Abstract
It is reported that an increase in aerobic bacteria, a lack of short-chain fatty acids (SCFAs), and immune disorders in the diverted colon are major causes of diversion colitis. However, the precise pathogenesis of this condition remains unclear. The aim of the present study was to examine the microbiota, intestinal SCFAs, and immunoglobulin A (IgA) in the diverted colon. Eight patients underwent operative procedures for colostomies. We assessed the diverted colon using endoscopy and obtained intestinal samples from the diverted colon and oral colon in these patients. We analyzed the microbiota and SCFAs of the intestinal samples. The bacterial communities were investigated using a 16S rRNA gene sequencing method. The microbiota demonstrated a change in the proportion of some species, especially Lactobacillus, which significantly decreased in the diverted colon at the genus level. We also showed that intestinal SCFA values were significantly decreased in the diverted colon. Furthermore, intestinal IgA levels were significantly increased in the diverted colon. This study was the first to show that intestinal SCFAs were significantly decreased and intestinal IgA was significantly increased in the diverted colon. Our data suggest that SCFAs affect the microbiota and may play an immunological role in diversion colitis.
Keywords: Diversion colitis, Microbiota, Short-chain fatty acids, Immunoglobulin A
Abbreviations: DC, diversion colitis; OA, organic acid; SCFAs, short-chain fatty acids; AA, acetic acid; BA, butyric acid; PA, propionic acid; IgA, immunoglobulin A
Highlights
-
•
Anaerobic bacteria like Lactobacillus significantly decreased in the diverted colon.
-
•
Fecal short-chain fatty acids significantly decreased in the diverted colon.
-
•
Acetic acid also decreased significantly in the diverted colon.
-
•
Fecal immunoglobulin A levels were significantly increased in the diverted colon.
1. Introduction
Diversion colitis (DC) was first described by Morson et al., in 1974 as nonspecific inflammation in the diverted colon [1]. Glotzer et al. termed this inflammation “diversion colitis” in 1981 [2]. A prospective study reported that almost all cases show colitis, as evidenced by endoscopic analyses, 3–36 months after a colostomy [3]. Most patients are asymptomatic; however, approximately one-third of patients may exhibit various symptoms of DC, such as abdominal discomfort, tenesmus, anorectal pain, mucous discharge, and rectal bleeding [4,5].
It has been reported that an increase in aerobic bacteria, a lack of short-chain fatty acids (SCFAs), and immune disorders of the diverted colon are major causes of DC [6]. However, the precise pathogenesis of this condition remains unclear. While there are a few reports on the microbiota in DC in humans, the relationship between the microbiota and SCFAs or immunoglobulin A (IgA) remains elusive.
The aim of the present study was to evaluate the intestinal microbiota, SCFAs, and IgA in the diverted colon and to elucidate the pathogenesis of DC.
2. Material and methods
2.1. Subjects and study protocol
Written informed consent was obtained from the patients for publication of the report and accompanying images. The study was reviewed and approved by the Institutional Review Board of Niigata University.
The subjects were 8 patients (65.9 ± 10.4 years) (male: female = 1:7), who had undergone stoma surgery within the previous 1–40 months. Five of these patients had a history of colon cancer, one had rectovaginal septum cancer, one had ovarian cancer, and one had a retroperitoneal abscess. These patients all underwent a colostomy (Table 1). None of the patients was administered with antibiotics or probiotics for one month leading up to the date of the colonoscopy.
Table 1.
Patient characteristics.
| Case (No) | Age (yrs) | Sex | Diagnosis | Operative procedure | Post-surgical treatment for primary disease (anti-cancer drugs or antibiotics) | Symptom | UCEIS | Postoperative period |
|---|---|---|---|---|---|---|---|---|
| 1 | 62 | M | Rectal Cancer | Low anterior resection + Colostomy | none | none | 1 | 10 M |
| 2 | 49 | F | Rectal Cancer | Low anterior resection + Colostomy | XELOX chemotherapy (6 M) | none | 2 | 6 M |
| 3 | 66 | F | Rectal Cancer | Low anterior resection + Colostomy | FOLFOX chemotherapy (6 M) | none | 2 | 6 M |
| 4 | 86 | F | Sigmoid colon Cancer | Low anterior resection + Colostomy | none | none | 1 | 1 M |
| 5 | 67 | F | Rectovaginal septum cancer | Transverse colostomy | Ceftriaxone (2 W) | mucous and<!--Soft-enter Run-on-- > bloody stool | 3 | 40 M |
| 6 | 74 | F | Ovarian Cancer | Transverse colostomy | Sulbactam/Cefoperazone (1 W) + Piperacillin/Tazobactam (1 W) | mucous stool | 3 | 18 M |
| 7 | 56 | F | Rectal Cancer | Low anterior resection + Colostomy | none | none | 1 | 1 M |
| 8 | 67 | F | Retroperitoneal abscess | Left hemicolectomy + Transverse colostomy | Piperacillin/Tazobactam (1 M) | mucous stool | 2 | 16 M |
We assessed the diverted colon endoscopically, and evaluated the severity using the ulcerative colitis endoscopic index of severity (UCEIS). We assessed the intestinal microbiota of the diverted colon and oral colon using a next-generation sequencer (Illumina MiSeq). We conducted 16S rRNA gene sequencing. The composition of intestinal microbiota was evaluated using quantitative insights into microbial ecology (QIIME), and β-diversity was measured using UniFrac-distances analysis.
We also measured the composition of intestinal organic acids, SCFA, and IgA through enzyme-linked immunosorbent assay (ELISA) in six patients.
2.2. Intestinal sample collection
Intestinal samples (approximately 100 mg) were suspended in 900 μL of guanidine thiocyanate solution (100 mM Tris–HCl [pH 9.0], 40 mM EDTA, and 4 M guanidine thiocyanate) and frozen at −80 °C until further analysis [7].
2.3. DNA preparations from intestinal samples
The collected samples were sent to the laboratory of Miyarisan Pharmaceutical Co., Ltd. and stored at −20 °C. DNA was extracted from collected intestinal samples using a glass bead extraction method and purified, according to a previously reported method [8]. The amount of DNA was determined using a Quanti Fluor dsDNA System and Quantus Fluorometer (Promega, Madison, WI, USA).
2.4. PCR amplification and analysis of 16S rRNA sequences
The V3–V4 region of the 16S rRNA gene was PCR amplified from stool DNA samples using a TaKaRa Ex Taq Hot Start PCR mixture (Takara Bio, Shiga, Japan). The primers used for PCR amplification were 341F and 785R, which contained the Illumina index and sequencing adapter overhangs [9]. PCR assays were performed using a TaKaRa PCR Thermal Cycler Dice Touch device (Takara Bio, Shiga, Japan) with the following parameters: initial denaturation at 98 °C for 30 s, followed by 35 cycles of 98 °C for 10 s and 60 °C for 30 s, with a final extension step at 72 °C for 5 min. The PCR products were purified and size selected using SPRIselect (Beckman Coulter, Brea, CA, USA). DNA concentrations were quantified with a QuantiFluor dsDNA System and Quantus Fluorometer (Promega, Madison, WI, USA), and equal amounts of purified PCR products were pooled for subsequent Illumina MiSeq sequencing. Sequencing was carried out with a Miseq Regent Kit V3 (600 cycles) (Illumina, San Diego, CA, USA), according to the manufacturer's instructions. Sequence processing and quality assessment were performed using the QIIME package (version 1.8.0) (http://qiime.org), an open source software created to address the problem of obtaining sequencing data from raw sequences for interpretation and database deposition [10]. To obtain an overall diversity analysis for subsequent comparative and statistical evaluations, we merged the Biological Observation Matrix (BIOM) tables provided by QIIME into a unique biom table using a script included in the QIIME package. Paired-end reads were merged using the Fastq-join script in ea-utils with the parameters m = 6 and P = 20, then quality filtered using QIIME's script split_libraries_fastq.py (r = 3, P = 0.75, q = 20, n = 0). De novo and reference-based chimera detection and removal were performed using USEARCH v6.1 with the Greengenes v13.8 database. Operational taxonomic units (OTUs) were chosen using an open reference OTU-picking pipeline against the 97% identity of the pre-clustered Greengenes v13.8 database using UCLUST. According to the manufacturer, a QIIME alpha diversity analysis script is used to perform rarefaction analysis by subsampling the OTUs biom table based on the minimum rarefaction depth value chosen by the user depending on the minimum number of sequences/samples obtained. For our subset, this value was 3957. Then, using different metrics, alpha diversity was computed for each rarefied OTU table. We used three non-phylogeny-based metrics: observed species, chao 1, and the Shannon index. After performing the rarefaction evaluation, the QIIME beta diversity analysis script was used to compute beta diversity with the rarefied OTUs table using different metrics. We used a non-phylogeny-based metric (Bray–Curtis metric) as well as phylogeny-based metrics (unweighted and weighted UniFrac) [11]. Finally, the script was used to obtain a distance metric to compute the principal coordinate analysis (PCoA) and convert it into plots for results visualization.
2.5. Analysis of intestinal organic acids
For determination of organic acids, 0.1 g of feces was placed in a 2.0 mL tube with zirconia beads and suspended in MilliQ. Samples were heated at 85 °C for 15 min, vortexed at 5 m/s for 45 s using FastPrep 24 5G (MP Biomedicals, CA, USA), and centrifuged at 15,350×g for 10 min. The supernatant was filtered through a 0.2 μm filter. Organic acids (acetic acid, propionic acid, butyric acid, iso-butyric acid, succinic acid, lactic acid, formic acid, valeric acid, and iso-valeric acid) in feces were measured using high-performance liquid chromatography (Prominence, SHIMADZU, Kyoto, Japan) using a post column reaction with a detector (CDD-10A, SHIMADZU, Kyoto, Japan), tandemly arranged two columns (Shim-pack SCR-102(H), 300 mm × 8 mm ID, SHIMADZU, Kyoto, Japan), and a guard column (Shim-pack SCR-102(H), 50 mm × 6 mm ID, SHIMADZU, Kyoto, Japan). The system was used with a mobile phase (5 mM p-toluenesulfonic acid) and a reaction solution (5 mM p-toluenesulfonic acid, 100 μM EDTA, and 20 mM Bis-Tris). The flow rate and oven temperature were 0.8 mL/min and 45 °C, respectively. The detector cell temperature was maintained at 48 °C.
2.6. Analysis of intestinal IgA
The quantification of intestinal IgA was performed using Cosmo Bio Co., Ltd. (Tokyo, Japan). Intestinal IgA content was quantified through ELISA using a mouse IgA ELISA Quantitation Set (Bethyl Laboratories, Inc., Montgomery, TX, USA). Approximately 50 mg of feces was used for IgA analysis, and the values were expressed as values per feces weight.
2.7. Statistical analysis
All quantitative data are expressed as mean ± SE. A two-sided Student's two-sample t-test was used for statistical analyses with the SPSS statistical package, version 24.0 (IBM SPSS Japan Inc., Tokyo, Japan). In this study, while values of P < 0.05 were considered statistically significant, if significance did not remain after correction using an optimized false discovery rate approach to account for false-positive results, presented as q values, they are instead described as tendencies. The significance of each PCoA plot was analyzed using permutational multivariate analysis of variance, a non-parametric test similar to analysis of variance that does not require the data to be normally distributed, and uses distance metrics to confirm the strength and statistical significance of sample groupings [12]. We used 999 Monte Carlo permutations in QIIME to assess statistical significance between group diversity metrics.
3. Results
3.1. Endoscopic evaluation and symptoms
Eight patients underwent stoma surgery within 1–40 months prior to this study. We performed an endoscopy for all patients and evaluated the inflammation of the diverted colon using the UCEIS. While the method for assessing inflammation of DC has not yet been established, it is often evaluated using the UCEIS score.
In all patients, mild DC (UCEIS = 1 to 3) was detected by colonoscopy (Table 1). Details of inflammation were UCEIS = 1 in 3 patients, UCEIS = 2 in 3 patients, and UCEIS = 3 in 2 patients. Only three patients presented with symptoms of mucous, bloody stool, and tenesmus, and all three had surgery over a year prior. Patients who had a longer period after surgery tended to have more colonic inflammation and were more symptomatic (Table 1).
3.2. Intestinal microbiota
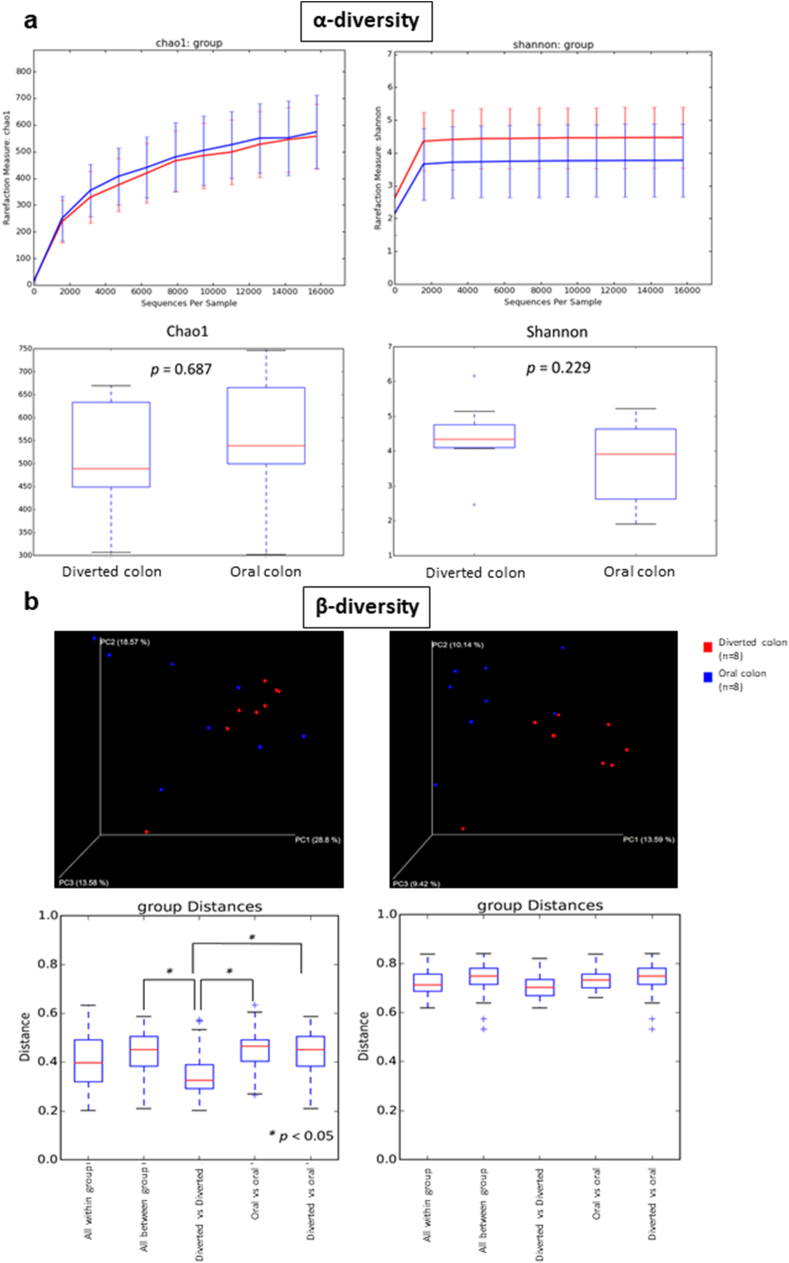
The α-diversity data showed that there was no significant difference (Fig. 1a) among the microbiota in the diverted colon and in the oral colon. Regarding the β-diversity, there was a significant difference between “diverted colon vs. diverted colon” and “oral colon vs. oral colon” and between “diverted colon vs. diverted colon” and “diverted colon vs. oral colon” (Fig. 1b). These results indicate that the difference in the composition ratio of the microbiota in the oral colon is larger than that in the diverted colon, and the difference in the composition ratio of the microbiota between these two groups was larger than that in the diverted colon group. The β-diversity data showed that the microbiota in the diverted colon formed a cluster, which was significantly different from the microbiota in the oral colon.
Fig. 1.
(a) Theα-diversity data of this study. Bars show the SD of the data. Data were analyzed using the Mann–Whitney U test (vs. Pre). (b) The β -diversity data of this study. Bars show the SD of the data. Data were analyzed using the Mann–Whitney U test (vs. Pre) followed by Benjamini–Hochberg (*: P < 0.05).
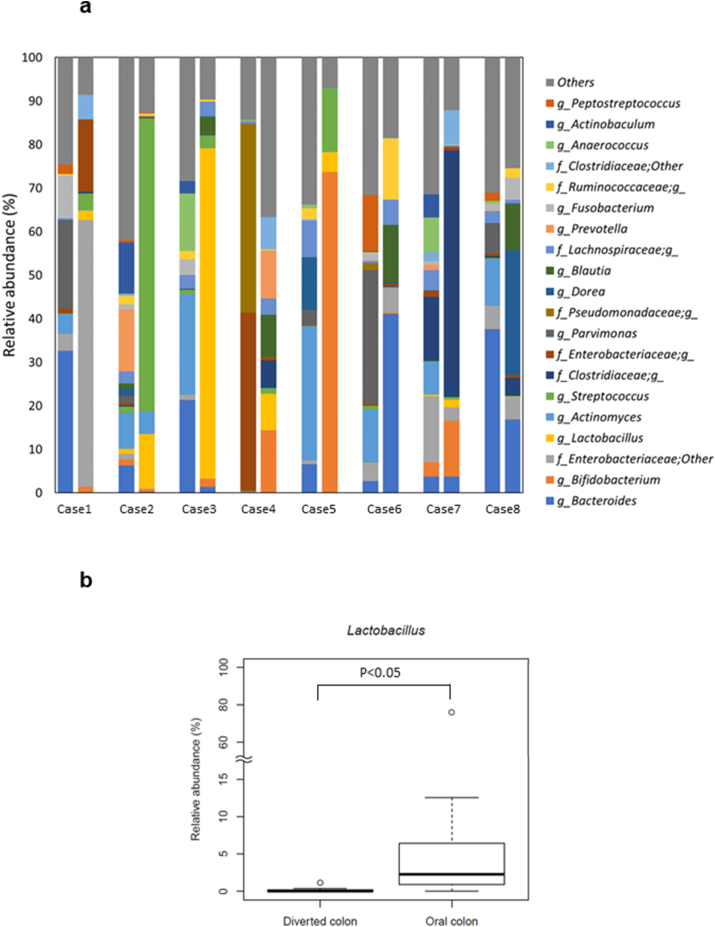
Regarding the intestinal microbiota at the genus level, the serial intestinal microbiota showed an increase in Actinomyces (P < 0.05), Anaerococcus (P < 0.05), Corynebacterium (P < 0.01), Peptoniphilus (P < 0.05), and Porphyromonas (P < 0.01), with significantly different intestinal organisms in the diverted colon compared to those in the oral colon (Fig. 2a). Moreover, we detected a decrease in Lactobacillus (P < 0.05) and Granulicatella (P < 0.05), which were significantly changed in the diverted colon compared to those in the oral colon (Fig. 2b).
Fig. 2.
(a) The analysis of the intestinal microbiota. The comparison of microbiota between the diverted colon and the oral colon of the patients, who underwent colostomy. (b) Significant differences were detected in the numbers of Lactobacillus (at genus level) (P < 0.05). Bars show the SD of the data. Data were analyzed using the Mann–Whitney U test (vs. Pre) followed by the Benjamini–Hochberg procedure (*P < 0.05, **p < 0.01).
3.3. Intestinal SCFA and IgA
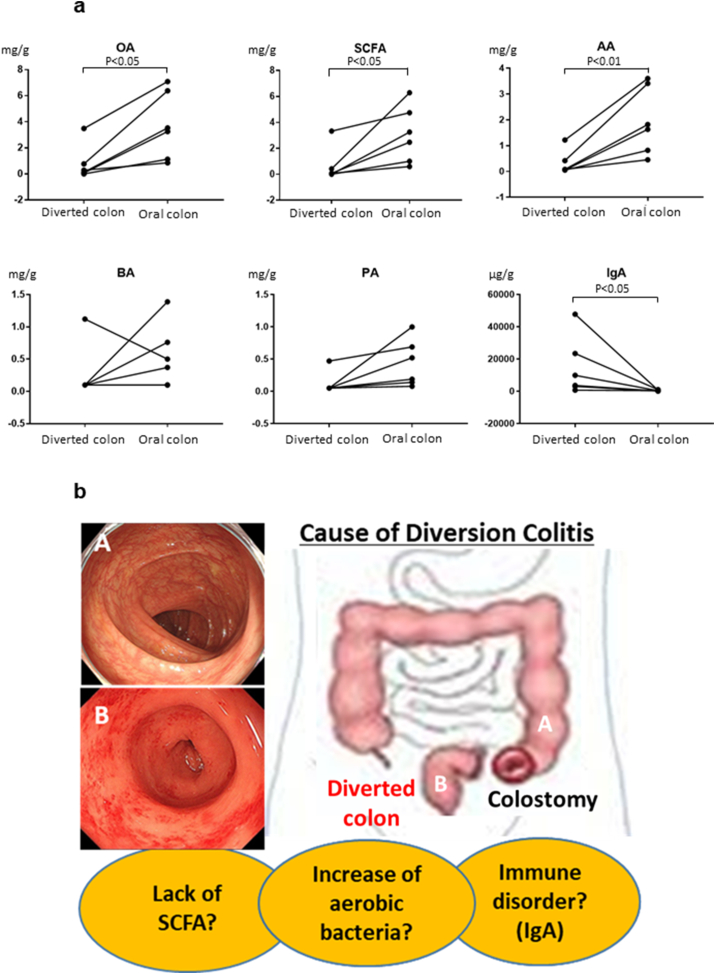
The comparison of intestinal organic acid (OA), SCFAs, and acetic acid (AA) in the diverted colon and the oral colon showed that there were significant differences in six patients, with averages (diverted colon vs oral colon, mean ± SD) of 0.79 ± 1.2 vs. 3.71 ± 2.4 (P < 0.05) for OA, 0.65 ± 1.2 vs. 3.06 ± 2.0 (P < 0.05) for SCFAs, and 0.45 ± 0.3 vs. 1.96 ± 1.2 (P < 0.01) for AA (Fig. 3a).
Fig. 3.
(a) The comparison of intestinal elements between the diverted colon and oral colon. Significant differences were detected for OA (P < 0.05), SCFAs (P < 0.05), AA (P < 0.01), and IgA (P < 0.05). OA; organic acid, SCFA; short-chain fatty acids, AA; acetic acid, BA; butyric acid, PA; propionic acid, IgA; immunoglobulin A. (b) Schematic of diversion colitis.
However, there were no significant differences in intestinal butyric acid (BA) and propionic acid (PA) values between these groups.
The comparison of intestinal IgA in the diverted colon and oral colon showed that there was a significant difference in six patients, with an average (diverted colon vs oral colon, mean ± SD) of 14821 ± 8221 vs. 520 ± 339 (P < 0.05) (Fig. 3a). Interestingly, the diverted colon had a higher IgA than the oral colon in all cases.
4. Discussion
The precise pathogenesis of DC remains unclear. It is considered that an increase in aerobic bacteria, a lack of SCFAs, and immune disorders in the diverted colon are major causes of DC. We have summarized the possible disease pathogenesis in Fig. 3b.
There are several reports on the microbiota in DC; Neut et al. showed a decrease in anaerobes of the diverted colon [13]. Se-Jin Baek et al. reported a decrease in anaerobes, notably in Lactobacillus and Bifidobacterium [14]. These reports are comparisons between patients with DC and normal subjects.
Almost all of our patients had mild inflammation; and therefore, we could not derive a correlation between the intensity of inflammation and the microbiota. Considering the diversity in the composition of the microbiota in different individuals, in our study, we instead compared the microbiota in the diverted colon with that in the oral colon of the same individual. We concluded that Lactobacillus was predominantly low in the diverted colon. Lactobacillus has been reported to promote regulatory T cell differentiation and suppress enteritis [15]; therefore, it is considered one of the causes of DC.
SCFAs are produced mainly through saccharolytic fermentation of carbohydrates that escape digestion and absorption in the small intestine [16]. The pathways of SCFA production are relatively well understood and have been recently described in detail [17]. However, to date, there have been no reports on SCFA measurements in DC. In this study, we showed that SCFAs were significantly reduced in the diverted colon. The complex metabolic cross-feeding relationships among the microbial populations is influenced by the environment, such as changes in the nutrient supply, particularly in complex environments, such as the human lower gut [18]. SCFAs are the major fuel source for the intestinal epithelium. Bacteria produce SCFAs as byproducts of carbohydrate fermentation in the colonic lumen, and SCFAs provide the primary energy source for colonic mucosal cells [19]. Their absence in the diverted colon may cause mucosal atrophy and inflammation.
IgA is the main antibody isotype secreted into the intestinal lumen, and it plays a critical role in defense against pathogens and in the maintenance of intestinal homeostasis [20]. However, how secreted IgA regulates the gut microbiota is not completely understood. It has been reported that intestinal IgA selectively binds harmful bacteria to eliminate them from the gut microbiota [21]. On the other hand, several reports have demonstrated that IgA binding could facilitate gut colonization by bound bacteria, rather than suppressing their growth [22,23]. There are no previous reports on intestinal IgA in DC. Here, we showed that intestinal IgA was increased in the diverted colon.
The fundamental treatment of DC is intestinal anastomosis. Therapeutic options other than surgical procedures include 5-aminosalicylic acids, glucocorticoids, antibiotics, SCFAs, and intestinal microbiota transplantation (FMT) [6,24]. The treatment of DC with SCFAs has shown some positive results, with Harig demonstrating improved symptoms and endoscopic inflammatory changes [24]. Komorowski et al. reported similar results in four patients with DC following SCFA irrigation [25]. In recent years, several studies on the usefulness of SCFAs, including butyrate, have been reported [26,27]. Cristina et al. [28] proposed that butyrate enemas might prevent the atrophy of the diverted colon/rectum, thus improving the recovery of tissue integrity. We also reported that the usefulness of FMT for DC and intestinal microbiota was important for homeostasis of the colon [29].
Our results suggest that SCFAs (including AA) affects the intestinal microbiota, which may play a role in the immunity of the diverted colon. This study was the first to show that intestinal SCFAs were significantly decreased and intestinal IgA was significantly increased in the diverted colon. Our results were also consistent with those of previous reports in which the proportion of Lactobacillus was significantly decreased in diverted colon. Our data suggest that SCFAs affect the microbiota and may play a role in improving DC. However, further studies, such as a comparison of the microbiota between patients with and without diversion colitis, on a large scale, are essential to elucidate the pathogenesis of DC.
Ethics approval and consent to participate
The study was reviewed and approved by the Institutional Review Board of Niigata University.
Consent for publication
Written informed consent was obtained from the patients for publication of the report and accompanying images.
Availability of data and materials
Not applicable.
Conflicts of interest
These authors disclose the following: Shuji Terai received research funding from Miyarisan Pharmaceutical Co., Ltd. The remaining authors have no conflicts of interest to disclose.
Funding
This work was supported by Takeda Japan Medical Office Funded Research Grant 2018, Tsukada Medical Funded Research Grant, Grant-in-Aid for Young Scientific Research (19K17393) from the Ministry of Education, Science, Technology, Sports, and Miyarisan Pharmaceutical Co., Ltd.
Author's contributions
KT, AT, TM, TY, YK, KT, HS, SI, KH, KM, YT, MN, YS, HK, and JY diagnosed and drafted the manuscript. AM, AM, KO, MT, TW, and ST analyzed data. All authors critically reviewed the manuscript and approved the final draft.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
The authors disclose the following: Shuji Terai received research funding from Miyarisan Pharmaceutical Co., Ltd. Asami Matsumoto, Ayaka Minemura, Kentaro Oka and Motomichi Takahashi are employees of Miyarisan Pharmaceutical Co., Ltd.The remaining authors disclose no conflicts.
Please note that all Biochemical and Biophysical Research Communications authors are required to report the following potential conflicts of interest with each submission. If applicable to your manuscript, please provide the necessary declaration in the box above.
-
(1)
All third-party financial support for the work in the submitted manuscript.
-
(2)
All financial relationships with any entities that could be viewed as relevant to the general area of the submitted manuscript.
-
(3)
All sources of revenue with relevance to the submitted work who made payments to you, or to your institution on your behalf, in the 36 months prior to submission.
-
(4)
Any other interactions with the sponsor of outside of the submitted work should also be reported. (5) Any relevant patents or copyrights (planned, pending, or issued).
-
(6)
Any other relationships or affiliations that may be perceived by readers to have influenced, or give the appearance of potentially influencing, what you wrote in the submitted work. As a general guideline, it is usually better to disclose a relationship than not.
Acknowledgements
The authors thank Yasutoshi Kuroki and Miyarisan Pharmaceutical Co., Ltd. for their cooperation.
Contributor Information
Atsunori Tsuchiya, Email: atsunori@med.niigata-u.ac.jp.
Shuji Terai, Email: terais@med.niigata-u.ac.jp.
References
- 1.Morson B.C., Dawson I.M.P. first ed. Blackwellfic Publications; London: 1972. Gastrointestinal Pathology. [Google Scholar]
- 2.Glotzer D.J., Glick M.E., Goldman H. Proctitis and colitis following diversion of the fecal stream. Gastroenterology. 1981;80:438–441. [PubMed] [Google Scholar]
- 3.Korelitz B.I., Cheskin L.J., Sohn N. Proctitis after fecal diversion in Crohn's disease and its elimination with reanastomosis: implications for surgical management. Report of four cases. Gastroenterology. 1984;87:710–713. [PubMed] [Google Scholar]
- 4.Szczepkowski M., Kobus A., Borycka K. How to treat diversion colitis? Current state of medical knowledge, own research and experience. Acta Chir. Iugosl. 2008;55:77–81. doi: 10.2298/aci0803077s. [DOI] [PubMed] [Google Scholar]
- 5.Ma C.K., Gottlieb C., Haas P.A. Diversion colitis: a clinicopathologic study of 21 cases. Hum. Pathol. 1990;21:429–436. doi: 10.1016/0046-8177(90)90206-k. [DOI] [PubMed] [Google Scholar]
- 6.Tominaga K., Kamimura K., Takahashi K. Diversion colitis and pouchitis: a mini-review. World J. Gastroenterol. 2018 Apr;24(16):1734–1747. doi: 10.3748/wjg.v24.i16.1734. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishimoto Y., Mizutani S., Nakajima Tet al. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut. 2016;65:1574–1575. doi: 10.1136/gutjnl-2016-311937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin J.S., Touyama M., Yamada S. Alteration of a human intestinal microbiota under extreme life environment in the Antarctica. Biol. Pharm. Bull. 2014;37:1899–1906. doi: 10.1248/bpb.b14-00397. [DOI] [PubMed] [Google Scholar]
- 9.Klindworth A., Pruesse E., Schweer T. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuczynski J., Stombaugh J., Walters W.A. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics. 2011;36:10.7.1–10.7.20. doi: 10.1002/0471250953.bi1007s36. (Chapter 10): Unit 10. 7:1 –28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks D.H., Beiko R.G. Measuring community similarity with phylogenetic networks. Mol. Biol. Evol. 2012;29:3947–3958. doi: 10.1093/molbev/mss200. [DOI] [PubMed] [Google Scholar]
- 12.Kelly B.J., Gross R., Bittinger K. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics. 2015;31:2461–2468. doi: 10.1093/bioinformatics/btv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neut C., Colombel J.F., Guillemot F. Impaired bacterial flora in human excluded colon. Gut. 1989;30:1094–1098. doi: 10.1136/gut.30.8.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baek S.J., Kim S.H., Lee C.K. Relationship between the severity of diversion colitis and the composition of colonic bacteria: a prospective study. Gut and Liver. 2014;8(2):170–176. doi: 10.5009/gnl.2014.8.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyagi Abdul Malik, Yu Mingcan, Trevor M. The microbial metabolite butyrate stimulates bone formation via T regulatory cell-mediated regulation of WNT10B expression. Immunity. 2018 Dec;49(6):1116–1131. doi: 10.1016/j.immuni.2018.10.013. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller T.L., Wolin M.J. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl. Environ. Microbiol. 1996;62:1589–1592. doi: 10.1128/aem.62.5.1589-1592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint H.J., Duncan S.H., Scott K.P. Links between diet, gut microbiota composition and gut metabolism. Proc. Nutr. Soc. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 18.Porcher E., Tenaillon O., Godelle B. From metabolism to polymorphism in bacterial populations: a theoretical study. Evolution. 2001;55:2181–2193. doi: 10.1111/j.0014-3820.2001.tb00734.x. [DOI] [PubMed] [Google Scholar]
- 19.Lim A.G., Langmead F.L., Feakins R.M. Diversion colitis: a trigger for ulcerative colitis in the in-stream colon? Gut. 1999;44:279–282. doi: 10.1136/gut.44.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandtzaeg P., Secretory IgA. Designed for anti-microbial defense. Front. Immunol. 2013;4:222. doi: 10.3389/fimmu.2013.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palm N.W., de Zoete M.R., Cullen T.W. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bollinger R.R., Everett M.L., Wahl S.D. Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type1pili. Mol. Immunol. 2006;43:378–387. doi: 10.1016/j.molimm.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Friman V., Adlerberth I., Connell H. Decreased expression of mannosespecific adhesins by Escherichia coli in the colonic microflora of immunoglobulin A-deficient individuals. Infect. Immun. 1996;64:2794–2798. doi: 10.1128/iai.64.7.2794-2798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harig J.M., Soergel K.H., Komorowski R.A. Treatment of diversion colitis with short-chain-fatty acid irrigation. N. Engl. J. Med. 1989;320:23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 25.Komorowski R.A. Histologic spectrum of diversion colitis. Am. J. Surg. Pathol. 1990;14:548–554. doi: 10.1097/00000478-199006000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Pal K., Tinalal S., Al Buainain H., Singh V.P. Diversion proctocolitis and response to treatment with short-chain fatty acids-a clinicopathological study in children. Indian J. Gastroenterol. 2015;34:292–299. doi: 10.1007/s12664-015-0577-0. [DOI] [PubMed] [Google Scholar]
- 27.Schauber J., Bark T., Jaramillo E., Katouli M., Sandstedt B., Svenberg T. Local short-chain fatty acids supplementation without beneficial effect on inflammation in excluded rectum. Scand. J. Gastroenterol. 2000;35:184–189. doi: 10.1080/003655200750024371. [DOI] [PubMed] [Google Scholar]
- 28.Luceri C., Femia A.P., Fazi M., Di Martino C., Zolfanelli F., Dolara P., Tonelli F. Effect of butyrate enemas on gene expression profiles and endoscopic/histopathological scores of diverted colorectal mucosa: a randomized trial. Dig. Liver Dis. 2016;48:27–33. doi: 10.1016/j.dld.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Tominaga K., Tsuchiya A., Yokoyama J., Terai S. How do you treat this diversion ileitis and pouchitis? Gut. 2019 Apr;68(4):593–758. doi: 10.1136/gutjnl-2017-315591. Epub 2018 Mar 27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.