Abstract
Purpose
Determining the optimal strategy to implement systemic treatment modalities has been challenging in triple-negative breast cancer (TNBC). We aim to investigate the role of microRNA-223 (miR-223) as prognostic factor and predictor of response toward chemotherapy in TNBC.
Patients and Methods
We retrospectively analyzed the association of pretreatment miR-223 expression with clinicopathologic characteristics and 36-month overall survival (OS) of 53 all stages TNBC patients. Tumor level of miR-223 was measured using real-time quantitative polymerase chain reaction (expressed in fold change). Cutoff value for miR-223 was determined by using receiver operating curve (ROC). Kaplan–Meier curve was used to perform survival analysis.
Results
The optimum cutoff value for miR-223 was 23.435 (AUC: 0.706, 95%CI: 0.565–0.848; p:0.01; sensitivity: 78.6%; specificity: 56%) and was used to categorize mir-223 expression into over- and underexpressed group. Overexpression of miR-223 was associated with increased expression of EGFR (69.7% vs 35%, p: 0.022) and lower 36-month OS (33.3% vs 70%; median OS±SE (months): 25.66±1.58 vs 30.23±1.99; log rank p<0.05). Worse survival is observed in miR-223 overexpressed group receiving platinum-based chemotherapy compared to miR-223 underexpressed group (mean OS (95%CI) months: 24.7 (20.3–29.1) vs 34.3 (31.2–37.4); p<0.01), while no significant difference observed in non-platinum containing regimen. No significant association was observed between miR-223 expression with other clinicopathologic characteristics.
Conclusion
Overexpression of miR-223 is associated with increased expression of EGFR, worse prognosis, and resistance toward platinum-based chemotherapy in Indonesian TNBC patients.
Keywords: miR-223, chemotherapy, prognosis, EGFR, TNBC
Introduction
Triple-negative breast cancer (TNBC), defined by the absence of estrogen (ER), progesterone (PR), and HER2 receptors, is associated with more aggressive disease characteristic, poor response toward therapy and shorter survival. Despite increased prominence of immunotherapy in TNBC, systemic chemotherapy is still the mainstay of treatment. Unfortunately, only 20% of all TNBC respond to systemic chemotherapy and little is understood behind the mechanism of chemotherapy resistance or susceptibility in these patients.1 Currently we are still unable to decide which patient might or might not benefit from receiving systemic chemotherapy.
MicroRNA-223 (miR-223) has pleiotropic effect in malignancy, meaning it can act both as an oncogenic-microRNA (oncomiR) or as a tumor suppressor, depending on the context.2 MicroRNA-223 is also known to contribute in resistance to chemotherapy in many different malignancies, including resistance to platinum- and anthracycline-based regimens, the two most commonly used regimens in breast cancer therapy.3,4 Evidence on the role of miR-223 in breast cancer, especially in TNBC, is still limited and showing conflicting results.5–8 Furthermore, most of these studies were preclinical studies limiting their implementation in clinical settings. We aim to investigate the role of miR-223 in Indonesian TNBC patients, including its prognostic role, its association with unfavorable clinical features, response toward chemotherapy, as well as association with other biomarkers, such as cytokeratin 5/6 (CK5/6), EGFR, and p53 mutant.
Patients and Methods
Patient Populations
This was a retrospective study conducted at Dr Sardjito Hospital, Yogyakarta, Indonesia. We consecutively included stage I–IV TNBC patients diagnosed between 2014 and 2017. Out of 272 detected patients, 219 were excluded due to incomplete data, leaving 53 patients included in this study. Clinical data was retrieved from medical records. This study has been approved by the IRB Ethics Committee Faculty of Medicine, Public Health, and Nursing, Gadjah Mada University/Dr Sardjito Hospital, Yogyakarta, Indonesia with approval numbers: KE/FK/0751/EC/2018 (first approval) and KE/0286/03/2020 (latest update).
Pathology Assessment
Tumor samples were obtained from formalin-fixed paraffin-embedded (FFPE) tissue stored at the Department of Anatomical Pathology, Dr Sardjito General Hospital, Yogyakarta, Indonesia; Waskhita Laboratory; Panti Rapih General Hospital; and CITO Laboratory.
Real-time Quantitative Polymerase Chain Reaction
Expression of miR-223 was determined by using real-time quantitative polymerase chain reaction (qRT-PCR). For preparation of RNA from FFPE samples, the (QIAGEN GmbH, Hilden, Germany) RNeasy FFPE kit was used according to manufacturer’s recommendations. Three sections of 10 µm FFPE thickness were used per preparation. For quantitative PCR (qPCR), RNA/sample was amplified using the (Genes Laboratories Inc., Seongnam, South Korea) NEXpro™ qRT-PCR Master (SYBR) (Cat. no. NexQ-7000) in (Bioneer, Daejeon, South Korea) a Step One Real Time PCR System (Bioneer ExicyclerTM 96 Real Time Quantitative Thermal Block). MicroRNA-223 forward primer, 5′-AGC CGT GTCAGTTTG TCA AAT-3′, and miR-223 reverse primer, 5′-GTGCAGGGTCCGAGG TC-3′. U6 was used as internal control. U6 forward primer, 5′-CTCGCTTCG GCAGCA CA-3′ and U6 reverse primer, 5′-AACGCTTCACGA ATTTGCGT-3′. GAPDH has been used for normalization of gene expression data. The cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles at 95°C for 20 seconds, annealing at 60°C for 40 seconds and extension at 72°C for 60 seconds.
Immunohistochemistry
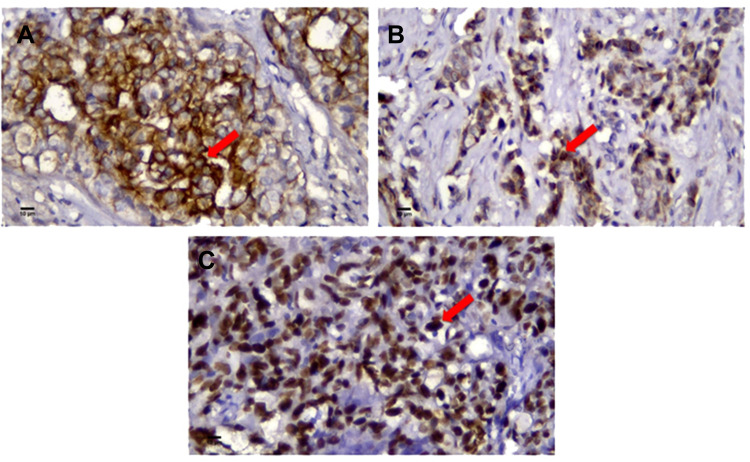
Block paraffin samples were cut 4 µm in thickness to analyze the expression of p53 mutant, EGFR and CK5/6 by immunohistochemistry method. Antibodies used in this study were (Abcam, Cambridge, USA) Clone Ab 32,049, rabbit mAb p53 mutant (dilution 1:50), (Novocastra, Newcastle, UK) Clone NCL-L-EGFR, mouse mAb EGFR (dilution 1:20), and (Cell Marque, Trappes, France) Clone D5 and 16B4, RTU, mouse mAb CK5/6, with chromogen DAB. In this study, the membranous expression of EGFR and CK5/6 in more than 10% of the tumor cells, nuclei expression of p53 mutant in more than 1% of the tumor cells were considered positive (Figure 1).9 The expression of those markers was examined by two senior pathologists independently. If there was any disagreement between the two pathologists, discussion was carried out and consensus had to be made between them.
Figure 1.
Immunohistochemistry of (A) CK5/6 (red arrow: positively stained in cytoplasmic membrane and cytoplasm of tumor cell). (B) EGFR (red arrow: positively stained in cytoplasmic membrane and cytoplasm of tumor cell). (C) p53 mutant (red arrow: positively stained in nucleus of tumor cell). Bar: 10 µm. Magnification: 400 times.
Statistical Analysis
Statistical analysis was performed using IBM SPSS version 24 software. Association between tumor subtype and clinicopathologic features was analyzed using chi-squared test or Fisher’s exact test if assumptions for chi-squared test were violated. All of the reported p-values are two-sided, with significance level set at p<0.05. Cutoff value for miR-223 expression was determined by using receiver operating curve (ROC) to categorize miR-223 expression into over- and underexpressed groups. Survival analysis was performed using Kaplan–Meier curve. Cutoff for overall survival (OS) analysis was set at 36 months.
Results
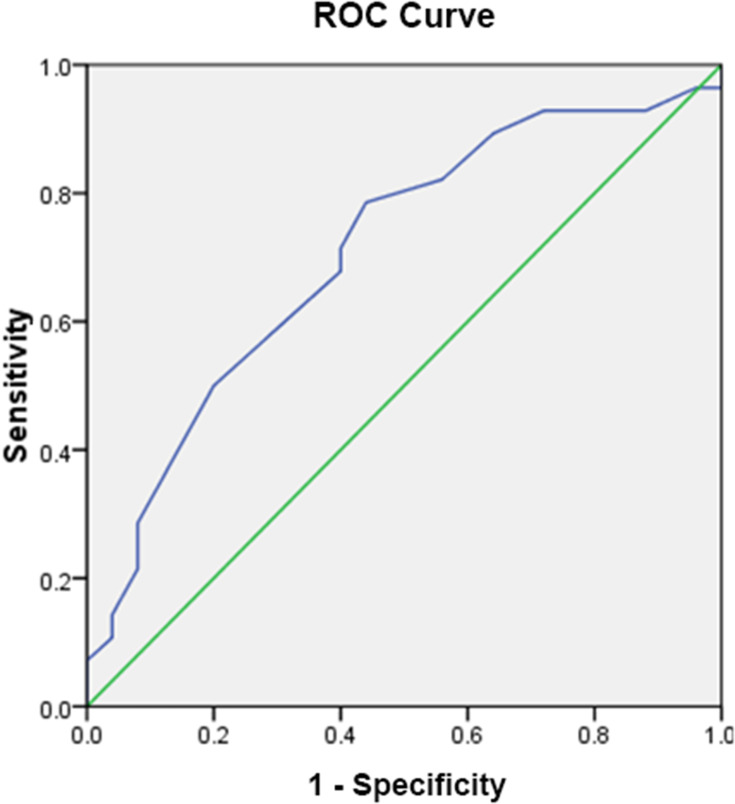
According to the ROC curve, the optimum cutoff value for miR-223 was 23.435 (AUC: 0.706, 95%CI: 0.565–0.848; p: 0.01; sensitivity: 78.6%; specificity: 56%). This cutoff was used to categorize miR-223 expression into over- and underexpressed groups (Figure 2, Table 1). Out of 53 included subjects, 20 were classified into miR-223 underexpressed subgroup and 33 were classified into the miR-223 overexpressed group. The mean age of the subjects was 53 years old, while the median age was 50.28. Subjects were divided into two groups according to its median value, 23 (43.4%) subjects were below 50 years old and 30 (56.6%) subjects more than or equal to 50 years old. The mean and median BMI in this study were 24. Thirty-four (64.2%) subjects had BMI <25 and 19 (35.8%) subjects had BMI ≥25. Seven (13.2%) subjects presented with early stage (I–IIA), 38 (71.7%) subjects with locally advanced stage (IIB-IIIC), while eight (15.1%) subjects with stage IV disease. Thirteen (24.5%) subjects presented with tumor size (T) <5 cm while 40 (75.5%) subjects with T ≥5 cm. Thirty-six (67.9%) subjects presented with no nodal involvement (N), while 17 (32.1%) subjects had at least one nodal metastasis. Distant metastasis was found in eight (15.1%) subjects. There was no significant difference in age, BMI, disease stage, tumor size, nodal status, as well as distant metastasis between miR-223 underexpressed and miR-223 overexpressed subgroup. Subjects in miR-223 overexpressed subgroup had higher frequency EGFR expression (69.7% vs 35%; p<0.05), while no significant difference in expression of p53 mutant (35% vs 45.5%; p:0.454) and CK5/6 (50% vs 60.6%; p:0.57) between the two subgroups (Table 2).
Figure 2.
ROC Curve of miR-223 to Predict 36-months OS with AUC of 0.706.
Table 1.
AUC Value of miR-223 ROC
| Area | Standard Error | p-value | 95%CI | |
|---|---|---|---|---|
| Lower Bound | Upper Bound | |||
| 0.706 | 0.072 | 0.010 | 0.565 | 0.8484 |
Table 2.
Clinicopathologic Profile of TNBC Patients with miR-223 Underexpression and Overexpression
| Parameter | miR-223 Underexpressed | miR-223 Overexpressed | p-value |
|---|---|---|---|
| (n=20) | (n=33) | ||
| EGFR | |||
| Positive | 7 | 23 | 0.013a |
| Negative | 13 | 10 | |
| CK5/6 | |||
| Positive | 10 | 20 | |
| Negative | 10 | 13 | 0.57 |
| p53 Mutant | |||
| Positive | 7 | 15 | |
| Negative | 13 | 18 | 0.454 |
| Stage | |||
| Early | 3 | 4 | |
| Locally- advanced | 16 | 22 | |
| Metastatic | 1 | 7 | 0.209 |
| Tumor size | |||
| <5 cm | 4 | 9 | |
| ≥5 cm | 16 | 24 | 0.555 |
| Nodal status | |||
| Positive | 12 | 24 | |
| Negative | 8 | 9 | 0.336 |
| Distant metastasis | |||
| Present | 1 | 7 | |
| Absent | 19 | 26 | 0.113 |
| Age | |||
| <50 y.o | 6 | 17 | |
| ≥50 y.o | 14 | 16 | 0.126 |
| BMI | |||
| <25 | 11 | 23 | |
| ≥25 | 9 | 10 | 0.279 |
| Chemotherapy regimen | |||
| Platinum | 9 | 14 | |
| Non-platinum | 11 | 17 | |
| Missing | 0 | 2 | 0.559 |
Note: aStatistically significant.
Abbreviations: CK5/6, cytokeratin 5/6; BMI, body mass index.
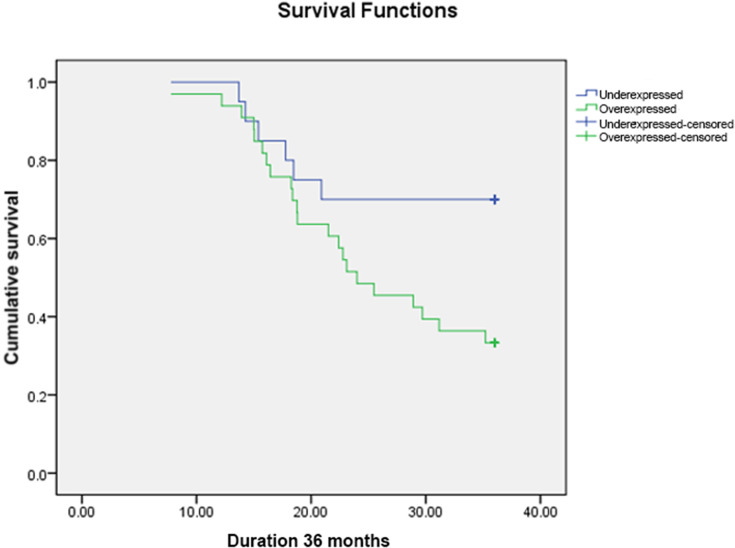
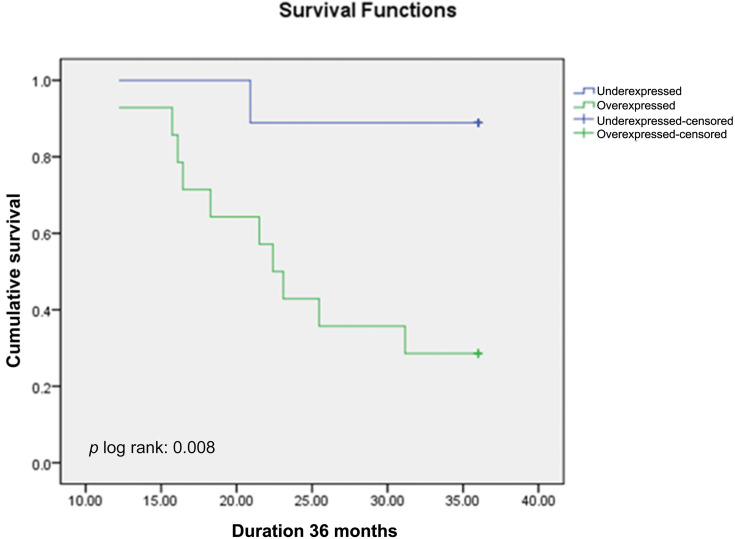
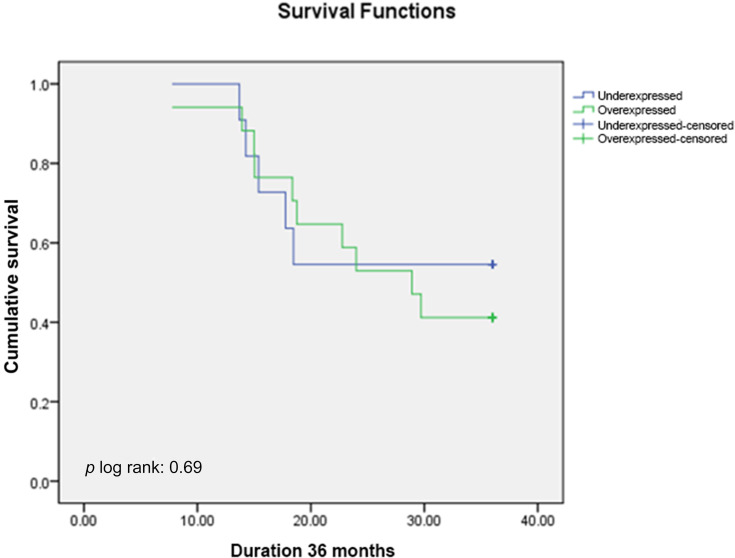
Out of 53 subjects included, 25 subjects (47.2%) were still alive at 36-months follow-up, while 28 subjects (52.8%) were deceased. Twenty-two out of 33 subjects (41.5%) in miR-223 overexpressed subgroup were deceased, which was worse compared to six out of 20 subjects (30%) in miR-223 underexpressed subgroup. Mean OS of miR-223 overexpressed subgroup was 25.7 months (95%CI: 22.6–28.7 months), which was significantly shorter compared to 30.2 months (95%CI: 26.3–34.1 months) in miR-223 underexpressed subgroup (p<0.05) (Figure 3, Table 3). The overall median OS of this study was 31.17 months. When assessing OS according to chemotherapy regimen, there was significant difference between miR-223 overexpressed and miR-223 underexpressed subgroup who received platinum containing regimen (mean OS (95%CI) months: 24.7 (20.3–29.1) vs 34.3 (31.2–37.4); p<0.01) (Figure 4), while there was no significant difference between the two subgroups who received non-platinum containing regimen (mean OS (95%CI) months: 26.2 (21.7–30.8) vs 26.8 (20.9–32.8); p:0.69) (Figure 5). Research data is available in supplementary file 1.
Figure 3.
Kaplan–Meier Overall Survival Curve of All TNBC Patients According to miR-223 Expression.
Table 3.
Mean OS of TNBC Patients According to miR-223 Expression
| miR-223 Expression | Mean OS (Month) | Standard Error (Month) | 95%CI | |
|---|---|---|---|---|
| Lower Bound (Month) | Upper Bound (Month) | |||
| Underexpressed | 30.225 | 1.997 | 26.311 | 34.139 |
| Overexpressed | 25.655 | 1.578 | 22.561 | 28.748 |
| Overall | 27.379 | 1.275 | 24.880 | 29.879 |
Abbreviation: OS, overall survival.
Figure 4.
Kaplan–Meier Overall Survival Curve of TNBC Patients Treated with Platinum-containing Regimen According to miR-223 Expression.
Figure 5.
Kaplan–Meier Overall Survival Curve of TNBC Patients Treated with Non-platinum-containing Regimen According to miR-223 Expression.
Discussion
In our cohort, overexpression of miR-223 was associated with worse prognosis (mean OS: 25.7 vs 30.2 months). Furthermore, EGFR was more frequently expressed in the miR-223 overexpressed group compared to the miR-223 underexpressed group (69.7% vs 35%; p<0.05). Expression of EGFR is generally associated with basal-likeness which represents more aggressive subtype of TNBC.10 Our finding differs from previous preclinical studies which observed miR-223 as tumor suppressor in TNBC cell lines.5,7 On the contrary, a retrospective clinical study observing serum level of circulating miRNA in 60 TNBC patients showed that miR-223 level is significantly elevated in relapsing group compared to nonrelapsing group, suggesting its role as oncomiR, which is consistent with our study.6
The prognostic role of miR-223 in our cohort was especially apparent in patients receiving platinum-based chemotherapy, while no survival difference was observed in patients receiving non-platinum chemotherapy regimen. This might suggest resistance to platinum as one of the underlying mechanisms. Research has shown that platinum-based chemotherapy results in better outcome in TNBC compared to non-TNBC.11,12 Mechanism behind the higher efficacy of this regimen is thought to be caused by homologous recombinant deficiency (HRD) which is a commonly observed characteristic in both BRCA mutated and sporadic-TNBC with “BRCAness” profile.13–16 However, molecular heterogeneity of TNBC causes varied response to chemotherapy amongst these patients. Roughly only 20% of all TNBC patients show excellent response, while the rest exhibit little to no benefit.1 Resistance to chemotherapy in TNBC, including to platinum-based regimen, has been observed in previous studies.17,18 The main mechanisms of chemotherapy resistance in TNBC are (1) ATP-binding cassette (ABC) transporter-mediated drug efflux; (2) cancer stem cells (CSC); (3) hypoxia; (4) avoidance of apoptosis; (5) various signaling pathways, including NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells), PTEN and PI3K-AKT-mTOR pathway, JAK/STAT pathway, and receptor tyrosine kinase; (6) mRNA (although evidence on miR-223 in this subject is lacking); and (7) TNBC heterogeneity.18 Although there is currently no evidence of platinum resistance associated with miR-223 in TNBC, such observations have been made in other types of malignancy. MicroRNA-223 has been reported to cause platinum resistance in non-small-cell lung cancer (NSCLC) through regulation of autophagy, and through deregulation of cell cycle in gastric cancer.3,19 A preclinical study to evaluate the mechanism of miR-223 causing platinum resistance through TNBC cell-line or a xenograft model can improve our understanding on this subject.
Determining the optimal strategy to implement systemic treatment modalities has been challenging in TNBC. Due to the heterogeneity of TNBC, there might not be a “one for all” solution. Instead, the more realistic approach is to identify and decipher the role of various biomarkers with strategic roles in the disease development and resistance/susceptibility towards various therapeutic modalities. Although we are yet unable to define the precise mechanism behind miR-223 as a contributor toward platinum resistance, we were able to define which group of patients would receive benefit from platinum-based chemotherapy and which group would not according to miR-223 expression. However, this is a small sample-sized retrospective study which ideally requires a verification cohort in a prospective study with sizable sample to validate its result.
Conclusion
In summary, miR-223 overexpression is associated with worse prognosis, increased expression of EGFR, and resistance to platinum-based chemotherapy in Indonesian TNBC patients.
Acknowledgments
The authors express gratitude to Sardjito General Hospital for providing the necessary assistance during data procurement for this publication.
Ethics Approval and Informed Consent
Written informed consent was obtained from the patients involved, including permission to use clinical data and reexamination of tissue specimen. This study was conducted in accordance with the Declaration of Helsinki. Ethical clearance was approved by the IRB Ethics Committee Faculty of Medicine, Public Health, and Nursing, Gadjah Mada University/Dr Sardjito Hospital, Yogyakarta, Indonesia with approval numbers: KE/FK/0751/EC/2018 (first approval) and KE/0286/03/2020 (latest update).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
The initial result of this study was presented at the 2020 ESMO Asia Virtual Congress as a poster presentation. The poster’s abstract was published in the Annals of Oncology: https://www.annalsofoncology.org/article/S0923-7534(20)42552-9/fulltext.
References
- 1.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi: 10.1200/JCO.2007.14.4147 [DOI] [PubMed] [Google Scholar]
- 2.Gao Y, Lin L, Li T, et al. The role of miRNA-223 in cancer: function, diagnosis and therapy. Gene. 2017;616:1–7. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Jin W, Jia H, et al. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. 2015;34(1):28. doi: 10.1186/s13046-015-0145-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding J, Zhao Z, Song J, et al. MiR-223 promotes the doxorubicin resistance of colorectal cancer cells via regulating epithelial-mesenchymal transition by targeting FBXW7. Acta Biochim Biophys Sin. 2018;50(6):597–604. doi: 10.1093/abbs/gmy040 [DOI] [PubMed] [Google Scholar]
- 5.Sun X, Li Y, Zheng M, et al. MicroRNA-223 increases the sensitivity of triple-negative breast cancer stem cells to trail-induced apoptosis by targeting HAX-1. PLoS One. 2016;11(9):e0162754. doi: 10.1371/journal.pone.0162754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahlberg KK, Bottai G, Naume B, et al. A serum microRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21(5):1207–1214. doi: 10.1158/1078-0432.CCR-14-2011 [DOI] [PubMed] [Google Scholar]
- 7.Yoshikawa M, Iinuma H, Umemoto Y, et al. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol Lett. 2018;15(6):9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citron F, Segatto I, Vinciguerra GL, et al. Downregulation of miR-223 expression is an early event during mammary transformation and confers resistance to cdk4/6 inhibitors in luminal breast cancer. Cancer Res. 2020;80(5):1064–1077. doi: 10.1158/0008-5472.CAN-19-1793 [DOI] [PubMed] [Google Scholar]
- 9.Maeda T, Nakanishi Y, Hirotani Y, et al. Immunohistochemical co-expression status of cytokeratin 5/6, androgen receptor, and p53 as prognostic factors of adjuvant chemotherapy for triple negative breast cancer. Med Mol Morphol. 2016;49:1:11–21. doi: 10.1007/s00795-015-0109-0 [DOI] [PubMed] [Google Scholar]
- 10.Changavi AA, Shashikala A, Ramji AS. Epidermal growth factor receptor expression in triple negative and nontriple negative breast carcinomas. J Lab Physicians. 2015;7(2):79–83. doi: 10.4103/0974-2727.163129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staudacher L, Cottu PH, Diéras V, et al. Platinum-based chemotherapy in metastatic triple-negative breast cancer: the Institut Curie experience. Ann Oncol. 2011;22(4):848–856. doi: 10.1093/annonc/mdq461 [DOI] [PubMed] [Google Scholar]
- 12.Koshy N, Quispe D, Shi R, et al. Cisplatin-gemcitabine therapy in metastatic breast cancer: improved outcome in triple negative breast cancer patients compared to non-triple negative patients. Breast. 2010;19(3):246–248. doi: 10.1016/j.breast.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 13.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 14.Belli C, Duso BA, Ferraro E, Curigliano G. Homologous recombination deficiency in triple negative breast cancer. Breast. 2019;45:15–21. doi: 10.1016/j.breast.2019.02.007 [DOI] [PubMed] [Google Scholar]
- 15.Tanino H, Kosaka Y, Nishimiya H, et al. BRCAness and prognosis in triple-negative breast cancer patients treated with neoadjuvant chemotherapy. PLoS One. 2016;11(12):e0165721. doi: 10.1371/journal.pone.0165721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oonk AMM, van Rijn C, Smits MM, et al. Clinical correlates of ‘BRCAness’ in triple-negative breast cancer of patients receiving adjuvant chemotherapy. Ann Oncol. 2012;23(9):2301–2305. doi: 10.1093/annonc/mdr621 [DOI] [PubMed] [Google Scholar]
- 17.Hill DP, Harper A, Malcolm J, et al. Cisplatin-resistant triple-negative breast cancer subtypes: multiple mechanisms of resistance. BMC Cancer. 2019;19(1):1039. doi: 10.1186/s12885-019-6278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells. 2019;8(9):957. doi: 10.3390/cells8090957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Chen J, Zhang S, et al. MiR-223 regulates autophagy associated with cisplatin resistance by targeting FBXW7 in human non-small cell lung cancer. Cancer Cell Int. 2020;20:258. doi: 10.1186/s12935-020-01284-x [DOI] [PMC free article] [PubMed] [Google Scholar]