Abstract
COVID-19 has broken out rapidly in nearly all countries worldwide, and has blossomed into a pandemic. Since the beginning of the spread of COVID-19, many scientists have been cooperating to study a vast array of old drugs and new clinical trial drugs to discover potent drugs with anti-COVID-19 activity, including antiviral drugs, antimalarial drugs, immunosuppressants, Chinese medicines, Mpro inhibitors, JAK inhibitors, etc. The most commonly used drugs are antiviral compounds, antimalarial drugs and JAK inhibitors. In this review, we summarize mainly the antimalarial drugs chloroquine and hydroxychloroquine, the antiviral drugs Favipiravir and Remdesivir, and JAK inhibitor Ruxolitinib, discussing their biological activities, clinical trials and synthesis progress.
Keywords: COVID-19, Remdesivir, Favipiravir, Chloroquine, Hydroxychloroquine, Ruxolitinib
Introduction
Coronavirus disease 2019 (COVID-19) has broken out rapidly in nearly all countries worldwide, and has blossomed into a pandemic. COVID-19 infections normally manifest with symptoms of high temperature, cough, myalgia, weakness, polypnea and other symptoms [1]. In grievous cases, it can also cause acute respiratory distress syndrome (ARDS), and result in fluids around in the lungs, eliciting infectious shock. Since the beginning of the COVID-19 spread, scientists have investigated cooperatively, examining abundant old drugs and new clinical treatments, such as Chinese medicines [2], vaccine development [3, 4], convalescent plasma [5–7], interferon-based therapies [8], monoclonal antibodies [9], cell-based therapies [10], immunopathology therapies [11] and small molecule drugs, aiming to discover drugs with potent anti-COVID-19 activity. However, the pathway to develop a new drug or vaccine usually takes more than 1 year or even 3–5 years. Monoclonal antibodies, cell-based therapies, interferon-based therapies, and immunopathology therapies are unacceptable for their high cost. Considering cost and time constraints, small molecule drugs, including existing drugs, e.g., those used to treat influenza, HBV, HCV, HIV, antimalarial and anti-filovirus drugs, have evoked great interest among researchers as they might allow more rapid development [12].
As traditional small molecule drugs used for treating malaria and certain autoimmune diseases, chloroquine (CLQ) and hydroxychloroquine (CLQ-OH) (Fig. 1) were demonstrated recently to exhibit certain activity against COVID-19 both in vitro and in vivo. Preliminary clinical results showed that CLQ has potential for use in the treatment of COVID-19 patients [13]. Compared with CLQ, CLQ-OH is more effective and has better safety properties in vitro [14]. Recently, the Gautret group [15] conducted a clinical trial using CLQ-OH in combination with Azithromycin to treat patients with COVID-19; the results showed that this was effective. But, afterwards, it was reported that use of these two antimalarial drugs may be fatal in some cases [16].
Fig. 1.
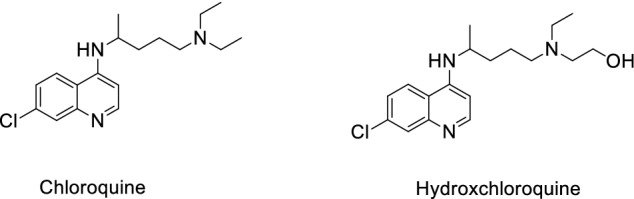
Structure of chloroquine (CLQ) and hydroxychloroquine (CLQ-OH)
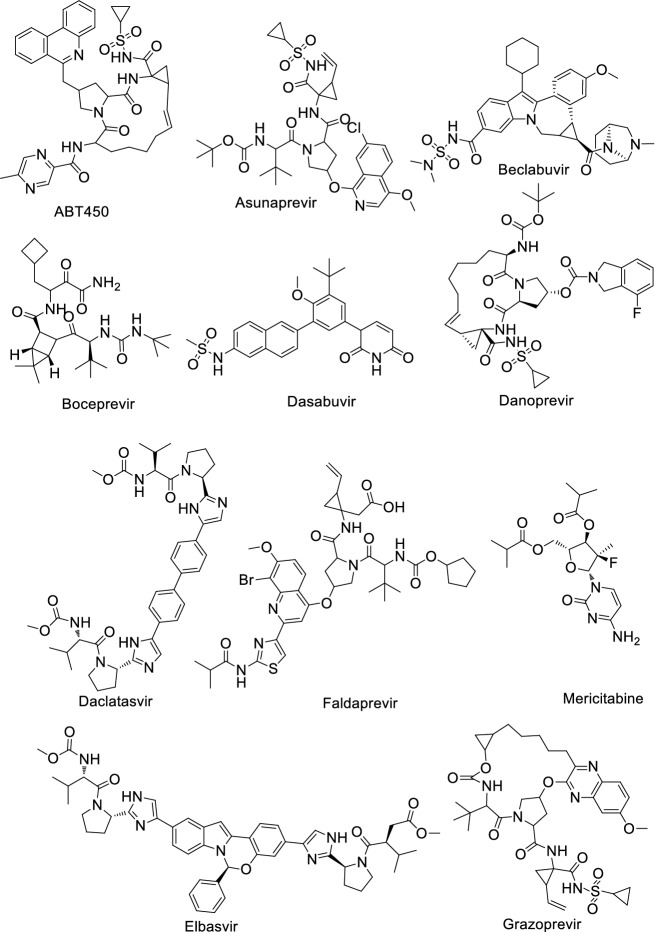
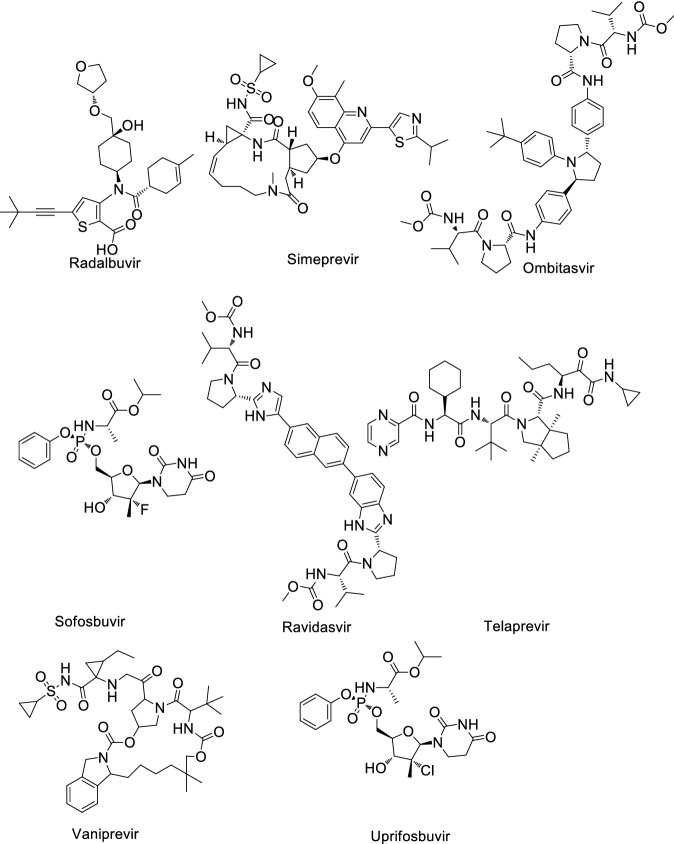
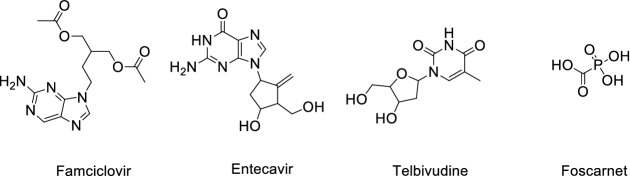
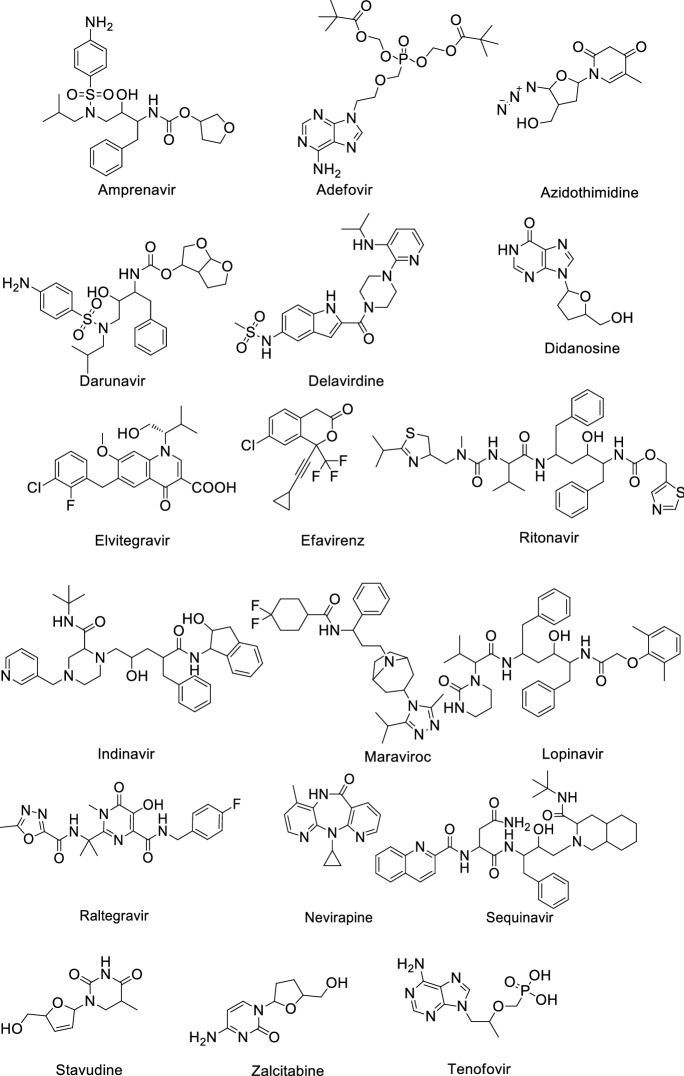
Shah et al. [17] reviewed a total of 61 antiviral drugs to screen efficient drugs against COVID-19 as shown in Tables 1, 2, 3, 4, 5, 6 and Figs. 2, 3, 4, 5, 6, 7. Some biological activities against COVID-19 are also listed based on literature reports [18–37].
Table 1.
Hepatitis C virus (HCV) antiviral agents against COVID-19
| Entry | Name | Biological activity against COVID-19 | Mechanism |
|---|---|---|---|
| 1 | ABT450 | ND | Inhibits NS3/4A serine protease |
| 2 | Asunaprevir | IC50: 53.9 ± 1 μM [18] | Protease inhibitor |
| 3 | Beclabuvir | ND | Inhibits NS5B protein |
| 4 | Boceprevir | IC50: 2.7 ± 0.05 μM [19] | NS3/4A protease inhibitor |
| 5 | Dasabuvir | ND | Inhibits the action of NS5B polymerase |
| 6 | Danoprevir | CC50: > 50 µM [20] | NS3/4A protease inhibitor |
| 7 | Daclatasvir | EC50: 0.8 µM [20] | NS5A inhibitor |
| 8 | Faldaprevir | ND | Protease inhibitor |
| 9 | Elbasvir | ND | NS5A inhibitor |
| 10 | Grazoprevir | Ki: 172.33 nM [21] | Blocks NS3 |
| 11 | Mericitabine | ND | Inhibitor of RdRp |
| 12 | Radalbuvir | ND | NS5B inhibitor |
| 13 | Simeprevir | EC50: 4.08 µM, CC50: 19.33 µM [22] | HCV protease inhibitor |
| 14 | Ombitasvir | ND | NS5A inhibitor |
| 15 | Sofosbuvir | EC50: 381 ± 34 µM [20] | Inhibits viral RNA synthesis by inhibiting NS5B protein |
| 16 | Ravidasvir | ND | NS5A inhibitor |
| 17 | Telaprevir | IC50: 10.7 ± 0.4 μM [18] | NS3/4a protease inhibitor |
| 18 | Velpatsvir | EC50: 0.77–2.74 µM [23] | NS5A protein inhibitor |
| 19 | Vedroprevir | ND | Inhibits HCV NS3 |
| 20 | Vaniprevir | ND | NS3/4A protease inhibitor |
| 21 | Uprifosbuvir | EC50: > 50 μM [24] | NS5B polymerase inhibitor |
ND, not determined, IC50 half maximal inhibitory concentration
Table 2.
Hepatitis B virus (HBV) antiviral agents against COVID-19
| Entry | Name | Biological activity against COVID-19 | Mechanism |
|---|---|---|---|
| 1 | Famciclovir | ND | Inhibits viral DNA polymerase |
| 2 | Entecavir | EC50: > 10 µM, CC50: > 50 µM [23] | Inhibits reverse transcription |
| 3 | Telbivudine | ND | Reverse transcriptase inhibitor |
| 4 | Foscarnet | ND | Viral DNA polymerases inhibitor |
Table 3.
Human immunodeficiency virus (HIV) antiviral agents against COVID-19
| Entry | Name | Biological activity against COVID-19 | Mechanism |
|---|---|---|---|
| 1 | Amprenavir | EC50: 31.32 μM, CC50: > 81 μM [25] | Protease inhibitor |
| 2 | Adefovir | ND | Reverse transcriptase inhibitor |
| 3 | Azidothimidine | ND | Inhibits reverse transcriptase |
| 4 | Darunavir | Kd: 57.30 nM (3CL protease), 6.09 nM (RdRp) and 46.16 nM (papain-like protease) [26] EC50 > 100 μM [27] | Inhibits HIV protease enzyme |
| 5 | Delavirdine | ND | Non-nucleoside reverse transcriptase inhibitor |
| 6 | Didanosine | ND | Nucleoside reverse transcriptase inhibitor |
| 7 | Elvitegravir | ND | Integrase inhibitor |
| 8 | Efavirenz | EC50: > 9.6 µM, CC50: 37.6 ± 10.7 μM [23] | Inhibits non-nucleoside reverse transcriptase enzyme |
| 9 | Ritonavir | C50: 8.63 µM, CC50: 74.11 µM [25] | HIV Protease inhibitor |
| 10 | Indinavir | EC50: 59.14 µM CC50: > 81 µM [25] | Protease inhibitor |
| 11 | Maraviroc | EC50: 2.7 µM [28] | C–C chemokine receptor type 5 allosteric modulator |
| 12 | Lopinavir | EC50: 5.73 µM, CC50: 74.44 µM [25] | Protease inhibitor |
| 13 | Raltegravir | ND | HIV-1 integrase inhibitor |
| 14 | Nevirapine | ND | Non-nucleoside reverse transcriptase inhibitor |
| 15 | Sequinavir | EC50: 8.83 µM, CC50: 44.43 µM [25] | Protease inhibitor |
| 16 | Stavudine | ND | Inhibits HIV reverse transcriptase enzyme |
| 17 | Zalcitabine | ND | Inhibits nucleoside reverse transcriptase |
| 18 | Tenofovir | EC50: 100 μM [29] | HIV-1 reverse transcriptase inhibitor |
Table 4.
Influenza antiviral agents against COVID-19
| Entry | Name | Active against | Biological activity against COVID-19 | Mechanism |
|---|---|---|---|---|
| 1 | Arbidol (Umifenovir) | Influenza | EC50: 4.11 μM [29] | Inhibits membrane fusion |
| 2 | Favipiravir | Influenza | EC50: 22.5 μM [30] | Inhibits viral RNA dependent RdRp |
| 3 | Amantadine | Influenza A | IC50 > 100 μM [31] | The influenza virus A-M2 proton channel agonist |
| 4 | Zanamivir | Influenza viruses | ND | Neuraminidase inhibitor |
| 5 | Oseltamivir | Influenza viruses A | EC50: > 100 μM [32] | Inhibits the neuraminidase enzyme |
Table 5.
Ebola antiviral agents against COVID-19
Table 6.
Other antiviral agents against COVID-19
| Entry | Name | Active against | Biological activity against COVID-19 | Mechanism |
|---|---|---|---|---|
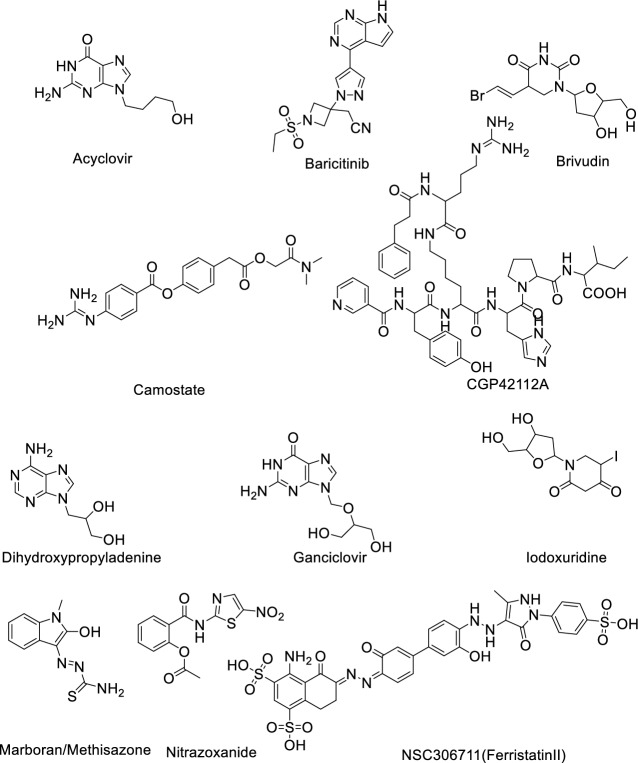
| 1 | Acyclovir | Cytomegalovirus infections | ND | Inhibits viral DNA polymerase |
| 2 | Baricitinib | Rheumatoid arthritis | IC50: 400–800 nM [34] | Inhibits Janus kinase |
| 3 | Brivudin | Herpes zoster | ND | Locks the action of DNA polymerases |
| 4 | Camostate | Pancreatitis | EC50: 107 nM [35] | Serine protease inhibitor |
| 5 | CGP42112A | Vasodilation and blood pressure reduction | ND | Angiotensin AT2 receptor agonist |
| 6 | Dihydroxy propyladenine | Herpes Virus | ND | Inhibits viral replication |
| 7 | Ganciclovir | Cytomegalovirus | ND | Inhibits viral DNA polymerases |
| 8 | Iodoxuridine | Herpes simplex virus | ND | Interferes viral DNA replication |
| 9 | Marboram/Methisazone | Small pox virus | ND | Inhibits mRNA and protein synthesis |
| 10 | Nitrazoxanide | Broad-spectrum antiviral | EC50: 2.21 μM [36] | Pyruvate:ferred oxinoxidoreductase (PFOR) enzyme |
| 11 | NSC306711 (Ferristatin II) | Flavivirus | ND | Degradation of Transferrin receptor-1 |
Fig. 2.
Structure of hepatitis C virus (HCV) antiviral agents against COVID-19
Fig. 3.
Structure of hepatitis B virus (HBV) antiviral agents against COVID-19
Fig. 4.
Structure of human immunodeficiency virus (HIV) antiviral agents against COVID-19
Fig. 5.
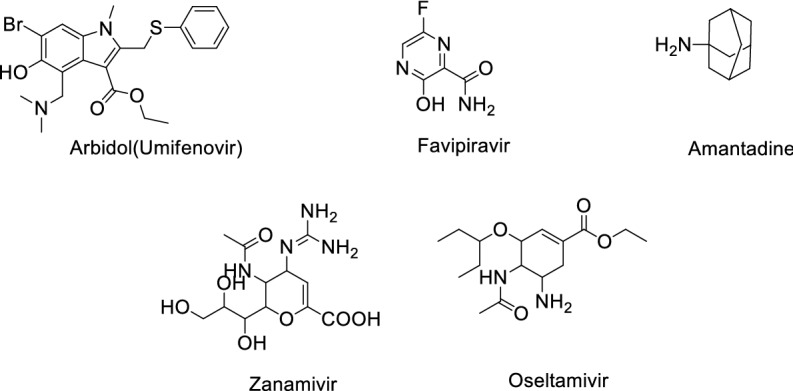
Structure of Influenza antiviral agents against COVID-19
Fig. 6.
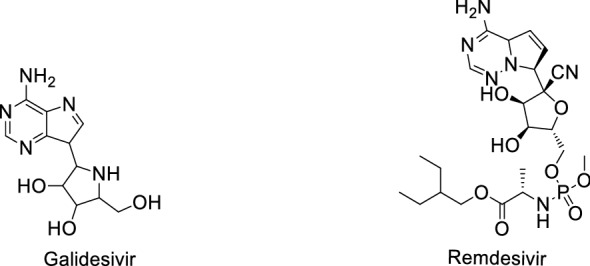
Structure of Ebola antiviral agents against COVID-19
Fig. 7.
Structure of other antiviral agents against COVID-19
Yan and Muller [37] provided a detailed analysis and described the use of the parent nucleoside of remdesivir, GS-441524 (Fig. 8), over remdesivir for the treatment of COVID-19, and appealed to Gilead Science to ditch GS-441524.
Fig. 8.
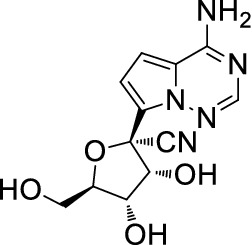
Structure of GS-441524 (remdesivir)
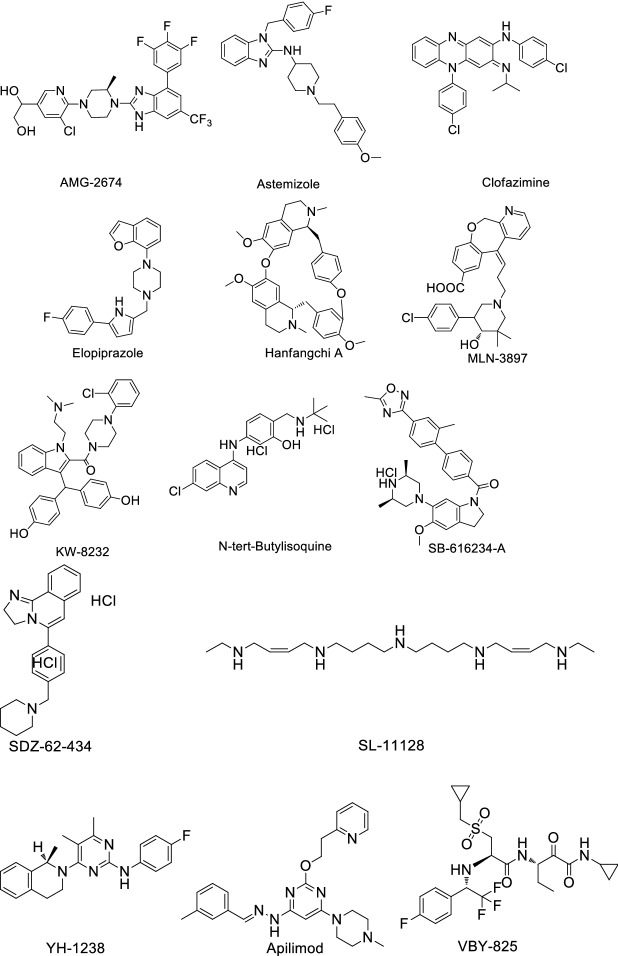
Riva et al. [38] identified 100 known drugs that can inhibit COVID-19 replication in mammalian cells, and found 21 compounds exhibiting effective dose response relationships with antiviral activity and confirmed their dose/activity relationships. Then, they found 13 compounds harboring EC50 values < 500 nM in at least one cell line (Table 7); for structures of these compounds see Fig. 9.
Table 7.
Drugs with known activity against COVID-19 in cell lines
| Entry | Drug name | EC50 value (µM) | Cell line |
|---|---|---|---|
| 1 | AMG2674a | 0.023 | Vero E6 cells |
| 2 | AMG2674 | 0.3 | 293T-ACE2 cells |
| 3 | AMG2674 | 0.41 | Hub-7-ACE2 cells |
| 4 | Astemizole | ~ 1.2 | Vero E6 cells |
| 5 | Astemizole | 0.87 | 293T-ACE2 cells |
| 6 | Astemizole | 1.3 | Hub-7-ACE2 cells |
| 7 | Clofazimine | 0.31 | Vero E6 cells |
| 8 | Clofazimine | ND | 293T-ACE2 cells |
| 9 | Clofazimine | 0.49 | Hub-7-ACE2 cells |
| 10 | Elopiprazole | 1.6 | Vero E6 cells |
| 11 | Elopiprazole | 0.13 | 293 T-ACE2 cells |
| 12 | Elopiprazole | ~ 2.7 | Hub-7-ACE2 cells |
| 13 | Hanfanychin A | ~ 1.2 | Vero E6 cells |
| 14 | Hanfanychin A | 0.56 | 293T-ACE2 cells |
| 15 | Hanfanychin A | 0.64 | Hub-7-ACE2 cells |
| 16 | MLN-3897 | 0.65 | Vero E6 cells |
| 17 | MLN-3897 | 0.41 | 293T-ACE2 cells |
| 18 | KW 8232 | ~ 1.2 | Vero E6 cells |
| 19 | KW 8232a | ~ 0.091 | 293T-ACE2 cells |
| 20 | KW 8232 | 1 | Hub-7-ACE2 cells |
| 21 | N-tert-Butylisoquine | ~ 1.2 | Vero E6 cells |
| 22 | N-tert-Butylisoquine | 0.29 | 293T-ACE2 cells |
| 23 | N-tert-Butylisoquine | 0.37 | Hub-7-ACE2 cells |
| 24 | SB 616234-A | ~ 1.2 | Vero E6 cells |
| 25 | SB 616,234-A | ~ 0.29 | 293 T-ACE2 cells |
| 26 | SB 616234-A | 0.64 | Hub-7-ACE2 cells |
| 27 | SDZ-62-434 | 0.63 | Vero E6 cells |
| 28 | SDZ-62-434 | 0.12 | 293T-ACE2 cells |
| 29 | SDZ-62-434 | 0.11 | Hub-7-ACE2 cells |
| 30 | SL-11128 | ~ 0.25 | Vero E6 cells |
| 31 | SL-11128 | ND | 293T-ACE2 cells |
| 32 | SL-11128 | ~ 2.5 | Hub-7-ACE2 cells |
| 33 | YH-1238 | ~ 0.95 | Vero E6 cells |
| 34 | YH-1238 | 1.1 | 293T-ACE2 cells |
| 35 | Apilimoda | 0.0203 | Vero E6 cells |
| 36 | Apilimoda | 0.012 | 293T-ACE2 cells |
| 37 | Apilimoda | 0.088 | Hub-7-ACE2 cells |
| 37 | VBY-825 | 0.3 | Vero E6 cells |
| 38 | VBY-825a | 0.071 | 293T-ACE2 cells |
| 39 | VBY-825a | 0.052 | Hub-7-ACE2 cells |
| 40 | ONO 5334 | 0.41 | 293T-ACE2 cells |
| 41 | ONO 5334a | 0.042 | Vero E6 cells |
| 42 | ONO 5334a | 0.078 | 293T-ACE2 cells |
| 43 | Z LVG CHN2 | 0.19 | Vero E6 cells |
| 44 | Z LVG CHN2a | 0.0011 | 293T-ACE2 cells |
| 45 | Z LVG CHN2a | 0.0069 | Hub-7-ACE2 cells |
| 46 | MDL 28170 | 0.22 | Vero E6 cells |
| 47 | MDL 28170a | 0.021 | 293T-ACE2 cells |
| 48 | MDL 28170a | 0.086 | Hub-7-ACE2 cells |
| 49 | DS-6930 | 0.36 | Vero E6 cells |
| 50 | R 82913 | 0.21 | Vero E6 cells |
aCompounds harboring EC50 values < 500 nM in at least one cell line.
Fig. 9.
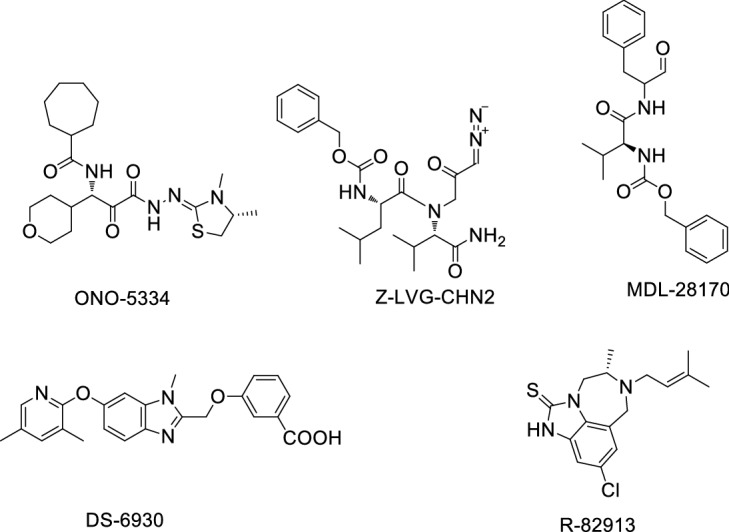
Compounds exhibiting good effectiveness in COVID-19 cell lines
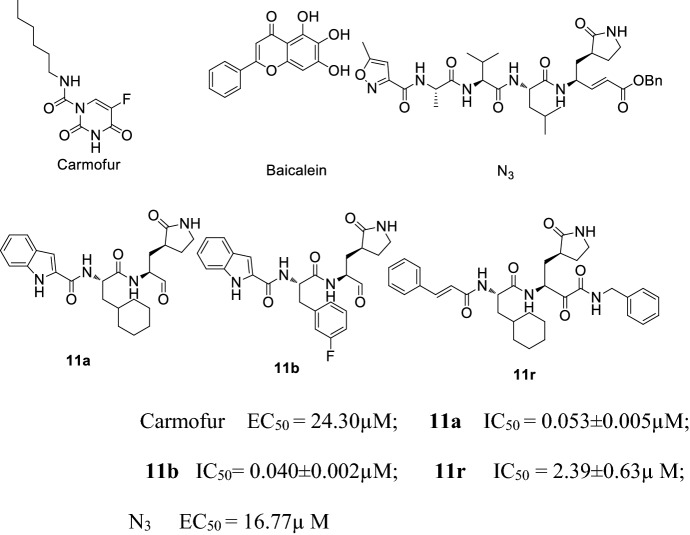
In addition to antimalarial and antiviral drugs, Mpro has also attracted the interest of researchers as a drug target for COVID-19 as it can mediate virus duplication and transcription. Jin and co-workers [39] reported the mechanism of Mpro inhibitor N3 via computer-aided drug design, and confirmed the crystal structure of COVID-19 Mpro and N3. Their results showed that N3 had the strongest antiviral COVID-19 effects at a concentration of 10 μM, the inhibition against COVID-19 EC50 value was 16.77 μM. They also discovered the crystal structure of the Mpro-Carmofur complex, confirming that Carmofur covalently links to the Cys145 residue via the carbonyl group, while the fatty acid group married with the hydrophobic S2 subsite of Mpro [40]. Su and co-workers [41] investigated the inhibition of COVID-19 Mpro by natural products derived from Chinese traditional medicines. They found that baicalin and baicalein showed non-covalent, non-peptidomimetic inhibition of COVID-19 Mpro, and had strong antiviral activities both in vitro and in a cell-based system. The in vitro study results and favorable safety data from clinical trials showed that baicalein has great potential to become a candidate for a much needed anti-coronaviral drug. Dai and co-workers [42] designed two Mpro inhibitors 11a and 11b, which showed perfect anti-COVID-19 activity. The structure–function relationship showed that the aldehyde group of the two compounds can covalently link to the Mpro Cys145 residue. Zhang and co-workers [43] reported the complex structure of COVID-19 Mpro and 11r, found that 11r showed excellent inhibitory activity and potent anti-COVID-19 activity. 11r could be used as a lead compound to develop potent inhibitors of COVID-19 Mpro (Fig. 10).
Fig. 10.
Structure and biological activity of Carmofur, Baicalein, 11a, 11b, 11r and N3
Vitner et al. [44] examined a Glucosyl Ceramide synthase (GCS) inhibitor GENZ-123346 (analogue of Cerdelga) and GENZ-66761 (structure unknown) for their antiviral effects on COVID-19. Both drugs can inhibit COVID-19 virus, and could be assessed further in preclinical and clinical trials (Fig. 11).
Fig. 11.
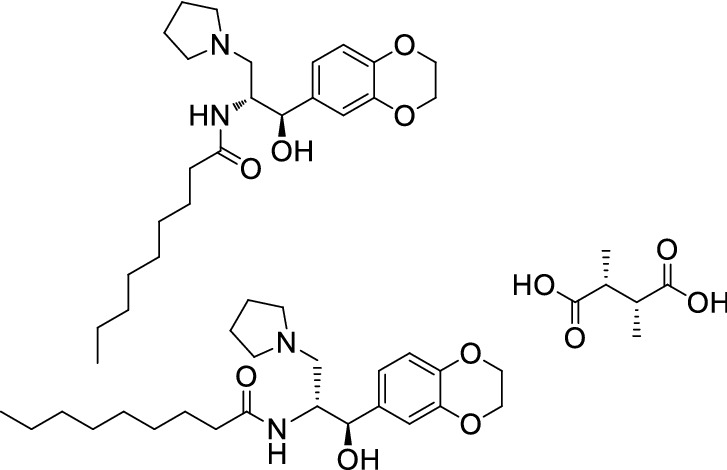
Structure of GENZ123346
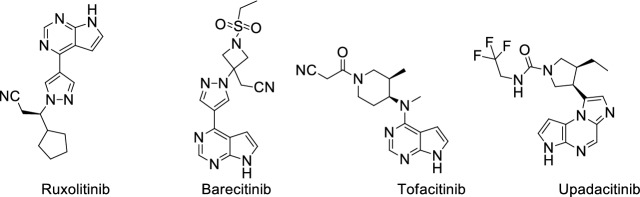
Cytokine storm is a driver of pathology and mortality in viral infections. In COVID-19-infected patients, cytokine storm increases the risk of death and other severe symptoms [45]. Plenty of COVID-19 patients with cytokine storm syndrome encounter a sharp respiratory function obstacle [46]. Secondary hemophagocytic lymphohistiocytosis (sHLH) is a hyper inflammatory syndrome characterized by noteworthy augmentation of cytokines, with multi-organ failure and high mortality rate [47]. COVID-19 patients with cytokine storm syndrome exhibit increased levels of several interleukins: IL-2, IL-6, IL-7, granulocyte colony-stimulating factor (G-CSF), interferon-gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α). The study showed that COVID-19 patients who died have higher plasma levels of ferritin and IL-6 [48]. Four United States Food and Drug Administration (FDA)-approved Janus kinase (JAK) inhibitors have proved useful for the treatment of COVID-19 [49]: Jakafi/Ruxolitinib; Xeljanz/Tofacitinib; Olumiant/Baricitinib; and Rinvoq/Upadacitinib (Fig. 12). Among these four JAK inhibitors, Ruxolitinib can significantly reduce the IL-6 and TNF-α level in spleen [50].
Fig. 12.
Structure of four Janus kinase (JAK) inhibitors
Of all the above-mentioned drugs that have potential activity in inhibiting COVID-19, CLQ, CLQ-OH, Favipiravir, Remdesivir and Ruxolitinib attracted our interest due to extensive research on these drugs worldwide.
Characteristics of COVID-19
Coronavirus is a positive sense, single-chain RNA virus [51]. Coronaviruses are classified as α-, β-, γ-, and δ-coronavirus. Only α-coronavirus and β-coronavirus can infect humans [52]. γ-Coronavirus and δ-coronavirus can affect humans indirectly through animals [53]. The coronavirus (severe acute respiratory syndrome coronavirus (SARS-CoV)-2) causing COVID-19 is a β-coronavirus and shares about 80% RNA sequence consistency with SARS-CoV [54]. The SARS-CoV-2 genome encodes four main non-structural proteins: helicase, Mpro, RNA-dependent papain-like protease, and RNA polymerase [55], which are absolutely necessary for the survival of SARS-CoV-2. Initial analyses found that the four main SARS-CoV-2 enzymes mentioned above are highly conserved [56], and the spike glycoprotein is indispensable for SARS-CoV-2 invading the host cell [57]. Similar to SARS-CoV, SARS-CoV-2 encodes a large spike protein with two domains (S1 and S2); SARS-CoV-2 virus binds and enters a host cell via this spike protein [58, 59] (Fig. 13).
Fig. 13.
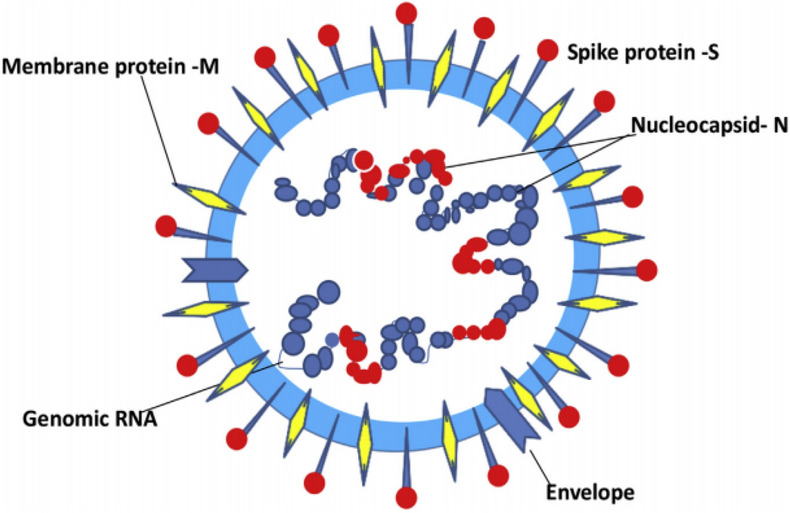
Schematic representation of a coronavirus virion. Image reproduced from Ref. [59] with permission from Elsevier
New research has shown that SARS-CoV-2 engages a receptor binding domain (RBD), binding to angiotensin-converting enzyme 2 (ACE2) to invade its human host cell [60] (Fig. 14).
Fig. 14.
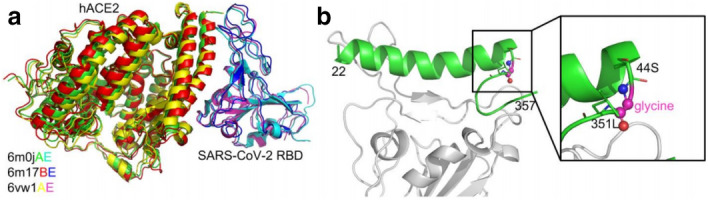
Comparison of severe acute respiratory syndrome coronavirus (SARS-CoV)-2 receptor binding domain (RBD)/human angiotensin-converting enzyme 2 (hACE2) complex structures (a) and the constructed SARS-CoV-2 RBD/hACE2 peptide complex (b). Image reproduced from Ref. [60] with permission from bioRxiv
Nevertheless, non-conserved mutations are highly accumulated in regions S1 and S2 which interact directly with ACE2. Mercurio et al. [61] performed a comparative in silico modeling analysis, and gained new insights into the spike protein of SARS-CoV-2. SARS-CoV-2 spike protein can interact with the ACE2 receptor on human cells at the RBD. This analysis can supply an ideal pipeline to identify characterized antibodies that might target the SARS-CoV-2 spike protein RBD to prevent interacting with human ACE2. Laurini et al. [62] reported an atomistic-based, reliable in silico structure of the viral transmembrane spike glycoprotein (S-protein)/ACE2 complex (Fig. 15), showing that residues D38, K31, E37, K353, Y41 on ACE2 and Q498, T500, R403 on the SARS-CoV-2 S-protein RBD are true hot spots to shaping and determining the stability of the relevant protein–protein interface. These results could be used in the structure-based design and development of neutralizing antibodies, vaccines, and protein–protein inhibitors against COVID-19.
Fig. 15.
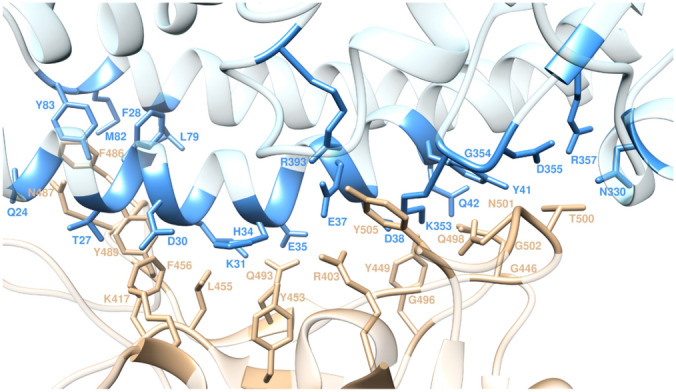
Structure of S-Protein/ACE2 complex. Image reproduced from Ref. [62] with permission from ACS
Monteil et al. [63] afforded a molecular explanation for the severe lung failure and death due to COVID-19, and proved that APN01 (human recombinant soluble ACE2, also named hrsACE2) can inhibit SARS-CoV-2 infections, which can be used for treatment of COVID-19 patients.
Chloroquine
CLQ is an old antimalarial drug used to treat malaria, amebiasis, rheumatoid, arthritis and lupus erythematosus syndrome. It inhibits the heme polymerase in malarial trophozoites via preventing heme conversion to hemozoin [64]. CLQ has strong antiviral effects on SARS-Cov infection [65] and Ebola in vitro [66], and is also able to inhibit influenza A virus replication in vitro [67]. Moreover, CLQ also has been shown to have some level of anti-HIV [68] and anti-H5N1 [69] activity.
Biological Activity of CLQ
CLQ can increase the pH value of host cell vacuoles. In lysosomes of the host cell, CLQ can change the catalytic activity of acidic hydrolases, affecting protein degradation, endosomal macromolecule composition, and post-translational modifications in Golgi [70]. In macrophages and antigen-presenting cells, an antirheumatic response is present that interrupts the immunological process [71]. Recent literature reports [72–74] assist our understanding of the three probable mechanisms of CLQ activity (Fig. 16).
Fig. 16.
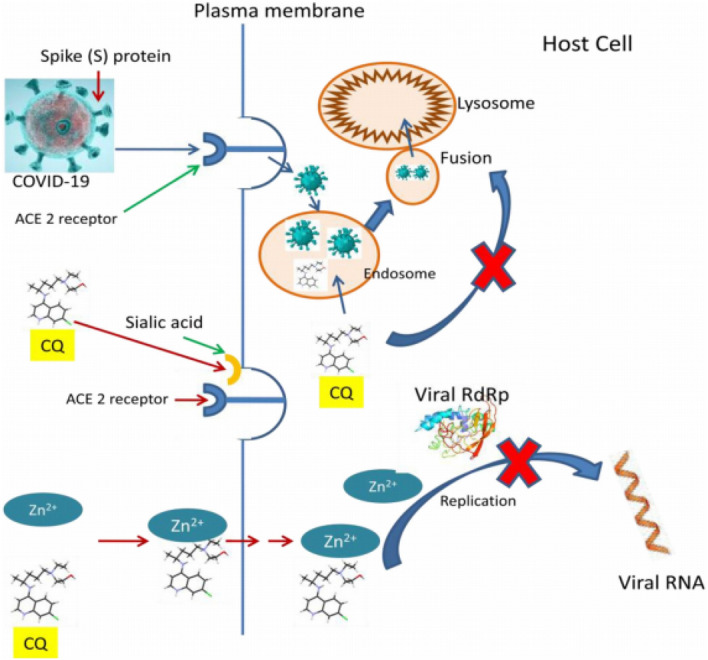
Proposed mechanism of action for CLQ. CQ CLQ, RdRp RNA dependent RNA polymerase. Image reproduced from Ref. [74] with permission from Elsevier
Gao et al. [13] found an EC50 value of 1.13 μM and a CC50 value greater than 100 mM for CLQ anti-COVID-19. Wang et al. [33] evaluated the efficiency of CLQ and six other drugs against SARS-CoV-2 in vitro, obtaining an EC50 value for CLQ of 1.13 μM and EC90 value of 6.90 μM in Vero E6 cells.
Clinical Trials of CLQ
Since the outbreak of COVID-19, CLQ has been used extensively to treat COVID-19 patients. Sun et al. [75] reported using CLQ phosphate at a dose of 500 mg to treat COVID-19 with a duration not exceeding 10 days in elderly patients; these authors summarized the main adverse reactions of CLQ in elderly patients: cardiac arrest, effects on skeletal musculoskeletal system, nerve irritability, medicated psychosis, granulocytopenia, aplastic anemia, thrombocytopenia, irreversible visual impairment, and others. Verscheijden et al. [76] established the best-evidence pediatric CLQ (free base) doses: 35 mg/kg, children 0–1 month, 47 mg/kg; children 1–6 months, 55 mg/kg; children 6 months to 12 years; and adolescents 44 mg/kg. Gao et al. [13] revealed that CLQ used in an intervention group had potent efficacy compared with a control group. Borba et al. [77] performed a parallel phase IIb clinical trial study to evaluate the safety and efficacy of high (0.6 g) and low (0.45 g) dose CLQ. Double-blinded and randomized participants were treated in intervention and control groups; the results suggested that the high dose of CLQ is hazardous to COVID-19 patients. Huang et al. [78] undertook a multicenter prospective observational study. Patients received a CLQ dose of 500 mg once or twice daily; patients treated with non-CLQ therapy were used as historical controls. In total, 197 patients used CLQ treatment, and 176 patients were treated as the historical control group. The duration of fever was clearly shortened. No serious adverse events were found in patients treated with CLQ. Patients treated with half dose CLQ experienced a lower rate of adverse events than with full dose. This study provides evidence for the safety and efficacy of CLQ in treatment of COVID-19 patients. Smit et al. [79] introduced pharmacokinetic and safety properties of CLQ for treatment of COVID-19, revealing that, for the use of CLQ to treatment of COVID-19 infection, early achievement of high ‘target’ concentrations is necessary, but, due to the high loading dose of CLQ and slow elimination, it can cause severe life-threatening toxicity and increased risk of mortality. Sinkeler [80] conducted a retrospective and observational study in a total of 397 patients aged over 18 years, found the CLQ gradually increased the QTc interval during treatment, which may result in ventricular tachycardia in patients. Sinkeler [80] suggested to measure the QTc interval before adjusting the dose of CLQ or withdrawing this potentially beneficial medication. While the pathogenesis of COVID-19 is still unknown, and because it does not show any anti SARS-Cov effect in an in vivo model, CLQ could be useless in treatment of COVID-19 patients and might even be harmful [81]; hence, before the pathogenesis of COVID-19 is known and the effect of CLQ is evaluated in clinical trials, it should be used cautiously.
Synthesis of CLQ
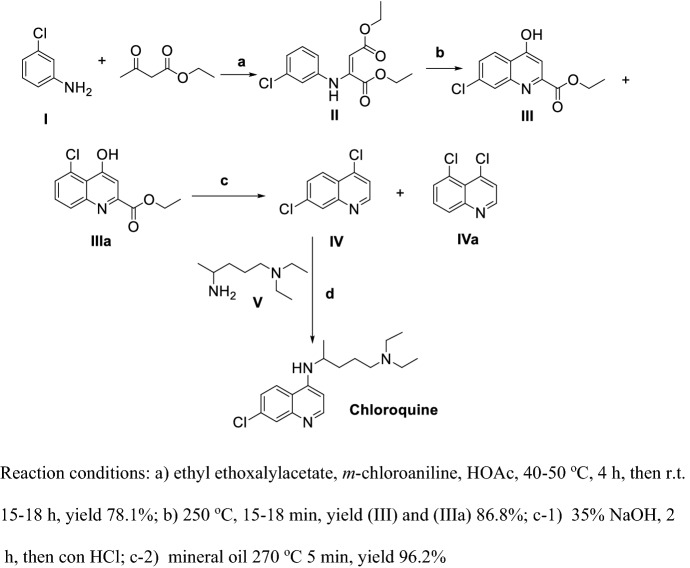
Surrey and Hammer [82] reported the first synthesis of CLQ (Scheme 1).
Scheme 1.
First generation synthesis of Chloroquine (CLQ)
Ethyl ethoxalylacetate reacting with m-chloroaniline (I) in the presence of HOAc under mild condition obtained II, then ring-closure at high temperature in a short time yields III and its isomer IIIa; III and IIIa were refluxed in strong base solution to obtain a mixture of IV and IVa, with a final coupling of IV and V at high temperature obtaining CLQ. This synthetic route was unacceptable due to steps (ii) and (iii) producing nearly half of an isomer byproduct and steps (ii) and (iv) proceeding at high temperature.
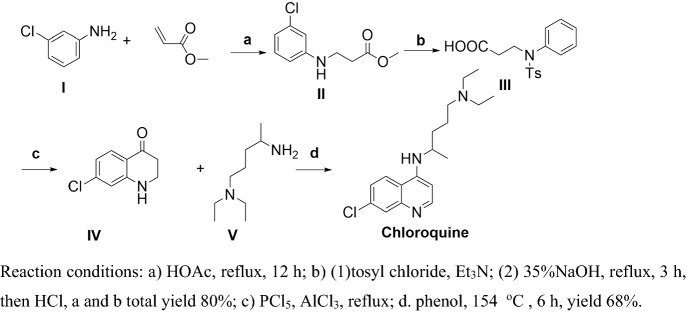
Jonnson and Buell [83] reported a second generation synthesis of CLQ (Scheme 2).
Scheme 2.
Compound II should be added an C-Cl at meta position
Methyl acrylate and m-chloroaniline (I) were refluxed in HOAc to obtain II, then II was protected by TsCl and ring-closure obtained IV. Finally, IV and V were refluxed in the presence of phenol to obtain CLQ. The total yield of this procedure is about 24.7%, which is higher than the first generation process, but, in step d, the reaction temperature is as high as more than 150 °C, at which temperature byproducts are easily produced, and the solvent phenol is harder to recover.
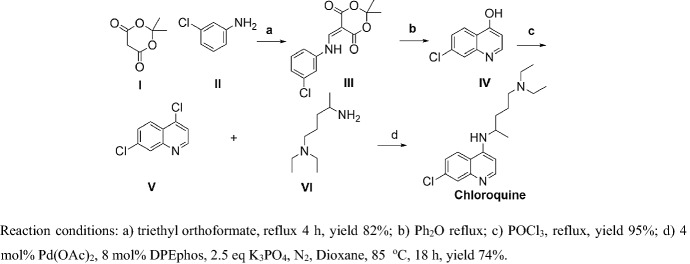
Margolis et al. [84] reported a third generation synthesis of CLQ (Scheme 3). I and II were refluxed without solvent and ring-closure obtained IV, then chlorination in the presence of POCl3 obtained V. Finally, V and VI coupling via Suzuki coupling obtained CLQ. The totally yield of this procedure is approximately 58% in mild conditions, which is suitable for large-scale production.
Scheme 3.
Third generation synthesis of CLQ
Hydroxychloroquine
Biological Activity of CLQ-OH
CLQ-OH is similar to CLQ, and. as an old antimalarial drug, also has potential effects on COVID-19 and lower toxicity than CLQ [85]. Moreover, it showed better anti-SARS-CoV-2 efficiency compared to CLQ in vitro [86]. Yao et al. [14] used physiologically-based pharmacokinetic models (PBPK) models to ensure the most efficacious concentrations of CLQ-OH and its safety profile, and found its EC50 value to be 0.72 μM in vitro. Fantini et al. [74] used an assembly of structure-molecular modeling methods and found that CLQ-OH can prevent binding of the SARS-CoV-2 spike to gangliosides, which is helpful in understanding the mechanism of CLQ-OH as an anti-COVID-19 drug (Figs. 17, 18).
Fig. 17.
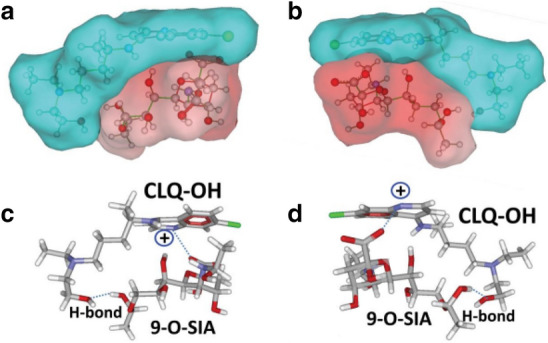
Molecular model of CLQ-OH interaction with sialic acids. Image reproduced from Ref. [74] with permission from Elsevier
Fig. 18.
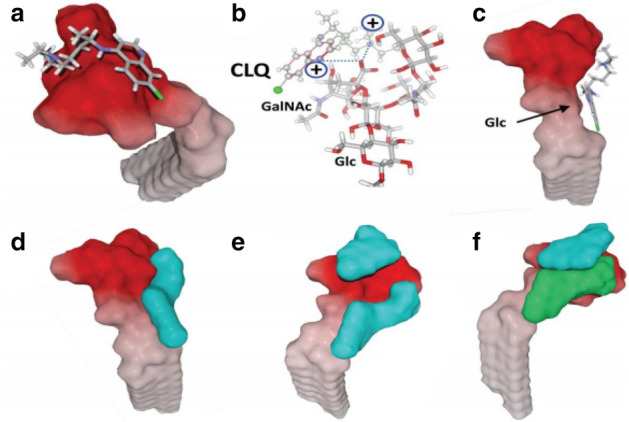
Molecular modeling simulations of CLQ and CLQ-OH binding to ganglioside. Image reproduced from Ref. [74] with permission from Elsevier
Zhang et al. [87] systematically evaluated the treatment of COVID-19 with CLQ and CLQ-OH for efficacy and safety, and asserted a potential mechanism involving COVID-19-induced injury of multiple organs as well as the pharmacological effects of CLQ-OH on COVID-19 (Fig. 19); the authors concluded that COVID-19 is actually a multisystem disease, with respiratory symptoms as the major clinical manifestation. Therefore, the treatment of COVID-19 with CLQ and CLQ-OH should be evaluated systematically, and patients should be monitored carefully for cardiovascular conditions to prevent lethal adverse events.
Fig. 19.
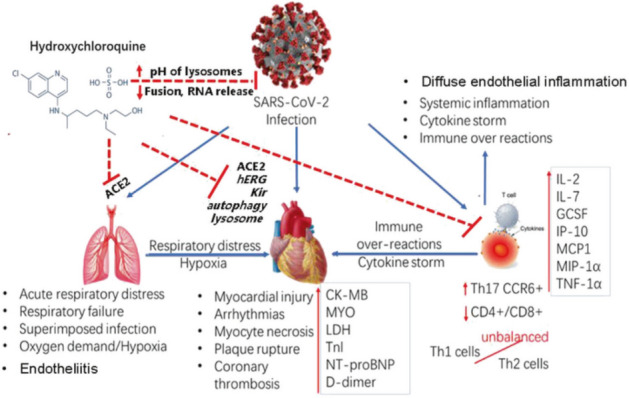
Potential mechanisms of SARS-COV-2-induced injury of multiple organs and pharmacological effects of CLQ-OH on COVID-19. Blue arrows indicate the actions of SARS-COV-2. ACE2 is key for SARS-COV-2 entering cells in human organs. Red dashed lines indicate the potential mechanisms of the therapeutic and toxic effects of CLQ-OH on SARS-COV-2 and organs in COVID-19 patients. Image reproduced from Ref. [87] with permission from Elsevier
Clinical Trials of CLQ-OH
Based on clinical studies [88], CLQ-OH is considered to be safer than CLQ. Gautret et al. [89] conducted a phase III clinical trial treating 80 mildly infected inpatients with CLQ-OH in combination with Azithromycin, demonstrating its therapeutic effect. Furthermore, the cost of this treatment is negligible. The suggested dose of CLQ-OH is 400 mg daily on the 1st day, then 200 mg daily for the next 3 days [90]. A total of 62 COVID-19 patients were assigned stochastically to the CLQ-OH (0.4 g daily, 0.2 g bid) and the control groups. After 5 days of treatment, patients in the CLQ-OH groups were clearly recovering in a shorter time [91]. To evaluate toxicity in patients who received high doses of CLQ-OH, a total of 63 patients were included [92], 58 females and 5 males. The mean dosage of CLQ-OH was 3.9 mg/kg, but 14 patient had doses higher than 5 mg/kg. A total of 36 patients were treated with CLQ-OH for more than 5 days. Only one (1.58%) patient exhibited CLQ-OH toxic retinopathy over a mean of 8 years treatment period. Patients on doses of > 5 mg/kg of CLQ-OH may be put at higher risk for retinal toxicity. Gautret et al. [15] used CLQ-OH, alone (0.6 g daily) or in combination with Azithromycin to treat COVID-19 patients in a small clinical trial. The results showed that Azithromycin combined with CLQ-OH was more efficient than CLQ-OH alone. A recent report [93] also confirmed significant efficacy of CLQ-OH in combination with Azithromycin. On the contrary, Sharma et al. [94] found opposite consequences in CLQ-OH combination with Azithromycin in a small, non-randomized clinical trial. Moreover, the risk of arrhythmia and prolonged QT interval was increased. Singh et al. [95] studied the efficacy of CLQ-OH in a treatment group compared with a control group in COVID-19, and concluded that there was no positive result in the CLQ-OH group, and that the death rate even increased in the CLQ-OH group compared to the control group. Gendelman et al. [96] performed a retrospective study based on a database including COVID-19 patients treated with CLQ-OH from February to March. A total sample of 14,520 were screened for COVID-19 infection, with 1317 being found positive. These findings raise doubts about the safety and efficacy of CLQ-OH against COVID-19 infection. Lauriola et al. [97] performed a retrospective study including 337 consecutive COVID-19 patients; 297 patients received CLQ-OH and azithromycin combination treatment, 17 patients CLQ-OH treatment alone and 63 patients did not recieve either of these two drugs due to contraindications. In this study, 146 patients died, including 102 in the combination treatment, 7 in the CLQ-OH treatment and 35 in the no treatment groups.
Synthesis of CLQ-OH
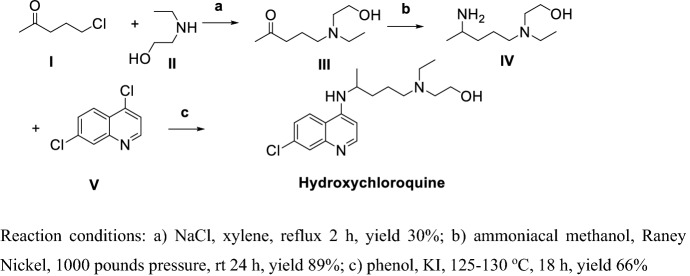
Alexander et al. [98] first reported the synthesis of CLQ-OH (Scheme 4).
Scheme 4.
Method for preparation of Hydroxychloroquine (CLQ-OH) by Alexander et al. [98]
I and II were refluxed in xylene to get III, then ammoniation and hydrogenation by Raney Nickel in high pressure obtained IV; finally, IV and V refluxed in phenol CLQ-OH. The overall yield of this procedure is as low as 17.6%, in step (b) intermediate (III) ammoniation and reduction by Raney Nickel requires high pressure, and in step (c) the solvent phenol is hard to recover. Hence, this method is unacceptable for large-scale production.
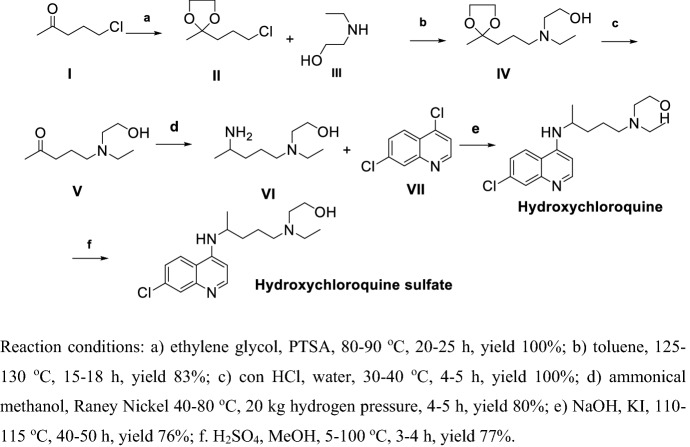
Ashok et al. [99] reported another synthetic route for the preparation of CLQ-OH (Scheme 5). Compound I was protected by ethylene glycol then reacted with III in refluxing toluene obtained IV, IV via deprotection, ammoniation and hydrogenation with Raney Nickel in high pressure to get VI, VI reacted with VII obtained CLQ-OH; finally, treatment of CLQ-OH with sulfuric acid yields CLQ-OH sulfate. The overall yield of this procedure is about approximately 40% via six steps, but in step (d), the Raney Nickel reduction of intermediate (IV) requires high pressure, which is a potential safety hazard.
Scheme 5.
Method for preparation of CLQ-OH by Ashok et al. [99]
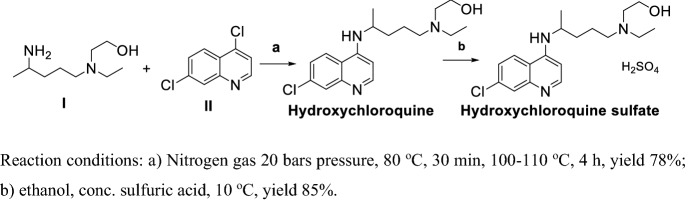
In 2010, Min et al. [100] reported a method for preparation of CLQ-OH (Scheme 6). I and II reacted in nitrogen conditions at high pressure obtained CLQ-OH with a yield of 78%; treatment sulfuric acid obtained CLQ-OH sulfate. This procedure led to CLQ-OH sulfate via two steps with an approximate total yield of 77%, but in step (a) the reaction proceeded in high pressure, which has safety implications.
Scheme 6.
Method for preparation of CLQ-OH by Min et al. [100]
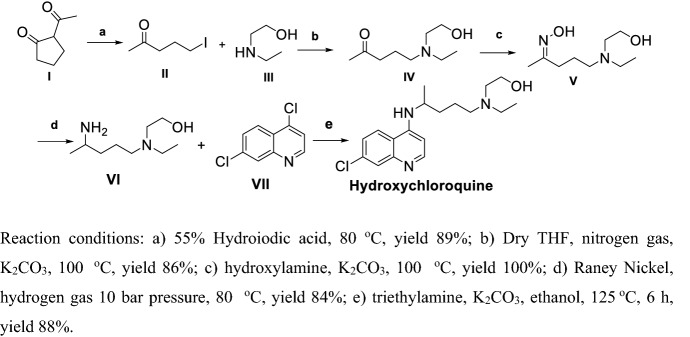
Yu et al. [101] and Frank et al. [102] used continuous-flow method for high-yield preparation of CLQ-OH (Scheme 7), which is not suitable for large-scale production. Iodization of I with hydroiodic acid and reaction with III in the presence of K2CO3 yielded IV, then oxime was obtained with hydroxylamine, and hydrogenation using Raney Nickel in high pressure obtained VI; finally, VI and VII reacted at high temperature in base conditions yielded CLQ-OH. This procedure yielded CLQ-OH in mild conditions, but the final step using ethanol as a solvent at high temperature is unacceptable, as ethanol volatilizes easily.
Scheme 7.
Preparation of CLQ-OH with a continuous-flow method
Favipiravir
Favipiravir (T-705) has been developed by Toyama Chemical Co., Ltd (Tokyo, Japan) as a broad spectrum antiviral drug against RNA viruses [103]. It shows good efficacy in the treatment of influenza infections [104] and pathogenic avian influenza A (H5N1). Favipiravir was also used as a potential drug for Ebola virus [105] and severe influenza [106], with EC50 values of 0.014–0.55 μg/mL to seasonal influenza and oseltamivir-resistant virus [107]. Favipiravir has an antiviral effect on SARS-CoV-2 by inducing a reduction in virus-induced cytopathic effect [108]. Observations show that Favipiravir induced the mutagenic effect responsible for the inhibition of replication; it was shown to act through lethal mutagenesis for several viruses, predominantly by competing with guanosine to cause transition mutations.
Biological Activity of Favipiravir
Favipiravir is a virus RdRp inhibitor. EC90 values for H5N1 resistance are in the 1.3–7.7 µM range [109]. Research in a P388D1 cell-based system [110] showed that Favipiravir can clearly inhibit the generation of TNF-α. Shirak et al. [111] established an influenza infection animal model to study the activity of Favipiravir, and proved that Favipiravir was significantly effective in alleviating influenza infection in mice. Janowski et al. [112] demonstrated that Favipiravir has EC50 values of 246 ± 76 µM (VA1), > 1000 µM (HAstV4) and 4.73 µM (IAV).
Clinical Trials of Favipiravir
Survival and virological characteristics were observed [113]. Randomized, multicenter phase II [114] and phase III [115] trials demonstrated clinical efficacy and safety of Favipiravir in Influenza. Lou [116] assessed the antiviral activity of Favipiravir and Baloxavir against COVID-19. Patients were randomized and assigned into three groups: Baloxavir group: dose 80 mg daily orally on Day 1 and Day 4, for patients still positive in virological test, given again on Day 7; Favipiravir group: The first dose 1.600 g or 2.2 g, followed by dose 0.6 g, 3 times daily; Control group: Continue existing antiviral treatment. The results showed that Favipiravir did not have any dramatic efficiency against COVID-19 even at high concentration. Cai et al. [117] conducted an Open-Label Controlled trial to test the efficacy of Favipiravir compared to Lopinavir (LPV)/Ritonavir (RTV) as anti-COVID-19 agents. Patients were randomly allocated to Favipiravir group (dose 1.6 g twice a day for the 1st day; dose 0.6 g twice a day for 2–14 days) and LPV/RTV group (dose 0.4 g/0.1 g twice a day); the results showed that the Favipiravir group patients exhibited higher efficacy than the LPV/RTV group.
Synthesis of Favipiravir
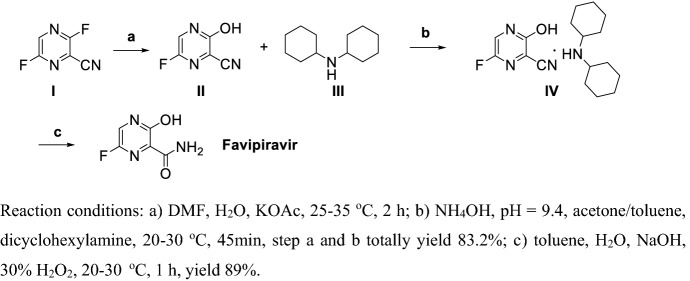
Takamatsu and Yonezawa [118] disclosed a method for producing Favipiravir and its intermediates in high yields (Scheme 8). Hydroxylation of I at room temperature in the presence of KOAc gives II. II reacted with III in the presence of ammonium hydroxide to give IV in 83.2% yield. Finally, cyan oxidation of IV by hydrogen peroxide in strong base solution gives Favipiravir in total yield of 74%. This procedure obtained Favipiravir via three steps under mild conditions. The process has the advantages of easy post-processing, low toxicity, and high yield for high purity of Favipiravir, and is suitable for large-scale production.
Scheme 8.
Synthesis of Favipiravir by Takamatsu and Yonezawa [118]
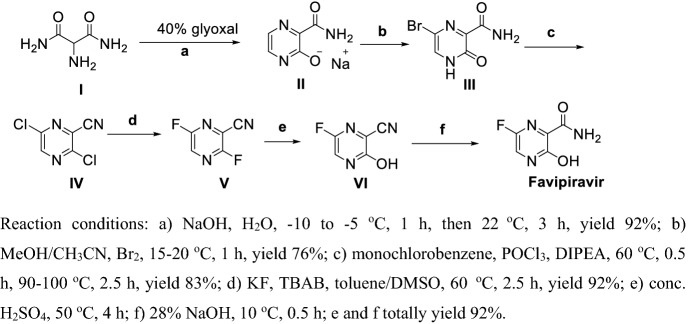
Hara et al. [119] reported a method for producing Favipiravir via six steps (Scheme 9), witha total yield of 53% under mild conditions. Compound I reacted with glyoxal in the presence of NaOH solution at lower temperature to obtain II, bromination, chlorination and dehydration obtained IV, fluoro-substitution in the presence of KF and tetra-n-butylammonium bromide (TBAB) gives V, hydrolyzation of 2-fluoro group of V in sulfuric acid and 3-cyan group of VI in NaOH solution obtained Favipiravir.
Scheme 9.
Preparation of Favipiravir by Hara et al. [119]
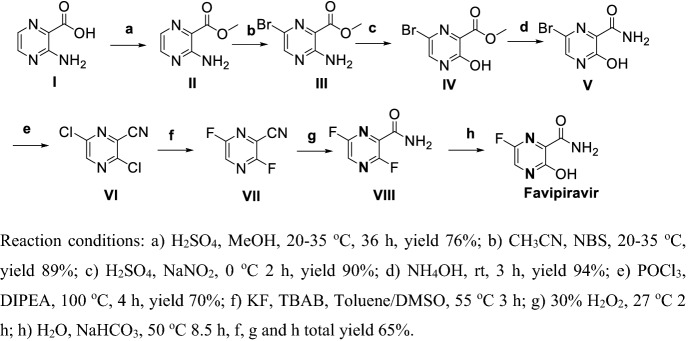
Liu et al. [120] reported a method for preparation of Favipiravir (Scheme 10) via 8 steps with a total yield of 24%. Esterification of I gave II, bromination by NBS obtained III, diazotization of III and hydrolyzation in the presence of ammonium hydroxide gave IV, ester group amidation obtained V, chlorination and dehydration obtained VI, fluoro-substitution chloro group of V in the presence of KF and TBAB got VII, hydrolyzation 3-cyan group of VII in the presence of hydrogen peroxide and 2-fluro group of VIII in weak base solution obtained Favipiravir. This procedure needs long steps and the diazotization reaction in step (c) has potential safety issues.
Scheme 10.
Preparation of Favipiravir by Liu et al. [120]
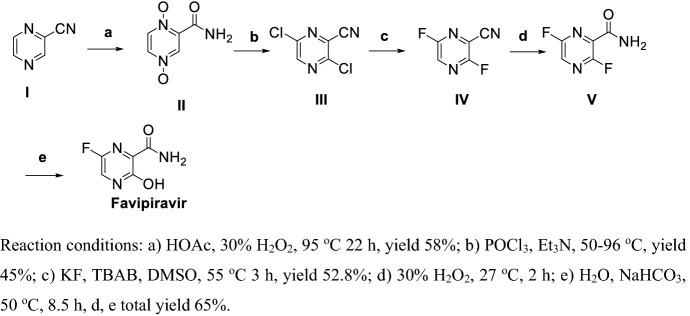
Li et al. [121] reported a method for preparation of Favipiravir (Scheme 11) via 4 steps with a total yield of less than 10%. Oxidation of I using hydrogen peroxide in the presence of HOAc gives II. Chlorination and dehydration of II obtained III, and then fluoro-substitution in presence of KF and TBAB gives IV. Hydrolyzation of 3-cyan group of IV in the presence of hydrogen peroxide gave V, followed by hydrolyzation of 2-fluoro group of V in weak base sulotion, obtained Favipiravir.
Scheme 11.
Preparation of Favipiravir by Li et al. [121]
Remdesivir
Remdesivir (Fig. 20) also named GS-5734, is a broad-spectrum antiviral RdRp inhibitor against MERS, Ebola, SARS and the like. Research showed an EC50 value of 0.77 μM in Vero E6 cells inhibiting COVID-19 [122]. Holshue [123] reported the first case of a US COVID-19 patient. After the COVID-19 spread worldwide in March, phase III clinical trials were launched in the US and other countries extensively. On 2 May, the FDA issued Remdesivir emergency use for COVID-19 treatment.
Fig. 20.
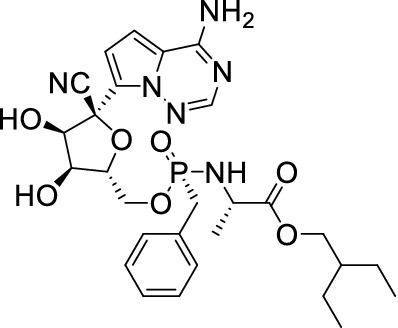
Structure of Remdesivir
History of Remdesivir
The HCV genome encodes two proteins, NS2 and NS5, that are important targets for drug design [124, 125]. The discovery of small molecules inhibiting virus replication has attracted more attention [25]. Abundant direct-acting antiviral (DAA) small molecules have been designed and tested in clinical trials over the last few decades. A number of DAAs have been proven to have anti-HCV activity. Nucleoside inhibitors (NIs) are the most outstanding due to their prominent therapeutic effect [126]. Most NIs used clinically are N-nucleosides, which have a 2′-C-Me in the sugar branch. The first 2′-C-Me branched N-nucleosides prepared in 1960s were used to treat HCV infection; their in vitro activity against HCV was test in the 2000s [127]. N-Nucleosides firstly metabolize into nucleoside triphosphates in cells, then link with the NS5B polymerase and insert into the viral RNA, inhibiting viral RNA extension and virus replication [128]. In 2012, Cho et al. [129] synthesized a few 2′-C-Me C-nucleosides targeting NS5B, and screened compound (1) as a HCV inhibitor (Fig. 21).
Fig. 21.
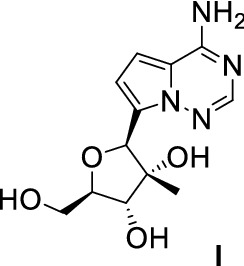
Structure and activity of (1) EC50 = 1.98 μM; CC50 = 85 μM (Huh-7); C50 = 0.31 μM
On the basis of compound (1), abundant analogues of compound (1) were synthesized to improve selectivity (Table 8) [130].
Table 8.
1′-substituted analogs of (1) in the NS5B enzyme assay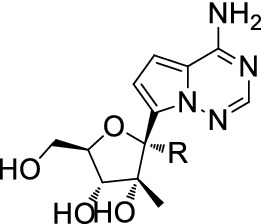
| Compound | R | EC50 (1b, μM) | IC50 (1b, μM) | mtRNA SNI (%) |
|---|---|---|---|---|
| 1 | H | 1.98 | 0.31 | 21 |
| 2 | Cyano | > 98 | 0.29 | 0.03 |
| 3 | Methyl | > 98 | 55 | 0.08 |
| 4 | Ethynyl | > 98 | > 200 | ND |
SNI Single nucleotide incorporation, ND not determined
Among these analogues of compound (1), it was found that compound (2) has perfect activity in the NS5B enzyme assay. Then compound (2) should be used as a lead nucleoside for further optimization (Table 9).
Table 9.
HCV replicon activity of phosphoramidate prodrugs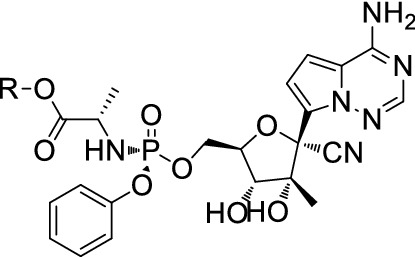
| Compound | R | EC50 (1b, μM) | CC50 (Huh-7, μM) |
|---|---|---|---|
| 5 | iso-Pr | 1.45 | > 89 |
| 6 | Me | 1.37 | > 89 |
| 7 | Et | 1.05 | > 89 |
| 8 | (S)-sec-Bu | 0.23 | > 89 |
| 9 | (R)-sec-Bu | 0.98 | > 89 |
| 10 | t-Bu | > 89 | > 89 |
| 11 | cyc-pentyl | 0.45 | > 89 |
| 12 | neo-pentyl | 0.18 | 85 |
| 13 | 2-EtBu | 0.44 | 35 |
| 14 | Bn | 0.33 | 63 |
In 2013, Cho et al. [131] prepared the first HCV inhibitor C-nucleoside GS-6620 (Fig. 22) with high efficiency in phase I clinical trials.
Fig. 22.
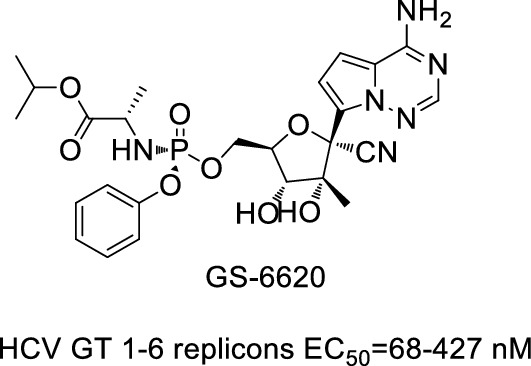
Structure and activity of GS-6620
In 2017, based on GS-6620, Siegel et al. [132] discovered and synthesized Remdesivir, and found it had good activity against Ebola virus.
Biological Activity of Remdesivir
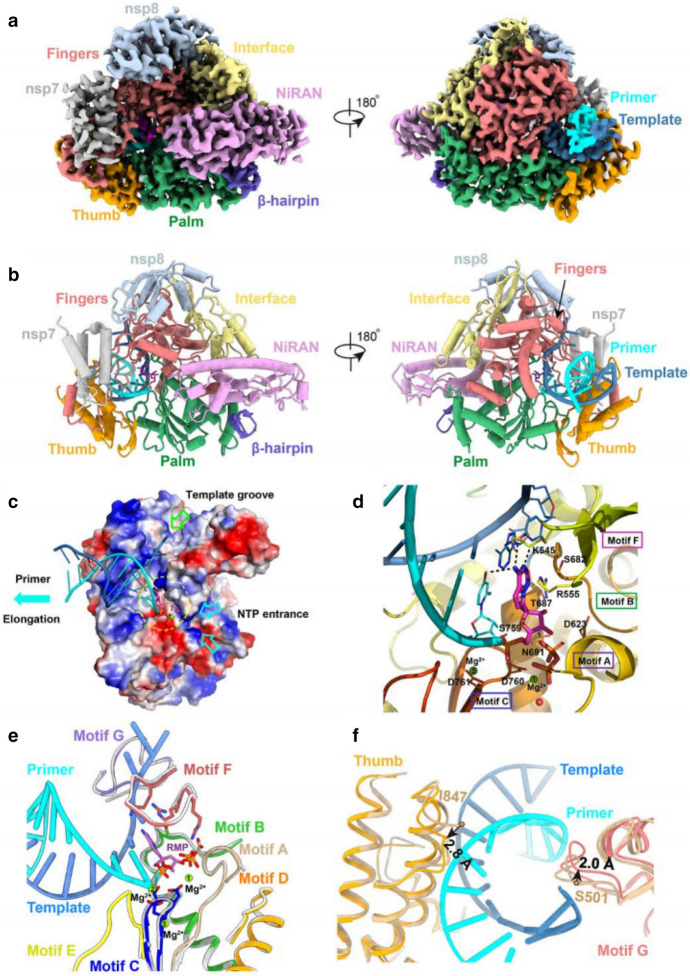
Remdesivir is RdRp inhibitor, suppressing virus genome replication [51]. RdRp controls the replication of virus RNA. Once Remdesivir metabolizes into the corresponding NTP, the latter competes with ATP for incorporation into the nascent RNA strand [133], leading to termination of virus RNA synthesis and prevention of the growth of RNA. Even though the virus can probe and delete C-nucleosides, resulting in tolerance, Remdesivir appears to be able to overcome this problem and maintain efficiency [134]. Yin et al. [135] reported the complex structure of Remdesivir and SARS-CoV-2 RdRp, where Remdesivir was covalently inserted into RdRp. This complex structure supplies a mechanism whereby Remdesivir inhibits SARS-CoV-2 replication, providing a platform for development of new drugs against COVID-19 (Fig. 23).
Fig. 23.
The complex structure of Remdesivir and SARS-CoV-2 RNA bound to RdRp complex. Image reproduced from Ref. [135] with permission from Wiley-VCH
Remdesivir has EC50 values about 0.07 μM for either SARS-CoV or MERS-CoV [136, 137]. Emmiede et al. [138] tested the efficiency of Remdesivir against MERS-CoV virus. The result showed that Remdesivir can clearly inhibit virus replication. Wang et al. [33] first examined the effect of Remdesivir against COVID-19 in Vero E6 cells and found an EC50 value 0.77 μM for Remdesivir inhibiting COVID-19. Elfiky et al. [139] targeted a few anti-polymerase drugs targeting RdRp, and found Remdesivir, Sofosbuvir, Galidesivir and Tenofovir as potent drugs against COVID-19. Choy et al. [140] reported the efficiency of Remdesivir and three other drugs against COVID-19 in Vero E6 cells, with EC50 values of 23.15 µM (Remdesivir), 26.63 µM (Lopinavir), 2.55 µM (Homorringtonine) and 0.46 µM (Emetine), respectively. Zhang et al. [141] found that the 1′-cyano group of Remdesivir has dual roles in inhibition of nucleotide addition and proofreading. Pruijssers et al. [142] reported that Remdesivir potently inhibits SARS-CoV-2 replication in human lung cells and primary human airway epithelial cultures (EC50 = 0.01 µM), in Vero E6 cells (EC50 = 1.65 µM), respectively. Wu et al. [143] found that Remdesivir and GS-441524 can inhibit cell proliferation and the expression of fibrotic markers (fibronectin, pSmad3, and aSMA) in NRK-49F and HK2 cells. Brandi et al. [144] used rhesus macaque model of COVID-19, and showed that Remdesivir can be used in transient lower respiratory tract disease.
Clinical Trials of Remdesivir
The first case of a COVID-19 patient inthe US appeared in January 2020 [90]; his condition improved after 8 days’ Remdesivir treatment, and no obvious adverse effect was observed during the treatment. Stephanie et al. [145] then described 12 US COVID-19 mild-to-moderate patients treated with Remdesivir, and all patients recovered at the end of this clinical trial. A randomized, controlled clinical trial was conducted [146] to investigate the safety and efficiency of Remdesivir against COVID-19 at the University of Nebraska Medical Center (UNMC). The clinical trial results showed no evidence that Remdesivir can improve clinical outcomes. A few phaseIII clinical trials have been conducted to evaluate the efficiency and safety of Remdesivir against mild and moderate COVID-19 patients [147–150]; the results do not support the efficacy and safety of Remdesivir. Grein et al. [151] reported 61 COVID-19 patients receiving compassionate use Remdesivir: 8 patients were not analyzed for various reasons, 22 patients were from the US, 22 patients from the EU or Canadian, 9 patients were Japanese. The results showed that 68% patients got well, 57% patients received mechanical ventilation, 47% patients were in recovery, and 13% patients died. Even though there were two absolutely contrary results in US and Chinese phase III clinical trials, on 2 May, the FDA authorized Remdesivir for compassionate use against COVID-19 for various reasons. Beige et al. [152] undertook a phase III trial using Remdesivir (dose 0.2 g daily in the first day, dose 0.1 g 2–10 days) in adult patients with COVID-19. A total of 1063 patients underwent randomization. The results, based on findings from analysis, showed that Remdesivir can shorten time to recovery. Goldman et al. [153] proceeded with a phase III clinical trial with Remdesivir (dose 0.2 g daily first day, 0.1 g the remaining days) against COVID-19 in 397 patients. Patients were allocated to two groups (10 days treatment and 5 days treatment). At baseline, prolonging treatment time did not improve clinical status.
Synthesis of Remdesivir
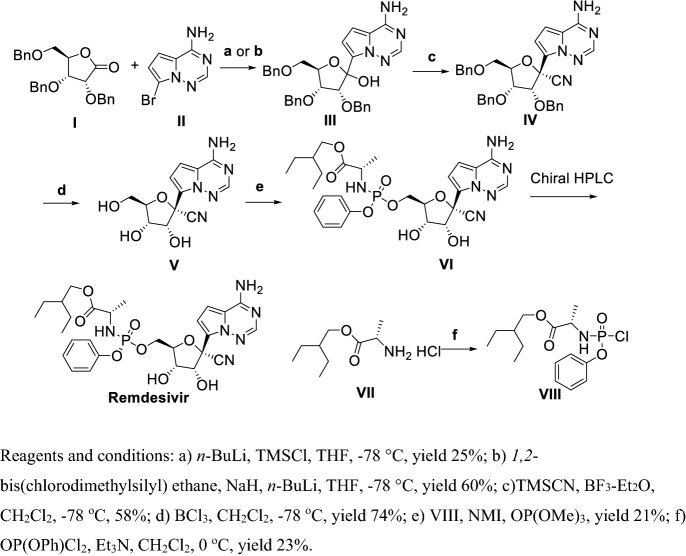
The first generation [154] for the synthesis of Remdesivir was as follows (Scheme 12). Coupling I and II in the presence of n-BuLi, and chlorotrimethylsilane (TMSCl) or 1,2-bis(chlorodimethylsilyl)ethane, NaH, and n-BuLi at ultralow temperature gave III, cyanation of III by TMSCN in the presence of Lewis acid BF3-Et2O at ultralow temperature obtained IV, benzyl deprotection using BCl3 gave V, V was reacted with VIII in the presence of NMI and OP(OMe)3 to give VI, with Remdesivir finally achieved by chiral HPLC. In the first generation for synthesizing of Remdesivir, Remdesivir was obtained in total yield of less than 2%. The first four steps were conducted at −78 ℃ and the β-anomer VII was separated by chromatography [155], which hindered this synthetic route from large-scale development.
Scheme 12.
First generation synthesis of Rendesivir
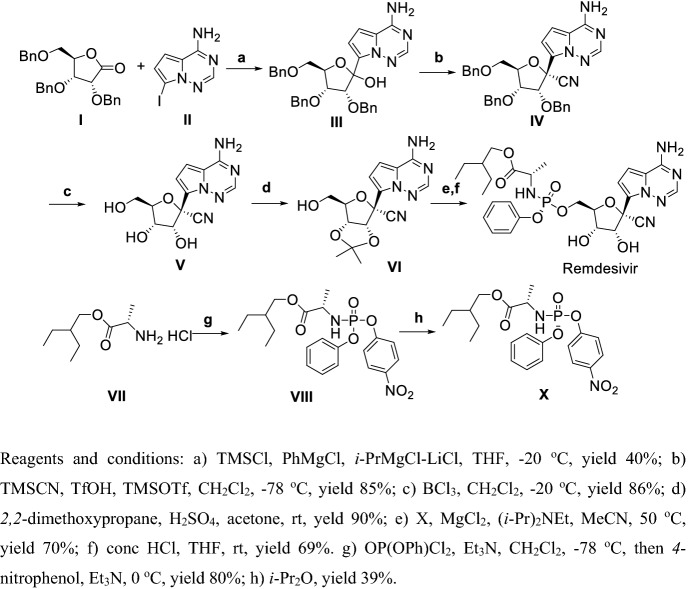
Second generation [138] Remdesivir was synthesized diastereoselectively via 7 steps in total 4% yield which is still unacceptable but suitable for large-scale production (Scheme 13). Compound I reacted with II in the presence of TMSCl and Grignard reagent at low temperature yielded III, then cyanation of III using TMSCN and TMSOTf in the presence of TfOH at ultralow temperature obtained IV, benzyl deprotection using BCl3 gave V, using 2,2-dimethoxypropane selectively protected two hydroxy of V to obtain VI, and then VI and X were reacted in the presence of DIPEA followed by deprotection to give Remdesivir.
Scheme 13.
Second generation synthesis of Remdesivir
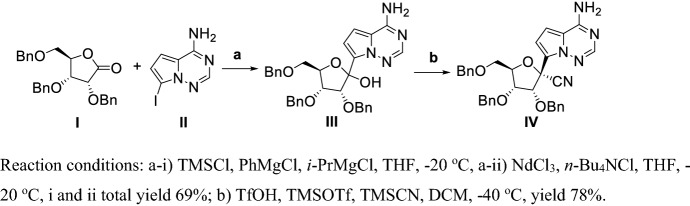
Vieira et al. [156] described a route to synthesize a key Remdesivir intermediate (IV) by using a continuous flow chemistry method, providing improved control over the reaction conditions and increasing the diastereoselectivity (Scheme 14). Coupling of I and II in the presence of TMSCl and Grignard reagent catalyzed by NdCl3 at low temperature obtained III in yields of 69%, and cyanation of III using TMSCN in the presence of TMSOTf and TfOH at lower temperatures obtained intermediate IV in yields of 78%. This synthetic route obtained the key intermediate IV in total yield 54% in two steps. But this process requires continuous flow conditions which is unacceptable in industrial production.
Scheme 14.
Large-scale cyanation process using continuous flow chemistry synthesis of Remdesivir key intermediate
Xue et al. [157] disclosed an improved methodology for the key C-glycosylation step for synthssis of Remdesivir using i-Pr2NH as a cost-effective additive in yield 75% (Scheme 15). The reaction was conducted in the presence of i-Pr2NH, and n-BuLi at ultralow temperature within 2 h using III as a protecting amino group of I. This procedure is unacceptable in large-scale production due to the ultralow temperature conditions.
Scheme 15.
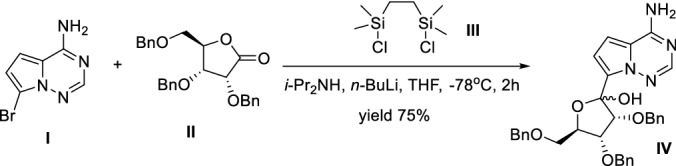
Improved methodology for the synthesis of Remdesivir key intermediate by Xue et al. [157]
Wang et al. [158] reported a gram-scale catalytic asymmetric synthesis of a key step of Remdisivir in high chiral purity and yield (Scheme 16). Compound I was reacted with II in the presence of 2,6-lutidine, and 4 Å MS catalyzed by chiral catalyst A at lower temperature gave III in 89% yield with chiral purity higher than 99%. III was then deprotected by 37% HCl to obtain Remdesivir at a yield of 73%. In this procedure, Remdesivir was synthesized asymmetrically in short steps in high yield under mild conditions at gram-scale, and is thus suitable for large-scale production.
Scheme 16.
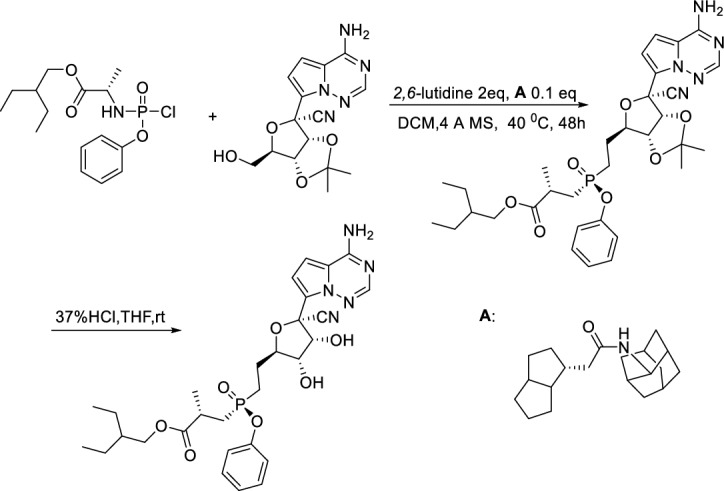
Catalytic asymmetric synthesis of a key step of Remdisivir by Wang et al. [158]
Ruxolitinib
Biological Activities of Ruxolitinib
Ruxolitinib is an FDA-approved targeting JAK1 and JAK2 inhibitor. It prevents the tyrosine phosphorylation of STAT1/3/5, which are downstream of cytokine receptors and drive T-ALL proliferation. Walker et al. [159] proved that Ruxolitinib and venetoclax worked synergistically to treat T-ALL in vitro, but were not effective in vivo. CXCR4-CXCL12 was implicated as the potential pathway that drives T-ALL infiltration into the central nervous system (CNS). By deleting the CXCR4 gene from T-ALL, they found prolonged survival in vivo with decreased neurologic clinical scores. Thus, T-ALL CNS infiltration should be blocked via CXCR4 inhibition. Ruxolitinib may be able to inhibit CXCR4 [160] (Fig. 24).
Fig. 24.
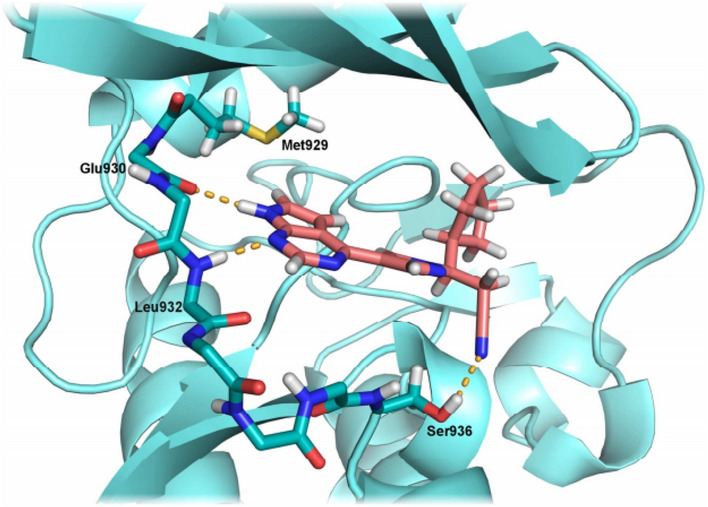
Ruxolitinib (1) docked into JAK2. Image reproduced from Ref. [160] with permission from Elsevier
The 7H-pyrrolo[2,3]pyrimidine core of Ruxolitinib links to the hinge area of Cxc Chemokin Receptor 4 (CXCR4) by hydrogen bond, the nitrile function of Ruxolitinib binds to Ser936 by hydrogen bond, the pyrazole ring linking the side chain with the hinge binding adenine mimic acts as a structural template. Since it is not involved in direct interactions with the enzyme, bioisosteric replacement with a triazole ring is a promising strategy to increase synthetic accessibility.
In in vitro experiments, Ruxolitinib was diluted into 50 μM in DMSO. In in vivo studies, Ruxolitinib dissolved in DMSO was added to 5% DMA in H2O. To survey the relevance of the JAK/STAT and BCL2 pathways on T-ALL proliferation and cell survival, Jurkat (mature T-ALL) and Loucy (early precursor T-ALL with high BCL2 expression) were assessed following treatment with Ruxolitinib. These cell lines were treated with different doses of Ruxolitinib over 72 h and assessed using a trypan blue exclusion assay and MTT (3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide) proliferation assay. Ruxolitinib decreased the survival and proliferation of both Jurkat and Loucy cell lines after 24, 48, and 72 h of treatment in 2.5 μM. Ruxolitinib failed to treat T-ALL in vivo because of leukemia CNS infiltration. In 2011, the FDA and European Food and Drug Administration (EFDA) approved Ruxolitinib first in class in treatment of myelofibrosis [161]. Tuttle et al. [162] used two mouse models to prove that interferon receptor genes are overexpressed in mice, and that JAK1 inhibitors can clearly restrain cytokine storm. Increased evidence proves that mortality with COVID-19 infections is caused mainly by the overexpression of a immune response to SARS-CoV-2, resulting in a cytokine storm and ARDS [163]. Many cytokines and chemokines involved in the cytokine storm employ JAKs for signal transduction. Cytokine analysis of COVID-19 patients shows that C-reactive protein (CRP) and interleukin (IL)-6 levels are significantly higher in patients who eventually died compared to those who survived [50]. Similar to some other mortal lung infections, the overexpression of immune response to the COVID-19 virus causes a cytokine storm, along with infiltration and activation of diverse immune cells, then generation secondary cell factors and chemotactic factors [164]. In this study, patients admitted to intensive care units (ICUs) showed significantly higher levels of IL-2, IL-10, IL-7, IP-10, MCP-1, MIP-1α, G-CSF, and TNF-α relative to non-ICU patients. All in all, these discoveries support the combination of antiviral treatment and targeted immunosuppression as a therapeutic program in COVID-19 [165].
Clinical Trials of Ruxolitinib
Several clinical trials have evaluated the safety and efficacy of Ruxolitinib of IL-6 and JAK/STAT signaling. Jung et al. [50] conducted a phase II clinical trial to evaluate Ruxolitinib treatment of myelofibrosis. Their results proved the efficiency of Ruxolitinib to treat myelofibrosis. Studies on Ruxolitinib treatment of sHLH patients showed encouraging results on overall survival [166]. Cao et al. [167] carried out a phaseII clinical trial to treat severe COVID-19 patients using Ruxolitinib. A total of 20 patients were distributed to the Ruxolitinib and standard of care (SoC) group, 21 patients were distributed to placebo group (SoC treatment) randomly. The results showed that the patients in Ruxolitinib and SoC group recovered faster than the placebo group. Most importantly, there were no deaths in the Ruxolitinib and SoC group.
Synthesis of Ruxolitinib
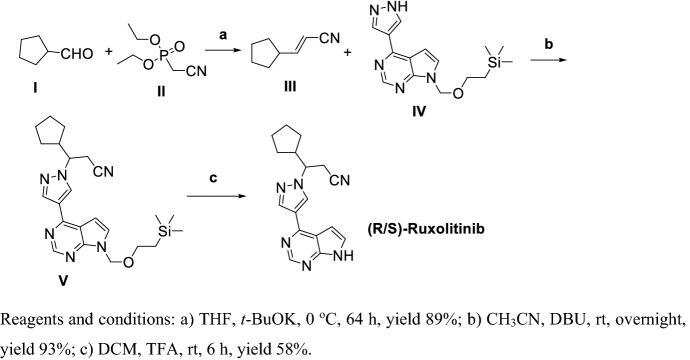
Rodgers et al. [168] synthesized racemic Ruxolitinib via three steps with a total yield of 48% (Scheme 17). Condensation of I with malonic acid in strong organic base condition obtained III, then III reacted with IV in the presence of 1,8-Diazabicyclo[5.4.0]undec-7-ene (DBU) gave V; finally, V was deprotected by TFA to racemate Ruxolitinib with a total yield of 48%.
Scheme 17.
Preparation of racemic Ruxolitinib by Rodgers et al. [168]
Haydl et al. [169] reported Rhodium-catalyzed asymmetric coupling of (I) with pyrazole derivatives (II) giving enantioenriched allylic pyrazoles, which can be used to synthesize the targeted drug Ruxolitinib (Scheme 18). They developed a Rhodium-catalyzed, enantioselective synthesis of (R)-Ruxolitinib in the presence of chiral ligand using cheap material I, II and V in high chiral purity and yield. Above all, there were only three reaction steps.
Scheme 18.
Gram-scale synthesis of (R)-Ruxolitinib by Haydl et al. [169]
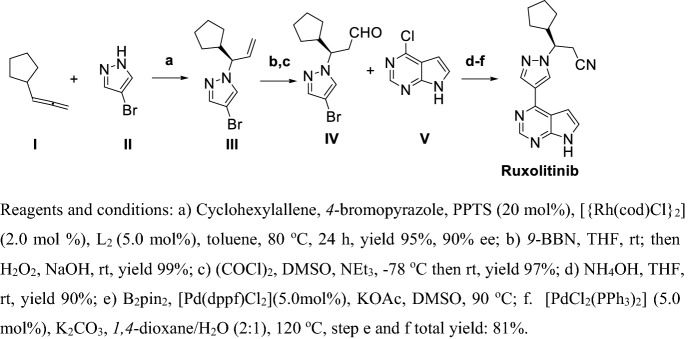
Deepshikha et al. [170] provided a method for preparation of Ruxolitinib and its phosphate giving a chiral purity of 99.96% but with a total yield as low as 5%, which is unacceptable (Scheme 19). In this synthetic route, I was protected by 2-(trimethylsilyl)ethoxymethyl chloride, then direct Suzuki coupling with III gave compound IV with a yield of 80.8% in two steps. IV reacted with V in base condition, deprotection with Lewis acid BF3-Et2O and treated with (+)-2,3-dibenzoyl-d-tartaric acid to obtain X with a yield as low as 7% in three steps. Finally, compound IV was chirally resolved and treated with phosphoric acid to give Ruxolitinib phosphate with yield 88% in two steps.
Scheme 19.
Preparation of Ruxolitinib and its phosphate by Deepshikha et al. [170]
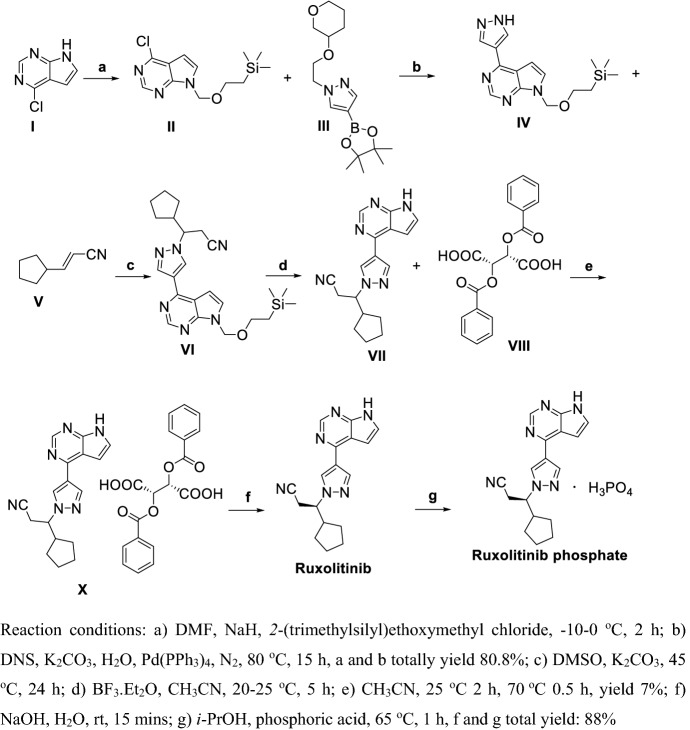
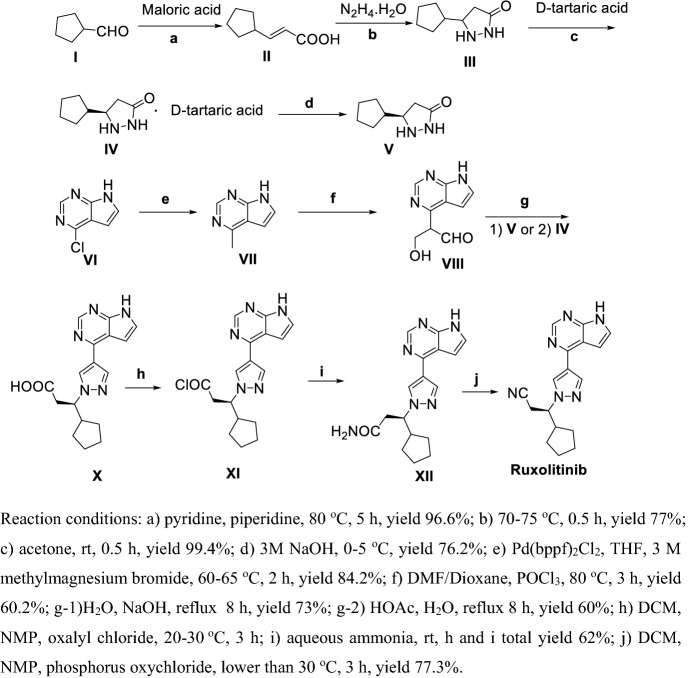
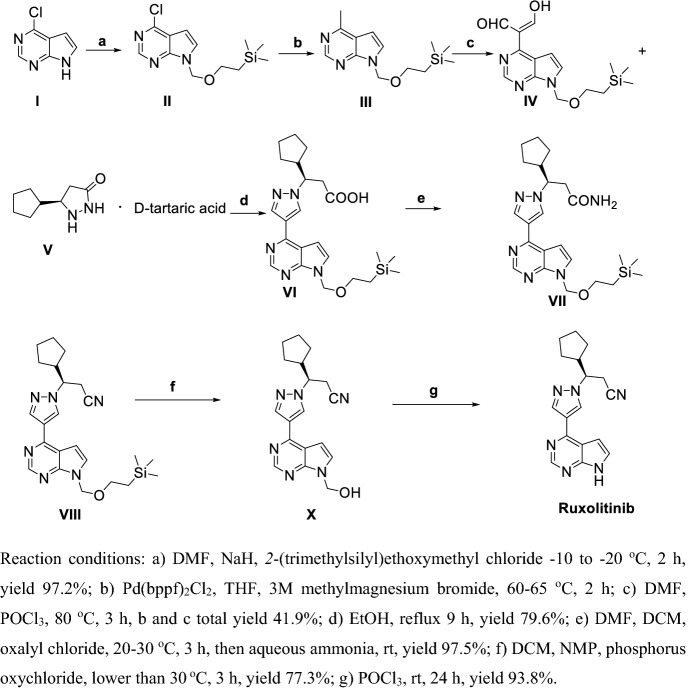
Zhang et al. [171] reported two methods for preparation of Ruxolitinib, which have characteristics such as high stereoselectivity, mild reaction conditions and convenient post treatment, avoiding use expensive asymmetric catalysts and suitable for industrial production (Schemes 20, 21). As shown in Scheme 20, condensation of I with malonic acid in weak organic base condition gave II, which directed ring-closure with hydrazine hydrate in quantitative yield to III. Chiral resolution of III by using d-tartaric acid then gave V. Methylation of VI using methylmagnesium bromide gave VII, then VIII was obtained via Vilsmeier–Haack reaction. VIII and IV or V refluxed in base or acid conditions obtained X, then acyl chlorination using oxalyl chloride and amidation obtained XII. Finally, dehydration of XII obtained Ruxolitinib.
Scheme 20.
Preparation of Ruxolitinib by Zhang et al. [171] synthetic route one
Scheme 21.
Preparation of Ruxolitinib by Zhang et al. [171] synthetic route 2
As shown in Scheme 21, compound I was protected by 2-(trimethylsilyl)ethoxymethyl chloride to II. Methylation of II using methylmagnesium bromide gave III, then IV was obtained via Vilsmeier–Haack reaction. V and IV or V were refluxed in ethanol to obtain VI, and then amidation and dehydration gave VII. Finally, deprotection of VII obtained Ruxolitinib.
Conclusions
After the spread of COVID-19 worldwide, tens of thousands medical scientists and pharmacologists have made great efforts to search for potent drugs that can inhibit COVID-19, and they successfully found a few drugs like CLQ, CLQ-OH, Favipiravir and Remdesivir that are useful in the treatment of COVID-19 patients. But, in clinical trials to date, CLQ and CLQ-OH may increase mortality due to their high doses (CLQ 500 mg dose, twice daily; CLQ-OH 400–600 mg dose). Favipiravir has high efficiency in treating Chinese patients, but it also needed a high dose (600 mg dose, 2–3 times daily); especially, there is a lack of clinical data to prove its efficacy and safety outside of China. Gilead Sciences conducted clinical trials with Remdesivir after the spread of COVID-19 worldwide, and the FDA authorized Remdesivir for compassionate use in treating COVID-19 patients in May 2020. To date, COVID-19 patients who die with cytokine storm syndrome have higher levels of IL-6 in plasma; four JAK inhibitors, Ruxolitinib, Tofacitinib, Baricitinib and Upadacitinib, have proved useful in the treatment of COVID-19 patients. Most importantly, there were no deaths using Ruxolitinib to treat COVID-19 patients in a phase II clinical trial.
As far as we know, there is no specific medicine for treatment of COVID-19 patients, but COVID-19 is still spreading rapidly worldwide. Combinations of antivirial drugs such as Remdesivir or GS-441524 and JAK inhibitor drugs may be a useful therapentic schedule to reduce mortality before a specific medicine appears.
In this review, we introduced lots of small molecules that exhibit potent activity in inhibiting COVID-19, especially CLQ, CLQ-OH, Favipiravir, Remdesivir and Ruxolitinib, presenting their biological activities, clinical trials and synthesis processes, which may help researchers to systematically understand the processes of these potential drugs.
Acknowledgments
The authors thank the Natural Science Foundation of China (21676076, 21878071, and 21971060), Hu-Xiang High Talent in Hunan Province (2018RS3042) and Recruitment Program of China (WQ20164300353) for financial support. The authors acknowledge Prof. Nobuaki Kambe (Osaka University) for helpful discussion.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ARDS
Acute respiratory distress syndrome
- BCL-2
β-Cell lymphoma 2
- CLQ-OH
Hydroxychloroquine
- CNS
Central nervous system
- COVID-19
Corona virus disease 2019
- CRP
Levels of C-reactive protein
- CXCR4
Cxc chemokin receptor 4
- CXCL-12
Cxc chemokin ligand 12
- DAAs
Direct-acting antivirals
- EC50
Concentration for 50% of maximal effect
- EC90
Concentration for 90% of maximal effect
- EFDA
European Food and Drug Administration
- FDA
Food and Drug Administration
- G-CSF
Granulocyte colony-stimulating factor
- hrs ACE2
Human recombinant soluble angiotensin-converting enzyme 2
- IC50
The half maximal inhibitory concentration
- ICU
Intensive care unit
- IL
Interleukins
- INF
Interferon
- JAK
Janus kinase
- MCP-1
Monocyte chemoattractant protein-1
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MIP-1α
Recombinant human chemokine CCL3
- PBPK
Physiologically-based pharmacokinetic models
- RBD
Receptor binding domain
- RdRp
RNA dependent RNA polymerase
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- STAT
Signal transducer and activator of transcription
- SHLH
Secondary hemophagocytic lymphohistiocytosis
- T-ALL
T cell acute lymphoblastic leukemia
- TNF
Tumor necrosis factor
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianbing Hu, Email: 378704627@qq.com.
Weijun Yang, Email: wjyang@hnu.edu.cn.
Renhua Qiu, Email: qiurh@qq.com, Email: renhuaqiu1@hnu.edu.cn.
References
- 1.Hussin AR, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao P, Wu S, Wu T, Deng Y, Zhang Q, Wang K, Zhang Y. The important role of polysaccharides from a traditional Chinese medicine-lung cleansing and detoxifying decoction against the COVID-19 pandemic. Carbohyd Polym. 2020 doi: 10.1016/j.carbpol.2020.116346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eriko PR. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect Dis Ther. 2020 doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhama K, Sharun K, Tiwari R, Dadar M, Malik YS, Singh KP, Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vacc Immunother. 2020 doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan K, Liu BD, Li CS, Zhang HJ, Yu T, Qu JM, Zhoug M, Chen L, Meng SL, Hu Y, Peng C, Yuan MC, Huang JY, Wang ZJ, Yu JH, Gao XX, Wang D, Yu XQ, Li L, Zhang JY, Wu X, Li B, Xu YP, Chen W, Peng Y, Hu YQ, Lin LZ, Liu XF, Huang SH, Zhou ZJ, Zhang LH, Wang Y, Zhang Z, Deng K, Xia ZW, Gong Q, Zhang W, Zheng XB, Liu Y, Yang HC, Zhou DB, Yu D, Hou JF, Shi ZL, Chen SJ, Chen Z, Zhang XX, Yang XM. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA. 2020;117(17):9490–9494. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Q, He Y. Challenges of convalescent plasma therapy on COVID-19. J Clin Virol. 2020;127:104358–104363. doi: 10.1016/j.jcv.2020.104358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. 2020;20(4):398–400. doi: 10.1016/S1473-3099(20)30141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, Zhou QQ, Li YZ, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Martin S, Zand MD. The potential for antibody-dependent enhancement of SARS-CoV-2 infection: translational implications for vaccine development. Infect Control Hosp Epidemiol. 2020 doi: 10.1017/cts.2020.39. [DOI] [Google Scholar]
- 10.He R, Lu Z, Zhang L, Fan T, Xiong R, Shen X, Feng H, Meng H, Lin W, Jiang W, Geng Q. The clinical course and its correlated immune status in COVID-19 pneumonia. J Clin Virol. 2020 doi: 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao XT. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020 doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lythgoe MP, Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol Sci. 2020 doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Tian Z, Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 14.Yao XT, Ye F, Zhang M, Cui C, Huang BY, Niu PH, Liu X, Zhao L, Dong ED, Song CL, Zhan SY, Lu RJ, Li HY, Tan WJ, Liu DY. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome main point: hydroxychloroquine was found to be more potent than chloroquine at inhibiting SARS-CoV-2 in vitro. Clin Infect Dis. 2020;2:1–25. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Dupont HT, Honoré S, Colson P, Chabriére E, Scola BL, Rolain JM, Brouqui P, Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;1:1. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Olano J, Howland MA, Su MK, Hoffman RS, Biary R. Toxicokinetics of hydroxychloroquine following a massive overdose. J Am Emerg Med. 2019;37(12):2264–2264. doi: 10.1016/j.ajem.2019.158387. [DOI] [PubMed] [Google Scholar]
- 17.Shah B, Modi P, Sagar SR. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anson BJ, Chapman ME, Lendy EK, Pshenychnyi S, Aquila RTD, Satchell KJF, Mesecar AD. Broad-spectrum inhibition of coronavirus main and papain-like proteases by HCV drugs. Res Square. 2020 doi: 10.21203/rs.3.rs-26344/v1. [DOI] [Google Scholar]
- 19.Liu SF, Lien CZ, Selvaraj P, Wang TT. Evaluation of 19 antiviral drugs against SARS-CoV-2 infection. BioRxiv. 2020 doi: 10.1101/2020.04.29.067983. [DOI] [Google Scholar]
- 20.Sacramento CQ, Fintelman-Rodrigues N, Temerozo JR, Dias SDSG, Ferreira AC, Mattos M, Pão CRR, Freitas CSD, Soares VC, Bozza FA, Bou-Habib DC, Bozza PT, Souza TML. The in vitro antiviral activity of the anti-hepatitis C virus (HCV) drugs daclatasvir and sofosbuvir against SARS-CoV-2. BioRxiv. 2020 doi: 10.1101/2020.06.15.153411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurung AB. In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep. 2020;21:100860–100871. doi: 10.1016/j.genrep.2020.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo HS, Hui KPY, Lai HM, Khan KS, Kaur S, Huang JZ, Li ZQ, Chan AKN, Cheung HHY, Ng KC, Ho JCW, Chen YW, Ma BW, Cheung PMH, Shin D, Wang KD, Lee MH, Selisko B, Eydoux C, Guillemot JC, Canard B, Wu KP, Liang PH, Dikic I, Zuo Z, Chan FKL, Hui DSC, Mok VCT, Wong KB, Aik WS, Chan MCW, Ng WL. Simeprevir potently suppresses SARS-CoV-2 replication and synergizes with remdesivir. BioRxiv. 2020 doi: 10.1101/2020.05.26.116020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie XP, Muruato AE, Zhang XW, Lokugamage KG, Fontes-Garfias CR, Zou J, Liu JY, Ren P, Balakrishnan M, Cihlar T, Tseng CTK, Makino S, Menachery VD, Bilello JP, Shi PY. A Nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. BioRxiv. 2020 doi: 10.1101/2020.06.22.165712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez S, Fernandez-Antunez C, Phama LC, Ryberga LA, Fenga S, Pedersena MS, Mikkelsena LS, Belouzardc S, Dubuissonc J, Gottweina JM, Fahnøe U, Bukh J. Efficient culture of SARS-CoV-2 in human hepatoma cells enhances viability of the virus in human lung cancer cell lines permitting the screening of antiviral compounds. BioRxiv. 2020 doi: 10.1101/2020.10.04.325316. [DOI] [Google Scholar]
- 25.Yamamoto N, Matsuyama S, Hoshino T, Yamamoto N. Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. BioRxiv. 2020 doi: 10.1101/2020.04.06.026476. [DOI] [Google Scholar]
- 26.Hu F, Jiang JX, Yin P (2020) Prediction of potential commercially inhibitors against SARS-CoV-2 by multi-task deep model. ARxiv [DOI] [PMC free article] [PubMed]
- 27.Meyer SD, Bojkova D, Cinatl J, Dammea EV, Buyck C, Loock MV, Woodfall B, Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int J Infect Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risner KH, Tieu KV, Wang YF, Bakovic A, Alem F, Bhalla N, Nathan S, Conway DE, Macklin P, Narayanan A. Maraviroc inhibits SARS-CoV-2 multiplication and S-protein mediated cell fusion in cell culture. BioRxiv. 2020 doi: 10.1101/2020.08.12.246389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Almeida SMVD, Soares JCS, Santos KLD, Alves JEF, Ribeirob AG, Jacob ÍTT, Ferreiraa CJDS, Santos JCD, Oliveirab JFD, Junior LBDC, Lima MDCAD. COVID-19 therapy: what weapons do we bring into battle? Bioorg Med Chem. 2020;28:115757–115782. doi: 10.1016/j.bmc.2020.115757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekins S, Mottin M, Ramos PRPS, Sousa BKP, Neves BJ, Foil DH, Zorn KM, Braga RC, Coffee M, Southan C, Puh AC, Andrade CH. Déjà Vu: stimulating open drug discovery for SARS-CoV-2. Drug Discov Today. 2020;25:928–941. doi: 10.1016/j.drudis.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodon J, Noguera-Julian M, Erkizia I, Valencia A, Guallar V, Carrillo J, Blanco J, Segalés J, Clotet B, Vergara-Alert J, Izquierdo-Useros N. Search for SARS-CoV-2 inhibitors in currently approved drugs to tackle COVID-19 pandemia. BioRxiv. 2020 doi: 10.1101/2020.04.23.055756. [DOI] [Google Scholar]
- 32.Wang DD, Huang JS, Yeung AWK, Tzvetkov NT, Horbánczuk JO, Willschke H, Gai ZB, Atanasov AG. The significance of natural product derivatives and traditional medicine for COVID-19. Processes. 2020;8:937–961. doi: 10.3390/pr8080937. [DOI] [Google Scholar]
- 33.Wang ML, Cao R, Zhang L, Yang X, Liu J, Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stebbing J, Krishnan V, Bono SD, Ottaviani S, Casalini G, Richardson PJ, Monteil V, Lauschke VM, Mirazimi A, Youhanna S, Tan YJ, Baldanti F, Sarasini A, Terres JAR, Nickoloff BJ, Higgs RE, Rocha G, Byers NL, Schlichting DE, Nirula A, Cardoso A, Corbellino M. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. 2020;12(8):12697–12731. doi: 10.15252/emmm.202012697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M, Hofmann-Winkler H, Smith JC, Krüger N, Sørensen LK, Søgaard OS, Hasselstrøm JB, Winkler M, Hempel T, Raich L, Olsson S, Yamazoe T, Yamatsuta K, Mizuno H, Ludwig S, Noé F, Sheltzer JM, Kjolby M, Pöhlmann S. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. BioRxiv. 2020 doi: 10.1101/2020.08.05.237651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amin SA, Jha T. Fight against novel coronavirus: a perspective of medicinal chemists. Eur J Med Chem. 2020;201:112559–112570. doi: 10.1016/j.ejmech.2020.112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan VC, Muller FL. Advantages of the parent nucleoside GS-441524 over remdesivir for Covid-19 treatment. ACS Med Chem Lett. 2020;11:1361–1366. doi: 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riva L, Yuan SF, Yin X, Martin-Sancho L, Matsunaga N, Pache L, Burgstaller-Muehlbacher S, Jesus PDD, Teriete P, Hull MV, Hang MW, Chan JFW, Cao JL, Poon VKM, Herbert KM, Cheng KY, Nguyen TTH, Rubanov A, Pu Y, Nguyen C, Choi A, Rathnasinghe R, Schotsaert M, Miorin L, Dejosez M, Zwaka TP, Sit KY, Martinez-Sobrido L, Liu WC, White KM, Chapman ME, Emma K, Lendy EK, Glynne RJ, Albrecht R, Ruppin E, Mesecar AD, Johnson JR, Benner C, Ren Sun R, Schultz PG, Su AI, García-Sastre A, Chatterjee AK, Yuen KY, Chanda SK. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020 doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin ZM, Du XY, Xu YC, Deng YQ, Liu MQ, Zhao Y, Zhang B, Li XF, Zhang LK, Peng C, Duan YK, Yu J, Wang L, Yang KL, LiuFJ JRD, Yang XL, You T, Liu XC, Yang XN, Bai F, Liu H, Liu X, Guddat LW, Xu WQ, Xiao GF, Qin CF, Shi ZL, Jiang HL, Rao ZH, Yang HT. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2019 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 40.Jin ZM, Zhao Y, Sun Y, Zhang B, Wang HF, Wu Y, Zhu Y, Zhu C, Hu TY, Du XY, Duan YK, Yu J, Yang XB, Yang XN, Yang KL, Liu X, Guddat LW, Xiao GF, Zhang LK, Yang HT, Rao ZH. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. BioRxiv. 2020 doi: 10.1101/2020.04.09.033233. [DOI] [PubMed] [Google Scholar]
- 41.Su HX, Yao S, Zhao WF, Li MJ, Liu J, Shang WJ, Xie H, Ke CQ, Gao MN, Yu KQ, Liu H, Shen JS, Tang W, Zhang LK, Zuo JP, Jiang HL, Bai F, Wu Y, Ye Y, Xu YC. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. BioRxiv. 2020 doi: 10.1101/2020.04.13.038687. [DOI] [Google Scholar]
- 42.Dai WH, Zhang B, Jiang XM, Su HX, Li J, Zhao Y, Xie X, Jin ZM, Peng JJ, Liu FJ, Li CP, Li Y, Bai F, Wang HF, Cheng X, Cen XB, Hu SL, Yang XN, Wang J, Liu X, Xiao GF, Jiang HL, Rao ZH, Zhang LK, Xu YC, Yang HT, Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang LL, Lin DZ, Sun XY, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitner EB, Avraham R, Achdout H, Tamir H, Agami A, Cherry L, Yahalom-Roen Y, Politi B, Erez N, Melamed S, Paran N, Israely T. Antiviral activity of glucosylceramide synthase inhibitors against SARS-CoV-2 and other RNA virus infections. BioRxiv. 2020 doi: 10.1101/2020.05.18.103283. [DOI] [Google Scholar]
- 45.He F, Deng Y, Li W. Coronavirus disease 2019 (COVID-19): what we know? J Med Virol. 2020 doi: 10.1002/jmv.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson HLH. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seguin A, Galicier L, Boutboul D, Lemiale V, Azoulay E. Pulmonary involvement in patients with hemophagocytic lymphohistiocytosis. Chest. 2016;149(5):1294–1301. doi: 10.1016/j.chest.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peterson D, Damsky W, King B. The use of Janus kinase inhibitors in the time of SARS-CoV-2. J Am Acad Dermatol. 2020;82(6):223–226. doi: 10.1016/j.jaad.2020.03.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung CW, Shih LY, Xiao ZJ, Jie J, Hou HA, Du X, Wang MC, Park S, Eom KS, Oritani K, Okamoto S, Tauchi T, Kim JS, Zhou DB, Saito S, Li JM, Handa H, Li JY, Ohishi K, Hou M, Wu DP, Takenaka K, Liu T, Hu Y, Amagasaki T, Ito K, Gopalakrishna P, Akashi K. Efficacy and safety of ruxolitinib in asian patients with myelofibrosis. Leuk Lymphoma. 2014;56(7):2067–2074. doi: 10.3109/10428194.2014.969260. [DOI] [PubMed] [Google Scholar]
- 51.Amirian ES, Levy JK. Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses. One Health. 2020;9:100128–100134. doi: 10.1016/j.onehlt.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paules CI, Marston HD, Fauci AS. Coronavirus infections more than just the common cold. J Am Med Assoc. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 53.Browna AJ, Wona JJ, Grahama RL, Dinnon KH, III, Sims AC, Feng JY, Cihlar T, Denisonc MR, Barica RS, Sheahan TP. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic delta-coronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2020;169:104541–104550. doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baranov PV, Henderson CM, Anderson CB, Gesteland RF, Atkins JF, Howard MT. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li GD, Clercq ED. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nature. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 56.Morse JS, Lalonde T, Shiqing X, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chem Bio Chem. 2020 doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen KY. Coronaviruses drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 59.Kaul D. An overview of coronaviruses including the SARS-2 coronavirus molecular biology, epidemiology and clinical implications. Curr Med Res Pract. 2020;10:54–64. doi: 10.1016/j.cmrp.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang XQ, Pearce R, Zhang Y. Computational design of peptides to block binding of the SARS-CoV-2 spike protein to human ACE2. BioRxiv. 2020 doi: 10.1101/2020.03.28.013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mercurio I, Tragni V, Busco F, Grassi AD, Pierri CL. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. Cell Mol Life Sci. 2020 doi: 10.1101/2020,04(17),pp.046185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laurini E, Marson D, Aulic S, Fermeglia M, Pricl S. Computational alanine scanning and structural analysis of the SARS-CoV-2 spike protein/angiotensin-converting enzyme 2 complex. ACS Nano. 2020;14:11821–11830. doi: 10.1021/acsnano.0c04674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Pozo CHD, Prosper F, Romero JP, Wirnsberger G, Zhang HB, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pooladanda V, Thatikonda S, Godugu C. The current understanding and potential therapeutic options to combat COVID-19. Life Sci. 2020;254:117765–117783. doi: 10.1016/j.lfs.2020.117765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vincent MJ, Bergeron E, Benjannet S, Erickson BR, Rollin PE, Ksiazek TG, Seidah NG, Nichol ST. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. J Virol. 2005;2:69–78. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dowall SD, Bosworth A, Watson R, Bewley K, Taylor I, Rayner E, Hunter L, Pearson G, Easterbrook L, Pitman J, Hewson R, Carroll MW. Chloroquine inhibited ebola virus replication in vitro but failed to protect against infection and disease in the in vivo guinea pig model. J Gen Virol. 2015;96:3484–3492. doi: 10.1099/jgv.0.000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ooi EE, Chew JSW, Loh JP, Chua RCS. In vitro inhibition of human influenza A virus replication by chloroquine. J Virol. 2006;3:39–41. doi: 10.1186/1743-422X-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Romanelli F, Smith KM, Hoven AD. Chloroquine and hydroxychloroquine as inhibitors of human immuno-deficiency virus (HIV-1) activity. Curr Pharm Des. 2004;10:2643–2648. doi: 10.2174/1381612043383791. [DOI] [PubMed] [Google Scholar]
- 69.Yan YW, Zou Z, Sun Y, Li X, Xu KF, Wei YQ, Jin NY, Jiang CY. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patil VM, Singhal S, Masand N. A Systematic review on use of aminoquinolines for the therapeutic management of COVID-19: efficacy, safety and clinical trials. Life Sci. 2020;254:117775. doi: 10.1016/j.lfs.2020.117775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt RLJ, Jutz S, Goldhahn K, Witzeneder N, Gerner MC, Trapin D, Greiner G, Hoermann G, Steiner G, Pickl WF, Burgmann H, Steinberger P, Ratzinger F, Schmetterer KG. Chloroquine inhibits human CD4+ T-cell activation by AP-1 signaling modulation. Sci Rep. 2017;7:42191–42204. doi: 10.1038/srep42191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, Nardo DD, Gohel TD, Emde M, Schmidlethner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-based network analysis reveals a spectrummodel of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Velthuis AJWT, van den Worm SHE, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11):1001176–1001186. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fantini J, Scala CD, Chahinian H, Yahi N. Structural and molecular modeling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55:105960–105967. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun XL, Li SQ, Li KX, Hu X. Pharmaceutical care of chloroquine phosphate in elderly patients with coronavirus pneumonia (COVID-19) Aging Med. 2020;3:98–101. doi: 10.1002/agm2.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verscheijden LFM, Van de Zanden TM, Van Bussel LPM, De Hoop-Sommen M, Russel FGM, Johnson TN, De Wildt SN. Chloroquine dosing recommendations for pediatric COVID-19 supported by modeling and simulation. Clin Pharmacol Ther. 2020;108(2):248–252. doi: 10.1002/cpt.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borba MGS, Val FFA, Sampaio VS, Alexandre MAA, Melo GC, Brito M, Mourao D, Brito-Sousa D, Guerra MVF, Hajjar LA, Pinto RC, Balieiro AAS, Naveca FG, Xavier MS, Salomao A, Siqueira AM, Schwarzbolt A, Croda JHR, Nogueira ML, Romero GAS, Bassat Q, Fontes CJ, Albuqueerque BCA, Daniel-Ribeiro CT, Monteiro WM, Lacerda MVG, CloroCovid-19 Team Chloroquine diphosphate in two different dosages as adjunctive therapy of hospitalized patients with severe respiratory syndrome in the context of coronavirus (SARS-CoV-2) infection: preliminary safety results of a randomized,double-blinded, phase IIb clinical trial (CloroCovid-19 Study) medRxiv. 2020 doi: 10.1101/2020.04.07.20056424. [DOI] [Google Scholar]
- 78.Huang MX, Li M, Xiao F, Liang JB, Pang PF, Tang TT, Liu SX, Chen BH, Shu JX, You YY, Li Y, Tang MW, Zhou JH, Jiang GM, Xiang JF, Hong WX, He SM, Wang ZQ, Feng JH, Lin CQ, Ye YN, Wu ZL, Li YC, Zhong B, Sun RL, Hong ZS, Liu J, Chen HL, Wang XH, Li ZH, Pei DQ, Tian L, Xia JY, Jiang SP, Zhong NS, Shan H. Preliminary evidence fom a multicenter prospective observational study of the safety and efficacy of chloroquine for the treatment of COVID-19. Natl Sci Rev. 2020;7(9):1428–1436. doi: 10.1093/nsr/nwaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smit C, Peeters MYM, Anker JNVD, Knibbe CAJ. Chloroquine for SARS-CoV-2: implications of its unique pharmacokinetic and safety properties. Clin Pharmacokinet. 2020;59:659–669. doi: 10.1007/s40262-020-00891-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sinkeler FS, BergerFA MHJ, Jansen MMPM. The risk of QTc-interval prolongation in COVID-19 patients treated with chloroquine. Netherlands Heart J. 2020;28:418–423. doi: 10.1007/s12471-020-01462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guastalegname M, Vallone A. Could chloroquine/hydroxychloroquine be harmful in coronavirus disease 2019 (COVID-19) treatment? Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Surrey AR, Hammer HF. Some 7-substituted 4-aminoquinoline derivatives. J Am Chem Soc. 1946;68:113–116. doi: 10.1021/ja01205a036. [DOI] [PubMed] [Google Scholar]
- 83.Jonnson WS, Buell BG. A new synthesis of chloroquine. J Am Chem Soc. 1952;74:4513–4516. doi: 10.1021/ja01138a014. [DOI] [Google Scholar]
- 84.Margolis BJ, Long KA, Laird DL, Ruble JC, Pulley SR. Assembly of 4-aminoquinolines via palladium catalysis: a mild and convenient alternative to SNAr methodology. J Org Chem. 2007;72:2232–2235. doi: 10.1021/jo062168u. [DOI] [PubMed] [Google Scholar]
- 85.Biot C, Daher W, Chavain N, Fandeur T, Khalife J, Dive D, De Clercq E. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J Med Chem. 2006;49(9):2845–2849. doi: 10.1021/jm0601856. [DOI] [PubMed] [Google Scholar]
- 86.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, Zhan S, Lu R, Li H, Tan W, Liu D. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71(15):732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang XL, Li ZM, Ye JT, Lu J, Ye LYLD, Zhang CX, Liu PQ, Duan DYD. Pharmacological and cardiovascular perspectives on the treatment of COVID-19 with chloroquine derivatives. Acta Pharmacol Sin. 2020;41:1377–1386. doi: 10.1038/s41401-020-00519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapoor A, Pandurangi U, Arora V, Gupta A, Jaswal A, Nabar A, Naik N, Namboodiri N, Vora A, Yadav R, Saxena A. Cardiovascular risks of Hydroxychloroquine in treatment and prophylaxis of COVID-19 patients: a scientific statement from the Indian Heart Rhythm Society. Ind Pac Electrophys J. 2020 doi: 10.1016/j.ipej.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Sevestre J, Mailhe M, Doudier B, Aubry C, Amrane S, Seng P, Hocquart M, Eldin C, Finance J, Vieira VE, Tissot-Dupont HT, Honoré S, Stein A, Million M, Colson P, Scola BL, Veit V, Jacquier A, Deharo JC, Drancourt M, Fournier PE, Rolain JM, Brouqui P, Raoult D. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycin in 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med Infect Dis. 2020;34:101663–101669. doi: 10.1016/j.tmaid.2020.101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, Xu QH, Sun ZY, Zhou L. Current targeted therapeutics against COVID-19: based on first-line experience in China. Pharmacol Res. 2020;157:104854–104860. doi: 10.1016/j.phrs.2020.104854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen Z, Hu J, Zhang Z, Jiang S, Han S, Yan D, Zhuang R, Hu B, Zhang Z. Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial. medRxiv. 2020 doi: 10.1101/2020.03.22.20040758. [DOI] [Google Scholar]
- 92.Adel FA, Shoughy SS, Tabbara KF. Hydroxychloroquine dosing and toxicity: a real-world experience in Saudi Arabia of 63 patients. Saudi J Ophthalmol. 2020 doi: 10.1016/j.sjopt.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Derendorf H. Excessive lysosomal ion-trapping of hydroxychloroquine and azithromycin. Int J Antimicrob Agents. 2020;55:106007–106011. doi: 10.1016/j.ijantimicag.2020.106007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Molina JM, Delaugerre C, Goff JL, Mela-Lima B, Ponscarme D, Goldwirt L, de Castro N. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Méd Malad Infect. 2020;50:382–387. doi: 10.1016/j.medmal.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sharma AN, Mesinkovska NA, Paravar T. Characterizing the adverse dermatologic effects of hydroxychloroquine: a systematic review. J Am Acad Dermatol. 2020 doi: 10.1016/j.jaad.2020.04.024. [DOI] [PubMed] [Google Scholar]
- 96.Singh AK, Singh A, Singh R, Misra A. Hydroxychloroquine in patients with COVID-19: a systematic review and meta-analysis. Diabetes Metab Syndr. 2020;14:589–596. doi: 10.1016/j.dsx.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lauriola M, Pani A, Ippoliti G, Mortara A, Milighetti S, Mazen M, Perseghin G, Pastori D, Grosso P, Scaglione F. Effect of combination therapy of hydroxychloroquine and azithromycin on mortality in COVID-19 patients. Clin Transl Sci. 2020 doi: 10.1111/cts.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ndelman O, Amital H, Bragazzi NL, Watad A, Chodick G. Continuous hydroxychloroquine or colchicine therapy does not prevent infection with SARS-CoV-2: insights from a large healthcare database analysis. Autoimmun Rev. 2020;19(7):102566–102569. doi: 10.1016/j.autrev.2020.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alexander R (1951) 7-Chloro-4-[5-(N-ethyl-N-2-hydroxyethylamino)-2-pentyl] aminoquinoline, its acid addition salts, and method of preparation. US2546658
- 100.Ashok K, Dhansukhlal VK, Dharmendra S, Sanjay N, Sanjay B, Atul J (2005) An improved process for the preparation of 7-chloro-4-(5-N-ethyl-N-2-hydroxyethylamine)-2-pentyl aminoquinoline and its intermediates. WO2005062723
- 101.Min YS, Cho HS, Mo KW (2010) New preparation of hydroxychloroquine. WO201002715
- 102.Yu E, Mangunuru HPR, Telang NS, Kong CJ, Verghese J, Gilliland SE, Ahmad S, Dominey RN, Gupton BF. High-yielding continuous-flow synthesis of antimalarial drug hydroxychloroquine. Beilstein J Org Chem. 2018;14:583–592. doi: 10.3762/bjoc.14.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Frank GB, Saeed A, Mangunure HPR, Telang NS (2019) High-yielding continuous flow synthesis of antimalarial drug hydroxychloroquine. WO2019165337 [DOI] [PMC free article] [PubMed]
- 104.Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir Res. 2013;100:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takahashi K, Furuta Y, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Shiraki K. In vitro and in vivo activities of T-705 and oseltamivir against influenzavirus. Antivir Chem Chemother. 2003;14:235–241. doi: 10.1177/095632020301400502. [DOI] [PubMed] [Google Scholar]
- 106.Takahashi K, Sakai-Tagawa Y, Shinya K, Sakabe S, Le QM, Kawaoka Y. T-705 (Favipiravir) activity against lethal H5N1 influenza A viruses. Proc Natl Acad Sci USA. 2010;107:882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bai CQ, Mu JS, Kargbo D, Song YB, Niu WK, Nie WM, Jiang JF. Clinical and virological characteristics of ebola virus disease patients treated with favipiravir (T-705)-Sierra Leone. Clin Infect Dis. 2014;63:1288–1294. doi: 10.1093/cid/ciw571. [DOI] [PubMed] [Google Scholar]
- 108.Cao B (2018) A pharmacokinetics study of favipiravir in patients with severe influenza. NCT03394209. https://clinicaltrials.gov/ct2/show/NCT03394209
- 109.Furuta Y, Takahashi K, Fukuda Y, Kuno M, Kamiyama T, Kozaki K, Shiraki K. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob Agents Chemother. 2002;46:977–981. doi: 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanaka T, Kamiyama T, Daikoku T, Takahashi K, Nomura N, Kurokawa M, Shiraki K. T-705 (favipiravir) suppresses tumor necrosis factor alpha production in response to influenza virus infection: a beneficial feature of T-705 as an anti-influenza drug. Acta Virol. 2017;61:48–55. doi: 10.4149/av_2017_01_48. [DOI] [PubMed] [Google Scholar]
- 111.Shiraki K, Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol Ther. 2020;209:107512–107526. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Janowski AB, Dudley H, Wang D. Antiviral activity of ribavirin and favipiravir against human astroviruses. J Clin Virol. 2019;123:104247–104258. doi: 10.1016/j.jcv.2019.104247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shannon A, Selisko B, Le NTT, Huchting J, Touret F, Piorkowski G, Fsttorini V, Ferron F, Decroly E, Meier C, Coutard B, Peersen O, Canard B. Favipiravir strikes the SARS-CoV-2 at its Achilles Heel, the RNA polymerase. BioRxiv. 2020 doi: 10.1101/2020.05.15.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dose-finding study of favipiravir in the treatment of uncomplicated influenza (2020) NCT01068912. https://clinicaltrials.gov/ct2/show/NCT01068912
- 115.Phase 3 Efficacy and safety study of favipiravir for treatment of uncomplicated in-fluenza in adults-T705. (2020) NCT02026349. https://clinicaltrials.gov/ct2/show/NCT02026349
- 116.Lou Y, Liu L, Yao HP, Hu XJ, Su JW, Xu KJ, Luo R, Yang X, He LJ, Lu XY, Zhao QW, Liang TB, Qiu YQ. Clinical outcomes and plasma concentrations of baloxavir marboxil and favipiravir in COVID-19 patients: an exploratory randomized, controlled trial. medRxiv. 2020 doi: 10.1101/2020.04.29.20085761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, Liao X, Gu Y, Cai Q, Yang Y, Shen C, Li X, Peng L, Huang D, Zhang J, Zhang S, Wang F, Liu J, Chen L, Chen S, Wang Z, Zhang Z, Cao R, Zhong W, Liu Y, Liu L. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020 doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takamatsu T, Yonezawa K (2010) Organic amine of 6-fluoro-3-hydroxy-2-pyrazinecarbonitrile and method for producing the same. US20100286394
- 119.Hara T, Norimatsu N, Kurushima H, Kano T (2011) Method for producing dichloropyrazine derivative. US20110275817
- 120.Liu FL, Li CQ (2017) A method for preparation favipiravir. CN106866553
- 121.Li MY (2017) A method for preparation favipiravir. CN107226794
- 122.Wang ML, Cao RY, Zhang LK, Yang YL, Liu J, Xu MY, Shi ZL, Hu ZH, Zhong W, Xiao GF. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Holshue ML, Chas DB, Lindquist S, Lofy KH, John W, Hollianne B, Christopher S, Keith E, Sara W, Ahmet T, George D, Amanda C, Fox LA, Patel A, Gerber S, Kin L, Tong SX, Lu XY, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zeuzem S, Berg T, Moeller B, Hinrichsen H, Mauss S, Wedemeyer H, Sarrazin C, Hueppe D, Zehnter E, Manns MP. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepat. 2009;16:75–90. doi: 10.1111/j.1365-2893.2008.01012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kronenberger B, Zeuzem S. New developments in HCV therapy. J Viral Hepat. 2012;19:48–51. doi: 10.1111/j.1365-2893.2011.01526.x. [DOI] [PubMed] [Google Scholar]
- 126.Sofia MJ. Nucleotide prodrugs for HCV therapy. Antivir Chem Chemother. 2011;22:23–49. doi: 10.3851/IMP1797. [DOI] [PubMed] [Google Scholar]
- 127.Carroll SS, Tomassini JE, Bosserman M, Getty K, Stahlhut MW, Eldrup AB, Bhat B, Hall D, Simcoe AL, LaFemina R, Rutkowski CA, Wolanski B, Yang ZC, Migliaccio G, Francesco RD, Kuo LC, MacCoss M, Olsen DB. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J Biol Chem. 2003;278:11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- 128.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, Francesco RD, Eldrup AB, Getty KL, Hou XL, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem. 2003;278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 129.Cho A, Zhang L, Xu J, Babusis D, Butler T, Lee R, Saunders OL, Wang T, Parrish J, Perry J, Feng JY, Ray AS, Kim CU. Synthesis and characterization of 2’-C-Me branched C-nucleosides as HCV polymerase inhibitors. Bioorg Med Chem Lett. 2012;22:4127–4132. doi: 10.1016/j.bmcl.2012.04.065. [DOI] [PubMed] [Google Scholar]
- 130.Cho A, Saunders OL, Butler T, Zhang L, Xu J, Vela JE, Feng JY, Ray AS, Kim CU. Synthesis and antiviral activity of a series of 1′-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg Med Chem Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cho A, Zhang LJ, Xu J, Lee R, Butler T, Metobo S, Aktoudianakis V, Lew W, Ye H, Clarke M, Doerffler E, Byun D, Wang T, Babusis D, Carey AC, German P, Sauer D, Zhong WD, Rossi S, Fenaux M, McHutchison JG, Perry J, Feng J, Ray AS, Kim CU. Discovery of the first C-nucleoside HCV polymerase inhibitor (GS-6620) with demonstrated antiviral response in HCV infected patients. J Med Chem. 2013;57(5):1812–1825. doi: 10.1021/jm400201a. [DOI] [PubMed] [Google Scholar]
- 132.Siegel D, Hui HC, Doerffler E, Clarke MO, Chun K, Zhang LJ, Neville S, Carra E, Lew W, Ross R, Wang Q, Wolfe L, Jordan R, Soloveva V, Knox J, Perry J, Perron M, Stray KM, Barauskas O, Feng JY, Xu YL, Lee G, Rheingold AL, Ray AS, Bannister R, Strickley R, Swaminathan S, Lee WA, Bavari S, Cihlar T, Lo MK, Warren TK, Mackman RL. Discovery and synthesis of a phosphoramidate prodrug of a pyrrolo[2,1-f][triazin-4-amino] adenine C-nucleoside(GS-5734) for the treatment of ebola and emerging viruses. J Med Chem. 2017;60(5):1648–1661. doi: 10.1021/acs.jmedchem.6b01594. [DOI] [PubMed] [Google Scholar]
- 133.Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chem Bio Chem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yin WC, Mao CY, Luan XD, Shen DD, Shen QY, Su HX, Wang XX, Zhou FL, Zhao WF, Gao MQ, Chang SH, Xie YC, Tian GH, Jiang HW, Tao SC, Shen JS, Jiang Y, Jiang HL, Xu YC, Zhang SY, Zhang Y, Xu HE. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. BioRxiv. 2020 doi: 10.1101/2020.04.08.032763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9(396):3653–3662. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, Ray AS, Cihlar T, Siegel D, Mackman RL, Clarke MO, Baric RS, Denison MR. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2):18–32. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wit ED, Feldmannb F, Cronin J, Jordanc R, Okumurad A, Thomas T, Scott D, Cihlar T, Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci USA. 2019 doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592–117597. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Choy KT, Wong YL, Kaewpreedee A, Sia P, Chen SF, Hui DY, Chu W, Chan W, Cheung PH, Huang P, Peiris X, Yen M. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir Res. 2020;178:104786–104790. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang L, Zhang D, Yuan CM, Wang XW, Li YF, Jia XL, Gao X, Yen HL, Cheung PPH, Huang XH. Role of 1’-ribose cyano substitution for remdesivir to effectively inhibit both nucleotide addition and proofreading in SARS-CoV-2 viral RNA replication. BioRxiv. 2020 doi: 10.1101/2020.04.27.063859. [DOI] [PubMed] [Google Scholar]
- 142.Pruijssers AJ, George AS, Schäfer A, Leist SR, Gralinksi LE, Dinnon KH, III, Yount BL, Agostini ML, Stevens LJ, Chappell JD, Lu XT, Hughes TM, Gully K, Martinez DR, Brown AJ, Graham RL, Perry JK, Pont VD, Pitts J, Ma B, Babusis D, Murakami E, Feng JY, Bilello JP, Porter DP, Cihlar T, Baric RS, Denison MR, Sheahan TP. Remdesivir potently inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020 doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu M, Xu L, Huang D, Yuan MJ, Ye CY. Remdesivir inhibits renal fibrosis in obstructed kidneys. bioRxiv. 2020 doi: 10.1101/2020.04.01.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Williamson BN, Feldmann F, Meade-White KBS, Porter DP, Schulz J, Doremalen NV, Leighton L, Yinda CK, Péreze-Péreze L, Okumura A, Lovaglio J, Hanley PW, Saturday G, Bosio CM, Anzick S, BAarbian K, Cihlar T, Martens C, Scott DP, Munster VJ, Wit ED, Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM, Abedi GR, Ahmed NS, Almendares O, Beer K, Ben-Aderet MA, Benowitz I, Biggs HM, Binder AM, Black SR, Bonin B, Bozio CH, Brown CM, Bruce H, Bryant-Genevier J, Budd A, Buell D, Bystritsky R, Cates J, Charles EM, Chatham-Stephens K, Chea N, Chiou H, Christians JM, Dawson P, DeSalvo T, Diaz G, Donahue M, Donovan S, Duca LM, Erichson K, Esona MD, Evans S, Falk J, Feldstein LR, Fricchione MJ, Gerber SI, Gunzenhauser JD, Harcourt J, Hunter JC, Kim L, Kamili S, Klos R, Layden JE, Livingston M, Lo K, Li Y, Malapati L, McGovern O, Robinson S, Robinson P, Rolfes MA, Routh JA, Rubin R, Rudman SL, Sakthivel SK, Scott S, Shepherd C, Shetty V, Smith EA, Smith S, Stierman B, Stoecher W, Sunenshine R, Sy-Santos R, Wang LJ, Watson JT, Westercamp M, Whitaker B, Wilkerson S, Wondruff RC, Wortham JM, Wu T, Xie A, Yousaf A, Zahn M, Zhang J. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.National Institutes of Health (2020) NIH Clinical Trial of Remdesivir to Treat COVID-19 Begins. https://www.nih.gov/news-events/news-releases/nih-clinical-trial-Remdesivir-treat-covid-19-begins
- 147.U.S.National Library of Medicine Clinical Trials Registry (2020) A trial of remdesivir in adults with mild and moderate COVID-19. NCT04252664. https://clinicaltrials.gov/ct2/show/NCT04252664
- 148.US National Library of Medicine Clinical Trials Registry (2020) Severe 2019-nCoV Remdesivir RCT. NCT04257656. https://clinicaltrials.gov/ct2/show/NCT04257656
- 149.Zhang Q, Wang Y, Qi C, Shen L, Li J. Clinical trial analysis of 2019-nCoV therapy registered in China. J Med Virol. 2020;92(6):540–545. doi: 10.1002/jmv.25733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang YM, Zhang DY, Du GH, Du RH, Zhao JP, Jin Y, Fu SZ, Gao L, Cheng ZS, Lu QF, Hu Y, Luo GW, Wang K, Lu Y, Li HD, Wang SZ, Ruan SN, Yang CQ, Mei CL, Wang Y, Ding D, Wu F, Tang X, Ye XZ, Ye YC, Liu B, Yang J, Yin W, Wang AL, Fan GH, Zhou F, Liu ZB, Gu XY, Xu JY, Shang LH, Zhang Y, Cao LJ, Guo TT, Wan Y, Qin H, Jiang YS, Jaki T, Hayden FG, Horby P, Cao B, Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Grein J, Ohmagari N, Diaz SG, Asperges DE, Castagna A, Feldt T, Green G, Green ML, Lescure FX, Nicastri E, Oda R, Yo K, Quiros-Roldan E, Studemeister A, Redinski J, Ahmed S, Bernett J, Chelliah D, Chen D, Chihara S, Cohen SH, Cunningham J, Monforte AD, Ismail S, Kato H, Lapadula G, Her EL, Maeno T, Majumder S, Massari M, Mora-Rillo M, Mutoh Y, Nguyen D, Verweij E, Zoufaly A, Osinusi AO, Dezure A, Zhao Y, Zhong L, Chokkalingam A, Elboudwarej E, Telep L, Mera R, Gaggar A, Myers RP, Brainard DM, Childs R, Flaniga T. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh M, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC. Remdesivir for the treatment of Covid-19 preliminary report. N Engl J Med. 2020 doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, Spinner CD, Galli M, Ahn MY, Nahass RG, Chen YS, SenGupta D, Hyland RH, Osinusi AO, Cao HY, Blair C, Wei XL, Gaggar A, Brainard DM, Towner WJ, Munoz J, Mullane KM, Marty FM, Tashima KT, Diaz G, Subramanian A. Remdesivir for 5 or 10 days in patients with severe Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Butler T, Cho A, Kim C, Saunders O, Zhang LJ, Parrish J (2011) 1'-Substituted carba-nucleoside analogs for antiviral treatment. EP2268642
- 155.Metobo SE, Xu J, Saunders OL, Butler T, Aktoudianakis E, Cho A, Kim CU. Practical synthesis of 1′-substituted tubercidin C-nucleoside. Tetrahedron Lett. 2012;53:484–486. doi: 10.1016/j.tetlet.2011.11.055. [DOI] [Google Scholar]
- 156.Vieira T, Stevens A, Chtchemelinine A, Gao D, Badalov P, Heumann L. Development of a large-scale cyanation process using continuous flow chemistry en route to the synthesis of remdesivir. Org Process Res Dev. 2020 doi: 10.1021/acs.oprd.0c00172. [DOI] [PubMed] [Google Scholar]
- 157.Xue F, Zhou XB, Zhou RJ, Zhou XH, Xiao D, Gu E, Guo XW, Xiang J, Wang K, Yang LK, Zhong W, Qin Y. Improvement of the C-glycosylation step for the synthesis of remdesivir. Org Process Res Dev. 2020;24:1772–1777. doi: 10.1021/acs.oprd.0c00310. [DOI] [PubMed] [Google Scholar]
- 158.Wang M, Zhang L, Huo XH, Zhang ZF, Yuan QJ, Li PP, Chen JZ, Zou YS, Wu ZX, Zhang WB. Catalytic asymmetric synthesis of the anti-COVID-19 drugremdesivir. Angew. Chem. Int. Ed. 2020 doi: 10.1002/anie.202011527. [DOI] [PubMed] [Google Scholar]
- 159.Walker KL, Kabakov SA, Zhu F, Bouchlaka MN, Olson SL, Cho MM, Quamine AE, Feils AS, Gavcovich TB, Rui LX, Capitini CM. Efficacy of JAK1/2 and BCL2 inhibition on human T cell acute lymphoblastic leukemia in vitro and in vivo. bioRxiv. 2020 doi: 10.1101/734913. [DOI] [Google Scholar]
- 160.Williams NK, Bamert RS, Patel O, Wang C, Walden PM, Wilks AF, Fantino E, Rossjohn J, Lucet IS. Dissecting specificity in the janus kinases: the structures of JAK specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol. 2009;387:219–232. doi: 10.1016/j.jmb.2009.01.041. [DOI] [PubMed] [Google Scholar]
- 161.Sonbol MB, Firwana B, Zarzour A, Morad M, Rana V, Tiu RV. Comprehensive review of JAK inhibitors in myeloproliferative neoplasms. Ther Adv Hematol. 2013;4:15–35. doi: 10.1177/2040620712461047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tuttle KD, Minter R, Waugh KA, Araya P, Ludwig M, Sempeck C, Smith K, Andrysik Z, Burchill MA, Tamburini BAJ, Orlicky DJ, Sullivan KD, Espinosa JM. JAK1 inhibition blocks lethal sterile immune responses: implications for COVID-19 therapy. BioRxiv. 2020 doi: 10.1101/2020.04.07.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y, Zhang L, Fan GH, Xu JY, Gu XY, Cheng ZS, Yu T, Xia JA, Wei Y, Wu WJ, Xie XL, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie JG, Wang GF, Jiang RM, Gao ZC, Jin Q, Wang JW, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:15–21. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Caocci G, Nasa GL. Could ruxolitinib be effective in patients with COVID-19 infection at risk of acute respiratory distress syndrome (ARDS)? Ann Hematol. 2020;99:1675–1676. doi: 10.1007/s00277-020-04067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cao Y, Wei J, Zou L, Jiang T, Wang G, Chen L, Huang L, Meng F, Huang L, Wang N, Zhou X, Luo H, Mao Z, Chen X, Xie J, Liu J, Cheng H, Zhao J, Huang G, Wang W, Zhou J. Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J Allergy Clin Immunol. 2020;146:137–146. doi: 10.1016/j.jaci.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rodgers JD, Shepard S, Maduskuie TP, Wang HS, Falahatpisheh N, Rafalski M, Arvanitis AG, Storace L, Jalluri RK, Fridman JS, Vaddi K (2007) Heteroaryl substituted pyrrolo[2,3-B]pyridines and pyrrolo[2,3-B]pyridines as janus kinese inhibitors. WO 2007070514
- 169.Haydl AM, Xu K, Breit B. Regio- and enantioselective synthesis of N-substituted pyrazoles by rhodium-catalyzed asymmetric addition to allenes. Angew Chem Int Ed. 2015;127:1–6. doi: 10.1002/ange.201501758. [DOI] [PubMed] [Google Scholar]
- 170.Deepshikha C, Chandra BD, Pranab C, Asok N, Mohan P (2016) Processes for the preparation of ruxolitinib phosphate. WO 2016035014.
- 171.Zhang XQ, Zhang AM, Zhou Z, Yang LL, Yao HD, Zhou XY, Wang HB (2018) Synthesis process of ruxolitinib. EP3398952