Abstract
Background
Brain-derived neurotrophic factor (BDNF) is a pro-survival factor in the brain that also regulates energy balance. BDNF loss-of-function point mutations are responsible for haploinsufficiency, causing severe early-onset obesity. Up to date, only a few studies have sequenced this gene to search for rare mutations related to obesity. In this study, we aimed to investigate the prevalence of BDNF variants in a cohort of adults with severe obesity from Brazil.
Material and Methods
This study comprised 201 adults with severe obesity (BMI ≥ 35.0 kg/m2) with onset during childhood- or adolescence/youth. As controls, 73 subjects with normal weight (18.5 ≤ BMI ≤ 24.9 kg/m2) were selected. The exclusion criteria were pregnancy, lactation, the use of medication to lose or gain weight, and the presence of symptoms suggestive of syndromic obesity (only for the case group). The coding region of the BDNF gene was screened by Sanger sequencing. Demographic, anthropometric, and blood pressure parameters were obtained from the participants as well as serum hormone and cytokines concentrations and biochemical values.
Results
As a result, three missense variants [p.(Thr2Ile), p.(Val66Met), and p.(Arg209Gln)] and four synonymous variants (p.Leu107=, p.Thr149=, p.Ala150=, and p.Ser213=) were identified. The p.(Arg209Gln) was predicted as pathogenic by all in silico algorithms used and was not observed in the control group. The individuals carrying the p.(Val66Met) mutated allele had higher waist circumference, HDL-cholesterol and MCP1 levels, and reduced risk of developing metabolic syndrome.
Conclusion
We observed that the common BDNF p.(Val66Met) variant has influenced waist circumference, HDL-cholesterol, and MCP1 levels. This polymorphism has also a protective effect on metabolic syndrome susceptibility. Additionally, we described for the first time a rare potentially pathogenic BDNF variant in a Brazilian patient with severe obesity and childhood-onset.
Keywords: BDNF, severe obesity, rare mutation, early-onset obesity, metabolic syndrome
Introduction
Obesity is characterized as an excessive accumulation of adipose tissue in the body. An individual is considered obese when having a body mass index (BMI, calculated by a ratio between weight [kilograms] to the squared height [meters]) equal to or higher than 30 kg/m2. This disease is an important public health problem, affecting high-, middle-, and low-income countries. Recent data estimated that there were more than 650 million adults and 340 million children and adolescents with obesity worldwide in 2016.1 In Brazil, 20.7% of women and 18.7% of men were obese in 2018.2 This disease has a massive impact on human wellbeing and is associated with the development of several metabolic-related disorders, including type 2 diabetes, hypertension, metabolic syndrome, cardiovascular diseases, and some types of cancers.1,3
Obesity is a complex disorder at both clinical and genetic levels.4,5 The common forms of obesity occur due to the interaction of environmental factors with multiple genetic variants of low or middle effect. However, rare monogenic non-syndromic forms can be caused by mutations with strong effects in a single gene. Currently, some genetic forms of obesity can be treated and novel therapeutic interventions are in development for some specific genetic types, highlighting the importance of determining the molecular contributions of those mutations.6–9 The number of known genes associated with monogenic and polygenic obesity has been growing in the last years, and overlaps between the two forms may occur.10 One of the genes associated with both monogenic and polygenic obesity is the brain-derived neurotrophic factor (BDNF).5,11,12
BDNF gene is localized on chromosome 11p14.1, spanning 4.77 kb. It encodes a 247-amino acid precursor protein, pre-pro-BDNF, that is then cleaved into pro-BDNF. Further, pro-BDNF can be proteolytically cleaved into mature BDNF. This secreted protein is widely expressed in the nervous system and plays an important role in neuronal proliferation, survival, and differentiation.13,14 It has been suggested that BDNF is also involved in energy balance and food intake, acting on the leptin-melanocortin pathway as a downstream target of MC4R signaling.15 Recently, Wang et al showed in mouse models that the deletion of BDNF from the paraventricular nucleus of hypothalamus significantly blunted the sympathetic innervation of adipose tissue and weight loss stimulated by exogenous leptin, implicating BDNF as an indirect but essential factor in leptin top-down signaling.16 In another report, Bdnf heterozygous mice developed aggressiveness and hyperphagia accompanied by increased weight gain and abnormalities in locomotor activity.17,18
In humans, BDNF haploinsufficiency has been related to severe early-onset obesity. Gray et al reported an 8-year-old proband with a de novo chromosomal inversion (46, XX, inv (11) (p13p15.3)), which disrupts one copy of BDNF. The patient exhibited reduced levels of serum BDNF, hyperphagia, severe obesity, impaired cognitive function, and hyperactivity.19 Harcourt et al have found a novel 2.0 Mb heterozygous deletion encompassing the BDNF in a mother and a child with obesity and development delay.20 In this context, complete functional loss of one BDNF allele seems to cause severe obesity and other neurodevelopmental abnormalities. Despite these results, data regarding point pathogenic mutations in this gene are still limited in the literature. Recently, Serra-Juhé et al have identified two potentially pathogenic mutations [p.(Ile231Val) and p.(Cys141Gly)] in patients with severe early-onset obesity and mild to severe hyperphagia from the Spanish population. None of them presented congenital and behavioral problems.12 In addition, Sonoyama et al have reported a missense variant [p.(Glu183Lys)] in a patient with severe obesity and moderately severe learning difficulties.21 Kleinendorst et al have also found two variants [c.133A>C; p.(Ser45Arg) and c.440G>A; p.(Trp147*)] with uncertain clinical significance (VUS) in patients with obesity.22 It is well established that BDNF is essential for regulating energy balance and food intake, and functional loss of one allele can cause severe obesity in humans. Since epidemiological studies of rare BDNF point mutations are scarce in the literature, we aimed to investigate the prevalence of BDNF variants in a cohort of patients with severe obesity from Rio de Janeiro, Brazil.
Subjects and Methods
Subjects and Sample
This cross-sectional observational study included 201 unrelated patients, aged 18 to 65 years from Rio de Janeiro State, Brazil. The inclusion criteria were severe obesity (BMI≥35 kg/m2) with childhood onset (n=122 individuals; 0–11 years) or adolescence/youth onset (n= 79 subjects; 12–21 years). The information about period of obesity-onset was self-reported. The exclusion criteria were pregnancy, lactation, the use of medication to lose weight, and the presence of symptoms suggestive of syndromic obesity (cognitive delay, dysmorphic characteristics, and organ-specific developmental abnormalities). All patients were candidates to undergo bariatric surgery in Brazil. All participants were recruited from a nongovernmental organization, called Rescue Group to Self-Esteem and Citizenship of the Obese (in Portuguese, “Grupo de Resgate à Autoestima e Cidadania do Obeso”). Additionally, we have selected 73 unrelated subjects with normal weight (18.5 ≤ BMI ≤ 24.9 kg/m2). The exclusion criteria were pregnancy, lactation, and the use of medication to lose or gain weight. These participants were volunteers from public hospitals in the same city. This study was performed according to the Declaration of Helsinki (1964) and was approved by the Ethics Committee of the Oswaldo Cruz Foundation (CAAE: 09225113.0.0000/Protocol no: 346.634) and all patients gave their written informed consent after the study had been explained.
Metabolic Phenotyping of Participants
All participants underwent a clinical examination, including anthropometric and biochemical parameters, as well as blood pressure measurements. All information was obtained, after an overnight fast, as previously described.23,24 Briefly, body weight, height, neck, waist, and hip circumference were measured. Waist circumference was measured at the midpoint between the iliac crest and the last costal arch. Hip circumference was evaluated at the level of the greater trochanters. Neck circumference was measured in the midway of the neck. BMI, body adiposity index (BAI), and waist to hip ratio (WHR) were calculated. BMI was calculated as weight in kilograms divided by the square of the height in meters. BAI is used to estimate the percentage of body fat, calculated using the formula: hip circumference/(height1.5)-18.25
The measurement of plasma glucose, total cholesterol (TC), high-density lipoprotein cholesterol (HDL-c), and triglyceride (TG) was performed by oxidase–peroxidase method (BioSystems). Low-density lipoprotein cholesterol (LDL-c) was calculated using Friedewald formula (LDL-c=TC−HDL-c−TG/5). Hemoglobin glycated and C-reactive protein (CRP) were measured using by latex agglutination method and turbidimetric inhibition immunoassay (TINIA), respectively. Blood pressure was measured with the individual seated using blood pressure monitor, indicated for patients with severe obesity.26
Suspicion of hypertension was considered when high blood pressure (140/90 mmHg) was observed or by the use of antihypertensive drug therapy.27 Metabolic syndrome was diagnosed by the presence of abnormalities in any three of the following five measurements: fasting glucose level (≥100 mg/dL or drug treatment for elevated glucose), triglycerides (≥150 mg/dL or drug treatment for elevated triglycerides), HDL-cholesterol (<40 mg/dL in men and <50 mg/dL in women or drug treatment for reduced HDL-cholesterol), waist circumference (≥102 cm in men and ≥88 cm in women) and blood pressure (≥130/85 mmHg or drug treatment for hypertension).28
Demographic Variables
Cigarette smoking status was defined as “never smoked” or “already smoked”, which included current smokers and ex-smokers. Exercise habit was classified as “yes”, if the patient has been practicing physical activities during the last month, or “no”. All information was collected by standardized questionnaires.
Hormone Analysis
Serum hormone and cytokines concentrations were determined in the group of patients with childhood-onset obesity in order to perform an extended characterization of their phenotype. Leptin, resistin, monocyte chemoattractant protein-1/CCL2 (MCP1), and plasminogen activator inhibitor-1 (PAI-1) were measured using Human Adipocyte Magnetic Bead on Bio-Plex 200 Multiplexing Analyzer System (Merck, Darmstadt, Germany). Assays were carried out in accordance with the manufacturer’s protocol.
Mutation Screening
Genomic DNA was isolated from peripheral blood using QIAamp Blood Kit (Qiagen, Valencia, CA, USA). The coding region of BDNF (ENST00000525528.1) was screened by Sanger automatic sequencing. Two sets of primers were designed using Primer3Plus software: BDNF-I forward primer, 5ʹ-GTCTGGTGCAGCTGGAGTTT-3ʹ; BDNF-I reverse primer, 5ʹ-CGCCGTTACCCACTCACTAA-3ʹ; BDNF-II forward primer, 5ʹ-CCTTTGGAGCCTCCTCTTCT-3ʹ; and BDNF-II reverse primer, 5ʹ-AAGTTGTGCGCAAATGACTG-3ʹ. Reactions were performed in a total volume of 25 μL, which included 10–20 ng of genomic DNA, 1 unit of Taq DNA Polymerase Class five, 1x Class five buffer with MgCl2, 0.2 mmol/L of each DNTP, and 0.3 pmoles of each primer. Amplification was carried out as follows: 95°C for 1 minutes, followed by 35 cycles of 94°C for 15 seconds, 61°C for 15 seconds, and 72°C for 1 minute and 30 seconds; then an elongation step of 72°C for 10 minutes.
After amplification, PCR products were visualized on 1.5% agarose gels, purified by ExoSAP-IT™ PCR Product Cleanup Reagent (ThermoFisher Scientific, Waltham, MA, USA) and sequenced using Big Dye Terminator kit v.3.1 (Applied Biosystems, Austin, TX, USA) on 3730xl DNA analyzer (Applied Biosystems). Reactions of sequencing were carried out in a total volume of 10 µL, including 10–40 ng of PCR products, 0.32 pmoles of primer, 1X sequencing buffer, and 1.0 µL Big Dye Terminator kit v.3.1. The sequencing of these products was performed as follows: 40 cycles of 94 ºC for 10 sec, 50 ºC for 5 sec, and 60 ºC for 4 min. The chromatograms were analyzed and aligned with the sequence provided by Ensembl database using BioEdit software. All genetic variations were confirmed by bidirectional sequencing of a second PCR.
Bioinformatic Tools
The BDNF genomic, transcript, and protein sequences were obtained from the Ensembl database (http://www.ensembl.org/) and the National Centre for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The reference sequence was ENST00000525528.1. UniProt database was used to obtain information on the protein structures (http://www.uniprot.org/). The pathogenicity of missense variants was analyzed using Polyphen (Polymorphism Phenotyping) (http://genetics.bwh.harvard.edu/pph2/), SIFT (Sorting Intolerant from Tolerant) (https://sift.bii.a-star.edu.sg/) and MutationTaster (http://www.mutationtaster.org/). The potential pathogenic variants were also interpreted according to current standards and guidelines.29 Identified variants were checked in public databases, such as PubMed, Clinvar, dbSNP (https://www.ncbi.nlm.nih.gov/), Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/), 1000 Genomes Project database (1000 genomes) (http://www.internationalgenome.org/) and Online Archive of Brazilian Mutations (ABraOM) (http://abraom.ib.usp.br/).
Statistical Analysis
Normality of quantitative variables was tested by Kolmogorov–Smirnov and Shapiro Wilk tests, in which all evaluations showed a non-normal distribution. Descriptive statistics were expressed as median (interquartile 25–75%) and number (percentage [%]). Mann–Whitney and χ2 tests were used to compare quantitative and qualitative variables, respectively. Association between polymorphism and qualitative variables was carried out by logistic regression analysis. The effect of polymorphisms on the quantitative parameters was tested by linear regression analysis after a logarithmic transformation of these variables. All linear and logistic regression analyses were adjusted for age and gender (model 1), and then adjusted for age, gender, exercises habits, smoking status, and BMI (model 2) [except for the body weight and BMI]. P-value of 0.05 was applied as a significance cutoff. Statistical analyses were performed using SPSS statistical package (IBM Corporation, Chicago, IL, USA).
Results
Characteristics of Study Population
This study included 201 unrelated Brazilian patients with severe obesity, stratified in childhood-onset obesity (60.7%) and adolescence/youth-onset obesity (39.3%). The clinical characteristics of the study population are shown in Table 1. We compared whether the period of obesity-onset influences on obesity-related traits. Our results showed an increased prevalence of metabolic syndrome in subjects with childhood-onset obesity (77.4% vs 63.1%; P= 0.049). No difference of anthropometric, biochemical, and blood pressure measurements as well as the presence of hypertension was found between the stratified groups.
Table 1.
Characteristics of the Study Population
| Variables | All | Childhood-Onset Obesity | Adolescence/Youth-Onset Obesity | P-value | |||
|---|---|---|---|---|---|---|---|
| n | Values | n | Values | n | Values | ||
| Age (years) | 201 | 36 (28; 43) | 122 | 37.0 (28; 45) | 79 | 34.0 (28.0; 40) | 0.190 |
| Gender | |||||||
| Female | 201 | 160 (79.6) | 122 | 97 (79.5) | 79 | 63 (79.7) | 0.967 |
| Male | 41 (20.4) | 25 (20.5) | 16 (20.3) | ||||
| Weight (kg) | 201 | 128.7 (122.9; 146.0) | 122 | 128.9 (114.0; 146.9) | 79 | 128.7 (110.6; 145.5) | 0.973 |
| Height (m) | 201 | 1.63 (1.58; 1.70) | 122 | 1.64 (1.58; 1.70) | 79 | 1.63 (1.58; 1.70) | 0.997 |
| BMI (kg/m2) | 201 | 47.3 (42.6; 53.0) | 122 | 47.0 (42.8; 52.6) | 79 | 48.0 (42.5; 55.0) | 0.723 |
| BAI | 199 | 50.3 (44.8; 55.6) | 122 | 49.2 (44.4; 55.7) | 77 | 50.7 (45.2; 54.6) | 0.472 |
| Waist circumference (cm) | 199 | 137.0 (126.0; 147.0) | 122 | 137.0 (126.0; 144.0) | 77 | 137.0 (123.5; 149.5) | 0.940 |
| Hip circumference (cm) | 199 | 142.0 (133.0; 152.0) | 122 | 141.8 (133.0; 151.2) | 77 | 143.0 (133.5; 154.0) | 0.492 |
| WHR | 199 | 0.96 (0.90; 1.00) | 122 | 0.97 (0.91; 1.00) | 77 | 0.95 (0.89; 0.99) | 0.195 |
| SBP (mm Hg) | 109 | 132.0 (119.0; 147.5) | 61 | 131.0 (121.0; 148.5) | 48 | 132.0 (115.2; 147.0) | 0.776 |
| DBP (mm Hg) | 109 | 87.0 (77.0; 95.5) | 61 | 88.0 (79.0; 97.5) | 48 | 84.0 (75.2; 94.2) | 0.568 |
| Glucose (mg/dL) | 149 | 96.0 (89.5; 108.0) | 85 | 97.0 (92.0; 105.5) | 64 | 94.5 (88.0; 112.0) | 0.598 |
| Cholesterol total (mg/dL) | 168 | 189.0 (165.0; 216.0) | 98 | 192.0 (162.2; 221.7) | 70 | 184.0 (166.5; 211.2) | 0.444 |
| HDL-cholesterol (mg/dL) | 168 | 46.0 (41.0; 52.7) | 98 | 44.5 (40.0; 52.2) | 70 | 48.0 (43.0; 53.2) | 0.186 |
| LDL-cholesterol (mg/dL) | 164 | 116.0 (96.2; 132.0) | 95 | 118.0 (97.0; 143.0) | 69 | 110.0 (95.0; 127.0) | 0.149 |
| Triglycerides (mg/dL) | 168 | 122.0 (89.2; 162.0) | 98 | 123.5 (84.7; 166.5) | 70 | 120.0 (91.5; 138.2) | 0.558 |
| Hemoglobin glycated (%) | 110 | 5.65 (5.07; 6.30) | 66 | 5.65 (5.10; 6.50) | 44 | 5.65 (5.00; 6.20) | 0.434 |
| CRP (mg/dL) | 109 | 1.05 (0.56; 1.58) | 66 | 1.08 (0.56; 1.64) | 43 | 0.85 (0.64; 1.52) | 0.685 |
| Leptin (pg/mL) | 96 | 2644.7 (2058.5; 3380.0) | 96 | 2644.7 (2058.5; 3380.0) | na | – | |
| MCP1 (pg/mL) | 96 | 246.3 (134.2; 366.8) | 96 | 246.3 (134.2; 366.8) | na | – | |
| PAI-1 (pg/mL) | 96 | 25,210.0 (1,7057.9; 31,640.4) | 96 | 25,210.0 (1,7057.9; 31,640.4) | na | – | |
| Resistin (pg/mL) | 96 | 8330.6 (5708.7; 10,843.6) | 96 | 8330.6 (5708.7; 10,843.6) | na | – | |
| Smoking status | |||||||
| Already smoked | 189 | 22 (11.6) | 118 | 15 (12.7) | 71 | 7 (9.9) | 0.554 |
| Never smoked | 167 (88.4) | 103 (87.3) | 64 (90.1) | ||||
| Physical activity practice | |||||||
| Yes | 191 | 48 (25.1) | 120 | 33 (27.5) | 71 | 15 (21.1) | 0.356 |
| No | 143 (74.9) | 87 (72.5) | 56 (78.9) | ||||
| Metabolic syndrome | |||||||
| Yes | 158 | 113 (71.5) | 93 | 72 (77.4) | 65 | 41 (63.1) | 0.033 |
| No | 45 (28.5) | 21 (22.6) | 24 (36.9) | ||||
| Hypertension | |||||||
| Yes | 193 | 128 (66.3) | 122 | 83 (68.0) | 71 | 45 (63.4) | 0.510 |
| No | 65 (33.7) | 39 (31.9) | 26 (36.6) | ||||
Notes: P-value for differences between childhood- and adolescence/youth-onset obesity. Bold font indicates statistical significance.
Abbreviations: BAI, body adiposity index; BMI, body mass index; CRP, C-reactive protein; DBP, diastolic blood pressure; HDL-cholesterol, high-density lipoprotein-cholesterol; LDL-cholesterol, low-density lipoprotein-cholesterol; MCP1, monocyte chemoattractant protein-1; na, not available; PAI-1, plasminogen activator inhibitor-1; SBP, systolic blood pressure; WHR, waist–hip ratio.
BDNF Mutations
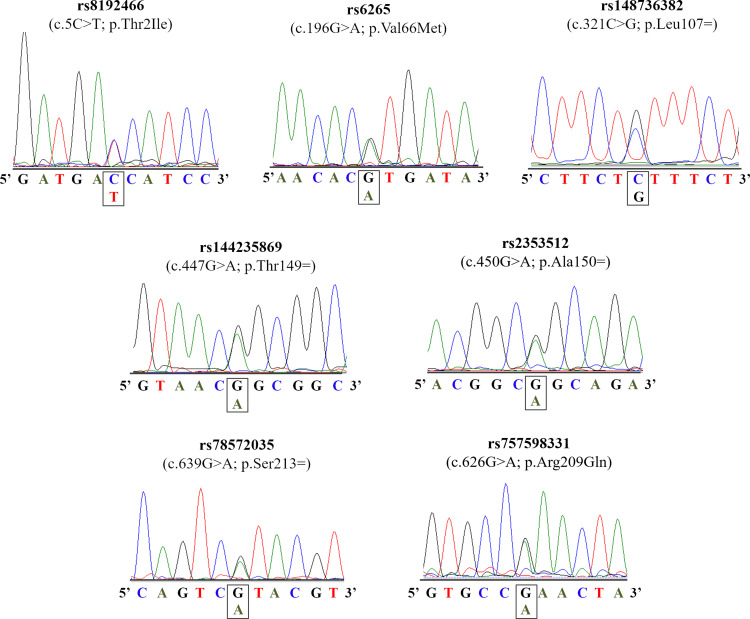
The coding region of BDNF gene was screened in our patients with severe obesity, leading to the identification of seven distinct variants (Table 2). Among these variations, three were missense [c.5C>T, p.(Thr2Ile); c.196G>A, p.(Val66Met); and c.626G>A, p.(Arg209Gln)] and four were synonymous (c.321C>T, p.Leu107=; c.447G>A, p.Thr149=; c.450G>A, p.Ala150=; and c.626G>A, p.Ser213=). The electropherogram of these variants is presented in Figure 1. All alterations, except for the p.Val66Met, were found only in the heterozygous state. Two subjects have two distinct genetic variants [p.(Val66Met)/p.Ser221= and p.(Val66Met)/p.Leu115=].
Table 2.
Summary of Variants Identified in the Coding Region of BDNF Gene
| Mutation Protein | cDNA | dbSNP | Type of Mutation | MAF GnomAD (%) | MAF GnomAD (%) | MAF 1000 Genomes (%) | MAF ABraOM (%) | MAF Our Study (%) | Polyphen-2 | SIFT | Mutation Taster | ClinVar | Onset of Obesity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exomes | Genomes | |||||||||||||
| p.(Thr2Ile) | c.5C>T | rs8192466 | Missense | 0.00126 | 0.001 | 0.001 | 0.001 | 0.0008 | 0.002 | Tolerated | Probably damaging | Disease causing | Uncertain significance | Childhood |
| p.(Val66Met) | c.196G>A | rs6265 | Missense | 0.1901 | 0.195 | 0.152 | 0.201 | 0.139 | 0.092 | Likely benign | Benign | Polymorphism | Benign | Childhood/adolescence/youth |
| p.Leu107= | c.321C>T | rs148736382 | Synonymous | 0.00002 | 0.0002 | 0.001 | 0.001 | NA | 0.005 | - | - | - | na | Childhood/adolescence/youth |
| p.Thr149= | c.447G>A | rs144235869 | Synonymous | 0.00012 | 0.00012 | 0.00016 | NA | 0.0008 | 0.002 | - | - | - | na | Adolescence/youth |
| p.Ala150= | c.450G>A | rs2353512 | Synonymous | 0.9987 | 0.999 | 0.996 | 0.005 | 0.0008 | 0.012 | - | - | - | na | Childhood/adolescence/youth |
| p.Ser213= | c.639G>A | rs78572035 | Synonymous | 0.0005 | 0.00049 | 0.001 | 0.001 | NA | 0.002 | - | - | - | Benign | Adolescence/youth |
| p.(Arg209Gln) | c.626G>A | rs757598331 | Missense | 0.00002 | 0.00001991 | NA | NA | NA | 0.002 | Deleterious | Probably damaging | Disease causing | na | Childhood |
Abbreviations: MAF, minor allele frequency; NA, not available.
Figure 1.
Electropherograms of the BDNF gene.
We also screened the coding region of BDNF in our control cohort, but we did not find the p.(Thr2Ile) or the p.(Arg209Gln) mutations. The p.(Val66Met) is a polymorphism and it was observed in our patient and in controls. Furthermore, we also observed the p.Thr149= and p.Ala150= variants in both groups. No other alteration was observed in our control group. We checked the presence of the missense variants in online mutation databases, and our results showed that the p.(Arg209Gln) variant is rare. This mutation was absent in 1000 Genomes Project and ABraOM, but it was presented in the gnomAD database with a very low allele frequency (MAF = 0.00001991). The p.(Thr2Ile) was found in the three databases with the minor allele frequency under 0.001.
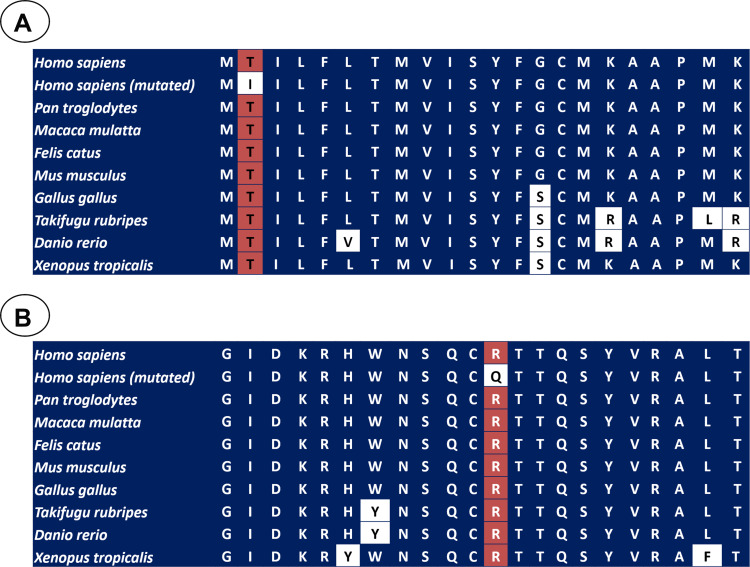
In silico analyses were carried out using three different algorithms in order to investigate the impact of the missense mutations on BDNF structure and function. The p.(Thr2Ile) variant was predicted to be “probably damaging” and “disease causing” by SIFT and Mutation Taster, respectively. Nevertheless, it was predicted as “tolerated” by PolyPhen. Additionally, the p.(Val66Met) alteration was predicted as non-disease causing by all the three softwares. The p.(Arg209Gln) mutation was predicted to be “deleterious”, “probably damaging” and “disease causing” by Polyphen, SIFT, and MutationTaster, respectively. The potential impact of this variant was interpreted according to current standards and guidelines, which classified this mutation as likely pathogenic.29 We also investigated the conservation of the BDNF sequence across different species, suggesting that the threonine at codon 2 [p.(Thr2Ile)] and arginine at codon 209 [p.(Arg209Gln)] are located in a highly conserved amino acid position (Figure 2A and B). Finally, the patients carrying p.(Thr2Ile) and p.(Arg209Gln) variants did not present other potentially deleterious variant in several candidate genes for non-syndromic monogenic obesity (LEP, MC4R, POMC, and MRAP2).
Figure 2.
A portion of the amino acid sequence of BDNF in diverse species, in which the position of p.Thr2Ile (A) and p.Arg209Gln (B) variants is showed in the red squares. The white squares showed the non-conserved position.
Clinical Features of the Patients with the Rare Variants
The patient carrying the BDNF p.(Thr2Ile) variant was a 35-year-old female patient, which reported severe early-onset obesity (childhood). Her body weight was 166.5 kg for 1.70 m of height with a BMI of 57.6 kg/m2. The waist circumference was 134 cm; hip circumference, 168 cm; WHR, 0.80; and BAI, 57.8. We measured biochemical parameters, in which fasting plasma glucose was 89 mg/dl, total cholesterol, 190 mg/dl; HDL-c, 50 mg/dl; LDL-c, 128 mg/dl; and TG, 61 mg/dl. We also measured the concentration of serum hormones and cytokines, leptin, 733.8 pg/mL; MCP1, 50.46 pg/mL; PAI-1, 10,280.98 pg/mL; and resistin, 1199.1 pg/mL. The patient had hypertension and was under treatment to control the blood pressure.
The p.(Arg209Gln) variant was detected in a 46-year-old female patient, who developed obesity during childhood. Her body weight was 102.7 kg for 1.64 m with a BMI of 38.2 kg/m2. On clinical examination, her waist circumference was 140 cm; hip circumference, 128 cm; WHR, 1.09; neck circumference, 37 cm; and BAI, 42.9. The blood pressure was 131/104 mmHg. We obtained biochemical parameters, in which fasting plasma glucose was 92 mg/dl; total cholesterol, 182 mg/dl; HDL-c, 40 mg/dl; LDL- c, 125 mg/dl; TG, 87 mg/dl; hemoglobin glycated, 5.2%; and CRP, 2.11 mg/dl. The serum concentration of hormones and cytokines was not available. The patient presented hypertension and metabolic syndrome (reduced HDL-cholesterol, increased waist circumference, and hypertension).
Genotype–Phenotype Analysis of Common Variants
Our case group was divided according to the period of obesity onset, stratified in childhood-onset obesity, and adolescence/youth-onset obesity. Firstly, we analyzed whether the BDNF p.(Val66Met) (rs6265) polymorphism influences the period of obesity onset; however, no association was found (P= 0.926) (Table 3).
Table 3.
Association of BDNF p.Val66Met Polymorphism and Obesity Related-Traits and Presence of Comorbidities
| Variables | BDNF p.Val66Met Polymorphism (rs6265) | P-valueδ | ||||
|---|---|---|---|---|---|---|
| n | Non-Carriers (GG) | n | Carriers (GA+AA) | P- value¥ | ||
| Weight (kg) | 166 | 128.0 (112.1; 146.0) | 35 | 131.0 (112.9; 145.5) | 0.749 | 0.857 |
| BMI (kg/m2) | 166 | 47.3 (42.7; 52.8) | 35 | 47.5 (42.3; 54.9) | 0.870 | 0.965 |
| BAI | 164 | 50.3 (44.8; 55.0) | 35 | 49.8 (44.5; 59.1) | 0.732 | 0.650 |
| Waist circumference (cm) | 164 | 135.5 (125.5; 145.5) | 35 | 139.0 (130.0; 152.0) | 0.161 | 0.043 |
| Hip circumference (cm) | 164 | 141.7 (133.0; 152.0) | 35 | 147.0 (135.0; 152.0) | 0.530 | 0.180 |
| WHR | 164 | 0.95 (0.90; 0.99) | 35 | 0.98 (0.93; 1.03) | 0.176 | 0.286 |
| SBP (mm Hg) | 90 | 132.0 (120.0; 149.0) | 19 | 126.0 (110.0; 140.0) | 0.683 | 0.935 |
| DBP (mm Hg) | 90 | 87.5 (78.0; 97.2) | 19 | 82.0 (72.0; 91.0) | 0.873 | 0.881 |
| Glucose (mg/dL) | 122 | 96.0 (89.7; 109.0) | 27 | 97.0 (89.0; 104.0) | 0.672 | 0.685 |
| Cholesterol total (mg/dL) | 137 | 190.0 (162.0; 217.0) | 31 | 187.0 (170.0; 214.0) | 0.538 | 0.476 |
| HDL-cholesterol (mg/dL) | 137 | 45.0 (40.0; 52.0) | 31 | 49.0 (44.0; 57.0) | 0.008 | 0.008 |
| LDL-cholesterol (mg/dL) | 134 | 116.0 (95.7; 132.2) | 30 | 116.0 (97.2; 133.7) | 0.563 | 0.456 |
| Triglycerides (mg/dL) | 137 | 125.0 (94.5; 167.0) | 31 | 106.0 (77.0; 135.0) | 0.177 | 0.128 |
| Hemoglobin glycated (%) | 88 | 5.60 (5.10; 6.27) | 22 | 5.80 (4.87; 7.15) | 0.379 | 0.345 |
| CRP (mg/dL) | 87 | 1.04 (0.56; 1.52) | 22 | 1.12 (0.55; 1.89) | 0.509 | 0.366 |
| Leptin (pg/mL) | 79 | 2645.4 (1903.4; 3354.6) | 17 | 2627.0 (2370.0; 3740.2) | 0.407 | 0.290 |
| MCP1 (pg/mL) | 79 | 239.8 (127.9; 361.5) | 17 | 327.1 (236.2; 437.1) | 0.015 | 0.041 |
| PAI-1 (pg/mL) | 79 | 24,940.2 (16,771.7; 31,045.5) | 17 | 34,083.5 (17,798.7; 39,676.8) | 0.104 | 0.252 |
| Resistin (pg/mL) | 79 | 7952.9 (5140.7; 10,504.1) | 17 | 10,488.3 (6901.7; 14,033.8) | 0.028 | 0.115 |
| Period of obesity onset | ||||||
| Childhood | 166 | 101 (60.8) | 35 | 21 (60.0) | 0.665 | 0.632 |
| Adolescence/youth | 65 (39.2) | 14 (40.0) | ||||
| Metabolic syndrome | ||||||
| Yes | 128 | 98 (76.6) | 30 | 16 (53.3) | 0.009 | 0.007 |
| No | 30 (23.4) | 14 (46.7) | ||||
| Hypertension | ||||||
| Yes | 158 | 106 (67.1) | 35 | 22 (62.8) | 0.545 | 0.569 |
| No | 52 (32.9) | 13 (37.1) | ||||
Notes: P-value for differences between BDNF p.Val66Met polymorphism carriers and non-carriers. P-value¥ was adjusted for age and gender (model 1). P-valueδ was adjusted for age, gender, exercises habits, smoking status and BMI (except for BMI and body weight) [model 2]. Bold font indicates statistical significance.
Abbreviations: BAI, body adiposity index; BMI, body mass index; CRP, C-reactive protein; DBP, diastolic blood pressure; HDL-cholesterol, high-density lipoprotein-cholesterol; LDL-cholesterol, low-density lipoprotein-cholesterol; MCP1, monocyte chemoattractant protein-1; na, not available; PAI-1, plasminogen activator inhibitor-1; SBP, systolic blood pressure; WHR, waist–hip ratio.
The effect of BDNF p.(Val66Met) polymorphism on anthropometric, biochemical, serum hormone and cytokines, and blood pressure measurements as well as the presence of comorbidities was also investigated in this study (Table 3). Our results showed that this polymorphism was associated with HDL-cholesterol, MCP1, and resistin levels. However, the association with resistin did not remain after correcting for covariates (model 2). We also observed that this polymorphism influenced waist circumference after adjusting for possible confounding variables (model 2). The results showed higher median of HDL-c among the carriers (49.0 [44.0; 57.0]; n= 31) when compared to non-carriers (45.0 [40.0; 52.0]; n= 137) (P= 0.008). We also observed a significant higher median of MCP1 among the carriers (327.1 [236.2; 437.1]; n= 17) than non-carriers (239.8 [127.9; 361.5]; n= 79) (P= 0.041). Moreover, the median of waist circumference was higher in individuals carrying the mutant allele (139.0 [130.0; 152.0]; n= 35) when compared to subjects carrying the wild-type allele (135.5 [125.5; 145.5]; n= 164) (P= 0.043).
We also observed that this polymorphism was associated with metabolic syndrome in patients with severe obesity. We found that the frequency of the wild-type genotype (GG) was significantly higher in patients with metabolic syndrome when compared to normal cases (76.6% vs 53.3%; P=0.009). This result remained significant after adjusting for the covariates (model 2). Furthermore, individuals with severe obesity carrying BDNF p.Val66Met (G) variant had a 3.4-fold increased risk of developing metabolic syndrome when compared to subjects carrying the mutant allele (GA and AA) (OR= 3.43 CI:[1.41–8.35]; P=0.007).
Discussion
In the present study, we screened the coding region of the BDNF gene in 201 unrelated Brazilian patients with severe obesity from the Southeast of Brazil. Patients developed this phenotype during childhood or adolescence/youth. As a result, a total of seven variants were identified, in which three were missense [p.(Thr2Ile), p.(Val66Met), and p.(Arg209Gln)] and four were synonymous variants (p.Leu107=, p.Thr149=, p.Ala150=, and p.Ser213=). The prevalence of BDNF variants in our sample was 21.9%. Only a few studies have screened the BDNF gene in a cohort of individuals with obesity or overweight in order to identify point mutations as the cause of this disorder. In our study, two patients carrying different rare potentially pathogenic variants [(p.(Thr2Ile) and p.(Arg209Gln)] were identified, and absent in the control group.
In the literature, Friedel et al have screened the coding region of BDNF in 183 children and adolescents with extreme obesity and in 187 students that were underweight from the German population. They identified the p.(Thr2Ile) variant in one patient with morbid obesity, who inherited this variation from the overweight mother.11 Zegers et al performed a mutation screening of BDNF in a large cohort from Belgium (554 children and adolescents with overweight or obesity and 561 subjects with normal weight). They have found the p.(Thr2Ile) variant in three subjects with obesity, but it was also present with comparable frequency in their control group.30 This variant was found in 1000 Genomes Project, ExAC database, and ABraOM. All these results suggest that the p.Thr2Ile is a rare, but a non-disease causing variant.
Furthermore, we identified a BDNF p.(Arg209Gln) missense variant in a 46-year-old female patient with severe obesity (BMI of 38.2 kg/m2), who developed this disease during childhood. The patient also showed hypertension and metabolic syndrome. Our in silico analyses predicted that this variant may have an impact on function, supporting a disease-causing effect. Moreover, BDNF p.(Arg209Gln) was interpreted as likely pathogenic by current standards and guidelines for variant classification.29 Additionally, this variant was absent in our normal-weight controls and in 1000 Genomes Project as well as in the Brazilian mutation database. This variant was observed in the gnomAD database with an extremely low allele frequency.
The arginine at codon 209 is positioned in a highly conserved position among different species from zebrafish up to human, suggesting that this region might be functionally important. According to UniProt database (P23560), the mutated arginine residue is present in the mature BDNF, supporting that it might have an effect in the protein activity. The patient was negative for any other variants in BDNF and in genes related or linked to monogenic obesity,24,31 and the pathogenic p.(Arg209Gln) mutation was the only one detected. All these results support that this variant is disease-causing for non-syndromic monogenic obesity. However, functional experiments are still needed in order to investigate the effect of the p.(Arg209Gln) variant on the function of mature BDNF and the development of obesity. Moreover, additional genetic studies with other genes associated to non-syndromic monogenic obesity should be performed in the future. These studies may allow us to identify other potentially pathogenic variants in our patients.
To the best of our knowledge, the p.(Arg209Gln) variant was not found in the literature. However, some studies have identified rare and potential pathogenic BDNF variants. Recently, Sonoyama et al have identified a missense variant [p.(Glu183Lys)] that disturbed the processing and secreting of the mature peptide. This variant was found in a 15-year-old girl with severe obesity and moderate learning difficulties, which was inherited from her father who also had severe obesity and learning difficulties.21 Serra-Juhé et al have found two predicted pathogenic variants [p.(Ile231Val) and p.(Cys141Gly)] in patients with severe early-onset obesity and mild to severe hyperphagia from the Spanish population. The patients did not exhibit congenital or behavioral problems. None of these variants were observed in the control group. Interestingly, the p.(Cys141Gly) variant was not observed in parental samples, suggesting that it occurred de novo in this participant.12
Finally, Kleinendorst et al have selected 1230 patients with obesity in order to screen 52 obesity-related genes. They identified two rare BDNF variants of unknown significance [c.133A>C; p.(Ser45Arg) and c.440G>A; p.(Trp147*)] in their patients with obesity. The p.(Ser45Arg) (c.133A>C) was found in a 48-year-old female patient and also in her overweight mother. The p.Trp147* (c.440G>A) was identified in a 39-year-old male participant and also in his daughter with obesity. However, his mother with normal weight also presented this variant.22 Further segregation analysis and/or functional studies are necessary to elucidate the pathogenicity of these genetic alterations.
Interestingly, BDNF patients could present some clinical features, including hyperactivity, impaired concentration and memory, reduced nociception, delayed speech and language, limited attention span, and pain sensation.5,32 However, Serra-Juhé et al have found obesity patients with pathogenic missense variants in BDNF which did not exhibit congenital and/or behavioral problems. Our study identified one female patient with potential deleterious variant, who did not present cognitive delay, dysmorphic characteristics, and organ-specific developmental abnormalities. Nevertheless, we suggest a future examination in order to investigate deeply the BDNF specific clinical features.
We also investigated the influence of BDNF p.(Val66Met) polymorphism on obesity-related traits as well as the presence of comorbidities. Our results showed higher levels of HDL-c, MCP1 and waist circumference in patients carrying the mutant allele (GA and AA genotypes). In addition, this polymorphism was associated with a lower prevalence of metabolic syndrome, suggesting a protective effect of this polymorphism. In contrast to our findings, Rana et al (2019) reported that this polymorphism (GA genotype) was associated with reduced HDL-c levels in males from Pakistani population. They also reported that this polymorphism is a risk factor for metabolic syndrome development (independent of BMI). However, they reported an increased risk of having abdominal obesity in female participants.33 Peng et al observed that individuals carrying the A allele had lower HDL-c than non-carriers in a long-lived population of Guangxi Province (China).34 In another study, Suriyaprom et al did not find any association between BDNF p.(Val66Met) polymorphism and risk of metabolic syndrome as well as HDL-c levels and waist circumference in Thai individuals.35 Reasons for the discrepancies of the results might be explained by differences in the genetic background between the cohorts and the selection criteria of the samples. Regarding MCP1 levels, this is the first study showing the association between p.(Val66Met) and the concentration of this chemokine. Therefore, more studies are necessary to mechanistically elucidate this result. The substitution of the valine for methionine at the 66th amino-acid residue in the pro-BDNF protein disturbs intracellular trafficking and decreases depolarization-induced secretion of BDNF without disturbing its baseline constitutive secretion.36,37 Despite these effects on protein secretion and function, several studies have been reporting that p.(Val66Met) polymorphism has a protective effect against obesity and metabolic-related traits,33,38,39 which is in accordance with our data showing an inverse association with the occurrence of metabolic syndrome. The mechanism for this protective effect is still unknown, but we show here that it is at least partially mediated by an increase in HDL-c levels. One possible hypothesis is that the diminished secretion of BDNF in specific activities might result in chronically higher constitutive basal secretion of BDNF, causing inhibition of food intake.40 Further functional studies are needed to investigate how this allele can have a protective effect.
Finally, we also identified that the prevalence of metabolic syndrome was higher in individuals with childhood-onset obesity when compared to whom developed during adolescence/youth. One possible explanation is that the individuals with early-onset had to cope with obesity for a longer period and this may have enhanced the chances of developing comorbidities, including metabolic syndrome. Another possibility may rely on the fact that early-onset obesity is often genetically inherited, and this genetic and epigenetic background may prime the individual for the development of comorbidities.
Our study has some limitations that must be considered when analyzing the results: (1) no family members were available, it was not possible to carry out the segregation analysis; (2) the period of obesity onset was self-reported; (3) functional analyses were not performed in order to investigate the impact of the identified variants on protein structure and function.
Conclusion
In summary, we observed that the BDNF common variant (p.Val66Met) has an influence on waist circumference, HDL-c, and MCP1 levels. Moreover, this polymorphism has a protective effect on metabolic syndrome development in patients with severe obesity. We also described, for the first time, a rare pathogenic mutation in BDNF gene that may cause non-syndromic monogenic obesity. Our results support the clinical utility of genetic testing to detect patients who might benefit from specific medical management, genetic counseling to the family and targeted therapeutic intervention.
Acknowledgments
The authors would like to thank the Nereida Proença da Fonseca and Edson Fernandes for their great technical assistance, Rosimere Lima for her excellent work with the participants and the Programa de Desenvolvimento Tecnológico em Insumos para Saúde (PDTIS) platform for multiplex and Sanger sequencing analyses. We are grateful to patients who kindly agreed to participate in this study. This work was supported by Oswaldo Cruz Foundation (FIOCRUZ, Rio de Janeiro - Brazil), National Council for Scientific and Technological Development (CNPq) and Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (FAPERJ).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.World Health Organization. Obesity and overweight. 2020. Available from: https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight. Accessed April22, 2020.
- 2.Vigitel B. Saúde Suplementar: Vigilaância De Fatores De Risco e Protecçaão Para Doenças Crônicas Por Inquérito Telefônico. Agência Nac Saúde Supl Ministério Da Saúde; 2018. [Google Scholar]
- 3.Garvey WT, Mechanick JI, Brett EM, et al.; Guidelines R of the AOCP. American association of clinical endocrinologists and American college of endocrinology comprehensive clinical practice guidelines for medical care of patients with obesity. Endocr Pract. 2016;22:1–203. doi: 10.4158/EP161365.GL [DOI] [PubMed] [Google Scholar]
- 4.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–442. doi: 10.1038/nrg2594 [DOI] [PubMed] [Google Scholar]
- 5.da Fonseca ACP, Mastronardi C, Johar A, Arcos-Burgos M, Paz-Filho G. Genetics of non-syndromic childhood obesity and the use of high-throughput DNA sequencing technologies. J Diabetes Complications. 2017;31:1549–1561. doi: 10.1016/j.jdiacomp.2017.04.026 [DOI] [PubMed] [Google Scholar]
- 6.Paz-Filho G, Mastronardi C, Delibasi T, Wong M, Licinio J. Congenital leptin deficiency: diagnosis and effects of leptin replacement therapy. Arq Bras Endocrinol Metabol. 2010;54:690–697. doi: 10.1590/S0004-27302010000800005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collet T, Dubern B, Mokrosinski J, et al. Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R de fi ciency. Mol Metab. 2017;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kühnen P, Clement K, Wiegand S, et al. Proopiomelanocortin deficiency treated with a melanocortin-4 receptor agonist. N Engl J Med. 2016;375:240–246. doi: 10.1056/NEJMoa1512693 [DOI] [PubMed] [Google Scholar]
- 9.Grandone A, Di Sessa A, Umano GR, Toraldo R, Del Giudice EM. New treatment modalities for obesity. Best Pract Res Clin Endocrinol Metab. 2018;32:535–549. doi: 10.1016/j.beem.2018.06.007 [DOI] [PubMed] [Google Scholar]
- 10.Pigeyre M, Yazdi FT, Kaur Y, Meyre D. Recent progress in genetics, epigenetics and metagenomics unveils the pathophysiology of human obesity. Clin Sci. 2016;130:943–986. [DOI] [PubMed] [Google Scholar]
- 11.Friedel S, Fontenla Horro F, Wermter AK, et al. Mutation screen of the brain derived neurotrophic factor gene (BDNF): identification of several genetic variants and association studies in patients with obesity, eating disorders, and attention‐deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;132:96–99. doi: 10.1002/ajmg.b.30090 [DOI] [PubMed] [Google Scholar]
- 12.Serra-Juhé C, Martos-Moreno GÁ, de Pieri FB, et al. Heterozygous rare genetic variants in non-syndromic early-onset obesity. Int J Obes. 2019;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tapia-Arancibia L, Rage F, Givalois L, Arancibia S. Physiology of BDNF: focus on hypothalamic function. Front Neuroendocrinol. 2004;25:77–107. doi: 10.1016/j.yfrne.2004.04.001 [DOI] [PubMed] [Google Scholar]
- 14.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B, Goulding EH, Zang K, et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003;6:736. doi: 10.1038/nn1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Loh KH, Wu M, et al. A leptin–BDNF pathway regulating sympathetic innervation of adipose tissue. Nature. 2020;583(7818):839–844. doi: 10.1038/s41586-020-2527-y [DOI] [PubMed] [Google Scholar]
- 17.Kernie SG, Liebl DJ, Parada LF. BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 2000;19:1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons WE, Mamounas LA, Ricaurte GA, et al. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci. 1999;96:15239–15244. doi: 10.1073/pnas.96.26.15239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray J, Yeo GSH, Cox JJ, et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55:3366–3371. doi: 10.2337/db06-0550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harcourt BE, Bullen DVR, Kao K, et al. Maternal inheritance of BDNF deletion, with phenotype of obesity and developmental delay in mother and child. Am J Med Genet A. 2018;176:194–200. doi: 10.1002/ajmg.a.38539 [DOI] [PubMed] [Google Scholar]
- 21.Sonoyama T, Stadler LKJ, Zhu M, et al. Human BDnf/trkB variants impair hippocampal synaptogenesis and associate with neurobehavioural abnormalities. Sci Rep. 2020;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleinendorst L, Massink MPG, Cooiman MI, et al. Genetic obesity: next-generation sequencing results of 1230 patients with obesity. J Med Genet. 2018;55:578–586. doi: 10.1136/jmedgenet-2018-105315 [DOI] [PubMed] [Google Scholar]
- 23.da Fonseca ACP, Abreu GM, Zembrzuski VM, et al. The association of the fat mass and obesity-associated gene (FTO) rs9939609 polymorphism and the severe obesity in a Brazilian population. Diabetes Metab Syndr Obes Targets Ther. 2019;12:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Fonseca ACP, de Medeiros Abreu G, Zembrzuski VM, et al. Identification of the MC4R start lost mutation in a morbidly obese Brazilian patient. Diabetes Metab Syndr Obes Targets Ther. 2019;12:257. doi: 10.2147/DMSO.S189455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergman RN, Stefanovski D, Buchanan TA, et al. Index of body adiposity. Obesity. 2011;19:1083–1089. doi: 10.1038/oby.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancia G, Fagard R, Narkiewicz K, et al. 2013 ESH/ESC guidelines for the management of arterial hypertension: the task force for the management of arterial hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC). Eur Heart J. 2013;34:2159–2219. [DOI] [PubMed] [Google Scholar]
- 27.Chobanian AV. National heart, lung, and blood institute; national high blood pressure education program coordinating committee. seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension. 2003;42:1206–1252. [DOI] [PubMed] [Google Scholar]
- 28.Grundy SM, Cleeman JI, Daniels SR, et al. Diagnosis and management of the metabolic syndrome: an American heart association/national heart, lung, and blood institute scientific statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404 [DOI] [PubMed] [Google Scholar]
- 29.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zegers D, Hendrickx R, Verrijken A, et al. Screening for genetic variants in BDNF that contribute to childhood obesity. Pediatr Obes. 2014;9:36–42. doi: 10.1111/j.2047-6310.2012.00131.x [DOI] [PubMed] [Google Scholar]
- 31.da Fonseca ACP, Abreu GM, Zembrzuski VM, et al. Study of LEP, MRAP2 and POMC genes as potential causes of severe obesity in Brazilian patients. Eat Weight Disord. 2020;1–10. [DOI] [PubMed] [Google Scholar]
- 32.Styne DM, Arslanian SA, Connor EL, et al. Pediatric obesity—assessment, treatment, and prevention: an endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2017;102:709–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rana S, Sultana A, Bhatti AA. Association of BDNF rs6265 and MC4R rs17782313 with metabolic syndrome in Pakistanis. J Biosci. 2019;44:95. doi: 10.1007/s12038-019-9915-1 [DOI] [PubMed] [Google Scholar]
- 34.Peng J-H, Liu C-W, Pan S-L, et al. Potential unfavorable impacts of BDNF Val66Met polymorphisms on metabolic risks in average population in a longevous area. BMC Geriatr. 2017;17:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suriyaprom K, Tungtrongchitr R, Thawnasom K. Measurement of the levels of leptin, BDNF associated with polymorphisms LEP G2548A, LEPR Gln223Arg and BDNF Val66Met in Thai with metabolic syndrome. Diabetol Metab Syndr. 2014;6:6. doi: 10.1186/1758-5996-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z-Y, Patel PD, Sant G, et al. Variant brain-derived neurotrophic factor (BDNF)(Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/S0092-8674(03)00035-7 [DOI] [PubMed] [Google Scholar]
- 38.Gunstad J, Schofield P, Paul RH, et al. BDNF Val66Met polymorphism is associated with body mass index in healthy adults. Neuropsychobiology. 2006;53:153–156. doi: 10.1159/000093341 [DOI] [PubMed] [Google Scholar]
- 39.Wu L, Xi B, Zhang M, et al. Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children. Diabetes. 2010;59:3085–3089. doi: 10.2337/db10-0273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han JC. Rare syndromes and common variants of the brain-derived neurotrophic factor gene in human obesity. Prog Mol Biol Transl Sci. 2016;140:75–95. [DOI] [PubMed] [Google Scholar]