Abstract
Several mutations act as driver mutations in breast cancer, including GATA3 mutations. Reports of the relation between GATA3 mutations and breast cancer prognosis remain conflicting. Also, the role of GATA3 germline mutations is not well studied. We hypothesize that different mutation types could have different effects. Also, this study aims to assess effect of GATA3 mutations on GATA3 protein function as a transcription factor, and target pathways affected. DNA from de novo breast cancer female patients was sequenced to detect exon 6 GATA3 mutation. Sequence analysis was performed along with clinical and prognostic parameters and disease-free survival. Public datasets were analyzed for differentially expressed genes and pathways with mutant GATA3 patients. Mutations in GATA3 exon 6 were detected in 56.1% of patients (including 2 novel, Lys368fs, Pro354Lys). Intronic mutations were significantly higher in long disease-free survival group, while frameshift mutations were significantly higher in short DFS group. Patients with tumor size ≥20 had significantly higher protein coding and lower intronic mutations compared to patients with tumor size <20 mm. Differential expression and pathway analysis showed that mutant GATA3 had lost its negative regulatory effect on several pathways such as: signaling by interleukins, regulation of TP53 expression, and RUNX3 regulated CDKN1A transcription pathway. PIK3CA, SKP1, FBP1, SMAD3, ANXA9 and CLSTN2 were positively correlated to wild-type GATA3 expression, but not mutant GATA3. Intronic germline mutations of GATA3 could be related to better prognosis, while protein coding GATA3 germline mutations could be related to unfavorable prognosis. GATA3 mutations lead to dysregulation of pathways related to immunity, breast cancer development, and metabolism.
Impact statement
GATA3 mutations are known to play an important role in breast cancer progression. The exact role and mechanisms of these mutations remain controversial as some studies suggest a relation to breast tumor growth, while others suggest a relation to longer survival. GATA3 germline mutations are not well studied in breast cancer. In this study, it was hypothesized that different types of GATA3 mutations could contribute to the breast cancer progression in different ways. GATA3 exon 6, which is important for GATA3 protein functions, was reported to have hotspots, and hence it was selected for study. Intronic GATA3 germline mutations were found to be related to favorable prognosis, while protein coding mutations were found to be related to unfavorable prognosis. Bioinformatics study of large publically available datasets showed that GATA3 mutations lead to dysregulation of pathways related to T-cells activation, inflammation, and breast cancer development.
Keywords: GATA3, breast cancer, driver genes, mutation, DFS, bioinformatics
Introduction
Breast cancer (BC) is the most prevalent cancer affecting women worldwide, and it is considered the leading cause of cancer-related death in women. Response to treatment is influenced by the genetic heterogeneity of the disease. In Egypt, BC is the most prevalent cancer among females, representing 21.6% of total cancer cases.1
Although GATA-binding protein 3 (GATA3) is one of the most repeatedly mutated genes in BC,2 the role of GATA3 mutations in BC is not yet fully understood. GATA3 is a zinc-finger transcription factor and key regulator of different pathways.3 Mutations in this gene, which encodes a protein crucial for breast development, occur in 15% of estrogen receptor-positive (ER+), or luminal-type, BC.2 Also, several GATA3 germline variants have been reported to affect treatment outcomes in BC.4 GATA3 functions as a tumor suppressor by prompting epithelial, and repressing mesenchymal fates.5 Binding of GATA3 chromatin is vital for the recruitment of other co-factors such as ERα and FOXA1 in BC cells.6 It was reported that the wild type allele expression could be maintained due to the heterozygous nature of the GATA3 mutations.7
Mutations in exons 5 and 6 have some hotspots, suggesting that they might be “gain of function” mutations. Whether GATA3 mutations are oncogenic drivers remains an open question. While some studies suggest they might support tumor growth,8 other studies linked them with longer survival2 and better response to hormonal therapy. Furthermore, loss of GATA3 expression is associated with poor prognosis.9 The roles of the other BC driver mutations such as those of PIK3CA and TP53 are well studied, unlike the role of GATA3, especially germline mutations.10
We hypothesize that different GATA3 germline mutation types could have different effects. Also, this study aims to assess both the effect of these mutations on GATA3 protein function as a transcription factor and also the target pathways affected.
Materials and methods
Participants
This study is a prospective observatory study that comprises 66 de novo BC Egyptian women. Diagnosis was confirmed histopathologically. Exclusion criteria were severe clinical symptoms, or recurrent cancer. Twenty age-matched, apparently healthy, females with comparable socioeconomics were enrolled as a control group. They were free from malignant and benign tumors.
Ethical approval
This study protocol was reviewed by the Institutional Review Board (IRB) of Al-Azhar University Hospital (Damietta) and approved by the Scientific Research Ethical Committee of Faculty of Pharmacy (Girls), Al-Azhar University, abiding by the Declaration of Helsinki. A prior written informed consent was obtained from each participant.
History and clinical investigations include age at diagnosis, tumor grade, histology, metastasis, size, lymph node metastasis, estrogen, progesterone and human epidermal growth factor receptor 2 (ER, PR, HER2 respectively) receptor statuses, and type of treatment. All BC patients were treated according to standard treatment protocols and followed up for 40 months. Disease-free survival (DFS) was recorded. DFS is the time elapsed between surgery and BC recurrence or metastasis.
Immunohistochemical staining for ER, PR, and HER-2 was performed on sections of 10% formalin-fixed paraffin embedded blocks for each patient using monoclonal antibodies: ER and PR (1D5, PgR 636, respectively, DAKO). HER2 antibody (HercepTest, DAKO) was used following the manufacturer’s instructions.
DNA extraction
Peripheral blood samples (5 mL) were collected from patients and controls; genomic DNA was extracted using a Quick- gDNA MiniPrep kit (Zymo Research Corporation, Valencia, CA; Cat. No. D3024) following the manufacturer's instructions.
PCR and sequencing
GATA 3 Exon 6 primers were designed using Primer Blast as follows: Forward primer: ATTAACAGACCCCTGACTATGAAGA, Reverse primer GAGGGGTGGTGTGGTCCAA. Cycling conditions on an ABI 9700 Thermal Cycler were: initial denaturation at 95°C for 2 min, 40 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, elongation at 72°C for 30 s, and final elongation at 72°C for 7 min.
After amplification, products were purified using a QIAquick PCR Purification Kit (Qiagen) and directly sequenced by Sanger sequencing on an ABI PRISM 3730 automated sequencer (Invitrogen) using the same primer. After sequence analysis by BLAST (reference GRCh38.p13), Mutation Finder11 and SIFT12 (RRID SCR: 012813) were used for sequencing results analysis.
Bioinformatics study
Co-expression analysis of BC datasets was performed at cBioportal13,14 (RRID SCR: 014555), while PANTHER15 (RRID SCR: 004869) and reactome16 (RRID SCR: 003485) were used for pathway analysis.
Datasets analyzed were Breast Invasive Carcinoma of TCGA PanCan 2018 (825 patients)17 and METABRIC (2509 primary breast tumors with 548 matched normal).18 The data that support the findings of this study are openly available in cBioportal at [https://www.cbioportal.org/datasets].
Statistical analysis
Statistical analysis includes Student’s T test, Z-test, and Pearson’s Chi square test.
Data were represented as mean (SD), P value <0.05 was considered statistically significant. In the bioinformatics study, the P-values were corrected for multiple comparisons (represented as q value) using the Benjamini Hochberg FDR correction procedure. Minimum significant FDR was 0.05.
Results
This study was comprised of 66 confirmed de novo BC cases with mean age 48.27 ± 10.18 years. The mean DFS of enrolled patients was 27.18 ± 11.59 months, ranging from 6 to 40 months (time of study) with a mortality rate of 4.5%.
Mutation analysis
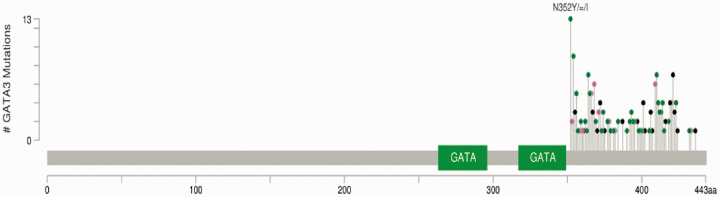
In BC patients, a total of 314 mutations in GATA3 exon 6 were detected in 56.1% of patients (37 patients) compared to none of the control group. Among the 314 mutations, there were 61 frameshift, 84 missense, 1 nonsense, 30 synonymous, and 138 intronic mutations (mostly at c.1051). The most frequent mutation in GATA3 exon 6 was a missense at c.1054 (Asn352) (Figure 1).
Figure 1.
Mutational landscape of GATA3 exon 6 in breast cancer study group. *Green lollipops represent Missense Mutations, black lollipops represent Truncating Mutations, and pink lollipops represent other types including Frameshift and Synonymous mutations. (A color version of this figure is available in the online journal.)
Frameshift mutations were: deletions (30/61), insertions (29/61), and indels (3/61). Most of them (33/61, 54%) were observed at threonine and lysine codons. Frameshift mutations were found to be significantly higher in association with short DFS (DFS ≤ 18 months, P value = 0.025), while intronic mutations were significantly higher in patients with a long DSF (DFS ≥ 24 months, P value = 0.003)
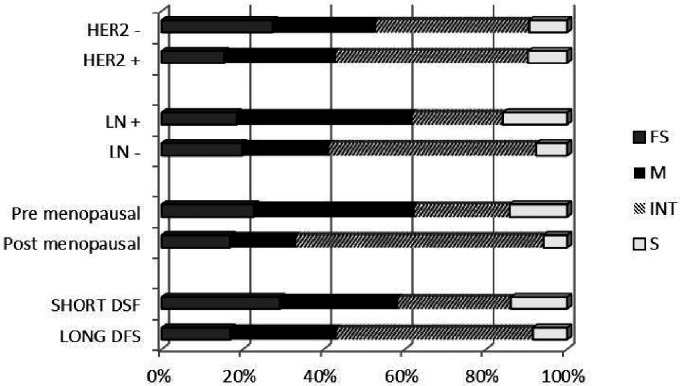
Regarding onset of diagnosis, missense and synonymous mutations were significantly higher in pre-menopause (P value = 0.0001 and 0.01 respectively), while intronic mutations were significantly higher in post menopause BC (P value = 0.00001).
Regarding lymph node (LN) involvement, missense and synonymous mutations were significantly higher in LN-positive patients, (P value = 0.0002 and 0.03 respectively), while intronic mutations are significantly lower (P value = 0.00001).
Missense and intronic mutations are significantly higher in HER2-positive cases (P value = 0.005 and 0.001 respectively) (Figure 2).
Figure 2.
Percentage of mutation types in different clinical subgroups. * FS: frameshift; M: missense; INT: intronic; S: synonymous; LN: lymph node.
Patients negative to ER had significantly higher intronic mutations compared to ER-positive patients (P value = 0.0002). No particular mutational pattern was observed in the TNBC group.
Patients with tumor size <20 mm had significantly lower protein coding mutations than patients with tumor size ≥20 mm (P value = 0.0001).
No significant correlation was observed between the mutational pattern and tumor stage or metastasis.
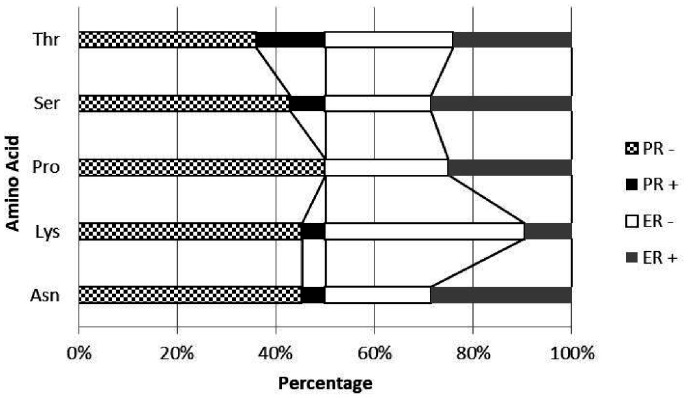
Analysis of the effect of nucleotide sequence change on the predicted amino acid sequence of GATA3 protein was performed using the Mutation Finder and SIFT prediction tool. Of the 176 protein coding mutations, there were 116 mutations at codons corresponding to asparagine (21), lysine (21), proline (28), serine (21), and threonine (25).
Mutations at asparagine codons are mostly missense at Asn352. The predicted amino acid change was mainly to tyrosine, which is predicted by SIFT to be tolerated (Seq Rep = 1).
There were several mutations at lysine codons especially at Lys368, and all were predicted by SIFT to be not tolerated (Seq Rep = 0.74). Frameshift mutations at Lys368 have not been reported previously. Mutations in lysine codons are significantly higher in ER-negative than ER-positive cases.
Mutations at proline codons were predominantly missense at Pro354 and frameshift Pro409. Pro354 is replaced by alanine, arginine, or lysine, and all were predicted by SIFT to be not tolerated (Seq Rep = 1). Pro354Lys has not been reported previously.
It was noticed that all mutations at proline codons were in PR-negative cases (Figure 3).
Figure 3.
Percentage of mutation at different amino acid codons in hormone receptors positive and negative subgroups.
Mutations at threonine codons were predominantly missense and frameshift at Thr364. Thr364 is replaced by asparagine or proline with frameshift, and all were predicted by SIFT to be not tolerated (Seq Rep = 0.98).
Mutations with lower SIFT predictive scores were ignored due to poor predictive value.
Results of bioinformatics analysis
Breast Invasive Carcinoma of TCGA PanCan 2018 (825 patients) and METABRIC (2509 primary breast tumors with 548 matched normal) were analyzed at cBioportal.
Co-expression analysis
The top 100 genes that significantly correlate with wild type and mutant GATA3 expression (either positive or negative Spearman correlation) in TCGA PanCancer 2018 and METABRIC datasets were compared. Comparison of q-values of differentially correlated gene sets showed the effect of GATA3 mutations on its expression and co-expressed genes revealing that mutant GATA3 have a co-expression profile different than that of wild-type GATA3. Besides, for some genes, the significance of correlation was lost in the mutant GATA3 subset (Table 1). These gene sets were analyzed with reactome to determine the related pathways (FDR ≤ 0.01) (Table 2).
Table 1.
q-valuea of genes negatively and positively correlated to GATA3 in patients with wild-type GATA3 and mutant GATA3.
|
Negatively correlated genes |
Positively correlated genes |
||||
|---|---|---|---|---|---|
| qa-wild-type GATA3 | qa- mutant GATA3 | qa-wild-type GATA3 | qa- mutant GATA3 | ||
| HSPA1A | 1.04E-09 | 0.185121 | SKP1 | 4.70E-177 | 0.055242 |
| CCNE1 | 7.99E-16 | 0.232416 | FBP1 | 1.16E-170 | 0.065526 |
| CCNB1b | 5.17E-06 | 0.647838 | SMAD3 | 1.13E-16 | 0.390394 |
| PTK2 | 2.71E-05 | 0.81302 | ANXA9 | 1.85E-173 | 0.494279 |
| COL6A1c | 0.08255 | 0.015705 | CLSTN2 | 3.04E-178 | 0.522665 |
| TP53 | 1.04E-09 | 0.535273 | |||
| PIK3CA | 9.00E-09 | 0.601376 | |||
| IGFBP2d | 1.61E-12 | 0.771508 | |||
aq value = P-value corrected for multiple comparisons using Benjamini Hochberg FDR correction procedure. Minimum significant FDR was 0.05.
bTurned to non-significant positive correlation.
cTurned from non-significant to significant.
dTurned to non-significant negative correlation.The bold characters shows non significant q-value.
Table 2.
Reactome pathways for genes which lost significance of correlation to GATA3 in mutant GATA3 subset.
|
Negatively correlated genes |
Positively correlated genes |
||||
|---|---|---|---|---|---|
| Pathway name | P-value | FDRa | Pathway name | P-value | FDRa |
| Regulation of TP53 Expression | 2.83e-06 | 9.21e-04 | Attenuation phase | 3.33e-15 | 4.56e-13 |
| RUNX3 regulated CDKN1ATranscription | 1.13e-05 | 0.002 | HSF1-dependent transactivation | 1.62e-14 | 1.10e-12 |
| Signaling by interleukins | 1.96e-05 | 0.002 | Regulation of HSF1-mediated heat shock response | 1.51e-12 | 6.79e-11 |
| Cellular response to heat stress | 5.21e-12 | 1.77e-10 | |||
| HSF1 activation | 1.95e-10 | 5.26e-09 | |||
| Cellular responses to stress | 2.73e-09 | 6.01e-08 | |||
| Cellular responses to externalStimuli | 3.49e-09 | 6.64e-08 | |||
| Polo-like kinase-mediated events | 1.70e-04 | 0.003 | |||
| G1/S transition | 2.39e-04 | 0.004 | |||
| Mitotic G1-G1/S phases | 3.63e-04 | 0.005 | |||
| G1/S-specific transcription | 5.89e-04 | 0.007 | |||
| Cell cycle, mitotic | 9.79e-04 | 0.011 | |||
| TP53 regulates transcription of cell cycle genes | 0.001 | 0.013 | |||
| HSP90 chaperone cycle for steroid hormone receptors (SHR) | 0.002 | 0.014 | |||
aMinimum significant FDR = 0.05.
Enrichment analysis
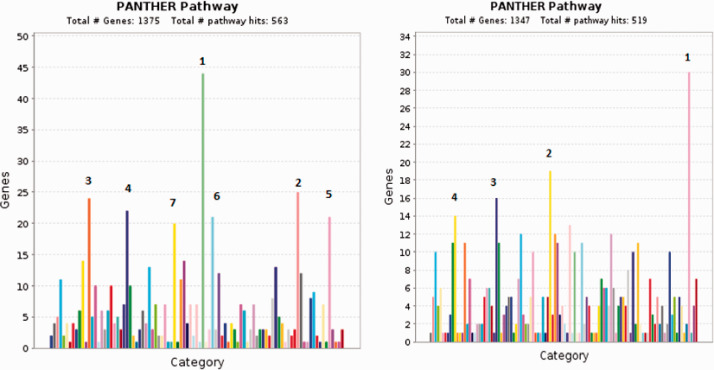
Comparison of mRNA levels in patients with mutant or wild type GATA3 in TCGA PanCan and METABRIC datasets for gene enrichment results in two gene sets of significantly over- or under- expressed genes in mutant-GATA3 patients. These two gene sets were analyzed using PANTHER pathway analysis. Under-expressed genes belong mainly to the following pathways: Inflammation mediated by chemokine and cytokine signaling pathway, T cell activation, angiogenesis, CCKR signaling map, Wnt signaling, Integrin signaling pathway, and gonadotropin-releasing hormone receptor pathway.
Over-expressed genes were found to belong to the following pathways: Wnt signaling, gonadotropin-releasing hormone receptor pathway, CCKR signaling map, and Alzheimer disease-presenilin pathway (Figure 4).
Figure 4.
Enrichment analysis for significantly differentially expressed genes in patients with GATA3 mutations compared to other BC patients. Left panel (under-expression): 1. Inflammation mediated by chemokine and cytokine signaling pathway, 2. T cell activation, 3. Angiogenesis, 4. CCKR signaling map, 5. Wnt signaling pathway, 6. Integrin signaling pathway, 7. Gonadotropin-releasing hormone receptor pathway. Right panel (over-expression): 1. Wnt signaling pathway, 2.Gonadotropin-releasing hormone receptor pathway, 3. CCKR signaling map, 4. Alzheimer disease-presenilin pathway. (A color version of this figure is available in the online journal.)
Discussion
Over two million BC new cases were recorded in 2018, accounting for about 25% cancer cases in women.19 GATA3 is expressed in the mammary glands playing an important role in mammary tissue proliferation. GATA3 protein binds and remodels inaccessible chromatin affecting cell development and differentiation. It also acts to open the chromatin at ER-responsive genes in the ducts.20 GATA3 contains two highly conserved zinc fingers with the carboxyl-terminal zinc finger responsible for DNA binding, hence important for its transcription factor functions.21 Being at the zinc finger domain necessary for DNA binding, mutations at exon 6 of GATA3 could affect the GATA3 functions.8 GATA3 germline mutations were not well studied in BC yet.
Correlation of germline mutation types with different clinical parameters were tested and could be interpreted as following:
Intronic mutations
Interestingly, more than half of the study participants harbored several GATA3 mutations, while none of the control group showed any mutations. Lack of Egyptian population germline mutation studies did not allow for comparison to the population. The 138 intronic mutations found were mostly at c.1051, which is a splicing mutation.22
Patients negative to ER had significantly higher intronic mutations compared to ER-positive patients. Previous studies reported that GATA3 levels were significantly lower in ER-negative tumors.23 This suggests that splicing mutations could decrease GATA3, affecting ER and its responsive genes.
Intronic mutations are significantly higher in HER2-positive patients. Since HER2 is related to more aggressive BC,24 the increased intronic splicing mutations in HER2-positive patients suggest that loss of GATA3 functions due to the splicing mutation could render the cells vulnerable to the oncogenic HER2.
On the other hand, intronic mutations were higher in correlation with favorable conditions such as long DFS, small tumor size, and lower in LN-positive cases. Patients positive to LN involvement are generally predicted to show poor prognosis.25 Patients with tumor size ≥20 mm had significantly higher protein coding mutations compared to patients with tumor size <20 mm.
Frameshift mutations
Frameshift mutations in the study group were mostly at threonine and lysine codons, including one novel mutation at Lys368 (predicted to be not tolerated). These mutations are predicted to impair the structure of the transcribed protein at the C-terminal. Frameshift mutations in exon 6 were reported to extend the ORF, potentially affecting protein stability.26 GATA3 mutations leading to protein extension could be providing a gain-of-function. This could explain the oncogenic character of mutant GATA3.27
Frameshift mutations were found to be significantly higher in patients with short DFS. Supportive evidence could be that more aggressive gene expression profiles were found in cells with mutations in the GATA3 protein DNA binding domain.8
Mutations at threonine codons were predominantly frameshift at Thr364. Mutations at this site are reported to cause loss of DNA binding in GATA3.28
Missense mutations
The aggressive gene expression profiles reported in patients with GATA3 exon 6 mutations in particular8 could explain the results of the current study that missense mutations and synonymous mutations were significantly higher in pre-menopause patients, while intron mutations were significantly higher in post-menopausal cases. Pre-menopausal onset of BC is related to a more aggressive form and a poor prognosis BC.29
Missense mutations are significantly higher in HER2-positive cases, which are usually associated with poor prognosis.24
Mutations in lysine codons were observed to be significantly higher in ER negative than in ER-positive cases. GATA3 is associated with ER expression and the expression level of GATA3 is strongly associated with estrogen receptor alpha (ERα).23 Hence, mutations in lysine codons could be affecting the GATA3 protein functions and expression, contributing to disease prognosis.
There were several missense mutations at Pro354 codon (including a novel one, Pro354Lys). All mutations at proline codons were in PR-negative cases. A previous study reported that mutation of one allele of the GATA3 second zinc finger leads to decreased expression of some genes, including PR.8 PR expression is a prognostic marker in BC, and lower expression of PR is associated with poor prognosis.8
Synonymous mutations
The study subjects were found to harbor several synonymous mutations. The change in nucleotide sequence could affect the transcription of the GATA3 gene through hindering the transcription factor binding, and epigenetic regulation, among others. Synonymous mutations are reported to be causal in several types of cancer.30
In this study, synonymous mutations were significantly higher in LN positive than LN-negative patients. Positive LN is usually considered a poor prognostic marker, while negative LN is controversial.31 These results suggest a correlation between GATA3 synonymous mutation and unfavorable prognosis of BC.
Previous studies had stated that GATA3 mutations largely affect its functionality. Several breast cancer cell lines such as MCF10A, which is a low-GATA3cell line and MCF7, which has a mutant GATA3 were studied. It was shown that cell lines with mutant GATA3 have a different expression profile of GATA3 target genes compared to cells with wild type GATA3.28
To inspect the role of GATA3 mutation in driving cancer, comparison of BC wild-type and mutant GATA3 subsets of TCGA PanCancer 2018 and METABRIC revealed a loss of correlation between GATA3 and some genes in the mutant GATA3 subset.
Regulation of the TP53 Expression pathway was negatively correlated with wild-type GATA3 expression, but not mutant GATA3. A simultaneous loss of wild type GATA3 expression and overexpression of TP53 in other type of cancer was previously reported.32 This could support the negative correlation between wild type GATA3 and TP53 found in the current study. Also, loss of this negative correlation in patients with mutant GATA3 suggests the mutation acts by a loss-of-function for this pathway. These data show that wild type GATA3 is required for TP53 expression and thus its tumor suppressor function. Also, the positive correlation between TP53 and wild type GATA3 expression is lost in patients with mutant GATA3. This adds evidence to the predicted loss-of-function due to GATA3 mutation.27
Likewise, signaling by interleukins pathway was negatively correlated with wild-type GATA3 expression, but not mutant GATA3. Loss of this negative correlation in mutant GATA3 cases suggests a potential mechanism of GATA3 mutation as a driver for BC through affecting immunity. It was previously reported that the production of IL-5, 9, and 13 needs GATA3. Also, IL-4 induction is sensitive to loss of GATA-3 developing Th2 cells.8
The RUNX3 regulated CDKN1A transcription pathway was negatively correlated with wild-type GATA3 expression, but not mutant GATA3. Loss of this negative correlation in mutant GATA3 cases suggests loss-of-function of the mutant protein. The expression of this gene is controlled by TP53.33 RUNX3 has important functions in the lymphocyte and myeloid lineages.34
Similarly, several heat shock pathways were negatively correlated with wild-type GATA3 expression, but not mutant GATA3. One of the key genes of this pathway, HSPA1A, correlates negatively with wild-type GATA3 expression but not mutant GATA3. Loss of the negative correlation in cases with GATA3 mutation suggests that GATA3 mutations could drive BC through loss of its effect on HSPA1A, rendering HSPA1A able to promote cancer progression.35
On the other hand, PIK3CA, SKP1, FBP1, SMAD3, ANXA9 and CLSTN2 were positively correlated with wild-type GATA3 expression, but not mutant GATA3. Loss of the negative correlation in mutant GATA3 cases suggests that GATA3 mutation could drive BC through mechanisms involving dysregulation of these genes. For example, PIK3CA (which is a known key gene in BC) was previously reported to be transcriptionally regulated by GATA3, likely contributing to its negative effect on PI3K-Akt signaling.36
Expression of FBP1, in addition to its crucial role in metabolism, negatively correlates with activity of the Wnt/β-Catenin pathway.37 Enrichment analysis of a mutant GATA3 subset revealed dysregulation of the Wnt signaling pathway. Several Wnt pathway genes are over-expressed (including proto-oncogenes like Wnt3, BCL9), and several Wnt pathway genes are under-expressed (mostly Wnt pathway modulators and inhibitors, including FBP1). These findings suggest that GATA3 mutation could contribute to BC progression through affecting the Wnt pathway. Also, ANXA9 expression was reported to be associated with aggressive BC.38
Enrichment analysis of mutant GATA3 subset transcriptome revealed under-expression of genes related to inflammation mediated by chemokine and cytokine signaling and T-cell activation pathway. GATA3 was previously reported to show a negative expression correlation with immune activities in BC including proliferation of T cells.39 GATA3 is critical for Th2 cell differentiation40 and controlling the development and function of B and thymic NK cells.41
Interestingly, enrichment analysis of mutant GATA3 subset transcriptome revealed a marked under-expression of genes related to angiogenesis. A previous study showed that downregulated genes in mutant GATA3 tumors are significantly associated with angiogenesis-related pathways.8 This explanation needs further study.
Conclusions
Intronic germline mutations of GATA3 could be related to a better prognosis (long DFS, smaller tumor size, LN negative, and post-menopause cases), while protein coding GATA3 germline mutations could be related to an unfavorable prognosis. GATA3 mutations lead to dysregulation of pathways related to immunity such as signaling by interleukins, T-cell activation, and inflammation mediated by chemokine and cytokine signaling pathways. GATA3 mutations could also lead to dysregulation of pathways related to BC development such as: regulation of TP53 expression and RUNX3 regulated CDKN1A transcription pathways. Similarly, GATA3 mutations affect genes related to BC development and metabolism such as PIK3CA, SKP1, FBP1, SMAD3, ANXA9, and CLSTN2.
Authors’ contributions
All authors participated in the study conceptualization, design, funding, analysis of the data and interpretation of results. IHI conducted experiments, conducted the bioinformatics study, HGA and FEMH conducted experiments, and drafted the manuscript. HSA supplied blood samples of breast cancer patients and clinical data (including tumor size, grade, and patients’ survival data). All authors reviewed and approved the manuscript.
Footnotes
Declaration OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval: This study protocol was reviewed by the Institutional Review Board (IRB) of Al-Azhar University Hospital (Damietta) and approved by the Scientific Research Ethical Committee of Faculty of Pharmacy (Girls), Al-Azhar University, Code no. 51/2015.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Iman H Ibrahim https://orcid.org/0000-0001-5752-2107
References
- 1.Ibrahim AS, Khaled HM, Mikhail NN, Baraka H, Kamel H. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol 2014; 2014:437971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira B, Chin SF, Rueda OM, Vollan HM, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut S, Tsui DW, Liu B, Dawson S, Abraham J, Northen H, Peden JF, Mukherjee A, Turashvili G, Green AR, McKinney S, Oloumi A, Shah SP, Rosenfeld N, Murphy L, Bentley DR, Ellis IO, Purushotham A, Pinder SE, Børresen-Dale A, Earl HM, Pharoah PD, Ross Mt Aparicio S, Caldas C. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nature Comm 2016; 7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shu J, Wu C, Wu Y, Li Z, Shao S, Zhao W, Tang XL, Yang H, Shen L, Zuo X, Yang W, Shi Y, Chi X, Zhang H, Gao G, Shu Y, Yuan K, He W, Tang C, Zhao Y, Deng H. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 2013; 153:963–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen V, Barlow WE, Yang JJ, Zhu Q, Liu S, Kwan ML, Ergas IJ, Roh JM, Hutchins LF, Kadlubar SA, Albain KS, Rae JM, Yeh I-T, Ravdin PM, Martino S, Lyss AP, Osborne CK, Hortobagyi GN, Kushi LH, Hayes DF, Ambrosone CB, Yao S. Germline genetic variants in GATA3 and breast cancer treatment outcomes in SWOG S8897 trial and the pathways study. Clin Breast Cancer 2019; 19:225–35.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol 2013; 15:201–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theodorou V, Stark R, Menon S, Carroll JS. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res 2013; 23:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerød A, Kåresen R, Oh DS, Dressler LG, Lønning PE, Strausberg R, Chanock SJ, Børresen-Dale A, Perou CM. Mutation of GATA3 in human breast tumors. Oncogene 2004; 23:7669–78 [DOI] [PubMed] [Google Scholar]
- 8.Takaku M, Grimm SA, Roberts JD, Chrysovergis K, Bennett BD, Myers PH, Perera L, Tucker CJ, Perou CM, Wade PA. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat Commun 2018; 9:1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Smissen WJ, Look MP, Liu J, Smissen WJ, Look MP, Sieuwerts AM, Smid M, Gelder ME, Foekens JA, Hollestelle A, Martens JW. GATA3 mRNA expression, but not mutation, associates with longer progression-free survival in ER-positive breast cancer patients treated with first-line tamoxifen for recurrent disease. Cancer Lett 2016; 376:104–9 [DOI] [PubMed] [Google Scholar]
- 10.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, Cortés MM, Fernandez-Lopez JC, Peng S, Ardlie KG, Auclair D, Bautista-Piña V, Duke F, Francis J, Jung J, Maffuz-Aziz A, Onofrio RC, Parkin M, Pho NH, Quintanar-Jurado V, Ramos AH, Rebollar-Vega RG, Rodriguez-Cuevas S, Romero-Cordoba SL, Schumacher SE, Stransky N, Thompson KM, Uribe-Figueroa L, Baselga J, Beroukhim R, Polyak K, Sgroi DC, Richardson AL, Jiménez-Sánchez G, Lander ES, Gabriel SB, Garraway LA, Golub TR, Melendez-Zajgla J, Toker A, Getz GA, Hidalgo-Miranda A, Meyerson M. Sequence Analysis of mutations and translocations across breast cancer subtypes. Nature 2012; 486:405–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hijikata A, Raju R, Keerthikumar S, Ramabadran S, Balakrishnan L, Ramadoss SK, Pandey A, Mohan S, Ohara O. Mutation@a glance: an integrative web application for analysing mutations from human genetic diseases. DNA Res 2010; 17:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sim N, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 2012; 40: W452–W7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne C, Heuer ML, Larsson E, Antipin Y, Reva BA, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross BE, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami EG, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman RD, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003; 13:2129–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos K, Cook J, Gillespie M, Haw R, Loney F, May B, Milacic M, Rothfels K, Sevilla C, Shamovsky V, Shorser S, Varusai TM, Weiser J, Wu G, Stein L, Hermjakob H, D'Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res 2019; 48:D498–D503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nawy T. A pan-cancer atlas. Nat Methods 2018; 15:407. [DOI] [PubMed] [Google Scholar]
- 18.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa SA, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green AR, Provenzano E, Wishart GC, Pinder SE, Watson PH, Markowetz F, Murphy LC, Ellis IO, Purushotham A, Børresen-Dale A, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012; 486:346–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 20.Kong SL, Li G, Loh SL, Sung WK, Liu ET. Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol 2011; 7:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krings G, Nystrom M, Mehdi I, Vohra P, Chen YY. Diagnostic utility and sensitivities of GATA3 antibodies in triple-negative breast cancer. Hum Pathol 2014; 45:2225–32 [DOI] [PubMed] [Google Scholar]
- 22.Belge H, Dahan K, Cambier JF, Benoît V, Morelle J, Bloch J, Vanhille P, Pirson Y, Demoulin N. Clinical and mutational spectrum of hypoparathyroidism, deafness and renal dysplasia syndrome. Nephrol Dial Transplant 2017; 32: 830–7 [DOI] [PubMed] [Google Scholar]
- 23.Ismail AM, Khalifa SE, Saied EM, El-Tamamy MM. Immunohistochemical study of GATA-3 expression versus estrogen and progesterone receptor in invasive mammary carcinomas. Kasr Al Ainy Med J 2018; 24:40–6 [Google Scholar]
- 24.Wang J, Xu B. Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Signal Transduct Target Ther 2019; 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67:7–30 [DOI] [PubMed] [Google Scholar]
- 26.Takaku M, Grimm SA, Wade PA. GATA3 in breast cancer: tumor suppressor or oncogene? Gene Expr 2015; 16:163–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mair B, Konopka T, Kerzendorfer C, Sleiman K, Salic S, Serra V, Muellner MK, Theodorou V, Nijman SM. Gain- and loss-of-Function mutations in the breast cancer gene GATA3 result in differential drug sensitivity. PLoS Genet 2016; 12:e1006279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali A, Christie PT, Grigorieva IV, Harding B, Esch HV, Ahmed SF, Bitner-Glindzicz MA, Blind E, Bloch C, Christin P, Clayton PP, Gécz J, Gilbert-Dussardier B, Guillén-Navarro E, Hackett A, Halac I, Hendy GN, Lalloo F, Mache C, Mughal Z, Ong AC, Rinat C, Shaw N, Smithson SF, Tolmie JD, Weill J, Nesbit MA, Thakker RV. Functional characterization of GATA3 mutations causing the hypoparathyroidism-deafness-renal (HDR) dysplasia syndrome: insight into mechanisms of DNA binding by the GATA3 transcription factor. Hum Mol Genet 2007; 16:265–75 [DOI] [PubMed] [Google Scholar]
- 29.Azim HA, Jr, Partridge AH. Biology of breast cancer in young women. Breast Cancer Res 2014; 16:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 2014; 156:1324–35 [DOI] [PubMed] [Google Scholar]
- 31.Öz B, Akcan A, Doğan S, Abdulrezzak Ü, Aslan D, Sözüer EM, Emek E, Akyüz M, Elmalı F, Ok E. Prediction of nonsentinel lymph node metastasis in breast cancer patients with one or two positive sentinel lymph nodes. Asian J Surg 2018; 41: 12–9 [DOI] [PubMed] [Google Scholar]
- 32.Goyal A, Zhang G, Yang B. Differential expression patterns of GATA3 in usual and differentiated types of vulvar intraepithelial neoplasia: potential diagnostic implications. Mod Pathol 2018; 31:1131–40 [DOI] [PubMed] [Google Scholar]
- 33.Kreis NN, Louwen F, Yuan J. The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers 2019; 11: 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebihara T, Seo W, Taniuchi I. Roles of RUNX complexes in immune cell development. Adv Exp Med Biol 2017; 962:395–413 [DOI] [PubMed] [Google Scholar]
- 35.Wu F-H, Yuan Y, Li D, Liao S, Yan B, Wei J, Zhou Y, Zhu J, Zhang G, Feng Z. Extracellular HSPA1A promotes the growth of hepatocarcinoma by augmenting tumor cell proliferation and apoptosis-resistance. Cancer Lett 2012; 317: 157–64 [DOI] [PubMed] [Google Scholar]
- 36.Nguyen AHT, Tremblay M, Haigh K, Koumakpayi IH, Paquet M, Pandolfi PP, Mes-Masson A, Saad F, Haigh JJ, Bouchard M. Gata3 antagonizes cancer progression in pten-deficient prostates. Hum Mol Genet 2013; 22:2400–10 [DOI] [PubMed] [Google Scholar]
- 37.Li K, Ying M, Feng D, Du J, Chen S, Dan B, Wang C, Wang Y. Fructose-1,6-bisphosphatase is a novel regulator of wnt/β-Catenin pathway in breast cancer. Biomed Pharmacother 2016; 84:1144–9 [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi N, Yamamoto H, Mimori K, Yamashita S, Miyazaki S, Nakagawa S, Ishii H, Noura S, Ohue M, Yano M, Doki Y, Mori M. ANXA9 gene expression in colorectal cancer: a novel marker for prognosis. Oncol Lett 2014; 8:2313–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Misumi I, Gu A, Curtis TA, Su L, Whitmire JK, Wan YY. GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat Immunol 2013; 14:714–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stern HM. Improving treatment of HER2-positive cancers: opportunities and challenges. Sci Transl Med 2012; 4: 127rv2. [DOI] [PubMed] [Google Scholar]
- 41.Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development 2018; 145:dev164384. [DOI] [PubMed] [Google Scholar]