Abstract
Background
This paper examines the expression, function, and molecular mechanism of long non-coding ribonucleic acid (lncRNA) ARAP1 antisense RNA 1 (ARAP1-AS1) in lung cancer. Specifically, it aims to clarify the molecular mechanism of lncRNA ARAP1-AS1 that affects the occurrence and development of lung cancer, and provide a theoretical basis and molecular targets for targeted therapy or early diagnosis of lung cancer.
Methods
Fluorescence quantitative detection of lncRNA ARAP1-AS1 expression in lung cancer tissues and cell lines, and methylthiazolyldiphenyl-tetrazolium (MTT), plate cloning experiment, and flow cytometry were used to detect the effect of knockdown of lncRNA ARAP1-AS1 on cell proliferation, clone formation, and the cell cycle, respectively. Western blotting was used to detect the expression of cell cycle-related proteins as well as the effect of knockdown of lncRNA ARAP1-AS1 on lung cancer. Cell proliferation was assessed by a nude mouse subcutaneous tumor formation experiment.
Results
LncRNA ARAP1-AS1 is highly expressed in lung cancer tissues and cells. Knockdown of LncRNA ARAP1-AS1 can significantly inhibit the proliferation and clonal formation of lung cancer cells and induce G0/G1 cell cycle arrest. Knockdown of ARAP1-AS1 can markedly inhibit the expression of cell cycle-related protein cyclin D1, but has no significant effect on the expression of cyclin-dependent kinase (CDK)4 and CDK6. Furthermore, knockdown of ARAP1-AS1 can also notably inhibit the growth of lung cancer cells and substantially reduce the expression of Ki-67 in tumor-bearing tissues in nude mice.
Conclusions
LncRNA ARAP1-AS1 is highly expressed in lung cancer. Knocking down of this gene can significantly inhibit cell proliferation in vitro and in vivo, and can also cause G0/G1 cell cycle arrest by inhibiting the expression of cyclin D1.
Keywords: Lung cancer, long non-coding ribonucleic acid ARAP1 antisense RNA 1 (lncRNA ARAP1-AS1), cell proliferation, cycle arrest, nude mice
Introduction
Presently, lung cancer has one of the highest malignancy-related morbidity and mortality rates worldwide, and exhibits an increasing trend every year (1). Owing to the substantial body of basic research in recent years, early diagnosis of lung cancer, minimally invasive surgery, and targeted therapy have been greatly improved; yet, the lung cancer death toll remains high (2,3). This is primarily due to the fact that there are no significant clinical symptoms in early-stage lung cancer (4,5). The majority of patients (approximately 75%) are already in advanced stages at initial diagnosis, and some even present with distant metastases. Studies have shown that deletion, mutation, and amplification of related genes can significantly promote the occurrence and development of lung cancer. For example, the deletion of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene, which encodes the auxin response factor (ARF) and p16 proteins to regulate the cell cycle, which encodes the ARF and p16 proteins to regulate the cell cycle as well as the activity of p53 and CDK4/6/pRB, respectively, and block the cell cycle, thereby inhibiting the occurrence and proliferation of tumors (6,7). In squamous cell and small cell carcinomas, some genes in the chromosomes, such as sex determining region Y-box2 (SOX2) and NK2 homeobox 1 gene (NKX2-1), also undergo copy number amplification (5). Mutations in oncogenes, such as Kirsten rat sarcoma 2 viral oncogene homolog (KRAS) and epidermal growth factor receptor (EGFR), will promote molecular activation as well as the excessive and disordered proliferation of tumor cells (8). The discovery of these mutations and the elucidation of lung cancer function have promoted the rapid development of targeted therapies, such as small molecule inhibitors targeting EGFR, which have significantly improved the survival of lung cancer patients (9,10). Therefore, exploring the key molecular targets that affect the regulation of lung cancer will further improve the understanding of the pathogenesis of lung cancer. At the same time, by exploring related molecules, powerful and effective diagnostic markers may be discovered for clinical preventive treatment.
In recent years, numerous studies have reported that some long-ignored ribonucleic acids (RNAs), such as non-coding RNAs, which do not possess the ability to encode. At present, it is established that approximately 93% of the sequences in the human genome can be transcribed into RNA, however only 2% of RNA can be translated into protein, with the remaining 98% being long non-coding RNAs (lncRNA) (11,12). These are RNA molecules that are transcribed from the cell genome with a length greater than 200 nucleotides (nt). They are involved in the regulation of the cell cycle as well as cell proliferation, differentiation, and apoptosis, and other important physiological processes (13,14). Recent studies have shown that there are also a large number of abnormally expressed lncRNAs in lung cancer tissues. These lncRNAs can either promote or inhibit the occurrence and development of lung cancer and are expected to become biomarkers for the early diagnosis of lung cancer (15,16).
Furthermore, studies have shown that the non-coding RNA, ARAP1 antisense RNA 1 (ARAP1-AS1), can markedly affect the occurrence and development of other tumors, including bladder, colorectal, and breast cancers (17-19). However, the expression level, biological function, and molecular function of lncRNA ARAP1-AS1 in lung cancer tissues are still unclear. This study aims to clarify the function and mechanism of this gene in lung cancer tissues, in vitro cell models, and in vivo nude mouse tumor formation experiments. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-3378).
Methods
Fifteen lung cancer patients who were treated and had their tumors surgically removed in our hospital from February 2018 to July 2019 were included in this study. The tumor tissue and matched normal tissue adjacent to the tumor (>5 cm from the edge of the cancer bed tissue) were taken from the patients during surgical resection. The patients did not undergo any treatment prior to surgery. All samples were taken and rinsed with phosphate-buffered saline (PBS) buffer immediately, and subsequently the samples were stored in liquid nitrogen for subsequent RNA extraction. All enrolled patients signed an informed consent form. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Soochow University (No. 20180945) and informed consent was taken from all the patients.
Human lung cancer cell lines (PC-9, A549, H226, H1975, and H1299 cells) and a normal human lung epithelial cell (BEAS-2B) were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The Cell Counting Kit 8 (CCK-8), crystal violet staining solution, and cell cycle detection kit were purchased from Beijing Biyuntian Biotechnology Co. Ltd (Beijing, China). The total RNA extraction kit, SYBR Green reagent, reverse transcription polymerase chain reaction (RT-PCR kit) (Takara, Japan), primary antibody rabbit anti-β-actin, cyclin D1, CDK4, CDK6, Ki-67, horseradish peroxidase-labeled secondary antibodies were purchased from Cell Signaling Technology (Shanghai, China). The Roswell Park Memorial Institute (RPMI)-1640 medium, Dulbecco's Modified Eagle Medium (DMEM) medium, fetal bovine serum, and double antibodies were purchased from Gibco (Shanghai, China).
RT-PCR method to detect the expression of lncRNA ARAP1-AS1
The tissue sample cells were collected, and the total RNA extraction kit was used to extract the RNA, strictly according to the manufacturer’s instructions. The reverse transcription kit was used to obtain complementary deoxyribonucleic acid (cDNA). SYBR Green dye method was used for the RT-PCR reaction. The ΔΔCt method was used to calculate the relative expression of lncRNA ARAP1-AS1; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene, and the lncRNA ARAP1-AS1 and GAPDH primer sequences were used as references (20).
Vector construction and transfection
Lung cancer cells with good growth status were inoculated in a 6-well plate. After the cells were cultured overnight, the fusion reached 80%. SuperFectin short hairpin RNA (shRNA) transfection reagent was used to combine shARAP1-AS1 (shRNA-1 and shRNA-2) with negative control shRNA (shNC) transfection cells, and the primer sequence was interfered with reference (19). After 48 hours, quantitative RT-PCR (qRT-PCR) and Western blotting were performed to detect transfection efficiency.
CCK-8 detects cell viability
Lung cancer cells in the logarithmic growth phase were collected after transfection. DMEM medium containing 10% fetal bovine serum was used to dilute the cells to a cell suspension of 5×104/mL. The cell suspension was then inoculated on a 96-well culture plate, and subsequently incubated in a cell culture incubator at 37 °C and 5% carbon dioxide (CO2). Each group had five parallel wells. After culturing for 24, 48, and 72 h, 10 µL of CCK-8 reagent was added. After culturing for a further 3 h in an incubator at 37 °C, a full-wavelength microplate reader was used to detect the optical density (OD) value of each well at 490 nm. The experiment was repeated three times, with the average of the absorbance values of five replicate wells taken, and the cell growth curve was drawn.
Clone formation experiment
Cells in the logarithmic growth phase were collected after transfection and digested into a single cell suspension with 0.25% trypsin and a pipette. They were then counted with a cell counting plate, inoculated into a 6-well plate (density: 600 cells/well), and gently rotated to disperse the cells evenly. The cells were then cultured in a CO2 incubator at 37 °C and 5% CO2. After 2 weeks, when visible colonies were formed, the culture medium was discarded and the cells were carefully immersed in PBS once. Subsequently, 1% paraformaldehyde was applied for 10 minutes to fix the cells. The fixative was then discarded, and the cells were washed with PBS buffer again. Crystal violet solution was then added for staining, and rinsed slowly under running water three times after staining. Pictures of the cells were then taken, and cell clusters of ≥50 cells were used as the number of clones to analyze the clone formation rate.
Flow cytometry to detect cell cycle
Lung cancer cells were collected 48 h after transfection and digested into a single cell suspension with 0.25% trypsin and a pipette. The cell pellet was collected by centrifugation, washed carefully with PBS once, and then pre-chilled 75% ethanol was used to fix for 12 h. After centrifugation, the cell pellet was washed with PBS buffer once, 50 µL of staining solution was added [ribonuclease (RNase) and propidium iodide (PI)], incubated in the dark for 30 minutes, and then flow cytometry was used to detect the distribution of the cell cycle.
Western blot detection
The lung cancer cells were collected after transfection, lysed on ice, and the protein concentration was determined by the Bicinchoninic acid disodium (BCA) protein quantification method. A sample amount of 50 µg was added to each well, and a 12% polyacrylamide gel was used for protein electrophoresis. The electrophoresis voltage was set to 90 and 120 V constant pressure. Following this, wet transfer to the PVDF membrane was performed; the membrane was diluted in PBS, and 5% skimmed milk powder was sealed at room temperature for 2 h. Incubate with primary antibodies β-actin (1:1,000), cyclin D1 (1:800), CDK4 (1:1,000), and CDK6 (1:800) at 4 °C overnight, and then 1:2,000 peroxidase-labeled was added. The secondary antibody was incubated for 2 h at room temperature, and after color development, it was placed in a gel imager (Beijing Hongtao Foundation Technology Development Co. LTD) to take pictures and perform image analysis.
Subcutaneous tumor formation experiment in nude mice
Female BALB/c nu/nu thymus-deficient nude mice were purchased from Shanghai Shrek Experimental Animal Center (Shanghai, China) (n=8, 4–6 weeks old, weighing approximately 18–20 g). They were kept in separate cages in SPF animal experiment centers, with four nude mice per group. The culture conditions in the animal room were as follows: alternating light (12 h) and dark (12 h); constant humidity and temperature; and follow-up experiments performed after observation and breeding for 1 week.
Animal experiments were performed under a project license (No. 201901293) granted by the ethics committee of the First Affiliated Hospital of Soochow University, in compliance with national or institutional guidelines for the care and use of animals.
Transfected cells with good growth status were collected. A single cell suspension was then made, and the cell density was adjusted to 1×107 cells/mL. Iodophor was used to disinfect the naked back, and then a disposable sterile syringe was used to draw 100 µL of cell suspension and inject it into the back of the nude mouse. The mice were reared under normal conditions for 30 days, and the tumor formation, feeding, drinking, and activity status were observed daily. At the same time, the nude mice were weighed and measured. At the end of the experiment, the mice were killed by cervical dislocation, and the transplanted tumor was stripped and fixed.
Statistical analysis
SPSS20.0 statistical software was used for data analysis. The data were expressed as mean ± standard deviation (SD). The t test or analysis of variance (ANOVA) were used for comparison between the groups, and P<0.05 was considered statistically significant.
Results
LncRNA ARAP1-AS1 is highly expressed in lung cancer tissues and cell lines
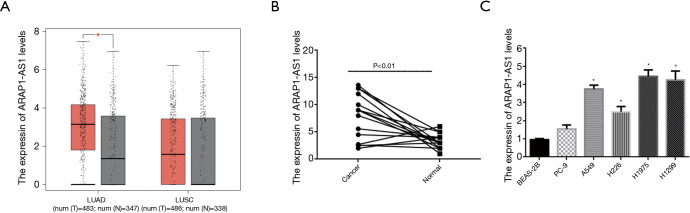
First, our analysis of lung cancer-related data in The Cancer Genome Atlas (TCGA) database found adenocarcinoma (483 cases and 347 cases adjacent to cancer) and squamous cell carcinoma (486 cases and 338 cases adjacent to cancer). The analysis results showed the following: LncRNA ArAP1-AS1 is highly expressed in adenocarcinoma cells, but has no significant change in squamous cell carcinoma (Figure 1A). Furthermore, qRT-PCR was used to detect the expression of lncRNA ARAP1-AS1 in cancerous and precancerous tissues. The results of fluorescence quantitative detection showed that the expression level of ARAP1-AS1 in cancer tissues was significantly higher compared with precancerous tissue (P<0.01) (Figure 1B).
Figure 1.
LncRNA ARAP1-AS1 is highly expressed in lung cancer tissues and cell lines. (A) TCGA database lncRNA ARAP1-AS1 transcription analysis, adenocarcinoma (LUAD): 483 cases of cancerous tissue and 347 cases of adjacent cancer; squamous cell carcinoma (LUSC): 486 cases of cancerous tissues and 338 cases adjacent to cancer. (B) Fluorescence quantitative PCR detection of lncRNA ARAP1-AS1 expression levels in 15 cases of lung cancer tissues and corresponding adjacent tissues. (C) Fluorescence quantitative PCR detection of lncRNA ARAP1-AS1 expression in different cell lines. *, P<0.05.
Therefore, in order to further study the specific biological function of lncRNA ARAP1-AS1 in lung cancer, we planned to use lung cancer cell lines. QRT-PCR was used to detect lncRNA ARAP1-AS1 in five lung cancer cell lines (PC-9, A549, H226, H1975, H1299) and a normal human lung epithelial cell (BEAS-2B). The results showed that the expression levels of lncRNA ARAP1-AS1 in various lung cancer cell lines differed substantially, with the highest expression levels in H1975 and H1299 cells, followed by A549 cells, H266 cells, and PC-9 cells. BEAS-2B cells exhibited the lowest expression (Figure 1C). Thus, we selected H1975 cells and H1299 cells with high expression of lncRNA ARAP1-AS1 for subsequent cell function research.
The effect of knocking down lncRNA ARAP1-AS1 on the proliferation of lung cancer cells
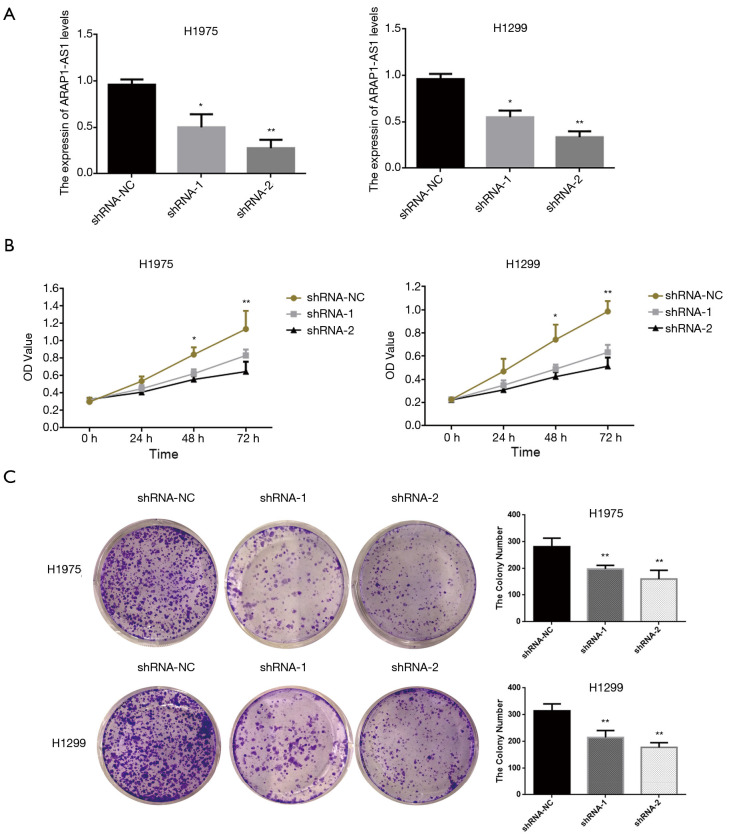
To further explore the specific biological functions of lncRNA ARAP1-AS1 in lung cancer cells, we constructed two lncRNA ARAP1-AS1 lentiviral interference vectors (shRNA-1 and shRNA-2) and a shRNA-control (shRNA-NC). H1975 and H1299 cells were stained. The results of qRT-PCR showed that the transfection of shRNA-1 and shRNA-2 interfering viruses that knock down lncRNA ARAP1-AS1 can significantly reduce the expression level of lncRNA ARAP1-AS1 in H1975 and H1299 cells compared with shRNA-NC (Figure 2A).
Figure 2.
The effect of lncRNA ARAP1-AS1 knockdown on the proliferation of lung cancer cells. (A) qRT-PCR detected the knockdown efficiency in lung cancer cells after transfection of shRNA-1, shRNA-2, and shRNA-NC; (B) CCK-8 detected the knockdown of lncRNA, and the effect of effect of ARAP1-AS1 on the proliferation ability of H1975 and H1299 lung cancer cells; (C) the clone formation experiment detected the clone formation ability of H1975 and H1299 lung cancer cells after the knocking down of lncRNA ARAP1-AS1 gene. Colonies were stained for 5 min with 0.5% crystal violet, then colonies were photographed with NIKON camera. *, P<0.05; **, P<0.01.
In order to further examine the effect of lncRNA ARAP1-AS1 knockdown on the proliferation of lung cancer cells (H1975 and H1299), CCK-8 was used to test cell viability. The results showed that shRNA-1 and shRNA-2 had a positive effect in 24 h compared with shRNA-group cells, and the proliferation of H1975 and H1299 cells was not significantly affected. As the culture time was extended to 48 h, shRNA-1 and shRNA-2 began to significantly inhibit the proliferation ability of the cells, which was particularly significant at 72 h (Figure 2B). The results of the clone formation experiment showed that transfection of lncRNA ARAP1-AS1 shRNA-1 and shRNA-2 could significantly inhibit the number of H1975 cells and H1299 clone formation compared with the shRNA-NC group (Figure 2C). This indicates that the knocking down of lncRNA ARAP1-AS1 can significantly inhibit the cloning ability of H1975 and H1299 lung cancer cells.
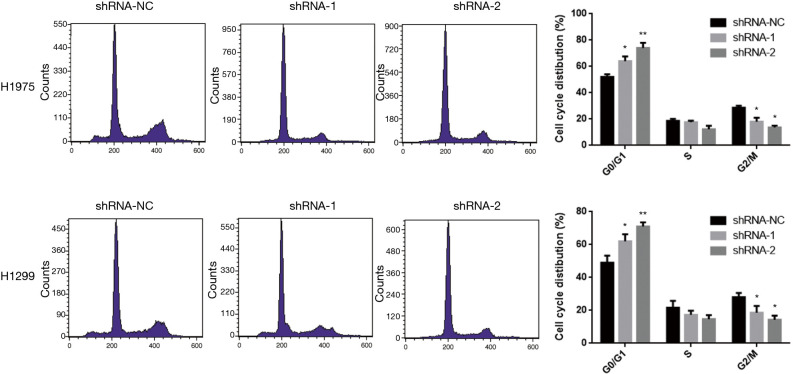
Knockdown of lncRNA ARAP1-AS1 can promote lung cancer G0/G1 cell cycle arrest
The clone formation experiment demonstrated that knocking down of lncRNA ARAP1-AS1 can inhibit the proliferation of lung cancer cells on the cloning plate. To further test whether knocking down of lncRNA ARAP1-AS1 affects the cell cycle, flow cytometry was used to detect the cell cycle. The analysis results show that, compared with the shRNA-NC group, transfection of lncRNA ARAP1-AS1 shRNA-1 and shRNA-2 can significantly promote H1975 and H1299 cells to arrest in the G0/G1 phase. The corresponding G2/M phase ratio was significant, while the ratio of S-phase cells decreased. This indicates that knocking down of lncRNA ARAP1-AS1 can significantly inhibit H1975 and H1299 lung cancer cell cycle arrest in the G0/G1 phase (Figure 3).
Figure 3.
Flow cytometry detection of the effects of knockdown of lncRNA ARAP1-AS1 on H1975 and H1299 lung cancer cell cycles. *, P<0.05; **, P<0.01.
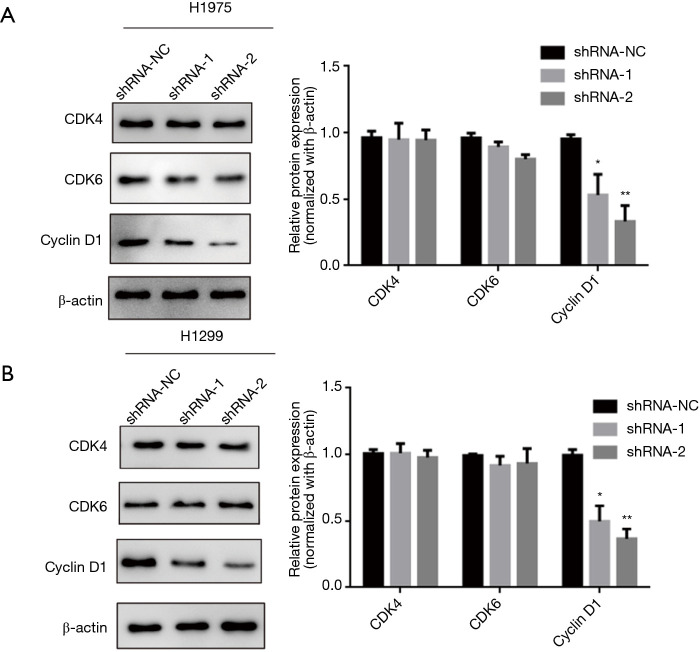
Knockdown of lncRNA ARAP1-AS1 can significantly inhibit the expression of cycle-related proteins
Based on the above results, it is clear that knocking down of lncRNA ARAP1-AS1 can significantly inhibit lung cancer cell cycle arrest in the G0/G1 phase. Therefore, western blotting was used to detect the expression of cycle-related proteins and the expression of related regulatory proteins in the G0/G1 phase. The results show that in H1975 cells, knocking down of lncRNA ARAP1-AS1 can significantly inhibit the expression of the cell cycle-related protein, cyclin D1, compared with the blank control group. Meanwhile, it has no significant effect on the expression of other proteins, such as CDK4, and the expression of CDK6 exhibited only a slight decrease, however the difference was not statistically significant (Figure 4).
Figure 4.
Western blot detection of the effect of knockdown of lncRNA ARAP1-AS1 on lung cancer H1299 cell cycle-related proteins. *, P<0.05; **, P<0.01.
The effect of knocking down of lncRNA ARAP1-AS1 on the growth of lung cancer cells in nude mice
The above experiment demonstrated at the cellular level that knockdown of lncRNA ARAP1-AS1 can inhibit the proliferation of lung cancer cells. In order to further examine the effect of knockdown of lncRNA ARAP1-AS1 on the growth of lung cancer cells in nude mice, we used a stable knockdown lentiviral vector (shRNA-2) to interfere with H1975 cells. After adding screening drugs, we obtained a stable knockdown cell line and inoculated it under the skin of male BALB/c nu/nu thymus-deficient nude mice (each group n=4). Each nude mouse was inoculated with viable cells: 1×106 cells; four nude mice in each group; measured with vernier calipers every 3 days. The tumor-bearing mice were weighed, and the long neck and short neck of the transplanted tumor were measured. The volume of the transplanted tumor mass was calculated according to the following formula: V=(a × b × b)/2; where V is the volume of the transplanted tumor mass, a is the long diameter, and b is the short diameter (Figure 5A). Knockdown of lncRNA ARAP1-AS1 can significantly inhibit the growth of H1975 cells in nude mice.
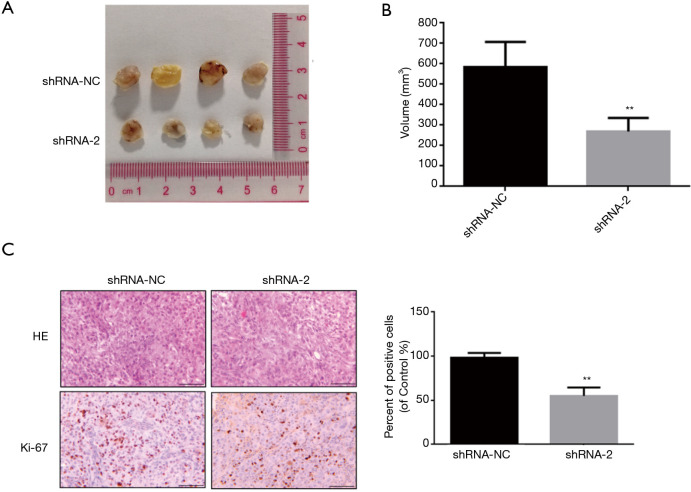
Figure 5.
The effect of knocking down of lncRNA ARAP1-AS1 on the growth of lung cancer cells (H1975) in nude mice. (A) Observations and measurements of the tumor-bearing tissue in nude mice; (B) analysis of the volume of tumor-bearing tissue in nude mice 30 days after inoculation; (C) HE stain detection of the expression of the proliferation molecule Ki-67 in tumor-bearing tumor tissues. Scale =100 µm; **, P<0.01.
After the nude mice were sacrificed, the tumor-bearing tissues were separated for measurement and photographs. The calculations based on the above formula showed that the tumor volume formed in vivo by H1975 cells in the blank control group (shRNA-NC) was 598.5±123.4 mm3, and after knockdown of LncRNA ARAP1, the volume of H1975 cells in the AS1 group was 269.1±113 mm3 (Figure 5B). Following statistical analysis and comparison of the data, it was found that the volume of H1975 cells in the lncRNA ARAP1-AS1 knockdown group was significantly lower than that of the blank control group, and the difference between the two was statistically significant (P<0.01).
Also, the immunohistochemistry results demonstrated that Ki-67 staining was darker in the nucleus of shRNA-NC group, while nucleus staining was lighter in the shRNA-2 group, indicating reduced expression of Ki-67 and slower cell proliferation. After statistical analysis of the positive cells, it was observed that the Ki-67 positive rate of tumor tissues in the shRNA-2 group was significantly lower than the shRNA-NC group (Figure 5C). This signifies that knocking down of lncRNA ARAP1-AS1 can significantly inhibit the proliferation of lung cancer cell in nude mice.
Discussion
Studies have shown that there are a large number of abnormally expressed lncRNAs in lung cancer tissues. These lncRNAs can either promote or inhibit the occurrence and development of lung cancer, and are expected to become biomarkers for the early diagnosis or treatment of lung cancer (15,16,21). Some genes can also perform physiological functions by regulating lncRNA. For example, Caveolin 1 (CAV1) can regulate the proliferation, migration, and invasion of lung cancer cells by regulating the expression of HOX transcript antisense RNA (HOTAIR), and the down-regulation of HOTAIR expression partially reverses the increase in cell survival and invasion ability induced by CAV1 (22). LncRNA not only participates in the regulation of tumorigenesis, but also regulates drug resistance-related processes. Silencing lncRNAs can effectively promote the cytotoxicity induced by gefitinib. H19 encapsulated in exosomes can be transferred to non-resistant cells, thereby inducing gefitinib resistance (23). In the present study, it was observed that lncRNA ARAP1-AS1 is highly expressed in lung cancer tissues and cell lines. Knockdown of lncRNA ARAP1-AS1 in high-expressing cell lines can significantly inhibit lung cancer cell proliferation and clone formation. Further research found that knocking down of lncRNA ARAP1-AS1 can also cause G0/G1 cell cycle arrest by inhibiting the expression of cyclin D1.
As is known, lncRNA does not possess the ability to transcribe and translate into functional proteins in cells, but it can regulate gene expression in a variety of ways (24). In recent years, lncRNA ARAP1-AS1 has been newly identified as a regulatory molecule, and is currently found to be highly expressed in colon, gastric, and cervical cancers, as well as being involved in the occurrence and development of tumors (18,20,25). LncRNA ARAP1-AS1 is highly expressed in colorectal cancer tissues and cell lines. Knockdown of lncRNA ARAP1-AS1 can significantly inhibit cell migration, invasion, and epithelial-mesenchymal transition (18). Moreover, lncRNA ARAP1-AS1 is also highly expressed in breast invasive carcinoma tissues, and the expression of lncRNA ARAP1-AS1 is significantly up-regulated in breast cancer cell lines. The down-regulation of lncRNA ARAP1-AS1 expression leads to inhibition of breast cancer cell proliferation, enhanced apoptosis, and blocked migration (19). In the present study, it was found that lncRNA ARAP1-AS1 was significantly up-regulated in lung cancer tissues and cells, and knocking down of lncRNA ARAP1-AS1 can significantly inhibit cell proliferation and clonal formation. Upon further study, it was observed that knocking down of lncRNA ARAP1-AS1 can also significantly promote G0/G1 cell cycle arrest.
Cell cycle arrest is a leading cause of slow cell growth, and the cell cycle is regulated by a variety of cycle control proteins (26). In order to assess the effect of knockdown of lncRNA ARAP1-AS1 on related cyclins, Western blotting was performed. The results showed that knockdown of lncRNA ARAP1-AS1 can significantly inhibit the expression of cyclin D1, but has no significant effect on the expression of CDK4 or CDK6. However, the mechanism through which lncRNA ARAP1-AS1 regulates the expression of cyclin D1 requires further research and exploration. Previous studies have shown that lncRNA ARAP1-AS1 is enriched in the cytoplasm of bladder cancer cells, suggesting that lncRNA ARAP1-AS1 may be used as a competing endogenous RNA (ceRNA) to regulate gene expression and biological processes. Further studies have reported that lncRNA ARAP1-AS1 acts as ceRNA to regulate microRNA (miR)-4735-3p/TCH2 (17). In breast cancer cells, lncRNA ARAP1-AS1 can sponge miR-2110 through the ceRNA mechanism to promote the expression of histone deacetylase 2 (HDAC2) in breast cancer (19). Another study has shown that knocking down of lncRNA ARAP1-AS1 inhibits the growth and spread of cervical cancer cells. Moreover, overexpression of lncRNA ARAP1-AS1 promotes the growth and spread of cervical cancer cells. Studies have shown lncRNA ARAP1-AS1 can directly interact with PSF (PTB associated splicing factor) to release Polypyrimidine tract-binding protein (PTB) and accelerate the internal ribosome entry site (IRES)-driven proto-oncogene c-Myc, thereby facilitating cervical cancer development and progression. In addition, c-Myc can transcriptionally activate lncRNA ARAP1-AS1 by directly binding to the E-box motif located on the promoter of lncRNA ARAP1-AS1 (20). Therefore, we infer that lncRNA ARAP1-AS1 may regulate the expression of downstream target genes through intermediate miRNA or other gene proteins in lung cancer tissues. In the next experiment, we will further reveal the specific molecular regulation mechanism of lncRNA ARAP1-AS1 in lung cancer and confirm its regulation of target genes.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: Scientific Research Fund for young and middle-aged of Wannan Medical College in 2020 (WK2020F15).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the First Affiliated Hospital of Soochow University (NO. 20180945) and informed consent was taken from all the patients. Animal experiments were performed under a project license (NO. 201901293) granted by the ethics committee of the First Affiliated Hospital of Soochow University, in compliance with national or institutional guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-3378
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-3378
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-3378). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Oberndorfer F, Müllauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol 2018;30:69-76. 10.1097/CCO.0000000000000429 [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Soudy H. Osimertinib in the treatment of leptomeningeal disease in T790M-negative, epidermal growth factor receptor-mutated non-small cell lung cancer: a case report. Chin Clin Oncol 2019;8:29. 10.21037/cco.2019.02.02 [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and Molecular Classification of Lung Cancer. Cancer Treat Res 2016;170:25-46. 10.1007/978-3-319-40389-2_2 [DOI] [PubMed] [Google Scholar]
- 4.Tu H, Wu M, Huang W, et al. Screening of potential biomarkers and their predictive value in early stage non-small cell lung cancer: a bioinformatics analysis. Transl Lung Cancer Res 2019;8:797-807. 10.21037/tlcr.2019.10.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med 2019;27:167-70. [PubMed] [Google Scholar]
- 6.Jeong EH, Lee TG, Ko YJ, et al. Anti-tumor effect of CDK inhibitors on CDKN2A-defective squamous cell lung cancer cells. Cell Oncol (Dordr) 2018;41:663-75. 10.1007/s13402-018-0404-6 [DOI] [PubMed] [Google Scholar]
- 7.Drak Alsibai K, Vacher S, Meseure D, et al. High Positive Correlations between ANRIL and p16-CDKN2A/p15-CDKN2B/p14-ARF Gene Cluster Overexpression in Multi-Tumor Types Suggest Deregulated Activation of an ANRIL-ARF Bidirectional Promoter. Noncoding RNA 2019;5:44. 10.3390/ncrna5030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YJ, Baek DS, Lee S, et al. Dual-targeting of EGFR and Neuropilin-1 attenuates resistance to EGFR-targeted antibody therapy in KRAS-mutant non-small cell lung cancer. Cancer Lett 2019;466:23-34. 10.1016/j.canlet.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 9.Zarredar H, Pashapour S, Ansarin K, et al. Combination therapy with KRAS siRNA and EGFR inhibitor AZD8931 suppresses lung cancer cell growth in vitro. J Cell Physiol 2019;234:1560-6. 10.1002/jcp.27021 [DOI] [PubMed] [Google Scholar]
- 10.Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:841-9. 10.1200/JCO.2017.74.7576 [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Zhu S, Meng N, et al. ncRNA-Encoded Peptides or Proteins and Cancer. Mol Ther 2019;27:1718-25. 10.1016/j.ymthe.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nigita G, Marceca GP, Tomasello L, et al. ncRNA Editing: Functional Characterization and Computational Resources. Methods Mol Biol 2019;1912:133-74. 10.1007/978-1-4939-8982-9_6 [DOI] [PubMed] [Google Scholar]
- 13.Peng WX, Koirala P, Mo YY. LncRNARNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thin KZ, Liu X, Feng X, et al. LncRNA-DANCR: A valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract 2018;214:801-5. 10.1016/j.prp.2018.04.003 [DOI] [PubMed] [Google Scholar]
- 15.Chen R, Li WX, Sun Y, et al. Comprehensive Analysis of LncRNARNA and mRNA Expression Profiles in Lung Cancer. Clin Lab 2017;63:313-20. 10.7754/Clin.Lab.2016.160812 [DOI] [PubMed] [Google Scholar]
- 16.Jiang N, Meng X, Mi H, et al. Circulating LncRNARNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin Chim Acta 2018;486:26-33. 10.1016/j.cca.2018.07.026 [DOI] [PubMed] [Google Scholar]
- 17.Teng J, Ai X, Jia Z, et al. Long non-coding RNA ARAP1-AS1 promotes the progression of bladder cancer by regulating miR-4735-3p/NOTCH2 axis. Cancer Biol Ther 2019;20:552-61. 10.1080/15384047.2018.1538613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Y, Gu B, Wang Y, et al. YY1-Induced Upregulation of Long Noncoding RNA ARAP1-AS1 Promotes Cell Migration and Invasion in Colorectal Cancer Through the Wnt/β-Catenin Signaling Pathway. Cancer Biother Radiopharm 2019;34:519-28. 10.1089/cbr.2018.2745 [DOI] [PubMed] [Google Scholar]
- 19.Lu C, Wang X, Zhao X, et al. Long non-coding RNA ARAP1-AS1 accelerates cell proliferation and migration in breast cancer through miR-2110/HDAC2/PLIN1 axis. Biosci Rep 2020;40:BSR20191764. 10.1042/BSR20191764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Wu D, Wang D. Long non-coding RNA ARAP1-AS1 promotes tumorigenesis and metastasis through facilitating proto-oncogene c-Myc translation via dissociating PSF/PTB dimer in cervical cancer. Cancer Med 2020;9:1855-66. 10.1002/cam4.2860 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Zhao Y, Feng C, Li Y, et al. LncRNARNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem 2019;460:1-8. 10.1007/s11010-019-03564-1 [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Yin NC, Liu H, et al. Cav-1 promote lung cancer cell proliferation and invasion through LncRNARNA HOTAIR. Gene 2018;641:335-40. 10.1016/j.gene.2017.10.070 [DOI] [PubMed] [Google Scholar]
- 23.Lei Y, Guo W, Chen B, et al. Tumor released LncRNARNA H19 promotes gefitinib resistance via packaging into exosomes in non small cell lung cancer. Oncol Rep 2018;40:3438-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jathar S, Kumar V, Srivastava J, et al. Technological Developments in LncRNARNA Biology. Adv Exp Med Biol 2017;1008:283-323. 10.1007/978-981-10-5203-3_10 [DOI] [PubMed] [Google Scholar]
- 25.Jiang YH, Zhu XY, Guo ZY, et al. Increased long non-coding RNA ARAP1-AS1 expression and its prognostic significance in human gastric cancer: a preliminary study. Eur Rev Med Pharmacol Sci 2020;24:1815-20. [DOI] [PubMed] [Google Scholar]
- 26.Dalton S. Linking the Cell Cycle to Cell Fate Decisions. Trends Cell Biol 2015;25:592-600. 10.1016/j.tcb.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as