardiol 2015;65:133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mohsin S, Khan M, Toko H, Bailey B, Cottage CT, Wallach K, Nag D, Lee A, Siddiqi S, Lan F, Fischer KM, Gude N, Quijada P, Avitabile D, Truffa S, Collins B, Dembitsky W, Wu JC, Sussman MA.. Human cardiac progenitor cells engineered with Pim-I kinase enhance myocardial repair. J Am Coll Cardiol 2012;60:1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao M-M, DePinho RA, Kost-Alimova M, Mourkioti F, Kraft P, Bernstein D, Protopopov A, Meeker AK, Blau HM, Day JW, Kustan J.. Role of telomere dysfunction in cardiac failure in Duchenne muscular dystrophy. Nat Cell Biol 2013;15:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katakura Y, Nakata E, Miura T, Shirahata S.. Transforming growth factor β triggers two independent-senescence programs in cancer cells. Biochem Biophys Res Commun 1999;255:110–115. [DOI] [PubMed] [Google Scholar]
- 26. Bernal A, Tusell L.. Telomeres: implications for cancer development. Int J Mol Sci 2018;19:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muraski JA, Fischer KM, Wu W, Cottage CT, Quijada P, Mason M, Din S, Gude N, Alvarez R, Rota M, Kajstura J, Wang Z, Schaefer E, Chen X, MacDonnel S, Magnuson N, Houser SR, Anversa P, Sussman MA.. Pim-1 kinase antagonizes aspects of myocardial hypertrophy and compensation to pathological pressure overload. Proc Natl Acad Sci U S A 2008;105:13889–13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khalil N. TGF-β: from latent to active. Microbes Infect 1999;1:1255–1263. [DOI] [PubMed] [Google Scholar]
- 29. Leri A, Franco S, Zacheo A, Barlucchi L, Chimenti S, Limana F, Nadal-Ginard B, Kajstura J, Anversa P, Blasco MA.. Ablation of telomerase and telomere loss leads to cardiac dilatation and heart failure associated with p53 upregulation. EMBO J 2003;22:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong LSM, Oeseburg H, Boer RD, Gilst WV, Veldhuisen DV, Harst PVD.. Telomere biology in cardiovascular disease: the TERC−/− mouse as a model for heart failure and ageing. Cardiovasc Res 2009;81:244–252. [DOI] [PubMed] [Google Scholar]
- 31. Lighthouse JK, Burke RM, Velasquez LS, Dirkx RA, Aiezza A, Moravec CS, Alexis JD, Rosenberg A, Small EM.. Exercise promotes a cardioprotective gene program in resident cardiac fibroblasts. JCI Insight 2019;4:e92098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao B, Liu L, Mao J, Zhang Z, Wang Q, Li Q.. PIM1 mediates epithelial-mesenchymal transition by targeting Smads and c-Myc in the nucleus and potentiates clear-cell renal-cell carcinoma oncogenesis article. Cell Death Dis 2018;9: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Warnecke-Eberz U, Bollschweiler E, Drebber U, Pohl A, Baldus SE, Hoelscher AH, Metzger R.. Frequent down-regulation of pim-1 mRNA expression in non-small cell lung cancer is associated with lymph node metastases. Oncol Rep 2008;20:619–624. [PubMed] [Google Scholar]
- 34. Reiser-Erkan C, Erkan M, Pan Z, Bekasi S, Giese NA, Streit S, Michalski CW, Friess H, Kleeff J.. Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in pancreatic ductal adenocarcinoma. Cancer Biol Ther 2008;7:1353–1360. [DOI] [PubMed] [Google Scholar]
- 35. Macdonald A, Campbell DG, Toth R, McLauchlan H, Hastie CJ, Arthur J.. Pim kinases phosphorylate multiple sites in Bad and promote 14-3-3 binding and dissociation from Bcl-XL. BMC Cell Biol 2006;7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y, Wang Z, Magnuson NS.. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res 2007;5:909–922. [DOI] [PubMed] [Google Scholar]
- 37. Mochizuki T, Kitanaka C, Noguchi K, Muramatsu T, Asai A, Kuchino Y.. Physical and functional interactions between pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem 1999;274:18659–18666. [DOI] [PubMed] [Google Scholar]
- 38. Conery AR, Cao Y, Thompson EA, Townsend CM, Ko TC, Luo K.. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-β-induced apoptosis. Nat Cell Biol 2004;6:366–372. [DOI] [PubMed] [Google Scholar]
- 39. Remy I, Montmarquette A, Michnick SW.. PKB/Akt modulates TGF-β signalling through a direct interaction with Smad3. Nat Cell Biol 2004;6:358–365. [DOI] [PubMed] [Google Scholar]
- 40. Sussman MA. Mitochondrial integrity: preservation through Akt/Pim-1 kinase signaling in the cardiomyocyte. Expert Rev Cardiovasc Ther 2009;7:929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.