Abstract
Emerging evidence has shown that circular RNAs (circRNAs) serve as a promising biomarker in different malignancies. Specifically, circ_001569 has been found to be upregulated in some types of human gastrointestinal cancer. In this study, we aimed to investigate clinical significances, diagnostic and prognostic values of circ_001569 in pancreatic cancer (PC), and identify its effects on the malignant behaviors of PC cells. The expression of circ_001569 was determined in 26 tissues samples and 97 plasma samples from PC patients by qRT-PCR. Relationship between circ_001569 expression and clinicopathological parameters was analyzed by Chi-square test. Diagnostic and prognostic values of circ_001569 were evaluated by ROC curves, Kaplan-Meier curves, and Cox regression analysis. The effects of circ_001569 on the proliferation, migration, invasion, and apoptosis of PC cells were assessed by MTT, wound healing, Transwell invasion assays, and flow cytometric analysis, respectively. Results showed that the expression of circ_001569 was upregulated in tissues and plasma of PC patients. High circ_001569 level was positively correlated with lymphatic metastasis, clinical stage, and venous invasion. Circ_001569 level was an independent prognostic indicator for overall survival rates of PC patients, and patients with high circ_001569 level had a poor prognosis. The AUC of circ_001569 was 0.716 (95% CI: 0.642-0.790) with a sensitivity and specificity of 62.76% and 74.29%, respectively. In vitro, circ_001569 silencing decreased cell proliferation, migration, and invasion, but promoted cell apoptosis of PC cells. Our data demonstrate that high circ_001569 level associates with tumor malignant behaviors, and may serve as a potential biomarker in the diagnosis and prognosis of PC.
Keywords: circular RNAs, circ_001569, pancreatic cancer, biomarker, survival rates
Introduction
Pancreatic cancer (PC) is one of the most malignancies and is the leading cause of cancer-associated mortalities worldwide, which characterized by rapid local invasion and early lymphatic metastasis.1,2 As a result of lack of early diagnosis, patients with PC have an extremely poor prognosis with the overall 5-year survival rates less than 5%.3 In recent years, although an increasing number of novel therapies has been applied in PC treatment, but limited progress has acquired owing to the low specificity and sensitivity.4 Thus, it is critical to identify novel and reliable biomarkers for the diagnosis and prognosis of PC. Simultaneously, a deeper understanding of the key factors associated the progression of PC will develop novel therapeutic target.
Circular RNAs (circRNAs) are closed-loop RNAs that are produced from pre-mRNAs during transcription and are initially proposed to be by-products of splicing or splicing errors with low abundance.5,6 Recently, circRNAs attract a great of attention due to their pivotal roles in malignant behaviors of cancer, including cell proliferation, differentiation, apoptosis, and metastasis.7,8 Additionally, growing evidence has suggested that circRNAs are often dysregulated in different malignancies, and can serve as potential biomarkers for the diagnosis and prognosis of cancers.9,10 In colorectal cancer, Yuan et al reported that circ_0026344 expression is downregulated in cancer tissues and predicts a poor overall survival rate.11 In PC, Jiang et al showed that circ_0001649 plays a tumor suppressor role and serves as a potential therapeutic target.12 In lung cancer, Zong et al demonstrated that circ_102231 is upregulated in tumor tissues and associates with advanced TNM stage, lymph node metastasis, and poor prognosis.13 Thus, above data support the significance of exploring circRNAs function in human cancers. However, PC-related circRNAs are still lacking and need to be further detected.
Circ_001569 is located on the plus strand of human chromosome 16q13.1, and aligns in a sense orientation to the ATP binding cassette subfamily C member 1 (ABCC1) and spans exons 33.14 ABCC1 gene is a member of the superfamily of ATP-binding cassette (ABC) transportersis, which is involved in multi-drug resistance.15 A previous study has shown that circ_001569 is upregulated in colorectal cancer tissues compared with normal tissues.16 Also, several papers reported that upregulation of circ_001569 predicts poor prognosis in non-small cell lung cancer and breast cancer.17,18 Till now, the clinical significances and biological roles of circ_001569 in PC have not yet been reported. Given that increased circ_001569 expression is common phenomenon in multiple cancers, it has been hypothesized that circ_001569 may associate with tumor development. In this study, we evaluated circ_001569 expression in 26 tissues samples and 97 plasma samples from PC patients, determine its diagnostic and prognostic values, and observed its effects on biological behaviors of PC cells. In general, our study provided critical evidences to prove that circ_001569 associates with malignant behaviors and may serve as a potential biomarker in the diagnosis and prognosis of PC.
Materials and Methods
PC Samples
Peripheral blood samples (each 5 ml) were collected from 26 patients with PC after admission at Hunan Provincial People’s Hospital from January 2017 to October 2018, and 26 paired cancer tissues and corresponding adjacent non-cancerous tissues were collected after pancreaticoduodenectomy surgery. None of the patients received radiotherapy chemotherapy, and/or other anticancer treatment before the surgery. These patients were followed up by telephone every 3 months after surgery. Clinical characteristics of these patients were shown in Table 1. Similarly, peripheral blood samples (each 5 ml) were acquired from another 71 PC patients and 71 healthy volunteers from January 2015 to April 2019 at our hospital. All samples were put into liquid-nitrogen immediately after collection and then stored at −80°C until further experiments. The study was approved by Institutional Review Board of Hunan Provincial People’s Hospital (No. 2015064) in accordance with the relevant guidelines and regulations, and written informed consent was obtained from each participant prior to enrollment.
Table 1.
The Association Between Circ_001569 Expression and Clinicopathological Parameters in Patients With PC.
| Parameters | No. of patients (n) | Circ_001569 | P | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ||||
| < 60 | 21 | 9 | 12 | 0.322 |
| ≥ 60 | 5 | 4 | 1 | |
| Gender | ||||
| Male | 10 | 6 | 4 | 0.688 |
| Female | 16 | 7 | 9 | |
| Pathological subtype | ||||
| PDAC | 19 | 11 | 8 | 0.467 |
| Mucinous carcinoma | 5 | 2 | 3 | |
| Acinar cell carcinoma | 2 | 0 | 2 | |
| Tumor diameter (cm) | ||||
| < 4 | 15 | 7 | 8 | 1.000 |
| ≥ 4 | 11 | 6 | 5 | |
| Tumor location | ||||
| Head | 12 | 8 | 4 | 0.238 |
| Body and tail | 14 | 5 | 9 | |
| Tumor differentiation | ||||
| Well/moderately | 19 | 10 | 9 | 1.000 |
| Poorly | 7 | 3 | 4 | |
| Venous invasion | ||||
| No | 20 | 13 | 7 | 0.015* |
| Yes | 6 | 0 | 6 | |
| Clinical stage | ||||
| I/II | 15 | 12 | 3 | 0.001* |
| III | 11 | 1 | 10 | |
| Lymphatic metastasis | ||||
| No | 14 | 10 | 4 | 0.047* |
| Yes | 12 | 3 | 9 | |
| CA19-9 level (U/ml) | ||||
| < 37 | 8 | 2 | 6 | 0.202 |
| ≥ 37 | 18 | 11 | 7 | |
PC: pancreatic cancer, PDAC: pancreatic ductal adenocarcinoma. *P < 0.05.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was obtained from human PC samples and cells using TRIzol™ reagent (Thermo Fisher Scientific, CA, USA), according to the manufacturer’s protocol. The cDNA was synthesized using PrimeScript RT Reagent Kit with gDNA Eraser from Beijing Takara Co., Ltd, China. qRT-PCR was performed using GoTaq qPCR Master Mix (Promega, Madison, USA) on an ABI StepOne™ Real-Time PCR System (Applied Biosystems, CA, USA) with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. The thermocycler programs were as follows: an initial step at 95°C for 20 min, followed by 40 cycles of denaturation at 98°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 45 s. Each reaction was performed in triplicate, and data were analyzed using the 2−ΔΔCt method. Primers were synthesized from Shanghai GenePharma Co., Ltd, China as follows: ABCC1, 5′-GTCGGGGCATATTCCTGGC-3′ (forward) and 5′-CTGAAGACTGAACTCCCTTCCT-3′ (reverse), GAPDH, 5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′ (reverse), and circ_001569, 5′-TCCCCTGAACATTCTCCCCAT-3′ (forward) and 5′-GAAAGCACTTGGTGAAGTCGG-3′ (reverse).
Cell culture
Human PC cell lines including Aspc-1, Bxpc-3, and Panc-1, and a normal pancreatic ductal epithelial cell line HPDE were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). These cells were cultured in RPMI1640 medium (Thermo Fisher Scientific, CA, USA) supplemented with 10% FBS (Gibco, CA, USA), and maintained in a humidified incubator with an atmosphere of 5% CO2 at 37°C.
Oligonucleotide and Cell Transfection
The small interfering RNA (siRNA) targeting circ_001569 (named si-circ_0015691 #1, #2, #3 and corresponding negative control (named NC#1, #2, #3) were purchased from Guangzhou RiboBio Co., Ltd, China. Cells were seeded in 6-well or 96-well plates overnight and then transfected using Lipofectamine 3000 (Thermo Fisher Scientific, CA, USA) for 24 h or 48 h at 37°C, according to the manufacturer’s protocol. After that, cells were harvested for subsequent analyses.
RNase R Digestion
To measure the stability of circ_001569, 5 µg of total RNA was treated with 3 U/µg of RNase R (Epicenter Biotechnologies, Shanghai, China) at 37°C for 15 min according to the manufacturer’s protocol. After that, circ_001569 and liner ABCC1 mRNA expression levels were assessed using qRT-PCR.
3-(4,5-dimethyl-2-thiazolyl)-2,5-Diphenyl-2-H-tetrazolium bromide (MTT) Assay
Cell growth was observed using MTT assay. Approximately 7 × 103 transfected cells were seeded in 96-well plates and cultured for another 1, 2, 3, and 4 days. Then, 16 µl of MTT reagent (Sigma-Aldrich, MO, USA) at 5 mg/ml concentration was added to the wells and incubated for another 4 h at 37°C. The culture medium was removed and 150 µl dimethyl sulfoxide (Sigma-Aldrich) was added to the wells. Each well was measured spectrophotometrically at 450 nm using a microplate reader (Molecular Devices, CA, USA).
Wound Healing Assay
Cell migration was assessed using wound healing assay. Approximately 5 × 106 transfected cells were seeded in 6-well plates and incubated overnight at 37°C. Then confluent cells were scratched with 100-µl pipette tips, and maintained with serum-free RPMI1640 medium in an cell incubator. Cells were photographed after 0 and 24 h after scratching, and the width of the wounds was calculated using an IX71 inverted microscope (Olympus, PA, USA).
Transwell Invasion Assay
Cell invasion was evaluated using Transwell invasion assay. Transwell chambers were coated with 50 µg reconstituted basement membrane matrix (Corning, Lowell, MA, USA), and approximately 2 × 104 transfected cells in 100 µl of serum-free RPMI1640 medium were seeded into the upper chambers. Bottom chambers were filled with 500 µl of RPMI1640 medium containing 15% FBS. After culturing for 24 h, cells on the inner membrane were removed. The cells in outer membrane was fixed with 4% paraformaldehyde for 20 min at 37°C and stained with 0.1% crystal violet solution for 10 min at 37°C. Images were observed under an IX71 inverted microscope and 5 fields were randomly selected to be photographed.
Flow Cytometric Assay
Cell apoptosis was detected using flow cytometric assay with Annexin V-FITC/PI Apoptosis Detection Kit (BD Biosciences, NJ, USA). After 48 h post-transfection, cells were collected and washed with cold PBS twice. The, approximately 1 × 106 cells were resuspended in 200 µl binding buffer, and added with 4 µl of annexin V-FITC reagent and 3 µl of propidium iodide for incubation for 30 min at 37°C in a dark room. Afterward, the stained cells were analyzed with a FACScan flow cytometer (BD Biosciences, NJ, USA), and apoptotic rate was calculated.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD), and all analyses were performed using SPSS 22.0 software (IBM Corporation, NY, USA). To compare the significance between 2 groups, student’s t-test was performed. One-way ANOVA followed by Dunnett’s post hoc test was applied to compare the differences among 3 groups. The clinical significances between circ_001569 expression and clinicopathologic parameters were analyzed by Chi-square test. Survival curves were obtained by using Kaplan-Meier method and compared by log-rank test. Univariate and multivariate survival analysis was performed using the Cox proportional hazards model. Receiver operating characteristic (ROC) curves were constructed to estimate the diagnostic value of circ_001569. P-value less than 0.05 was considered statistically significant.
Results
Circ_001569 Is Highly Expressed in Tissues and Plasma From PC Patients
To explore the expression pattern of circ_001569 in PC, we first analyzed GSE69362 microarray in gene expression omnibus (GEO) database,19 and highly expression levels of circ_001569 were found in cancer tissues (Figure 1A, P < 0.05). Similarly, the expression of circ_001569 was determined in 26 paired cancer tissues and corresponding adjacent non-cancerous tissues using qRT-PCR. As shown in Figure 1B, the levels of circ_001569 were markedly upregulated in cancer tissues compared to the non-cancerous tissues (P < 0.01). Further, compared to plasma of 71 healthy volunteers, circ_001569 expression was significantly increased in plasma of 97 PC patients (Figure 1C, P < 0.001). Additional, circ_001569 was present at higher levels in PC cell lines (Aspc-1, Bxpc-3, and Panc-1) compared to HPDE (Figure 1D, P < 0.05). Based on the higher levels of circ_001569 in Aspc-1 and Bxpc-3 cells than that in Panc-1 cell line, the 2 cell lines were selected for the subsequent experiments.
Figure 1.
The expression pattern of circ_001569 in PC. A, Relative expression levels of circ_001569 in GSE69362 microarray. B, Relative circ_001569 expression of 26 paired cancer tissues and corresponding adjacent non-cancerous tissues was measured by qRT-PCR. C, Relative circ_001569 expression in plasma of 71 healthy volunteers and 97 PC patients. D, Analysis for circ_001569 levels in human PC cell lines (Aspc-1, Bxpc-3, and Panc-1) and a normal pancreatic ductal epithelial cell line HPDE. PC: pancreatic cancer, qRT-PCR: quantitative real-time PCR, SD: standard deviation. Data were expressed as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.01.
To evaluate the stability of circ_001569 in Aspc-1 and Bxpc-3 cells, the cells were treated with 3 U/µg of ribonuclease R (RNase R). Results of qRT-PCR indicated that the resistance of circ_001569 to RNase R degradation was higher than liner ABCC1 in both Aspc-1 and Bxpc-3 cells (Figure 2A and B, P < 0.01).
Figure 2.
Circ_001569 as a potential biomarker in the diagnosis and prognosis of PC patients. A and B, qRT-PCR analysis of circ_001569 and liner ABCC1 gene after treatment of RNase R in both Aspc-1 and Bxpc-3 cells. C, Kaplan-Meier survival analysis and log-rank test were used to assess circ_001569 expression and PC patients’ overall survival. D, ROC curves of circ_001569 in discriminating patients with PC patients from healthy volunteers. ABCC1: ATP binding cassette subfamily C member 1, ROC: receiver operating characteristic. **P < 0.01.
Highly Expression of Circ_001569 Associates With Adverse Clinicopathological Factors and Predicts a Poor Prognosis in PC Patients
Based on the median value of circ_001569 expression in cancer tissues, 26 patients with PC were classified into 2 groups, including high expression of circ_001569 expression group (n = 13) and low circ_001569 expression group (n = 13). Subsequently, by Chi-square test, we found high circ_001569 level was positively correlated with lymphatic metastasis (P = 0.047), clinical stage (P = 0.001), and venous invasion (P = 0.015) (Table 1). However, there was no significant differences in age, gender, tumor diameter, tumor location, tumor differentiation, pathological subtype, and CA19-9 level.
Among the 26 patients with PC, 15 patients survival times shorter than 12 months and 11 patients survival times longer than 12 months, with an average survival time of 19.52 ± 2.06 months. Kaplan-Meier survival analysis of prognosis revealed that patients with high circ_001569 expression had shorter overall survival rates than those patients with lower circ_001569 expression (Figure 2C, P = 0.028). Moreover, multivariate Cox regression analyses validated that circ_001569 was an independent prognostic indicator for overall survival rates of PC patients (Table 2, P = 0.042).
Table 2.
Univariate and Multivariate Cox Proportional Hazards Model Analysis of Prognostic Factors for Overall Survival in Patients With PC.
| Parameters | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age (< 60 vs. ≥ 60) | 0.714 (0.452-1.302) | 0.820 | NA | NA |
| Gender (male vs. female) | 0.821 (0.589-1.577) | 0.501 | NA | NA |
| Pathological subtype (PDAC vs. mucinous carcinoma/acinar cell carcinoma) | 1.306 (0.811-2.960) | 0.175 | NA | NA |
| Tumor diameter (< 4 vs. ≥ 4) | 1.122 (0.759-2.518) | 0.464 | NA | NA |
| Tumor location (head vs. body and tail) | 1.297 (0.773-2.848) | 0.362 | NA | NA |
| Tumor differentiation (Well/moderately vs. poorly) | 1.744 (0.922-3.158) | 0.079 | NA | NA |
| CA19-9 level (< 37 vs. ≥ 37) | 0.869 (0.470-1.721) | 0.627 | NA | NA |
| Venous invasion (no vs. yes) | 1.930 (0.953-3.862) | 0.061 | NA | NA |
| Lymphatic metastasis (no vs. yes) | 2.728 (1.509-5.516) | 0.034* | 1.945 (0.970-3.480) | 0.073 |
| Clinical stage (I/II vs. III) | 2.184 (0.997-3.905) | 0.057 | NA | NA |
| Circ_001569 level (low vs. high) | 2.952 (1.821-7.778) | 0.028* | 2.256 (1.132-4.836) | 0.042* |
HR: hazard ratio, CI: confidence interval. *P < 0.05.
Circ_001569 Serves as a Potential Biomarker for Diagnosis of PC Patients
To investigate the diagnostic value of circ_001569 in PC, the ROC curve analysis was conducted. As shown in Figure 2D, plasma circ_001569 could efficiently discriminate PC patients from healthy volunteers (P < 0.001), with an area under curve (AUC) of 0.716 (95% CI: 0.642-0.790), sensitivity was 62.76%, and specificity was 74.29%.
Circ_001569 Promotes Cell Proliferation and Arrested Cell Apoptosis of PC Cells
To investigate the biological roles of circ_001569 in PC, Aspc-1 and Bxpc-3 cells were used for loss-of-function experiments. circ_001569 levels were significantly decreased in both Aspc-1 and Bxpc-3 cells following transfection with si-circ_0015691 #1 compared with NC #1-transfected cells (Figure 3A, P < 0.01). However, si-circ_0015691 #2 could only effectively silence circ_0015691 expression in Bxpc-3 cells (Figure 3B, P < 0.05), and si-circ_0015691 #3 could only effectively inhibit circ_0015691 expression in Aspc-1 cells (Figure 3C, P < 0.05). Therefore, the si-circ_0015691 #1 and NC #1 were chose and used for subsequent experiments.
Figure 3.
Circ_001569 promotes cell proliferation and arrested cell apoptosis of PC cells. A-C, Comparing the efficiency of circ_001569 silencing in Aspc-1 and Bxpc-3 cells by qRT-PCR analysis after transfection of si-circ_0015691 #1, 2#, 3# or NC #1, 2#, 3#. D and E, MTT assay was used to detect cell proliferative ability of Aspc-1 and Bxpc-3 cells after circ_001569 knockdown. F, Flow cytometric analysis showed that downregulation of circ_001569 increases cell apoptotic rates of Aspc-1 and Bxpc-3 cells. MTT: 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide, NC: negative control. *P < 0.05, **P < 0.01.
MTT assay showed that knockdown of circ_001569 markedly repressed cell proliferation of Aspc-1 and Bxpc-3 cells (Figure 3D and E, P < 0.001). Moreover, to evaluate whether the anti-proliferative effect of circ_001569 knockdown on PC cells resulted from an alteration of cell apoptosis, flow cytometric assay was applied. Results confirmed that increased cell apoptotic rates were found in Aspc-1 and Bxpc-3 cells after silencing of circ_0015691 (Figure 3F, P < 0.01).
Circ_001569 Facilitates Cell Migration and Invasion of PC Cells
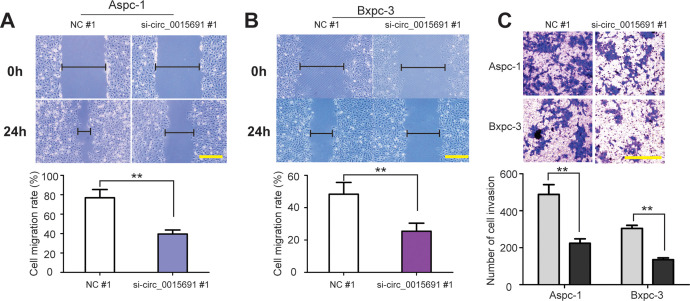
What’s more, by wound healing assay, we found that knockdown of circ_001569 slowed lower cell migration abilities of Aspc-1 and Bxpc-3 cells (Figure 4A and B, P < 0.01). Similar effect was observed in Transwell invasion assay, indicating that silencing of circ_001569 decreased the number of invaded cells (Figure 4C, P < 0.01). Taken together, upregulation of circ_001569 associates with the progression of PC.
Figure 4.
Circ_001569 facilitates cell migration and invasion of PC. A and B, Representative images of wound healing and cell migration rate after 0 h and 24 h scratching in Aspc-1 and Bxpc-3 cells transfected with si-circ_0015691 #1 or NC #1. C, Images of cell invasion and cell counts in Aspc-1 and Bxpc-3 cells by Transwell invasion assay after circ_001569 silencing. Scale bar = 100 µm. **P < 0.01.
Discussion
Currently, identifying novel biomarkers for diagnosis and prognosis as well as detection of promising therapeutic targets for the treatment of PC are urgently needed.20,21 Several circRNAs-mediated clinical significances and functions have been reported in PC.22 For example, Yang et al demonstrated that circ_LDLRAD3 as a biomarker in diagnosis of PC.23 Chen et al showed that circ_100782 modulates PC cell proliferation via IL6/STAT3 signaling pathway.24 Qu et al demonstrated that circ_RHOT1 expression is upregulated and facilitates cell proliferation and invasion in PC.25 In addition, Yao et al revealed that downregulation of circ_LDLRAD3 represses PC progression via miR-137-3p/PTN axis.26 In our study, we found that upregulation of circ_001569 may serve as a potential biomarker for the diagnosis and prognosis of PC, and knockdown of circ_001569 exerts a inhibitory role on malignant behaviors of PC cells.
A recent study reveals that circ_001569 expression is upregulated in colorectal cancer.16 Also, some reports have demonstrated that high circ_001569 level predicts poor prognosis in non-small cell lung cancer, breast cancer and hepatocellular carcinoma.17,18,27 Identify with these discoveries, a markedly increase of circ_001569 expression was shown in tissues and plasma from PC patients during our study. At the same time, we compared the expression of circ_001569 in 3 PC cell lines and a normal pancreatic ductal epithelial cell lines HPDE, and showed that circ_001569 levels in PC cells were also significantly higher than that in HPDE cells. Then, we investigated the clinicopathologic significances of circ_001569 in PC patients. Statistical analyses demonstrated that high circ_001569 level was positively correlated with lymphatic metastasis, clinical stage, and venous invasion. These data strong implied that upregulation of circ_001569 may play an key role in the progression of PC.
To our knowledge, early effective diagnosis could improve the therapeutic effect and prognosis of PC patients. A large number of studies had reported that circRNAs serve as potential biomarkers in plasma of patients with cancer.9,10 Then, we detected the prognostic value of circ_001569 in PC. Kaplan-Meier analysis showed that the patients with high expression of circ_001569 had shorter overall survival rates after pancreaticoduodenectomy surgery compared with patients with low circ_001569 expression. Further multivariate Cox analysis verified that circ_001569 serves as an independent prognostic factor for PC patients. Moreover, the diagnostic value of miR-18a-5p was also been assessed according to ROC analysis. From the ROC curves, the high diagnostic accuracy of circ_001569 was demonstrated, indicating that circ_001569 could distinguish PC patients from healthy volunteers with high sensitivity and specificity. However, in our study, tumor diameter, lymphatic metastasis and clinical stage had no related with prognosis of PC patients. The possible explaination might be caused by the small sample size of this research. Therefore, these data must be treated and interpreted with caution, and a larger cohort of PC patients should be recruited to further validate the clinical values of circ_001569 in the future.
Currently, increasing proofs gave the persuasion to the oncogene role of circ_001569 in malignant tumors. For example, Liu et al suggested that upregulation of circ_001569 promotes cell growth and metastasis in hepatocellular carcinoma by sponging miR-411-5p and miR-432-5p.27 Similarly, Xie et al reported that circ_001569 acts as a positive regulator in cell proliferation and invasion of colorectal cancer.16 Consistent with the above studies, we found circ_001569 silencing decreased cell proliferation, migration, and invasion. In addition, flow cytometric assay for cell apoptosis demonstrated that circ_001569 knockdown induced PC cell apoptosis. These data demonstrated that upregulation of circ_001569 facilitates the progression of PC. However, due to the limitation of time and funds, we have only detected the role of circ_001569 in PC cells in vitro. Further researches are needed to focus on in vivo study and underlying molecular mechanism to further confirm support the findings in our study.
In summary, our study revealed that circ_001569 is upregulated in tissues and plasma of PC patients and predicted a poor overall survival. Circ_001569 has a high diagnostic value for PC and facilitates cell malignant behaviors. Therefore, circ_001569 may be a promising biomarker in the diagnosis and prognosis of PC.
Footnotes
Authors’ Note: N.Z. contributed to the study design, manuscript writing and revision. X.S. contributed to sample collection and experiment. Y.C. and J.L. contributed to experiment. H.H. and C.L. contributed to data analysis. All authors read and approved the final manuscript. The study was approved by Institutional Review Board of Hunan Provincial People’s Hospital (No. 2015064) in accordance with the relevant guidelines and regulations, and written informed consent was obtained from each participant prior to enrollment.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Project of Improving the Diagnosis and Treatment Capacity of Hepatobiliary, Pancreas and Intestine diseases in Hunan Province (Xiangwei [2019] No. 118), and the Youth PhD Fund of Hunan Provincial People’s Hospital (BSJJ201808).
ORCID iD: Ning Zhou  https://orcid.org/0000-0002-7416-1494
https://orcid.org/0000-0002-7416-1494
References
- 1. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. [DOI] [PubMed] [Google Scholar]
- 2. Dimastromatteo J, Houghton JL, Lewis JS, Kelly KA. Challenges of pancreatic cancer. Cancer J. 2015;21(3):188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 4. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15(6):333–348. [DOI] [PubMed] [Google Scholar]
- 5. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. [DOI] [PubMed] [Google Scholar]
- 9. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. [DOI] [PubMed] [Google Scholar]
- 10. Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP, Yang M. Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol. 2018;101:531–538. [DOI] [PubMed] [Google Scholar]
- 11. Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503(2):870–875. [DOI] [PubMed] [Google Scholar]
- 12. Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88–93. [DOI] [PubMed] [Google Scholar]
- 13. Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644. [DOI] [PubMed] [Google Scholar]
- 14. Sun J, Lian M, Ma H, et al. Competing endogenous RNA network analysis of CD274, IL10 and FOXP3 coexpression in laryngeal squamous cell carcinoma. Mol Med Rep. 2018;17(3):3859–3869. [DOI] [PubMed] [Google Scholar]
- 15. Li S, Zhang W, Yin X, et al. Mouse ATP-Binding Cassette (ABC) transporters conferring multi-drug resistance. Anticancer Agents Med Chem. 2015;15(4):423–432. [PubMed] [Google Scholar]
- 16. Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding L, Yao W, Lu J, Gong J, Zhang X. Upregulation of circ_001569 predicts poor prognosis and promotes cell proliferation in non-small cell lung cancer by regulating the Wnt/beta-catenin pathway. Oncol Lett. 2018;16(1):453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu JH, Wang Y, Xu D. Hsa_circ_001569 is an unfavorable prognostic factor and promotes cell proliferation and metastasis by modulating PI3K-AKT pathway in breast cancer. Cancer Biomark. 2019;25(2):193–201. [DOI] [PubMed] [Google Scholar]
- 19. Qu S, Song W, Yang X, et al. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Z, Qin W, Sun Y. Contribution of biomarkers for pancreatic cancer-associated new-onset diabetes to pancreatic cancer screening. Pathol Res Pract. 2018;214(12):1923–1928. [DOI] [PubMed] [Google Scholar]
- 21. Kunovsky L, Tesarikova P, Kala Z, et al. The use of biomarkers in early diagnostics of pancreatic cancer. Can J Gastroenterol Hepatol. 2018;2018:5389820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xing C, Ye H, Wang W, et al. Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif Cells Nanomed Biotechnol. 2019;47(1):3920–3928. [DOI] [PubMed] [Google Scholar]
- 23. Yang F, Liu DY, Guo JT, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qu S, Hao X, Song W, et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. 2019;11(1):53–63. [DOI] [PubMed] [Google Scholar]
- 26. Yao J, Zhang C, Chen Y, Gao S. Downregulation of circular RNA circ-LDLRAD3 suppresses pancreatic cancer progression through miR-137-3p/PTN axis. Life Sci. 2019;239:116871. [DOI] [PubMed] [Google Scholar]
- 27. Liu H, Xue L, Song C, Liu F, Jiang T, Yang X. Overexpression of circular RNA circ_001569 indicates poor prognosis in hepatocellular carcinoma and promotes cell growth and metastasis by sponging miR-411-5p and miR-432-5p. Biochem Biophys Res Commun. 2018;503(4):2659–2665. [DOI] [PubMed] [Google Scholar]