Abstract
The mechanisms of inflammation in bone and joint tissue are complex and involve long non-coding RNAs (lncRNAs), which play an important role in this process. The aim of the present study was to screen out differentially expressed genes in human osteoblasts stimulated by inflammation, and to further explore the mechanisms underlying inflammatory responses and the functional activity of human osteoblasts through bioinformatics methods and in vitro experiments. For this purpose, MG63 cells were stimulated with various concentrations of lipopolysaccharide (LPS) for different periods of time to construct an optimal inflammatory model and RNA sequencing was then performed on these cells. The levels of nuclear enriched abundant transcript 1 (NEAT1), various inflammatory factors, Nod-like receptor protein 3 (NLRP3) protein and osteogenesis-related proteins, as well as the levels of cell apoptosis- and cell cycle-related markers were measured in MG63 cells stimulated with LPS, transfected with NEAT1 overexpression plasmid and treated with bexarotene by western blot analysis, RT-qPCR, immunofluorescence, FISH, TEM and flow cytometry. There were 427 differentially expressed genes in the LPS-stimulated MG63 cells, in which NEAT1 was significantly downregulated. LPS upregulated the expression of inflammatory cytokines and NLRP3, inhibited the expression of autophagy-related and osteogenesis-related proteins, promoted apoptosis and altered the cell cycle, which was partially inhibited by NEAT1 overexpression and promoted by bexarotene. LPS stimulated inflammation in the MG63 cells and inhibited the retinoid X receptor (RXR)-α to downregulate the expression of NEAT1 and decrease levels of autophagy, which promoted the activation of NLRP3 and the release of inflammatory factors, and impaired the functional activity of osteoblasts, thus promoting the development of inflammation.
Keywords: nuclear enriched abundant transcript 1, osteoblast, inflammation, autophagy
Introduction
Bone is a unique type of tissue that undergoes continuous modelling and remodeling, a process that is accomplished by 2 major cell types: Bone-forming osteoblasts and bone-resorbing osteoclasts (1). Osteoblasts and osteoclasts are involved in the complex and precise network of signaling pathways that regulate this process (2-4). However, the mechanisms underlying osteoblastogenesis, which is a complex process, remain to be elucidated. However, several molecules and signaling pathways, such as the Wnt signaling pathway, BMP-2/Smad/Runx2 pathway and AMPK pathway (5-7), have been found to regulate the maturation, differentiation and function of pre-osteoblasts and osteoblasts. The differentiation and function of osteoclasts are mainly regulated by the RANKL/RANK/OPG pathway (8-11). Various skeletal diseases occur when the balance between bone resorption and formation is lost.
Inflammation is one of the major reasons for this imbalance and is manifested as impaired bone formation, as well as excessive bone degradation (1,12). Inflammatory cytokines and other messenger molecules produced by activated cells are delivered to target cells of bone tissue. Lipopolysaccharide (LPS), a main component of Gram-negative bacterial membranes, is commonly used as a stimulator of the inflammatory response in cell experiments and has been demonstrated to suppress osteoblastic differentiation (13-20), increase osteoclast activity (21) and induce bone loss (22,23). Recent studies have demonstrated that LPS-activated autophagy negatively regulates Wnt signaling via the autophagic degradation of dishevelled 2 (Dvl2), which plays an important role in osteoclastogenesis from pre-osteoclasts (24). Accordingly, exploration at the genetic level seems necessary for elucidating the mechanisms that mediate the inflammatory process in skeletal disease.
Long non-coding RNAs (lncRNAs) are non-coding RNAs (ncRNAs) of >200 bp in length, which have been reported to be involved in a variety of biological functions, such as the regulation of gene expression through epigenetic regulation, transcriptional regulation and post-transcriptional regulation (25,26). Recent research has indicated that several lncRNAs expressed in macrophages and monocytes mediate pro-inflammatory and anti-inflammatory processes, cell differentiation and survival (27). Previous studies have demonstrated that lncRNAs may regulate osteogenic differentiation by interacting with miRNAs or specific pathways (28-30). Additionally, several lncRNAs involved in bone mineral homeostasis and osteoclastogenesis have been found in monocytes of osteoporotic patients (31).
Nuclear enriched abundant transcript 1 (NEAT1) is a classic lncRNA that is specifically located in paraspeckles and functions as an essential structural determinant by interacting with members of the Drosophila behavior human splicing (DBHS) family of proteins (32-35). Recently, emerging evidence has suggested that NEAT1 is intricately associated with inflammation. First, NEAT1 has been identified as an inflammatory regulator in human lupus, sepsis and atherosclerosis (36-40). The expression levels of several chemokines and cytokines are significantly linked with the status of NEAT1. Additionally, NEAT1 has been found to promote the formation of Nod-like receptor protein 3 (NLRP3), which leads to enhanced pro-caspase-1 processing, caspase-1 activation and interleukin (IL)-1β maturation (41). In the present study, novel mechanisms of NEAT1 were explored in LPS-induced inflammation in osteoblasts via activating autophagy and suppressing the NLRP3 inflammasome, which may provide novel insight for the therapeutic application of lncRNAs in inflammatory diseases.
Materials and methods
Cells and cell culture
The human osteosarcoma cell line, MG63, donated by the State Key Laboratory of Oral Disease, Sichuan University was used to establish the osteoblast model. A total of 1×106 cells were seeded in each 10-cm dish and cultured with 8 ml α-MEM medium containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 incubator. Passaging was achieved using 2 ml trypsin containing 0.25% EDTA (HyClone; Cytiva) when cells were fused to 80%.
Cell stimulation
To construct an optimal inflammatory model, MG63 cells were separately stimulated with 0.5, 1 and 2 µg/ml Porphyromonas gingivalis-derived LPS (LPS-PG; Invitrogen; Thermo Fisher Scientific, Inc.) for 3 h and then examined by reverse transcription-quantitative PCR (RT-qPCR). The MG63 were then stimulated with LPS-PG at the optimal drug concentration for 2, 3, 4, 5 and 6 h separately and examined by RT-qPCR to identify the optimal treatment time.
To determine the optimal drug concentration of retinoid X receptor (RXR)-α agonist, bexarotene, the MG63 cells were separately treated with 0, 0.1, 0.3, 0.9 and 2.7 µg/ml bexarotene (Cell Signaling Technology, Inc.) for 24 h and then examined by RT-qPCR. Bafilomycin A1 (Baf A1) and MCC950 were obtained from Selleck Chem. Co. Ltd. To inhibit autophagy, cells were incubated with 200 nM Baf A1 for 4 h. The cells were also incubated with 1 µM MCC950 for 3 h to selectively inhibit NLRP3.
NEAT1 overexpression
The NEAT1 overexpression plasmid was constructed by GeneCopoeia, and the null vector was used as a control. The heat shock method was used for the transformation of the plasmid DNA into E. coli. In brief, following a 30 min of incubation on ice, a mixture of 10 µl of competent bacteria and 2 µl of plasmid were placed at 42°C for 90 sec and placed back on ice for 2 min. A total of 2 ml of Luria-Bertani (LB) medium (Thermo Fisher Scientific, Inc.) was added to an agar plate containing ampicillin and 200 µl of the mixture was added onto the LB. The plates were incubated at 37°C overnight. Recombinant plasmid DNA was isolated using a E.Z.N.A.® Endo-Free Plasmid Midi kit (Omega Bio-tek) according to the manufacturer's instructions. Agarose gel electrophoresis was conducted for the selection of the plasmid DNA. Sequencing of the DNA was performed as follows by TSINGKE Biological Technology. The MG63 cells were seeded at a density of 1×105 cells each well into a 6-well plate for transfection. After 24 h, the cells were 70 to 90% confluent and the medium were replaced with an antibiotic-free and serum-free medium. A total of 300 µl of mixture of the plasmid DNA and Lipo2000™ transfection reagent (Beyotime Institute of Biotechnology) were added to each well and the cells were incubated for 6 h at 37°C in a 5% CO2 incubator. The medium was then replaced with α-MEM medium containing 10% FBS. After 24 to 48 h, the success of transfection was determined by RT-qPCR and using a fluorescence microscope (Olympus Corporation).
Western blot analysis
Total protein was extracted using a whole cell lysis assay (Nanjing KeyGen Biotech. Co., Ltd.) according to the manufacturer's protocols. The total protein concentration was determined by BCA protein assay (Nanjing KeyGen Biotech. Co., Ltd.). A total of 4 V of protein sample were mixed with 1 V of 5X protein loading buffer (Sigma-Aldrich; Merck KGaA), incubated for 5 min in boiling water and stored at -20°C. Western blot analysis was performed following a standard protocol. A total of 20 µg of protein lysate was subjected to 7.5% or 10% sodium dodecyl sulfate polyacrylamide gels (Epizyme, Inc.) and transferred onto polyvinylidene difluoride (PVDF) membranes (KeyGen BioTech). The membranes were blocked with 5% non-fat dried milk in TBST (Kelong Chemical Co.) for 1 h at room temperature, and subsequently probed with specific primary antibodies [anti-GAPDH (mouse, 60004-1-Ig), anti-β-actin (rabbit, 66009-1-Ig), anti-Unc-51 like autophagy activating kinase (ULK1; rabbit, 20986-1-AP), anti-p62 (rabbit, 18420-1-AP) (all from Proteintech Group, Inc. and diluted at 1:500), anti-apoptosis-associated speck-like protein containing a CARD (ASC; rabbit, ab151700; Abcam), anti-caspase-1 (rabbit, ab179515; Abcam), anti-NLRP3 (rabbit, ab270449; Abcam) diluted at 1:1,000, anti-p-ULK-1 (rabbit, #14202; Cell Signaling Technology, Inc.) diluted at 1:500, anti-interleukin (IL)-1β (rabbit; Abcam, ab234437 for full-length IL-1β and ab9722 for cleaved IL-1β) at 1:200), caspase-3 (rabbit, #9662; Cell Signaling Technology, Inc.) diluted at 1:500, cytochrome c (cyto c, rabbit, 10993-1-AP) diluted at 1:200, LC3 (rabbit, 14600-1-AP) at 1:200, osteopontin (OPN; rabbit, 22952-1-AP) diluted at 1:500, poly(ADP-ribose) polymerase (PARP; rabbit, 13371-1-AP) diluted at 1:500, Bax (rabbit, 50599-2-Ig) diluted at 1:200, collagen type I (Col-I; rabbit, 14695-1-AP) diluted at 1:500 (all from Proteintech Group, Inc.) at 4°C overnight]. Goat anti-mouse IgG antibody or goat anti-rabbit IgG antibody or donkey anti-goat IgG antibody (AS09-602 and AS10-1427; AmyJet Scientific, Inc.) were used as the secondary antibodies with a 1:200 dilution at room temperature for 1-2 h. The signal was detected using an ECL kit (EMD Millipore) with a Bio-Rad Geldoc EZ instrument (Bio-Rad Laboraroties, Inc.), according to the manufacturer's instructions. ImageJ 1.7 soft-ware was used to quantify the density and size of the blots, and statistical analysis was performed using the Graphpad Prism 7.0 package.
Agarose gel electrophoresis
A total of 500 ml of 50X TAE buffer was prepared with 121 g of Tris base (Sigma-Aldrich; Merck KGaA), 28.55 ml of acetic acid and 50 ml of 0.5 ml/l EDTA. Subsequently, 50 ml of 50X TAE buffer was diluted to 500 ml of 5X TAE buffer. Agarose (0.4 g) was then added into 40 ml of 5X TAE buffer, and the mixture was then heated in a microwave oven followed by the addition of 4 drops of Genecolor (Bio-Gene Technology Ltd.) to prepare the agarose gel. A total of 3 µl of DNA samples were loaded into each well with 1 µl of loading buffer. Electrophoresis was run at 120 V and 50 mA for 20 min. The fragments of DNA were visualized under UV light.
RT-qPCR
According to the manufacturer's instructions, RNA was extracted from the cultured cells using an RNA-Quick Purification kit (Yishan Biotechnology, Co. Ltd.). Reverse transcription was accomplished using the PrimeScriptTM RT reagent kit with a gDNA eraser (Takara Bio, Inc.), and samples were immediately stored at −80°C. Sequences of the primers of GAPDH, IL-1β, IL-6, NEAT1-1, NEAT1-2, Runx2, Col-I, and osteocalcin in the MG63 cells were designed and synthesized by TSINGKE Biological Technology as shown in Table I. The thermocycling was set following the manufacturer's instructions: The cDNA synthesis reaction mix was incubated at 37°C for 15 min, and followed by 85°C for 5 sec. Then the reaction was terminated at 4°C. The qPCR reaction mix was incubated at 95°C for 30 sec (stage 1 for 1 cycle), at 95°C for 5 sec and at 60°C for 30 sec (stage 2 for 40 cycles), followed by dissociation stage. Quantitative PCR was performed on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) with 25 µl reaction mix. The relative expression of a gene of interest was calculated using the 2−ΔΔCq method (42) and normalized against an internal control (GAPDH).
Table I.
Sequences of primers used in RT-qPCR analysis.
| Gene | Primer |
|---|---|
| GAPDH | |
| F | 5′-ACAACTTTGGTATCGTGGAAGG-3 |
| R | 5′-GCCATCACGCCACAGTTTC-3′ |
| IL-1β | |
| F | 5′-TGTGAAATGCCACCTTTTGA-3′ |
| R | 5′-TGAGTGATACTGCCTGCCTG-3′ |
| IL-6 | |
| F | 5′-AGCCACTCACCTCTTCAGAAC-3′ |
| R | 5′-GCCTCTTTGCTGCTTTCACAC-3′ |
| NEAT1-1 | |
| F | 5′-AGCTGCGTCTATTGAATTGGTAAAGTAA-3′ |
| R | 5′-GACAGAAAGATCCCAACGATAAAAATAA-3′ |
| NEAT1-2 | |
| F | 5′-GTCTTTCCATCCACTCACGTCTATTT-3′ |
| R | 5′-GTACTCTGTGATGGGGTAGTCAGTCAG-3′ |
| Runx2 | |
| F | 5′-TGGTTACTGTCATGGCGGGTA-3′ |
| R | 5′-TCTCAGATCGTTGAACCTTGCTA-3′ |
| Col-I | |
| F | 5′-AGGGACACAGAGGTTTCAGT-3′ |
| R | 5′-AGCACCATCATTTCCACGAG-3′ |
| Osteocalcin | |
| F | 5′-CTCACACTCCTCGCCCTATTG-3′ |
| R | 5-GCTTGGACACAAAGGCTGCAC-3 |
F, forward; R, reverse; NEAT1, nuclear enriched abundant transcript 1; IL, interleukin; Col-I, collagen type I.
RNA sequencing and bioinformatics analysis
The purity, quantity and integrity of RNA was measured using a NanoDrop™One/OneC spectrophotometer (Thermo Fisher Scientific, Inc.), a Qubit™ RNA HSAssay kit (Thermo Fisher Scientific, Inc.) and an Agilent 4200 TapeStation (Agilent Technologies, Inc.) separately. Libraries were generated by PCR amplification, purified by AmPure XP magnetic beads, and quantified using a Kapa qPCR and Agilent 4200 TapeStation. The PCR amplification mix was incubated in the following progress: 1 cycle 98°C for 30 sec; 15 cycles 98°C for 10 sec, 60°C for 30 sec, 72°C for 30 sec; 1 cycle 72°C for 10 min; hold at 4°C. Libraries were pooled and sequenced on an Illumina HiSeq4000 platform. The expression profiles of each lncRNAs and coding-genes were quantified and normalized by the DESeq method, and the differential expressed genes among each group were calculated by FDR (false discovery rate) and a Student's t-test. The significant differentially expressed genes (DEGs) were recognized with an FDR <0.01 (or adjusted P-value) and a ≥2-fold change in expression. The DEGs were enriched by their Gene Ontology (GO) or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with the GSEA method, as well as the lncRNA annotation, Encyclopedia of DNA Elements (ENCODE) database, and mapping to Reactome database (43) using the clusterProfiler package (44).
Immunofluorescence
Cells in 6-well plates were selected to be transfected with GFP-LC3 plasmid (Addgene, #21073) based on group design. MG63 cells were grown in α-MEM medium containing 10% FBS, cells at sub-confluency were transfected with the indicated cDNAs using Lipo3000 reagent (Thermal Fisher Scientific, Inc.). Cells were analyzed at 24-48 h following transfection. Mock transfection was performed using the empty vector. First, the medium was discarded. The cells were then washed twice with PBS for 5 min each time and fixed using 4% paraformaldehyde for 15 min. After washing in cold PBS twice, the cells were permeabilized using 0.5% Triton X-100 in PBS at room temperature for 20 min. After rinsing with PBS 3 times, the cells were blocked with 5% goat serum (Sigma-Aldrich; Merck KGaA) in PBS for 30 min at room temperature and incubated with 30 µl of the indicated primary antibodies [anti-caspase-1 (rabbit, 22915-1-AP; Proteintech Group, Inc.), anti-ASC (rabbit, ab155970; Abcam), anti-NLRP3 (rabbit, ab270449; Abcam) and LC3-II (rabbit, 18725-1-AP; Proteintech Group, Inc.)] in a wet box at 4°C overnight. After reheating at room temperature for 30 min and washing 3 times with PBS, the cells were incubated with goat anti-rabbit IgG antibody conjugated with Alexa Fluor 488, Alexa Fluor 594 or Cy3 (AS09-608, AS11-1814 and AS11-1815; AmyJet Scientific, Inc.) at a dilution of 1:100 at 37°C for 1 h, and 100 µl of Hoechst stain solution or DAPI were added for 5 min, and the cells were again rinsed with PBS 3 times. A blocking solution containing an anti-fluorescent quencher was used for mounting. Images of the cells were captured under a fluorescence microscope (Olympus Corporation).
Fluorescent in situ hybridization (FISH)
A fluorescence in situ hybridization kit (Guangzhou RiboBio Co., Ltd.) was used, according to the manufacturer's instructions. The methods used for fixation and permeabilization were the same as those used for immunofluorescence (described above). The cells were incubated with 200 µl of prehybridization solution at 37°C for 30 min and were then incubated with 20 µl of DNA probe/hybridization buffer at 37°C overnight. The following steps were performed in the dark: Each well was washed sequentially with washing solution I, II, III and PBS, while blocking, staining and observation of the cells was performed using the same methods as those described above for immunofluorescence.
Transmission electron microscopy (TEM)
The cells were collected using a cell scraper, centrifugated at 100 × g for 5 min at room temperature and washed twice with PBS. The cells were fixed using 2.5% glutaraldehyde at 4°C overnight and seeded to a formvarstabilized carbon support films, negatively stained with phosphotungstic acid solution (2% w/v), and then dried naturally for TEM analysis. The intracellular morphology of MG63 cells was analyzed using a transmission electron microscopy (TEM, H-600; Hitachi, Ltd.).
Cell cycle analysis and apoptosis assay
The cells were dissociated using trypsin without EDTA and the suspension was centrifuged at 1,000 × g for 5 min at room temperature. The cells were gently washed with PBS twice and again centrifugated at 1,000 × g for 5 min at room temperature. The supernatant was discarded, and the cells were collected and detected on a BD FACSCanto-II flowcytometry instrument (BD Biosciences). For the cell cycle assay, the cells were resus-pended and fixed using cold 70% (v/v) ethanol at 4°C for at least 30 min. The ethanol used was discarded through centrifugation at 1,000 × g for 5 min at 4°C. After washing with PBS twice, the cells were incubated with 1 ml of PI staining solution in the dark at room temperature for 20 min and the positive cells were then detected. For the cell apoptosis assay, the cells were resuspended using a 1X binding buffer. The mixture was incubated with 10 µl of PI staining solution and 5 µl of Annexin V-FITC solution (C1062L, Annexin V-FITC/PI dual staining kit; Beyotime Institute of Biotechnology, Inc.) in the dark at room temperature for 5 min. Subsequently, 400 µl of 1X of a binding buffer was added to each tube and the positive cells were then detected.
Statistical analysis
All experiments were performed at least 3 times and all results are reported as the means ± standard deviation (SD). Statistical analyses were performed using an independent Student's t-test, one-way ANOVA with the Bonferroni post hoc test and Pearson's correlation analysis on GraphPad Prism 7.0 software or R packages. P-values of <0.05 were considered to indicate a statistically significant difference.
Results
Expression of NEAT1 is decreased following LPS stimulation
In order to construct an optimal inflammatory model, MG63 cells were stimulated with various concentrations of LPS-PG for different periods of time and the results of RT-qPCR revealed that following stimulation with 0.5, 1 and 2 µg/ml LPS-PG for 3 h, the expression levels of IL-6 and IL-1β were upregulated in the MG63 cells, as shown in Fig. S1A. The levels of IL-6 and IL-1β were significantly increased in the 1 and 2 µg/ml groups compared with the control group, although the difference was not significant between these 2 groups. Thereafter, the levels of inflammatory factors were detected at different time points. The results revealed that the levels of IL-6 and IL-1β were significantly upregulated within a short time duration (2 and 3 h); however, they decreased along with the increase in treatment duration (5 and 6 h) (Fig. S1B). Therefore, stimulation with 1 µg/ml LPS-PG for 2 h was confirmed to produce the optimal MG63 inflammatory model (Fig. S1C), and these conditions were used to treat the cells used in further RNA-seq experiments.
In order to assess LPS-induced changes in RNA expression in MG63 cells, high-throughput sequencing of RNAs from MG63 cells with and without LPS stimulation was conducted. Over 14,000 genes were identified (Table II) and 427 genes were found to be differentially expressed in the MG63 cells following LPS stimulation, compared with that of MG63 cells without stimulation (Fig. 1A and B). Among these genes, 241 genes were upregulated (Log2 fold change >0), while 186 were downregulated (Log2 fold change <0). The top 10 genes with the most significant differences in abundance (upregulated and downregulated) between the MG63 cells with or without LPS stimulation are presented in Table III.
Table II.
Results of comparison of reads to reference genomes.
| Sample | Total clean reads | Total mapped reads | Total mapping ratio (%) | Uniq mapping ratio (%) | Total gene number |
|---|---|---|---|---|---|
| Control-1 | 41,449,747 | 30,129,821 | 72.69 | 48.49 | 14029 |
| Control-2 | 41,153,924 | 30,375,711 | 73.81 | 49.21 | 14068 |
| Control-3 | 40,884,000 | 30,556,702 | 74.74 | 49.93 | 14034 |
| LPS-1 | 41,325,764 | 30,386,834 | 73.53 | 48.91 | 14205 |
| LPS-2 | 40,855,494 | 30,800,957 | 75.39 | 50.22 | 14430 |
| LPS-3 | 41,469,689 | 31,060,797 | 74.90 | 49.91 | 14444 |
Figure 1.
Bioinformatics analysis of RNA-Seq results in LPS-stimulated MG63 cells. (A and B) Differential expression level of RNAs in a heatmap and volcano plot. (A) Red color indicates relative overexpression, while blue color indicates relative underexpression. (B) Red indicates upregulated, green indicates downregulated, and black indicates non-statistically significant altered genes. The horizontal dashed line indicates a statistical threshold corresponding to an adjusted P-value of <0.05; x-axis, mRNA log2 fold change; y-axis, P-value in negative log10 scale. (C and D) Enriched pathway in KEGG analysis. (E) Functional genes network in reactome analysis. Red indicates upregulated, and blue indicates downregulated genes. Main types of functional biological processes are the cell cycle, MAPK activation and ncRNA processing. (F) Each line in the middle of the illustration represents a gene and its rank in GSEA enrichment analysis. The enrichment score indicates the enrichment level of genes in the target gene lists. NES, normalized enrichment score; LPS, lipopolysaccharide.
Table III.
Top 10 differential expressed up- and downregulated genes.
| Gene name | Gene description | Log2 fold change |
|---|---|---|
| Upregulated genees | ||
| CXCL8 | C-X-C motif chemokine ligand 8 | 3.831031313 |
| LIF | LIF, interleukin 6 family cytokine | 3.434091582 |
| EGR1 | Early growth response 1 | 2.949483692 |
| BIRC3 | Baculoviral IAP repeat containing 3 | 2.869020403 |
| SERPINE1 | Serpin family E member 1 | 2.767710691 |
| KRTAP1-5 | Keratin associated protein 1-5 | 2.641349766 |
| CCL2 | C-C motif chemokine ligand 2 | 2.582271496 |
| EDN1 | Endothelin 1 | 2.571977567 |
| CXCL3 | C-X-C motif chemokine ligand 3 | 2.512770455 |
| GADD45B | Growth arrest and DNA damage inducible beta | 2.263219387 |
| Downregulated genes | ||
| TXNIP | Thioredoxin interacting protein | −2.166372864 |
| TEF | TEF, PAR bZIP transcription factor | −1.727267478 |
| SESN3 | Sestrin 3 | −1.679757426 |
| SNAI2 | Snail family transcriptional repressor 2 | −1.672595539 |
| PPP1R3C | Protein phosphatase 1 regulatory subunit 3C | −1.659475556 |
| ARRDC3 | Arrestin domain containing 3 | −1.620649654 |
| C10orf10 | Chromosome 10 open reading frame 10 | −1.617743297 |
| HIST1H3G | Histone cluster 1 H3 family member g | −1.563950632 |
| NEURL1B | Neuralized E3 ubiquitin protein ligase 1B | −1.554898668 |
| MN1 | MN1 proto-oncogene, transcriptional regulator | −1.497164046 |
The results of the GO enrichment analysis and Reactome Pathway Database analysis indicated that the cell cycle, MAPK pathway and ncRNA processing may play a key role in inflammation induced by LPS (Fig. 1C-E). Further results obtained through the GO analysis are shown in Fig. S2.
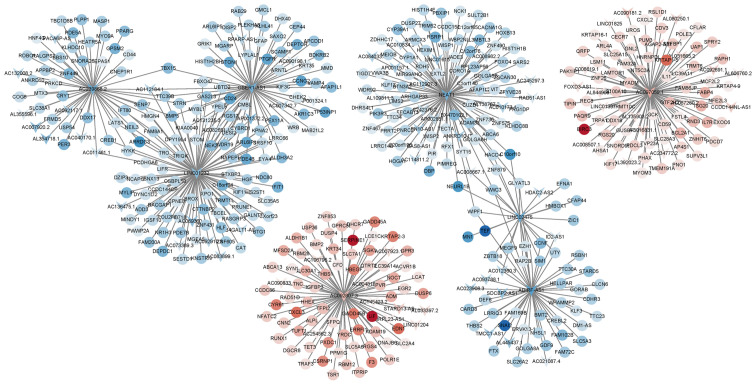
After analysis using the ENCODE database, 10 differentially expressed lncRNAs were identified and these are listed in Table IV, among which NEAT1, LINC02475, OSER-AS1, AC097059.1 and AC092807.3 exhibited a high co-expression association with multiple mRNAs (Fig. 2). Consistent with these results, the RNA expression levels of the NEAT1 transcripts (NEAT1-1 and NEAT1-2) decreased significantly in the LPS-stimulated MG63 cells, as shown through immunofluorescence analysis and RT-qPCR (Fig. 3A and B).
Table IV.
Differential expressed lncRNAs.
| lncRNA | Gene description | Log2 fold change |
|---|---|---|
| NEAT1 | Nuclear paraspeckle assembly transcript 1 | −1.217166677 |
| LINC02475 | Long intergenic non-protein coding RNA 2475 | −1.216221753 |
| OSER1-AS1 | OSER1 antisense RNA 1 (head to head) | −1.164111533 |
| PSMB8-AS1 | PSMB8 antisense RNA 1 (head to head) | −1.12672866 |
| AC048341.3 | −1.092688339 | |
| AP001372.2 | −1.022974571 | |
| AC083843.2 | −0.924327576 | |
| AC097059.1 | 1.415095278 | |
| AC092807.3 | 1.131243985 | |
| SNHG15 | Small nucleolar RNA host gene 15 | 0.982590385 |
Figure 2.
lncRNA-mRNA co-expression network. Arrows and circles represent lncRNAs and mRNAs, respectively. Red color indicates upregulation, while blue color indicates downregulation. The deeper the color, the more significant the difference. lncRNAs and mRNAs connected by lines have co-expression association. The absolute value of Pearson's correlation coefficient was >0.99.
Figure 3.
Expression of NEAT1 and inflammation-related proteins in LPS-stimulated MG63 (LPS) vs. normal MG63 cells (controls). (A) mRNA expression levels of NEAT1-1 and NEAT1-2 examined by RT-qPCR. (B) Differential expression of NEAT1 was shown by immunofluorescence. Alexa Fluor 594 (red) with DAPI nuclear counterstain (blue). Scale bar, 20 µM. (C) Levels of proteins related to NOD-like pathway were measured by western blot analysis. LPS, lipopolysaccharide; NEAT1, nuclear enriched abundant transcript 1. (**P<0.01, ***P<0.001).
Furthermore, combining the results of the KEGG pathway analysis and gene set enrichment analysis (GESA), revealed that the NOD-like receptor signaling pathway was significantly activated, indicating that it may be a potential key pathway for the mediation of inflammatory responses in the current inflammatory model (Fig. 1F).
NEAT1 suppresses LPS-induced inflammatory responses via the NOD-like pathway
In order to examine the effects of NEAT1 expression on the activation of the NOD-like pathway, MG63 cells were successfully transfected with NEAT1 overexpression plasmid (Fig. S3). The expression levels of IL-1β precursors and splicers, caspase-1 precursors and splicers, caspase-3 precursors and splicers, and NLRP3 were found to be upregulated following LPS stimulation and were downregulated by NEAT1 overexpression (Fig. 3C), although the expression levels of cytochrome c were not markedly altered, the above-mentioned results still suggest that NEAT1 can inhibit the expression of NLRP3 and the maturation of IL-1β and caspase-1.
Overexpression of NEAT1 promotes autophagy and further reduces the activation of the NLPR3 inflammasome in MG63 cells
Previous studies have theoretically found that autophagy may be involved in the control of IL-1β secretion by targeting pro-IL-1β for lysosomal degradation or by regulating the activation of the NLRP3 inflammasome or by other potential mechanisms (45,46). The present study explored autophagy induction in MG63 cells in response to LPS stimulation or NEAT1 overexpression. Following transfection with NEAT1 overexpression plasmid, the LPS-stimulated MG63 cells secreted increased levels of ULK1, p-ULK1 and LC3, and a decreased level of p62, indicating that autophagy was inhibited in response to LPS, but was promoted when the cells overexpressed NEAT1 (Fig. 4B). Furthermore, the increased expression of GFP-labeled-LC3 was observed on the autophagosome membrane in NEAT1-overexpressing MG63 cells, while decreased levels were observed in LPS-stimulated MG63 cells (Fig. 4A). In addition, an increased number of autophagosomes in NEAT1-overexpressing cells was observed through TEM (Fig. 4D).
Figure 4.
NEAT1 inhibits the activation of the NLRP3 inflammasome through autophagy. (A) GFP-labeled LC3 (green) was shown by immunofluorescence in cells subjected to different treatments. The position of the nucleus is shown in blue color. Scale bar, 6 µM. (B and C) Expression levels of ULK1, p-ULK1, LC3, p62, NLRP3, OPN and GAPDH were measured in each group by western blot analysis. (D) The autophagosome vacuoles are shown by TEM, and the number of autophagosomes increased in MG63 cells overexpressing NEAT1. Scale bar, 5 µM. LPS, lipopolysaccharide; NEAT1, nuclear enriched abundant transcript 1; NLRP3, Nod-like receptor protein 3; ULK1, Unc-51 like autophagy activating kinase; LC3, light chain 3; OPN, osteopontin.
The assembly of an inflammasome complex has been shown to be required for the activation of caspase-1 and the processing of pro-IL-1β (47). Combining previous results that demonstrated that the expression of NLRP3 was upregulated by LPS and downregulated by NEAT1, in the present study, the autophagy inhibitor, Baf A1 (200 nM for 4 h of incubation), was applied on the cells in the different groups, and western blot analysis of LC3, p62, NLRP3 and OPN expression was then performed. As shown in Fig. 4C, BafA1 markedly decreased the expression of LC3-II and increased the expression of p62, demonstrating that autophagy may be inhibited by Baf A1. The expression of NLRP3 increased following the application of Baf A1 on NEAT1-overexpressing cells stimulated with LPS, and the changes in the expression of OPN in each group were in contrast to those of NLRP3. These results suggested that the inhibitory effect of NEAT1 on NLRP3 was dependent on the suppression of the cellular autophagy process.
Overexpression of NEAT1 affects the cell cycle and impairs the osteogenic function of MG63 cells
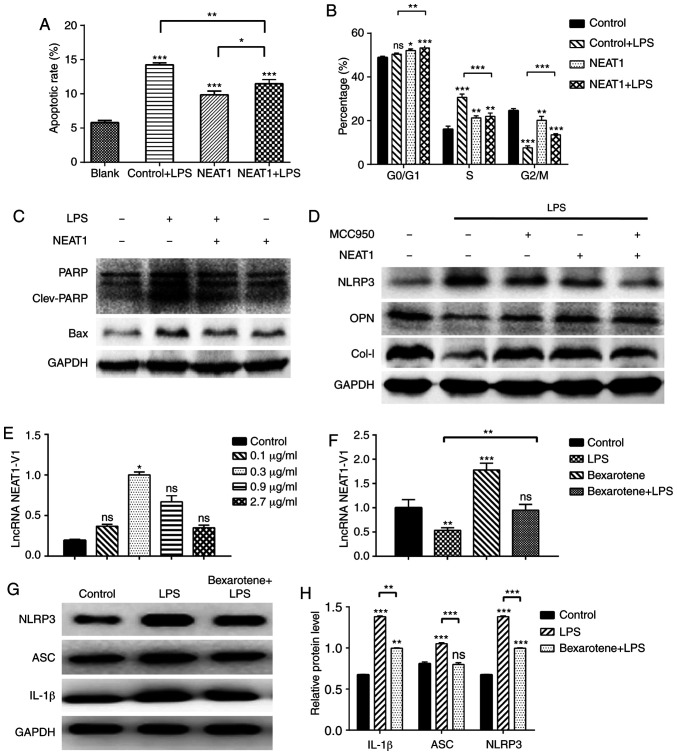
Considering the potential of cell cycle involvement in inflammation, based on the RNA-seq results and the association between inflammasomes, autophagy and cell death pathways involved in the inflammatory process, flow cytometry was performed on the MG63 cells following the different treatments to compare the apoptotic ratio and phases of the cell cycle. The results revealed that the apoptotic ratio increased significantly in the LPS-stimulated cells, but was attenuated in the NEAT1-overexpressing cells (Figs. 5A and S4). Additionally, NEAT1 decreased the levels of the apoptosis-related proteins, PARP and Bax, as shown through the results of western blot analysis (Fig. 5C). The changes in the cell cycle of the LPS-stimulated MG63 cells were mainly reflected by the increase in the proportion of cells at the S phase and the decrease in the proportion of cells at the G2/M phase, compared with the untreated group. The overexpression of NEAT1 increased the proportion of cells at the G0/G1 phase to varying degrees. Compared with the LPS group, the proportion of cells at the S phase decreased, and the proportion of cells at the G2/M phase increased in the NEAT1 + LPS group, indicating that the overexpression of NEAT1 inhibited the cell cycle progression of MG63 cells that were in an inflammatory state (Figs. 5B and S5).
Figure 5.
(A and D) Differential expression of NEAT1 and NLRP3 affects the cell cycle and osteogenic function of MG63 cells. (E-H) Bexatotene significantly increased expression of NEAT1 and decreased expression of inflammasome-related proteins. (A and B) Different apoptotic rate of apoptosis and cell cycle of MG63 cells without or with LPS stimulation, NEAT1 overexpression or both were examined by flow cytometry; (C and D, and G and H) Levels of various proteins were detected by western blot analysis in MG63 cells subjected to different treatments. (E and F) mRNA levels of NEAT1-1 were measured in MG63 cells treated with (E) various concentrations of bexarotene and (F) with different treatment combinations by RT-qPCR (*P<0.05, **P<0.01, ***P<0.001; ns, no significant difference). LPS, lipopolysaccharide; NEAT1, nuclear enriched abundant transcript 1; NLRP3, Nod-like receptor protein 3.
In addition, the levels of osteogenesis-related proteins were detected in the MG63 cells. The LPS-stimulated cells secreted lower levels of OPN and Col-I, and the overexpression of NEAT1 significantly reversed this effect. The novel selective NLRP3 inhibitor, MCC950 (1 µM for 3 h of incubation), exerted the same effect on the expression of OPN and Col-I, as that exerted by the NEAT1 overexpression plasmid; however, no synergistic effect was observed (Fig. 5D). It was hypothesized that NEAT1 and MCC950 may function in a similar manner.
RXR-α agonist reverses the LPS-induced loss of osteogenic factors by regulating NEAT1 expression
In order to determine whether RXR-α is an upstream transcription factor of NEAT1 that can regulate NEAT1 expression, the response of MG63 cells to various concentrations of bexarotene (0.1-2.7 µg/ml for 24 h of incubation), an RXR-α agonist, was observed. All 4 concentrations of bexarotene used increased the expression of NEAT1 and the concentration of 0.3 µg/ml was found to be optimal (Fig. 5E). As shown in Fig. 5F, bexarotene also effectively increased the expression of NEAT1 in MG63 cells under the inflammatory condition. In addition, the protein expression levels of IL-1β, ASC and NLRP3 increased (Fig. 5G and H), and the mRNA expression levels of osteocalcin, COL-I and Runx2 decreased (Fig. 6A) in response to bexarotene. These results suggested that bexarotene reversed the inflammatory process and osteogenesis-related gene expression induced by NEAT1 in MG63 cells. Additionally, it was found that RXR-α may be located upstream of NEAT1 and could positively regulate NEAT1 expression, further abrogating the biological functional damage of osteoblasts induced by inflammation.
Figure 6.
(A) Bexarotene effects the expression of osteogenic genes. The relative mRNA levels of Runx2, collagen-1 and osteocalcin were examined in each group by RT-qPCR; (B) NEAT1 directly interacted with caspase-1. The MG63 cellular lysates with or without LPS incubation were incubated with anti-casp1 antibody or normal IgG for RIP. The NEAT1 levels in immuno-precipitants were analyzed by RT-qPCR and the protein levels were detected by western blot analysis (*P<0.05, **P<0.01, ***P<0.001; ns, no significant difference). LPS, lipopolysaccharide; NEAT1, nuclear enriched abundant transcript 1; Col-I, collagen type I.
Given that there was a recent report indicating that lncRNA Neat1 (murine form) interacted with the NLRP3 inflamma-some in murine BMDMs and functioned as an inflammation stimulator (41), the present study determined the interactions between NEAT1 and caspase-1 by using RIP (RNA immuno-precipitated) from the MG63 cellular lysates with or without LPS stimulation (Fig. 6B). The results suggested that NEAT1 was significantly enriched in caspase-1 immunoprecipitants in the control group; however, the NEAT1 levels did not exhibit any difference in the immunoprecipitants of caspase-1 and IgG in the LPS-stimulated group (Fig. 6B).
To explore the mechanisms through which bexarotene may regulate NEAT1, immunofluorescence and FISH techniques were further combined to observe the levels of NEAT1, caspase-1, ASC, LC3-II and NLRP3 expression in MG63 cells. The results revealed that NEAT1 was widely distributed in the nucleus and cytoplasm. Following stimulation with LPS for 2 h, NEAT1 accumulated in the nucleus, while there was also an evident increase in the size of the paraspeckle. Moreover, the level of caspase-1 tended to increase in the nucleus (Fig. 7A), and the increased expression of ASC was observed (Fig. 7B) in the LPS-stimulated MG63 cells. However, treatment with 0.3 µg/ml bexarotene decreased the LPS-induced activation of the NLRP3 inflammasome, and promoted NEAT1 expression and autophagosome accumulation (Fig. 7C). Taken together, it was hypothesized that the regulation of inflammasomes by NEAT1 may be associated with the cytoplasmic transport of related proteins and the post-transcriptional regulation of proteins, for which the mechanisms of action remain to be proven through further experiments.
Figure 7.
Bexarotene upregulates the expression of NEAT1 and inhibits the activation of the inflammasome. The fluorescence laser confocal microscopic images of NEAT1 and co-stained with (A) caspase-1 and (B) ASC and NLRP3 (yellow) by using immunofluorescence and FISH. Scale bar, 6 µM. Yellow arrows indicate the position of paraspeckles, purple arrows indicate the potential position of the inflammasome. Cy3, red; Alexa Fluor 488, green. (C) Co-staining of NEAT1, NLRP3 and LC3-II by immunofluorescence and FISH. Yellow arrows indicate the fusion of NLRP3 inflammasome and autophagosomes. LPS, lipopolysaccharide; NEAT1, nuclear enriched abundant transcript 1; ASC, apoptosis-associated speck-like protein containing a CARD; NLRP3, Nod-like receptor protein 3; LC3, light chain 3.
Discussion
Previous studies have suggested that the metabolism of bone tissue and inflammation are regulated by various lncRNAs (47,48). In the present study, RNA-seq analysis was performed on MG63 cells and it was found that NEAT1, a key lncRNA, was downregulated in response to LPS. NEAT1 suppressed the downstream inflammatory process and impaired osteoblastic function by promoting autophagy, which inhibited the activation of NLRP3 and downregulated the expression levels of caspase-1 and IL-1β. In addition, the finding that bexarotene increased NEAT1 expression, decreased the expression of inflammatory cytokines and restored osteogenic activity in LPS-stimulated MG63 cells indicated that RXR-α may be a positive regulator of NEAT1. As far as is known, this is the first study to reveal the role of NEAT1 in the inflammation process of bone tissue.
In the present study, it was first found that the expression of NEAT1 was downregulated by LPS. However, the mechanism of NEAT1 expression regulation is complex, which includes gene mutations, copy number alterations, transcription factors, DNA methylation, miRNA and RNA-binding proteins. Several research studies have found that the expression of NEAT1 is increased in prostate cancer, gastric cancer, hepatocellular carcinoma, papillary renal-cell carcinoma and clear cell renal cell carcinoma (49-52), suggesting that NEAT1 may potentially be involved in the process of carcinogenesis in certain tumors. However, the expression of NEAT1 has been found to be decreased in multiple myeloma and leukemia (53-55), indicating that NEAT1 may also function as a tumor suppressor gene in certain other tumors. Therefore, it is currently considered that NEAT1 plays differential roles in different cells. NEAT1 downregulation is generally considered to be regulated by the p53-dependent DNA damage response mechanism. p53 is a cell pressure sensor that responds to signals, such as DNA damage and oncogene expression, as well as mediates cell cycle regulation and apoptosis by regulating hundreds of target genes. As a transcription factor of NEAT1, p53 induces paraspeckle formation by regulating the expression of NEAT1, thereby decreasing DNA damage-induced cell death under oncogene-induced replication stress. In turn, paraspeckles inhibit replication-related DNA damage and p53 activation, which is another mechanism through which p53 maintains genomic integrity (56,57). Apart from p53, NEAT1 expression can also be upregulated by Oct4, HIF-2α, RXR-α and Runx1, and downregulated by BRCA1 (58-62).
RXRs, a family of nuclear receptors, are the primary receptors and mediators of retinoid effects (63). There are 3 isotypes: α, β and γ. The role RXRs in bone metabolism and inflammation remains controversial due to multiple combinations of its isotypes and experimental treatment conditions. The 3 RXR isotypes and their heterodimer partners are widely expressed in the osteoblast lineage, while only RXR-α and RXR-β are expressed in bone marrow myeloid cells (osteoclast progenitors) (64,65). It is also widely accepted that RXRs can modulate osteoclast and osteoblast formation, and function at several levels of cell differentiation and activation. For example, a previous study demonstrated that bexarotene, a selective agonist of RXRs, increased bone turnover in rats (66). On the other hand, Wang et al found that LPS inhibited RXR function and the reduction of its pathway-associated proteins (67). Of note, bexarotene upregulates NEAT1, which inhibits apoptosis and inflammation, thereby resulting in better functional recovery in mice following traumatic brain injury (60). Combined with the results of the present study, RXR-α has been found to be the most probable upstream regulator of NEAT1.
Apart from upstream of NEAT1, downstream is also crucial to understanding its mechanisms, such as the noteworthy targets: Inflammasomes, particularly the NLRP3 inflammasome, which are vital members of the innate immune system (68-71). NLRP3 receives extracellular stimulation and then interacts with ASC, which recruits and activates pro-caspase-1, inducing the maturation, activation and secretion of IL-1β and IL-18, which improve the inflammatory process (72). Several diseases have been proven to be associated with NLRP3 inflammasome activation dysfunction and NLRP3 agonists are a feasible target option for the alleviation of these diseases. However, none of the NLRP3 agonists, which are structurally and functionally diverse, can directly bind to NLRP3. Afonina et al(73) promoted the hypothesis that all NLRP3 agonists may act via a common intermediate, with which they communicate through the induction of membrane damage, potassium efflux and elevation of intracellular calcium. Interestingly, NEAT1 has been demonstrated to enhance the activation of NLRP3 and caspase-1, the processing of pro-IL-1β and the secretion of mature IL-1β in mouse macrophages, and these results are different from the present results (41). It is hypothesized that different cell types or different transcription factors produce differential effects, and that macrophages can differentiate into osteoclasts, which exert different biological effects from that of osteoblasts (Fig. 8).
Figure 8.
Potential mechanisms of NEAT1 in LPS-induced inflammation in MG63 cells. LPS, lipopolysaccharide; NEAT1, nuclear enriched abundant transcript 1; NLRP3, Nod-like receptor protein 3.
The present study also wished to determine the mechanisms through which NEAT1 regulates inflammasomes. The results in the literature published on this topic were summarized and it was found that autophagy can inhibit inflammasomes, and it was then conjectured that activated autophagosomes may act as the mediator between NEAT1 and NLRP3. This has been demonstrated in a number of studies, indicating that NEAT1 promotes autophagy in various cell types (74,75). Moreover, Cao et al(76) discussed the interactions between autophagy and the NLRP3 inflammasome in detail. The mechanism of autophagy inhibition in NLRP3 inflammasomes, which has been observed mostly in macrophages, may be related to the reduction in ASC activity, the phosphorylation of NLRP3 and the clearance of mitochondrial ROS. On the contrary, autophagy may also positively regulate the activation of the NLRP3 inflammasome by enhancing the activation of caspase-1 through a Atg5-dependent non-classical pathway in yeast under starvation conditions.
In order to further understand the function of NEAT1, researchers can perform experiments in vivo or investigate the role of NEAT1 in inflammation induced by hypoxia. A previous study demonstrated that NEAT1-induced para-speckle formation was dependent on HIF-2α expression. Moreover, osteoclasts are another important cell type for bone metabolism, and the KEGG analysis performed in the present study revealed the involvement of osteoclast-differentiation-related-genes, indicating that osteoclasts may be a potential cell target.
In conclusion, the present study clearly demonstrated that NEAT1 suppressed the downstream inflammatory process and impaired osteoblastic function by promoting autophagy and downregulating the expression levels of caspase 1 and IL-1β via the RXR-α/NEAT1/NLRP3 axis. It would be of great interest to further verify the exact mechanism of NEAT1 function in the current inflammatory model and investigate potential inflammasome inhibitors in order to identify potential therapeutic targets.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The authors are grateful for the financial support from the National Natural Science Foundation of China (grant nos. 81870804 and 81571009).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WD, MW, RS and DB conceived and designed the study. MW, PW and JWe performed the experiments. WD, MW, JWa, SC, XX and YH analyzed the data and prepared the figures. WD drafted and wrote the initial manuscript. WD and RS reviewed and revised the manuscript. All authors discussed the results and commented on the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Redlich K, Smolen JS. Inflammatory bone loss: Pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012;11:234–250. doi: 10.1038/nrd3669. [DOI] [PubMed] [Google Scholar]
- 2.Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- 3.Zaidi M. Skeletal remodeling in health and disease. Nat Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 4.Matsuo K, Irie N. Osteoclast-osteoblast communication. Arch Biochem Biophys. 2008;473:201–209. doi: 10.1016/j.abb.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Karner CM, Long F. Wnt signaling and cellular metabolism in osteoblasts. Cell Mol Life Sci. 2017;74:1649–1657. doi: 10.1007/s00018-016-2425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komori T. Regulation of proliferation, differentiation and Functions of osteoblasts by Runx2. Histochem Cell Biol. 2019;20:1694. doi: 10.3390/ijms20071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdallah BM, Jafari A, Zaher W, Qiu W, Kassem M. Skeletal (stromal) stem cells: An update on intracellular signaling path-ways controlling osteoblast differentiation. Bone. 2015;70:28–36. doi: 10.1016/j.bone.2014.07.028. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa N, Kinosaki M, Yamaguchi K, Shima N, Yasuda H, Yano K, Morinaga T, Higashio K. RANK is the essential signaling receptor for osteoclast differentiation factor in osteoclastogenesis. Biochem Biophys Res Commun. 1998;253:395–400. doi: 10.1006/bbrc.1998.9788. [DOI] [PubMed] [Google Scholar]
- 9.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi Y, Udagawa N, Takahashi N. Action of RANKL and OPG for osteoclastogenesis. Crit Rev Eukaryot Gene Expr. 2009;19:61–72. doi: 10.1615/CritRevEukarGeneExpr.v19.i1.30. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Walter TS, Huang P, Zhang S, Zhu X, Wu Y, Wedderburn LR, Tang P, Owens RJ, Stuart DI, et al. Structural and functional insights of RANKL-RANK interaction and signaling. J Immunol. 2010;184:6910–6919. doi: 10.4049/jimmunol.0904033. [DOI] [PubMed] [Google Scholar]
- 12.Mundy GR. Osteoporosis and inflammation. Nutr Rev. 2007;65:S147–151. doi: 10.1301/nr.2007.dec.S147-S151. [DOI] [PubMed] [Google Scholar]
- 13.Kadono H, Kido J, Kataoka M, Yamauchi N, Nagata T. Inhibition of osteoblastic cell differentiation by lipopolysaccha-ride extract from Porphyromonas gingivalis. Infect Immun. 1999;67:2841–2846. doi: 10.1128/IAI.67.6.2841-2846.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandow K, Maeda A, Kakimoto K, Kusuyama J, Shamoto M, Ohnishi T, Matsuguchi T. Molecular mechanisms of the inhibitory effect of lipopolysaccharide (LPS) on osteoblast differ-entiation. Biochem Biophys Res Commun. 2010;402:755–761. doi: 10.1016/j.bbrc.2010.10.103. [DOI] [PubMed] [Google Scholar]
- 15.Daigang L, Jining Q, Jinlai L, Pengfei W, Chuan S, Liangku H, Ding T, Zhe S, Wei W, Zhong L, Kun Z. LPS-stimulated inflammation inhibits BMP-9-induced osteoblastic differentiation through crosstalk between BMP/MAPK and Smad signaling. Exp Cell Res. 2016;341:54–60. doi: 10.1016/j.yexcr.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Huang RL, Yuan Y, Zou GM, Liu G, Tu J, Li Q. LPS-stimulated inflammatory environment inhibits BMP-2-induced osteoblastic differentiation through crosstalk between TLR4/MyD88/NF-κB and BMP/Smad signaling. Stem Cells Dev. 2014;23:277–289. doi: 10.1089/scd.2013.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Quan J, Long W, Chen H, Wang R, Guo J, Lin X, Mai S. LL-37 inhibits LPS-induced inflammation and stimulates the osteogenic differentiation of BMSCs via P2X7 receptor and MAPK signaling pathway. Exp Cell Res. 2018;372:178–187. doi: 10.1016/j.yexcr.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 18.Yao Z, Yang Z, Chen F, Jiang Y, Fu C, Wang Y, Lu R, Wu H. Autophagy is essential for the endothelial differentiation of breast cancer stemlike cells. Int J Mol Med. 2020;45:255–264. doi: 10.3892/ijmm.2019.4399. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Yang Y, Li X, Zhang H, Gang Y, Bai L. Alterations of autophagy in knee cartilage by treatment with treadmill exercise in a rat osteoarthritis model. Int J Mol Med. 2019;43:336–344. doi: 10.3892/ijmm.2018.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang M, Feng C, Zhang Y, Liu C, Li B, Zhu Q, Huang B, Zhou Y. Autophagy protects nucleus pulposus cells from cyclic mechanical tensioninduced apoptosis. Int J Mol Med. 2019;44:750–758. doi: 10.3892/ijmm.2019.4212. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Wang S, Zhang P, Said-Al-Naief N, Michalek SM, Feng X. Molecular mechanism of the bifunctional role of lipopolysaccharide in osteoclastogenesis. J Biol Chem. 2009;284:12512–12523. doi: 10.1074/jbc.M809789200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orcel P, Feuga M, Bielakoff J, De Vernejoul MC. Local bone injections of LPS and M-CSF increase bone resorption by different pathways in vivo in rats. Am J Physiol. 1993;264:E391–E397. doi: 10.1152/ajpendo.1993.264.3.E391. [DOI] [PubMed] [Google Scholar]
- 23.Chiang CY, Kyritsis G, Graves DT, Amar S. Interleukin-1 and tumor necrosis factor activities partially account for calvarial bone resorption induced by local injection of lipopolysaccharide. Infect Immun. 1999;67:4231–4236. doi: 10.1128/IAI.67.8.4231-4236.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Yang Y, Bao J, Wang Z, Xia M, Dai A, Tan J, Zhou L, Wu Y, Sun W. Autophagy negative-regulating Wnt signaling enhanced inflammatory osteoclastogenesis from Pre-OCs in vitro. Biomed Pharmacother. 2020;126:110093. doi: 10.1016/j.biopha.2020.110093. [DOI] [PubMed] [Google Scholar]
- 25.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 27.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18:962–972. doi: 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7:e2327. doi: 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin C, Jia L, Huang Y, Zheng Y, Du N, Liu Y, Zhou Y. Inhibition of lncRNA MIR31HG promotes osteogenic differentiation of human adipose-derived stem cells. Stem Cells. 2016;34:2707–2720. doi: 10.1002/stem.2439. [DOI] [PubMed] [Google Scholar]
- 30.He S, Yang S, Zhang Y, Li X, Gao D, Zhong Y, Cao L, Ma H, Liu Y, Li G, et al. LncRNA ODIR1 inhibits osteogenic differentiation of hUC-MSCs through the FBXO25/H2BK120ub/H3K4me3/OSX axis. Cell Death Dis. 2019;10:947. doi: 10.1038/s41419-019-2148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Wang XQ, Liu XT, Guo R, Zhang RD. Integrative analysis reveals key mRNAs and lncRNAs in monocytes of osteoporotic patients. Math Biosci Eng. 2019;16:5947–5971. doi: 10.3934/mbe.2019298. [DOI] [PubMed] [Google Scholar]
- 32.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31:4020–4034. doi: 10.1038/emboj.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Hao T, Zhang J, Zhang M, Sun P, Wu L. MicroRNA-592 suppresses the malignant phenotypes of thyroid cancer by regulating lncRNA NEAT1 and downregulating NOVA1. Int J Mol Med. 2019;44:1172–1182. doi: 10.3892/ijmm.2019.4278. [DOI] [PubMed] [Google Scholar]
- 35.Liu R, Tang A, Wang X, Chen X, Zhao L, Xiao Z, Shen S. Inhibition of lncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int J Mol Med. 2018;42:2903–2913. doi: 10.3892/ijmm.2018.3829. [DOI] [PubMed] [Google Scholar]
- 36.Zhang F, Wu L, Qian J, Qu B, Xia S, La T, Wu Y, Ma J, Zeng J, Guo Q, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 37.He F, Zhang C, Huang Q. Long noncoding RNA nuclear enriched abundant transcript 1/miRNA-124 axis correlates with increased disease risk, elevated inflammation, deteriorative disease condition, and predicts decreased survival of sepsis. Medicine (Baltimore) 2019;98:e16470. doi: 10.1097/MD.0000000000016470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Q, Huang C, Luo Y, He F, Zhang R. Circulating lncRNA NEAT1 correlates with increased risk, elevated severity and unfavorable prognosis in sepsis patients. Am J Emerg Med. 2018;36:1659–1663. doi: 10.1016/j.ajem.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Chen DD, Hui LL, Zhang XC, Chang Q. NEAT1 contributes to ox-LDL-induced inflammation and oxidative stress in macro-phages through inhibiting miR-128. J Cell Biochem. 2018 Sep 11; doi: 10.1002/jcb.27541. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 40.Huang-Fu N, Cheng JS, Wang Y, Li ZW, Wang SH. Neat1 regulates oxidized low-density lipoprotein-induced inflammation and lipid uptake in macrophages via paraspeckle formation. Mol Med Rep. 2018;17:3092–3098. doi: 10.3892/mmr.2017.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun. 2019;10:1495. doi: 10.1038/s41467-019-09482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 43.Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, et al. Reactome: A knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Claude-Taupin A, Bissa B, Jia J, Gu Y, Deretic V. Role of autophagy in IL-1β export and release from cells. Seminars in cell & developmental biology. 2018;83:36–41. doi: 10.1016/j.semcdb.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krakauer T. Inflammasomes, autophagy, and cell death: The trinity of innate host defense against intracellular bacteria. Mediators Inflamm. 2019;2019:2471215. doi: 10.1155/2019/2471215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao C, Chen J, Fan F, Long Y, Tang S, Jiang C, Wang J, Xu Y, Xu Y. RIPK2-mediated autophagy and negatively regulated ROS-NLRP3 inflammasome signaling in GMCs stimulated with high glucose. Mediators Inflamm. 2019;2019:6207563. doi: 10.1155/2019/6207563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S, Shuch BM, Gerstein MB. Whole-genome analysis of papillary kidney cancer finds significant noncoding alterations. PLoS Genetics. 2017;13:e1006685. doi: 10.1371/journal.pgen.1006685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ning L, Li Z, Wei D, Chen H, Yang C. LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial-mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomark. 2017;19:75–83. doi: 10.3233/CBM-160376. [DOI] [PubMed] [Google Scholar]
- 53.Taiana E, Favasuli V, Ronchetti D, Todoerti K, Pelizzoni F, Manzoni M, Barbieri M, Fabris S, Silvestris I, Gallo Cantafio ME, et al. Long non-coding RNA NEAT1 targeting impairs the DNA repair machinery and triggers anti-tumor activity in multiple myeloma. Leukemia. 2020;34:234–244. doi: 10.1038/s41375-019-0542-5. [DOI] [PubMed] [Google Scholar]
- 54.Zeng C, Liu S, Lu S, Yu X, Lai J, Wu Y, Chen S, Wang L, Yu Z, Luo G, Li Y. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol Cancer. 2018;17:130. doi: 10.1186/s12943-018-0884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, Chen S, Li Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22:861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 57.Mello SS, Sinow C, Raj N, Attardi LD. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017;31:1095–1108. doi: 10.1101/gad.284661.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16:104. doi: 10.1186/s12943-017-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng X, Zhang Y, Liu Y, Fang L, Li L, Sun J, Pan Z, Xin W, Huang P. HIF-2α activated lncRNA NEAT1 promotes hepa-tocellular carcinoma cell invasion and metastasis by affecting the epithelial-mesenchymal transition. J Cell Biochem. 2018;119:3247–3256. doi: 10.1002/jcb.26481. [DOI] [PubMed] [Google Scholar]
- 60.Zhong J, Jiang L, Huang Z, Zhang H, Cheng C, Liu H, He J, Wu J, Darwazeh R, Wu Y, Sun X. The long non-coding RNA Neat1 is an important mediator of the therapeutic effect of bexarotene on traumatic brain injury in mice. Brain Behav Immun. 2017;65:183–194. doi: 10.1016/j.bbi.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Barutcu AR, Hong D, Lajoie BR, McCord RP, van Wijnen AJ, Lian JB, Stein JL, Dekker J, Imbalzano AN, Stein GS. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim Biophys Acta. 2016;1859:1389–1397. doi: 10.1016/j.bbagrm.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lo PK, Zhang Y, Wolfson B, Gernapudi R, Yao Y, Duru N, Zhou Q. Dysregulation of the BRCA1/long non-coding RNA NEAT1 signaling axis contributes to breast tumorigenesis. Oncotarget. 2016;7:65067–65089. doi: 10.18632/oncotarget.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henning P, Conaway HH, Lerner UH. Retinoid receptors in bone and their role in bone remodeling. Front Endocrinol (Lausanne) 2015;6:31. doi: 10.3389/fendo.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menéndez-Gutiérrez MP, Ricote M. The multi-faceted role of retinoid X receptor in bone remodeling. Cell Mol Life Sci. 2017;74:2135–2149. doi: 10.1007/s00018-017-2458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Menéndez-Gutiérrez MP, Rőszer T, Fuentes L, Núñez V, Escolano A, Redondo JM, De Clerck N, Metzger D, Valledor AF, Ricote M. Retinoid X receptors orchestrate osteoclast differentiation and postnatal bone remodeling. J Clin Invest. 2015;125:809–823. doi: 10.1172/JCI77186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nowak B, Matuszewska A, Filipiak J, Nikodem A, Merwid-Ląd A, Pieśniewska M, Fereniec-Gołębiewska L, Kwiatkowska J, Szeląg A. The influence of bexarotene, a selective agonist of the retinoid receptor X (RXR), and tazarotene, a selective agonist of the retinoid acid receptor (RAR), on bone metabolism in rats. Adv Med Sci. 2016;61:85–89. doi: 10.1016/j.advms.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Moser AH, Shigenaga JK, Grunfeld C, Feingold KR. Downregulation of liver X receptor-alpha in mouse kidney and HK-2 proximal tubular cells by LPS and cytokines. J Lipid Res. 2005;46:2377–2387. doi: 10.1194/jlr.M500134-JLR200. [DOI] [PubMed] [Google Scholar]
- 68.Huang Y, Wang H, Hao Y, Lin H, Dong M, Ye J, Song L, Wang Y, Li Q, Shan B, et al. Myeloid PTEN promotes chemotherapy-induced NLRP3-inflammasome activation and antitumour immunity. Nat Cell Biol. 2020;22:716–727. doi: 10.1038/s41556-020-0510-3. [DOI] [PubMed] [Google Scholar]
- 69.He H, Jiang H, Chen Y, Ye J, Wang A, Wang C, Liu Q, Liang G, Deng X, Jiang W, Zhou R. Oridonin is a covalent NLRP3 inhibitor with strong anti-inflammasome activity. Nat Commun. 2018;9:2550. doi: 10.1038/s41467-018-04947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao X, Zhang C, Xing Y, Xue G, Zhang Q, Pan F, Wu G, Hu Y, Guo Q, Lu A, et al. Remodelling of the gut microbiota by hyperactive NLRP3 induces regulatory T cells to maintain homeostasis. Nat Commun. 2017;8:1896. doi: 10.1038/s41467-017-01917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Jiang W, Yan Y, Gong T, Han J, Tian Z, Zhou R. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat Immunol. 2014;15:1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- 72.Hoss F, Rodriguez-Alcazar JF, Latz E. Assembly and regulation of ASC specks. Cell Mol Life Sci. 2017;74:1211–1229. doi: 10.1007/s00018-016-2396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Afonina IS, Zhong Z, Karin M, Beyaert R. Limiting inflammation-the negative regulation of NF-kappaB and the NLRP3 inflammasome. Nat Immunol. 2017;18:861–869. doi: 10.1038/ni.3772. [DOI] [PubMed] [Google Scholar]
- 74.Kong Y, Huang T, Zhang H, Zhang Q, Ren J, Guo X, Fan H, Liu L. The lncRNA NEAT1/miR-29b/Atg9a axis regulates IGFBPrP1-induced autophagy and activation of mouse hepatic stellate cells. Life Sci. 2019;237:116902. doi: 10.1016/j.lfs.2019.116902. [DOI] [PubMed] [Google Scholar]
- 75.Liu F, Ai FY, Zhang DC, Tian L, Yang ZY, Liu SJ. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2020;9:1079–1091. doi: 10.1002/cam4.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao Z, Wang Y, Long Z, He G. Interaction between autophagy and the NLRP3 inflammasome. Acta Biochim Biophys Sin (Shanghai) 2019;51:1087–1095. doi: 10.1093/abbs/gmz098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.