Summary
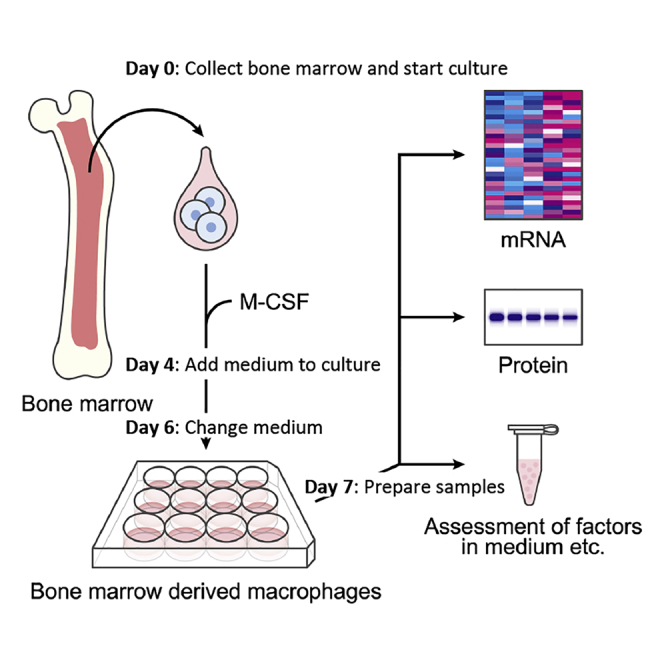
The assessment of macrophage function has been a topic of intense discussion due to multiple subtypes. This protocol describes the collection of bone marrow cells from the femur and tibia of mice, differentiation into bone marrow-derived macrophages (BMDM cells), and sampling from cultures. This protocol focuses on the efficient preparation of BMDM cells, providing a way to assess the function of macrophages.
For complete details on the use and execution of this protocol, please refer to Toda et al. (2020).
Subject areas: Cell isolation, Immunology, Model Organisms, Cell Differentiation
Graphical Abstract
Highlights
-
•
Fast and efficient collection of bone marrow from mice
-
•
Differentiation into BMDM that are >90% positive for CD11b and F4/80
-
•
Culture of BMDM for in vitro functional analysis of RNA and protein
-
•
Cells can be assessed without assuming polarization to M1/M2 subtype
The assessment of macrophage function has been a topic of intense discussion due to multiple subtypes. This protocol describes the collection of bone marrow cells from the femur and tibia of mice, differentiation into bone marrow-derived macrophages (BMDM cells), and sampling from cultures. This protocol focuses on the efficient preparation of BMDM cells, providing a way to assess the function of macrophages.
Before you begin
Prepare mice
Note: Mice should be available sufficiently before the in vitro experiment. Although we have not experienced marked differences in the viability of harvested cells among differently aged mice between 6–12 weeks of age or between male/female mice, the mice used should be of the same age and sex in the same experiment.
Mice used in this protocol can be fasted or fed ad libitum at time of collection of bone marrow cells. Cell viability should be evaluated in relation to wild type mice in mice with defects in hematopoietic cells.
Prepare reagents and equipment
Timing: 30 min
Note: 1 week before in vitro experiment. 1 h before collection of bone marrow cells.
-
1.
Add 10% FBS (heat inactivated) and 1% penicillin/streptomycin to DMEM high glucose (DMEM 10% FBS) and store at 4°C until preparation of the bone marrow culture medium.
-
2.
Keep DMEM (10% FBS) at 37°C. Add CSF-1 to obtain bone marrow culture medium after collection of bone marrow cells. Freshly prepare before harvesting bone marrow (Zhang et al., 2008).
-
3.
Prepare ice cold PBS (10 mL/ mouse) in 10 mL syringe with 23G needle, culture plate (one 12 well plate/ mouse), scissors, and forceps.
-
4.
Bring mice on site after preparation.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE Rat Anti-CD11b | BD | 553311 |
| APC anti-mouse F4/80 antibody | BioLegend | 123116 |
| 7-AAD | BD | 559925 |
| Phospho-Akt (Ser473) Rabbit mAb | Cell Signaling | 9271 |
| Phospho mTOR antibody | Cell Signaling | 2971 |
| ECL anti-rabbit IgG horseradish peroxidase lined whole antibody | GE | NA934-1ML |
| GAPDH rabbit monoclonal antibody | Cell Signaling | 2118 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant mouse IL-13 | R and D | 413-ML-005 |
| Recombinant mouse CSF-1 | Gibco | PMC2044 |
| DMEM (high glucose) | Gibco | 5796 |
| Stain buffer | BD | 554656 |
| FBS | Biowest | S1650 |
| Penicillin streptomycin | Gibco | 10378016 |
| PBS tablets | Takara | T900 |
| 0.05% Trypsin-EDTA | Gibco | 25300-054 |
| Cell lysis buffer | CST | 9803 |
| Complete protease inhibitor cocktail | Roche | 11697498001 |
| 4× Laemmli sample buffer | Bio-Rad | 1610747 |
| Experimental models: organisms/strains | ||
| C57BL6/J | Jackson Laboratory | 664 |
| Oligonucleotides | ||
| IL-10 Fw GCTCTTACTGACTGGCATGAG | N/A | N/A |
| IL-10 Rv CGCAGCTCTAGGAGCATGTG | N/A | N/A |
| TNF-α Fw CTGAACTTCGGGGTGATCGG | N/A | N/A |
| TNF-α Rv GGCTTGTCACTCGAATTTTGAGA | N/A | N/A |
| Arg1 Fw CTCCAAGCCAAAGTCCTTAGAG | N/A | N/A |
| Arg1 Rv GGAGCTGTCATTAGGGACATCA | N/A | N/A |
| Software and algorithms | ||
| Prism 7 | GraphPad | N/A |
| MS Excel | Microsoft | N/A |
| Photoshop CC | Adobe | N/A |
| Other | ||
| Cyclophillin probe | Applied Biosystems | 4316034 |
| Cell strainer (100 μm) | Falcon | 352360 |
| Lysing buffer | BD | 555899 |
| Cell scraper | IWAKI | 9000-220-CS |
| RNeasy mini kit | QIAGEN | 74106 |
| Round bottom polystyrene tubes | Falcon | 352058 |
Materials and equipment
| Bone marrow culture medium | Final concentration (mM or μM) |
|---|---|
| DMEM | N/A |
| FBS | 10% |
| Penicillin streptomycin | 1% |
| M-CSF1 | 1 ng/mL (test multiple concentrations) |
Step-by-step method details
Collection of bone marrow cells
Timing: 30 min
Note: Day 0
This step prepares the bone marrow cells including progenitors which will differentiate into bone marrow-derived macrophages after culture. The time it takes to start of culture in the bone marrow culture medium should be minimal to maximize cell yield and to avoid contamination.
-
1.
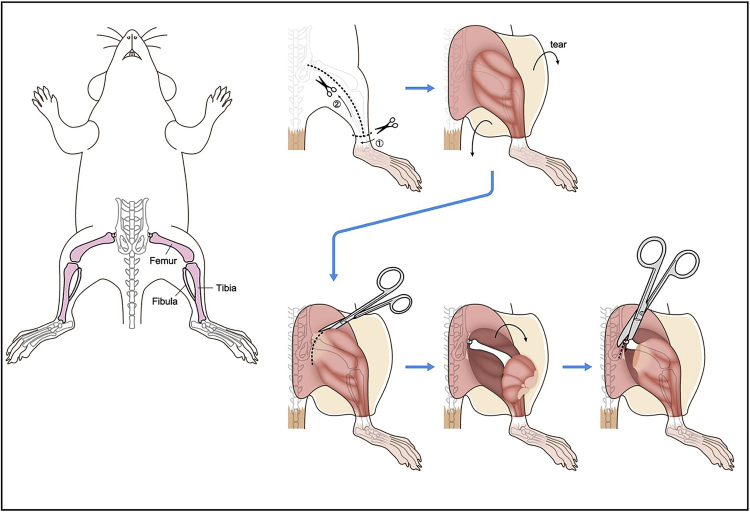
Euthanize mouse by an appropriate method approved by the supervising institution. Appropriately remove excess blood in the following steps.
-
2.
Soak with 70% ethanol. (Also see Troubleshooting problem 1)
-
3.
Remove skin and expose lower limb.
-
4.
Cut major muscles near the base of the lower limb to expose the hip joint.
-
5.
Cut femur near the base (Figure 1).
-
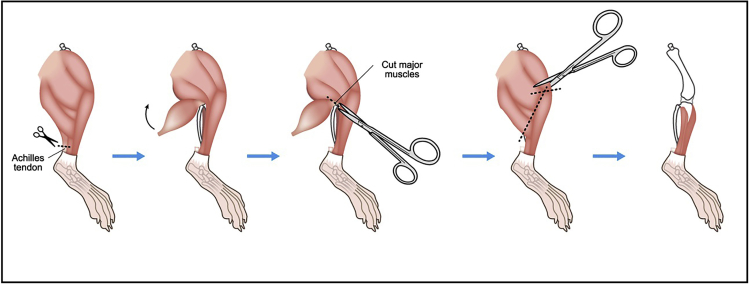
6.
Cut Achilles tendon and remove major muscles around tibia.
-
7.
Remove major muscles around femur (Figure 2).
-
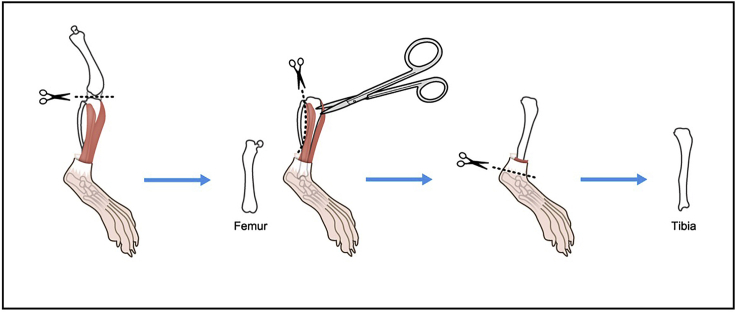
8.
Cut at knee joint.
-
9.
Remove major muscles around tibia, and remove fibula (Figure 3).
-
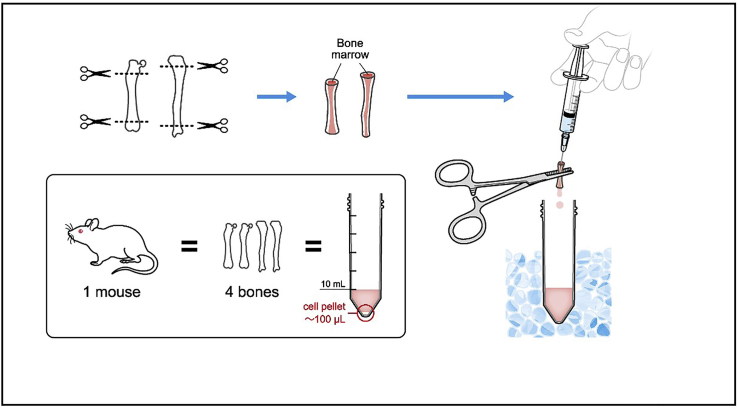
10.
Remove the epiphyses of femur and tibia so that the bone marrow can be accessed from the ends with a 23G needle. This can be confirmed by poking the ends with a needle (Also see Troubleshooting problem 2).
-
11.
Wash the bone marrow into a 50 mL tube by slowly injecting approximately 2–3 mL PBS per bone. The bone marrow will come out from the other end. Make sure the PBS does not backflow, to avoid contamination. Keep the bone marrow on ice.
-
12.
Repeat with other lower limb (Figure 4).
Optional: For collection of bone marrow from 5 mice or more, or if collection takes more than 1 h, use DMEM without FBS for washing bone marrow.
-
13.
Centrifuge bone marrow suspension at 200 × g, 5 min at 4°C.
-
14.
Decant or aspirate PBS.
Optional: To remove erythrocytes resuspend in 1× lyse buffer on ice, and let sit for 3 min. Add 10 mL DMEM (10% FBS 1% penicillin streptomycin) and centrifuge suspension at 200 × g, 5 min at 4°C, and repeat if necessary.
Figure 1.
Illustration describing steps 1–5.
Figure 2.
Illustration describing steps 6 and 7.
Figure 3.
Illustration describing steps 8 and 9.
Figure 4.
Illustration describing steps 10–12.
Differentiation into bone marrow-derived macrophages
-
15.
Resuspend bone marrow (approx. 1 × 107 live cells per mouse) in 1 mL bone marrow culture medium, and filter through a 100 μm cell strainer.
-
16.
Pipette suspension (approx. 1 × 107 live cells) into total volume of bone marrow culture medium (approx. 12 mL for 12 well plate). 6 well plates and 24 well plates can also be used.
-
17.
Add bone marrow culture medium to wells (1 mL/well for 12 well plate). Culture at 37°C 5% CO2.
-
18.
Add half this volume of bone marrow culture medium on day 4 (e.g., 500 μL/well for 12 well plate). Adherent cells should show 1–2 protrusions at low magnification.
Preparation for in vitro experiment
Timing: 10 min
Note: Day 6
This step describes the preparation for in vitro stimulation using BMDMs. FBS contains signaling peptides etc. that may obscure, for example, detection of phosphorylation in response to stimulation of interest. Therefore, a period of serum-free culture before stimulation is recommended in such experiments. Overnight culture in serum-free medium does not have a marked impact on cell viability, but baseline Akt phosphorylation is suppressed which makes detection of phosphorylation in response to in vitro stimulation clearer.
-
19.
Wash cells with sterile PBS, and change to DMEM 10% FBS, and continue culture at 37°C 5% CO2.
Optional: For detection of phosphorylation signals etc., change to DMEM without FBS at this point.
Timing: 10 min
Note: Day 7 (2 h before in vitro experiment)
-
20.
Change medium to DMEM without FBS if not already changed.
Sampling from bone marrow-derived macrophage cultures
Timing: 30 min to several hours
This step describes sampling from BMDM cultures. To minimize altered signaling events, or gene expression, sampling directly from the culture plate is recommended.
-
21.
Change medium to medium containing stimulation of interest.
-
22.
After stimulation, wash cells twice with PBS, and aspirate.
-
23.
For creating a cell suspension, add 0.05% trypsin-EDTA (approx. 100 μL/well for 12 well plate) and incubate at 37°C for 3 min. Add DMEM (10% FBS, 1 mL/well for 12 well plate), dislodge the cells using a cell scraper, and transfer to a 5 mL round bottom tube.
-
24.
Centrifuge at 200 × g, 4°C for 3 min.
-
25.
Suspend cells in 1 mL FACS buffer for counting, and subsequent analysis.
-
26.
If sampling of protein or RNA takes place later, freeze the washed culture plate with liquid nitrogen, and keep in −80°C freezer. In the following steps, add extraction buffer directly to the wells, and pipette repeatedly until no adherent cells remain. Optionally, a cell scraper may be used to dislodge the cells.
-
27.
For extracting protein samples for western blot, add cell lysis buffer supplemented with 1× protease cocktail to the wells (approx. 100 μL/well for a 12 well plate), and transfer to a 1.5 mL sampling tube.
-
28.
Sonicate for 5 min at 4°C, and vortex.
-
29.
Centrifuge for 10 min at 10,000 × g and transfer supernatant to a 1.5 mL sampling tube. Avoid aspirating or contacting the sediment (Trouble shooting problem 3). 100–200 μg of protein is typically extracted.
-
30.
Add 1/3 volume of 4× Laemmli buffer, and boil for 10 min.
-
31.
Keep at −20°C until SDS-PAGE.
-
32.
For extracting RNA using RNeasy mini kit, add 350 μL buffer RLT to the wells. Final elution should use 50–100 μL TE. 5–10 μg RNA is typically extracted. Keep at −80°C until gene expression analysis.
Expected outcomes
From three male C57BL6/J mice at 8 weeks of age weighing 24.7 ± 0.8 g, (1.6 ± 0.2) × 107 live bone marrow cells were isolated. During culture, (1.2 ± 0.1) × 105 cells were adherent at day 4, and (1.7 ± 0.1) × 105 cells were adherent at day 7. Examples of gene expresion analyses using FACS, RT-PCR, and western blot are shown (Figures 5, 6, and 7).
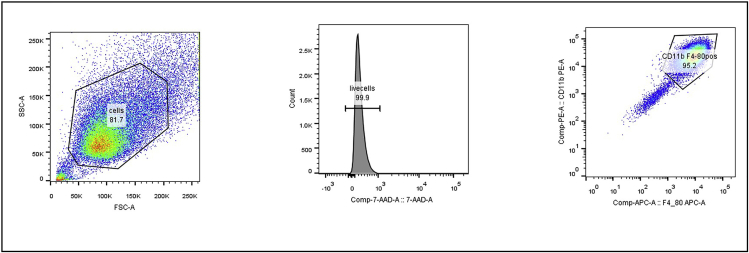
Figure 5.
FACS analysis of BMDM cell suspension
Over 90% of the cells were positive for CD11b and F4/80, which are markers for murine macrophages (Wynn et al., 2013).
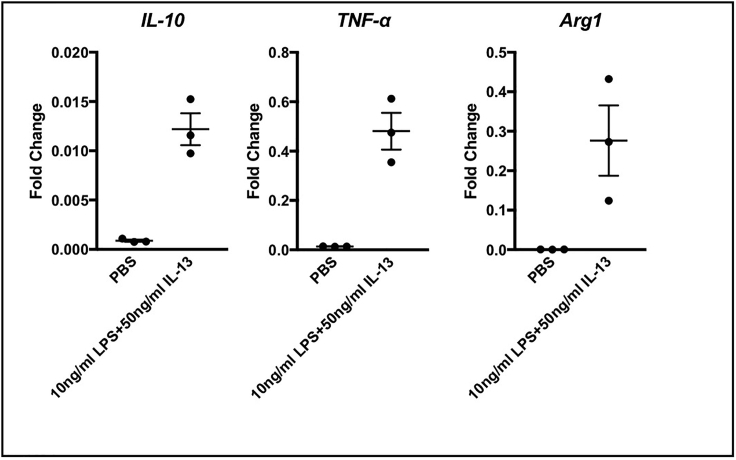
Figure 6.
Gene expression assessed by RT-PCR using RNA extracted from BMDM cultures stimulated overnight with 10 ng/mL LPS and 50 ng/mL IL-13
Genes known to be expressed by M1 and M2 macrophages could be detected (Martinez and Gordon, 2014; Wynn et al., 2013).
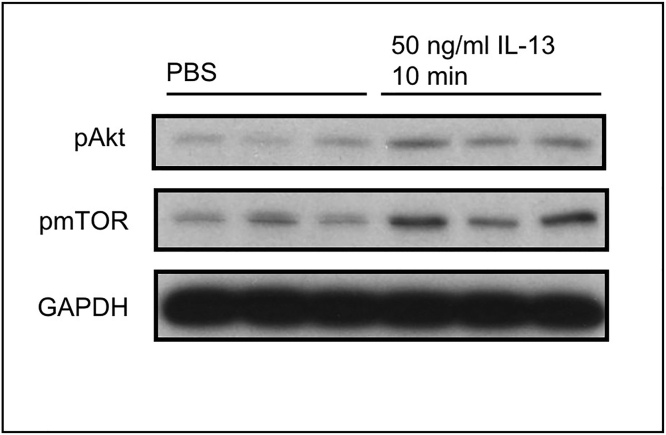
Figure 7.
Akt phosphorylation assessed by western blot of protein extracted from BMDM cultures stimulated with 50 ng/mL IL-13
Signaling in response to IL-13 has been described by others (Brown Lobbins et al., 2018; Jiang et al., 2000).
Limitations
The cultures are viable for at least 3–4 days after differentiation, but have little or no replicating potential after differentiation, and therefore the in vitro experiment should conclude without passage.
Although the BMDM cultures generated using this protocol are useful for analyzing genes known to be expressed by multiple macrophage subtypes, they cannot be used to assess different macrophage functions between tissues (Toda et al., 2020; Weischenfeldt and Porse, 2008). Additionally, this protocol only covers the assessment of BMDM cells before polarization into different macrophage subsets as described by others (Wynn et al., 2013). The cells may be further cultured with IFN-γ, IL-4 etc. to assess the function of polarized macrophage subsets although one should be aware that functions of such strongly polarized macrophages only limitedly reflect the in vivo variety of macrophages (Murray et al., 2014). As assessment of macrophage function using unpolarized BMDM cultures generated as described in this protocol provides a view of cellular function without narrowing the scope of the analysis to a certain polarization, it provides candidates of cellular functions some of which in combination with in vivo analyses may subsequently prove to be of importance in physiological or pathological conditions. The BMDM cultures described in this protocol, as in all immune cell cultures do not completely reflect the variety of immune cells in vivo, and future development and utilization of single-cell techniques may be key to further analysis of immune cell function in high spatiotemporal resolution.
Troubleshooting
Problem 1
Generated cells are sparse.
Potential solution
BMDM cultures generated should be approximately 75% confluent. If cells are sparser, the amount of collected bone marrow may be inadequate perhaps due to cutting the edges of the femur and tibia too much. The edges of the bone should be cut ∼0.5 mm at a time to minimize loss. Additionally, some mouse models may have smaller amounts of bone marrow cells than others, and may require isolation of more bone marrow to acquire the same amount of BMDM cells as wild type mice.
Problem 2
Generated cells are not viable.
Potential solution
BMDM cultures will become nonadherent when contaminated. Mice should be thoroughly soaked with 70% ethanol before exposing the bone marrow. Feces and fur are inevitably close to the site of dissection, and exposed bone must not touch them. If possible, the isolation of bone marrow may be done in sterile conditions.
Problem 3
Western blot resulted in bands with too much background.
Potential solution
As the volume of the lysate is typically <100 μL, this may occur when the supernatant of the lysate after centrifugation is inadequately aspirated, particularly when the sediment is aspirated. The steps following sonication should be done with care.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kohjiro Ueki (uekik@ri.ncgm.go.jp).
Materials availability
This protocol does not include unique materials.
Data and code availability
This protocol does not include data sets.
Acknowledgments
This work has been funded by Japan Society for the Promotion of Science grants 16K15488 (to K.U.) and 17K16141 and 19K17954 (to G.T.); Japan Agency for Medical Research and Development grant 16fk0210204h0002 (to K.U.); and grants from the Japan Diabetes Foundation, the MSD Life Science Foundation, and the Kanae Foundation for the Promotion of Medical Science (to G.T.). Hiroko Uchida holds the copyright of the graphical abstract and schemes. Please contact "me@uchidahiroko.com" for secondary use of these graphics.
Author contributions
G.T. performed the experiments and wrote the protocol. T.Y., T.K., and K.U. helped in planning the experiments.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Gotaro Toda, Email: gtoda-tky@umin.ac.jp.
Kohjiro Ueki, Email: uekik@ri.ncgm.go.jp.
References
- Brown Lobbins M.L., Shivakumar B.R., Postlethwaite A.E., Hasty K.A. Chronic exposure of interleukin-13 suppress the induction of matrix metalloproteinase-1 by tumour necrosis factor α in normal and scleroderma dermal fibroblasts through protein kinase B/Akt. Clin. Exp. Immunol. 2018;191:84–95. doi: 10.1111/cei.13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Harris M.B., Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J. Allergy Clin. Immunol. 2000;105:1063–1070. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]
- Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda G., Soeda K., Okazaki Y., Kobayashi N., Masuda Y., Arakawa N., Suwanai H., Masamoto Y., Izumida Y., Kamei N. Insulin- and lipopolysaccharide-mediated signaling in adipose tissue macrophages regulates postprandial glycemia through Akt-mTOR activation. Mol. Cell. 2020;79:43–53.e4. doi: 10.1016/j.molcel.2020.04.033. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J., Porse B. Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc. 2008;2008 doi: 10.1101/pdb.prot5080. pdb.prot5080. [DOI] [PubMed] [Google Scholar]
- Wynn T.A., Chawla A., Pollard J.W. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 2008;Chapter 14 doi: 10.1002/0471142735.im1401s83. Unit 14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol does not include data sets.