Abstract
Methods to assess environmental exposure to hazardous chemicals have primarily focused on quantification of individual chemicals, although chemicals often occur in mixtures, presenting challenges to the traditional risk characterization framework. Sampling sites in a defined geographic region provide an opportunity to characterize chemical contaminants, with spatial interpolation as a tool to provide estimates for non-sampled sites. At the same time, the use of in vitro bioactivity measurements has been shown to be informative for rapid risk-based decisions. In this study, we measured in vitro bioactivity in 39 surface soil samples collected immediately after flooding associated with Hurricane Harvey in Texas in a residential area known to be inundated with polycyclic aromatic hydrocarbon (PAH) contaminants. Bioactivity data were from a number of functional and toxicity assays in 5 human cell types, such as induced pluripotent stem cell-derived hepatocytes, cardiomyocytes, neurons, and endothelial cells, as well as human umbilical vein endothelial cells. Data on concentrations of PAH in these samples were also available and the combination of data sources offered a unique opportunity to assess the joint spatial variation of PAH components and bioactivity. We found significant evidence of spatial correlation of a subset of PAH contaminants and of cell-based phenotypes. In addition, we show that the cell-based bioactivity data can be used to predict environmental concentrations for several PAH contaminants, as well as overall PAH summaries and cancer risk. This study’s impact lies in its demonstration that cell-based profiling can be used for rapid hazard screening of environmental samples by anchoring the bioassays to concentrations of PAH. This work sets the stage for identification of the areas of concern and direct quantitative risk characterization based on bioactivity data, thereby providing an important supplement to traditional individual chemical analyses by shedding light on constituents that may be missed from targeted chemical monitoring.
Keywords: polycyclic aromatic hydrocarbons, mixtures toxicology, in vitro models, iPSC, humanized models, human risk assessment, risk assessment, new approach methods, bioactivity, spatial analysis
Environmental samples from contaminated sites contain complex mixtures of chemicals and may pose concern to both human health and the environment (Escher et al., 2020; Stehle and Schulz, 2015). The regulatory authorities in the United States (U.S. EPA, 1986) and Europe (Backhaus et al., 2010; Brack et al., 2019; Kortenkamp and Faust, 2018) are tasked with the evaluation of the mixtures; however, multiple challenges with the current approaches they rely upon have been widely acknowledged. Traditional methods for assessment of environmental exposures focus on the few individual chemicals that were detected in environmental samples, an approach that can underestimate the risks both because chemicals may be missed due to their not being analyzed for, as well as because interactions among the components in a mixture may complicate attempts at dose reconstruction (Kortenkamp and Faust, 2018). Several regulatory agencies are considering amendments to the traditional risk characterization frameworks to mixtures (Bopp et al., 2019; ECHA, 2017; More et al., 2019) to address the knowledge gaps in regulatory science with respect to quantitative characterization of the effects by the mixtures of unknown, or yet to be characterized, chemical composition.
To better characterize potential hazards of complex environmental mixtures, novel approaches based on chemical analysis methods (Hollender et al., 2017) and biological assays (Fang et al., 2020; Judson et al., 2010) have been proposed. Recent advances in analytical chemistry assays and their application to the analysis of environmental samples contribute greatly to the opportunities to reconstruct exposure to complex mixtures (Patel, 2017; Rager et al., 2016). Both targeted and untargeted approaches have demonstrated that environmental and human samples may contain hundreds to thousands of chemicals (Rappaport, 2018; Sille et al., 2020); however, this complexity presents a formidable challenge to confident identification and quantitation of the constituent chemicals. Even with the most contemporary high-resolution analytical techniques, only partial characterization of the chemicals in complex environmental samples is attainable.
A complementary approach for hazard characterization of complex substances or mixtures is the use of in vitro methods that can evaluate the effects of the whole substance, rather than its individual constituents (Escher et al., 2020). Examples over the last decade include applications to hazard identification of oil dispersant formulations (Judson et al., 2010), environmental samples (Blackwell et al., 2017, 2019; Escher et al., 2018; Horzmann et al., 2017; Neale et al., 2020), petroleum substances (Grimm et al., 2016; Kamelia et al., 2019), and botanicals (Catlin et al., 2018). The high-throughput format of in vitro assays allows for rapid testing, and it has been suggested that additions of in vitro bioactivity data to mixture risk assessment may hold promise in reducing uncertainties (Drakvik et al., 2020; Ginsberg et al., 2019). It has also been posited that cell-based bioassays can be used in support of environmental quality standards (Escher et al., 2018).
Most studies that used cell-based bioassays to evaluate the effects of environmental mixtures take advantage of readily-available cancer cell lines and rely on cell viability or reporter assays (Alimba et al., 2016; Blackwell et al., 2019; Fang et al., 2020; Neale et al., 2017). Seldom are primary or induced pluripotent stem cell (iPSC)-derived human cell types and functional phenotypes used. Therefore, this study used a compendium of human cell lines from different organs to test bioactivity of a set of soil samples collected from a residential area in Texas with reported contamination from polycyclic aromatic hydrocarbon (PAH)-containing substances during Hurricane Harvey-associated flooding (Horney et al., 2018; Stone et al., 2019). The potential for a small set of in vitro models to inform rapid risk-based decision-making for environmental chemicals was recently demonstrated (Chen et al., 2020). Here, we show that PAH concentrations and in vitro bioactivity in environmental samples were spatially correlated for only a subset of cell-based phenotypes; however, in vitro bioactivity data can be used to predict environmental concentrations and cancer risk from PAH contaminants.
MATERIALS AND METHODS
Chemicals and biologicals
Dimethyl sulfoxide (DMSO, cell-culture grade, ≥99%) was purchased from Santa Cruz Biotechnology (Santa Cruz, California). Cyclohexane (HPLC grade) was obtained from Fisher Scientific (Waltham, Massachusetts). Reference compounds that served as positive controls for each cell type (Supplementary Table 1) were purchased from Sigma-Aldrich (St Louis, Missouri). Hoechst 33342, MitoTracker Orange CMTMRos, and Calcein Green AM were obtained from Life Technologies (Grand Island, New York). Four types of human iPSC-derived cells (iCell hepatocytes 2.0, catalog no. C1023; iCell neurons, catalog no. C1008; iCell cardiomyocytes, catalog no. CMC-100-010-001; and iCell endothelial cells, catalog no. C1023) used in these studies were from Fujifilm Cellular Dynamics (Madison, Wisconsin). Pooled human umbilical vein endothelial cells (HUVEC, catalog no. CC-2519A) were from Lonza (Walkersville, Maryland). Cell-specific media and supplements were purchased from the same vendor as the cells. Rationale for cell selection, metabolic competency of the iCell hepatocyte model, and the justification for selected phenotypes in each cell type are detailed elsewhere (Chen et al., 2020; Grimm et al., 2015; Iwata et al., 2017; Sirenko et al., 2014a,b).
Environmental sample collection and extraction
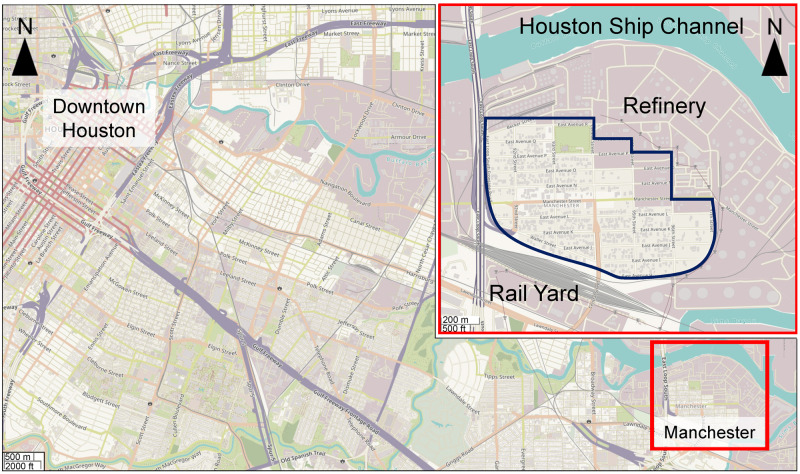
Surface soil samples were collected from a residential area in Manchester, Texas, which is a neighborhood in the greater Houston region (Figure 1). This area was selected for sampling because it is known to be contaminated with PAHs (Bera et al., 2019; Sansom et al., 2018, 2020; Stone et al., 2019). Samples were collected on September 1, 2017, immediately after the area became accessible following Hurricane Harvey landfall. Soil was taken from the top 2–3 cm depth using a metal shovel and deposited into Fisherbrand Certified Clean Clear Glass Straight-Sided Jars (250 ml, catalog no. 11704299; Fisher Scientific, Waltham, Massachusetts). The longitude and latitude of each sample location were recorded and all samples were transported to the laboratory in an ice-filled chest and stored at −80°C until extractions.
Figure 1.
Geographical map of the study area of Manchester neighborhood in Houston, Texas. A map of South-East section of the greater Houston area showing both downtown (top left) and Manchester (red box, bottom right) areas. Inset is a zoom-in of the Manchester neighborhood (blue outline) and surroundings that include a major petrochemical refinery (North-East), an inter-state highway (West), and a rail yard (South). Background and inset maps are from ESRI/OpenStreetMap. Map resolutions are indicated in the bottom left corners.
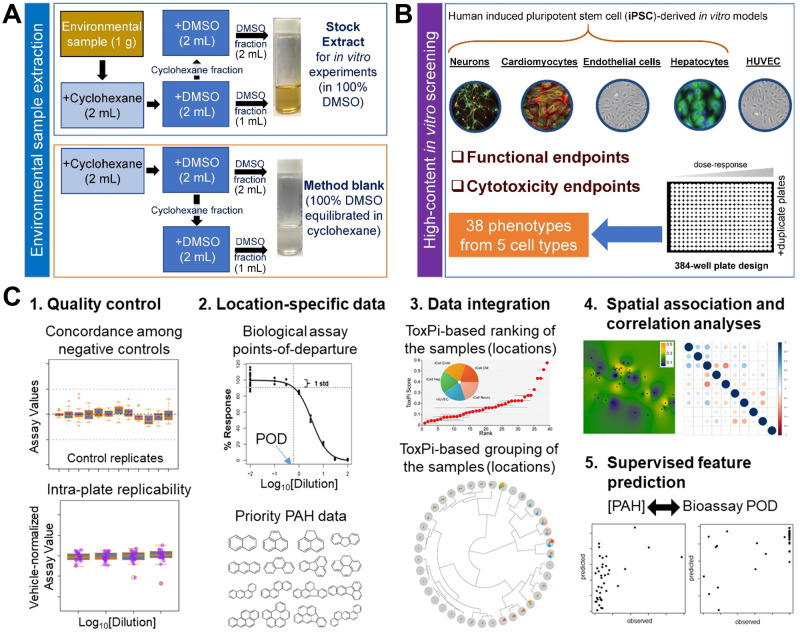
Prior to extraction, soil samples were freeze dried (Malcolm, 1968). The extraction procedure was designed to concentrate the “biologically active” fraction (polycyclic aromatics, but also other polar constituents) of each environmental sample. Samples were extracted (Figure 2A) with cyclohexane and DMSO using a procedure based on the IP346 method (CONCAWE, 1994). Specifically, 1 g of each sample was decanted into a 15-ml conical-bottom disposable plastic tube (Corning, Vernon Hills, Illinois) and mixed with 2 ml of cyclohexane and 2 ml of DMSO pre-equilibrated with cyclohexane at 10:1 ratio. Tubes were vortexed for 1 min and centrifuged for 5 min at 4700 rpm. A 2 ml of DMSO layer was removed and placed into a clean 5-ml glass vial (Lab Products, Houston, Texas). Additional amount of 2 ml of pre-equilibrated DMSO was added to the tube with the sample and the sample was vortexed and centrifuged as detailed above. The DMSO layer (2 ml) was removed and combined with the first DMSO fraction. This sample was used as a stock solution of each sample for subsequent in vitro experiments. In addition, we prepared a “method blank” sample using the procedure detailed above but without addition of a soil sample. This sample contained 100% DMSO with trace amounts of cyclohexane and was used as a “vehicle” sample throughout all in vitro experiments.
Figure 2.
Overall experimental design of the study. A, A schematic diagram of the extraction procedure for environmental soil samples. B, Bioactivity data collection overview. In vitro experiments were performed in 384-well plates using 5 human cell types. C, Data analysis workflow. Quality control (QC) was used to filter assay/cell line combinations to ensure high concordance among controls and high intra- and inter-plate reproducibility. For the assays passing QC, points of departure (POD) were estimated using logistic (Hill) function curve fitting, and overall and cell-type-specific measures of bioactivity computed across the assays. Analysis of bioactivity was further grounded in comparisons to polycyclic aromatic hydrocarbon (PAH) data on the same samples. Data were integrated using ToxPi approach. Spatial association and correlations between biological and PAH data were evaluated. Finally, trained (supervised) models to “predict” the PAH data from bioactivity or vice versa were constructed.
Cell culture experiments
All cell types (Figure 2B) were cultured in 384-well flat bottom plates (iCell Hepatocytes, catalog no. 356667, Corning; iCell Neurons, catalog no. 781946, Greiner Bio-One, Monroe, North Carolina; iCell Cardiomyocytes, catalog no. 3764, Corning; iCell Endothelial cells and HUVECs, catalog no. 353962, Corning) in the media as recommended by the manufacturers (Fujifilm Cellular Dynamics or Lonza). Cells were cultured without treatment for a period of time required to achieve functional capacity. Cell plating density and other culture conditions for each of these cell types have been previously detailed (Grimm et al., 2015; Iwata et al., 2017; Sirenko et al., 2014a,b). Each environmental sample’s stock extract in 100% DMSO was used to prepare 10× serial dilutions with cell culture grade DMSO. A master test plate was prepared to contain 308 experimental wells. All outer wells of the 384-well plates were filled with 75 µl of sterile distilled water to enhance temperature balance for the entire plate and were not used in the experiments. In the master plate, experimental wells were filled with one serial dilution (four 10× dilutions) of each of environmental sample extracts, “method blanks,” or pure DMSO. Three environmental extracts were placed on the master plate twice to enable examination of intra-plate reproducibility. Remaining wells were kept unfilled for cell-specific positive control chemicals and media-only wells. The master plate was sealed with aluminum film and stored at −80°C until used. Copies of a master plate were prepared for use in each in vitro experiment to avoid freeze thawing.
On the day of an experiment for a specific cell type, the master plate was removed from the freezer and placed at room temperature. Content of each well was diluted 100-fold with warm cell culture medium corresponding to the cell type under investigation to yield 4× working solution in 1% DMSO. Positive control chemicals (in 4× concentrations) and cell culture medium were added to the designated empty wells. Next, 12.5 µl (for cardiomyocytes) or 25 µl (for all other cell types) of each well on the working plate was transferred to the plates with cells using 384-well automatic dispenser. The final concentration of DMSO in all assay wells (except for media-only wells) following addition of the test substances was 0.25% (v/v). This amount of DMSO was without effects in all cell types used in these studies (Grimm et al., 2015; Iwata et al., 2017; Sirenko et al., 2014a,b). The environmental sample extracts were assayed in the final dilution of 400–400 000× from the stock solution. All experiments included inter-plate replicates because two identical plates were screened for each cell type.
Cytotoxicity and functional phenotype assays
For each cell line, a number of phenotypes (Supplementary Table 2) were evaluated using high-content or kinetic imaging. A total of 38 phenotypes from 5 tested cell types, including cytotoxicity and functional readouts, were used in subsequent analyses. At the end of the exposure period, cells were stained with different fluorescent dyes and imaged as detailed in previous studies (Grimm et al., 2015; Iwata et al., 2017; Sirenko et al., 2014a,b). Images were processed using the Multi-Wavelength Cell Scorning, Neurite Outgrowth, or Angiogenesis Tube Formation application modules in MetaXpress (Molecular Devices, San Jose, California) software and quantitative data were extracted for concentration-response modeling. Briefly, effects on the mitochondrial integrity and intensity of iCell hepatocytes, and neurite outgrowth of iCell neurons were measured using high-content imaging (ImageXpress Micro Confocal, Molecular Devices). Calcium flux reflecting the contract beating of iCell cardiomyocytes was determined by FLIPR tetra (Molecular Devices) instrument using EarlyTox Cardiotoxicity Kit (Molecular Devices). Effects on angiogenesis of both iCell endothelial cells and HUVECs were measured by 3D cell culture using extracellular gel matrix followed by high-content imaging (ImageXpress Micro Confocal, Molecular Devices).
Assay quality controls
The overall workflow of data processing and analysis is detailed in Figure 2C. Data quality in this study was evaluated using previously established protocols (Grimm et al., 2015). All cell responses were normalized to the vehicle control (0.25% “method blank”-treated wells). Overall quality control criteria were established to evaluate each cell-based assay based on 3 parameters (Supplementary Tables 3 and 4): (1) lack of a statistically significant difference between negative controls, (2) lower than 20% coefficient of variation (% CV) for the negative controls, and (3) confidence that positive control chemicals displayed expected effects in each cell type (expected direction of the effect and comparison of the EC50 of the positive controls to those in previously published methods).
Concentration-response modeling
Vehicle control-scaled data for each treatment were fitted to a curve with a nonlinear logistic function to determine point-of-departure (POD) values, defined as the dilutions at which the fitted curve exceeded one standard deviation above or below the mean of vehicle-treated controls, using R software-based script as reported previously (Sirenko et al., 2017). The choice of one standard deviation “benchmark response” was based on the U.S. EPA guidance for dose-response modeling and determination of the POD (U.S. EPA, 2012), as well as empirical testing of various thresholds as detailed in Sirenko et al. (2017), which showed that a choice of one standard deviation generates consistently high classification accuracy.
Data integration in ToxPi and clustering analyses
POD values generated from concentration-response modeling of each phenotype in tested cell types (Supplementary Table 5) were converted into toxicological priority index (ToxPi) scores (Reif et al., 2013), which were inversely scaled from 0 to 1, with 0 representing the highest POD value in a given dataset (ie, the lowest observed bioactivity) and 1 representing the lowest POD value (ie, the highest observed bioactivity). The scaled POD values were then used as quantitative inputs in ToxPi Graphical User Interface (Marvel et al., 2018) for data integration and visualization of bioactivity profiling. For the clustering, tested environmental samples were grouped based on the similarity between the biological profiling from each cell type in an unsupervised analysis, without prior knowledge of sample categories.
Spatial association of the bioactivity and PAH concentration data
For each sampling location, geographic distances were calculated from GPS coordinates using the geosphere package in R. Spatial interpolation was performed using inverse distance weighting in using the gstat package in R with idp = 3. Test of spatial association for bioactivity or PAH data used the standard Mantel (1967) approach for space-time association, with values for the biological and chemical features taking the place of the “time” dimension, and geographical distances calculated using latitude/longitude coordinates. This approach compares matrices of geographical distances to squared feature differences for all pairs of sampling sites normalized according to the methods in Zhou et al. (2013). For global tests using all biological or chemical features, distance matrices using all paired samples (i, j) were calculated using 1 − ρij, where ρij is the Spearman correlation of all features. The test statistic is the summed element-wise product of the two distance matrices, and rejects the null hypothesis for large values, corresponding to evidence of spatial correlation. Each test was implemented in R v.3.6.1 and p values were obtained using 10 000 permutations, and padj were derived from multiple testing correction using Benjamini-Hochberg computation (Benjamini and Hochberg, 1995) using the p.adjust function in R.
Both bioactivity data (ToxPi scores for each cell type) and chemical concentrations of PAHs in these samples (Sansom et al., 2020) were used for these analyses (Supplementary Table 6). Concentration of PAHs in these environmental samples was measured by Geochemical and Environmental Research Groups at Texas A&M University. The priority 16 PAHs, which have been designated high priority pollutants by the U.S. EPA (Keith, 2015), as well as the total PAH concentrations were analyzed by gas chromatography (HP5890, Hewlett Packard, Wilmington, Delaware) with mass spectrometry detection (Agilent 5972, Agilent Technologies, Santa Clara, California) in selected ion monitoring mode.
Hazard index calculation and cancer risk assessment based on PAHs concentrations
We characterized the non-cancer and cancer risk (Supplementary Table 7) associated with each sample as follows using the U.S. EPA Regional Screening Level Soil Screening Levels (SSL) for residential soil (U.S. EPA, 2020). For non-cancer risk levels for each sample, we calculated (Supplementary Table 8) the hazard index (HI) by summing the ratios between the measured soil concentration Ck for PAH k (converted to mg/kg) and the corresponding non-cancer SSLnc,k:
This calculation is based on the individual PAH non-cancer SSLnc,k corresponding to a hazard quotient of 1. Several PAHs did not have SSLs, so they were not included in the calculation. For cancer risk, we converted each PAH concentration to benzo[a]pyrene (BaP)-equivalents using the Toxic Equivalency Factors (TEFs) from Nisbet and LaGoy (1992), CBaPeq,k = Ck TEFk, and then calculated the cancer risk using the cancer SSLc, BaP for BaP (Supplementary Table 8):
This calculation is based on the individual PAH cancer SSLnc,k corresponding to a cancer risk of 10−6. Similar results were obtained when using alternative TEFs (U.S. EPA, 1993, 2010) (Supplementary Table 8).
Prediction between chemical and biological profiling
For prediction of individual chemical features from the collection of biological features, and individual biological features from the collection of chemical features, ordinary linear regression performs extremely poor due to the large number of prediction features compared to the sample size. Penalized ridge regression is a useful alternative, and we used the multivariate nature of the prediction (eg, multiple chemicals simultaneously) to offer further improvements in a unified model. Briefly, one can envision the chemical concentration data as a multidimensional readout Y with n = 39 rows and 19 columns for cancer risk and chemicals (include PAH aggregate values) and a predictor matrix X with 39 rows and 39 columns (including the intercept unit column) for biological features. Prior to fitting, all data columns were centered and scaled to unit variance for comparability and to ensure no predictor dominated simply due to scale differences. For tuning parameter λ, is a coefficient matrix, with final prediction . λ was evaluated on a grid such that log10(λ) varied uniformly from −1.0 to 6.0 in increments of 0.1. Evaluations were performed using leave-one-out cross-validation, ie, prediction for elements of Y from the ith sample used coefficients obtained after removing the ith sample, to avoid overfitting. Selection of the tuning parameter was performed to give minimum mean-squared prediction error. Final predictions were returned to the original Y scale by multiplying each column by the original standard deviation and adding the original mean. The entire procedure was then run again to predict biological features by reversing the assignment of X and Y matrices.
RESULTS
A recent longitudinal study that assessed exposure to PAHs among residents of Manchester, an environmental justice neighborhood located in the East End of Houston, Texas (Figure 1), showed evidence of redistribution of PAHs due to extreme flooding associated with Hurricane Harvey in 2017 (Horney et al., 2018; Stone et al., 2019). We sampled a total of 39 locations across the whole neighborhood; surface soil samples were collected immediately after the flooding receded. Because of the large number of potential sources of PAH in and around Manchester, and previous reports of considerable gradients of PAH concentrations among these samples, we processed (Figure 2A) the soils using a procedure that is designed to extract carcinogenic PAHs (ASTM International, 2014; CONCAWE, 1994). Specifically, this method preferentially extracts PAH that are toxicologically relevant, those with >3 rings, naked or partially alkylated (Carrillo et al., 2019).
To profile the bioactivity of the environmental samples, we used a targeted set of human cell-based models and phenotypes (Figure 2B) that can be used to assign compounds to chemical classes. The quantitative estimates of in vitro effects from these cells/phenotypes can serve as a conservative surrogate for regulatory in vivo POD (Chen et al., 2020). The data were analyzed using a multi-stage workflow (Figure 2C) that included quality assurance, concentration-response analysis, integration of the data from multiple cell types/phenotypes, spatial and correlation analyses of both bioactivity and PAH data, and supervised feature prediction between bioactivity and PAH datasets.
ToxPi and Clustering Analyses of Bioactivity in Environmental Soil Samples
We used dilution series-derived POD data from 38 phenotypes in 5 cell types (Supplementary Table 5) to compute an overall bioactivity ToxPi score for each of the 39 tested environmental samples (Figure 3A). Most of the samples exhibited little to no bioactivity in most phenotypes, as can be seen from low ToxPi values for about 75% of all samples analyzed. Only a handful of samples were bioactive, as signified by a sharp increase in the ToxPi scores. Interestingly, the sensitivity analysis, showed by the confidence interval (95%) whiskers for each red dot, demonstrated that high ToxPi samples’ rank was largely invariable, whereas the low-ranked samples’ confidence intervals were wide and largely overlapping. Figure 3B shows the ToxPi profiles and their clustering for each sample. The bioactive samples showed effects in several cell types, primarily in endothelial cells and iCell cardiomyocytes. Clustering of the ToxPi profiles for each sample showed that several clusters of very similar bioactivity were present.
Figure 3.
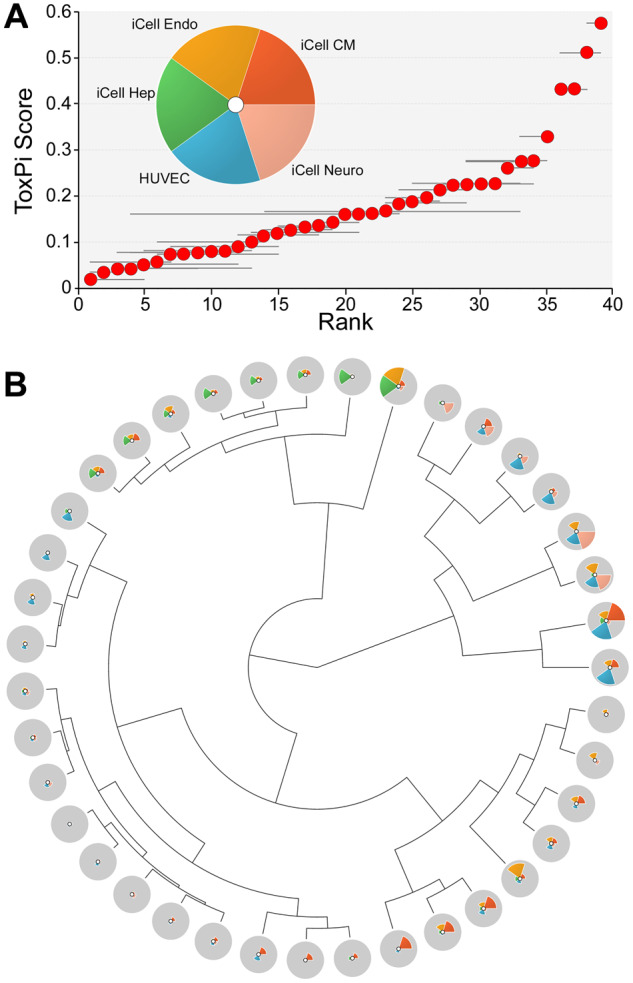
Bioactivity-based ranking of the sampling locations based on the data from 5 human cell types. A, The Toxicological Priority Index (ToxPi) approach was used to combine data across cell types (pie chart inset) and rank them based on the combined ToxPi score. Horizontal whisker represents a resampling-based confidence interval (95%) on the rank of each sampling location (red dots). B, Clustering (Ward’s D method) of the sampling locations using ToxPi scores. ToxPi radial plots were the same as those shown in panel (A).
Spatial Association of Bioactivity, PAH Concentration Data, and Risk Characterization
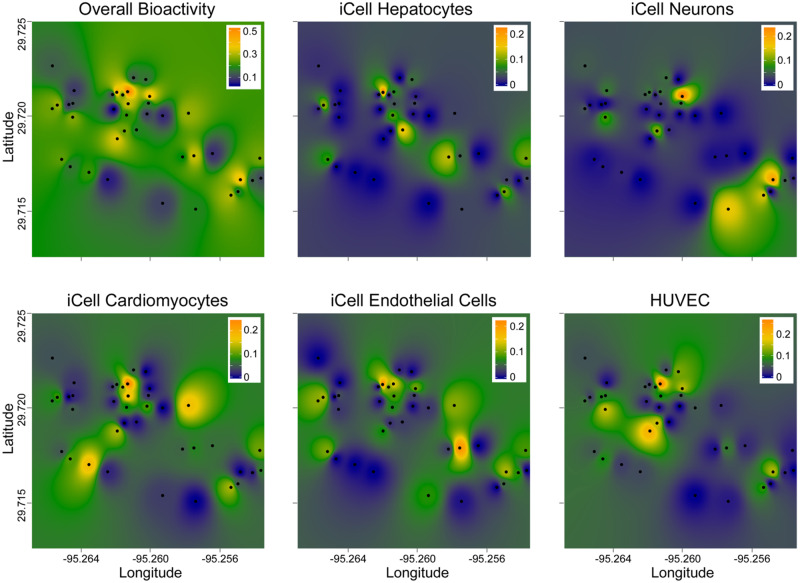
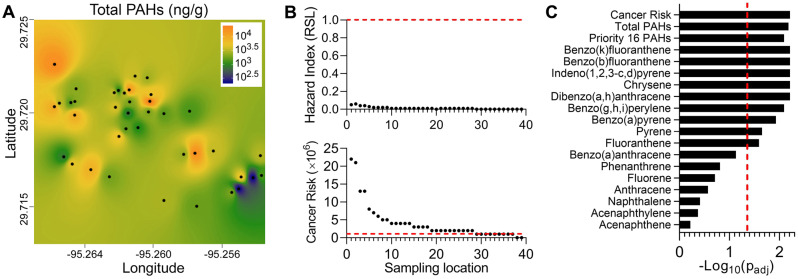
Next, we tested if spatial association was significant for bioactivity profiles. First, we mapped the overall bioactivity ToxPi scores, or scores for each cell type separately, for each location (Figure 4). Clusters of bioactivity were evident; however, the signatures of the individual cell types were quite distinct, similarly to our previous finding that each of tested cell types contributed independently to the utility of this overall in vitro model (Chen et al., 2020). Although a number of tested locations had consistently low bioactivity across the whole panel, several locations appeared to be hot spots identified by this analysis. Next, we used a statistical test of spatial association (a modified version of Mantel [1967]) to determine whether physical proximity among sampling sites was associated with the similarity of the bioactivity. Upon stringent false discovery rate correction procedures, no bioactivity phenotype individually, or in aggregate, reached significance (Supplementary Table 6). Similar analyses were performed using PAH concentrations in the same samples (Figure 5). Several clear “hot spots” were apparent for both total PAH (Figure 5A) and the individual PAHs (data not shown).
Figure 4.
Interpolation of the spatial patterns in bioactivity of the samples. Sampling locations are identified as black dots and the ToxPi integrated bioactivity (on a scale from 0 = lowest effect [dark blue], to 1 = highest effect [orange]) was used to create the maps (see Materials and Methods) that visualize ToxPi values as a color gradient (see the legends in each graph). The maps show overall bioactivity based on all 5 cell types (top left), or bioactivity in each of the cell types individually (see labels for each map for cell identifier).
Figure 5.
Polycyclic aromatic hydrocarbons (PAH) levels in the studied sample locations. A, Interpolation of the spatial patterns in total concentration of PAH (ng/g soil). Sampling locations are identified as black dots and the cumulative PAH concentrations were used to create the maps (see Materials and Methods) that visualize PAH levels as a color gradient (see the legend inset for concentration/color). B, Screening-level risk characterization for non-cancer (upper panel) and cancer (lower panel) risks, based on EPA Soil Screening Levels. Horizontal dashed lines denote screening levels of potential concern, based on a non-cancer Hazard Index = 1 and a cancer risk of 10−6. C, p values (log10 scale) for spatial correlation/persistence for cancer risks (based on PAH TEF), individual, or cumulative concentrations of 16 priority PAH, or total PAH. These p values were derived using a modification of a standard space-time correlation method as described in text. Shown are adjusted p values for each parameter and a vertical dotted line represents padj = .05 (false discovery rate) threshold.
As shown in Figure 5B and Supplementary Table 8, the non-cancer risks associated with these measured PAH concentrations are well below the levels of concern denoted by HI = 1. For cancer, however, the calculated cancer risks for many samples are above the commonly used screening level threshold of 1 × 10−6, though still within EPA’s generally acceptable risk range of 10−4 to 10−6 (U.S., 2011). In addition, a statistical test of spatial association for PAH data (Figure 5C, Supplementary Table 6) showed that most of the substances, as well as their cumulative values and PAH-derived cancer risk factor, were highly significantly co-located, even when stringent false discovery rate correction procedures were applied.
Prediction Between Chemical and Biological Profiling
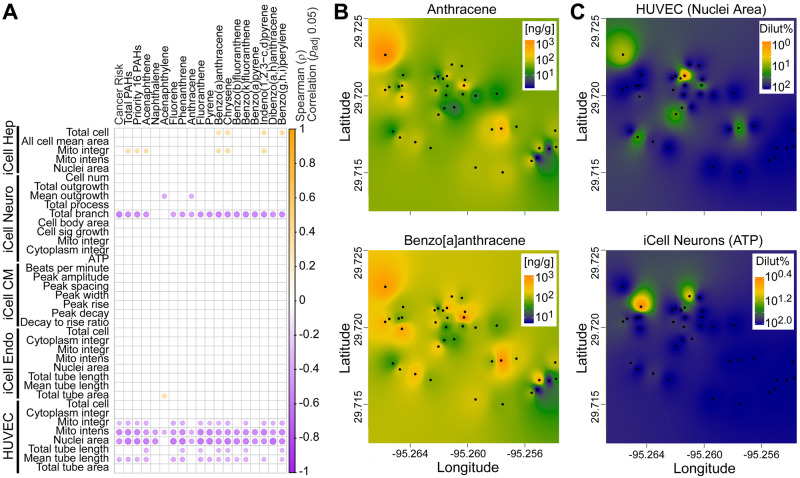
Next, we tested if overall in vitro bioactivity correlated with PAH-derived non-cancer (ie, HI) or cancer risk values for each sampling locations. Highly significant positive correlation was observed for both HI (r = 0.45, p < .01) and cancer risk (r = 0.48, p < .005) when samples 102 and 49 were removed, as these had the highest HI and cancer risk values. Next, we sought to determine what individual in vitro bioactivity phenotypes and soil PAH concentrations correlated (Figure 6). Most of the phenotypes (all of the phenotypes in iCell endothelial cells and iCell cardiomyocytes) did not correlate significantly with PAH values after adjustment for multiple comparisons (Figure 6A); however, several in vitro phenotypes showed strong negative correlations—most of the phenotypes in HUVECs and total branch phenotype in iCell neurons. Negative correlation for the individual phenotypes is expected as it indicates that higher PAH concentration indicate higher potency (ie, lower POD). Interestingly, in iCell hepatocytes, several PAHs were positively associated with cell viability and mitochondrial integrity, also concordant with the expected relationship between PAHs and these effects in hepatocytes. Figure 6B shows examples of two PAHs, anthracene and benzo[a]anthracene, which showed somewhat different geographical distribution of the “hot spots”; however, their concentrations were highly correlated with the environmental sample-induced effects HUVEC nuclei area phenotype (Figure 6C, top). We also show a geographical distribution of bioactivity in iCell neurons ATP phenotype (Figure 6C, bottom), as an example of a phenotype that did no correlate with PAH concentrations in environmental samples.
Figure 6.
Correlation analysis between PAH content and bioactivity of the soil samples. A, Spearman correlation of all bioactivity phenotypes with cancer risk, total, 16 priority or individual PAH concentrations. Significant (padj < .05) correlations are shown as dots that are colored based on the ρ value as indicated in the color bar. B, Interpolation of the spatial patterns in concentrations of anthracene and benzo[a]anthracene (ng/g soil) as representative PAH. Sampling locations are identified as black dots and the cumulative PAH concentrations were used to create the maps (see Materials and Methods) that visualize PAH levels as a color gradient (see the legend inset for concentration/color). C, Interpolation of the spatial patterns in bioactivity of soil samples for the HUVEC (nuclei area) and iCell Neurons (ATP) as representative phenotypes. Sampling locations are identified as black dots and the effective concentrations (as % dilution of the soil extract) were used to create the maps (see Materials and Methods) that visualize bioactivity as a color gradient (see the legend inset for effective concentration/color).
Because of the strong correlation among the PAH concentrations and some bioactivity phenotypes (Figure 6, Supplementary Table 6), we tested whether in vitro bioactivity data can be used collectively to infer PAH concentrations in these environmental samples, or vice versa. This question is relevant because both in vitro analyses and analytical chemistry assays are time consuming and if these data streams are predictive of each other, considerable time and resource savings can be achieved by prioritizing sample analyses.
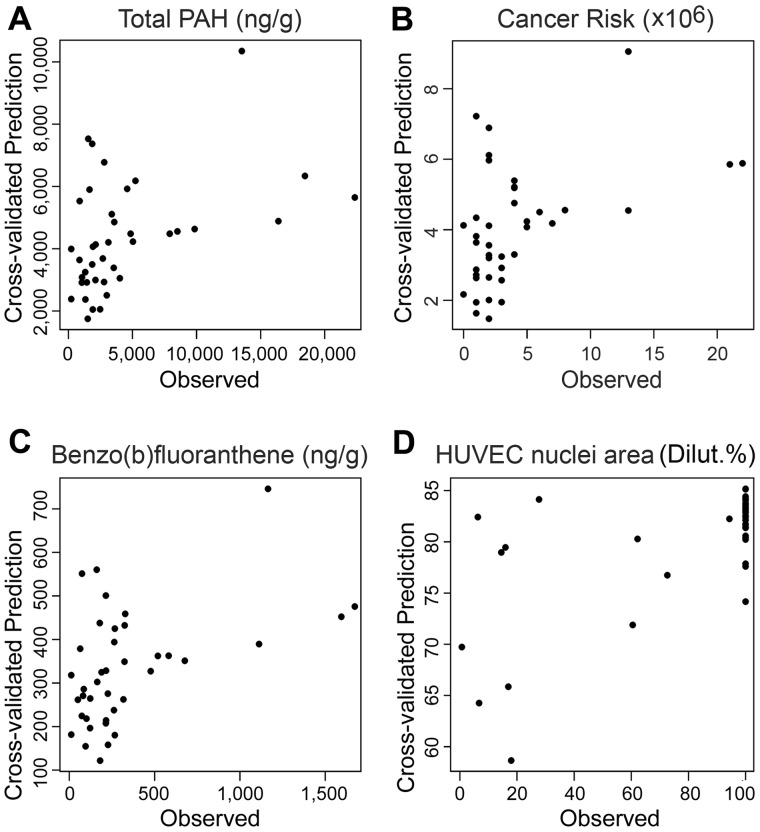
Using a regression model with rigorous cross-validation, we found that bioactivity data were highly predictive of the PAH concentrations, both for many individual priority PAHs, their summary measures, and the cancer risk (Table 1). Figure 7 shows representative examples of the relationships between observed and predicted values. Because the individual PAHs, their sum, and the cancer risk values are highly correlated, it is not surprising that similar patterns exist between observed and predicted values (Figs. 7A–C). It is noteworthy, however, that due to the nature of regression-based predictions, predicted values are “shrunken” estimates (toward a common mean) with less variation than the actual data. For predictions of cancer risk and PAH content, the most informative in vitro phenotypes were HUVEC nuclei area and mitochondria intensity, and total branch length in iCell neurons, see Supplementary Table 9 for all the predictor coefficients for all summary and individual priority PAHs, and their relative ranks. For HUVEC nuclei area and iCell neurons total branch length, the result may not be unexpected, as the pairwise correlations of these quantities with total PAH and cancer risk as observed in Figure 6A were of high significance. However, the high rank of iCell hepatocyte nuclei area as a highly informative predictor was not apparent from the pairwise correlations, and points to the advantage of using a multivariate regression prediction model in this context.
Table 1.
Cross-Validated Prediction of PAH Levels From All In Vitro Bioactivity Data, and of Bioactivity Quantification From All PAH Levels
| Parameter | r a | padj b | ρ a | padj b |
|---|---|---|---|---|
| Predicting PAH levels from in vitro bioactivity dataa | ||||
| Cancer risk | 0.47 | <.01 | 0.46 | <.01 |
| Total PAHs | 0.44 | <.01 | 0.48 | <.01 |
| Priority 16 PAHs | 0.42 | <.01 | 0.43 | <.05 |
| Benzo(k)fluoranthene | 0.48 | <.01 | 0.42 | <.05 |
| Benzo(b)fluoranthene | 0.48 | <.01 | 0.41 | <.05 |
| Indeno(1,2,3,-c, d)pyrene | 0.45 | <.01 | 0.48 | <.01 |
| Chrysene | 0.45 | <.01 | 0.46 | <.01 |
| Dibenzo(a, h)anthracene | 0.45 | <.01 | 0.42 | <.05 |
| Benzo(g, h, i)perylene | 0.42 | <.01 | 0.46 | <.01 |
| Benzo(a)pyrene | 0.40 | <.05 | 0.37 | <.05 |
| Pyrene | 0.36 | <.05 | 0.41 | <.05 |
| Fluoranthene | 0.34 | <.05 | 0.37 | <.05 |
| Benzo(a)anthracene | 0.27 | n.s. | 0.36 | <.05 |
| Fluorene | 0.17 | n.s. | 0.30 | <.05 |
| Predicting in vitro bioactivity data from PAH levels | ||||
| HUVEC nuclei area | 0.65 | <.001 | 0.54 | <.01 |
| iCell Neurons mean outgrowth | 0.44 | <.05 | 0.12 | n.s. |
| HUVEC mitochondria intensity | 0.39 | n.s. | 0.47 | <.05 |
| iCell neurons total branch | 0.31 | n.s. | 0.49 | <.05 |
Correlation (Pearson r or Spearman ρ) of predicted response values using multivariate ridge regression prediction compared to actual response values.
Associated padj values expressed as false discovery-adjusted using the Benjamini-Hochberg method applied to all responses (only significant results using padj < .05 shown).
Figure 7.
Illustrative cross-validated regression predicted values versus actual values, for predicting (A) total PAH concentrations, (B) cancer risk and (C) benzo(b)fluranthene from bioactivity measurements, and (D) HUVEC nuclei areas from the measured PAH concentrations. See correlation coefficients and p values for the correlations shown here listed in Table 1.
Predictions of in vitro bioactivity from PAH concentrations were less informative, with only 4 of 38 phenotypes having multiple testing-corrected significant correlations (Table 1) between observed and predicted values. Even though the correlations were significant for at least some phenotypes, the ranges of predicted bioactivity values were far narrower than those of the actual effects (Figure 7D), indicating that such predictions are difficult to interpret with respect to the potential range of hazards among real environmental samples. Supplementary Table 10 shows all of the regression predictor coefficients for various PAHs, and their relative ranks in predicting the bioactivity measures. Overall, the predictor coefficients were far smaller than those in case of predicting PAH concentrations from bioactivity.
DISCUSSION
Many have suggested the potential utility of cell-based in vitro bioassays for addressing the potential human and ecological health hazard of complex mixtures (Blackwell et al., 2017, 2019; Drakvik et al., 2020; Escher et al., 2020; Hayes et al., 2020; Kassotis et al., 2016; Neale et al., 2020). Large-scale in vitro toxicity screening programs such as Tox21 or ToxCast focus largely on the first-pass testing for individual chemical compounds and some complex substance formulations (Catlin et al., 2018; Judson et al., 2010), and have yet to be widely applied in the evaluation of complex environmental mixtures (Kassotis et al., 2016). In the past two decades, dozens of studies used various cell types, derived from both mammalian and aquatic species and prokaryotes, have been used to study sediment, soil, and water samples. Most often these studies examined general cytotoxicity, effects on the DNA (various genotoxicity and mutagenicity endpoints), as well as activation of various hormone and metabolism-related receptors. Concentration-response relationships were routinely evaluated and it was demonstrated that bioactivity can be used not only in comparative analysis of the relative potency among samples, but also to derive quantitative estimates of hazard (Blackwell et al., 2017, 2019; Escher et al., 2014, 2015, 2018; Jia et al., 2015). Overall, these studies established a body of evidence that a battery of in vitro bioassays can be used to support decision-making based on the bioactivity of the actual environmental samples.
Our work builds on this empirical foundation and shows that human iPSC-derived cells may not only be used to rank environmental samples with respect to potential human health concerns, but they also introduce additional valuable information through the analysis of cell function phenotypes. We found that among a large number of samples collected in a relatively confined geographical area with equal potential of PAH contamination associated with the proximity of numerous point sources (Stone et al., 2019), only some locations indicated a potential concern, information that could serve as a rationale for follow-up analyses with additional assays and models. Interestingly, we found that depending on the cell type, the “hot spots” varied. This finding is commensurate with evidence that certain cell types and phenotypes are differentially affected by various chemicals (Chen et al., 2020; Grimm et al., 2020). Although a screening-level risk characterization based on PAH concentrations indicated little concern for non-cancer effects, a possible concern was identified for some samples for PAH-related cancer risks, which in many cases exceed the screening level risk of 10−6. Similarly, while the calculated risk levels were still within the “generally acceptable” range used by EPA, because only PAHs were measured, the cumulative effects of other, unmeasured toxicants (which are surely present in this neighborhood) are not accounted for. In such a situation, it is common under EPA guidelines to strive for the “lower end” of the risk range and the bioactivity data may provide important clues on the types of hazards that may be present and also the level of concern for follow-up analyses.
A number of previous studies examined spatial relationships in bioactivity between sampling locations, or tested for the strength of association between bioactivity and chemical contamination in a spatial dimension. A study of 41 surface soil samples from Tianjin, China used a suite of in vitro cell bioassays focused on nuclear receptors and genotoxicity endpoints to examine the spatial clustering of the bioassay data (Xiao et al., 2006). This study found that the geographic distribution of aryl hydrocarbon receptor (AhR)-agonism and genotoxic bioactivity exhibited strong positive spatial correlation; however, the geographic distribution of pro-estrogenic bioactivity was markedly different from that of AhR-agonists effects. An example of a study that correlated in vitro and analytical data from environmental samples is a publication by Leusch et al. (2010) who compared the responses of 5 bioassays designed to measure estrogenic activity and chemical analysis on water samples (ground and river water, and raw and treated sewage). The authors showed that the bioassays that were robust in terms of assay sensitivity and reproducibility were well-correlated with the data from chemical assays. An example of a study that looked at both spatial and bioactivity-chemical analysis correlations is the work of Jung et al. (2012) who used 21 sediment samples from Masan Bay, Korea, to identify several “hot spots” of bioactivity (estrogen- and dioxin-responsive receptor assays). The authors also used spatial correlation analysis between organochlorine pesticides, polychlorinated biphenyls, dioxins and alkylphenols and their biological effects to pinpoint the sources, such as sewage treatment and industrial outfall, of environmental hazards. Recent examples of studies in the United States and Europe demonstrate that bioassay-based analysis of environmental mixtures for detecting biological effects should be combined with the analysis of a wide range of chemical contaminants to ascertain additional risks that may not be evident from the chemical analyses alone (Blackwell et al., 2017, 2019; Konig et al., 2017; Neale et al., 2020).
These studies established an important foundation for examining the relationships between exposure and bioactivity-derived hazard and for quantifying these relationships bioanalytical equivalent concentrations (Jahnke et al., 2018) and exposure-activity ratios (Blackwell et al., 2017). Our study provides strong additional evidence of such relationships. It is noteworthy that our study showed that while there was an overall significant positive correlation between bioactivity and PAH-associated HI and cancer risk, the correlation coefficients were only about 0.5. A similar finding was reported in a study of rain events impact on the chemical pollution in river water where the measured chemicals explained only a small fraction (<8%) of the in vitro biological effects (Neale et al., 2020). These data show indicate that bioactivity, while valuable information, may not be sufficient for evaluating certain chemical-specific risks. These results are not altogether surprising, as certain endpoints, such as cancer and immunotoxicity, are known to be poorly covered by currently available in vitro assays. Thus, we reason that for environmental monitoring, high bioactivity scores may be able to identify “hot spots” or areas of concern for the follow-up investigation, but that low bioactivity scores are not sufficient to rule out potential risk.
Another interesting corollary to the datasets that combine measurements of chemical contamination and bioactivity on the same samples is the possibility of using one or the other as predictors. Previously, Leusch et al. (2010) calculated a predicted estrogenicity for environmental water samples by multiplying the concentration of each chemical as determined by standard chemical methods with the relative potency for each individual compound. This report concluded, based on dose reconstruction from the individual chemical concentrations, that there was a good agreement between the predicted and measured estrogenicity; however, this study only attempted prediction of hazard for one type of hazard (ie, estrogenicity) and only through dose reconstruction. In this respect our study offers several additional advances. The correlations between PAH measurements and bioactivity levels show that a relatively small proportion of bioactivity measurements are substantially correlated with PAH levels in environmental samples. However, these correlations are sufficiently large (correlation >0.5) such that summary PAH levels and cancer risk values can be predicted with reasonable accuracy from bioactivity measurements. Interestingly, we found that a reverse prediction, from PAH concentrations to bioactivity, was not as informative. This finding reflects the potential indication that other compounds in the samples may have contributed to the overall bioactivity. It is also possible that poorer prediction of bioactivity may simply be due to an imbalance in the number of PAHs versus the number of bioactivity phenotypes. Ideally, it would be possible to obtain quantitative understanding of the contributions of individual measured substances to overall bioactivity, such as utilizing exposure-activity ratios based on bioactivity data on individual substances. Although some success in this regard have been made for water contamination (Blackwell et al., 2017, 2019; Neale et al., 2020), such efforts are more challenging in the case of soil contamination due to differences in extraction and bioavailability when comparing soil concentrations with concentrations in vitro media (Luo et al., 2020). Future studies are needed to better understand differential extraction efficiency and bioavailability of compounds of interest in order to make more confident comparisons across matrices. Overall, these findings provide additional important evidence as a proof of concept for the use of bioactivity as an approximate chemical concentration surrogate, although additional data should be generated to refine these findings. In addition, the ability to generalize beyond the range of concentrations observed is unknown, and we emphasize that aspects of our data structure and PAH content may be specific to the Manchester neighborhood sampled here.
This study has important limitations. First, the chemical comparisons and environmental sample extraction methods were focused on PAH contamination and as such provide an over-simplified representation of the chemical complexity of the environmental samples, especially after a major natural disaster. Although this chemical class was the most natural choice because of previous reports of PAH contamination in this area (Horney et al., 2018; Stone et al., 2019) and geographical proximity of the relevant point sources, additional chemicals need to be considered in future studies. For example, we found that bioactivity “hot spots” varied among cell types indicating that other contaminants may also be present and additional chemical analyses need to be performed. Second, our risk characterization and comparisons to HI and cancer slope factors were equally restricted to PAH-derived values, which may have reduced our ability to observe true relationships between chemical concentrations and bioactivity. Third, we emphasize that, by focusing on a single neighborhood, the range of variation in PAH may have been limited in comparison to other areas. Thus, under a wider sampling regime it is likely that the observed spatial relationships would have been significant for a larger number of individual chemical and/or bioactivity components. Finally, we note that the large number of cell-based phenotypes and measured contaminants relative to the sample size necessitated the use of penalized regression as a prediction tool, which can provide biased estimation in order to achieve higher prediction accuracy. Follow-up studies focusing on only a select few cell-based assays, informed by this and previous studies, as well as a larger pool of assays from ToxCast/Tox21 (Paul Friedman et al., 2020) might be required in order to provide unbiased estimation of the precise relationships between bioactivity and PAH contaminant concentrations.
In summary, this study explored the use of a small compendium of human cell lines representing multiple potential target tissues for bioactivity-based prioritization in the context of environmental monitoring. Using samples with suspected PAH contamination in a community in a greater Houston area that experienced massive flooding associated with Hurricane Harvey, we found joint spatial variation of PAH components and bioactivity, with different cell-types exhibiting largely distinct spatial patterns of activity. In addition, we found that the cell-based bioactivity data correlate with, and can be used to predict environmental concentrations for several PAH contaminants, as well as overall PAH summaries and cancer risk. However, several high concentration outliers in terms of PAH contamination were not well predicted by bioactivity, possibly due to the need for broader coverage of biological space in the cell-based assays. Overall, these results suggest that human cell-based assays, data that can be procured within weeks after a contamination event, can provide useful information for rapid decision-making in emergency situations, supplementing traditional targeted chemical monitoring with human effects-based monitoring so as to identify possible “hot spots” that warrant additional investigation for their potential to increase human health risk.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank students, staff, and faculty at Texas A&M Superfund Research Center for collecting, transporting and processing the environmental samples used in these studies.
FUNDING
National Institute of Environmental Health Sciences (P42 ES027704, P30 ES029067);a cooperative agreement with the United States Environmental Protection Agency (STAR RD83580201). The views expressed in this manuscript do not reflect those of the funding agencies. The use of specific commercial products in this work does not constitute endorsement by the funding agencies.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Alimba C. G., Gandhi D., Sivanesan S., Bhanarkar M. D., Naoghare P. K., Bakare A. A., Krishnamurthi K. (2016). Chemical characterization of simulated landfill soil leachates from Nigeria and India and their cytotoxicity and DNA damage inductions on three human cell lines. Chemosphere 164, 469–479. [DOI] [PubMed] [Google Scholar]
- ASTM International. (2014). Standard test method for determining carcinogenic potential of virgin base oils in metalworking fluids. E1687-10, West Conshohocken, PA.
- Backhaus T., Blanck H., Faust M. (2010). Hazard and risk assessment of chemical mixtures under REACH state of the art, gaps and options for improvement Swedish Chemicals Agency, Stockholm, Sweden. Report No. PM 3/10. www.kemi.se.
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300. [Google Scholar]
- Bera G., Camargo K., Sericano J. L., Liu Y., Sweet S. T., Horney J., Jun M., Chiu W., Rusyn I., Wade T. L., et al. (2019). Baseline data for distribution of contaminants by natural disasters: Results from a residential Houston neighborhood during Hurricane Harvey flooding. Heliyon 5, e02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell B. R., Ankley G. T., Bradley P. M., Houck K. A., Makarov S. S., Medvedev A. V., Swintek J., Villeneuve D. L. (2019). Potential toxicity of complex mixtures in surface waters from a nationwide survey of United States streams: Identifying in vitro bioactivities and causative chemicals. Environ. Sci. Technol. 53, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell B. R., Ankley G. T., Corsi S. R., DeCicco L. A., Houck K. A., Judson R. S., Li S., Martin M. T., Murphy E., Schroeder A. L., et al. (2017). An “EAR” on environmental surveillance and monitoring: A case study on the use of exposure-activity ratios (EARs) to prioritize sites, chemicals, and bioactivities of concern in Great Lakes waters. Environ. Sci. Technol. 51, 8713–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp S. K., Kienzler A., Richarz A. N., van der Linden S. C., Paini A., Parissis N., Worth A. P. (2019). Regulatory assessment and risk management of chemical mixtures: Challenges and ways forward. Crit. Rev. Toxicol. 49, 174–189. [DOI] [PubMed] [Google Scholar]
- Brack W., Ait Aissa S., Backhaus T., Dulio V., Escher B. I., Faust M., Hilscherova K., Hollender J., Hollert H., Muller C., et al. (2019). Effect-based methods are key. The European Collaborative Project SOLUTIONS recommends integrating effect-based methods for diagnosis and monitoring of water quality. Environ. Sci. Eur. 31, 1–6. [Google Scholar]
- Carrillo J. C., van der Wiel A., Danneels D., Kral O., Boogaard P. J. (2019). The selective determination of potentially carcinogenic polycyclic aromatic compounds in lubricant base oils by the DMSO extraction method IP346 and its correlation to mouse skin painting carcinogenicity assays. Regul. Toxicol. Pharmacol. 106, 316–333. [DOI] [PubMed] [Google Scholar]
- Catlin N. R., Collins B. J., Auerbach S. S., Ferguson S. S., Harnly J. M., Gennings C., Waidyanatha S., Rice G. E., Smith-Roe S. L., Witt K. L., et al. (2018). How similar is similar enough? A sufficient similarity case study with Ginkgo biloba extract. Food Chem. Toxicol. 118, 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liu Y., Wright F. A., Chiu W. A., Rusyn I. (2020). Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs. ALTEX 37, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONCAWE. (1994). The use of the dimethyl sulphoxide (DMSO) extract by the IP 346 method as an indicator of the carcinogenicity of lubricant base oils and distillate aromatic extracts, Brussels, Belgium. Report No. 94/51.
- Drakvik E., Altenburger R., Aoki Y., Backhaus T., Bahadori T., Barouki R., Brack W., Cronin M. T. D., Demeneix B., Hougaard Bennekou S., et al. (2020). Statement on advancing the assessment of chemical mixtures and their risks for human health and the environment. Environ. Int. 134, 105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECHA. (2017). Read-Across Assessment Framework (RAAF) - Considerations on multi-constituent substances and UVCBs. European Chemical Agency, Helsinki, Finland.
- Escher B. I., Aït-Aïssa S., Behnisch P. A., Brack W., Brion F., Brouwer A., Buchinger S., Crawford S. E., Du Pasquier D., Hamers T., et al. (2018). Effect-based trigger values for in vitro and in vivo bioassays performed on surface water extracts supporting the environmental quality standards (EQS) of the European Water Framework Directive. Sci. Total. Environ. 628-629, 748–765. [DOI] [PubMed] [Google Scholar]
- Escher B. I., Allinson M., Altenburger R., Bain P. A., Balaguer P., Busch W., Crago J., Denslow N. D., Dopp E., Hilscherova K., et al. (2014). Benchmarking organic micropollutants in wastewater, recycled water and drinking water with in vitro bioassays. Environ. Sci. Technol. 48, 1940–1956. [DOI] [PubMed] [Google Scholar]
- Escher B. I., Neale P. A., Leusch F. D. (2015). Effect-based trigger values for in vitro bioassays: Reading across from existing water quality guideline values. Water Res. 81, 137–148. [DOI] [PubMed] [Google Scholar]
- Escher B. I., Stapleton H. M., Schymanski E. L. (2020). Tracking complex mixtures of chemicals in our changing environment. Science 367, 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W., Peng Y., Yan L., Xia P., Zhang X. (2020). A tiered approach for screening and assessment of environmental mixtures by omics and in vitro assays. Environ. Sci. Technol. 54, 7430–7439. [DOI] [PubMed] [Google Scholar]
- Ginsberg G. L., Pullen Fedinick K., Solomon G. M., Elliott K. C., Vandenberg J. J., Barone S. Jr,, Bucher J. R. (2019). New toxicology tools and the emerging paradigm shift in environmental health decision-making. Environ. Health Perspect. 127, 125002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm F. A., Iwata Y., Sirenko O., Bittner M., Rusyn I. (2015). High-content assay multiplexing for toxicity screening in induced pluripotent stem cell-derived cardiomyocytes and hepatocytes. Assay Drug Dev. Technol. 13, 529–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm F. A., Iwata Y., Sirenko O., Chappell G. A., Wright F. A., Reif D. M., Braisted J., Gerhold D. L., Yeakley J. M., Shepard P., et al. (2016). A chemical-biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives. Green Chem. 18, 4407–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm F. A., Klaren W. D., Li X., Lehmler H. J., Karmakar M., Robertson L. W., Chiu W. A., Rusyn I. (2020). Cardiovascular effects of polychlorinated biphenyls and their major metabolites. Environ. Health Perspect. 128, 077008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes A. W., Muriana A., Alzualde A., Fernandez D. B., Iskandar A., Peitsch M. C., Kuczaj A., Hoeng J. (2020). Alternatives to animal use in risk assessment of mixtures. Int. J. Toxicol. 39, 165–172. [DOI] [PubMed] [Google Scholar]
- Hollender J., Schymanski E. L., Singer H. P., Ferguson P. L. (2017). Nontarget screening with high resolution mass spectrometry in the environment: Ready to go? Environ. Sci. Technol. 51, 11505–11512. [DOI] [PubMed] [Google Scholar]
- Horney J. A., Casillas G. A., Baker E., Stone K. W., Kirsch K. R., Camargo K., Wade T. L., McDonald T. J. (2018). Comparing residential contamination in a Houston environmental justice neighborhood before and after Hurricane Harvey. PLoS One 13, e0192660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horzmann K. A., de Perre C., Lee L. S., Whelton A. J., Freeman J. L. (2017). Comparative analytical and toxicological assessment of methylcyclohexanemethanol (MCHM) mixtures associated with the Elk River chemical spill. Chemosphere 188, 599–607. [DOI] [PubMed] [Google Scholar]
- Iwata Y., Klaren W. D., Lebakken C. S., Grimm F. A., Rusyn I. (2017). High-content assay multiplexing for vascular toxicity screening in induced pluripotent stem cell-derived endothelial cells and human umbilical vein endothelial cells. Assay Drug Dev. Technol. 15, 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke A., Sobek A., Bergmann M., Braunig J., Landmann M., Schafer S., Escher B. I. (2018). Emerging investigator series: Effect-based characterization of mixtures of environmental pollutants in diverse sediments. Environ. Sci. Process Impacts 20, 1667–1679. [DOI] [PubMed] [Google Scholar]
- Jia A., Escher B. I., Leusch F. D., Tang J. Y., Prochazka E., Dong B., Snyder E. M., Snyder S. A. (2015). In vitro bioassays to evaluate complex chemical mixtures in recycled water. Water Res. 80, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S., Martin M. T., Reif D. M., Houck K. A., Knudsen T. B., Rotroff D. M., Xia M., Sakamuru S., Huang R., Shinn P., et al. (2010). Analysis of eight oil spill dispersants using rapid, in vitro tests for endocrine and other biological activity. Environ. Sci. Technol. 44, 5979–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. H., Hong S. H., Yim U. H., Ha S. Y., Shim W. J., Kannan N. (2012). Multiple in vitro bioassay approach in sediment toxicity evaluation: Masan Bay, Korea. Bull. Environ Contam. Toxicol. 89, 32–37. [DOI] [PubMed] [Google Scholar]
- Kamelia L., de Haan L., Ketelslegers H. B., Rietjens I., Boogaard P. J. (2019). In vitro prenatal developmental toxicity induced by some petroleum substances is mediated by their 3- to 7-ring PAH constituent with a potential role for the aryl hydrocarbon receptor (AhR). Toxicol. Lett. 315, 64–76. [DOI] [PubMed] [Google Scholar]
- Kassotis C. D., Tillitt D. E., Lin C. H., McElroy J. A., Nagel S. C. (2016). Endocrine-disrupting chemicals and oil and natural gas operations: Potential environmental contamination and recommendations to assess complex environmental mixtures. Environ. Health Perspect. 124, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith L. H. (2015). The source of US EPA's sixteen PAH priority pollutants. Polycycl. Aromat. Comp. 35, 147–160. [Google Scholar]
- Konig M., Escher B. I., Neale P. A., Krauss M., Hilscherova K., Novak J., Teodorovic I., Schulze T., Seidensticker S., Kamal Hashmi M. A., et al. (2017). Impact of untreated wastewater on a major European river evaluated with a combination of in vitro bioassays and chemical analysis. Environ. Pollut. 220, 1220–1230. [DOI] [PubMed] [Google Scholar]
- Kortenkamp A., Faust M. (2018). Regulate to reduce chemical mixture risk. Science 361, 224–226. [DOI] [PubMed] [Google Scholar]
- Leusch F. D., de Jager C., Levi Y., Lim R., Puijker L., Sacher F., Tremblay L. A., Wilson V. S., Chapman H. F. (2010). Comparison of five in vitro bioassays to measure estrogenic activity in environmental waters. Environ. Sci. Technol. 44, 3853–3860. [DOI] [PubMed] [Google Scholar]
- Luo Y. S., Ferguson K. C., Rusyn I., Chiu W. A. (2020). In vitro bioavailability of the hydrocarbon fractions of dimethyl sulfoxide extracts of petroleum substances. Toxicol. Sci. 174, 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcolm R. (1968). Freeze-drying of organic matter, clays, and other earth materials. US Geol. Surv. Prof. Pap. 600-C, C211–C216. [Google Scholar]
- Mantel N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220. [PubMed] [Google Scholar]
- Marvel S. W., To K., Grimm F. A., Wright F. A., Rusyn I., Reif D. M. (2018). ToxPi graphical user interface 2.0: Dynamic exploration, visualization, and sharing of integrated data models. BMC Bioinf. 19, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- More S., Bampidis V., Benford D., Boesten J., Bragard C., Halldorsson T., Hernandez‐Jerez A., Hougaard‐Bennekou S., Koutsoumanis K., Naegeli H., et al. (2019). Genotoxicity assessment of chemical mixtures. EFSA J. 17, e05519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale P. A., Altenburger R., Ait-Aissa S., Brion F., Busch W., Umbuzeiro G. D., Denison M. S., Du Pasquier D., Hilscherova K., Hollert H., et al. (2017). Development of a bioanalytical test battery for water quality monitoring: Fingerprinting identified micropollutants and their contribution to effects in surface water. Water Res. 123, 734–750. [DOI] [PubMed] [Google Scholar]
- Neale P. A., Braun G., Brack W., Carmona E., Gunold R., Konig M., Krauss M., Liebmann L., Liess M., Link M., et al. (2020). Assessing the mixture effects in in vitro bioassays of chemicals occurring in small agricultural streams during rain events. Environ. Sci. Technol. 54, 8280–8290. [DOI] [PubMed] [Google Scholar]
- Nisbet I. C., LaGoy P. K. (1992). Toxic equivalency factors (TEFs) for polycyclic aromatic hydrocarbons (PAHs). Regul. Toxicol. Pharmacol. 16, 290–300. [DOI] [PubMed] [Google Scholar]
- Patel C. J. (2017). Analytic complexity and challenges in identifying mixtures of exposures associated with phenotypes in the exposome era. Curr. Epidemiol. Rep. 4, 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul Friedman K., Gagne M., Loo L. H., Karamertzanis P., Netzeva T., Sobanski T., Franzosa J. A., Richard A. M., Lougee R. R., Gissi A., et al. (2020). Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol. Sci. 173, 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager J. E., Strynar M. J., Liang S., McMahen R. L., Richard A. M., Grulke C. M., Wambaugh J. F., Isaacs K. K., Judson R., Williams A. J., et al. (2016). Linking high resolution mass spectrometry data with exposure and toxicity forecasts to advance high-throughput environmental monitoring. Environ. Int. 88, 269–280. [DOI] [PubMed] [Google Scholar]
- Rappaport S. M. (2018). Redefining environmental exposure for disease etiology. NPJ Syst. Biol. Appl. 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif D. M., Sypa M., Lock E. F., Wright F. A., Wilson A., Cathey T., Judson R. R., Rusyn I. (2013). ToxPi GUI: An interactive visualization tool for transparent integration of data from diverse sources of evidence. Bioinformatics 29, 402–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom G. T., Kirsch K. R., Casillas G. A., Camargo K., Wade T. L., Knap A. H., Baker E. S., Horney J. A. (2020). Spatial distribution of polycyclic aromatic hydrocarbons contaminants after Hurricane Harvey in a Houston neighborhood. Submitted. [DOI] [PMC free article] [PubMed]
- Sansom G. T., Kirsch K. R., Stone K. W., McDonald T. J., Horney J. A. (2018). Domestic exposures to polycyclic aromatic hydrocarbons in a Houston, Texas. Environ. Justice Neighborhood. Environ Justice 11, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sille F. C. M., Karakitsios S., Kleensang A., Koehler K., Maertens A., Miller G. W., Prasse C., Quiros-Alcala L., Ramachandran G., Rappaport S. M., et al. (2020). The exposome - A new approach for risk assessment. Altex 37, 3–23. [DOI] [PubMed] [Google Scholar]
- Sirenko O., Grimm F. A., Ryan K. R., Iwata Y., Chiu W. A., Parham F., Wignall J. A., Anson B., Cromwell E. F., Behl M., et al. (2017). In vitro cardiotoxicity assessment of environmental chemicals using an organotypic human induced pluripotent stem cell-derived model. Toxicol. Appl. Pharmacol. 322, 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O., Hesley J., Rusyn I., Cromwell E. F. (2014. a). High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev. Technol. 12, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirenko O., Hesley J., Rusyn I., Cromwell E. F. (2014. b). High-content high-throughput assays for characterizing the viability and morphology of human iPSC-derived neuronal cultures. Assay Drug Dev. Technol. 12, 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle S., Schulz R. (2015). Agricultural insecticides threaten surface waters at the global scale. Proc. Natl. Acad. Sci. USA 112, 5750–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone K. W., Casillas G. A., Karaye I., Camargo K., McDonald T. J., Horney J. A. (2019). Using spatial analysis to examine potential sources of polycyclic aromatic hydrocarbons in an environmental justice community after Hurricane Harvey. Environ. Justice 12, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. (2011). 40 CFR 300.430 - Remedial Investigation/Feasibility Study and Selection of Remedy. Washington, DC.
- U.S. EPA. (1986). Guidelines for the health risk assessment of chemical mixtures. Fed. Reg. 51, 34014–34025. [Google Scholar]
- U.S. EPA. (1993). Provisional Guidance for quantitative risk assessment of polycyclic aromatic hydrocarbons (PAH). U.S. Environmental Protection Agency, Office of Research and Development, Office of Health and Environmental Assessment, Washington, DC.
- U.S. EPA. (2010). Development of a relative potency factor (Rpf) approach for polycyclic aromatic hydrocarbon (PAH) mixtures (external review draft). U.S. Environmental Protection Agency, Washington, DC.
- U.S. EPA. (2012). Benchmark Dose Technical Guidance.
- U.S. EPA. (2020). Regional Screening Levels RSLS Generic Tables. Available at: https://www.epa.gov/risk/regional-screening-levels-rsls-generic-tables. Accessed July 1, 2020.
- Xiao R., Wang Z., Wang C., Yu G. (2006). Soil screening for identifying ecological risk stressors using a battery of in vitro cell bioassays. Chemosphere 64, 71–78. [DOI] [PubMed] [Google Scholar]
- Zhou Y. H., Mayhew G., Sun Z., Xu X., Zou F., Wright F. A. (2013). Space time clustering and the permutation moments of quadratic form. Stat 2, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.