Abstract
The necessity to decrease our fossil energy dependence requests bioprocesses based on biomass degradation. Cellobiose is the main product released by cellulases when acting on the major plant cell wall polysaccharide constituent, the cellulose. Escherichia coli, one of the most common model organisms for the academy and the industry, is unable to metabolize this disaccharide. In this context, the remodeling of E. coli to catabolize cellobiose should thus constitute an important progress for the design of such applications. Here, we developed a robust E. coli strain able to metabolize cellobiose by integration of a small set of modifications in its genome. Contrary to previous studies that use adaptative evolution to achieve some growth on this sugar by reactivating E. coli cryptic operons coding for cellobiose metabolism, we identified easily insertable modifications impacting the cellobiose import (expression of a gene coding a truncated variant of the maltoporin LamB, modification of the expression of lacY encoding the lactose permease) and its intracellular degradation (genomic insertion of a gene encoding either a cytosolic β-glucosidase or a cellobiose phosphorylase). Taken together, our results provide an easily transferable set of mutations that confers to E. coli an efficient growth phenotype on cellobiose (doubling time of 2.2 h in aerobiosis) without any prior adaptation.
Keywords: β-Glucosidase, Cellobiose phosphorylase, LacI, LamB, Escherichia coli, Cellobiose metabolism
Highlights
-
•
Production of a cytosolic β-glucosidase or cellobiose phosphorylase allows E. coli to metabolize cellobiose.
-
•
Optimization of cellobiose uptake through E. coli enveloppe to sustain an efficient growth on this carbon source.
-
•
Genetic engineering of E. coli can provide the capacity of an efficient cellobiose catabolism.
1. Introduction
For several years now, the ability to degrade the inexpensive, very abundant and renewable lignocellulosic material has grown a massive interest since it constitutes a solution to overcome the problems arisen from the consumption of fossil energy. Hydrolysis of the major plant biomass component, the cellulose, by enzymatic cocktails mainly generates the disaccharide cellobiose (Cb), composed of 2 glucosyl residues (Parisutham et al., 2017; Shi and Weimer, 1996). Escherichia coli is unable to utilize Cb, but this microorganism is the most well-known and characterized bacterium, grows rapidly, and can easily be modified thanks to the numerous available genetic tools. This model bacterium is therefore an attractive microorganism for industrial applications including the biofuel production sector. Different strategies were thus formerly applied to enable E. coli to catabolize this disaccharide such as the reactivation of the cryptic operons cel, bgl, chb or asc but only limited growth on this carbon source could be achieved (Hall and Xu, 1992; Kricker and Hall, 1984; Parisutham and Lee, 2011, 2015; Parker and Hall, 1990). The secretion or cell surface exposure of β-glucosidases, an enzyme cleaving the β-1,4 glycosidic bond in Cb (Aso et al., 2019; Gao et al., 2015; Luo et al., 2014; Muñoz-Gutiérrez et al., 2012; Soma et al., 2012), has also been used to extracellularly hydrolyze this sugar into two glucose units in sufficient quantity to support the growth (Aso et al., 2019; Ma et al., 2011; Singhania et al., 2013).
In E. coli, glucose is then imported in the cytoplasm via the phosphotransferase system (PTS) known to directly control the uptake and utilization of various carbohydrates (Tchieu et al., 2001). In short, the PTS triggers a sugar catabolic repression that allows E. coli to quickly adapt to its preferred energy and carbon source: glucose. However, this fast adaptation is a burden for the development of lignocellulolytic based biotechnological applications since the use of other fermentable sugars released in the medium during the plant cell wall degradation process (arabinose, galactose or xylose) is likely to be hampered (Park et al., 2012).
To alleviate this PTS induced catabolic repression, the expression of a cytosolic β-glucosidase or cellobiose phosphorylase (Cb phosphorylase) could be envisioned to generate intracellular glucose (de Groeve et al., 2011; Parisutham et al., 2017). In this respect, the Cb phosphorylase produces one glucose and one α-glucose 1-P (Fosses et al., 2017), the latter being converted into glucose 6-P by the α-phosphoglucomutase (PGM) which directly enters the glycolytic pathway, thus allowing to save one ATP molecule. The major limitation of this strategy is the import of the Cb. Previous studies related to E. coli strains producing either a β-glucosidase or a Cb phosphorylase reported an interaction of the inner membrane lactose permease LacY encoded by the lac operon with Cb and that a ΔlacY mutant strain producing a Cb phosphorylase was found unable to grow on this sugar (Collins et al., 1989; Sekar et al., 2012), suggesting the involvement of this permease in Cb uptake.
In the present study, by combining genomic editing with directed evolution of E. coli followed by genome sequencing, we have designed a robust strain of E. coli engineered by markerless chromosomic modifications to metabolize Cb by 1/genomic integration of genes coding for a β-glucosidase or a Cb phosphorylase allowing the intracellular use of Cb, 2/insertion of specific mutations favorizing the uptake of Cb (either by production of a modified maltoporin LamB and/or by tuning the expression level of the lacY permease). We have thus defined a set of modifications that could be genetically transferred in any E. coli strain to provide an optimized Cb utilization phenotype without prior adaptation. We also propose a model of Cb catabolism of the engineered E. coli strains synthesizing either the β-glucosidase or the Cb phosphorylase.
2. Materials and methods
2.1. Bacterial strains and plasmids
The strains Ruminococcus albus F40, Zymomonas mobilis 4 (ATCC 31821), Ruminiclostridium cellulolyticum H10 (ATCC 35319) and Escherichia coli MG1655 were used in this study. All the primers used in this study are listed in Supplemental Table 1. All the engineered E. coli strains and plasmids used are reported in Supplemental Table 2.
2.2. Construction of pET28-bglA and pET28-bglAA781S expression vectors
The sequence of the β-glucosidase encoding gene bglA of R. albus was amplified by PCR using the genomic DNA of R. albus as the template and the primers rbglA F and rbglA R (Suppl. Table 1). The amplicon was cloned between the NcoI and XhoI restriction sites into the pET28a(+) generating the pET28-bglA vector coding for the recombinant protein rBglA bearing 6 histidine residues at its C-terminus. The resulting plasmid was introduced in E. coli NEB 5-α competent cells (NEB, Ipswich, MA, Suppl. Table 2). Recombinant clones were identified by PCR and the insert subsequently verified by sequencing.
The pET28-bglAA781S vector coding for the variant rBglAA781S in which the residue Ala781 is replaced by Ser was constructed by the one-step PCR method (Ansaldi et al., 1996) using plasmid pET28-bglA as a template and the two overlapping divergent primers: bglA A781S F and bglA A781S R (Suppl. Table 1). The DpnI treated PCR product was transferred to E. coli NEB 5-α competent (NEB, Suppl. Table 2). Clones were analyzed as described above.
2.3. Strains construction
2.3.1. Construction of the integrative vectors containing bglA or cbpA expression cassette
A multi-cloning site (MCS) containing the restriction sites: BamHI, BglII, XhoI and SphI, was generated by the annealing of the oligonucleotides pKD3-MCS F and pKD3-MCS R (Mounts et al., 1989; Suppl. Table 1). This double strand DNA was then cloned in the SphI-linearized close to the cat selection marker gene (coding for the chloramphenicol resistance) in pKD3 vector (Datsenko and Wanner, 2000). The resulting pKD3-MCS vector was subsequently introduced in competent E. coli BW25141 (pir+) cells (Suppl. Table 2). Transformants were selected on LB solid medium containing 34 μg/mL of chloramphenicol at 37 °C. The sequence of plasmids extracted from liquid cultures of clones was verified with the primer seq-MCS F (Suppl. Table 1).
The cassette “PpdcZm-bglA-TgforZm” containing the coding sequence of the gene bglA from R. albus fused to the promoter Ppdc from Z. mobilis (PpdcZm) and the terminator Tgfor from Z. mobilis (TgforZm), was amplified by PCR using the primers BamHI bglA F and XhoI bglA R (Suppl. Table 1) and the vector pJIR300-RA as template (courtesy of L. Dedieu, Ohmiya et al., 1991). The “PpdcZm-bglA-TgforZm” cassette was cloned between the BamHI and XhoI sites of the pKD3-MCS vector to generate the pKD3-bglA plasmid. After transformation of competent E. coli BW25141 cells (Suppl. Table 2), the corresponding plasmid was extracted and the sequence verified as described above.
The coding sequence of the gene cbpA was amplified using the primers NdeI cbpA F and XbaI cbpA R (Suppl. Table 1) and the genomic DNA of R. cellulolyticum H10 as template. The internal NdeI restriction sequence of the cbpA amplicon was then submitted to site directed mutagenesis by overlapping PCR using the primers cbpA mut NdeI F and cbpA mut NdeI R (Suppl. Table 1). The corresponding PCR fragment was subcloned between the NdeI and XbaI in pKD3-bglA to create the pKD3-cbpA vector containing the cassette “PpdcZm-cbpA-TgforZm”. The corresponding plasmid was extracted and its sequence verified as described above.
2.3.2. Genomic integration of bglA and cbpA between the ybfL and ybfD loci in E. coli MG1655 strain
“PpdcZm-bglA-TgforZm cat” or the “PpdcZm-cbpA-TgforZm cat” cassettes were integrated in the intergenic region between the loci ybfL and ybfD, located at 34 min in E. coli MG1655 (Suppl. Table 2) genome, using the method described by Datsenko and Wanner (2000). PCR fragments containing the “PpdcZm-bglA-TgforZm cat” or the “PpdcZm-cbpA-TgforZm cat” cassettes were produced respectively using the pKD3-bglA or pKD3-cbpA as templates and the primers pKD3-ybfD F and pKD3-ybfD R (Suppl. Table 1). Electrotransformation of E. coli MG1655 pKD46 (Suppl. Table 2) by the purified PCR product was then carried out using an Eporator device (Eppendorf, Hamburg, Germany) to generate the chromosomic mutations zde::bglA or zde::cbpA according to the genetic nomenclature for an insertion in a bacterial genome at the non-coding position located at 34 min (Maloy et al., 1994). Integrants were analyzed by PCR using external primers verif ybfD F/verif ybfD R (Suppl. Table 1). After curing of the pKD46 plasmid at 42 °C, pCP20 electrotransformed clones were thermally selected to generate markerless mutations and obtain the strains CbB (Cellobiose using BglA) clones 2 and 3 (CbB2 and CbB3, Suppl. Table 2), harboring the mutation zde::bglA, and CbCzero (Cellobiose using CbpA) clones 1 and 2 (CbCzero1 and CbCzero2, Suppl. Table 2), harboring the mutation zde::cbpA, respectively.
2.3.3. Construction of the E. coli strain CbB3 ΔlacI
The strain MG1655 ΔlacI (Suppl. Table 2) was generated by integration of a cat cassette in lacI using the method of Datsenko and Wanner (2000) as described above. The cat cassette was synthetized using the pKD3 vector as the template and the primers lacI-cat F/lacI-cat R (Suppl. Table 1). Integrants were selected on LB solid medium containing 17 μg/mL of chloramphenicol at 30 °C and analyzed by PCR using the pair of external primers seq lacI F/seq Plac R (Suppl. Table 1) and pKD46 plasmid was cured at 42 °C. Following the method of Baba and coworkers (Baba et al., 2006), the resulting ΔlacI::cat deletion was transduced in the strain CbB3 (Suppl. Table 2). Transductants were selected on LB solid medium containing 17 μg/mL of chloramphenicol at 37 °C and analyzed by PCR using the pair of external primers seq lacI F/seq Plac R (Suppl. Table 1).
2.3.4. Construction of the strain CbC
The zde::PpdcZm-cbpA-TgforZm cat mutation was transduced from CbCzero1 strain (Suppl. Table 2; Baba et al., 2006) to the evolved strain CbB3-evo1 (Suppl. Table 2, see below “Directed evolution of E. coli strains on Cb and genome sequencing”). The resulting strains CbC clones 2 and 3 (CbC2 and CbC3, Suppl. Table 2) displaying the mutation ΔbglA::cbpA, in which the gene cbpA was transduced in place of the bglA gene, was verified by PCR using external primers verif ybfD F/verif ybfD R (Suppl. Table 1).
2.3.5. Replacements of lac operon promoter and PpdcZm in E. coli CbB and CbC genomes
The Plac inducible promoter controlling the lacZYA operon was replaced by the PacpP or PtonB promoters. Similarly, the PpdcZm promoter controlling the expression of the genome-integrated bglA or cbpA genes was replaced by the Pgmk promoter from E. coli. First, PacpP, PtonB and Pgmk promoters’ sequences were amplified from E. coli MG1655 (Suppl. Table 2) genomic DNA using the pairs of primers cat-PacpP F/PacpP-lacZ R, cat-PtonB F/PtonB-lacZ R and cat-Pgmk F/bglA Pgmk R or cat-Pgmk F/cbpA Pgmk R, respectively (Suppl. Table 1). Second, “FRT-cat-FRT” cassettes were amplified using the pKD3 template and the pairs of primers Plac-cat F/cat-PacpP R, Plac-cat F/cat-PtonB R or cat-ybfD F/cat-Pgmk R (Suppl. Table 1). Finally, the corresponding purified PCR fragments were combined by overlapping PCR using the pairs of primers Plac-cat F/PacpP-lacZ R or Plac-cat F/PtonB-lacZ R, and cat-ybfD F/bglA Pgmk R or cat-ybfD F/cbpA Pgmk R, respectively (Suppl. Table 1). “FRT-cat-FRT – promoter” amplicons were then integrated into the genome of E. coli MG1655 (Suppl. Table 2) as described above (Datsenko and Wanner, 2000) to generate the strains CbB3 PacpP::lac, CbB3 PtonB::lac, CbB3-evo1-Pgmk, and CbB3-evo1-Pgmk, respectively (Suppl. Table 2). Integrants were verified by PCR using the pairs of external primers seq lacI F/seq Plac R or verif ybfD F/verif bglA R or verif ybfD F/verif cbpA R (Suppl. Table 1). The pKD46 plasmid was then cured at 42 °C, before markerless mutations were obtained as described above using the pCP20 vector (Datsenko and Wanner, 2000).
2.3.6. Construction and transduction of marked loci lamBΔ363-446::cat and yfeNS121P::kan
PCR fragments were synthetized using the vectors pKD3 (Cmr) and pKD4 (Kmr) as templates with primers lamB mut-Cter F/lamB mut-Cter R or yfeN-kan F/yfeN-kan R respectively (Suppl. Table 1) to generate specific resistance cassettes flanked by the intergenic region downstream lamBΔ363-446 (named lamB∗ mutation) or yfeNS121P loci. The strain CbC2-evo1 (Suppl. Table 2) previously transformed by pKD46 vector was separately electrotransformed with these PCR fragments to mark the mutated loci using the method of Datsenko and Wanner (2000). Integrants were selected on LB solid medium containing 17 μg/mL of chloramphenicol or 35 μg/mL of kanamycin at 37 °C and analyzed by PCR using the external primers seq lamB F/seq lamB R or seq yfeN F/seq yfeN R. Then, pKD46 plasmid was cured at 42 °C. The resulting mutations lamBΔ363-446::cat or yfeNS121P::kan were transduced into CbC2 and CbB3-evo1 strains (Suppl. Table 2; Baba et al., 2006).
2.3.7. Construction of the strain CbB3-evo1 ΔlamB
Following the method developed by Baba and coworkers (Baba et al., 2006), the mutation ΔlamB::kan was transduced from the strain of the Keio collection (Baba et al., 2006) into CbB3-evo1 strain (Suppl. Table 2). Transductants were selected on LB solid medium containing 35 μg/mL of kanamycin at 37 °C and analyzed by PCR with the pair of external primers seq lamB F/verif lamB R (Suppl. Table 1).
2.3.8. Genetic engineering of the E. coli strain able to metabolize the Cb
Mutation lamBΔ363-446::cat was transduced into the engineered strain CbB3 PacpP::lac (Suppl. Table 2; Baba et al., 2006), leading to CbB3 PacpP::lac lamB∗. The strain with markerless mutation was obtained using the pCP20 vector as described above (Datsenko and Wanner, 2000).
2.4. Construction of the reporter plasmid pUA-Ppdc
The sequence of the PpdcZm promoter from Z. mobilis was amplified by PCR using pKD3-bglA plasmid as a template and the primers Ppdc-zm XhoI F and Ppdc-zm BamHI R (Suppl. Table 1). The amplicon was cloned between the XhoI and BamHI restriction sites of the pUA-66 (Zaslaver et al., 2006) generating the pUA-Ppdc reporter vector coding for the transcriptional fusion PpdcZm-gfp.
The resulting plasmid was introduced in E. coli NEB 5-α competent cells (Suppl. Table 2). Transformants were analyzed by PCR and positive clones were subsequently verified by sequencing.
2.5. Growth in M9 minimum medium
Overnight cultures of E. coli strains were performed under shaking (150 rpm) at 37 °C in LB medium supplemented with the appropriate antibiotics. 200 μL of preculture were then used to inoculate aerobically 20 mL of M9 liquid medium supplemented with 0.5% w/v Cb (M9 Cb) or 0.2% w/v glucose (M9 Glu) under shaking (150 rpm) at 37 °C, and growth was monitored at 600 nm. For anoxic cultures, M9 medium was boiled under argon flow, and aliquoted in sealed 50 mL serum bottle, prior to sterilization. Other compounds (MgSO4, CaCl2, Thiamine, FeSO4) including glucose (5 g/L) or cellobiose (5 g/L) were subsequently added anaerobically. Cultures were inoculated anaerobically using 200 μL of overnight cultures in LB medium supplemented with the appropriate antibiotics.
2.6. Directed evolution of E. coli strains on Cb and genome sequencing
Cb utilization directed evolution of strains CbB2 and CbB3 (zde::bglA, Suppl. Table 2), CbCzero1 and CbCzero2 (zde::cbpA, Suppl. Table 2) and CbC2 and CbC3 (CbB3-evo1 ΔbglA::cbpA, Suppl. Table 2) were performed in M9 Cb liquid medium at 37 °C as described above. When the OD600nm reached a value of 1, subcultures were performed in fresh M9 Cb medium. Clones from the corresponding subcultures grown until the stationary phase were first isolated on M9 Cb solid medium at 37 °C, and secondly on LB plate at 37 °C. PCR were performed on the evolved largest colonies CbB2-evo1, CbB2-evo2, CbB3-evo1 and CbB3-evo2 (Suppl. Table 2) to analyze the sequence of specific gene targets as: lacI, Plac, PpdcZm, lacY, ompF, ompC, micF. Amplicons from PCR using respectively the primer pairs verif micF F/verif ompC R, verif ompF F/verif ompF R, seq lacI F/seq Plac R, seq lacY F/seq lacY R (Suppl. Table 1), were sequenced. Isolated clones from strains CbB3-evo 1 and CbC2-evo1 were also subcultured in LB medium at 37 °C under shaking (150 rpm) for genomic DNA extraction using Wizard genomic DNA preparation kit (Promega, Madison, WI) to perform whole genome sequencing (GATC-Eurofins, Luxembourg city, Luxembourg).
2.7. Promoter activity assay
Cultures of MG1655 transformed with pUA66, pUA-Plac, pUA-PacpP, pUA-PtonB, pUA-Pgmk, pUA-PmscI and pUA-PrrnB from the plasmid GFP reporter collection of Zaslaver and coworkers (Zaslaver et al., 2006) and pUA-Ppdc were performed in triplicate on microreader plate in 200 μL M9 medium supplemented with 100 μg/mL of ampicillin, 0.2% casamino acids and 0.2% glucose at 37 °C. OD600nm and GFP fluorescence (excitation at 488 nm and emission at 521 nm) were measured every 10 min with a Spark 10 M device (Tecan, Männedorf, Switzerland). Alternatively, the growth and the GFP fluorescence of CbB3-evo1 transformed with the same vectors and cultured in 50 mL M9 supplemented with 100 μg/mL of ampicillin, 0.2% casamino acids and either 0.2% glucose or 0.5% cellobiose, at 37 °C were monitored similarly.
2.8. Production and purification of rBglA and rBglAA781S
E. coli BL21(DE3) (NEB, Suppl. Table 2) transformed with the vectors pET28-bglA or pET28-bglAA781S, were grown at 37 °C under shaking (150 rpm) in 2 x 1 L of LB liquid medium supplemented with 50 μg/mL of kanamycin. 200 μM of isopropyl-thio-β-D-galactoside (Euromedex, France) were added at OD600nm 1 and the growth temperature was switched to 16 °C overnight. Cells were harvested by centrifugation (10 min at 4 °C and 3,000 g) and resuspended in 50 mL of 30 mM Tris-HCl pH 8.0, 5 mM Imidazole, containing few mg of DNAseI (Roche, Mannheim, Germany). Cells were broken using a French Press (Stansted Fluid Power Ltd, Harlow, UK) and the crude extract was subsequently centrifuged (15,000 g, 10 min, 4 °C) to remove cell debris, prior to loading on 3 mL of Ni-NTA resin (Qiagen, Hilden, Germany) equilibrated in 30 mM Tris-HCl, 5 mM Imidazole pH 8.0. The proteins of interest were eluted using 30 mM Tris-HCl, 100 mM Imidazole pH 8.0, buffer. Finally, the purified enzymes were buffer exchanged against 10 mM Tris-HCL pH 8.0 and concentrated by ultrafiltration using Vivaspin20 (cut off 30 kDa, Sartorius, Göttingen, Germany), and stored at −80 °C. The concentration of the proteins was estimated using the absorbance at 280 nm and the absorption coefficient calculated with the program ProtParam tool (www.expasy.org/tools/protparam.html).
2.9. Determination of the kinetic parameters of rBglA and rBglAA781S
The kinetic parameters of rBglA and rBglAA781S were determined on Cb as follows. Enzymes (1 μM) were incubated at 37 °C with Cb at concentrations of 0.8, 1, 1.25, 1.67, 2.5, 5, 10, 20, 50, and 250 mM in 20 mM Hepes pH 7.0 and 5 mM CaCl2. After 5 and 10 min of incubation, aliquots (1–10 μL) were pipetted and diluted in a final volume of 250 μL in 0.1 M NaOH. Samples were analyzed by High Pressure Anion Exchange Chromatography coupled with Pulsed Amperometric Detection (HPAEC-PAD) (Thermofisher, Waltham, MA). Quantification of glucose and Cb was performed by applying 25 μL of diluted samples to a Dionex CarboPac PA1 column (4 x 250 mm) (Thermofisher) and the corresponding guard column (4 x 50 mm) at 30 °C. Sugars were eluted with the buffers 0.1 M NaOH and 0.5 M sodium acetate +0.1 M NaOH as the eluents A and B, respectively. The following multi-step procedure was used: isocratic separation (5 min, 95% A + 5% B), separation gradient (8 min, 10–37% B), column wash (2 min, 99% B) and subsequent column equilibration (2.5 min, 95% A + 5% B). The flow rate was kept at 1 mL/min in all cases. Injection of samples containing glucose and Cb (Sigma-Aldrich, Saint-Louis, MI) at known concentrations ranging from 4 to 100 μM were used for quantification.
2.10. Determination of CbpA and BglA activities in crude extracts
Fifty-mL overnight cultures of CbB3-evo1, CbB3-evo1-Pgmk, CbC2-evo1, CbC2-evo1-Pgmk and MG1655 (Suppl. Table 2) in lysogenic broth were centrifuged at 7,000 g for 10 min at 4 °C. The cell-containing pellet was resuspended in 8 mL of 50 mM KHPO4 buffer (pH 7.0) supplemented with few mg of DNAse I, and broken in a French press. The crude extract was centrifuged at 15,000 g for 10 min at 4 °C, and the supernatant was concentrated by ultrafiltration on Vivaspin20 (cut off 30 kDa) to 1 mL. The activity of BglA was monitored by incubating 10 μL of 20-fold diluted sample of CbB3-evo1, CbB3-evo1-Pgmk concentrated crude extracts, or 10 μL of MG1655 non-diluted concentrated crude extract with 990 μL of p-nitrophenyl-β-D-glucoside (pNP-Glu) at 1 g/L in 50 mM KHPO4 buffer (pH 7.0) at 37 °C. The activity was determined by monitoring the release of p-nitrophenol at 400 nm as previously described (Ravachol et al., 2016). The Cb phosphorylase activity was determined by incubating 20 μL of CbC2-evo1, CbC2-evo1-Pgmk or MG1655 crude extract with 980 μL of 10 mM Cb in 50 mM KHPO4 buffer (pH 6.2) containing 0.01% NaN3 (w/v) at 37 °C. At specific time points, 200 μL were pipetted and mixed with 50 μL of 0.025 M H2SO4. Twenty-five μL of samples were the applied on an (300 x 7.8 mm) Aminex HPX87H column (Bio-rad, Hercules, CA) preceded by the corresponding (30 x 4.6 mm) guard column at 65 °C. Sugars were eluted using the eluent 5 mM H2SO4 (isochratic separation for 15 min), and detection was performed using a refractive index detector (Iota, Marseille, France). Injections of glucose and Cb at known concentrations (ranging from 0.5 to 10 mM) were used to determine the amount of consumed Cb and released glucose.
2.11. Sugar consumption by CbB3 PacpP::lac lamB∗ and MG1655 strains, and release of fermentation products in anaerobiosis
The consumption of glucose and Cb by CbB3 PacpP::lac lamB∗ (Suppl. Table 2) and MG1655 strains under aerobic conditions was followed by pipetting 1-mL culture samples at specific time points. Samples were centrifuged 10 min at 4 °C at 15,000 g, and 200 μL of supernatant were mixed with 50 μL of 0.025 M H2SO4 prior to analyses on Aminex HPX87H as described above.
For anaerobic condition, the same procedure was followed except that the isochratic separation was extended to 25 min, and in addition to glucose and Cb, injection of succinate, lactate, formate, acetate and ethanol at known concentration (0.5–30 mM) were performed to quantify both the sugar consumption and the release of fermentation products.
3. Results and discussion
3.1. Selection of the Cb degrading enzymes and determination of the catalytic properties of the β-glucosidase (BglA) of Ruminococcus albus
In this study, we aimed to create an E. coli strain able to intracellularly catabolize Cb. Two major types of enzymes can cleave the stable β-1,4 glycosidic bond linking the two glucosyl residues in Cb: the β-glucosidases and the Cb phosphorylases, that release two glucoses or one glucose and one α-glucose 1-phosphate, respectively (de Groeve et al., 2011; Fosses et al., 2017; Kim et al., 2018; Parisutham et al., 2017; Wilson, 2009). We selected 1/the β-glucosidase BglA from R. albus F40 (Ohmiya et al., 1991) belonging to the Glycoside Hydrolase 3 family (Lombard et al., 2014) and whose plasmid-based overproduction in E. coli allowed its growth in M9 Cb (L. Dedieu, personal communication), and 2/the efficient Cb phosphorylase CbpA from R. cellulolyticum H10 belonging to the Glycoside Hydrolase family 74 (Lombard et al., 2014) that was previously characterized (Fosses et al., 2017), and known to be the most active enzyme of R. cellulolyticum on Cb (Liu et al., 2010, 2019). In order to precise the enzymatic properties of BglA, its encoding gene was therefore cloned in a pET28 vector which was subsequently used to transform the strain BL21(DE3). Recombinant protein rBglA was purified to homogeneity and kcat and KM values of 1045.3 ± 22.6 min−1 and 49.32 ± 2.81 mM were obtained on Cb (at pH 7.0 and 37 °C), respectively. The Cb phosphorylase CbpA was formerly purified and characterized, providing kcat and KM values of 1458.4 ± 108.0 min−1 and 2.85 ± 0.47 mM, respectively (Fosses et al., 2017). Both Cb degrading enzymes thus exhibit catalytic velocities in the same range, but rBglA displays a 17-fold higher KM for the disaccharide, thus leading to a 24-fold lower catalytic efficiency (kcat/KM) compared with CbpA (Suppl. Table 3).
Both enzymes were assayed without a priori to generate a Cb-utilization phenotype to E. coli since no preliminary information were available concerning the Cb intracellular concentration and fluxes. It is however worth considering that rBglA hydrolyses Cb into two glucose residues whereas CbpA generates one glucose and one α-glucose 1-P, thereby allowing the saving of one ATP molecule when the α-glucose 1-P is converted into glucose 6-P by the α-phosphoglucomutase (PGM) of E. coli (Lu and Kleckner, 1994) and directly enters the glycolytic pathway.
3.2. Effect of the genomic integration of bglA or cbpA genes in E. coli: metabolism of the Cb and growth
As stated above, the expression of the bglA gene of R. albus cloned in the pJRD300Ra expression vector sustained a limited growth of the E. coli recipient strain on M9 minimal medium supplemented with 0.5% Cb. To overcome any problems due to vector instability (Hershberger and Rosteck, 1991; Jakupciak and Wells, 1999; Patient and Summers, 1993; Standley et al., 2019), we chose to integrate the expression “PpdcZm-bglA- TgforZm” cassette in the E. coli MG1655 chromosome. In this cassette bglA expression is under the control of the strong promoter Ppdc from Z. mobilis strain 4 and the terminator Tgfor from Z. mobilis strain 4 (Reynen et al., 1990; Seo et al., 2005). We successfully integrated this cassette between the loci ybfL and ybfD in the intergenic region zde (Maloy et al., 1994), a position allowing foreign DNA integration up to 4 kb (Espeli et al., 2008; Lesterlin et al., 2012). The generated strain harboring the mutation zde::bglA is hereafter named CbB (Cellobiose using BglA). Corresponding isolated clones identified as E. coli CbB2 and CbB3 were selected. Similarly, E. coli strain harboring the mutation zde::cbpA was obtained by the integration of the “PpdcZm-cbpA-TgforZm” cassette between the same loci, and named CbCzero (Cellobiose using CbpA). Corresponding isolated clones CbCzero1 and CbCzero2 strains were selected.
Upon sequencing of all integrated regions, one mutation was found in the bglA gene leading to the replacement of the Ala 781 by a Ser residue. The mutated gene was cloned in a pET28 vector, and the corresponding modified rBglAA781S was produced and purified using the same procedure as the wild type (WT) rBglA. The kinetic parameters of rBglAA781S were established on Cb as formerly described and both kcat (1231.0 ± 18.3 min−1) and KM (53.99 ± 2.09 mM) were similar to that of the WT rBglA (Suppl. Table 3). Therefore, this mutation leading to a “neutral” aminoacid substitution in rBglA in E. coli CbB2 and CbB3 strains was kept in this following strain engineering experiments.
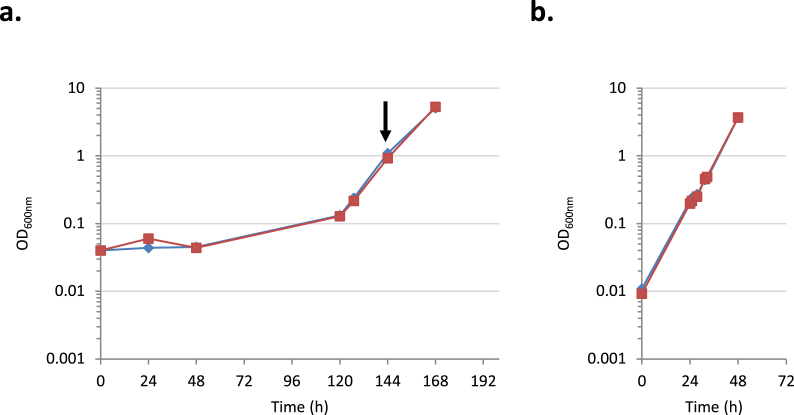
The ability of each strain to use the disaccharide as the sole carbon source was then evaluated on M9 minimum medium supplemented with 0.5% Cb. Despite the presence of cbpA, no growth was observed for E. coli CbCzero1 and CbCzero2 upon 15 days of cultivation (data not shown) whereas E. coli CbB2 and CbB3 displayed a growth after a 5-day lag phase (Fig. 1a) but no lag phase was observed upon direct subculture in the same fresh minimum medium supplemented with 0.5% Cb (Fig. 1b) suggesting that an adaptation event occurred. These results indicate that the integration of bglA or cbpA genes in E. coli genome is not sufficient alone to directly induce a growth on Cb, and that an in vivo evolution step conferred the capacity to grow on Cb-based medium to E. coli CbB2 and CbB3. Colonies of evolved clones were isolated by plating dilutions of each liquid cultures on M9-Cb solid medium. The 4 largest colonies named E. coli CbB2-evo1, CbB2-evo2 and CbB3-evo1, CbB3-evo2 were stored for further studies. Among them, the clone CbB3-evo1 which generated the largest colonies on M9-Cb plates, was therefore selected for subsequent analyses and engineering.
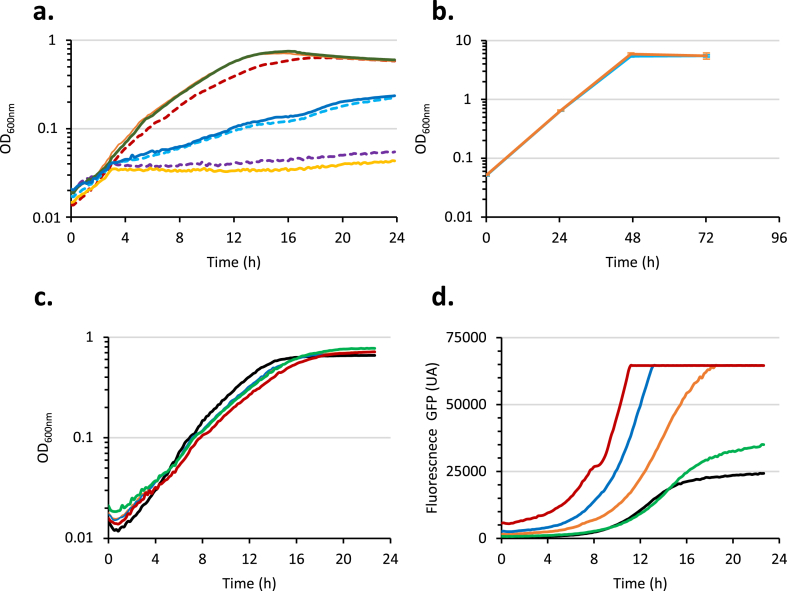
Fig. 1.
Growth of CbB strains in Cb-based M9 medium.a. CbB2 (blue curve) and CbB3 (red curve) were cultured in M9 medium containing 0.5% Cb at 37 °C. They were sub-cultured when OD600nm reached 1 (black arrow). b. Sub-cultures of CbB2 (blue curve) and CbB3 (red curve) grown in M9 medium containing 0.5% Cb at 37 °C. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Identification of the mutation in lacI responsible of the Cb growth phenotype of the CbB3-evo1 strain
To identify the beneficial traits implicated in the adaptation of the CbB2-evo1, CbB2-evo2, CbB3-evo1 and CbB3-evo2 clones to grow on Cb, we firstly performed conventional PCR-based sequencing on specific loci. We targeted 1/the PpdcZm promoter sequence that we introduced to control the expression of bglA, 2/various regions of the E. coli lac operon coding for genes previously proposed to be involved in Cb uptake in adapted strain (Collins et al., 1989; Sekar et al., 2012; Vilar et al., 2003) such as the PlacZ promoter of the lac operon (Donovan et al., 1996), lacI coding the repressor of the expression of lac operon (Lewis, 2005), lacY coding the lactose permease proposed to import the Cb (Guan and Kaback, 2006; Kaback, 2005), and 3/ompC and ompF, coding porins of the outer membrane known to leave sugars enter into the periplasm (Prilipov et al., 1998).
Only lacI was found to be affected by different types of mutation in the 4 clones we tested, and this exclusive event was confirmed by the complete genome resequencing of E. coli CbB2-evo1 and CbB3-evo1. The various modifications identified on LacI structure are mapped in Supplemental Fig. 1. One single-nucleotide polymorphism (SNP) impacts the helix-turn-helix motif implicated in the interaction with the lacO operator (Val 23 changed into Met in the clone CbB3-evo2). We also detected modifications potentially altering the structure of the tetramerization region of LacI: two SNPs changing the Glu 181 into Tyr and Asp 247 into Ala for clones CbB2-evo2 and CbB2-evo1 respectively, and 4-base pair insertion generating a premature stop codon and leading to the production of the truncated LacIΔ198-331 (hereafter called LacI∗) for the clone CbB3-evo1. None of these mutations were ever reported in the literature concerning LacI variants (Barker et al., 1998; Bell et al., 2001; Miura-Onai et al., 1995; Tang et al., 2006; Xu and Matthews, 2009).
All these mutations suggest that the Cb growth phenotype upon adaptation results from a modification of the lac operon expression engendered by LacI variants presumably exhibiting decreased affinity for the lacO operators or altered tetramerization properties, thereby hampering the regulation activity of these modified repressors (Goodson et al., 2013). We hypothesized that the impaired binding of LacI drastically modifies the expression of the lac operon leading to an increased synthesis of the encoded proteins, as the inner membrane permease LacY previously proposed to prone to transport Cb into the cytosol (Collins et al., 1989; Sekar et al., 2012). In former studies (Beckwith, 1967; Desai et al., 2014; Donovan et al., 1996; Ibrahim et al., 2017; Mueller-Hill et al., 1964; Yang et al., 2015), the role of LacY in the Cb import was not investigated as IPTG was used as an inducer for the expression of Cb catabolic enzymes, which indeed also triggered the expression of the lac operon.
3.4. Tuning the expression of the lac operon impacts the Cb metabolism and the growth of the resulting modified E. coli
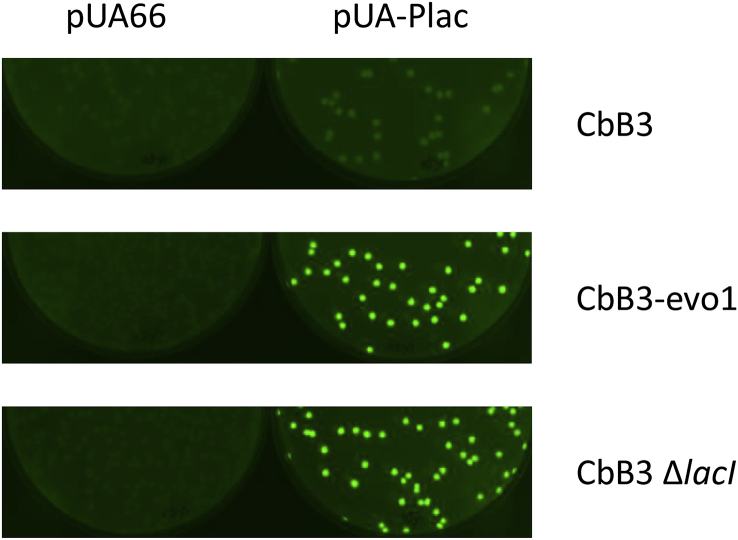
To verify that the mutations in lacI increased the expression of the lac operon, we firstly used pUA-Plac from the plasmid reporter library of Zaslaver et al. (2006) containing the Plac-gfp transcriptional fusion to investigate the repression activity of the LacI∗ variant we identified. E. coli CbB3 expressing lacI WT, CbB3-evo1 expressing lacI∗ and a newly constructed strain CbB 3ΔlacI, in which the gene lacI has been fully deleted, were transformed with the pUA66 (carrying for the gfp gene without any promoter) or the pUA-Plac. Fluorescence images of the corresponding transformed clones on LB plates are shown in Fig. 2.
Fig. 2.
Expression of thelacoperon in CbB and CbB evo strains revealed by Plac-gfpreporter. Fluorescence signal of the colonies grown on LB plate read Typhoon FLA9500 imager for CbB3, CbB3-evo1 and CbB3-ΔlacI, transformed with vectors pUA66 (gfp without any promoter) or pUA-Plac (Plac-gfp).
As expected, all pUA66 harboring clones were not fluorescent. Unsurprisingly, a high fluorescence signal was observed for the non-evolved E. coli CbB3 ΔlacI pUA-Plac clones because of the non-repression of the gfp gene in the absence of LacI repressor. The same pattern of fluorescence was observed for the evolved E. coli CbB3-evo1 pUA-Plac clones indicating that the LacI∗ is less active than the WT variant. The non-evolved E. coli CbB3 pUA-Plac did not display any GFP emission since the expression of the gfp gene is not induced due to the presence of the LacI WT repressor. Consequently, these data suggest that the repression of the lac operon in the E. coli CbB3-evo1 strain is significantly reduced, allowing the strain to produce the inner membrane LacY permease in a suitable quantity to favor the efficient uptake of Cb. Nevertheless, the CbB3 ΔlacI, strain was found unable to grow on both Cb and lactose (Suppl. Fig. 2b and c), suggesting that a total de-repression of the lac operon induces an overproduction and an aggregation of LacY, as formerly reported (Geerstma et al., 2008), thereby decreasing the amount of functional permease in the inner membrane. This observation also suggests that in CbB3-evo1 strain, the C-terminal truncation of LacI does not trigger a full de-repression of the lac operon.
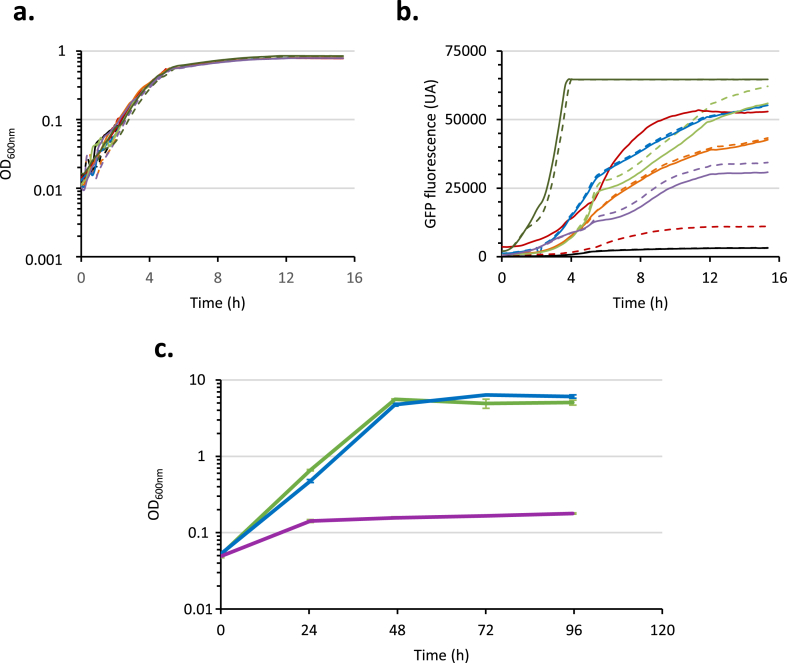
However, to secure the import of Cb through the inner membrane, these results prompted us to replace the Plac promoter by another promoter of E. coli. We selected a set of constitutive promoters displaying various promoter activity: Pgmk < PtonB < PmscS < PacpP << PrrnB (rRNA promoter known as one of the strongest promoters of E. coli) according to Zaslaver and coworkers (Zaslaver et al, 2006, 2009). We searched among these constitutive promoters which could mimic the strength of the Plac promoter like in the CbB3-evo1 strain harboring the LacI∗ variant. E. coli CbB3 and CbB3-evo1 were then transformed with the plasmids: pUA66 (no promoter -gfp) as a negative control, pUA-pPlac (Plac-gfp) as a comparative reference, pUA-pPacpP (PacpP-gfp), pUA-pPgmk (Pgmk-gfp), pUA-pPmscS (PmscS-gfp), pUA-pPtonB (PtonB-gfp), and pUA-PrrnB (PrrnB-gfp) from the E. coli gfp reporter plasmids library (Zaslaver et al, 2006, 2009). The corresponding strains were tested for their capacity to grow in M9 containing 0.2% casamino acids and 0.2% glucose at 37 °C and no growth defect was observed for any of the tested strains (Fig. 3a). As expected, a significant difference in the fluorescence profiles was observed for the strains E. coli CbB3 and CbB3-evo1 both bearing the pUA-Plac (Fig. 3b, red curves) due to the de-repression effect of the LacI∗ in the evolved strain. The selected promoters did not display altered activity in presence of LacI WT or its variant LacI∗ (Fig. 3b, dotted and filled curves, respectively). The strongest PrrnB promoter (Fig. 3b, dark green curves) exhibits a much higher constitutive activity than the LacI∗ de-repressed Plac one. In contrast, both PacpP (blue curves) and PmscS (green curves) promoters displayed an activity similar to the LacI∗ de-repressed Plac promoter (filled red curve) whereas Pgmk (orange curves) and PtonB (purple curves) promoters exhibit a weaker activity.
Fig. 3.
Promoter activities in CbB3 and CbB3-evo1 strains grown on glucose (a and b) and growth of CbB3 PacpP::lacand CbB3 PtonB::lacstrains in Cb-based M9 medium (c). Cells (a and b) were cultured in M9 0.2% CASA 0.2% glucose at 37 °C, and growth and fluorescence signal were monitored in a Tecan Spark plate reader. a. Optical density at 600 nm. b. Fluorescence level of GFP (excitation 488 nm / emission 521 nm). CbB3 (dotted line) or CbB3-evo1 (filled line) transformed by pUA66 (black), pUA-Plac (red), pUA-PacpP (blue), pUA-Pgmk (orange), pUA-PmscS (light green), pUA-PtonB (purple), or pUA-PrrnB (dark green). The mean of the data obtained on three culture replicates is shown. c. CbB3-evo1 (green), CbB3 PacpP::lac (blue), and CbB3 PtonB::lac (purple) strains were cultured in M9 medium containing 0.5% Cb at 37 °C. The mean of the data obtained on three culture replicates is shown. Bars indicate the standard deviations. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Based on these observations, we replaced the chromosomic Plac promoter by an equivalent (PacpP) or a weaker (PtonB) promoter in the E. coli CbB 3 strain to test the impact of the promoter strength on the cell capacity to use Cb for its growth. Growth curves on Cb-based medium of the corresponding strains CbB3 PacpP::lac and CbB3 PtonB::lac are reported in Fig. 3c. Only the PacpP promoter in CbB3 PacpP::lac strain was able to restore a growth phenotype similar to that of the evolved E. coli CbB3-evo1 in M9-Cb (Fig. 3c, blue and green curves, respectively) with a comparable doubling time (Table 1).
Table 1.
Generation time of CbB and CbC strains in M9 based Cb medium.
| Strain |
Cb catabolic enzyme |
Doubling time (h) |
||
|---|---|---|---|---|
| CbB3 | BglA | ND | ||
| CbB3-evo1 | BglA | 7.03 | +/− | 0.61 |
| CbCzero1 | CbpA | ND | ||
| CbC2 | CbpA | ND | ||
| CbC2-evo1 | CbpA | 2.58 | +/− | 0.06 |
| CbC2-evo2 | CbpA | 2.39 | +/− | 0.07 |
| CbC2-evo3 | CbpA | 2.57 | +/− | 0.05 |
| CbC2-evo4 | CbpA | 2.49 | +/− | 0.09 |
| CbC2-evo5 | CbpA | 2.52 | +/− | 0.03 |
| CbC2-evo6 | CbpA | 2.58 | +/− | 0.02 |
| CbC2 lamB∗ | CbpA | 2.62 | +/− | 0.11 |
| CbC2 yfeNS121P | CbpA | ND | ||
| CbB3-evo1 lamB∗ | BglA | 2.48 | +/− | 0.03 |
| CbB3-evo1 yfeNS121P | BglA | 7.20 | +/− | 0.79 |
| CbB3-evo1 ΔlamB | BglA | 6.94 | +/− | 0.38 |
| CbB3-evo1-Pgmk | BglA | 19.59 | +/− | 2.30 |
| CbC2-evo1-Pgmk | CbpA | 49.19 | +/− | 4.07 |
| CbB3 PacpP::lac | BglA | 6.94 | +/− | 0.53 |
| CbB3 PtonB::lac | BglA | ND | ||
| CbB3 PacpP::lac lamB∗ | BglA | 2.20 | +/− | 0.05 |
The mean of the data obtained on three culture replicates is shown.
ND: not determined.
Altogether, these data confirm that the control of the lac operon expression is the keystone of the evolution of the E. coli CbB3 strain toward the E. coli CbB3-evo1 strain able to use Cb. They also demonstrate that the replacement of Plac by PacpP is sufficient to establish a Cb growth phenotype in a non-evolved strain.
3.5. Identification of other complementary mutations that improve the Cb growth phenotype of the modified E. coli
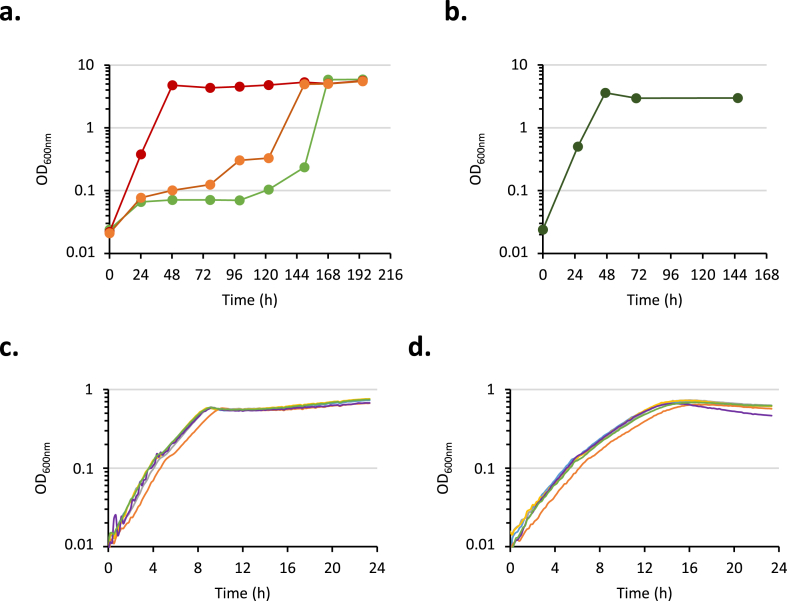
Considering that CbpA displays higher catalytic efficiency than BglA and allows to save one ATP molecule per metabolized Cb, that should thus provide a benefit in terms of metabolism, we replaced the integrated gene bglA by cbpA in the E. coli CbB3-evo1 strain. The cassette “PpdcZm-cbpA-TgforZm cat” was transduced into CbB3-evo1 to generate the strain CbC, displaying the mutation ΔbglA::cbpA, and clones CbC2 and CbC3 were studied. In contrast to CbCzero1 or 2 (see above), these genetically reengineered strains started to grow after 5 days of culture in M9-Cb medium (Fig. 4a, orange and green curves) and subsequent subcultures did not display any lag phase (Fig. 4b, red curve) suggesting that another evolution event occurred. From CbC2 adapted culture, several clones called CbC2-evo1 to CbC2-evo6 were isolated. They all exhibited similar growths in M9-Glucose (Fig. 4c) and -Cb (Fig. 4d) minimum media, with a doubling time ranging from 2.39 to 2.58 h (Table 1). CbC2-evo1 displays a more than two-fold reduced generation time compared to CbB3-evo1 strains (Table 1) demonstrating that the growth of CbC2-evo1 is significantly improved compared to the initial CbB3-evo1 strain (Suppl. Fig. 3, 6a and b).
Fig. 4.
Growth of CbC strains in Cb-based M9 medium. a. CbC2 (orange curve), CbC3 (green curve), and CbB3-evo1 (red curve) strains were cultured in M9 medium supplemented with 0.5% Cb at 37 °C. b. CbC2 was sub-cultured when initial culture reached the stationary phase. Sub-culture (dark green curve) was performed in M9 containing 0.5% Cb at 37 °C. c. Culture of isolated colonies CbC2-evo1 (blue), CbC2-evo2 (orange), CbC2-evo3 (grey), CbC2-evo4 (yellow), CbC2-evo5 (purple), and CbC2-evo6 (green) in M9 0.2% glucose at 37 °C and d. in M9 containing 0.5% Cb at 37 °C. Growth was monitored in Tecan Spark plate reader, of isolated colonies CbC2-evo1 (blue), CbC2-evo2 (orange), CbC2-evo3 (grey), CbC2-evo4 (yellow), CbC2-evo5 (purple), or CbC2-evo6 (green). The mean of the data obtained on three culture replicates (c. and d.) is shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Genome sequencing of CbC2-evo1 revealed the presence of two additional mutations in loci yfeN and lamB. Both mutations were confirmed by Sanger sequencing of the target loci. YfeN is a putative outer membrane protein belonging to the Tsx nucleoside transporter family (Ye and Van der Berg, 2004). The SNP mutation leads to the replacement of the Serine residue at position 121 by a Proline that could modify the tertiary structure of this porin. LamB, also known as Lambda phage receptor (Berkane et al., 2006; Chatterjee and Rothenberg, 2012), is the maltose outer membrane porin involved in the import of maltodextrins in the periplasm (Charbit, 2003; Klebba, 2002; Mulvihill et al., 2019). Mutation observed in lamB (hereafter named lamB∗) resulted from the insertion of the natural transposable element IS2D (Gonçalves et al., 2014; Saedler and Heiss, 1973). The modification of the ORF sequence at lamB locus led to the addition after the 362nd residue of four codons (Arg-Leu-Ala-Pro) and a premature termination codon thus resulting in the production of a truncated LamB (Δ363–446) whose 83 C-terminal residues are missing.
3.6. A mutation in lamB gene modifying the structure of the porin allows the strain to grow faster on Cb
To assess the impact of these mutations, yfeNS121P and lamB∗ were marked by integration of kan and cat resistance genes respectively, and were subsequently individually transduced into non-evolved CbC2 strain and CbB3-evo1 strain. Growth was followed on Cb-based medium (Fig. 5a). The transduction of lamB∗ mutation was found sufficient to allow the strain CbC2 lamB∗ to grow on M9 Cb without any adaptation step (orange curve). In contrast, the mutation yfeNS121P does not provide any benefit to both strains (yellow and blue curves). Surprisingly, CbB3-evo1 carrying lamB∗ mutation is able to grow as fast as CbC2-evo1, exhibiting a doubling time reduced to 2.5 h (Table 1).
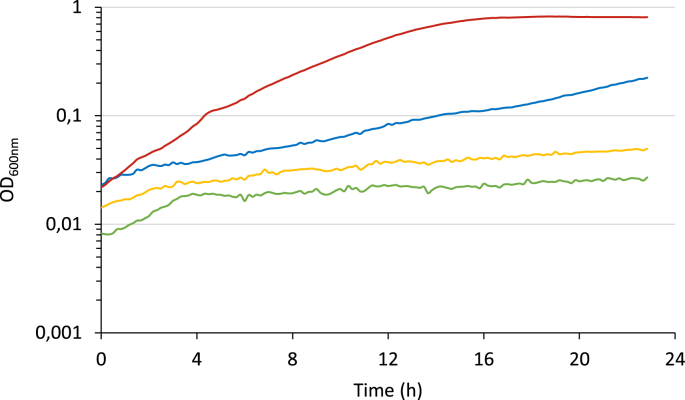
Fig. 5.
Growth of strains carryinglamBmutations in Cb-based M9 medium (a and b)and promoter activities in CbB3-evo1lamB∗strain grown on Cb (c and d). a. Culture in M9 containing 0.5% Cb at 37 °C of the strains CbB3evo1 (blue, dotted line), CbB3evo1 yfeNS121P (blue), CbB3evo1 lamB∗ (green), CbC2 (purple, dotted line), CbC2evo1 (red, dotted line), CbC2 yfeNS121P (yellow), and CbC2 lamB∗ (orange). The growth was monitored in Tecan Spark plate reader, and the data show the mean of three culture replicates. b. Culture in M9 containing 0.5% Cb at 37 °C of the strains CbB3-evo1 (blue) and CbB3-evo1 DlamB (orange). The mean of the data obtained on three culture replicates is shown.c. and d. The strains CbB3-evo1 lamB∗ transformed with pUA66 (black), pUA-Plac (red), pUA-PacpP (blue), pUA-Pgmk (orange), pUA-Ppdc (green) were grown in M9 containing 0.5% Cb at 37 °C. Growth and fluorescence signal were monitored in Tecan Spark plate reader. The mean of the data obtained on three culture replicates is shown. c. Optical density at 600 nm. d. Fluorescence level of GFP (excitation 488 nm / emission 521 nm). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
LamB barrels are assembled into trimers in the outer membrane (Baldwin et al., 2011; Luckey et al., 1991; Vos-Scheperkeuter and Witholt, 1984). The mutation engenders a deletion of 83 residues encompassing the three last β-sheets (Δ363–446) at the C-terminal extremity (Suppl. Fig. 4a) which contains the key residues Trp383 and Trp445 (Trp358 and Trp420 in the mature LamB) involved in the direct interaction with maltose, and allowing the specific transport of the disaccharide trough the membrane (Nikaido, 2003). Indeed, it is well known that LamB porin allows the diffusion of several other disaccharides displaying α-linkage (maltose, trehalose, melibiose, Suppl. Fig. 4b) but restricts the diffusion of disaccharides harboring β-linkage (cellobiose, sucrose, lactose) (Luckey and Nikaido, 1980). This Δ363–446 truncation should lead to a reshaping of the internal channel (Suppl. Fig. 4b and c) of LamB as illustrated by the predicted structure of the truncated LamB∗ based on the Omp-Pst2 from Providencia stuartii (Suppl. Fig. 4c), another outer membrane protein belonging to the porin superfamily as LamB. Omp-Pst2 is the closest porin, sharing 30.8% similarity with the truncated LamB∗ in the same range of size (Suppl. Fig. 5). Overall, the in-depth modification of the inner channel and the missing recognition residues might explain the ability of Cb to diffuse through LamB∗.
We then compared the capacity of the various Cb-metabolizing strains to grow on limited available Cb. The strains CbB3-evo1, CbC2-evo1, and CbB3-evo1 lamB∗ were grown on M9 medium containing a range of Cb concentrations from 0.025% to 1% (Suppl. Fig. 6). The CbB3-evo1 strain is still able to grow at a minimal concentration of 0.1% Cb, whereas the CbC2-evo1 strain displays a significant growth at only 0.05% Cb. Nevertheless, the CbB3-evo1 lamB∗ strain is the most efficient as it can grow, thanks to the lamB∗ mutation at only 0.025% Cb. These results indicate that the production of the truncated LamB favors the growth of the strains (CbC2-evo1 and CbB3-evo1 lamB∗), especially at low Cb concentration, compared to the strain displaying the full-length LamB porin (CbB3-evo1).
Since LamB∗ variant as found in CbC2-evo1 strain provides a better growth phenotype on Cb to both CbC2 and CbB3-evo1 lamB∗strains compared to CbB3-evo1, we investigated if in the initial strain CbB3-evo1, the native porin LamB is involved in the transport of Cb through the outer membrane. For this study, we constructed the strain CbB3-evo1 ΔlamB which does not produce LamB and its growth was compared to that of the strain CbB3-evo1 on Cb-based medium (Fig. 5b). Both strains exhibit similar growth, suggesting that Cb diffused through the outer membrane in the periplasm by another transport system in the strain CbB3-evo1. Thus, truncation of LamB improves the growth in the strain CbB3-evo1 lamB∗ by providing an additional Cb-transport system to the periplasm.
3.7. A stronger expression of bglA or cbpA is deleterious for the growth on Cb
To improve the Cb based growth capacity of the engineered strains, we first investigated the strength of PpdcZm promoter in E. coli and the effect of the replacement of this promoter controlling the expression of bglA and cbpA by stronger ones. The CbB3-evo1 lamB∗ strain was selected to monitor the promoter activity during the growth on Cb. This strain was transformed with pUA vectors containing PacpP, Plac, Pgmk (Zaslaver et al., 2006) or PpdcZm promoters, the latter being cloned in a pUA66 upstream of the gene encoding the GFP. The activity of the promoters was then measured on cells grown in M9 medium containing Cb (Fig. 5c and d). Compared to PacpP (blue curve), Plac (red curve), or Pgmk (orange curve), the fluorescence level reached with the PpdcZm promoter is very low depicting a weaker strength of this promoter.
Based on these observations, the PpdcZm promoter regulating the expression of bglA and cbpA was replaced by Pgmk in the strains CbB3-evo1 and CbC2-evo1, respectively. The growth of the resulting strains was afterwards examined on Cb-based medium (Fig. 6). CbB3-evo1-Pgmk (in yellow) and CbC2-evo1-Pgmk (in green) exhibited a very limited growth compared to CbB3-evo1 (blue curve) and CbC2-evo1 (red curve). In the case of CbC2-evo1-Pgmk, the doubling time was increased up to 33 h. Clearly, the replacement of PpdcZm promoter by the stronger promoter Pgmk severely affected the growth of both strains. This observation indicates that the use of a stronger promoter is detrimental and that PpdcZm induces a suitable expression of the genes encoding the heterologous Cb-degrading enzymes. Analyses of the activity of crude extracts of strains CbB3-evo1, CbB3-evo1-Pgmk, CbC2-evo1 and CbC2-evo1-PgmK, revealed that the BglA activity on pNP-Glu in CbB3-evo1-Pgmk extract (2,445 ± 148 iu/mL) was reduced by 70% compared with CbB3-evo1 extract (7,907 ± 317 iu/mL), whereas the CbpA activity on Cb measured in CbC2-evo1-PgmK extract (8.5 ± 0.4 iu/mL) was more than 100-fold lower than that observed for CbC2-evo1 extract (1,240 ± 32 iu/mL). These drastic diminutions observed for both CbB3-evo1-Pgmk and CbC2-evo1-PgmK are consistent with their impaired growth on Cb-based medium (Fig. 6), and may suggest that a large fraction of the heterologous Cb-degrading enzymes aggregates in these modified strains.
Fig. 6.
Growth of CbB3-evo1-Pgmk and CbC2-evo1-Pgmk strains in Cb-based M9medium. CbB3-evo1 (blue curve) CbC2-evo1 (red curve), CbB3-evo1-Pgmk (yellow curve), and CbC2-evo1-Pgmk (green curve) strains were cultured in M9 supplemented with 0.5% Cb at 37°C. OD at 600 nm was monitored in a Tecan Spark plate reader. The mean of the data obtained on three culture replicates is shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.8. PpdcZm exhibits a promoter activity in the range of the majority of promoters of E. coli
Since the PpdcZm promoter provides an appropriate expression of bglA or cbpA genes to sustain the growth on Cb-medium, we investigated its promoter strength among E. coli promoters. First, based on the culture of CbB3-evo1 lamB∗ pUA-PacpP, pUA-Pgmk or pUA-Ppdc grown on M9 Cb 0.5% (Fig. 5c and d), the promoter activity (PA) of PpdcZm, Pgmk and PacpP was calculated using the formula of Zaslaver and coworkers (PA = ) (Zaslaver et al., 2006) and their maximum of activity (maxPA) was extracted from the PA profiles for all the promoters (Suppl. Fig. 7a). Since PA were measured from a fluorescence quantification that strongly depends on the sensitivity of the device used, maxPA values were arbitrarily normalized to the PacpP maxPA to facilitate the comparison with the previously reported data (Zaslaver et al, 2006, 2009). Pgmk displays here a maxPA corresponding to 51% of the PacpP maxPA whereas the maxPA of PpdcZm drops to 21% (Suppl. Fig. 7a). These results are consistent with already published data from the E. coli promoter database, in which Pgmk displays a maxPA of 50% compared to PacpP (Suppl. Table 4). The maxPA of the 1916 promoters from the complete E. coli promoter database were also normalized with respect to PacpP maxPA (Suppl. Fig. 7b). It appears that 2/3 of the promoters (1208 promoters) have maxPA lower or similar to that of PpdcZm (21% of PacpP maxPA) whereas only 124 promoters exhibit similar or higher maxPA compared to Pgmk (50% of PacpP maxPA). Thus, PpdcZm promoter seems to exhibit an activity in E. coli in the range of the majority of promoters found in this bacterium.
3.9. Genome editing of E. coli MG1655 can provide an efficient Cb metabolism
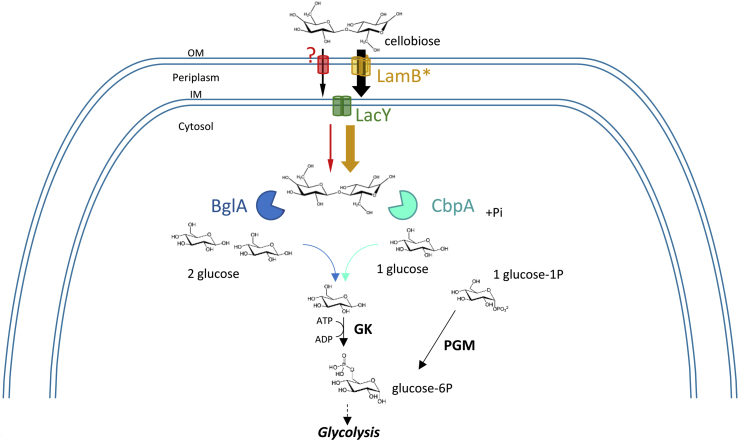
Altogether our data provide a list of genetic modifications enabling to convert the E. coli strain MG1655 into a bacterium able to efficiently catabolize and use Cb as the sole carbon source. As summarized in Supplemental Fig. 3 and in the Cb metabolism model depicted in the Fig. 7, the following mutations were thus inserted in the wild type MG1655 genome by P1 phage transduction: zde::bglA to provide a metabolic enzyme cleaving Cb into glucose (CbB3 strain, Suppl. Table 2); ΔPlac::PacpP to allow a constitutive expression of the lac operon and to provide a sufficient amount of LacY permease in the inner membrane (CbB3 PacpP::lac strain, Suppl. Table 2); lamB∗ to generate a truncated form of the outer membrane LamB porin that efficiently uptakes Cb (CbB3 PacpP::lac lamB∗ strain, Suppl. Table 2).
Fig. 7.
Model of Cb uptake and metabolism by engineered E. coli strains. LamB∗, truncated E. coli LamB (Δ363-446) porin; LacY, E. coli lactose permease; BglA, Ruminococcus albus β-glucosidase A; CbpA, Ruminiclostridium cellulolyticum Cb phosphorylase A; GK, E. coli glucokinase; PGM, E. coli phosphoglucomutase; IM, inner membrane; OM, outer membrane.
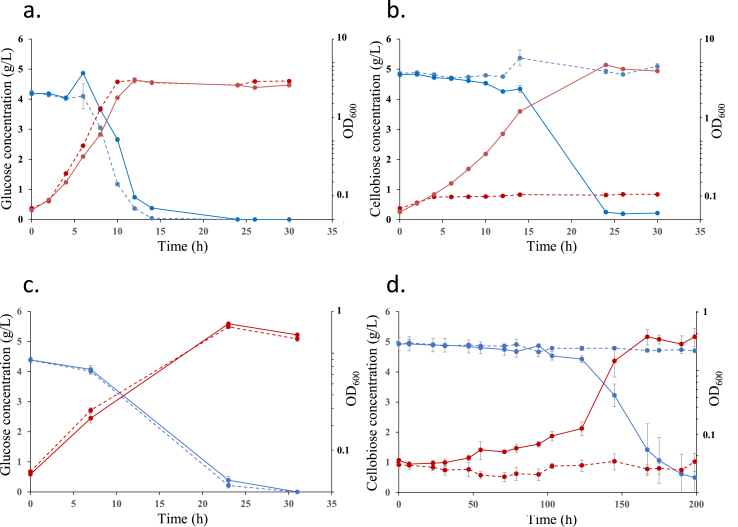
The resulting strain CbB3 PacpP::lac lamB∗ was able to grow aerobically without any lag phase on M9 Cb medium, displayed a doubling time of 2.2 h and reached a final OD600nm of 5 (Fig. 8b). This strain is also able to ferment cellobiose anaerobically (Fig. 8 d, and Suppl. Fig. 8) and exhibits a doubling time of 12 h on Cb under anoxic condition.
Fig. 8.
Growth of CbB3PacpP::lac lamb∗and MG1655 strains in aerobic and anaerobic conditions on glucose and cellobiose. The growth and sugar consumption under aerobic conditions on glucose and cellobiose are shown is a. and b., respectively. The growth and sugar consumption under anaerobic conditions on glucose and Cb are shown in c. and d., respectively. The growth (right axes) was monitored at 600 nm (red curves), and the sugar concentration (left axes) was determined by HPLC using an Aminex HPX87H column (blue curves). CbB3 PacpP ::lac lamb∗ and MG1655 strains are represented by solid and dotted lines, respectively. The data show the mean of three biological replicates, and bars represent the standard deviations. Fermentations products related to figures c. and d., are reported in supplemental Fig. 8a and b, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions
E. coli is not able to grow directly on Cb-based medium but previous studies showed that the strain can evolve to metabolize Cb by reactivating the cryptic operons chb (Chitibiose utilization loci, cryptic for Cb utilization) and asc (encoding a β-glucosidase and a PTS transporter for Cb) followed by 30 days of Cb-based adaptative evolution (Parisutham and Lee, 2011, 2015). Nevertheless, the formerly engineered strain had a limited growth on Cb, only exhibiting a maximal OD600nm = 1.6 and a generation time around 4 h when grown on minimum medium at a concentration of 0.4% Cb. In the present study, we have developed robust E. coli strains to provide an efficient organism able to grow on Cb as single carbon source. The best strain we obtained (CbB3 PacpP::lac lamB∗) reached on M9 0.5% Cb at 37 °C a final OD600nm = 5 in aerobiosis and 0.6 in anaerobiosis (Fig. 8b) and displayed respectively a doubling time of 2.2 h in aerobiosis (Table 1) and around 12 h in anaerobiosis (Fig. 8d). This capacity to efficiently catabolize Cb was attained by chromosomic insertion of a β-glucosidase that cleaves β-1,4 linkage to produce two glucose molecules as well as of a Cb phosphorylase that produces one glucose and one α-glucose 1-P by phosphorolysis. We have also unraveled mutations in two distinct genes improving the uptake of Cb: a lamB mutant gene encoding a truncated outer membrane maltoporin (LamB∗) and a lacI mutant gene encoding a dysfunctional LacI repressor leading to an upregulation of the lac operon and an appropriated production of the LacY permease. Furthermore, we engineered a constitutive lac operon expressing LacY under the control of PacpP promoter, without prior adaptation or involvement of LacI, a key repressor whose activity is used to control the expression of genes in many biotechnological applications (Ahmad et al., 2018; Gopal and Kumar, 2013; Gupta and Shukla, 2017; Hayat et al., 2018). Such a robust strain able to metabolize cellobiose should be valuable for future bioprocess engineering such as fermentation of saccharified cellulosic wastes into biomolecules of interest (biofuels, commodity chemicals…).
Authors contribution
RB: Conceptualization, Investigation, Formal analysis, Visualization, Writing - Original Draft NV: Conceptualization, Investigation, Formal analysis, Writing - Original Draft - Review & Editing NF: Writing - Review & Editing CT: Writing - Review & Editing SP: Writing - Review & Editing HPF: Conceptualization, Investigation, HPAEC-PAD and HPLC-RI analyses, Funding acquisition, Supervision, Resources, Writing - Review & Editing.
Declaration of competing interest
None.
Acknowledgements
The authors thank Luc Dedieu for providing pJIR215-bglA plasmid and sharing data about the use of this vector in E. coli. They also thank Mireille Ansaldi for providing pKD3, pKD4, pKD46 and pCP20 plasmids and their storage strains, BW25141 λpir strain and P1vir bacteriophage lysate for transduction. Emmanuelle Bouveret is also thanked for providing pUA vectors and advices for promoter activity assays. Isabelle Meynial-Salles, Philippe Soucaille, Pascale de Philip and Sandrine Pagès are thanked for helpful discussions. This study was supported by the grant ANR-14-CE05-0019-01 from the “Agence Nationale de la Recherche”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2020.e00157.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmad I., Nawaz N., Darwesh N.M., Ur Rahman S., Mustafa M.Z., Khan S.B., Patching S.G. Overcoming challenges for amplified expression of recombinant proteins using Escherichia coli. Protein Expr. Purif. 2018;144:12–18. doi: 10.1016/j.pep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Ansaldi M., Lepelletier M., Méjean V. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal. Biochem. 1996;234:110–111. doi: 10.1006/abio.1996.0060. [DOI] [PubMed] [Google Scholar]
- Aso Y., Tsubaki M., Dang Long B.H., Murakami R., Nagata K., Okano H., Phuong Dung N.T., Ohara H. Continuous production of d-lactic acid from cellobiose in cell recycle fermentation using β-glucosidase-displaying Escherichia coli. J. Biosci. Bioeng. 2019;127:441–446. doi: 10.1016/j.jbiosc.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin V., Bhatia M., Luckey M. Folding studies of purified LamB protein, the maltoporin from the Escherichia coli outer membrane: trimer dissociation can be separated from unfolding. Biochim. Biophys. Acta. 2011;1808:2206–2213. doi: 10.1016/j.bbamem.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker A., Fickert R., Oehler S., Müller-hill B. Operator search by mutant Lac repressors. J. Mol. Biol. 1998;278:549–558. doi: 10.1006/jmbi.1998.1729. [DOI] [PubMed] [Google Scholar]
- Beckwith J.R. Regulation of the lac operon. Recent studies on the regulation of lactose metabolism in Escherichia coli support the operon model. Science. 1967;156:597–604. doi: 10.1126/science.156.3775.597. [DOI] [PubMed] [Google Scholar]
- Bell C.E., Barry J., Matthews K.S., Lewis M. Structure of a variant of lac repressor with increased thermostability and decreased affinity for operator. J. Mol. Biol. 2001;313 doi: 10.1006/jmbi.2001.5041. [DOI] [PubMed] [Google Scholar]
- Berkane E., Orlik F., Stegmeier J.F., Charbit A., Winterhalter M., Benz R. Interaction of bacteriophage lambda with its cell surface receptor: an in vitro study of binding of the viral tail protein gpJ to LamB (Maltoporin) Biochemistry. 2006;45:2708–2720. doi: 10.1021/bi051800v. [DOI] [PubMed] [Google Scholar]
- Charbit A. Maltodextrin transport through lamb. Front. Biosci. 2003;8:s265–274. doi: 10.2741/1046. [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Rothenberg E. Interaction of bacteriophage λ with its E. coli receptor, LamB. Viruses. 2012;4:3162–3178. doi: 10.3390/v4113162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.C., Permuth S.F., Brooker R.J. Isolation and characterization of lactose permease mutants with an enhanced recognition of maltose and diminished recognition of cellobiose. J. Biol. Chem. 1989;264:14698–14703. [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groeve M.R.M., Desmet T., Soetaert W. Engineering of cellobiose phosphorylase for glycoside synthesis. J. Biotechnol. 2011;156:253–260. doi: 10.1016/j.jbiotec.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Desai S.H., Rabinovitch-Deere C.A., Tashiro Y., Atsumi S. Isobutanol production from cellobiose in Escherichia coli. Appl. Microbiol. Biotechnol. 2014;98:3727–3736. doi: 10.1007/s00253-013-5504-7. [DOI] [PubMed] [Google Scholar]
- Donovan R.S., Robinson C.W., Glick B.R. Review: optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter. J. Ind. Microbiol. 1996;16:145–154. doi: 10.1007/BF01569997. [DOI] [PubMed] [Google Scholar]
- Espeli O., Mercier R., Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 2008;68:1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- Fosses A., Maté M., Franche N., Liu N., Denis Y., Borne R., de Philip P., Fierobe H.-P., Perret S. A seven-gene cluster in Ruminiclostridium cellulolyticum is essential for signalization, uptake and catabolism of the degradation products of cellulose hydrolysis. Biotechnol. Biofuels. 2017;10:250. doi: 10.1186/s13068-017-0933-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Luan Y., Wang Q., Liang Q., Qi Q. Construction of cellulose-utilizing Escherichia coli based on a secretable cellulase. Microb. Cell Factories. 2015;14:159. doi: 10.1186/s12934-015-0349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerstma E.R., Groeneveld M., Slotboom D.-J., Poolman B. Quality control of overexpressed membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5722–5727. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves G.A.L., Oliveira P.H., Gomes A.G., Prather K.L.J., Lewis L.A., Prazeres D.M.F., Monteiro G.A. Evidence that the insertion events of IS2 transposition are biased towards abrupt compositional shifts in target DNA and modulated by a diverse set of culture parameters. Appl. Microbiol. Biotechnol. 2014;98:6609–6619. doi: 10.1007/s00253-014-5695-6. [DOI] [PubMed] [Google Scholar]
- Goodson K.A., Wang Z., Haeusler A.R., Kahn J.D., English D.S. LacI-DNA-IPTG loops: equilibria among conformations by single-molecule FRET. J. Phys. Chem. B. 2013;117 doi: 10.1021/jp308930c. [DOI] [PubMed] [Google Scholar]
- Gopal G.J., Kumar A. Strategies for the production of recombinant protein in Escherichia coli. Protein J. 2013;32:419–425. doi: 10.1007/s10930-013-9502-5. [DOI] [PubMed] [Google Scholar]
- Guan L., Kaback H.R. Lessons from lactose permease. Annu. Rev. Biophys. Biomol. Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.K., Shukla P. Microbial platform technology for recombinant antibody fragment production: a review. Crit. Rev. Microbiol. 2017;43:31–42. doi: 10.3109/1040841X.2016.1150959. [DOI] [PubMed] [Google Scholar]
- Hall B.G., Xu L. Nucleotide sequence, function, activation, and evolution of the cryptic asc operon of Escherichia coli K12. Mol. Biol. Evol. 1992;9:688–706. doi: 10.1093/oxfordjournals.molbev.a040753. [DOI] [PubMed] [Google Scholar]
- Hayat S.M.G., Farahani N., Golichenari B., Sahebkar A. Recombinant protein expression in Escherichia coli (E. coli): what we need to know. Curr. Pharmaceut. Des. 2018;24:718–725. doi: 10.2174/1381612824666180131121940. [DOI] [PubMed] [Google Scholar]
- Hershberger C.L., Rosteck P.R. Stability of recombinant plasmids for production of heterologous proteins in Escherichia coli. Bioprocess Technol. (N. Y.) 1991;13:3–16. [PubMed] [Google Scholar]
- Ibrahim E., Jones K.D., Taylor K.E., Hosseney E.N., Mills P.L., Escudero J.M. Molecular and biochemical characterization of recombinant cel12B, cel8C, and peh28 overexpressed in Escherichia coli and their potential in biofuel production. Biotechnol. Biofuels. 2017;10 doi: 10.1186/s13068-017-0732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakupciak J.P., Wells R.D. Genetic instabilities in (CTG.CAG) repeats occur by recombination. J. Biol. Chem. 1999;274:23468–23479. doi: 10.1074/jbc.274.33.23468. [DOI] [PubMed] [Google Scholar]
- Kaback H.R. Structure and mechanism of the lactose permease. C. R. Biol. 2005;328:557–567. doi: 10.1016/j.crvi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Kim S.-K., Himmel M.E., Bomble Y.J., Westpheling J. Expression of a cellobiose phosphorylase from Thermotoga maritima in Caldicellulosiruptor bescii improves the phosphorolytic pathway and results in a dramatic increase in cellulolytic activity. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02348-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba P.E. Mechanism of maltodextrin transport through LamB. Res. Microbiol. 2002;153:417–424. doi: 10.1016/s0923-2508(02)01340-2. [DOI] [PubMed] [Google Scholar]
- Kricker M., Hall B.G. Directed evolution of cellobiose utilization in Escherichia coli K12. Mol. Biol. Evol. 1984;1:171–182. doi: 10.1093/oxfordjournals.molbev.a040310. [DOI] [PubMed] [Google Scholar]
- Lesterlin C., Gigant E., Boccard F., Espéli O. Sister chromatid interactions in bacteria revealed by a site-specific recombination assay. EMBO J. 2012;31:3468–3479. doi: 10.1038/emboj.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. The lac repressor. C. R. Biol. 2005;328:521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Liu N., Fosses A., Kampik C., Parsiegla G., Denis Y., Vita N., Fierobe H.-P., Perret S. In vitro and in vivo exploration of the cellobiose and cellodextrin phosphorylases panel in Ruminiclostridium cellulolyticum: implication for cellulose catabolism. Biotechnol. Biofuels. 2019;12:208. doi: 10.1186/s13068-019-1549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Bevan D.R., Zhang Y.-H.P. The family 1 glycoside hydrolase from Clostridium cellulolyticum H10 is a cellodextrin glucohydrolase. Appl. Biochem. Biotechnol. 2010;161:264–273. doi: 10.1007/s12010-009-8782-x. [DOI] [PubMed] [Google Scholar]
- Lu M., Kleckner N. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 1994;176:5847–5851. doi: 10.1128/jb.176.18.5847-5851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey M., Ling R., Dose A., Malloy B. Role of a disulfide bond in the thermal stability of the LamB protein trimer in Escherichia coli outer membrane. J. Biol. Chem. 1991;266:1866–1871. [PubMed] [Google Scholar]
- Luckey M., Nikaido H. Specificity of diffusion channels produced by lambda phage receptor protein of Escherichia coli. Proc. Natl. Acad. Sci. Unit. States Am. 1980;77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Zhang Y., Bao J. Extracellular secretion of β-glucosidase in ethanologenic E. coli enhances ethanol fermentation of cellobiose. Appl. Biochem. Biotechnol. 2014;174:772–783. doi: 10.1007/s12010-014-1108-7. [DOI] [PubMed] [Google Scholar]
- Ma L., Zhang J., Zou G., Wang C., Zhou Z. Improvement of cellulase activity in Trichoderma reesei by heterologous expression of a beta-glucosidase gene from Penicillium decumbens. Enzym. Microb. Technol. 2011;49:366–371. doi: 10.1016/j.enzmictec.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Maloy S.R., Cronan J.E., Freifelder D. Jones and Bartlett Publishers; Boston: 1994. Microbial Genetics. [Google Scholar]
- Miura-Onai S., Yabuta M., Ohsuye K. Mutational study at Ser300 position of the Escherichia coli lactose repressor. Biochem. Biophys. Res. Commun. 1995;209:126–130. doi: 10.1006/bbrc.1995.1479. [DOI] [PubMed] [Google Scholar]
- Mounts P., Wu T.C., Peden K. Method for cloning single-stranded oligonucleotides in a plasmid vector. Biotechniques. 1989;7:356–359. [PubMed] [Google Scholar]
- Mueller-Hill B., Rickenberg H.V., Wallenfels K. Specificity of the induction of the enzymes of the lac operon in Escherichia coli. J. Mol. Biol. 1964;10:303–318. doi: 10.1016/s0022-2836(64)80049-8. [DOI] [PubMed] [Google Scholar]
- Mulvihill E., Pfreundschuh M., Thoma J., Ritzmann N., Müller D.J. High-resolution imaging of maltoporin LamB while quantifying the free-energy landscape and asymmetry of sugar binding. Nano Lett. 2019;19:6442–6453. doi: 10.1021/acs.nanolett.9b02674. [DOI] [PubMed] [Google Scholar]
- Muñoz-Gutiérrez I., Oropeza R., Gosset G., Martinez A. Cell surface display of a β-glucosidase employing the type V secretion system on ethanologenic Escherichia coli for the fermentation of cellobiose to ethanol. J. Ind. Microbiol. Biotechnol. 2012;39:1141–1152. doi: 10.1007/s10295-012-1122-0. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya K., Takano M., Shimizu S. Cloning of a β-glucosidase gene from Ruminococcus albus and its expression in Escherichia coli. Ann. N. Y. Acad. Sci. 1991;646:41–52. doi: 10.1111/j.1749-6632.1991.tb18562.x. [DOI] [PubMed] [Google Scholar]
- Parisutham V., Chandran S.-P., Mukhopadhyay A., Lee S.K., Keasling J.D. Intracellular cellobiose metabolism and its applications in lignocellulose-based biorefineries. Bioresour. Technol. 2017;239:496–506. doi: 10.1016/j.biortech.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Parisutham V., Lee S.K. Novel functions and regulation of cryptic cellobiose operons in Escherichia coli. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0131928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisutham V., Lee S.K. Engineering Escherichia coli for efficient cellobiose utilization. Appl. Microbiol. Biotechnol. 2011;92:125–132. doi: 10.1007/s00253-011-3434-9. [DOI] [PubMed] [Google Scholar]
- Park J.M., Vinuselvi P., Lee S.K. The mechanism of sugar-mediated catabolite repression of the propionate catabolic genes in Escherichia coli. Gene. 2012;504:116–121. doi: 10.1016/j.gene.2012.04.074. [DOI] [PubMed] [Google Scholar]
- Parker L.L., Hall B.G. Mechanisms of activation of the cryptic cel operon of Escherichia coli K12. Genetics. 1990;124:473–482. doi: 10.1093/genetics/124.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patient M.E., Summers D.K. ColE1 multimer formation triggers inhibition of Escherichia coli cell division. Mol. Microbiol. 1993;9:1089–1095. doi: 10.1111/j.1365-2958.1993.tb01238.x. [DOI] [PubMed] [Google Scholar]
- Prilipov A., Phale P.S., Koebnik R., Widmer C., Rosenbusch J.P. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 1998;180:3388–3392. doi: 10.1128/jb.180.13.3388-3392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravachol J., de Philip P., Borne R., Mansuelle P., Maté M.J., Perret S., Fierobe H.-P. Mechanisms involved in xyloglucan catabolism by the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Sci. Rep. 2016;6:22770. doi: 10.1038/srep22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynen M., Reipen I., Sahm H., Sprenger G.A. Construction of expression vectors for the gram-negative bacterium Zymomonas mobilis. Mol. Gen. Genet. 1990;223:335–341. doi: 10.1007/BF00265073. [DOI] [PubMed] [Google Scholar]
- Saedler H., Heiss B. Multiple copies of the insertion-DNA sequences IS1 and IS2 in the chromosome of E. coli K-12. Mol. Gen. Genet. 1973;122:267–277. doi: 10.1007/BF00278602. [DOI] [PubMed] [Google Scholar]
- Sekar R., Shin H.-D., Chen R. Engineering Escherichia coliCells for cellobiose assimilation through a phosphorolytic mechanism. Appl. Environ. Microbiol. 2012;78 doi: 10.1128/AEM.06693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.-S., Chong H., Park H.S., Yoon K.-O., Jung C., Kim J.J., Hong J.H., Kim H., Kim J.-H., Kil J.-I., Park C.J., Oh H.-M., Lee J.-S., Jin S.-J., Um H.-W., Lee H.-J., Oh S.-J., Kim J.Y., Kang H.L., Lee S.Y., Lee K.J., Kang H.S. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2005;23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Weimer P.J. Utilization of individual cellodextrins by three predominant ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 1996;62:1084–1088. doi: 10.1128/aem.62.3.1084-1088.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhania R.R., Patel A.K., Sukumaran R.K., Larroche C., Pandey A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour. Technol. 2013;127:500–507. doi: 10.1016/j.biortech.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Soma Y., Inokuma K., Tanaka T., Ogino C., Kondo A., Okamoto M., Hanai T. Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system. J. Biosci. Bioeng. 2012;114:80–85. doi: 10.1016/j.jbiosc.2012.02.019. [DOI] [PubMed] [Google Scholar]
- Standley M.S., Million-Weaver S., Alexander D.L., Hu S., Camps M. Genetic control of ColE1 plasmid stability that is independent of plasmid copy number regulation. Curr. Genet. 2019;65:179–192. doi: 10.1007/s00294-018-0858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.L., Wang S.C., Wang T., Zhao S.G., Wu Y.J., Wu L.J., Yu Z.L. Mutational spectrum of the lacI gene in Escherichia coli K12 induced by low-energy ion beam. Mutat. Res. 2006;602:163–169. doi: 10.1016/j.mrfmmm.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Tchieu J.H., Norris V., Edwards J.S., Saier M.H. The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001;3:329–346. [PubMed] [Google Scholar]
- Vilar J.M.G., Guet C.C., Leibler S. Modeling network dynamics: the lac operon, a case study. J. Cell Biol. 2003;161:471–476. doi: 10.1083/jcb.200301125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos-Scheperkeuter G.H., Witholt B. Assembly pathway of newly synthesized LamB protein an outer membrane protein of Escherichia coli K-12. J. Mol. Biol. 1984;175:511–528. doi: 10.1016/0022-2836(84)90182-7. [DOI] [PubMed] [Google Scholar]
- Wilson D.B. Cellulases and biofuels. Curr. Opin. Biotechnol. 2009;20:295–299. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Xu J., Matthews K.S. Flexibility in the inducer binding region is crucial for allostery in the Escherichia coli lactose repressor. Biochemistry. 2009;48:4988–4998. doi: 10.1021/bi9002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Yang X., Li Z., Du C., Wang J., Li S. Overexpression and characterization of a glucose-tolerant β-glucosidase from T. aotearoense with high specific activity for cellobiose. Appl. Microbiol. Biotechnol. 2015;99:8903–8915. doi: 10.1007/s00253-015-6619-9. [DOI] [PubMed] [Google Scholar]
- Ye Jiqing, Van der Berg Bert. Crystal structure of the bacterial nucleoside transporter Tsx. EMBO J. 2004;23(16):3187–3195. doi: 10.1038/sj.emboj.7600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A., Bren A., Ronen M., Itzkovitz S., Kikoin I., Shavit S., Liebermeister W., Surette M.G., Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Zaslaver A., Kaplan S., Bren A., Jinich A., Mayo A., Dekel E., Alon U., Itzkovitz S. Invariant distribution of promoter activities in Escherichia coli. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.