Abstract
Osteosarcoma (OS) is one of the most aggressive malignancies, accompanied by an elevated incidence and a decreased rate of healing. Recently, several long non-coding RNAs (lncRNAs) have been reported to be involved in OS progression. Although tumor suppressor candidate 7 (TUSC7) was reported as a novel lncRNA, little is known about its biological functions in OS. The present study was designed to explore whether TUSC7 was involved in the pathological development of OS using various methods, including hematoxylin and eosin staining, Cell Counting Kit-8 assay, colony formation assay and Transwell assay. The present study revealed that TUSC7 expression was downregulated in OS tissues and cell lines compared with in normal tissues and cell lines. Functionally, the current results revealed that overexpression of TUSC7 inhibited OS cell proliferation, migration and invasion, while promoting apoptosis in vitro and in vivo. Next, the subcellular distribution of TUSC7 was examined by nuclear/cytoplasmic RNA fractionation and reverse transcription-quantitative PCR. Mechanistic studies revealed that TUSC7 exerted its role by sponging microRNA (miR)-181a in OS cell lines. Ras association domain family member 6 (RASSF6) was confirmed as a target gene of miR-181a, and the expression levels of RASSF6 were negatively regulated by miR-181a. Additionally, the results of rescue experiments suggested that overexpression of miR-181a neutralized the inhibitory effects of TUSC7 overexpression on OS cells. Overall, the present study demonstrated that the tumor suppressor role of TUSC7 in OS progression was mediated through the miR-181a/RASSF6 axis, which may represent a new therapeutic target for OS.
Keywords: osteosarcoma, long non-coding RNA tumor suppressor candidate 7, microRNA-181a, Ras association domain family member 6
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor that affects children, adolescents and young adults (1,2). With the application of chemotherapeutics, a combination of surgical resection and multi-chemotherapy has become a standard clinical treatment strategy for almost all patients with OS, which has significantly improved patient survival (3). Despite extensive advances achieved in OS therapy, the overall survival rate of patients with distant metastasis remains poor (4,5). To date, the biological characteristics of OS are well understood, but it is urgent to explore the mechanisms of OS progression, which may contribute to the development of effective strategies for the diagnosis, treatment and prognosis of patients with OS.
Long non-coding RNAs (lncRNAs) belong to the non-coding RNA family and are generally comprised of ~200 nucleotides (6,7). Abnormal expression levels of lncRNAs contribute to tumor initiation, growth and metastasis (8-11). Recently, several lncRNAs have been reported to be involved in OS progression, such as ODRUL (12), LINC01278 (13) and lncRNA DLEU1 (14). However, the functions of lncRNAs in OS require further investigation. Tumor suppressor candidate 7 (TUSC7) is a lncRNA that has been reported to be a cancer suppressor gene in numerous types of human cancer, such as colorectal cancer (15), pancreatic carcinoma (16) and esophageal squamous cell carcinoma (17). Recently, TUSC7 has been also identified as a tumor suppressor in OS (18). However, the regulatory mechanisms of TUSC7 in OS require additional investigation.
In addition to lncRNAs, microRNAs (miRNAs/miRs) are another group of non-coding but short-length (<25 nucleotides) RNAs. Previous studies have demonstrated that miR-181a serves important roles in the development and progression of OS (19-21). In recent years, the discovery of crosstalk between lncRNAs and miRNAs has revealed a new mechanism of protein-coding gene modulation (22,23). Specifically, it has been demonstrated that lncRNAs work with miRNAs by acting as competitive endogenous RNAs (ceRNAs) or as miRNA sponges (24,25). Accordingly, the present study hypothesized a regulatory mechanism for lncRNA-miRNA-mRNA and investigated its functional roles in OS.
The present study investigated TUSC7 expression in OS and the vital role of TUSC7 in the proliferation, migration and invasion of OS both in vitro and in vivo.
Materials and methods
Clinical OS specimens
A total of 45 pairs of OS tissue samples and adjacent normal tissue samples (<2 cm from tumor) were collected between November 2018 and October 2019 from patients diagnosed with OS at the Affiliated Hospital of Inner Mongolia Medical University (Hohhot, China). There were 34 males and 11 females, with a median age of 19.4 years (age range, 10-25 years). The pathological diagnoses of OS were confirmed by two independent pathologists. Patients receiving chemotherapy or radiotherapy treatment were excluded. Written informed consent was provided by all patients with OS enrolled in the study. The present study was approved by the Ethics Committee of the Affiliated Hospital of Inner Mongolia Medical University (approval no. Y K D2017142). All tissue samples were immediately frozen in liquid nitrogen and then stored at −80°C until further use.
Cell culture
Human OS cell lines (Saos2, U2OS, MG63 and 143B) and a human osteoblast cell line (hFOB 1.19) were purchased from the American Type Culture Collection. All cell lines were cultured in DMEM supplemented with 10% FBS and 1% antibiotics (penicillin, 100 IU/ml and streptomycin, 10 mg/ml; all Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5% CO2.
Cell transfection
For transient transfection, cells were seeded into 6- or 96-well plates (5×106 or 104 cells/well, respectively). U2OS and MG63 cells were transfected with 2 µg pcDNA3.1-TUSC7 (TUSC7 overexpression vector), 2 µg pcDNA3.1-NC (empty vector), 100 nM non-targeting negative control (NC)-mimic or 100 nM miR-181a mimic. The synthesized sequences were 5′-AAC AUU CAA CGC UGU CGG UGA GU-3′ for the miR-181a mimic and 5′-UUC UCC GAA CG UGU CAC GUT T-3′ for the NC-mimc. In the present study, pcDNA3.1-TUSC7, pcDNA3.1-NC, 2 µg small interfering (si)-RASSF6 inhibitor (si-RASSF6), si-NC, miR-181a mimic and NC-mimic were provided by Shanghai GenePharma Co., Ltd. Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection according to the manufacturer's protocol. The RASSF6 siRNA target sequence was 5′-GAC CCA GAU UCC UAU GUC U-3′, while the si-NC target sequence was 5′-UUC UCC GAA CGU GUC ACG UTT-3′. After transfection for 2 days at the room temperature, the cells were collected for subsequent use.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from tissues and cells was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. RNA (1 µg) was reverse transcribed to cDNA using the Prime Script RT Master Mix kit (Takara Bio, Inc.) according to the manufacturer's protocol. After RT, qPCR analysis was conducted using SYBR Premix Ex Taq™ II (Takara Bio, Inc.) on a StepOnePlus™ Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR protocol was set at 95°C for 10 min, followed by 40 cycles at 95°C for 10 sec and 60°C for 1 min. The 2−ΔΔCq method (26) was used to calculate relative mRNA expression. The expression levels of TUSC7 and RASSF6 were normal-ized to those of GAPDH, while miR-181a expression was normalized to that of U6. The following gene-specific primers were used: TUSC7 forward, 5′-CAC TGC CTA TGT GCA CGA CT-3′ and reverse, 5′-AGA GTC CGG CAA GAA GAA CA-3′; miR-181a forward, 5′-ACA CTC CAG CTG GGA ACA TTC AAC GCT GTC G-3′ and reverse, 5′-GGT GTC GTG GAG TCG GCA ATT CAG TTG AG-3′; RASSF6 forward, 5′-AGG CCA GAC AGC TCT GAT GT-3′ and reverse, 5′-AGG CCA GAC AGC TCT GAT GT-3′; U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCAC GAA TTT GCG T-3′; GAPDH forward, 5′-TGA CTT CAA CAG CGA CAC CCA-3′ and reverse, 5′-CAC CCT GTT GCT GTA GCC AAA-3′; and β-actin forward, 5′-CTC CAT CCT GGC CTC GCT GT-3′ and reverse, 5′-GCT GTC ACC TTC ACC GTT CC-3′.
Western blot analysis
Total proteins in tissues and cells were lysed using RIPA lysate buffer with protease inhibitor cocktail (Roche Applied Science). The protein concentration was quantified using the BCA assay. SDS-PAGE (10%) was conducted to isolate 20 µg/lane of protein and transfer it to PVDF membranes (EMD Millipore). After blocking with 5% skimmed milk solution at room temperature for 1 h, membranes were incubated overnight with primary antibodies at 4°C and then incubated with HRP-conjugated secondary antibody (cat. no. A8419; 1:5,000; Sigma-Aldrich; Merck KGaA) at room temperature for 1 h. All primary anti-bodies were purchased from Abcam, including anti-RASSF6 (cat. no. ab220111; 1:1,000), anti-Bax (cat. no. ab32503; 1:1,000), anti-Bcl2 (cat. no. ab182858; 1:2,000), anti-Caspase-3 (cat. no. ab13847; 1:500), anti-Caspase-8 (cat. no. ab32397; 1:500) and anti-GAPDH (cat. no. ab9485, 1:2,500). Target protein levels were normalized to GAPDH, which served as the control. Protein expression was measured using enhanced chemiluminescence (EMD Millipore), and the software used for densitometry was ImageJ v1.8.0 (National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 (Beyotime Institute of Biotechnology) assays were conducted according to the manufacturer's protocol to assess cell proliferation. U2OS and MG63 cells (104 cells/well) were seeded into 96-well plates and transfected with TUSC7 overexpression vector or control vector, or miR-181a mimic or NC-mimic, as aforementioned. Cells were incubated with 10% CCK-8 solution added to each well in the dark for 2 h at 37°C. Proliferation rates were determined at 24, 48 and 72 h after transfection. The optical density (OD) of cells was measured using an ultraviolet spectrophotometer (ELX800; BioTek Instruments, Inc.; Agilent Technologies, Inc.) at a wavelength of 450 nm.
Colony formation assay
After transfection, cells were incubated in 6-well plates (500 cells/well) for colony formation assays. After 2 weeks of culture at 37°C, colonies were fixed with 10% formaldehyde at room temperature for 15 min and stained with 0.1% crystal violet solution at room temperature for 10 min. A colony was defined as containing >50 cells. The colony number in each well was counted and imaged under a light microscope (magnification, ×20; Olympus Corporation).
Transwell assay
The migratory and invasive abilities of U2OS and MG63 cells were estimated using Transwell assays. For the Transwell migration assay, transfected U2OS and MG63 cells (1×105) were seeded into the upper chamber of 8-µm pore size membranes (Merck KGaA) containing serum-free medium. For the Transwell invasion assay, U2OS and MG63 cells (2×105) were added in serum-free medium to the upper chambers pre-coated with diluted Matrigel® (1:5; BD Biosciences) at 37°C for 5 h. A total of 500 µl medium containing 10% FBS was added into the lower chamber. After incubation at 37°C for 48 h, cells on the upper membrane were removed using a cotton swab, and cells that had traversed the membrane were stained using 0.1% crystal violet at room temperature for 10 min and counted under a light microscope (magnification, ×100; Olympus Corporation) in five randomly chosen microscopic fields. The number of cells that entered the lower chamber reflected the migratory or invasive ability of tumor cells.
Apoptosis assay
Flow cytometry analysis was used to evaluate apoptosis. Briefly, U2OS and MG63 cells (2×105 cells/well) were seeded into 6-well plates, and after transfection for 48 h, cells were treated with Annexin V-FITC (5 µl) and PI (5 µl) using an Annexin V-FITC Apoptosis Detection kit (Invitrogen; Thermo Fisher Scientific, Inc.). The apoptosis rate was detected using a CytoFLEX flow cytometry (cat. no. C02945; Beckman Coulter, Inc.) following the manufacturer's protocol and analyzed using ModFit LT v5.0 (Verity Software House, Inc.).
Dual luciferase reporter assay
Firstly, the starBase data-base (http://starbase.sysu.edu.cn/) was used to predict the binding sites between miR-181a and TUSC7 or RASSF6, and luciferase reporter assay was used to confirm this association. For the luciferase reporter assay, sequences of TUSC7 and RASSF6 containing wild-type (WT) binding sites to miR-181a or mutant (Mut) sites were amplified and cloned into the luciferase reporter vector pGL3 (Promega Corporation), called TUSC7-WT and RASSF6-WT, and TUSC7-Mut and RASSF6-Mut, respectively. Cells were transfected with TUSC7-WT/Mut or RASSF6-WT/Mut, as well as with miR-181a mimic or NC-mimic using Lipofectamine 2000 as aforementioned. After ~48 h, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation) following the manufacturer's protocol. Renilla luciferase activity was used as an internal reference.
Nuclear/cytoplasmic RNA fractionation
The cytoplasmic/nuclear fraction isolation assay was performed using a PARIS kit (Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. After purification and DNase I treatment, RNA from the isolated nuclear and cytoplasmic fractions was reverse transcribed, and qPCR was used to evaluate the relative expression levels of TUSC7 and GAPDH in each sample, as aforementioned.
Hematoxylin and eosin (H&E) staining
Samples were collected from mouse subcutaneous tumors. First, samples were fixed in 4% paraformaldehyde solution at room temperature for 48 h, embedded in paraffin and transversely cut into 5-µm-thick sections. Second, paraffin sections were deparaffinized using xylene I and II for 10 min and rehydrated using a descending alcohol series (100, 90, 80 and 70% alcohol for 5 min each, followed by washing with water for 5 min). Finally, slices were subjected to H&E staining at room temperature for 15 min and observed under a light microscope (magnification, ×100; Olympus Corporation).
Immunohistochemistry
Mouse subcutaneous tumors were removed and fixed in 4% formaldehyde at room temperature for 48 h, embedded in paraffin and transversely cut into 5-µm-thick sections. Subsequently, sections were washed three times with 0.1 M PBS after deparaffinization using xylene I and II for 10 min and rehydration as aforementioned, and blocked with blocking buffer (Dual Endogenous Enzyme Block; Dako; Agilent Technologies, Inc.) at room temperature for 5 min. Sections were incubated with goat anti-Ki67 primary antibody (cat. no. ab15580; 1:100; Abcam) for 24 h at 4°C, followed by incubation with an HRP-conjugated rabbit anti-goat IgG secondary antibody (cat. no. ab6741; 1:500; Abcam) for 30 min at room temperature. Finally, sections were stained with 3,3-diaminobenzidine (Wuhan Servicebio Technology Co., Ltd.) and visualized using a light microscope (magnification, ×100; Olympus Corporation).
In vivo mouse xenograft tumor assay
A total of 30 male BALB/c nude mice (weight, 18-20 g; age, 4-6 weeks) from Beijing Vital River Laboratory Animal Technology Co., Ltd., were kept in a 12-h day/night cycle and in a temperature-controlled room (temperature, 18-22°C; humidity, 45-65%) with free standard food and tap water. There were 6 mice in each group. Each experiment was repeated 3 times. Animal experiments were approved by the Animal Experimentation Ethics Committee of the Affiliated Hospital of Inner Mongolia Medical University and conducted following the Guide for the Care and Use of Laboratory Animals (27). U2OS cells stably expressing TUSC7 or control vector were propagated, and 1×106 cells/100 µl culture medium were subcutaneously inoculated into the right side of the posterior flank in nude mice. Animals were sacrificed 5 weeks post-inoculation by intraperitoneal injection of pentobarbital sodium (100 mg/kg). Tumors were surgically dissected and weighed.
Statistical analysis
All data were collected from at least three independent experiments and are reported as the mean ± SD. Statistical significance between normal and tumor tissues were analyzed using a paired Student's t-test, while other comparisons between two groups were analyzed using an unpaired Student's t-test. One-way ANOVA followed by Tukey's post-hoc test was used for multiple comparisons using SPSS version 21.0 software (IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
TUSC7 expression is significantly downregulated in OS tissues and cell lines
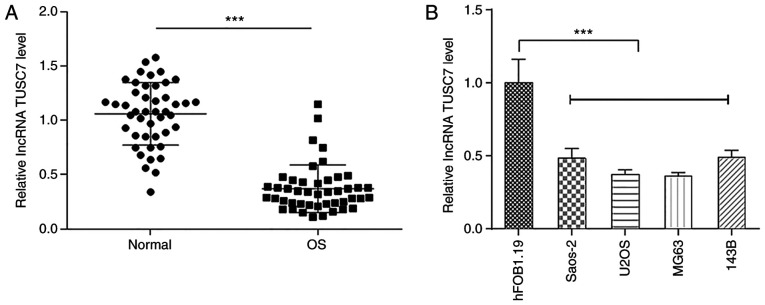
To investigate the role of TUSC7 in OS progression, the expression levels of TUSC7 in OS and normal bone tissues were measured by RT-qPCR. Results demonstrated that TUSC7 expression was significantly lower in OS tissues compared with in normal bone tissues (Fig. 1A). In addition, TUSC7 expression was significantly decreased in OS cell lines compared with in the human osteoblast hFOB1.19 cell line (Fig. 1B), especially in U2OS and MG63 cells, which were therefore used for subsequent experiments.
Figure 1.
TUSC7 expression is downregulated in OS tissues and cell lines. (A) Reverse transcription-quantitative PCR showing the expression levels of TUSC7 in (A) OS and normal tissues, and (B) four OS cell lines and the normal osteoblast hFOB1.19 cell line. Data are shown as the mean ± SD. ***P<0.001. OS, osteosarcoma; lncRNA TUSC7, long non-coding RNA tumor suppressor candidate 7.
TUSC7 overexpression inhibits the progression of OS
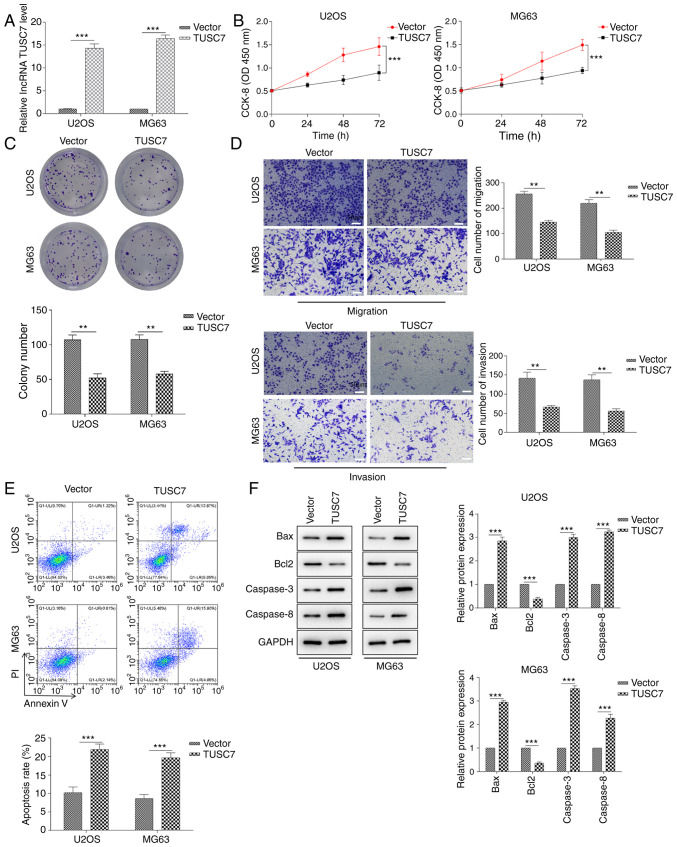
To explore the functional roles of TUSC7 in OS cell proliferation, migration, invasion and apoptosis, U2OS and MG63 cells were transfected with TUSC7-overexpressing plasmid or control vector. RT-qPCR assays revealed that the expression levels of TUSC7 in U2OS and MG63 cells were significantly increased in response to TUSC7 transfection (Fig. 2A). Cell proliferation was measured using CCK-8 and colony formation assays. The results demonstrated that cell proliferation rate and colony formation ability in the TUSC7 group were significantly lower compared with those in the control vector group (Fig. 2B and C). Additionally, the results of the Transwell assay revealed that migration and invasion of U2OS and MG63 cells were significantly decreased after TUSC7 overexpression (Fig. 2D). By contrast, the rate of early and late apoptotic cells in the TUSC7 transfection group was significantly increased (Fig. 2E). In addition, the protein expression levels of the anti-apoptotic protein Bcl2 were significantly decreased, while those of the pro-apoptotic proteins Bax, Caspase-3 and Caspase-8 were significantly increased in the TUSC7 group (Fig. 2F). The current results indicated that TUSC7 overexpression significantly inhibited proliferation, migration and invasion, and promoted apoptosis in U2OS and MG63 cells.
Figure 2.
TUSC7 overexpression inhibits the progression of OS. (A) Reverse transcription-quantitative PCR analysis of TUSC7 expression. (B) CCK-8 and (C) colony formation assays were performed to examine the cell proliferation rate of U2OS and MG63 cells after overexpression of TUSC7. (D) Transwell assays were used to evaluate the effects of TUSC7 overexpression on cell migration and invasion in U2OS and MG63 cells (scale bar, 50 µm). (E) Flow cytometry assays were used to analyze the effects of TUSC7 on apoptosis. (F) Western blotting was performed to measure the protein expression levels of several biomarkers of apoptosis, such as Bcl2, Bax, Caspase-3 and Caspase-8. Data are presented as the mean ± SD. **P<0.01 and ***P<0.001. CCK-8, Cell Counting Kit-8; OS, osteosarcoma; lncRNA TUSC7, long non-coding RNA tumor suppressor candidate 7.
TUSC7 competitively binds to miR-181a and suppresses its expression in OS cell lines
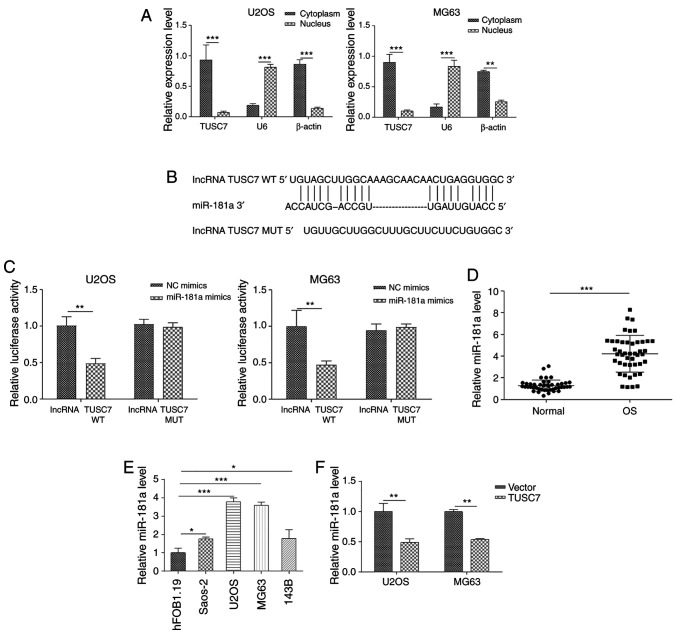
To examine the role of TUSC7 in OS progression, the subcellular localization of TUSC7 in U2OS and MG63 cells was analyzed. RT-qPCR revealed that TUSC7 was primarily localized in the cytoplasm (Fig. 3A), indicating its role in post-transcriptional regulation of gene expression. Previous studies have proven that lncRNA expression in the cytoplasm is vital for pathophysiological processes and partially occurs by sponging miRNAs (28,29). Since the promoting role of miR-181a in OS progression is well known and its expression levels have been reported to be aberrantly elevated in OS tissues and cells (20), the present study examined whether TUSC7 inhibited OS progression by sponging miR-181a expression. First, it was revealed that miR-181a was directly bound to the 3′-untranslated regions (UTR) of TUSC7 using the star-Base database (Fig. 3B). Subsequently, a luciferase reporter vector ligated with TUSC7-WT and TUSC7-Mut sequences was constructed. The results revealed that luciferase activities of TUSC7-WT, but not TUSC7-Mut, were significantly decreased following transfection with miR-181a mimics (Fig. 3C). In addition, miR-181a expression was significantly increased in both OS tissues and cell lines (Fig. 3D and E). As shown in Fig. 3F, the expression levels of miR-181a were significantly downregulated by TUSC7 overexpression compared with the control vector. In summary, the present results indicated that TUSC7 regulated OS progression by binding to miR-181a.
Figure 3.
TUSC7 competitively binds miR-181a and suppresses its expression in OS cell lines. (A) RT-qPCR showing the distribution of TUSC7 in the cytoplasm and nucleus of U2OS and MG63 cells. (B) WT and Mut sequences of TUSC7 3′-untranslated region and miR-181a. (C) Luciferase assays in U2OS and MG63 cells transfected with TUSC7 WT or Mut luciferase reporter plasmid, with miR-181a or NC mimics. Expression levels of miR-181a in (D) OS tissues and (E) OS cell lines detected by RT-qPCR. (F) Expression levels of miR-181a detected by RT-qPCR. Data are presented as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. WT, wild-type; Mut, mutant; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; OS, osteosarcoma; lncRNA TUSC7, long non-coding RNA tumor suppressor candidate 7.
RASSF6 is a direct target of miR-181a in OS cell lines
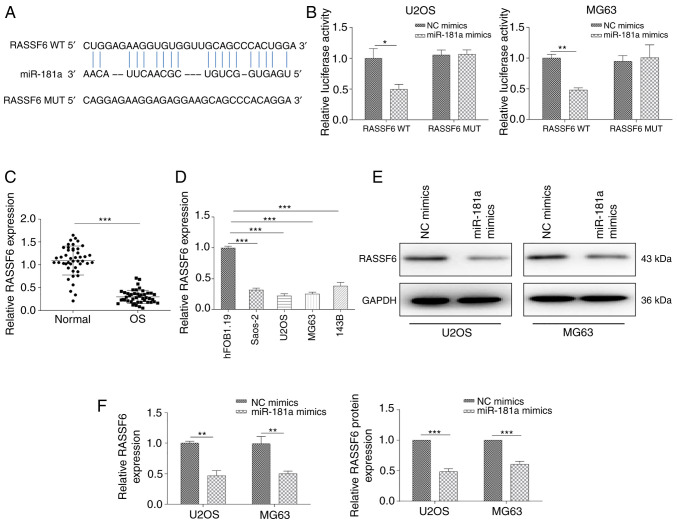
Having determined the potential competitive mechanism between miR-181a and TUSC7, the present study attempted to identify critical direct targets underlying the mechanistic contribution of miR-181a to OS progression. The predicted binding site of miR-181a at the 3′-UTR of RASSF6 was determined using the starBase online database. The potential binding sequences are shown in Fig. 4A. A dual-luciferase reporter assay was further conducted to validate direct binding of the 3′-UTR of RASSF6 mRNA with miR-181a. The results revealed that miR-181a mimics significantly suppressed luciferase activity of the WT 3′-UTR of RASSF6, but not of the Mut 3′-UTR of RASSF6 (Fig. 4B). In addition, RT-qPCR assays demonstrated that RASSF6 expression was significantly decreased in OS tissues and cell lines (Fig. 4C and D). RASSF6 expression at both the protein and mRNA levels was significantly suppressed after transfection with the miR-181a mimic (Fig. 4E and F). In summary, the current findings suggested that RASSF6 may act as a down-stream effector of miR-181a in OS progression.
Figure 4.
RASSF6 is a direct target of miR-181a in OS cell lines. (A) Putative binding sites between miR-181a and RASSF6 obtained from the starBase database. (B) Relative luciferase activities of U2OS and MG63 cells after co-transfection with WT or Mut RASSF6 reporter plasmid and miR-181a or NC mimics. RT-qPCR analysis of RASSF6 expression in (C) OS tissues and (D) OS cell lines. RASSF6 expression assessed via (E) western blotting and (F) RT-qPCR following transfection with miR-181a or NC mimics. Data are presented as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. WT, wild-type; Mut, mutant; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; OS, osteosarcoma; RASSF6, Ras association domain family member 6.
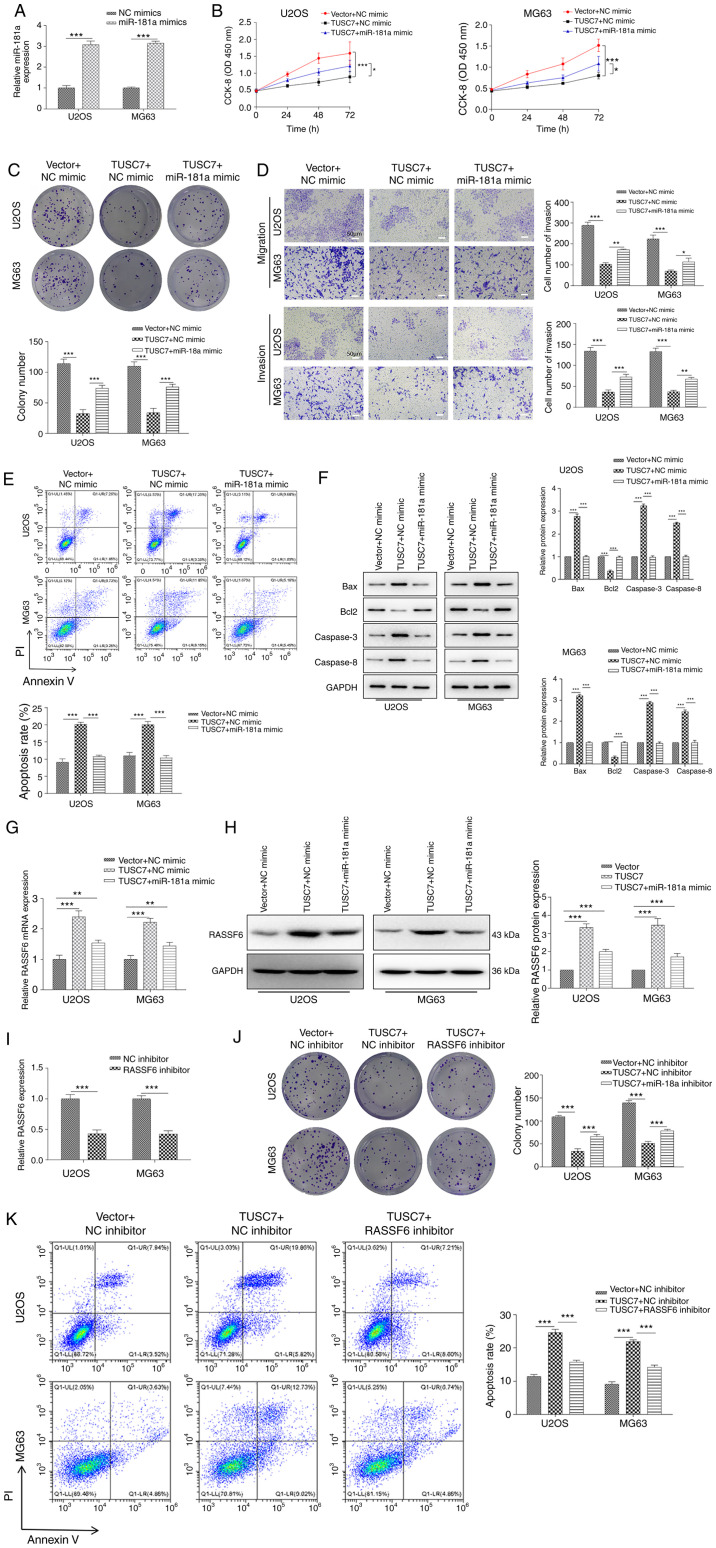
TUSC7/miR-181a/RASSF6 regulates the progression of OS
The present study demonstrated that TUSC7 served as an endogenous sponge for miR-181a in OS. Thus, whether TUSC7 inhibited OS cell proliferation, migration and invasion by inhibiting miR-181a expression was investigated. As shown in Fig. 5A, miR-181a expression was significantly increased in both U2OS and MG63 cells after transfection with the miR-181a mimic compared with the NC mimic. CCK-8 and colony formation assays demonstrated that miR-181a significantly reversed TUSC7-induced inhibition of cell proliferation in cells co-transfected with miR-181a mimic and TUSC7 vector (Fig. 5B and C). Additionally, Transwell assay revealed that TUSC7 overexpression significantly decreased cell migration and invasion, which was rescued by the addition of miR-181a (Fig. 5D). Furthermore, flow cytometry assays demonstrated that miR-181a overexpression rescued TUSC7-induced apoptosis in OS cells (Fig. 5E) and reversed TUSC7-induced decreases in Bcl2 expression and increases in Bax, Caspase-3 and Caspase-8 expression, as shown by western blotting (Fig. 5F). RT-qPCR and western blot assays revealed that the expression levels of RASSF6 were significantly increased following TUSC7 overexpression, and this effect was significantly reversed by the addition of the miR-181a mimic (Fig. 5G and H).
Figure 5.
TUSC7/miR-181a/RASSF6 regulates the progression of OS. (A) Relative miR-181a expression measured via RT-qPCR in miR-181a mimics-transfected U2OS and MG63 cells. Proliferation of U2OS and MG63 cells co-transfected with TUSC7 and miR-181a mimics identified by (B) CCK-8 and (C) colony formation assays. (D) Migration and invasion of U2OS and MG63 cells co-transfected with TUSC7 and miR-181a mimics measured by Transwell assays (scale bar, 50 µm). (E) Apoptosis of OS cells determined by flow cytometry in TUSC7- and miR-181a mimics-transfected U2OS and MG63 cells. (F) Expression levels of the apoptosis-associated proteins Bcl2, Bax, Caspase-3 and Caspase-8 detected via western blot assays in TUSC7- and miR-181a mimics-transfected U2OS and MG63 cells. (G) RT-qPCR and (H) western blot assays of the expression levels of RASSF6 in TUSC7- and miR-181a mimics-transfected U2OS and MG63 cells. (I) Relative RASSF6 expression measured by RT-qPCR in RASSF6 inhibitor-treated U2OS and MG63 cells. (J) Proliferation of U2OS and MG63 cells co-transfected with TUSC7 and RASSF6 inhibitor detected by colony formation assay. (K) Apoptosis of OS cells determined by flow cytometry in TUSC7- and RASSF6 inhibitor-transfected U2OS and MG63 cells. Data are presented as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. CCK-8, Cell Counting Kit-8; OD, optical density; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; miR, microRNA; OS, osteosarcoma; RASSF6, Ras association domain family member 6; TUSC7, tumor suppressor candidate 7.
In addition, to further detect the functions of RASSF6 on the effects induced by TUSC7, a RASSF6 inhibitor was used to treat OS cells. As shown in Fig. 5I, the RASSF6 inhibitor significantly attenuated mRNA expression levels of RASSF6 in U2OS and MG63 cells compared with the NC inhibitor. Colony formation assays revealed that the RASSF6 inhibitor significantly reversed the TUSC7-induced decrease in cell proliferation (Fig. 5J). Flow cytometry demonstrated that TUSC7 overexpression significantly increased apoptosis, while the RASSF6 inhibitor significantly reversed the effects of TUSC7 (Fig. 5K). The current data illustrated that TUSC7 directly regulated RASSF6 expression by competitively binding to miR-181a as a miRNA sponge.
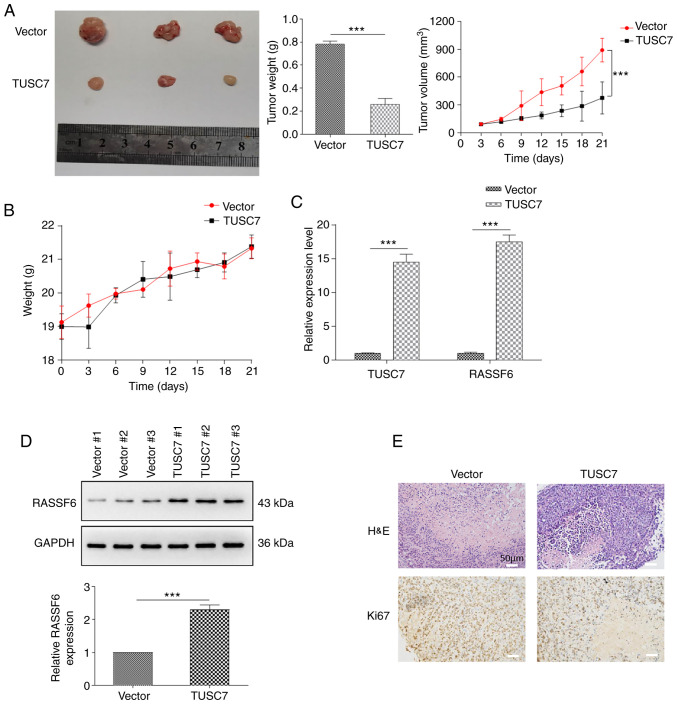
TUSC7 overexpression inhibits tumor growth in vivo
To further explore the roles of TUSC7 in OS tumorigenesis, U2OS cells were transfected with control or TUSC7 over-expression vectors for in vivo analysis of tumor growth. Cells were subcutaneously injected into BALB/c nude mice, which were euthanized for tumor collection after 5 weeks. Consistent with the in vitro results, overexpression of TUSC7 resulted in significantly smaller tumors (Fig. 6A). Moreover, tumor volumes of TUSC7-overexpressing mice were significantly smaller compared with those in the control vector group, while there was no significant difference between the body weights of TUSC7-overexpressing mice and the control vector group (Fig. 6A and B). In addition, RASSF6 expression at both the protein and mRNA levels was significantly increased in response to overexpression of TUSC7 (Fig. 6C and D). Additionally, H&E and Ki-67 staining revealed that the TUSC7-overexpressing group exhibited a decrease in proliferating cells (Fig. 6E). Overall, the present results demonstrated that TUSC7 negatively influenced OS progression.
Figure 6.
TUSC7 overexpression inhibits tumor growth in vivo. Tumor growth in experimental BALB/c nude mice after injection with U2OS cells transfected with control vector or TUSC7 overexpression vector. (A) Tumor volume and weight, and (B) mouse weight from U2OS cells after transfection with TUSC7 overexpression vector or control vector. (C) mRNA expression levels of RASSF6 determined by reverse transcription-quantitative PCR analysis. (D) Protein levels of RASSF6 assessed by western blotting (n=3). (E) Representative images of H&E and Ki67 staining (scale bar, 50 µm). Data are presented as the mean ± SD. ***P<0.001. H&E, hematoxylin and eosin; RASSF6, Ras association domain family member 6; TUSC7, tumor suppressor candidate 7.
Discussion
lncRNAs and miRNAs are reportedly dysregulated in numerous types of cancer, including OS (12,30,31). lncRNAs act as carcinogenic factors or tumor suppressor factors by regulating different mRNAs by sponging them (32,33). Several lncRNAs have been reported to be involved in OS progression (18,34,35). For example, as oncogenes, lncRNA PGM5-AS1 promotes epithelial-mesenchymal transition, invasion and metastasis of OS cells by impairing miR-140-5p-mediated fibrillin-1 inhibition (34). On the other hand, lncRNAs can act as tumor suppressors. lncRNA CEBPA-AS1 expression is decreased in OS tissues and cell lines, and overexpression of CEBPA-AS1 inhibits proliferation and migration while enhancing apoptosis in OS cells through the miR-10b-5p/nuclear receptor corepressor 2/Notch signaling pathway (35). lncRNA TUSC7 has been previously demonstrated to be a tumor suppressor in OS (18). The present study confirmed that TUSC7 was expressed at low levels in OS tissues and cell lines, while overexpression of TUSC7 inhibited proliferation and invasion, and promoted apoptosis in OS cells.
Functionally, the current study revealed that overexpression of TUSC7 inhibited cell proliferation and migration, while promoting apoptosis both in vitro and in vivo. Numerous studies have indicated that lncRNAs function as ceRNAs by competitively binding miRNAs, eliminating the inhibition of miRNAs on their target gene transcripts (36,37). The present study demonstrated that TUSC7 was primarily localized in the cytoplasm, and a luciferase reporter assay revealed that TUSC7 competitively bound to miR-181a. miR-181a has previously been reported to significantly promote proliferation and inhibit apoptosis in OS cells (19). However, to the best of our knowledge, there are no studies on the role of lncRNAs on miR-181a or its mechanism in OS. In the current study, RT-qPCR assays revealed that miR-181a expression was significantly increased in OS tissues and cell lines, and was negatively associated with TUSC7 expression.
Multiple members of the RASSF exhibit anticancer effects (38). RASSF6 is a novel tumor suppressor that serves an important role in the pathogenesis of various types of cancer (39,40). For instance, miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signaling activation (41). Moreover, miR-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer (42). However, the roles of RASSF6 and its regulatory mechanism in OS remain unclear. To explore the association between miRNA and RASSF6, a luciferase reporter assay was used to identify miR-181a-specific binding to RASSF6. Moreover, RASSF6 expression was downregulated in OS tissues and cell lines, and was negatively regulated by miR-181a. The current data suggested that RASSF6 was a direct target gene of miR-181a.
Additionally, a critical finding of the present study was that overexpression of miR-181a rescued the inhibitory effect of TUSC7 on OS cell proliferation, migration and invasion. Moreover, overexpression of miR-181a significantly decreased the expression levels of RASSF6 induced by TUSC7 over-expression. The current data indicated that TUSC7 directly regulated RASSF6 expression by competitively binding to miR-181a as a miRNA sponge.
In conclusion, the present study defined a novel role for TUSC7 as a tumor suppressor in OS development by sponging miR-181a, leading to RASSF6 upregulation. Overall, TUSC7 may represent a potential therapeutic and prognostic target for OS, although its clinical value should be further consolidated in future studies.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Scientific Foundation of China (grant no. Bai891018).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
RB, AZ and WL conceived the study. XC, NW and YW wrote the manuscript and performed the experiments. LS, HX and LW analyzed the data and designed the figures. SC and YY contributed to the resources, interpreted the data and critically revised the manuscript. All authors reviewed the paper and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the Affiliated Hospital of Inner Mongolia Medical University (approval no. Y K D2017142; Hohhot, China). All procedures involving human participants were performed in accordance with the ethical standards of the Institutional and National Research Committee and with the Declaration of Helsinki. Written informed consent was provided by all patients enrolled in the study. All mice were treated according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health, and animal experiments were approved by the Animal Experimentation Ethics Committee of the Affiliated Hospital of Inner Mongolia Medical University (approval no. Y K D2017142).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: Current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwamoto Y, Tanaka K, Isu K, Kawai A, Tatezaki S, Ishii T, Kushida K, Beppu Y, Usui M, Tateishi A, et al. Multiinstitutional phase II study of neoadjuvant chemotherapy for osteosarcoma (NECO study) in Japan: NECO-93J and NECO-95J. J Orthop Sci. 2009;14:397–404. doi: 10.1007/s00776-009-1347-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17:301–307. [PubMed] [Google Scholar]
- 5.Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: Morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand. 2017;59:71. doi: 10.1186/s13028-017-0341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Yu X, Shen J. Long non-coding RNAs: Emerging players in osteosarcoma. Tumour Biol. 2016;37:2811–2816. doi: 10.1007/s13277-015-4749-4. [DOI] [PubMed] [Google Scholar]
- 9.Xu S, Gong Y, Yin Y, Xing H, Zhang N. The multiple function of long noncoding RNAs in osteosarcoma progression, drug resistance and prognosis. Biomed Pharmacother. 2020;127:110141. doi: 10.1016/j.biopha.2020.110141. [DOI] [PubMed] [Google Scholar]
- 10.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, Kon OL, Lee CG. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019;79:5131–5139. doi: 10.1158/0008-5472.CAN-19-0255. [DOI] [PubMed] [Google Scholar]
- 12.Zhu KP, Ma XL, Zhang CL. LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 axis. Mol Ther. 2017;25:2383–2393. doi: 10.1016/j.ymthe.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu Z, Li S. Long noncoding RNA LINC01278 favors the progression of osteosarcoma via modulating miR-133a-3p/PTHR1 signaling. J Cell Physiol. 2020 Jan 29; doi: 10.1002/jcp.29582. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Zhang C, Wang X. Long noncoding RNA DLEU1 aggravates osteosarcoma carcinogenesis via regulating the miR-671-5p/DDX5 axis. Artif Cells Nanomed Biotechnol. 2019;47:3322–3328. doi: 10.1080/21691401.2019.1648285. [DOI] [PubMed] [Google Scholar]
- 15.Ren W, Chen S, Liu G, Wang X, Ye H, Xi Y. TUSC7 acts as a tumor suppressor in colorectal cancer. Am J Transl Res. 2017;9:4026–4035. [PMC free article] [PubMed] [Google Scholar]
- 16.Yue L, Guo J. LncRNA TUSC7 suppresses pancreatic carcinoma progression by modulating miR-371a-5p expression. J Cell Physiol. 2019 doi: 10.1002/jcp.28248. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 17.Chang ZW, Jia YX, Zhang WJ, Song LJ, Gao M, Li MJ, Zhao RH, Li J, Zhong YL, Sun QZ, Qin YR. LncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal squamous cell carcinoma by competitively regulating DESC1. J Exp Clin Cancer Res. 2018;37:56. doi: 10.1186/s13046-018-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong M, Jing R. Long non-coding RNA TUSC7 suppresses osteosarcoma by targeting miR-211. Biosci Rep. 2019;39:BSR20190291. doi: 10.1042/BSR20190291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu ZJ, Huang P, Chong YX, Kang LX, Huang X, Zhu ZX, Nie L. MicroRNA-181a promotes proliferation and inhibits apoptosis by suppressing CFIm25 in osteosarcoma. Mol Med Rep. 2016;14:4271–4278. doi: 10.3892/mmr.2016.5741. [DOI] [PubMed] [Google Scholar]
- 20.Ba Z, Gu L, Hao S, Wang X, Cheng Z, Nie G. Downregulation of lncRNA CASC2 facilitates osteosarcoma growth and invasion through miR-181a. Cell Prolif. 2018;51:e12409. doi: 10.1111/cpr.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savary G, Dewaeles E, Diazzi S, Buscot M, Nottet N, Fassy J, Courcot E, Henaoui IS, Lemaire J, Martis N, et al. The long noncoding RNA DNM3OS Is a reservoir of FibromiRs with major functions in lung fibroblast response to TGF-β and pulmonary fibrosis. Am J Respir Crit Care Med. 2019;200:184–198. doi: 10.1164/rccm.201807-1237OC. [DOI] [PubMed] [Google Scholar]
- 23.Xiao G, Yao J, Kong D, Ye C, Chen R, Li L, Zeng T, Wang L, Zhang W, Shi X, et al. The long noncoding RNA TTTY15, which is located on the Y chromosome, promotes prostate cancer progression by sponging let-7. Eur Urol. 2019;76:315–326. doi: 10.1016/j.eururo.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467–484. doi: 10.1007/s00018-017-2626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Urrutia E, Bustamante Montes LP, Ladron de Guevara Cervantes D, Perez-Plasencia C, Campos-Parra AD. Crosstalk between long non-coding RNAs, Micro-RNAs and mRNAs: Deciphering molecular mechanisms of master regulators in cancer. Front Oncol. 2019;9:669. doi: 10.3389/fonc.2019.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.National Research Council (US): Committee for the Update of the Guide for the Care and Use of Laboratory Animals . Guide for the Care and Use of Laboratory Animals. 8th edition. National Academies Press; Washington, DC: 2011. [Google Scholar]
- 28.Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y, Li X, Yang H. LINC00612 functions as a ceRNA for miR-214-5p to promote the proliferation and invasion of osteosarcoma in vitro and in vivo. Exp Cell Res. 2020;392:112012. doi: 10.1016/j.yexcr.2020.112012. [DOI] [PubMed] [Google Scholar]
- 30.Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8:1015. doi: 10.3390/cells8091015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong P, Xiong Y, Yue J, Xu D, Ihira K, Konno Y, Kobayashi N, Todo Y, Watari H. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res. 2019;38:295. doi: 10.1186/s13046-019-1306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang YR, Xu J, Wei W, Kang-Sheng G. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10:252. doi: 10.1038/s41419-019-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Liu P, Gao H, Wang X, Yan M. Long non-coding RNA PGM5-AS1 promotes epithelial-mesenchymal transition, invasion and metastasis of osteosarcoma cells by impairing miR-140-5p-mediated FBN1 inhibition. Mol Oncol. 2020;14:2660–2677. doi: 10.1002/1878-0261.12711. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Xia P, Gu R, Zhang W, Sun YF. lncRNA CEBPA-AS1 over-expression inhibits proliferation and migration and stimulates apoptosis of OS cells via notch signaling. Mol Ther Nucleic Acids. 2020;19:1470–1481. doi: 10.1016/j.omtn.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Huang G, Wang M, Li X, Wu J, Chen S, Du N, Li K, Wang J, Xu C, Ren H, et al. TUSC7 suppression of Notch activation through sponging MiR-146 recapitulated the asymmetric cell division in lung adenocarcinoma stem cells. Life Sci. 2019;232:116630. doi: 10.1016/j.lfs.2019.116630. [DOI] [PubMed] [Google Scholar]
- 37.Duan X, Wu Y, Zhang Z, Lu Z. Identification and analysis of dysregulated lncRNA and associated ceRNA in the pathogenesis of keloid. Ann Transl Med. 2020;8:222. doi: 10.21037/atm.2020.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: Biological functions and regulation. FEBS Lett. 2014;588:2671–2684. doi: 10.1016/j.febslet.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 39.Iwasa H, Jiang X, Hata Y. RASSF6; the putative tumor suppressor of the RASSF family. Cancers (Basel) 2015;7:2415–2426. doi: 10.3390/cancers7040899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu N, Si M, Yang N, Jing Y, Fu Y, Zhao X, Lin Z, Yang G. Overexpression of RAS-association domain family 6 (RASSF6) inhibits proliferation and tumorigenesis in hepatocellular carcinoma cells. Oncol Res. 2017;25:1001–1008. doi: 10.3727/096504016X14796039599926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi Y, Zhang D, Jiang W, Weng J, Zhou C, Huang K, Tang H, Yu Y, Liu X, Cui W, et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:11–22. doi: 10.1016/j.canlet.2016.12.033. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Yan B, Zhang P, Liu S, Li Q, Yang J, Yang F, Chen E. MiR-496-promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J Cell Physiol. 2020;235:1469–1479. doi: 10.1002/jcp.29066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.