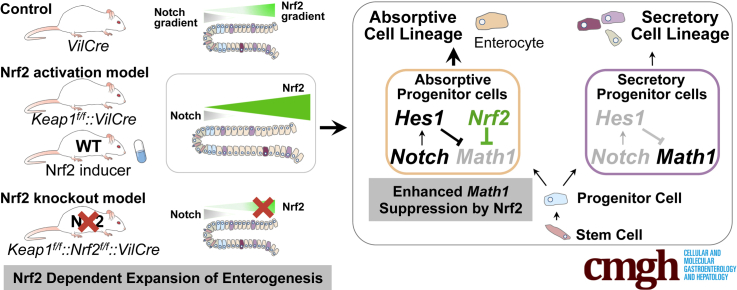
Abstract
Background & Aims
Notch signaling coordinates cell differentiation processes in the intestinal epithelium. The transcription factor Nrf2 orchestrates defense mechanisms by regulating cellular redox homeostasis, which, as shown previously in murine liver, can be amplified through signaling crosstalk with the Notch pathway. However, interplay between these 2 signaling pathways in the gut is unknown.
Methods
Mice modified genetically to amplify Nrf2 in the intestinal epithelium (Keap1f/f::VilCre) were generated as well as pharmacological activation of Nrf2 and subjected to phenotypic and cell lineage analyses. Cell lines were used for reporter gene assays together with Nrf2 overexpression to study transcriptional regulation of the Notch downstream effector.
Results
Constitutive activation of Nrf2 signaling caused increased intestinal length along with expanded cell number and thickness of enterocytes without any alterations of secretory lineage, outcomes abrogated by concomitant disruption of Nrf2. The Nrf2 and Notch pathways in epithelium showed inverse spatial profiles, where Nrf2 activity in crypts was lower than villi. In progenitor cells of Keap1f/f::VilCre mice, Notch downstream effector Math1, which regulates a differentiation balance of cell lineage through lateral inhibition, showed suppressed expression. In vitro results demonstrated Nrf2 negatively regulated Math1, where 6 antioxidant response elements located in the regulatory regions contributed to this repression.
Conclusions
Activation of Nrf2 perturbed the dialog of the Notch cascade though negative regulation of Math1 in progenitor cells, leading to enhanced enterogenesis. The crosstalk between the Nrf2 and Notch pathways could be critical for fine-tuning intestinal homeostasis and point to new approaches for the pharmacological management of absorptive deficiencies.
Keywords: Signaling Crosstalk, Progenitor Cells, Intestinal Homeostasis
Abbreviations used in this paper: ARE, antioxidant response element; CDDO-Im, oleanolic acid 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole; cDNA, complementary DNA; GFP, green fluorescent protein; Hes1, hairy and enhancer of split-1; Keap1, Kelch-like ECH-associated protein 1; LCM, laser capture microdissection; Math1, mouse atonal homolog 1; mRNA, messenger RNA; Nrf2, NF-E2-related factor 2; PCR, polymerase chain reaction
Graphical abstract
Summary.
Activation of Nrf2 signaling in the intestinal epithelium leads to lengthened intestines and expansion of enterogenesis, which is mediated through suppression of Notch downstream effector Math1 in enterocyte progenitor cells. This study indicates the facilitation of intestinal homeostasis by Nrf2.
The small intestine is one of the fastest self-renewing tissues in the mammalian body.1 The rapid rate of cell proliferation is driven by multipotent intestinal epithelial stem cells, which are located at the base of the crypts and produce transit-amplifying progenitors.2 The progenitor cells differentiate into one of the specialized intestinal epithelial cell types, which are categorized into 2 distinct cell lineages; absorptive (enterocyte) and secretory (goblet, Paneth, and enteroendocrine).3 Homeostasis within the stem cell compartment and matured intestinal cells is orchestrated by several signaling pathways.4 Notch and its downstream effectors are pivotal during the multiple processes involved in the maintenance of stem cells, the proliferation of progenitor cells, and the balance of differentiation toward distinct cell lineages.5,6 In the crypt compartment, progenitor cells are committed either to a secretory or absorptive lineage based on the expression of Notch downstream effectors, mouse atonal homolog 1 (Math1) or hairy and enhancer of split-1 (Hes1).7,8 Transgenic mice expressing the Notch intracellular domain constitutively upregulate Hes1 expression, increase the number of proliferating cells and exhibit loss of all secretory cells.5 Hes1-null mice are embryonic lethal, but intestines from these animals show an increase in secretory cells and a decrease in absorptive enterocytes concomitant with elevated expression of Math1.7,9 Embryos from Math1 null mice8 and mice with an intestine-specific deletion of Math110 show loss of the secretory cell lineage, wherein almost all epithelial cells have characteristics of intact absorptive enterocytes. These studies, supported by mouse transgenic technology, powerfully support a Notch-mediated lateral inhibitory mechanism, wherein Notch signaling silences Math1 expressing progenitor cells through direct transcriptional regulation of Hes1 and Math1-negative cells differentiate to enterocytes, whereas Math1-positive cells adopt the secretory fate.11 Taken together, Notch signaling and its downstream effectors are the master molecular players of cell fate commitment in the intestine.
However, signaling crosstalk between the core signaling module, Notch, and other pathways that could influence intestinal homeostasis are still largely unknown. We previously demonstrated the biological significance of signaling crosstalk between the Nrf2 (NF-E2 p45-related factor 2; encoded by Nfe2l2) and Notch pathways in mouse liver, where cross transcriptional regulation between the 2 transcription factors is important in liver development and function.12,13 Nrf2 plays a critical role in the maintenance of cellular redox and metabolic homeostasis, as well as the regulation of inflammation and cellular detoxication pathways. The contributions of the Nrf2 pathway to organismal homeostasis has raised intense attention toward targeting its clinical promise.14 Many phytochemicals (eg, sulforaphane, curcumin) found in commonly consumed foods also activate Nrf2 signaling.15 Under unstressed conditions, Keap1 (kelch-like ECH-associated protein 1), which is an adaptor protein for Cullin3-based ubiquitin E3 ligase, represses transcriptional activity of Nrf2 by ubiquitination and subsequent degradation of Nrf2 through the proteasome pathway.16, 17, 18 Upon exposure to oxidative or electrophilic stress, Keap1 inactivation allows Nrf2 to bind to antioxidant response elements (AREs), activating a battery of Nrf2 targeted genes related with cellular redox homeostasis.19,20 Among 300 transcriptional targets21 are cellular defense enzymes, such as NAD(P)H quinone oxidoreductase 1 (Nqo1) and catalytic and regulatory subunits of glutamate-cysteine ligase (Gclc and Gclm, respectively).22,23
Given these observations, it was hypothesized that the Nrf2 pathway engages with the Notch signaling cascade in the small intestine. Either genetic- or pharmacological- activation of Nrf2 within the intestinal epithelial cells evoked expansion of enterogenesis, leading to larger and increased numbers of absorptive cells. Interestingly, the activities of Nrf2 and Notch pathways in the epithelial cells were detected in reciprocal gradients along the crypt-villus axis in wild-type mice. In the progenitor cells from Nrf2 activated mice, expression of 1 important Notch downstream effector, Math1, was suppressed significantly without changing the expression of a potent Math1 suppresser, Hes1. In vitro results demonstrated the negative transcriptional regulation of Math1 by Nrf2 through AREs in the Math1 promoter. Thus, activation of Nrf2 in the progenitor cells through genetic or epigenetic dysfunction or by drugs and food phytochemicals could perturb the signaling network in the crypt through Math1 negative regulation, which might contribute to enterocyte lineage-specific expansion.
Results
Genetic Activation of Nrf2 Caused Longer Small Intestines and Taller Villi
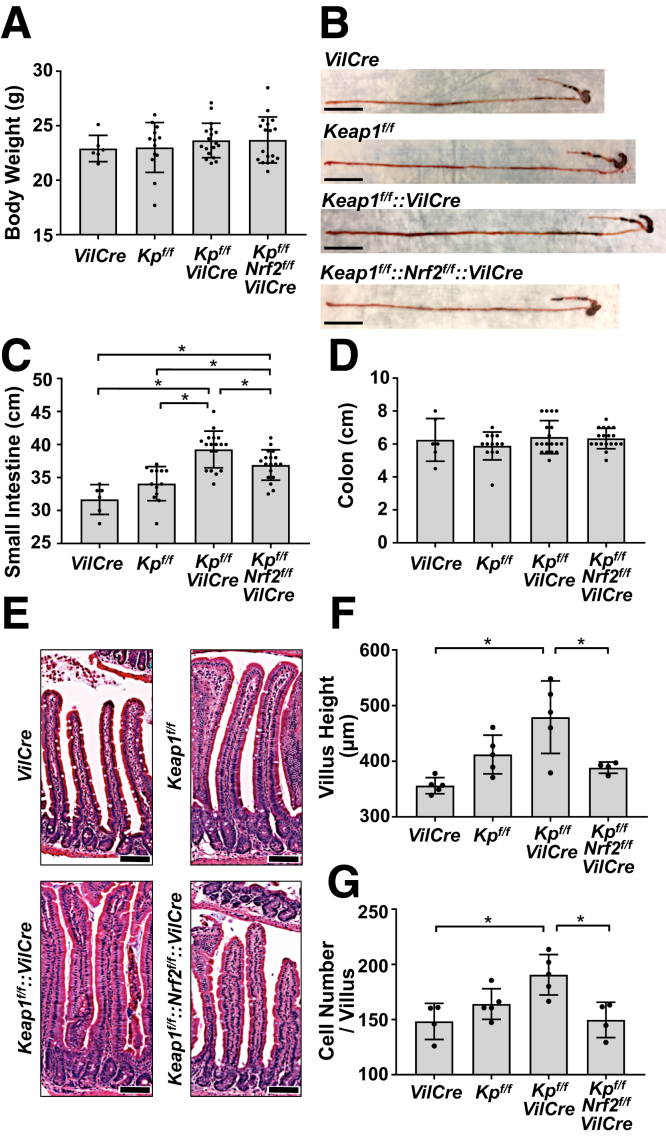
To investigate the association between Nrf2 and the Notch signaling cascade in intestinal epithelial cells, Keap1f/f mice were crossed with VilCre mice (Keap1f/f::VilCre) to establish constitutive activation of Nrf2, where Nrf2 is activated within the intestinal epithelial cells by genetic disruption of the Keap1 gene,24 leading to diminished proteolysis of nascent Nrf2 and amplified signaling. Three genotypes of mice were investigated along with Keap1f/f::VilCre mice: VilCre control mice, Keap1f/f mice (which function as systemic knockdown of Keap1 because of hypomorphic Keap1 gene alleles),25 and Keap1f/f::Nrf2f/f::VilCre mice, wherein the Nrf2 gene was concomitantly disrupted. Body weights of the 4 genotypes of mice did not differ (Figure 1A). Surprisingly, Keap1f/f::VilCre mice exhibited significantly longer small intestines compared with either VilCre or Keap1f/f (Figure 1B and C). Although the small intestines of Keap1f/f::Nrf2f/f::VilCre were longer than in VilCre mice, they were nonetheless significantly shorter than those of Keap1f/f::VilCre, indicating the elongation of entire small intestine observed in Keap1f/f::VilCre was partially canceled by Nrf2 knockout. Even though VilCre is expressed in epithelial cells throughout the entire intestinal tract including the colon,26 the length of colon did not change between the 4 genotypes (Figure 1D).
Figure 1.
Genetic activation of Nrf2 caused longer small intestines and taller villi. Male 8-week-old VilCre, Keap1f/f (Kpf/f), Keap1f/f::VilCre (Kpf/f-VilCre) and Keap1f/f::Nrf2f/f::VilCre (Kpf/f-Nrf2f/f-VilCre) mice were used. (A) Body weights (n = 6–18). (B) Gross morphology of the small intestines. Scale bar = 5 cm. Length of (C) small intestines and (D) colons (n = 6–18). (E) Hematoxylin and eosin staining of longitudinal sections of small intestines. Scale bar = 100 μm. Quantification of (F) villus height and (G) epithelial cell number (n = 4–5). Values are mean ± SD throughout. ∗P < .05 using 1-way analysis of variance.
In addition to intestinal elongation, the Keap1f/f::VilCre exhibited dramatically increased height of villi compared with VilCre control. This enhancement did not occur in Keap1f/f::Nrf2f/f::VilCre (Figure 1E and F), again confirming a role for Nrf2 in these phenotypes. Villus height in Keap1f/f mice trended upward, but did not reach statistical difference from the control animals. The depth of crypts did not show any changes across the 4 genotypes (data not shown). In accordance with increased villus height, the numbers of epithelial cells in villi were significantly elevated in Keap1f/f::VilCre, which again was not observed in Keap1f/f::Nrf2f/f::VilCre (Figure 1G). It is likely that the increased number of epithelial cells contributed to the expanded villus height observed in Keap1f/f::VilCre mice.
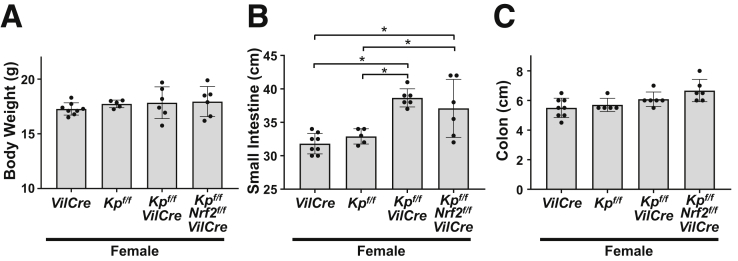
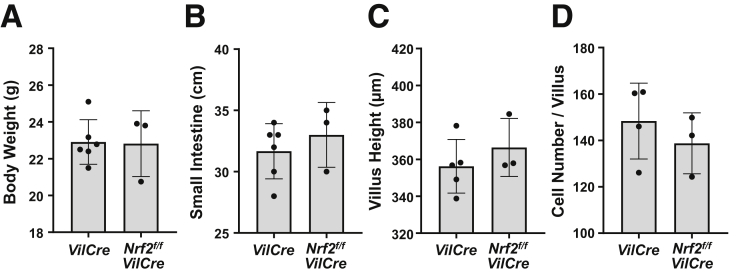
As seen with male mice, elongation of small intestines was consistently observed in female Keap1f/f::VilCre mice without any changes in body weight and colon length (Figure 2A–C). Male mice were analyzed in further studies on the phenotype and underlying mechanisms. Nrf2f/f::VilCre mice were also investigated as a possible control for the comparative analysis with Keap1f/f::Nrf2f/f::VilCre mice; they demonstrated equivalent results with VilCre mice in measures of body weights, length of small intestine, height of villus, and cell number in the villus (Figure 3A–D). Hence, VilCre mice were analyzed as a representative control throughout the study, which is focused on gain-of-function responses to Nrf2 signaling.
Figure 2.
Activation of Nrf2 caused longer small intestines in female mice. Female 8-week-old VilCre, Keap1f/f (Kpf/f), Keap1f/f::VilCre (Kpf/f-VilCre) and Keap1f/f::Nrf2f/f::VilCre (Kpf/f-Nrf2f/f-VilCre) mice were used for the following data. (A) Body weights (n = 5–8). Length of (B) small intestines and (C) colons (n = 5–8). Values are mean ± SD throughout. ∗P < .05 using 1-way analysis of variance.
Figure 3.
Genetic disruption of Nrf2 itself did not alter the length of small intestine or villus height. Comparisons of VilCre (8 weeks old, n = 5–6) and Nrf2f/f::VilCre (8 weeks old, n = 3) genotypes as controls were conducted in male mice for (A) body weight, (B) length of small intestine, (C) villus height, and (D) cell number per villus. No statistical differences by Student’s t test. Values are mean ± SD throughout.
Pharmacological Activation of Nrf2 Signaling Did Not Cause Longer Small Intestines But Increased the Height of Villi
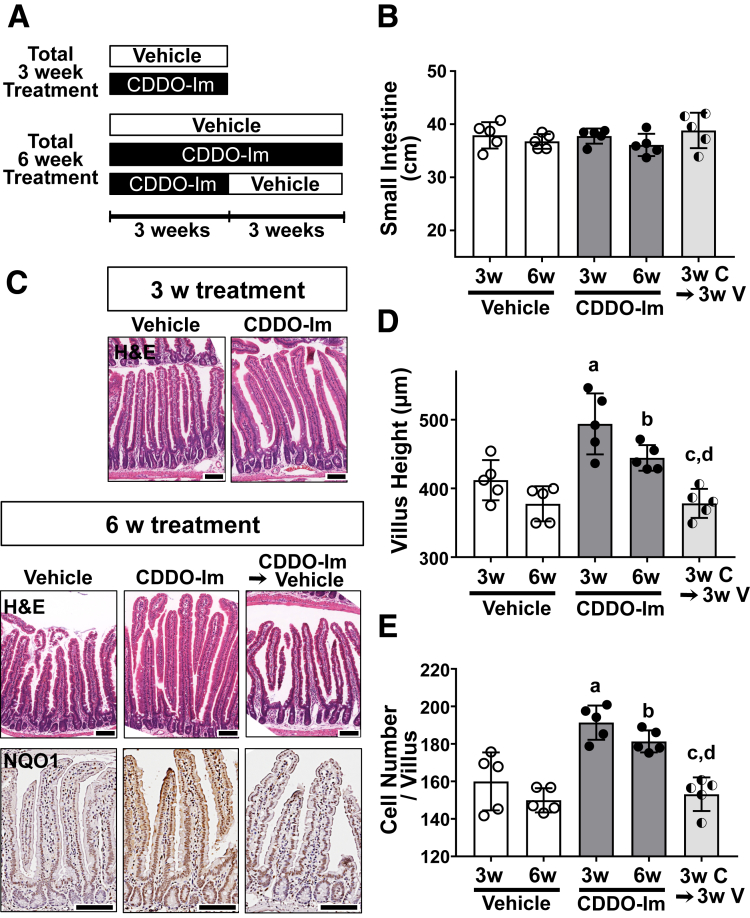
In addition to the genetic models, pharmacological activation of Nrf2 signaling was employed in wild-type mice by treatment with the potent Nrf2 inducer, CDDO-Im (oleanolic acid 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole).27 Mice were orally administered CDDO-Im or vehicle every other day for either 3 or 6 weeks (Figure 4A). To examine possible reversibility of effects of CDDO-Im, mice were also treated with CDDO-Im for 3 weeks, followed by 3 weeks of vehicle treatment. The length of the small intestines did not show any changes by intervention (Figure 4B). However, both 3 and 6 weeks of CDDO-Im administration led to statistically increased heights of villi compared with vehicle (Figure 4C and D). This effect was reversible as 3 weeks of CDDO-Im administration followed by 3 weeks of vehicle treatment restored villus height to basal levels (Figure 4C and D). Immunohistochemistry of NQO1 showed the administration of CDDO-Im indeed activated Nrf2 signaling in the epithelial cells, which disappeared too with cessation of CDDO-Im administration (Figure 4C, lower panels). The number of intestinal epithelium cells per villus was increased significantly by CDDO-Im, but that too was reversible (Figure 4E).
Figure 4.
Pharmacological activation of Nrf2 caused taller villi. (A) Schema displaying treatment groups. Wild-type mice (male, 6 weeks old) were treated with CDDO-Im (C) or vehicle (V), 3 times a week for 3 or 6 weeks. (B) Length of small intestines (n = 5). (C) Hematoxylin and eosin staining in the small intestinal sections of mice treated for 3 weeks (3w) or 6 weeks (6w) (upper and middle panels), and immunohistochemistry for NQO1 (lower panels). Scale bar = 100 μm. Quantification of average (D) villus height and (E) epithelial cell number (n = 5). a,bP < .05 vs 3w or 6w vehicle, respectively; c,dP < .05 vs 3w or 6w CDDO-Im, respectively, using 1-way analysis of variance. Values are mean ± SD throughout.
Activation of Nrf2 Signaling Expanded Enterogenesis in the Small Intestine
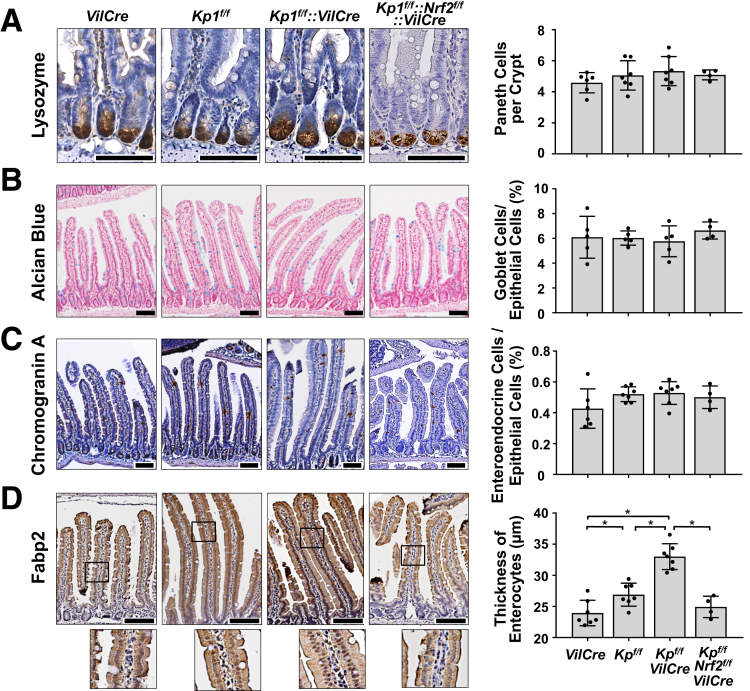
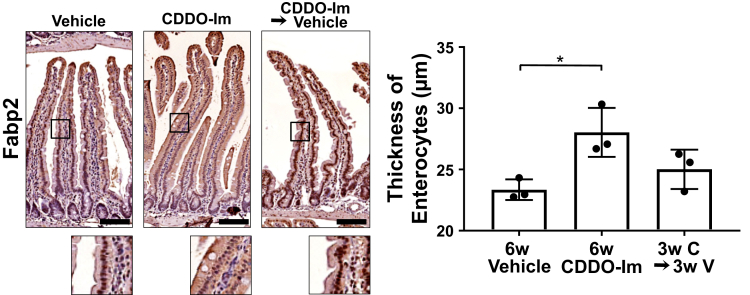
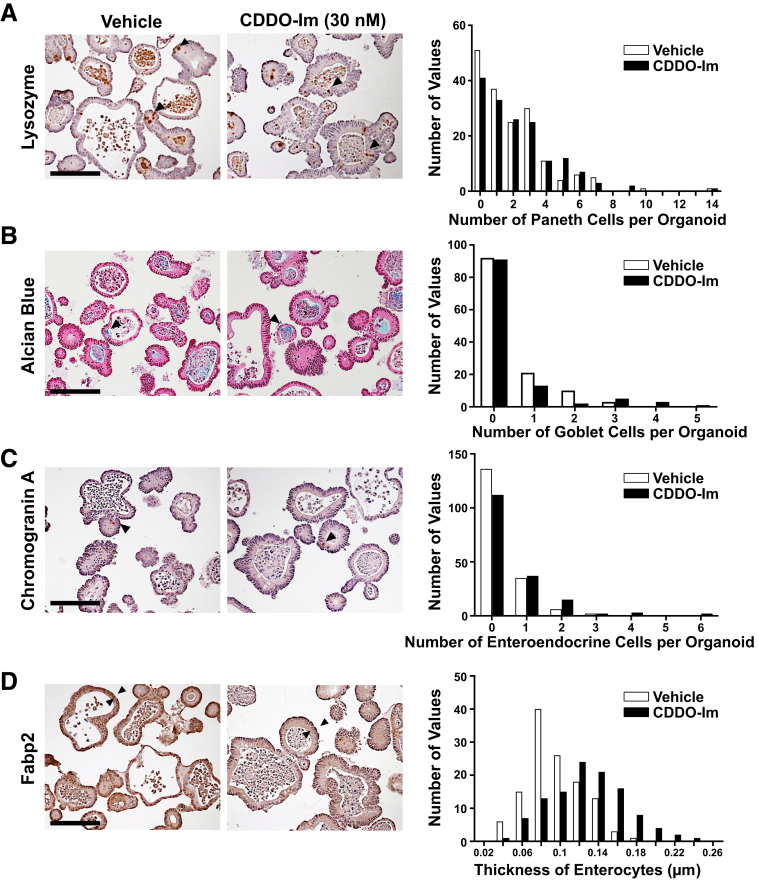
To identify which types of cells are affected by Nrf2 activation within the elongated villi, cell lineage was analyzed. The number of lysozyme positive-Paneth cells in the crypt did not change between the 4 genotypes of mice (Figure 5A). The percentage of Alcian blue–positive goblet cells and Chromogranin A–positive enteroendocrine cells per total epithelial cells did not show any differences across the examined genotypes (Figure 5B and C). Interestingly, the enterocytes of the Keap1f/f::VilCre mice showed morphological alterations; the thickness of Fabp2 positive-enterocytes was dramatically increased in Keap1f/f::VilCre compared with that of VilCre or Keap1f/f, and was completely attenuated in the Keap1f/f::Nrf2f/f::VilCre (Figure 5D). Increased thickness of enterocytes was also observed in the mice treated with CDDO-Im, and partially reversed back to the basal level by cessation of CDDO-Im administration (Figure 6).
Figure 5.
Activation of Nrf2 selectively expanded enterogenesis in the small intestine. Male 8-week-old VilCre, Keap1f/f (Kpf/f), Keap1f/f::VilCre (Kpf/f-VilCre) and Keap1f/f::Nrf2f/f::VilCre (Kpf/f-Nrf2f/f-VilCre) mice were used (n = 5–7). (A) Number of Paneth cells per crypt, identified by immunostaining for lysozyme. (B) Percentage of goblet cells and (C) enteroendocrine cells per total epithetical cells, identified by Alcian blue staining and immunostaining for chromogranin A, respectively. (D) Thickness of enterocytes, identified by immunostaining for Fabp2. Scale bar = 100 μm. Values are mean ± SD, ∗P < .05 using 1-way analysis of variance.
Figure 6.
Pharmacological activation of Nrf2 increased the thickness of enterocytes. Enterocytes were identified by immunostaining for Fabp2 and the graph shows the thickness of Fabp2 positive cells in the mice treated with vehicle or CDDO-Im for total 6 weeks, or 3 weeks of CDDO-Im followed by 3 weeks of vehicle treatment (n = 3). Values are mean ± SD, ∗P < .05 using 1-way analysis of variance. Scale bar = 100 μm.
Activation of Nrf2 Signaling Expanded Enterogenesis in Intestinal Organoids
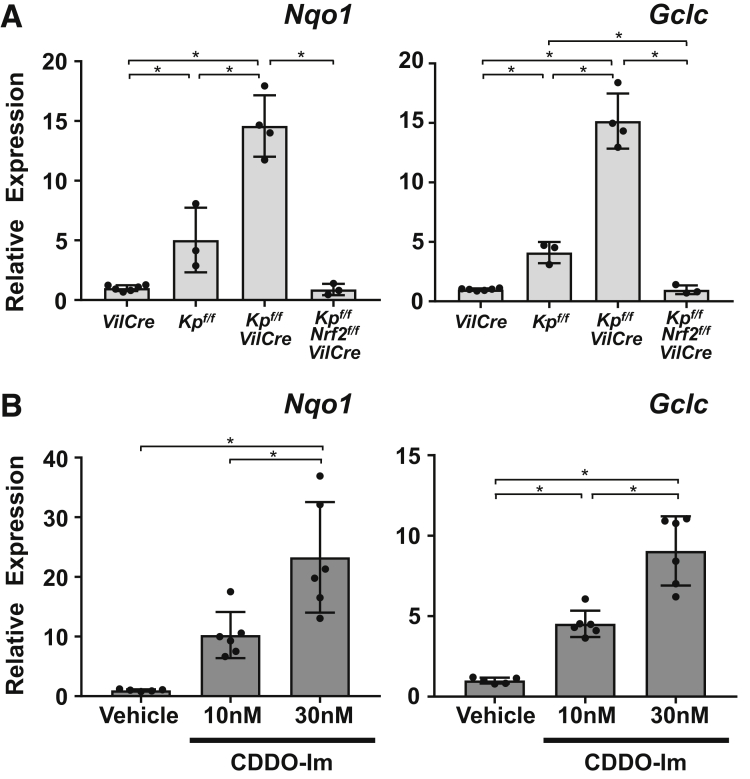
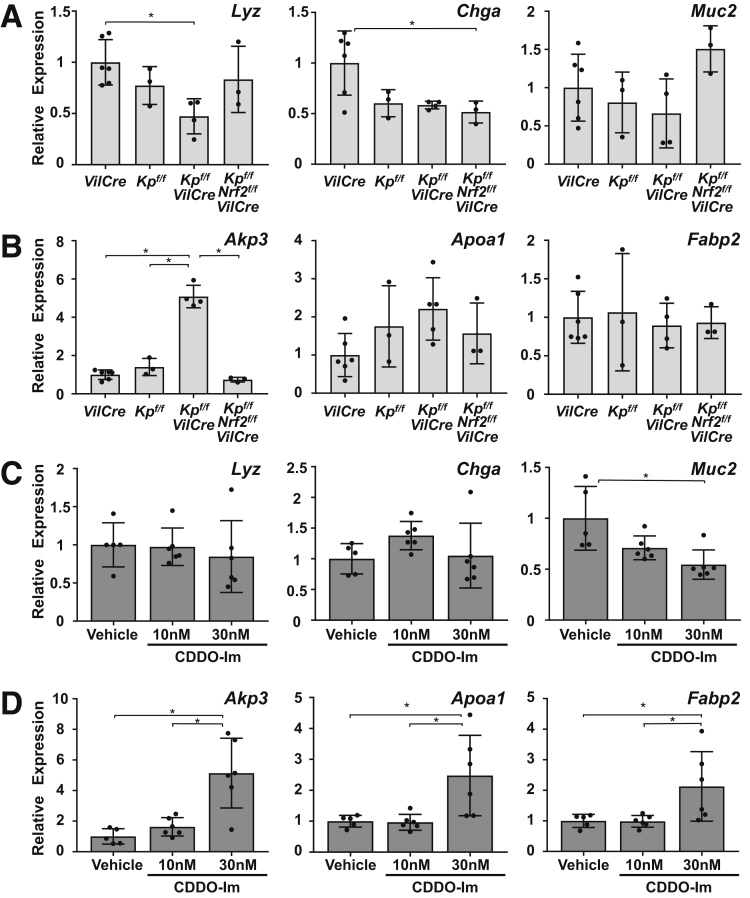
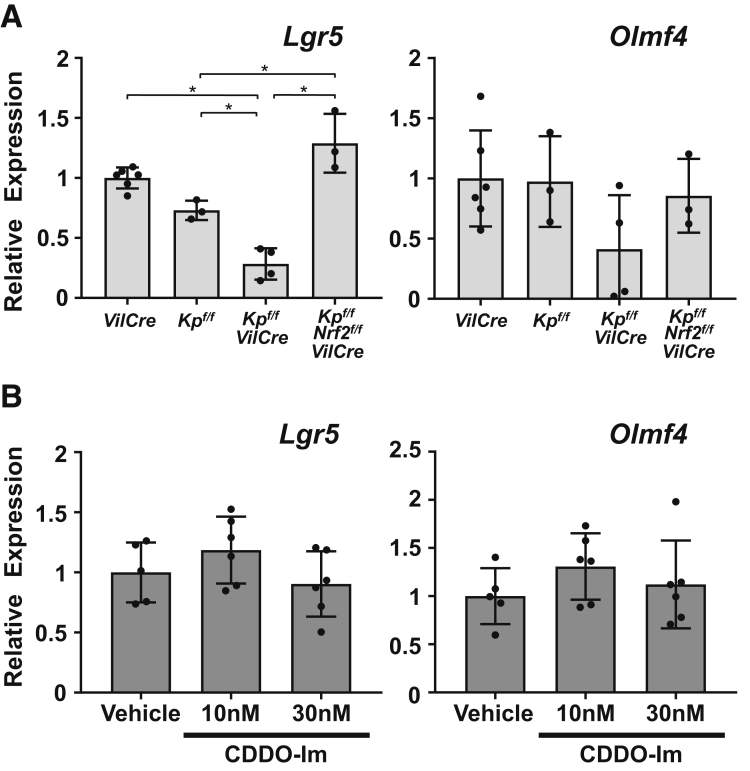
Intestinal organoid cultures were prepared from the 4 genotypes of mice and transcript levels of cell lineage markers were examined. The organoids derived from the small intestine of Keap1f/f::VilCre mice showed significant elevation of Nrf2 target genes (Nqo1 and Gclc) compared with that of VilCre-derived organoids, which was also confirmed in the wild-type background organoids treated by CDDO-Im (Figure 7A and B). Keap1f/f::Nrf2f/f::VilCre organoids did not show any elevation of these gene transcripts. As secretory cell lineage markers, messenger RNA (mRNA) expression of Lyz (Paneth cell), Chga (enteroendocrine cell), and Muc2 (goblet cell) were examined: Lyz expression was decreased in Keap1f/f::VilCre compared with VilCre control (Figure 8A, left panel); Chga showed lower expression in Keap1f/f::Nrf2f/f::VilCre compared with VilCre, but VilCre, Keap1f/f and Keap1f/f::VilCre did not show any differences (Figure 8A, middle panel); and no differences in Muc2 expression were observed in the organoids derived from the 4 genotypes of mice (Figure 8A, right panel). Lyz and Chga expression did not show significant changes following CDDO-Im treatment (Figure 8C, left and middle panels) while Muc2 expression decreased in the organoids treated with 30 nM of CDDO-Im (Figure 8C, right panel). In the same way, enterocytes of the absorptive cell lineage were examined by 3 markers. Organoids from Keap1f/f::VilCre showed significant elevation of Akp3 expression compared with that of VilCre, which did not occur in Keap1f/f::Nrf2f/f::VilCre organoids (Figure 8B, left panel). Apoa1 showed an increasing trend in the Keap1f/f::VilCre organoids and Fabp2 did not show substantial changes (Figure 8B, middle and right panels). The wild-type mouse organoids treated with 30 nM of CDDO-Im showed significant elevation of these 3 enterocyte markers, which was not observed with 10 nM of CDDO-Im (Figure 8D). Histological examination supported these results, demonstrating that the organoids with thicker enterocytes were observed more frequently with CDDO-Im than vehicle (Figure 9D); there was significant difference of average thickness between the 2 groups (P < .0001, data not shown) without changing the number of secretory cells per organoid (Figure 9A–C). The expression of stem cell markers demonstrated that the organoids from Keap1f/f::VilCre mice exhibited decreased Lgr5 expression compared with Keap1f/f and VilCre, which was canceled in Keap1f/f::Nrf2f/f::VilCre organoids, whereas Olmf4 did not show statistical differences between the 4 genotypes (Figure 10A). Neither of the stem cell markers changed with CDDO-Im treatment (Figure 10B). The inconsistent results on stem cell markers imply that activation of Nrf2 does not affect stem cell lineage, but rather indicates that Nrf2 may weakly affect Lgr5 gene expression in Keap1f/f::VilCre organoids.
Figure 7.
Activation of Nrf2 signaling in intestinal organoids. (A, B) mRNA expression levels of Nrf2 target genes (Nqo1 and Gclc) in intestinal organoids prepared from (A) VilCre, Keap1f/f (Kpf/f), Keap1f/f::VilCre (Kpf/f-VilCre), and Keap1f/f::Nrf2f/f::VilCre (Kpf/f-Nrf2f/f-VilCre) mice (8 weeks old, n = 5–6) and (B) wild-type background organoids treated by vehicle or CDDO-Im (n = 5–6). Intestinal organoids were prepared from 8-weeks-old wild-type mice, and the culture was maintained with medium adding CDDO-Im over 5 days with every other day of medium change. Data were normalized to Actb, and the expression level of VilCre or vehicle treatment was set as 1. Values are mean ± SD, ∗P < .05 using 1-way analysis of variance.
Figure 8.
Activation of Nrf2 expanded enterogenesis in the intestinal organoids. mRNA expression levels of (A) secretary lineage markers (Lyz, Chga, and Muc2) and (B) enterocyte markers (Akp3, Apoa1, and Fabp2) in organoids derived from the small intestine of VilCre, Keap1f/f (Kpf/f), Keap1f/f::VilCre (Kpf/f-VilCre), and Keap1f/f::Nrf2f/f::VilCre (Kpf/f-Nrf2f/f-VilCre) mice (8 weeks old, n = 5–6). The cells were collected after 5 days of culture. mRNA expression levels of (C) secretary lineage markers and (D) enterocyte markers in the wild-type background intestinal organoids treated with CDDO-Im over 5 days. The cells were collected on culture day 5 (n = 5–6). Data were normalized to Actb and expression levels in VilCre or vehicle treatment were set as 1. Values are mean ± SD, ∗P < .05 using 1-way analysis of variance.
Figure 9.
Activation of Nrf2 signaling increased the thickness of enterocytes in intestinal organoids. The intestinal organoids derived from wild-type mice were treated with either vehicle or CDDO-Im (30 nM) for 5 days. The frequency distribution of the number of (A) Paneth cells, (B) goblet cells, and (C) enteroendocrine cells per organoid were identified by immunostaining for lysozyme, Alcian blue staining, and immunostaining for Chromogranin A, respectively. (D) The frequency distribution of the thickness of enterocytes in organoids was identified by immunostaining for Fabp2. Three independent experiments were performed. Scale bar = 10 μm.
Figure 10.
Nrf2 signaling and expression of stem cell markers in intestinal organoids. (A, B) mRNA expression levels of intestinal stem cell markers (Lgr5 and Olmf4) in intestinal organoids prepared from (A) VilCre, Keap1f/f, Keap1f/f::VilCre and Keap1f/f::Nrf2f/f::VilCre mice (8 weeks old, n = 5–6) and (B) wild-type background organoids treated with vehicle or CDDO-Im for 5 days (n = 5–6). Data were normalized to Actb, and the expression level of VilCre or vehicle treatment was set as 1. Values are mean ± SD, ∗P < .05 using 1-way analysis of variance.
Cumulatively, the results achieved by genetic and pharmacologic approaches indicate that maximal activation of Nrf2 expands the enterocyte lineage in the intestinal organoids. By contrast, the cell types belong to the secretory lineages did not show any enhancement with largely null changes in the expression of their respective cell markers.
Nrf2 Signaling and Math1 Expression in Crypt Compartments of the Small Intestine
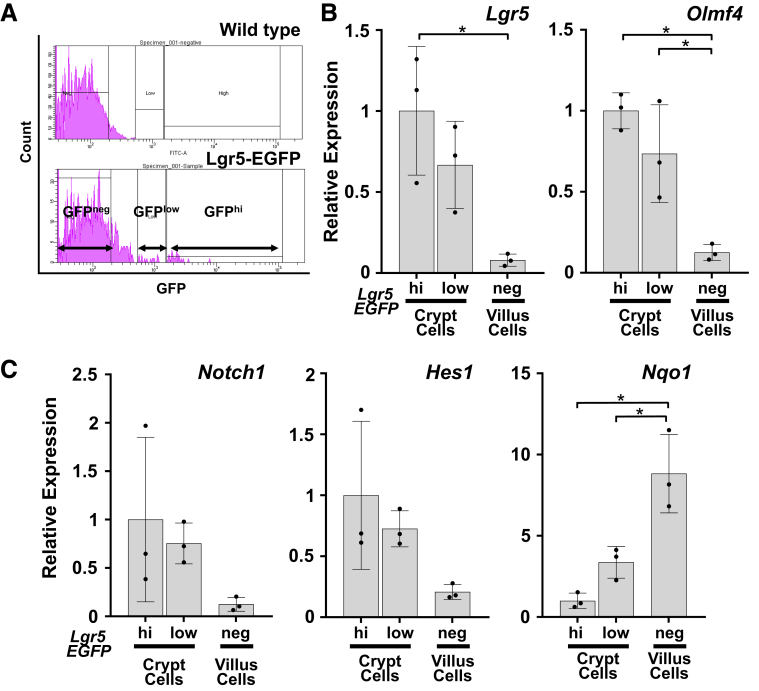
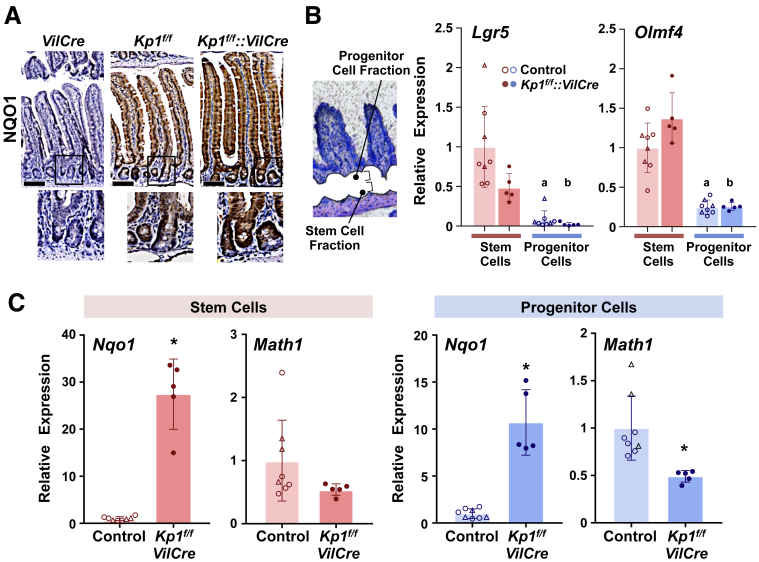
To assess the molecular mechanism underlying the enterocyte lineage-specific expansion evoked by Nrf2 activation, potential crosstalk between Nrf2 and the Notch downstream pathway was approached. Expression profiles of pathway-related genes along the crypt-villus axis were determined. Hence, Lgr5-EGFP-CreER reporter mice were employed to study each cell fraction, wherein enrichment was confirmed using stem cell markers (Figure 11A and B). Notch1 and Hes1 mRNA expression showed a clear increasing trend in the crypt cells compared with that in the villi (Figure 11C, left and middle panels), as supported by a previous report.28 Interestingly, Nqo1 expression was significantly lower in the crypt cells than in the villi, following an inverse gradation along the crypt-villus axis compared with the spatial profile of the Notch pathway related genes (Figure 11C, right panel). Although the gradation profile of Nqo1 expression in the Lgr5-EGFP-CreER reporter mice was not fully recapitulated by immunohistochemistry of NQO1 in the intestinal sections of VilCre mice (Figure 12A, left panel), which was likely due to a limitation of its sensitivity at the antibody dilution used, Keap1f/f::VilCre showed notable activation of Nrf2 within almost all epithelial cells except for cells populating the very bottom of the crypts (Figure 12A, right panel). Keap1f/f showed similar results, albeit with milder staining than seen in Keap1f/f::VilCre.
Figure 11.
Distribution of Nrf2 and Notch signaling along the crypt-villus axis in the wild-type mice. (A) The fluorescence-activated cell sorting histogram of wild-type background control mouse (upper panel) and Lgr5-EGFP-CreER reporter mouse (lower panel). The 3 distinct enhanced GFP (EGFP)–expressing cell fractions from Lgr5-EGFP-CreER reporter mice were sorted on the basis of EGFP intensities, indicated by either EGFP negative (GFPneg), EGFP low (GFPlow), or EGFP high (GFPhi) cell fractions. GFPneg cells are identified as villus cells and GFPlow and GFPhi cells are identified as crypt cells. (B) mRNA expression levels of stem cell markers and (C) Notch1, Hes1, and Nqo1 in each cell fraction. Data were normalized to the geometric mean value of 3 most stably expressed housekeeping genes across the 3 cell fractions: Tbp, Ppib, and B2m, according to the stability analysis. The expression level in Lgr5-EGFPneg cell fraction was set as 1. Values are ± SD, ∗P < .05 using 1-way analysis of variance.
Figure 12.
Nrf2 signaling and Math1 gene expression in intestinal crypts. (A) Immunohistochemistry for NQO1 in intestinal sections from VilCre, Keap1f/f (Kpf/f), and Keap1f/f::VilCre (Kpf/f-VilCre) mice (8 weeks old). Scale bar = 100 μm. (B) Blight-field image of intestinal sections after laser dissection and mRNA expression of stem cell markers in the microdissected fragments derived from Lgr5-EGFP-CreER background control (8 weeks old, VilCre::Lgr5-EGFP-CreER [n = 3, open triangles] and Lgr5-EGFP-CreER [n = 5, open circles]) and Keap1f/f::VilCre (Kpf/f-VilCre) mice (8 weeks old, Keap1f/f::VilCre::Lgr5-EGFP-CreER [n = 5, closed circles]). Stem cell fractions were obtained using indirect immunohistochemistry for GFP on a section cut from Lgr5-EGFP-CreER background mice adjacent to those used for LCM. Crypt cells excluding Lgr5-GFP–positive stem cells were captured as progenitor cell fractions. mRNA expression data were normalized to the geometric mean value of the 3 most stably expressed housekeeping genes across examined 4 groups (stem and progenitor cells from control and Keap1f/f::VilCre): Tbp, Ppib, and Actb, according to stability analysis. The expression level in the stem cell fraction of control mice was set as 1. Mean ± SD, a,b,P < .05 vs stem cells in either control or Keap1f/f::VilCre, respectively using 2-way analysis of variance. (C) mRNA expression of Nqo1 and Math1 in stem cells (left panels) and progenitor cells (right panels) from Lgr5-EGFP background control (VilCre::Lgr5-EGFP-CreER and Lgr5-EGFP-CreER) and Keap1f/f::VilCre (Kpf/f-VilCre) mice. Data were normalized to the geometric mean value of the 3 most stably expressed housekeeping genes across examined 2 groups (stem cells from control and Keap1f/f::VilCre [left panels] and progenitor cells from control and Keap1f/f::VilCre [right panels]): Tbp, Actb, and Hprt, according to stability analysis. The expression level in the stem cell fraction from control mice was set as 1. Values are mean ± SD, ∗P < .05 using Student’s T-test.
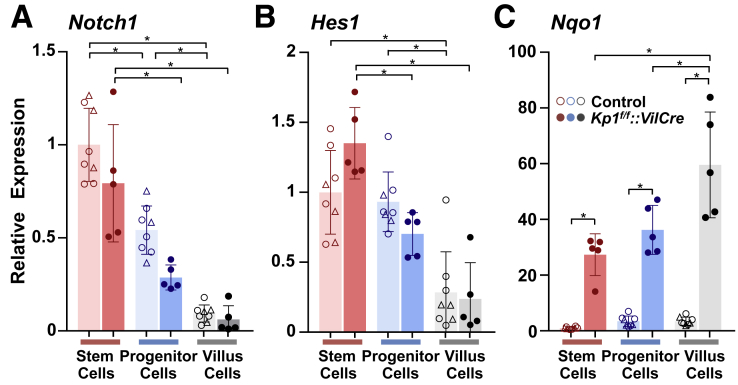
Given that enterocyte lineage-specific expansion emerged from Nrf2 activation, 1 of the downstream effectors of Notch signaling, Math1, known to regulate the differentiation balance between secretory and absorptive lineages through a lateral inhibitory mechanism, was identified as a probable candidate. Math1 is expressed in mature secretory cells facilitating maintenance of their differentiated phenotype in addition to the crypt cells that initiate this pathway of cell differentiation.8,29 To focus on potential interaction of Nrf2 signaling toward Math1 that could mediate alteration of enterogenesis, the spatial profiles of Math1 and Nrf2 signaling within the crypt compartment were examined. The stem and progenitor cells were collected by laser capture microdissection (LCM). Based on their location along the crypt-villus axis, crypt compartments were easily identified, and stem cell fractions were obtained using indirect immunohistochemistry for green fluorescent protein (GFP) on a section cut from Lgr5-EGFP-CreER background mice adjacent to those used for LCM. As the progenitor cells remained associated with the stem cell, crypt cells excluding Lgr5-GFP–positive stem cells were captured as progenitor fractions (Figure 12B). The enrichment of the captured segment was confirmed by expression of stem cell markers in both Lgr5-EGFP-CreER background control and Keap1f/f::VilCre mice (Figure 12B). Nqo1 expression was significantly higher in Lgr5-EGFP-CreER background Keap1f/f::VilCre mice than control mice in both stem and progenitor cell fractions (Figure 12C). Importantly, in the progenitor cells, mRNA expression of Math1 was significantly lower in Keap1f/f::VilCre than control mice (Figure 12C). Although a decreasing trend of Math1 in Keap1f/f::VilCre was observed in the stem fraction as well, the differences did not reach statistical significance.
Expression profiles of the Nrf2 and Notch pathway-related genes in the wild-type background Lgr5-EGFP-CreER mice shown in Figure 11 were confirmed in Keap1f/f::VilCre mice as demonstrated by LCM analysis, wherein Notch1 and Hes1 mRNA expression was highest in the stem cells, moderate in the progenitor cells and lowest in the villi (Figure 13A and B), and there were no significant differences in Notch1 and Hes1 mRNA expression between the 2 genotypes. On the other hand, Nqo1 expression was lowest in the stem cells and highest in the villus with dramatic elevation of Nqo1 expression in all cell fractions between Keap1f/f::VilCre and control mice (Figure 13C). Hes1 is a well-studied negative regulator of Math1 in several types of cells.30,31 Given the data showing comparable expression of Hes1 mRNA in control and Keap1f/f::VilCre mice, it is plausible that the decreased expression of Math1 in the progenitor cells of Keap1f/f::VilCre mice is uniquely caused by activation of Nrf2 signaling, and is not simply an indirect regulation mediated through an intermediary factor such as Hes1.
Figure 13.
Distribution of Nrf2 and Notch signaling along the crypt-villus axis in Nrf2 activated mice. The mRNA expression of (A) Notch1, (B) Hes1, and (C) Nqo1 in the laser capture microdissected cell fractions originating from Lgr5-EGFP-CreER background control (8 weeks old, VilCre::Lgr5-EGFP-CreER [n = 3, open triangles] and Lgr5-EGFP-CreER [n = 5, open circles]) and Keap1f/f::VilCre mice (8 weeks old, Keap1f/f::VilCre::Lgr5-EGFP-CreER [n = 5, closed circles]). Data were normalized to the geometric mean value of the 3 most stably expressed housekeeping genes examined across the 6 groups (stem, progenitor, and villus cells from control and Keap1f/f::VilCre): Tbp, Ppib, and Hprt according to the stability analysis. The expression level in the stem cell fraction of control mice was set as 1. Mean ± SD, ∗P < .05 using 2-way analysis of variance.
Nrf2-ARE Signaling Negatively Regulated Math1 Expression
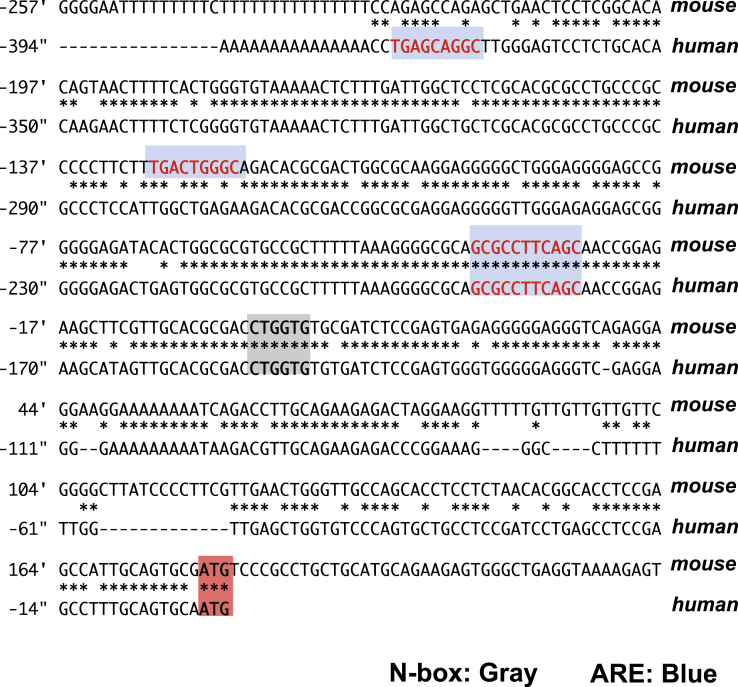
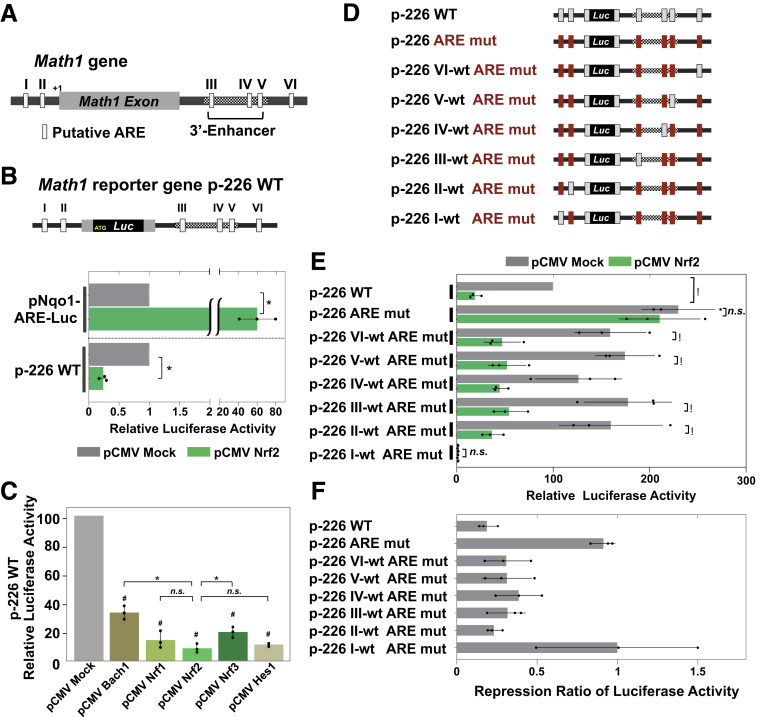
Given the data showing Keap1f/f::VilCre mice exhibit decreased expression of Math1 in their progenitor cells, a direct transcriptional regulation of Math1 by Nrf2 was hypothesized. The proximal promoter region of Atonal homologue 1 (ATOH1) gene in human and Math1 in mice is highly conserved between the 2 species; both possess ARE sequences (Figure 14). AREs are also identified in the downstream region of the Math1 gene, and 3 of them are located in the 3′ enhancer region (Figure 15A).32 To investigate whether these ARE sequences contribute to a functional cis element for Math1 transcriptional regulation by Nrf2, a Math1 luciferase reporter construct (p-226 WT) was produced (Figure 15B). As a positive control, it was confirmed that Nrf2 overexpression in Hepa1-6 cells led to significantly higher luciferase activity than that of control vector with the Nqo1-ARE reporter construct (pNqo1-ARE-Luc). Importantly, the Math1 reporter showed significantly lower luciferase activity than the basal level with Nrf2 overexpression (Figure 15B), indicating Nrf2 directly affects Math1 expression and AREs in Math1 regulatory region might contribute to the suppression of its expression. To investigate the specificity of the Nrf2-ARE pathway for Math1 transcriptional regulation, the Math1 reporter was examined with multiple transcriptional effectors (Figure 15C). Significant suppression of luciferase activity from the basal level was observed with all examined effectors; notably though, Nrf2 showed stronger suppression than Nrf3 or Bach1 (BTB and CNC homology1, basic leucine zipper transcription factor 1), while suppression was comparative to that with Nrf1 and Hes1. Further analyses adopting serial reporter constructs bearing point mutations designed to isolate single wild-type AREs were performed in order to identify the responsible AREs for Math1 transcriptional repression (Figure 15D). Nrf2 overexpression into Hepa1-6 cells resulted in attenuated luciferase activity with Math1 reporter construct harboring wild-type AREs (p-266 WT). Interestingly, the reporter construct with point mutations in all 6 AREs (p-266 ARE mut) significantly elevated the basal level of luciferase activity together with loss of Nrf2 responsiveness, highlighting an overall role of the AREs in repressing Math1. The constructs with a series of 5 mutated AREs while each harboring 1 wild-type ARE designated as p-266 VI-wt ARE mut, p-266 V-wt ARE mut, p-266 III-wt ARE mut, p-266 II-wt ARE mut, respectively, showed comparable levels of basal luciferase activity, which did not differ from the basal luciferase activity observed in p-266 WT construct. Thus, any of these wild-type AREs could support the suppression of Math1. Of particular interest, the construct harboring 5 point mutations but maintaining wild-type ARE I (p-266 I-wt ARE mut) dramatically lowered the basal luciferase activity; and Nrf2 did not evoke any further change (Figure 15E). Clearly, ARE I is dominantly, but not uniquely, responsible for Math1 repression. To simplify estimates of the responsiveness toward Nrf2 overexpression, the suppression ratios of pCMV mock to pCMV Nrf2 of luciferase activities are indicated in Figure 15F. These data highlight the molecular basis of transcriptional regulation of Math1 by Nrf2, reinforcing the involvement of Nrf2-ARE signaling in selectively modulating the Notch cascade in the murine small intestine.
Figure 14.
Sequence conservation of the Math1 promoter region between mouse and human. The proximal promoter sequence of the human ATHOH1 and Math1 gene, featuring AREs (highlighted in blue), an N-box (highlighted in gray), and translation start site (highlighted in red).
Figure 15.
Nrf2-ARE signaling negatively regulated Math1 expression. (A) Map of the mouse Math1 promoter and 3′ enhancer site (transcription start site [+1]) indicating the distribution of putative ARE motifs. AREs are indicated as I, II, III, IV, V, and VI. (B) Diagram of the p-226 WT Math1-ARE-Luc reporter construct (p-226 WT) and the relative luciferase activities of pNqo1-ARE-Luc reporter and p-226 WT in Hepa1-6 cells. In both analyses, the luciferase activities with pCMV Mock were set at 1.0. The dashed horizontal line indicates that the data obtained from 2 different promoters were not directly compared. ∗P < .05 using Student’s t test. (C) Luciferase activities of p-226 WT Math1-ARE-Luc reporter with multiple effectors in Hepa1-6 cells. The luciferase activities with pCMV Mock were set at 100, and the relative luciferase activities are shown. #P < .05 vs pCMV Mock control, ∗P < .05 for indicated comparison, using 1-way analysis of variance. (D) Diagram of serial point mutations of the p226-WT Math1-ARE-Luc reporter. The intact AREs (gray) and AREs having a point mutation (red) are indicated. (E) Luciferase activities in Hepa1-6 cells with serial point mutation reporter. The luciferase activities of Math1-ARE-Luc reporter (p-226 WT) driven by pCMV Mock were set at a value of 100, and the relative luciferase activities of each construct were shown. ∗P < .05 using 1-way analysis of variance. (F) The repression ratio of luciferase activity with each serial point mutation construct. The luciferase activities with pCMV Mock for each reporter were set at a value of 1.0, and the relative luciferase activities were indicated. Luciferase activities were normalized by the Renilla luciferase activity from a cotransfected reporter vector throughout. Three independent experiments were performed for each assay. Values are mean ± SD.
Discussion
This study presents new insights into how interactions between Nrf2 and Notch signaling within the intestinal crypts affect enterogenesis, leading to increased numbers and larger enterocytes. These 2 features of the phenotype of the Keap1f/f::VilCre mice are notable in that both outcomes are observed in the genetic and pharmacological models. In the former case, enterocyte enhancement is eliminated by Nrf2 disruption. Given that the pharmacodynamic action of CDDO-Im is blunted in Nrf2 knockout mice,33 these elements of the phenotype are strictly Nrf2 dependent. A third element of the phenotype is the increased length of the small intestine in Keap1f/f::VilCre mice. The underlying mechanism is not clear in that concomitant disruption of Nrf2 did not completely abrogate this aspect of the phenotype, unlike the effects on enterocyte number and size. Different too, the 6-week subchronic treatment of wild-type mice with CDDO-Im did not evoke intestinal elongation but did expand villus length. The lack of pharmacological effect for this phenotype could be a simple reflection of short duration or late onset relative to the genetic model. Alternatively, Nrf2 earlier in life affects other signaling cues guiding intestinal length or point of growth stasis.
The in vivo and in vitro approaches indicate that the constitutive activation of Nrf2 signaling in the progenitor cells perturbs the signaling network through negative regulation of Math1. Inasmuch as a lateral inhibition mechanism driven by the Notch/Math1 pathway regulates the balance of cell lineage differentiation, it is proposed that the disproportionate crosstalk between Nrf2 and the Notch/Math1 cascade in the crypt contributes to the selective expansion of enterogenesis in the Keap1f/f::VilCre mice.
Supporting our interpretation, the intestinal progenitors alter their cell fate determination to become only absorptive enterocytes in the absence of Math1.8,10 Given the central role of Notch signaling during the differentiation of intestinal epithelium, crosstalk between the Nrf2 and Notch downstream effectors could be critical for fine-tuning intestinal homeostasis.
Our earlier studies demonstrated the functional significance of positive signaling crosstalk between Nrf2 and Notch in the murine liver.12,13 This crosstalk facilitated liver regeneration and resistance to xenobiotic toxicities. By contrast, in the small intestine, the Nrf2 and Notch pathways exhibit a reciprocal manner of expression along the crypt-villus axis. This spatial expression profile of 2 signaling activities is unique enough to suggest a novel signaling crosstalk in the small intestine. Math1 is a key player for controlling intestinal cell fate commitment, where Hes1 is a negative regulator of Math1 within the crypts.8,30,31 As Hes1 expression was not changed by Nrf2 activation in the progenitor cells, it appears Hes1 does not mediate the perturbed signaling network evoked by Nrf2 activation. Rather this outcome suggests a direct association between Nrf2 and Math1. Indeed, our results using cell-based reporter constructs unveiled that Nrf2 transcriptionally contributes to repression of Math1. The transcriptional importance of the Math1 enhancer region has been shown in mammalian neural tubes, in which the zinc-finger transcription factor binds the Math1 enhancer region, repressing Math1 along with suppressed activity of the Math1 enhancer.34 We did not probe the detailed mechanism underlying Nrf2-mediated transcriptional regulation of Math1. However, our results did demonstrate that AREs located in the Math1 promoter regions, especially ARE I, as well as the 3′ enhancer region, are responsible for the negative regulation of Math1. Conceivable mechanisms of Math1 silencing by Nrf2 could be approached with potential targets such as corepressors or heterodimers of Math1,35,36 or molecular events including enhancer activity and an autoregulation pathway of Math1.34
The functional consequences of expanded enterogenesis through Nrf2 signaling remains unclear. Morphological changes of the gut are observed in animal models and pathological disorders, where various factors such as aging, diet, and inflammation affect villus height, crypt depth, the structure of tight junctions, and microvilli of enterocytes.37,38 In humans, short bowel syndrome is a highly morbid condition in which small intestinal length is inadequate for proper nutrient absorption. Structural and functional changes during intestinal adaptation are necessary to compensate for the sudden loss of digestive and absorptive capacity after massive intestinal resection.39 Pharmacologic therapy for short bowel syndrome is under evaluation. Teduglutide, a recombinant analogue of glucagon-like peptide-2 appears to be a helpful adjunct to promote bowel adaptation through increases in absorptive capacity due to its associated increase in villus height and crypt depth as well as its slowing effect on motility.40 A broader examination of the effects of clinically useful Nrf2 inducers,41 both drugs and food-based phytochemicals, on absorptive capacity and other intestinal functions is warranted.
While Nrf2 activation in the intestinal epithelium causes a specific expansion of the absorptive lineage without affecting secretory cells, a molecular target existing throughout the population of epithelial cells cannot fully approach the phenotype observed in Keap1f/f::VilCre. This point notwithstanding, Nrf2 does exert effects on mature intestinal cells. Pharmacological intervention with the triterpenoid bardoxolone methyl enhances signaling of DNA-damage response and protects colonic epithelium from ionizing radiation.42 Activation of Nrf2 by endogenous factors, such as bacterial metabolites, enhances intestinal epithetical barrier along with anti-inflammatory effects.43 The natural compound urolithin A enhances the gut barrier function by inducing tight-junction proteins through activation of aryl hydrocarbon receptor and Nrf2-dependent pathways.44 These findings indicate another aspect for a physiological role of Nrf2 in the gut, wherein activation of Nrf2 might be important for host defense. Nrf2 also plays a major role in the regulation of bile acid homeostasis in the liver and intestine by affecting expression of regulators of bile acid synthesis, bile acid, and organic solute transporters.45 Regarding an enterocyte-specific alteration by Nrf2, it is interesting to note that the intestinal alkaline phosphatase (Alp) gene, which was used as an enterocyte marker, has multiple AREs in its gene regulatory region (data not shown). Two ALP isoenzymes, dIALP (Akp3) and gIALP (Alpi or Akp6), are expressed in the intestine. Indeed, Akp3 (Figure 8B and D) and Alpi (data not shown) showed Nrf2-dependent elevation of mRNA in the intestinal organoids, suggesting a possible transcriptional regulation by Nrf2. Intestinal ALP regulates lipid transport and inflammatory responses in enterocytes by dephosphorylation of toxic microbial ligands. Thus, Nrf2-mediated Alp expression may be indicative of the functional involvement of Nrf2 in the matured enterocyte. The effect of Nrf2 activation in mature villus cells requires further study.
In conclusion, Nrf2 signaling mediates a dialogue in equilibrium signaling within the crypt with Notch downstream effectors such as Math1, which in turn constitute the essential molecular basis for cell lineage homeostasis in the intestinal epithelium. The conspicuous phenotype of Nrf2 activation in mice demonstrated that constitutive facilitation of Nrf2 perturbs the Notch signaling pathway in the progenitor cells via Math1 transcriptional regulation, leading to an expansion of enterogenesis. These findings provide the new insights into the role of Nrf2 in intestinal epithelial cells, broaden our understanding of the biological functions of Nrf2, and point to new approaches for the management of absorptive deficiencies.
Materials and Methods
Mice
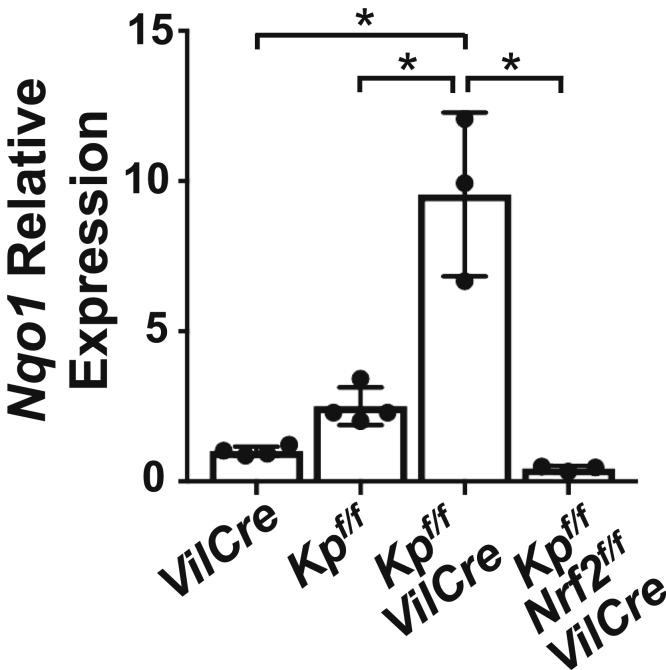
Keap1flox/flox (Keap1f/f) and Nrf2flox/flox (Nrf2f/f) mice were used to generate conditional knockout mice, as described previously.24,46 The Villin-Cre (VilCre, No.004586) and Lgr5-EGFP-CreER (No. 008875) mouse lines were supplied from the Jackson Laboratory (Bar Harbor, ME). All mice used in the experiments were albino C57BL/6J background (B6(Cg)-Tyrc-2J/J), male, and 8–10 weeks old, unless indicated otherwise. The breeding performed in this study was as follows; Keap1f/f::VilCre mice were crossed with Keap1f/f, and Keap1f/f::Nrf2f/f::VilCre mice were mated with Keap1f/f::Nrf2f/f mice to obtain Keap1f/f, Keap1f/f::VilCre, and Keap1f/f::Nrf2f/f::VilCre mice. VilCre mice were mated with wild-type mice, and age- and sex- matched mice were used as control animals. The intestinal mucosal tissue of Keap1f/f::VilCre mice showed significant elevation of a canonical Nrf2 target gene, Nqo1 compared with VilCre control and Keap1f/f (Figure 16), conforming constitutive activation of Nrf2 in the intestinal epithelial cells. Increased expression of Nqo1 was abrogated in Keap1f/f::Nrf2f/f::VilCre, wherein the Nrf2 gene was concomitantly disrupted. Keap1f/f mice, which function as systemic knockdown of Keap1 independent of Cre recombinase expression because of hypomorphic Keap1 gene alleles,25 showed milder elevation of Nqo1 expression than Keap1f/f::VilCre. To get the desired genotype of Lgr5-EGFP-CreER background mice, Lgr5-EGFP-CreER::Keap1f/f were crossed with Keap1f/f::VilCre mice and Lgr5-EGFP-CreER were mated with VilCre mice to obtain Lgr5-EGFP-CreER::Keap1f/f::VilCre, Lgr5-EGFP-CreER::VilCre, and Lgr5-EGFP-CreER mice. All mice were housed in a pathogen-free animal facility and handled according to the guidelines of the National Institutes of Health. All animal experiments were approved by the Institutional Animal Care and Use Committees at the University of Pittsburgh or the Fred Hutchinson Cancer Research Center.
Figure 16.
Genetic model of Nrf2 activation in intestinal epithelial cells. The mRNA expression levels of Nqo1 in small intestinal mucosa from VilCre, Keap1f/f (Kpf/f), Keap1f/f::VilCre (Kpf/f-VilCre), and Keap1f/f::Nrf2f/f::VilCre (Kpf/f-Nrf2f/f-VilCre) mice (8 weeks old, male, n = 3–4). Values are mean ± SD throughout. ∗P < .05 using 1-way analysis of variance.
CDDO-Im Treatment
Male albino C57BL/6J background (B6(Cg)-Tyrc-2J/J) wild type mice (6 weeks old) were orally administered CDDO-Im (30 μmol/kg of body weight; generously provided by Dr. Michael Sporn, Geisel School of Medicine at Dartmouth, Hanover, NH) in 10% dimethyl sulfoxide, 10% Cremophor-EL, and phosphate-buffered saline as a vehicle or vehicle alone, at a set time during the light cycle. At 24 hours after the last treatment, tissue was collected.
Immunostaining
Isolated intestines were placed in 4% paraformaldehyde at 4°C overnight and then cut length wise, rolled, and paraffin embedded. Immunohistochemistry and Alcian blue staining was carried out using antibodies or regents listed in Table 1. Imaging data were collected using Nikon E800 (Nikon, Tokyo, Japan) and APERIO AT (Leica, Wetzlar, Germany) for digital scanning. Morphometric analysis and cell counts were performed using HALO software (Indica Labs, Albuquerque, NM). For measurement of length of villi, depth of crypt, and cell number per villus, the hematoxylin and eosin–stained sections were imaged and a total 20 of well-orientated villi and crypts were randomly chosen from 4 regions of whole small intestines, which were defined as the posterior and anterior regions from either between duodenum and jejunum or between jejunum and ileum. For lysozyme-positive cells, a total of 20 adequately sectioned crypts were selected from 4 different areas of the whole small intestine and the average number of positive cells per crypt were determined per mouse. The percent of Chromogranin A–positive cells and Alcian blue–positive cells were determined by examining a total of 4500–7500 epithelial cells from at least 45 villi selected from 4 different regions of whole small intestine per mouse. The adequately sectioned villi were selected from 4 different regions of whole small intestine, and the thickness of Fabp2 positive enterocytes populated in the middle position of the villus was examined. The average thickness was determined from at least 40 enterocytes per mouse.
Table 1.
Antibodies and Staining Solution
| Antibodies | Company | Catalog No. | Dilution |
|---|---|---|---|
| Rabbit polyclonal anti-Chromogranin A | Abcam | ab15160 | 1:500 |
| Rabbit monoclonal anti-Lysozyme | Abcam | ab108508 | 1:1,000 |
| Goat polyclonal anti-FABP2 | NOVUS Biologicals | NB100-59746 | 1:1,000 |
| Goat polyclonal anti-NQO1 | Abcam | ab2346 | 1:500 |
| Goat polyclonal anti-GFP | Abcam | ab6673 | 1:100 |
| Alcian Blue Stain Kit | Abcam | ab150662 | - |
Organoid Culture
Mouse organoids were established and maintained as previously described,47 with modification. Approximately 30% length of the whole small intestine from each proximal and distal region was harvested and the isolated crypts were cultured in a mixture of Matrigel (BD Biosciences, Franklin Lakes, NJ) and medium (IntestCult Organoid Growth Medium, mouse; STEMCELL, Vancouver, Canada). For pharmacological activation, the culture was maintained with medium adding 10 nM or 30 nM of CDDO-Im over 5 days with every other day of medium-change (days 0, 2, 4).
Fluorescence-Activated Cell Sorting Analysis and Sorting
Fluorescence-activated cell sorting isolation of single LGR5+ cells from Lgr5-EGFP-CreER mice was performed as previously described48 with modifications. Propidium iodide solution was used for exclusion of dead cells, and CD31 (BioLegend, San Diego, CA; #102417) and CD45 (BioLegend; #103113)–positive cells were excluded from the propidium iodide–negative cells to gate out endothelial cells and lymphocytes, respectively. The sorting of Lgr5-EGFP+ cells was performed with a FACSAria II cell sorter (BD Biosciences).
Laser Capture Microdissection
Tissues for microdissection were obtained from Lgr5-EGFP-CreER background mice. Approximately 10 cm of intestinal segments from each proximal and distal part of small intestine were dissected and harvested specimens were flushed with ice-cold phosphate-buffered saline and cut lengthwise, rolled, and embedded into optimal cutting temperature compound, following a freezing process using a metal block placed in a Styrofoam container containing liquid nitrogen without fixation. Sections (10 μm) were cut and mounted on polyethylene naphthalate membrane–coated slides (Applied Biosystems, Foster City, CA). A total 850,000–950,000 μm2 area was microdissected by a UV laser-based unit LMD6 (Leica) from a total of 6 serial sections (3 sections each from proximal and distal intestine) per mouse with the following parameters: cut energy 25 (1–60), aperture 10 (1–45), cut speed 30 (1–100) with 40× objective.
RNA Preparation and Quantitative Polymerase Chain Reaction
The mucosal tissue, organoids and sorting cells were homogenized in Trizol (Thermo Fisher Scientific, Waltham, MA), and total RNA was extracted as previously reported.49 For laser capture samples, RNAqueous-Micro Total RNA Isolation kit (Ambion, Austin, TX) was used. Complementary DNA (cDNA) was synthesized using the qScript system (Quanta Biosciences, Beverly, MA) and quantitative polymerase chain reaction (PCR) was performed by QuantStudio 7 (Applied Biosystems) with TaqMan Fast Advanced Master Mix (Applied Biosystems). For organoids and microdissected samples, a multiplex PCR preamplification of specific cDNA targets and endogenous control was performed using TaqMan PreAmp Master Mix (Applied Biosystems) following the manufacturer’s instructions. The primers (Applied Biosystems) are shown in Table 2.
Table 2.
Primer List for Quantitative Polymerase Chain Reaction
| Gene Name | Systematic Name |
|---|---|
| Lyz | Mm00727183_s1 |
| Chga | Mm00514341_m1 |
| Muc2 | Mm00458299_m1 |
| Akp3 | Mm00475847_g1 |
| Apoa1 | Mm00437569_m1 |
| Fabp2 | Mm00433188_m1 |
| Lgr5 | Mm00438890_m1 |
| Olmf4 | Mm01320260_m1 |
| Nqo1 | Mm01253561_m1 |
| Math1 | Mm01181529_s1 |
| Gclc | Mm00802655_m1 |
| Actb | Mm01205647_g1 |
| Hprt | Mm00446968_m1 |
| B2m | Mm00437762_m1 |
| Tbp | Mm01277042_m1 |
| Ppib | Mm00478295_m1 |
| Gusb | Mm01197698_m1 |
| Notch1 | Mm00435245_m1 |
| Hes1 | Mm00468601_m1 |
Stability Analysis of Housekeeping Gene From Cell Fractions
For quantitative PCR analysis of intestinal cell fractions, a systematic study was performed to identify suitable reference genes across the cell fractions, considering the substantial biological differences between stem, progenitor, and mature villus cells.50,51 The expressions of a total of 6 housekeeping genes (Actb, Hprt, B2m, Tbp, Ppib, and Gusb) were examined and associated software52 was used to identify the 3 most stably expressed genes across the examined groups. The geometric mean of 3 reference genes was used for normalization of relative value of mRNA expression for further study.53
Cell Culture
Hepa1-6 cells were cultured in D-MEM (Gibco, Waltham, MA) supplement with 10% fetal bovine serum (Gibco) in 37°C, 5% CO2 incubator.
Expression Vectors and Its Control Vector
Mouse CNC family genes (Nrf1, Nrf2, Nrf3, and Bach1) and Hes1 cDNA were inserted in pcDNA3 (Invitrogen, Waltham, MA). Mouse Nrf1 and Bach1 cDNA were purchased from Origene (Rockville, MD) (MC206760 and MC203298, respectively). Mouse Nrf3 cDNA was prepared from pCMV3xFLAG mNrf3 (gift from Dr. Akira Kobayashi, Doshisha University, Kyoto, Japan) by HindIII and BamHI reaction and inserted into a pcDNA3 vector. Mouse Hes1 cDNA was isolated from pCI Hes1 (provided from Dr. Ryoichiro Kageyama, Kyoto University, Kyoto, Japan) by EcoRI digestion and transferred into pcDNA3 vector. pCMV Mock was produced from pcDNA3 by eliminating multiple cloning sites through self-ligation following HindIII, XbaI, and T4-polymerase reaction.
Construction of Math1 Reporter Genes
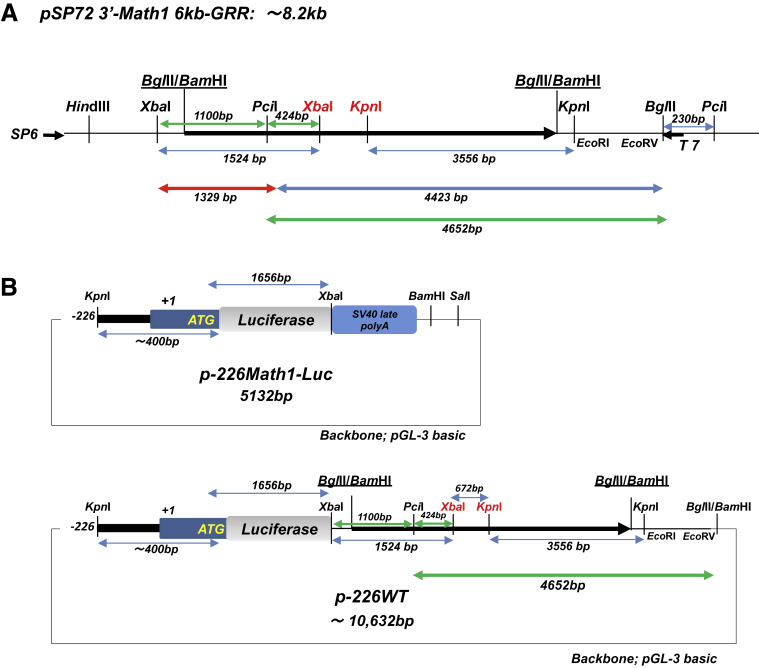
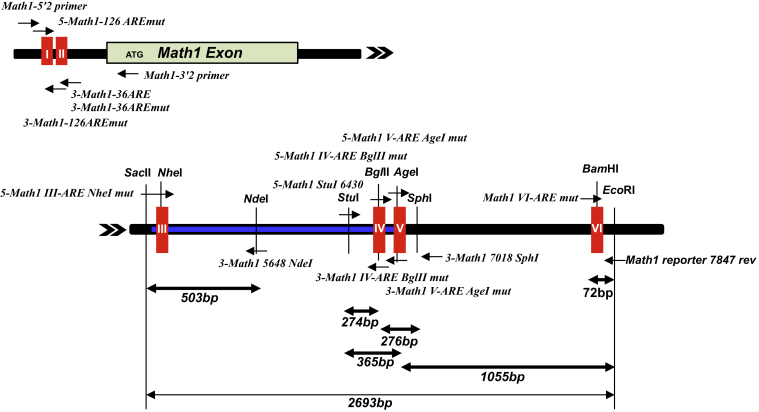
Math1 5′-proximal promoter region was cloned by nested PCR using 2 primer sets (Math1 5′1, Math1 3′1 and Math1 5′2, Math1 3′2) (Table 3) from C57BL6 mouse tail genomic DNA. Math1 reporter constructs were prepared as shown in Figure 17. The target PCR products directly cloned between the KpnI and NcoI sites of pGL-3 basic (Promega). Consequently, ATG of the Math1 gene functioned as the initiation codon of Firefly luciferase reporter gene (p-226 Math1 GL3). Tg#6 Math1-LacZ transgenic vector was a kind gift from Dr. Jane E. Johnson (UT Southwestern Medical Center, Houston, TX).32 This vector contains ∼6 kb downstream of the Math1 coding exon and 3′-gene regulatory region (6kb-GRR). 6kb-GRR was isolated from Tg#6 by BglII reaction and subcloned into pSP72 (Promega, Madison, WI) BamHI site once to create the useful unique restriction sites (pSP72 3′-Math1 6kb-GRR). To construct p-226 WT, 4.6-kb fragment isolated from pSP72 3′-Math1 6kb-GRR by BglII, PciI, and HindIII reaction and 1.3-kb fragment from pSP72 3′-Math1 6kb-GRR by XbaI and PciI reaction were inserted between XbaI and BamHI sites of p-226 Math1 GL3 (Figure 17). To construct putative ARE mutants and each possible ARE reconstituted mutant reporter gene, specific primers were prepared for PCR (Table 3) and locations displayed in Figure 18. The essential fragments produced by PCR were subcloned into the Litmus 28 vector (New England Biolabs, Ipswich, MA), and then confirmed by sequencing. Each appropriate mutant fragment was replaced by recombination using the restriction enzyme sites shown in Figure 18, and consequently the target reporter genes were established.
Table 3.
Primer List Used to Generate Math1 Reporter Constructs
| Primers used in Math1 reporter construction | |
| Math1-5'1 primer | TAGGCGCGTCCAGCTTGGCCGGG |
| Math1-3′1 primer | AGGTGGCTGTGGCGTCAGCGGCGGG |
| Math1-5'2 primer | TTTGGTACCAGAGCCAGAGCTGAACTCCTCGG |
| Math1-3′2 primer | GGGACCATGGCACTGCAATGGCTCGGAGGTG |
| Primers used in Math1 ARE mutants reporter construction | |
| 5-Math1-126AREmut | TTCTTTGcCcGGGCAGACACGCGACTGGCGCAAGG |
| 3-Math1-126AREmut | TCGCGTGTCTGCCCgGgCAAAGAAGGGG |
| 3-Math1-36AREmut | CGAAGCTTCTCCGGTTGCgGAAGGCtCTGC |
| 3-Math1-36ARE | CGAAGCTTCTCCGGTTGCTGAAGGCGCTGC |
| 5-Math1 StuI 6430 | TACGTTTAGTCTTCTCTGCACCCCAGGCCTAGTGTCTCCCC |
| 5-Math1 III-ARE NheI mut | TTTGCTGCCGCGGTGAGCAGAGCTTCCACTTCACCTCTCTGgcTagcCAACTCCTGTTCGCCCTTCTCAGAATGG |
| 5-Math1 IV-ARE BglII mut | AGCACGgGagatctGCTGGTGAGCGCACTCGCTTTCAGGCCGC |
| 5-Math1 V-ARE AgeI mut | TCCCCGGCaccggtAACCTCGGCCTCCTCCTCGTAG |
| 3-Math1 5648 NdeI | GTCAGCATGGCAGGAGTGGTCTGGCATATGGG |
| 3-Math1 7018 SphI | CCTCATCCCCTCCCCTAGGCTTTGCTTGACTC |
| 3-Math1 IV-ARE BglII mut | CGCTCACCAGCagatctCcCGTGCTCCGCTCCAGACGCTCCGCACC |
| 3-Math1 V-ARE AgeI mut | GAGGTTaccggtGCCGGGGAGGGTGACGACGGTGTT |
| Math1 reporter 7847 rev | ATGAATTCGAGCTCGGTACCCGGGGATCTCTCCTAAAAC |
| Math1 VI-ARE mut | ATTCCCTAGGCCCCATCgGAtCCAtCCATTAAGGAGCAGACATC |
| Math1 VI-ARE WT | ATTCCCTAGGCCCCATCTGACCCAGCCATTAAGGAGCAGACATC |
| Primers for sequence confirmation | |
| Math1 GL3-5048 | CCTTATCACCAGATCAGGGAGCCCGCC |
| Math1 GL3-5006 | CACAGCCAGAAAATACCAATGTCCATGGCTGCAC |
ARE, antioxidant response element.
Figure 17.
Construction of Math1 reporter gene. (A) The schema of pSP72 3′-Math1 6kb-GRR construct containing the 3′-gene regulatory region of Math1 (6kb-GRR). (B) To construct p-226 WT, a 4.6-kb fragment isolated from pSP72 3′-Math1 6kb-GRR by BglII, PciI, and HindIII reaction and a 1.3-kb fragment by XbaI and PciI reaction were inserted between XbaI and BamHI sites of p-226 Math1 GL3.
Figure 18.
Construction of ARE mutant Math1 reporter genes. Each appropriate mutant fragment was replaced by recombination using adequate restriction enzyme sites shown in the schema, with subsequent establishment of target reporter genes.
Statistical Analysis
GraphPad Prism 7 software (GraphPad Software, San Diego, CA) was used for statistical analysis of datasets. Quantitative data are presented as mean ± SD.
Acknowledgments
The authors thank Dr Jane E. Johnson (UT Southwestern Medical Center) for providing Math1-LacZ transgenic vector, Dr Akira Kobayashi (Doshisha University) for providing pCMV3xFLAG mNrf3 construct, Dr Motoko Koyama (Fred Hutchinson Cancer Research Center) for advice about experiments using Lgr5-EGFP-CreER mice, Dr Dionysios V. Chartoumpekis (University of Patras) for providing advice regarding normalization strategies for gene expression, the Department of Pathology, University of Washington, for assistance with histology samples. This research was also supported by the Experimental Histopathology and Flow Cytometry Shared Resource of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704).
CRediT Authorship Contributions
Yoko Yagishita, DDS, PHD (Conceptualization: Equal; Formal analysis: Lead; Investigation: Equal; Methodology: Equal; Validation: Equal; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Melissa L. McCallum, MS (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Thomas W. Kensler, PhD (Conceptualization: Equal; Funding acquisition: Lead; Investigation: Supporting; Project administration: Lead; Resources: Equal; Supervision: Equal; Validation: Lead; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Lead)
Nobunao Wakabayashi, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Resources: Equal; Supervision: Equal; Validation: Equal; Visualization: Equal; Writing – original draft: Supporting; Writing – review & editing: Equal)
Footnotes
Conflicts of Interest The authors disclose no conflicts.
Funding This work was supported by the National Institutes of Health (R35 CA197222 [to Thomas W. Kensler]) and the Washington State Andy Hill CARE Fund (to Thomas W. Kensler), Japan Society for the Promotion of Science Overseas Research Fellowships (JSPS 201860777 [to Yoko Yagishita]).
References
- 1.Heath J.P. Epithelial cell migration in the intestine. Cell Biol Int. 1996;20:139–146. doi: 10.1006/cbir.1996.0018. [DOI] [PubMed] [Google Scholar]
- 2.Radtke F., Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 3.van der Flier L.G., Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 4.Crosnier C., Stamataki D., Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7:349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- 5.Stanger B.Z., Datar R., Murtaugh L.C., Melton D.A. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci U S A. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 7.Jensen J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q., Bermingham N.A., Finegold M.J., Zoghbi H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K., Fukui H., Kayahara T., Sawada M., Seno H., Hiai H., Kageyama R., Okano H., Chiba T. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem Biophys Res Commun. 2005;328:348–352. doi: 10.1016/j.bbrc.2004.12.174. [DOI] [PubMed] [Google Scholar]
- 10.Shroyer N.F., Helmrath M.A., Wang V.Y., Antalffy B., Henning S.J., Zoghbi H.Y. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 11.van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 12.Wakabayashi N., Shin S., Slocum S.L., Agoston E.S., Wakabayashi J., Kwak M.K., Misra V., Biswal S., Yamamoto M., Kensler T.W. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakabayashi N., Skoko J.J., Chartoumpekis D.V., Kimura S., Slocum S.L., Noda K., Palliyaguru D.L., Fujimuro M., Boley P.A., Tanaka Y., Shigemura N., Biswal S., Yamamoto M., Kensler T.W. Notch-Nrf2 axis: regulation of Nrf2 gene expression and cytoprotection by notch signaling. Mol Cell Biol. 2014;34:653–663. doi: 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto M., Kensler T.W., Motohashi H. The KEAP1-NRF2 System: a Thiol-Based Sensor-Effector Apparatus for Maintaining Redox Homeostasis. Physiol Rev. 2018;98:1169–1203. doi: 10.1152/physrev.00023.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D.D., Chapman E. The role of natural products in revealing NRF2 function. Nat Prod Rep. 2020;37:797–826. doi: 10.1039/c9np00061e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh K., Wakabayashi N., Katoh Y., Ishii T., O'Connor T., Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D.R., Harada T., Engel J.D., Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 19.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi M., Li L., Iwamoto N., Nakajima-Takagi Y., Kaneko H., Nakayama Y., Eguchi M., Wada Y., Kumagai Y., Yamamoto M. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 22.Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J Biol Chem. 2000;275:16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 23.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., Yamamoto M., Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 24.Okawa H., Motohashi H., Kobayashi A., Aburatani H., Kensler T.W., Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 25.Taguchi K., Maher J.M., Suzuki T., Kawatani Y., Motohashi H., Yamamoto M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol Cell Biol. 2010;30:3016–3026. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.el Marjou F., Janssen K.P., Chang B.H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 27.Liby K., Hock T., Yore M.M., Suh N., Place A.E., Risingsong R., Williams C.R., Royce D.B., Honda T., Honda Y., Gribble G.W., Hill-Kapturczak N., Agarwal A., Sporn M.B. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65:4789–4798. doi: 10.1158/0008-5472.CAN-04-4539. [DOI] [PubMed] [Google Scholar]
- 28.Fre S., Hannezo E., Sale S., Huyghe M., Lafkas D., Kissel H., Louvi A., Greve J., Louvard D., Artavanis-Tsakonas S. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akazawa C., Ishibashi M., Shimizu C., Nakanishi S., Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 31.Zheng J.L., Shou J., Guillemot F., Kageyama R., Gao W.Q. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127:4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- 32.Helms A.W., Abney A.L., Ben-Arie N., Zoghbi H.Y., Johnson J.E. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- 33.Yates M.S., Tauchi M., Katsuoka F., Flanders K.C., Liby K.T., Honda T., Gribble G.W., Johnson D.A., Johnson J.A., Burton N.C., Guilarte T.R., Yamamoto M., Sporn M.B., Kensler T.W. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6:154–162. doi: 10.1158/1535-7163.MCT-06-0516. [DOI] [PubMed] [Google Scholar]
- 34.Ebert P.J., Timmer J.R., Nakada Y., Helms A.W., Parab P.B., Liu Y., Hunsaker T.L., Johnson J.E. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation. Development. 2003;130:1949–1959. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- 35.Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 36.McLarren K.W., Lo R., Grbavec D., Thirunavukkarasu K., Karsenty G., Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem. 2000;275:530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 37.Cani P.D., Possemiers S., Van de Wiele T., Guiot Y., Everard A., Rottier O., Geurts L., Naslain D., Neyrinck A., Lambert D.M., Muccioli G.G., Delzenne N.M. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP–2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Secondulfo M., Iafusco D., Carratu R., deMagistris L., Sapone A., Generoso M., Mezzogiomo A., Sasso F.C., Carteni M., De Rosa R., Prisco F., Esposito V. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis. 2004;36:35–45. doi: 10.1016/j.dld.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 39.McMellen M.E., Wakeman D., Longshore S.W., McDuffie L.A., Warner B.W. Growth factors: possible roles for clinical management of the short bowel syndrome. Semin Pediatr Surg. 2010;19:35–43. doi: 10.1053/j.sempedsurg.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouch J.D., Dunn J.C. New insights and interventions for short bowel syndrome. Curr Pediatr Rep. 2017;5:1–5. doi: 10.1007/s40124-017-0119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat Rev Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 42.Kim S.B., Pandita R.K., Eskiocak U., Ly P., Kaisani A., Kumar R., Cornelius C., Wright W.E., Pandita T.K., Shay J.W. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. Proc Natl Acad Sci U S A. 2012;109:E2949–E2955. doi: 10.1073/pnas.1207718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wlodarska M., Luo C., Kolde R., d'Hennezel E., Annand J.W., Heim C.E., Krastel P., Schmitt E.K., Omar A.S., Creasey E.A., Garner A.L., Mohammadi S., O'Connell D.J., Abubucker S., Arthur T.D., Franzosa E.A., Huttenhower C., Murphy L.O., Haiser H.J., Vlamakis H., Porter J.A., Xavier R.J. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe. 2017;22:25–37.e6. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh R., Chandrashekharappa S., Bodduluri S.R., Baby B.V., Hegde B., Kotla N.G., Hiwale A.A., Saiyed T., Patel P., Vijay-Kumar M., Langille M.G.I., Douglas G.M., Cheng X., Rouchka E.C., Waigel S.J., Dryden G.W., Alatassi H., Zhang H.G., Haribabu B., Vemula P.K., Jala V.R. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10:89. doi: 10.1038/s41467-018-07859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerachayaphorn J., Mennone A., Soroka C.J., Harry K., Hagey L.R., Kensler T.W., Boyer J.L. Nuclear factor-E2-related factor 2 is a major determinant of bile acid homeostasis in the liver and intestine. Am J Physiol Gastrointest Liver Physiol. 2012;302:G925–G936. doi: 10.1152/ajpgi.00263.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong X., Thimmulappa R., Craciun F., Harvey C., Singh A., Kombairaju P., Reddy S.P., Remick D., Biswal S. Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am J Respir Crit Care Med. 2011;184:928–938. doi: 10.1164/rccm.201102-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 48.Magness S.T., Puthoff B.J., Crissey M.A., Dunn J., Henning S.J., Houchen C., Kaddis J.S., Kuo C.J., Li L., Lynch J., Martin M.G., May R., Niland J.C., Olack B., Qian D., Stelzner M., Swain J.R., Wang F., Wang J., Wang X., Yan K., Yu J., Wong M.H. A multicenter study to standardize reporting and analyses of fluorescence-activated cell-sorted murine intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2013;305:G542–G551. doi: 10.1152/ajpgi.00481.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yagishita Y., Fukutomi T., Sugawara A., Kawamura H., Takahashi T., Pi J., Uruno A., Yamamoto M. Nrf2 protects pancreatic β-cells from oxidative and nitrosative stress in diabetic model mice. Diabetes. 2014;63:605–618. doi: 10.2337/db13-0909. [DOI] [PubMed] [Google Scholar]
- 50.Wang F., Wang J., Liu D., Su Y. Normalizing genes for real-time polymerase chain reaction in epithelial and nonepithelial cells of mouse small intestine. Anal Biochem. 2010;399:211–217. doi: 10.1016/j.ab.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 51.Sirakov M., Borra M., Cambuli F.M., Plateroti M. Defining suitable reference genes for RT-qPCR analysis on intestinal epithelial cells. Mol Biotechnol. 2013;54:930–938. doi: 10.1007/s12033-012-9643-3. [DOI] [PubMed] [Google Scholar]
- 52.Andersen C.L., Jensen J.L., Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 53.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]