Abstract
Previous studies have reported that long non-coding (lnc) RNA FGD5-antisense 1 (FGD5-AS1) promotes tumor proliferation, migration and invasion. Therefore, the present study aimed to elucidate the biological role and underlying molecular mechanisms of FGD5-AS1 in cisplatin (DDP) resistance of lung adenocarcinoma (LAD) cells. The results demonstrated that FGD5-AS1 was highly expressed in DDP-resistant LAD tissues and cells. Knockdown of FGD5-AS1 decreased the proliferative, migratory and invasive abilities of DDP-resistant LAD cells. Moreover, it was identified that FGD5-AS1 acted as a molecular sponge for microRNA (miR)-142, and FGD5-AS1 enhanced the resistance of A549/DDP cells to DDP by directly interacting with miR-142. Programmed cell death 1 ligand 1 (PD-L1) was also found to be a key effector of the FGD5-AS1/miR-142 axis to regulate the chemoresistance of DDP-resistant LAD cells. In conclusion, the present study demonstrated that FGD5-AS1 increased DDP resistance of LAD via the miR-142/PD-L1 axis, which may offer a novel treatment strategy for patients with DDP-resistant LAD.
Keywords: FGD5-antisense 1, microRNA-142, programmed cell death 1 ligand 1, cisplatin, lung adenocarcinoma
Introduction
Lung cancer is one of the most common malignant tumor types worldwide, and non-small cell lung cancer (NSCLC) accounts for ~80% of all lung cancer types (1). NSCLC includes adenocarcinoma and squamous cell carcinoma, among which lung adenocarcinoma (LAD) is the most common pathological type (2). In addition to surgical treatment, chemotherapy is an effective method to improve the survival rate of patients with LAD (3). However, chemotherapeutic resistance is a major barrier to chemotherapy failure. For instance, drug-resistant tumor cells continue to metastasize to the distal end, can proliferate quickly and have a strong invasive ability. Therefore, it is important to understand the molecular mechanism of cisplatin (DDP) resistance in LAD and to identify novel targets to prevent the occurrence of DDP resistance.
Previous studies have reported that long non-coding RNA (lncRNA) could be involved in the development of tumors. For instance, Yang et al (4) revealed that lncRNA MIR4435-2HG was upregulated and promoted the tumorigenesis of NSCLC, while Zhao et al (5) observed that GMDS-AS1 inhibited the proliferation of LAD cells and enhanced cell apoptosis. Moreover, the underlying mechanism of lncRNAs in multiple tumor chemoresistance is a hot topic of research. It has been demonstrated that some lncRNAs promote DDP-resistance in multiple cancer types. For example, Hu et al (6) discovered that colon cancer associated transcript 1 (CCAT1) enhanced DDP resistance in esophageal cancer via the microRNA (miRNA/miR)-142/PLK1/BUBR axis. Furthermore, Yan et al (7) reported that nuclear enriched abundant transcription 1 decreased the sensitivity of anaplastic thyroid carcinoma cells to DDP via the miR-9/c-Jun-amino-terminal kinase-inter-acting protein 4 axis. Therefore, understanding the molecular mechanism of lncRNA in DDP-resistant LAD could help identify novel therapeutic targets. lncRNA FGD5-antisense 1 (FGD5-AS1) is considered as a potential target in the treatment of various types of cancer, including colorectal cancer, periodontitis and kidney carcinoma (8-10). However, the role of FGD5-AS1 in DDP-resistant LAD remains unknown.
miRNAs have biological characteristics and targeted specificity, and have the potential to be a marker for tumor treatment and prognosis (11,12). Liang et al (13) revealed that miR-485-5p suppressed papillary thyroid cancer development by modulating Raf1. Moreover, Fan et al (14) suggested that miR-1281 expression was significantly downregulated in breast cancer tissues, which was associated with the pathological stage and poor prognosis of patients with breast cancer. These characteristics of miRNA provide novel strategies for the treatment of various malignant tumors.
The present study aimed to elucidate the biological role and underlying molecular mechanisms of FGD5-AS1 in DDP-resistant LAD cells. It was demonstrated that FGD5-AS1 enhanced the resistance of LAD cells to DDP via the miR-142/programmed cell death 1 ligand 1 (PD-L1) axis, and thus may provide potential therapeutic strategies for LAD treatment.
Materials and methods
Tumor tissues
A total of 46 LAD tissues, including 25 DDP-resistant LAD tissues and 21 DDP-sensitive LAD tissues, were obtained from 46 patients with LAD with a mean age of 47 years (age range, 33-64 years; 31 male patients and 15 female patients) between April 2015 and August 2017 at The Third Affiliated Hospital of Soochow University (Changzhou, China). All patients agreed to cooperate with the present research and provided signed informed consent. The research was approved by the Ethics Committee of The Third Affiliated Hospital of Soochow University. Fresh tissues were immediately frozen in liquid nitrogen and stored at -80°C.
Cell culture
Human LAD cell lines (A549 and HCC827) were purchased from the American Type Culture Collection. All cell lines were cultured in RPMI0-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) and supplemented with 10% FBS (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in a humidified chamber with 5% CO2. To establish DDP-resistant LAD cells (A549/DDP and HCC827/DDP), A549 and HCC827 cells were incubated at room temperature with increasing concentrations of DDP (0.5-10 µg/ml; Sigma Aldrich; Merck KGaA) for >6 months.
Cell transfection
Short hairpin (sh)-RNA targeting FGD5-AS1 (sh-FGD5-AS1; 5′-GCA AUG AUG CGC CAC UAG AUU G-3′) or PD-L1 (sh-PD-L1; 5′-AGC AAA CUG CAC GCC CAG CUG C-3′), miR-142 mimics (5′-CAU AAA GUA GAA AGC ACU ACU-3′), miR-142 inhibitor (5′-AGT AGT GCT CCT TTC TAC TTT ATG-3′) and their negative control (NC mimic, 5′-GGU UCG UAC GUA CAC UGU UCA-3′; NC inhibitor, 5′-CCA UCA GUC CCA AAU CCA-3′) group were purchased from Shanghai GenePharma Co., Ltd.. To overexpress FGD5-AS1, FGD5-AS1 was subcloned into pcDNA3.1 by Shanghai GenePharma Co., Ltd., and pcDNA3.1 served as the control. A549 and HCC827 cells (1×106) were transfected with sh-FGD5-AS1 (35 nM), sh-PD-L1 (35 nM), sh-NC (35 nM), miR-142 mimics (35 nM), miR-142 inhibitor (35 nM), NC mimic (35 nM), NC inhibitor (35 nM), pcDNA4.1 (15 nM) or FGD5-AS1 (15 nM) using Lipofectamine® 2000 reagent (Invitrogen, Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The subsequent experiments were performed 48 h after transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from LAD tissues and cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA was reverse-transcribed to cDNA using the PrimeScript™ RT Reagent Kit (Takara Biotechnology Co., Ltd.) at 37°C for 15 min. RT-qPCR was conducted using SYBR-Green PCR Master Mix kit (Takara Biotechnology Co., Ltd.) with the following thermocycling conditions: Initial denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 20 sec, and subsequently the final extension at 72°C for 5 min. GAPDH or U6 was used as an internal control. The expression levels of genes were calculated using the 2−ΔΔCq method (15). The primer sequences were as follows: FGD5-AS1 forward, 5′-AGA AGC GGA GGG GTG AAA AT-3′ and reverse, 5′-CCG CCT TAT AGT TGG CCC TC-3′; miR-142-5p forward, 5′-GGC CCA TAA AGT AGA AAG C-3′ and reverse, 5′-TTT GGC ACT AGC ACA TT-3′; PD-L1 forward, 5′-TAG AAT TCA TGA GGA TAT TTG CTG TCT T-3′ and reverse, 5′-TAG GAT CCT TAC GTC TCC TCC AAA TGT G-3′; GAPDH forward, 5′-AAC GGA TTT GGT CGT ATT G-3′ and reverse, 5′-GGA AGA TGG TGA TGG GAT T-3′; and U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′ and reverse, 5′-AAC GCT TCA CGA ATT TGC GT-3′.
Cell Counting Kit (CCK)-8 assay
To determine the IC50 value, the cells (1×104 cells/well) were seeded into 96-well plates at 37°C and 5% CO2. The cells were treated with (0, 2, 4, 6, 8 and 10 µg/ml) DDP for 48 h in the medium with 10% FBS. For the detection of cell viability, the transfected A549/DDP and HCC827/DDP cells were treated with 4 µg/ml DDP for 48 h. Then, cells were incubated with 10 µl CCK-8 reagent (Beyotime Institute of Biotechnology) for another 2 h. The absorbance at 450 nm wavelength was observed on a micro-plate reader (Thermo Fisher Scientific, Inc.). The IC50 value was calculated using the relative survival curve.
RNA immunoprecipitation (RIP) assay
A RIP assay was performed using the EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (cat. no. 17-701; EMD Millipore). Cell lysate was prepared from 1.5×107 cells and the cell lysate was centrifuged at 513 × g for 15 min at 4°C. The supernatant was incubated in 100 µl RIP buffer containing magnetic beads conjugated with the Ago2 antibody (cat. no. ab32381; 1:1,000; Abcam) or IgG antibody (cat. no. ab150077; 1:1,000; Abcam). Subsequently, the enrichment of FGD5-AS1 and miR-142 was determined via RT-qPCR.
Transwell assay
The migratory and invasive abilities were assessed using Transwell chambers (8.0 µm pore size; EMD Millipore) and Matrigel (Corning, Inc.). For the migration assay, transfected cells (1×105) were placed in the upper chamber containing 200 µl RPMI-1640 medium, and 600 µl RPMI-1640 medium containing 10% FBS was added in the lower chamber. The cells were cultured for 48 h, and the cells in the lower chamber were fixed with methanol and stained with crystal violet (Sigma-Aldrich; Merck KGaA) both for 20 min at room temperature. For the invasion assay, the insert membranes were coated with Matrigel for 1 h at room temperature and then cultured under the same conditions as aforementioned. Migrated and invaded cells were counted under a light microscope (magnification, ×200) and imaged.
Luciferase reporter assay
StarBase version 2.0 (http://star-base.sysu.edu.cn) was used to predict the binding sites between FGD5-AS1 and miR-142, and TargetScan 7.2 (http://www.targetscan.org) was used to predict the binding sites between miR-142 and 3′-untranslated region (UTR) of PD-L1. A luciferase reporter assay was performed to verify the binding ability between RNAs. The wild-type (WT) and mutant-type (Mut) of the FGD5-AS1 sequences (or PD-L1 3′-UTR) were cloned into firefly luciferase gene reporter vectors pmirGLO (Promega Corporation). These constructed vectors were co-transfected with NC mimics or miR-142 mimics into LAD cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the relative luciferase activity was determined using the Dual-Luciferase® Reporter Assay System (Promega Corporation) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase (Promega Corporation) gene activity.
Western blot analysis
Proteins were extracted from transfected LAD cells using RIPA buffer (Thermo Fisher Scientific, Inc.). Protein concentration was measured using the bicinchoninic acid assay (Beyotime Institute of Biotechnology). A total of 10 µg protein/lane were separated using 10% SDS-PAGE (EMD Millipore), and then transferred to PVDF membranes (Bio-Rad Laboratories, Inc.). After blocking with 5% skimmed milk for 2 h at room temperature, membranes were probed with primary antibodies against PD-L1 (1:1,000; cat. no. ab205921; Abcam) and anti-GAPDH (1:1,000; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.) overnight at 4°C. Subsequently, membranes were incubated with horse-radish peroxidase-conjugated goat anti-mouse IgG (cat. no. ab205719) and goat anti-rabbit IgG (cat. no. ab205718) secondary antibodies (1:1,000; Abcam) at room temperature for 2 h. The protein bands were visualized using an enhanced chemiluminescence kit (Bio-Rad Laboratories, Inc.) and semi-quantified by densitometric analysis of protein signals using ImageJ (version 1.49; National Institutes of Health). GAPDH served as the loading control.
Statistical analysis
SPSS 22.0 (IBM Corp.) was used for statistical analysis and each experiment was repeated ≥3 times. Comparisons of parameters between two groups were analyzed using a paired Student's t-test. Comparisons among multiple groups were performed using one-way ANOVA followed by Tukey's test. Pearson's correlation analysis was used for analyzing the correlation between FGD5-AS1 and miR-142. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA FGD5-AS1 is upregulated in DDP-resistant LAD tissues and cells
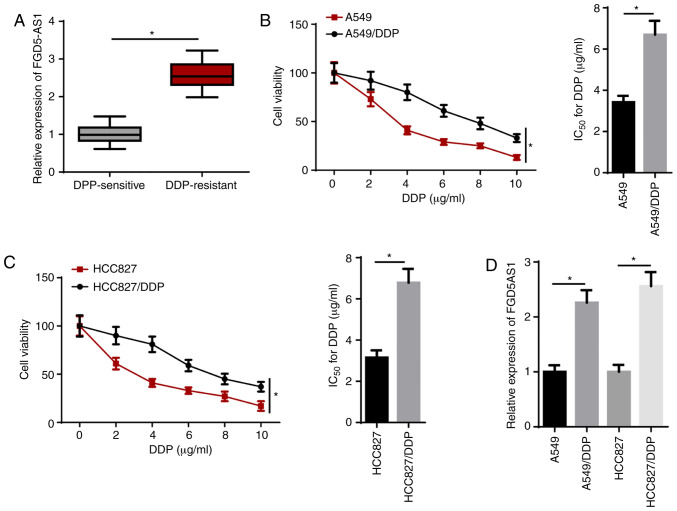
RT-qPCR results demonstrated that FGD5-AS1 expression was significantly upregulated in DDP-resistant LAD tissues compared with that in DDP-sensitive LAD tissues (Fig. 1A). To investigate whether DDP-resistant LAD cells were successfully established, the IC50 of DDP was measured using a CCK-8 assay. The results indicated that the IC50 of DDP in DDP-resistant LAD (A549/DDP and HCC827/DDP) cells was significantly enhanced compared with their parental cells (Fig. 1B and C). In addition, RT-qPCR identified that FGD5-AS1 expression was significantly upregulated in A549/DDP and HCC827/DDP cells compared with their counterparts (Fig. 1D). These results suggested that FGD5-AS1 was highly expressed in DDP-resistant LAD tissues and cells.
Figure 1.
lncRNA FGD5-AS1 is upregulated in DDP-resistant LAD tissues and cells. (A) RT-qPCR showed the relative FGD5-AS1 expression in DDP-sensitive (n=21) and DDP-resistant (n=25) LAD tissues. (B and C) Cell Counting Kit-8 assay showed the viability and IC50 of LAD or DDP-resistant LAD cells. (D) RT-qPCR showed the relative FGD5-AS1 expression in A549, A549/DDP, HCC827 and HCC827/DDP cells. The data are presented as the mean ± SD. *P<0.05. lncRNA, long non-coding RNA; FGD5-AS1, FGD5-antisense 1; DDP, cisplatin; LAD, lung adenocarcinoma; RT-qPCR, reverse transcription-quantitative PCR.
FGD5-AS1 knockdown decreases DDP resistance in DDP-resistant LAD cells
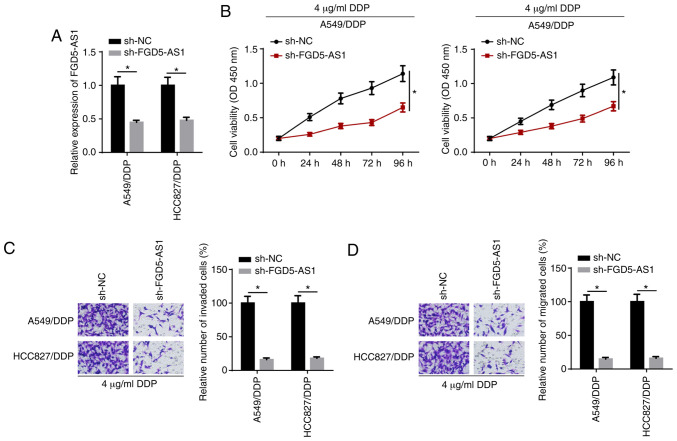
To evaluate the role of FGD5-AS1 in DDP-resistant LAD cells, sh-FGD5-AS1 was transfected into HCC827/DDP and A549/DDP cells. RT-qPCR confirmed that the expression of FGD5-AS1 decreased in the FGD5-AS1 knockdown group compared with the control group (Fig. 2A). CCK-8 assay results demonstrated that cell viability was suppressed after the knockdown of FGD5-AS1 in DDP-resistant LAD cells treated with 4 µg/ml DDP (Fig. 2B). Moreover, FGD5-AS1 knockdown decreased the invasion and migration of HCC827/DDP and A549/DDP cells treated with 4 µg/ml DDP (Fig. 2C and D). Thus, the results indicated that FGD5-AS1 knockdown enhanced the DDP sensitivity of DDP-resistant LAD cells.
Figure 2.
FGD5-AS1 knockdown decreases DDP resistance in DDP-resistant LAD cells. (A) Reverse transcription-quantitative PCR showed the relative FGD5-AS1 expression in A549/DDP and HCC827/DDP cells transfected with sh-NC and sh-FGD5-AS1. (B) Cell viability was determined by a Cell Counting Kit-8 assay in DDP-resistant LAD cells treated with 4 µg/ml DDP. (C) Cell invasion and (D) migration were evaluated by a Transwell assay (magnification, ×200). The data are presented as the mean ± SD. *P<0.05. FGD5-AS1, FGD5-antisense 1; DDP, cisplatin; LAD, lung adenocarcinoma; sh-, short hairpin RNA; NC, negative control.
FGD5-AS1 interacts with miR-142 in LAD cells
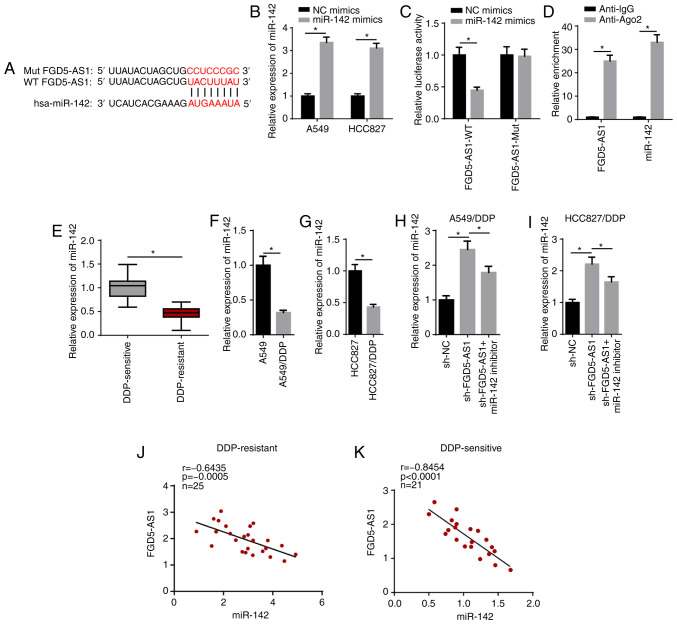
lncRNAs have been reported to promote cancer progression by acting as competing endogenous RNA (ceRNA). Using StarBase, miR-142 was predicted as a candidate target of FGD5-AS1 (Fig. 3A). RT-qPCR showed that miR-142 expression was significantly increased in A549 and HCC827 cells transfected with miR-142 mimics (Fig. 3B). Subsequently, the luciferase reporter assay results identified that overexpression of miR-142 significantly weakened the luciferase activity of the FGD5-AS1-WT group, while it had no effect on the FGD5-AS1-Mut reporter group (Fig. 3C). The RIP assay revealed that miR-142 and FGD5-AS1 were significantly enriched with the anti-ago2 antibody compared with anti-IgG, which further indicated that FGD5-AS1 may be associated with miR-142 (Fig. 3D).
Figure 3.
FGD5-AS1 interacts with miR-142 in LAD cells. (A) Bioinformatics prediction of the binding site of miR-142 on FGD5-AS1. (B) RT-qPCR showed the expression of miR-142 in A549 and HCC827 cells transfected with NC mimics and miR-142 mimics. The interaction between miR-142 and FGD5-AS1 was verified by (C) luciferase reporter and (D) RNA immunoprecipitation assays. (E) RT-qPCR showed the relative miR-142 expression in DDP-sensitive (n=21) and DDP-resistant (n=25) LAD tissues. RT-qPCR showed the relative miR-142 expression in (F) A549 and A549/DDP cells, and (G) HCC827 and HCC827/DDP cells. RT-qPCR showed that FGD5-AS1 knockdown upregulated miR-142 expression in (H) A549/DDP and (I) HCC827/DDP cells, whereas miR-142 reversed this effect. (J and K) The correlation between FGD5-AS1 and miR-142 in DDP-sensitive (n=21) and DDP-resistant (n=25) LAD tissues was assessed by Pearson's correlation analysis. The data are presented as the mean ± SD. *P<0.05. FGD5-AS1, FGD5-antisense 1; DDP, cisplatin; LAD, lung adenocarcinoma; NC, negative control; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; WT, wild-type; Mut, mutant; sh-, short hairpin RNA.
The RT-qPCR results identified that miR-142 expression was significantly upregulated in DDP-sensitive tissues compared with that in DDP-resistant LAD tissues (Fig. 3E). Moreover, RT-qPCR indicated a decrease in miR-142 expression in A549/DDP and HCC827/DDP cells (Fig. 3F and G). It was found that transfection with the miR-142 inhibitor abolished the promotive effect of sh-FGD5-AS1 on miR-142 expression in DDP-resistant LAD cells (Fig. 3H and I). In addition, miR-142 expression was negatively correlated with FGD5-AS1 expression in DDP-resistant (Fig. 3J) and DDP-sensitive LAD tissues (Fig. 3K). Taken together, the results suggested that FGD5-AS1 functioned as a sponge for miR-142, and FGD5-AS1 inhibited miR-142 expression in DDP-resistant LAD cells.
FGD5-AS1 enhances DDP resistance via miR-142 in DDP-resistant LAD cells
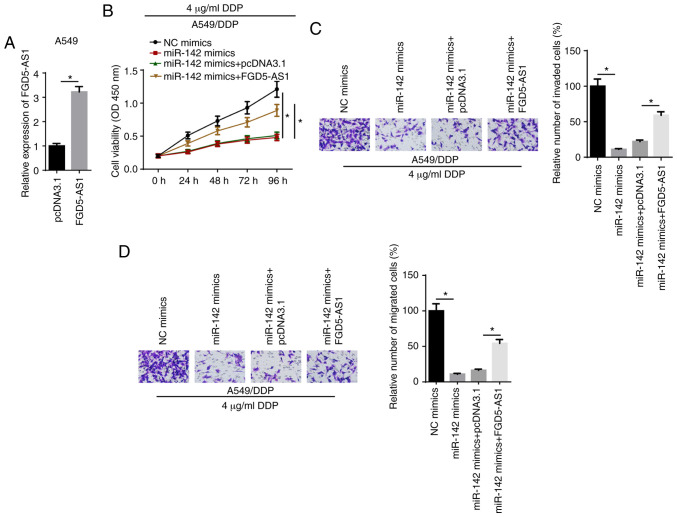
Rescue experiments were performed by transfecting A549/DDP cells with NC mimics, miR-142 mimics, miR-142 mimics + pcDNA3.1 or miR-142 mimics + FGD5-AS1. RT-qPCR showed that FGD-AS1 expression was upregulated in A549 cells transfected with the FDG5-AS1 overexpression plasmid (Fig. 4A). CCK-8 results identified that miR-142 mimics decreased the viability of A549/DDP cells treated with 4 µg/ml DDP compared with the NC mimics group, which was reversed by FGD5-AS1 overexpression (Fig. 4B). Moreover, Transwell assay results indicated that miR-142-attenuated invasion and migration of A549/DDP cells treated with 4 µg/ml DDP were reversed by the overexpression of FGD5-AS1 (Fig. 4C and D). These data demonstrated that FGD5-AS1 increased DDP resistance by modulating miR-142.
Figure 4.
FGD5-AS1 enhances DDP resistance via miR-142 in DDP-resistant lung adenocarcinoma cells. (A) Reverse transcription-quantitative PCR showed the expression of FGD5-AS1 in A549 cells transfected with FGD5-AS1 overexpression plasmid. (B) A Cell Counting Kit-8 assay showed the viability of A549/DDP cells transfected with NC mimics, miR-142 mimics, miR-142 mimics + pcDNA3.1 and miR-142 mimics + FGD5-AS1. Transwell assays showed the (C) invasion and (D) migration of A549/DDP cells treated with DDP in different groups (magnification, ×200). The data are presented as the mean ± SD. *P<0.05. FGD5-AS1, FGD5-antisense 1; DDP, cisplatin; NC, negative control; miR, microRNA.
PD-L1 is directly targeted by miR-142
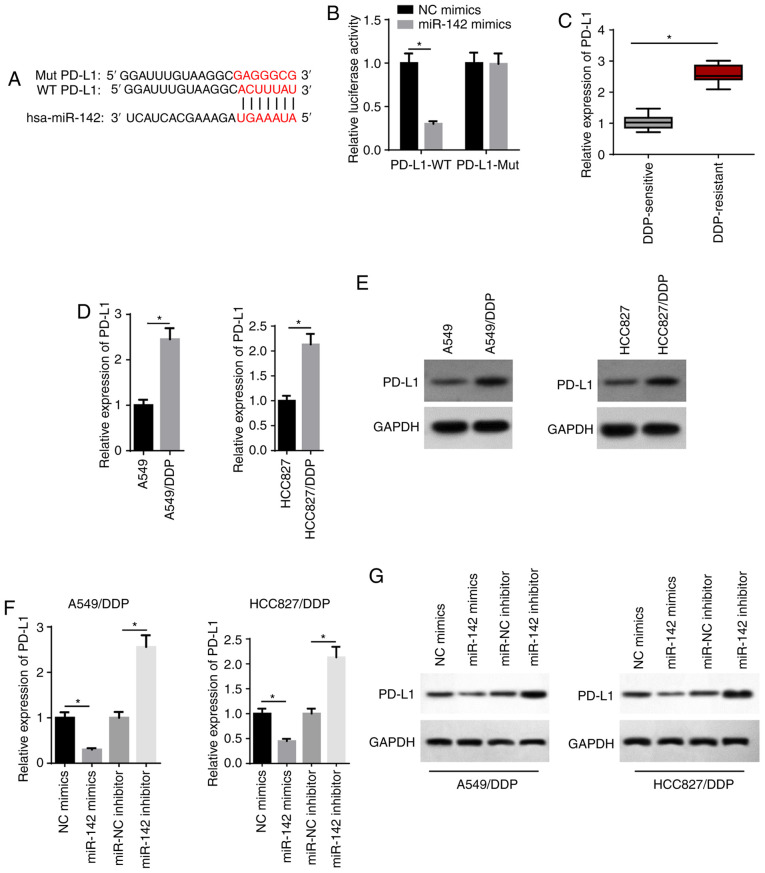
TargetScan analysis was performed and it was found that miR-142 possessed a binding site for PD-L1 (Fig. 5A). miR-142 mimics significantly suppressed the relative luciferase activity of the PD-L1-WT group, but had no effect on the PD-L1-Mut group (Fig. 5B). In addition, RT-qPCR results identified that PD-L1 expression was significantly increased in DDP-resistant LAD tissues (Fig. 5C). RT-qPCR and western blot assays also revealed that the mRNA and protein expression levels of PD-L1 were significantly increased in A549/DDP and HCC827/DDP cells (Fig. 5D and E). It was found that miR-142 mimics decreased PD-L1 expression, whereas transfection with the miR-142 inhibitor increased PD-L1 expression (Fig. 5F and G). Thus, miR-142 may exert its biological function via PD-L1.
Figure 5.
PD-L1 is directly targeted by miR-142. (A) Bioinformatics prediction of the binding site of miR-142 on PD-L1. (B) The interaction between miR-142 and PD-L1 was verified by a luciferase reporter assay. (C) RT-qPCR showed the relative PD-L1 expression in DDP-sensitive (n=21) and DDP-resistant (n=25) lung adenocarcinoma tissues. (D) RT-qPCR and (E) western blotting showed the relative PD-L1 expression in A549, A549/DDP, HCC827 and HCC827/DDP cells. (F) RT-qPCR and (G) western blotting showed the relative PD-L1 expression in A549/DDP and HCC827/DDP cells transfected with NC mimics, miR-142 mimics, miR-NC inhibitor and miR-142 inhibitor. The data are presented as the mean ± SD. *P<0.05. PD-L1, programmed cell death 1 ligand 1; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; DDP, cisplatin; NC, negative control; WT, wild-type; Mut, mutant.
FGD5-AS1 decreases the chemosensitivity of DDP-resistant LAD cells via the miR-142/PD-L1 axis
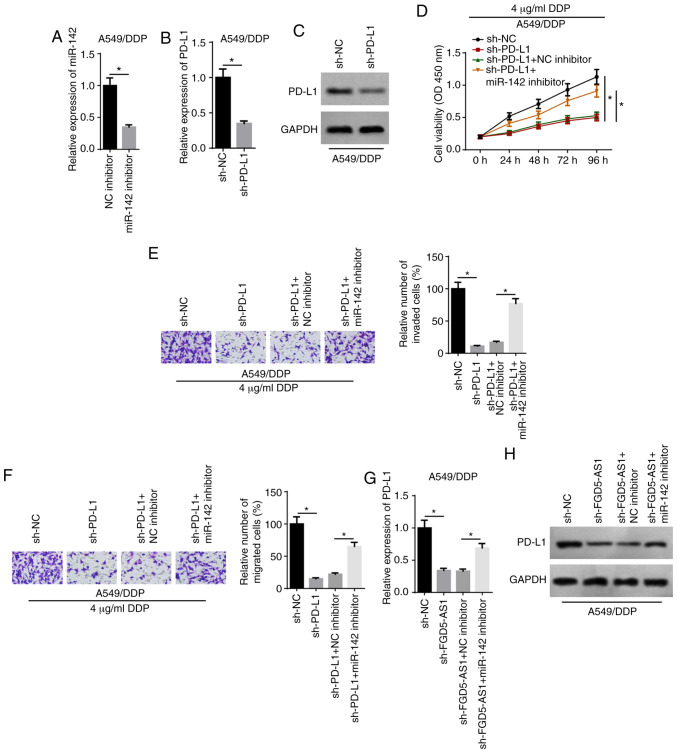
RT-qPCR analysis demonstrated that the expression of miR-142 was decreased in A549/DDP cells following transfection with the miR-142 inhibitor (Fig. 6A). Moreover, RT-qPCR and western blot assays indicated that the expression of PD-L1 was downregulated in A549/DDP cells after PD-L1 knockdown (Fig. 6B and C). PD-L1 knockdown significantly inhibited the viability of A549/DDP cells treated with 4 µg/ml DDP compared with the sh-NC group, whereas transfection with the miR-142 inhibitor reversed the effect (Fig. 6D). Moreover, rescue experiments suggested that the miR-142 inhibitor abolished the inhibitory effect of PD-L1 knockdown on the invasion and migration of A549/DDP cells treated with 4 µg/ml DDP (Fig. 6E and F). It was also observed that FGD5-AS1 knockdown downregulated the expression of PD-L1, whereas the miR-142 inhibitor partially reversed this expression (Fig. 6G and H). Collectively, these data supported the hypothesis that FGD5-AS1 promoted LAD cell viability, invasion and migration, and DDP resistance via the miR-142/PD-L1 axis.
Figure 6.
FGD5-AS1 decreases the chemosensitivity of DDP-resistant lung adenocarcinoma cells via the miR-142/PD-L1 axis. (A) RT-qPCR showed the relative miR-142 expression in A549/DDP cells transfected with NC inhibitor and miR-142 inhibitor. (B) RT-qPCR and (C) western blotting showed the relative PD-L1 expression in A549/DDP cells transfected with sh-NC and sh-PD-L1. (D) Cell Counting Kit-8 assay showed the viability of DDP-treated A549/DDP cells transfected with sh-NC, sh-PD-L1, sh-PD-L1 + NC inhibitor, sh-PD-L1 + miR-142 inhibitor. Transwell assays showed the (E) invasion and (F) migration of DDP-treated A549/DDP cells in different transfection groups (magnification, ×200). (G and H) RT-qPCR and western blotting showed PD-L1 expression in A549/DDP cells transfected with sh-NC, sh-FGD5-AS1, shFGD5-AS1 + NC inhibitor, shFGD5-AS1 + miR-142 inhibitor. The data are presented as the mean ± SD. *P<0.05. FGD5-AS1, FGD5-antisense 1; DDP, cisplatin; PD-L1, programmed cell death 1 ligand 1; miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; sh-, short hairpin RNA.
Discussion
LAD is a malignant tumor type that is a threat to human health worldwide (16). In addition to surgical treatment, DDP-based chemotherapy is the primary therapeutic strategy for LAD cancer therapy. As a result of DDP resistance in tumor cells, chemotherapy suffers from the bottleneck phenomenon, whereby tumor cell populations decrease for a brief period of time but then quickly repopulate. It was previously reported that lncRNA serves a vital regulatory role in the chemo-resistance of various cancer types (17). The present study demonstrated that lncRNA FGD5-AS1 was highly expressed in DDP-resistant LAD cells compared with parental cells, and FGD5-AS1 knockdown decreased the DDP resistance of A549/DDP and HCC827/DDP cells.
A ceRNA network exerts its regulatory function in human cancer, including LAD (18,19). For example, Wang et al (20) revealed that CCAT1 knockdown promoted chemical sensitivity in DDP-induced ovarian cancer cells by sponging downstream miR-454, while Qu et al (21) observed that LINC00461 promoted DDP resistance of rectal cancer by targeting miR-593. Similarly, Wu et al (22) found that lncRNA MIAT modulated proliferation and promoted DDP resistance in NSCLC cells by targeting miR-184. The present study indicated that FGD5-AS1 could function as a ceRNA to inhibit miR-142 expression.
miR-142 has been identified to exert an inhibitory effect in several cancer types, including squamous cell carcinoma, gastric cancer and ovarian cancer (23-25). In addition, it has been demonstrated that miR-142 inhibits chemoresistance of various tumors, such as ovarian cancer and osteosarcoma (26,27). The present results suggested that miR-142 was downregulated in LAD, and the overexpression of miR-142 contributed to chemosensitivity in DDP-resistant LAD cells.
PD-L1 may promote tumor cell proliferation and differentiation by creating an imbalance between immune cells and the cellular environment (28,29). PD-L1 is also reported to be involved in the regulation of tumor development. For example, PD-L1 is upregulated in human osteosarcoma tissues and increases cell invasion (30). Moreover, high expression of PD-L1 affects the survival and prognosis of patients with ovarian cancer (31). Zuo et al (32) reported that PD-L1 silencing suppressed chemoresistance of DDP-resistant ovarian cancer cells to DDP, as evidenced by inhibited proliferation, G1-phase cell cycle arrest and promotion of apoptosis. To the best of our knowledge, the present study provided the first evidence that PD-L1 could directly interact with miR-142 via a specific binding site, and that PD-L1 knockdown suppressed tumor progression and enhanced chemosensitivity of DDP-resistant LAD cells.
However, some limitations remain to be addressed in future studies. Other miRNAs or downstream effectors that are crucial to FGD5-AS1-regulated DDP resistance of LAD cells need to be identified in the future. In addition, the expression of FGD5-AS1 was detected using limited sample sizes. Therefore, further studies are required to confirm the expression of FGD5-AS1 in a large sample size and to further assess the clinical significance.
In conclusion, the present results suggested that the FGD5-AS1/miR-142/PD-L1 axis contributed to DDP resistance in LAD. These findings provided a novel insight into the issue of DDP resistance.
Acknowledgments
Not applicable.
Funding
This study was funded by the Scientific Research Project of Wuxi Municipal Health Commission (grant no. M202009).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
FZ and XiaonanS designed the present study. RN and XiaoliangS performed the experiments, analyzed the data and prepared the figures. FZ and XiaonanS drafted the initial manuscript. XiaonanS reviewed and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The research was approved by the Ethics Committee of The Third Affiliated Hospital of Soochow University (Changzhou, China) and all patients provided signed informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tang H, Han X, Li M, Li T, Hao Y. Linc00221 modulates cisplatin resistance in non-small-cell lung cancer via sponging miR-519a. Biochimie. 2019;162:134–143. doi: 10.1016/j.biochi.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Huang T, Li J, Zhang C, Hong Q, Jiang D, Ye M, Duan S. Distinguishing lung adenocarcinoma from lung squamous cell carcinoma by two hypomethylated and three hypermethylated genes: A meta-analysis. PLoS One. 2016;11:e0149088. doi: 10.1371/journal.pone.0149088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu R, Mao Y, Chen K, He W, Shi W, Han Y. The long noncoding RNA ANRIL acts as an oncogene and contributes to paclitaxel resistance of lung adenocarcinoma A549 cells. Oncotarget. 2017;8:39177–39184. doi: 10.18632/oncotarget.16640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang M, He X, Huang X, Wang J, He Y, Wei L. LncRNA MIR4435-2HG-mediated upregulation of TGF-β1 promotes migration and proliferation of nonsmall cell lung cancer cells. Environ Toxicol. 2020;35:582–590. doi: 10.1002/tox.22893. [DOI] [PubMed] [Google Scholar]
- 5.Zhao M, Xin XF, Zhang JY, Dai W, Lv TF, Song Y. LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by regulating miR-96-5p/CYLD signaling. Cancer Med. 2020;9:1196–1208. doi: 10.1002/cam4.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu M, Zhang Q, Tian XH, Wang JL, Niu YX, Li G. lncRNA CCAT1 is a biomarker for the proliferation and drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1 axis. Mol Carcinog. 2019;58:2207–2217. doi: 10.1002/mc.23109. [DOI] [PubMed] [Google Scholar]
- 7.Yan P, Su Z, Zhang Z, Gao T. LncRNA NEAT1 enhances the resistance of anaplastic thyroid carcinoma cells to cisplatin by sponging miR-9-5p and regulating SPAG9 expression. Int J Oncol. 2019;55:988–1002. doi: 10.3892/ijo.2019.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Ju H, Zhao T, Yu S, Li P, Jia J, Li N, Jing X, Tan B, Li Y. hsa_circ_0092306 targeting miR-197-3p promotes gastric cancer development by regulating PRKCB in MKN-45 cells. Mol Ther Nucleic Acids. 2019;18:617–626. doi: 10.1016/j.omtn.2019.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H, Lan Z, Li Q, Li Y. Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-κB pathway. Artif Cells Nanomed Biotechnol. 2019;47:2098–2106. doi: 10.1080/21691401.2019.1620256. [DOI] [PubMed] [Google Scholar]
- 10.Zhu H, Lu J, Zhao H, Chen Z, Cui Q, Lin Z, Wang X, Wang J, Dong H, Wang S, Tan J. Functional long noncoding RNAs (lncRNAs) in clear cell kidney carcinoma revealed by reconstruction and comprehensive analysis of the lncRNA-miRNA-mRNA regulatory network. Med Sci Monit. 2018;24:8250–8263. doi: 10.12659/MSM.910773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Liu X, Pan B, Hu X, Zhu Y, Su Y, Guo Z, Zhang G, Xu M, Xu X, et al. Serum exosomal miR-122 as a potential diagnostic and prognostic biomarker of colorectal cancer with liver metastasis. J Cancer. 2020;11:630–637. doi: 10.7150/jca.33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin Y, Zhou X, Huang C, Li L, Liu H, Liang N, Chen Y, Ma D, Han Z, Xu X, et al. Serum miR-342-3p is a novel diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Pathol. 2018;11:2742–2748. [PMC free article] [PubMed] [Google Scholar]
- 13.Liang ZX, Liu HS, Wang FW, Xiong L, Zhou C, Hu T, He XW, Wu XJ, Xie D, Wu XR, Lan P. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10:829. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan LY, Shi KY, Xu D, Ren LP, Yang P, Zhang L, Wang F, Shao GL. LncRNA GIHCG regulates microRNA-1281 and promotes malignant progression of breast cancer. Eur Rev Med Pharmacol Sci. 2019;23:10842–10850. doi: 10.26355/eurrev_201912_19788. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Yang QS, Li B, Xu G, Yang SQ, Wang P, Tang HH, Liu YY. Long noncoding RNA LINC00483/microRNA-144 regulates radiosensitivity and epithelial-mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J Cell Physiol. 2019;234:11805–11821. doi: 10.1002/jcp.27886. [DOI] [PubMed] [Google Scholar]
- 17.Xiao J, Lv Y, Jin F, Liu Y, Ma Y, Xiong Y, Liu L, Zhang S, Sun Y, Tipoe GL, et al. LncRNA HANR promotes tumorigenesis and increase of chemoresistance in hepatocellular carcinoma. Cell Physiol Biochem. 2017;43:1926–1938. doi: 10.1159/000484116. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Xu C, Gao Y, Wang Y, Ding Z, Zhang Y, Shen W, Zheng Y, Wan Y. A novel long non-coding RNA, MSTRG.51053.2 regulates cisplatin resistance by sponging the miR-432-5p in non-small cell lung cancer cells. Front Oncol. 2020;10:215. doi: 10.3389/fonc.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng F, Xu R. CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145-5p/ABCC1 axis. Biomed Pharmacother. 2020;124:109828. doi: 10.1016/j.biopha.2020.109828. [DOI] [PubMed] [Google Scholar]
- 20.Wang DY, Li N, Cui YL. Long non-coding RNA CCAT1 sponges miR-454 to promote chemoresistance of ovarian cancer cells to cisplatin by regulation of surviving. Cancer Res Treat. 2020;52:798–814. doi: 10.4143/crt.2019.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu W, Huang W, Yang F, Ju H, Zhu G. Long noncoding RNA LINC00461 mediates cisplatin resistance of rectal cancer via miR-593-5p/CCND1 axis. Biomed Pharmacother. 2020;124:109740. doi: 10.1016/j.biopha.2019.109740. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Liu C, Zhang Z. Knockdown of lncRNA MIAT inhibits proliferation and cisplatin resistance in non-small cell lung cancer cells by increasing miR-184 expression. Oncol Lett. 2020;19:533–541. doi: 10.3892/ol.2019.11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai D, Feng XD, Zhu WQ, Bao YN. LncRNA BLACAT1 regulates the viability, migration and invasion of oral squamous cell carcinoma cells by targeting miR-142-5p. Eur Rev Med Pharmacol Sci. 2019;23:10313–10323. doi: 10.26355/eurrev_201912_19669. [DOI] [PubMed] [Google Scholar]
- 24.Li M, Cai O, Tan S. LOXL1-AS1 drives the progression of gastric cancer via regulating miR-142-5p/PIK3CA axis. Onco Targets Ther. 2019;12:11345–11357. doi: 10.2147/OTT.S223702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Liu H, Chen R, Kang F, Lai H, Wang Y. KCNQ1OT1 promotes ovarian cancer progression via modulating MIR-142-5p/CAPN10 axis. Mol Genet Genomic Med. 2020;8:e1077. doi: 10.1002/mgg3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Chen W, Jin Y, Xue R, Su J, Mu Z, Li J, Jiang S. miR-142-5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Biochem Pharmacol. 2019;161:98–112. doi: 10.1016/j.bcp.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang L, Zhao G, Ji S, Yuan Q, Zhou H. Downregulated long non-coding RNA MSC-AS1 inhibits osteosarcoma progression and increases sensitivity to cisplatin by binding to MicroRNA-142. Med Sci Monit. 2020;26:e921594. doi: 10.12659/MSM.921594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalim M, Iqbal Khan MS, Zhan J. Programmed cell death ligand-1: A dynamic immune checkpoint in cancer therapy. Chem Biol Drug Des. 2020;95:552–566. doi: 10.1111/cbdd.13677. [DOI] [PubMed] [Google Scholar]
- 29.Yang M, Hu Z, Yue R, Yang L, Zhang B, Chen Y. The efficacy and safety of qiming granule for dry eye disease: A systematic review and meta-analysis. Front Pharmacol. 2020;11:580. doi: 10.3389/fphar.2020.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Chou X, Zhuang M, Zhu C, Hu Y, Cheng D, Liu Z. LINC00657 activates PD-L1 to promote osteosarcoma metastasis via miR-106a. J Cell Biochem. 2020;121:4188–4195. doi: 10.1002/jcb.29574. [DOI] [PubMed] [Google Scholar]
- 31.Xue C, Xu Y, Ye W, Xie Q, Gao H, Xu B, Zhang D, Jiang J. Expression of PD-L1 in ovarian cancer and its synergistic antitumor effect with PARP inhibitor. Gynecol Oncol. 2020;157:222–233. doi: 10.1016/j.ygyno.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Zuo Y, Zheng W, Liu J, Tang Q, Wang SS, Yang XS. MiR-34a-5p/PD-L1 axis regulates cisplatin chemoresistance of ovarian cancer cells. Neoplasma. 2020;67:93–101. doi: 10.4149/neo_2019_190202N106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.