Abstract
As a flavonoid, baicalein exhibits remarkable anti-cancer roles in several cancers. However, the factors regulating the antitumorigenic roles of baicalein in cervical cancer remain undefined. Here, we revealed that long noncoding RNA SNHG1 is implicated in the tumor-suppressive roles of baicalein. Functional assays demonstrated that ectopic expression of SNHG1 attenuates the roles of baicalein in repressing cervical cancer cell viability, inducing apoptosis, and repressing migration. SNHG1 silencing promotes the tumor-suppressive roles of baicalein in cervical cancer cell viability, apoptosis, and migration. Xenograft assays showed that SNHG1 reverses the tumor-suppressive roles of baicalein in repressing cervical cancer growth in vivo. Mechanistic investigations revealed that SNHG1 directly binds miR-3127-5p and up-regulates FZD4, a target of miR-3127-5p. Via regulating miR-3127-5p/FZD4, SNHG1 activates Wnt/β-catenin signaling. Moreover, SNHG1 reverses the repressive role of baicalein on Wnt/β-catenin signaling. The effect of SNHG1 on the antitumorigenic process of baicalein was abolished by Wnt/β-catenin signaling inhibitor ICG-001. Together, our observations demonstrated that SNHG1 represses the tumor-suppressive roles of baicalein in cervical cancer through regulating miR-3127-5p/FZD4/Wnt/β-catenin axis, and suggested that targeting SNHG1 represents a potential strategy to enhance the tumor-suppressive roles of baicalein in cervical cancer.
Impact statement
Baicalein exhibits anti-cancer roles in several cancers. However, the factors influencing the antitumorigenic efficiencies of baicalein in CC remain largely unclear. Here, we provide convincing evidences that lncRNA SNHG1 attenuates the tumor-suppressive roles of baicalein in CC cell viability, apoptosis, migration, and CC tumor growth. This study further demonstrates that the influences of SNHG1 in the antitumorigenic process of baicalein are achieved through modulating the miR-3127-5p/FZD4Wnt/β-catenin axis. SNHG1 attenuates the repressive role of baicalein on Wnt/β-catenin. Therefore, SNHG1 is a novel modulator of the tumor-suppressive roles of baicalein and SNHG1 represents a therapeutic intervention target to reinforce the tumor-suppressive roles of baicalein in CC.
Keywords: Long noncoding RNA, SNHG1, baicalein, Wnt/β-catenin signaling, competing endogenous RNA, cervical cancer
Introduction
Cervical cancer (CC) ranks fourth in the incidence and mortality of women malignancies, with 569,847 estimated new diagnoses and 311,365 estimated deaths globally in 2018.1 Surgery and radiochemotherapy are still the most common treatments against CC.2 The prognosis of CC is unsatisfactory with five-year survival rate of only about 65% in most countries.3
Recently, many nature products have exhibited potential anti-cancer roles via various mechanisms.4 Baicalein is a flavonoid drug, which is acquired from the root of Scutellaria baicalensis.5–7 Increasing evidences demonstrate that baicalein exerts potential anti-cancer roles in thyroid cancer, cervical cancer, hepatocellular carcinoma, breast cancer, pancreatic cancer, osteosarcoma, and so on.8–12 The molecular mechanisms of baicalein are complex.13 Many critical molecules and signaling pathways were modulated by baicalein, including p53, c-Myc, PTEN, TGF-β, NF-κB, Wnt/β-catenin, and so on.14–18 However, the molecules responsible for the tumor-suppressive roles of baicalein are largely unknown. Changing the expression of these molecules may modulate the tumor-suppressive roles of baicalein.
Great advances in genomic and transcriptomic sequencings led to the identification of >58,000 long noncoding RNAs (lncRNAs) in cells.19 Nevertheless, the number of human protein-encoding-gene is ∼21,000.19 LncRNAs are a group of novel long transcripts not translated into proteins.20,21 Many lncRNAs exhibit dysregulated expressions and important roles in many malignancies, including cervical cancer.22–28 LncRNA C5orf66-AS1 is highly expressed and drives cell proliferation in cervical cancer.29 lncRNA799 is highly expressed and promotes metastasis of cervical cancer.30 SNHG16 promotes cervical cancer tumorigenesis.31 LncRNA CRNDE promotes growth and metastasis of cervical cancer.32 LncRNA SNHG1 is a multi-functional lncRNA, which plays critical roles in various cancers.33 SNHG1 is reported to have pro-tumorigenic functions in acute myeloid leukemia, colorectal cancer, laryngeal cancer, breast cancer, and so on.34–39 Nevertheless, the roles of SNHG1 in the tumor-suppressive processes of baicalein in CC are still unknown.
Here, the effects of SNHG1 during the tumor-suppressive processes of baicalein were studied using functional experiments. Our findings revealed that SNHG1 represses the antitumorigenic roles of baicalein in CC. Further mechanistic studies revealed that SNHG1 binds miR-3127-5p and functions as a microRNA sponge. Via agglutinating miR-3127-5p, SNHG1 up-regulates FZD4, a target of miR-3127-5p. Through up-regulating FZD4, SNHG1 further activates Wnt/β-catenin signaling. Collectively, these data presented that SNHG1 represses the antitumorigenic roles of baicalein in CC through modulating miR-3127-5p/FZD4/Wnt/β-catenin axis.
Materials and methods
Cell culture and treatment
CC cell lines HeLa and SiHa were acquired from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Eagle’s Minimum Essential Medium (Invitrogen) added with 10% FBS (Invitrogen) in standard conditions. Where indicated, baicalein (Selleck) and ICG-001 (Selleck) were added for indicated time to stimulate cells. Baicalein and ICG-001 were both dissolved in DMSO.
Quantitative real-time polymerase chain reaction
The isolated RNA was subjected to reverse transcription using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara). qRT-PCR was undertaken using the standard SYBR Green protocol on ABI 7500 Real-time PCR system (Applied Biosystems). Primer sequences were: for SNHG1, 5′-AATGTCTTATTGGGCTCCTG-3′ (forward) and 5′-CTGTAACGCTGGCTTTGC-3′ (reverse); for FZD4, 5′-CAACTTTCACACCGCTCATC-3′ (forward) and 5′-GGCAAATCCAAATTCCTTCAG-3′ (reverse); for cyclin D1, 5′-ACAACTTCCTGTCCTACTACCG-3′ (forward) and 5′-TCCTCCTCCTCTTCCTCCTC-3′ (reverse); for c-Myc, 5′-GGGCTTTATCTAACTCGCTGTA-3′ (forward) and 5′-GCTATGGGCAAAGTTTCGTG-3′ (reverse); for β-actin, 5′-GGGAAATCGTGCGTGACATTAAG-3′ (forward) and 5′-TGTGTTGGCGTACAGGTCTTTG-3′ (reverse). For miR-3127-5p and miR-122-5p quantitation, qRT-PCR was undertaken by TaqMan microRNA assay (Applied Biosystems) as described above.
Northern blot
Northern blot was conducted with the NorthernMax Kit (Ambion). Briefly, 10 µg of total RNA was loaded to formaldehyde gel electrophoresis, followed by being transferred onto Biodyne Nylon membranes (Pall). Biotin-16-dUTP (Roche)-labeled SNHG1 cDNA probe was prepared using PCR with the primers 5′-TACTGGTGAAGGAATGGG-3′ (forward) and 5′-GAAGTGGAGTTATGGGAAG-3′ (reverse). After 60 min of prehybridization in ULTRAhyb™-Oligo buffer (Ambion), the membranes were hybridized for 12 h at 68°C in ULTRAhyb™-Oligo buffer with SNHG1 cDNA probe. After three washes, the membranes were scanned on Odyssey infrared scanner (Li-Cor).
Vectors construction and transfection
SNHG1 was PCR-amplified using TransTaq® DNA Polymerase High Fidelity (TransGen Biotech) with the primers 5′-GGGGTACCCTCATTTTTCTACTGCTCG-3′ (forward) and 5′-CGGGATCCAAACAGGACTATGTAATCAATC-3′ (reverse). The PCR product was cloned into Kpn I and BamH I sites of pcDNA3.1(+) vector (Invitrogen) to synthesize SNHG1 expression vector pcDNA3.1-SNHG1. In addition, the PCR product was cloned into Kpn I and BamH I sites of pSPT19 (Promega) to synthesize SNHG1 transcription vector pSPT19-SNHG1. miR-3127-5p binding site mutated SNHG1 expression vector pcDNA3.1-SNHG1-mut and transcription vector pSPT19-SNHG1-mut were synthesized by GenScript (Nanjing, China). Two independent oligonucleotides specifically repressing SNHG1 were cloned into the shRNA expression vector pGPU6/Neo (GenePharma), termed as shSNHG1-1 and shSNHG1-2. A scrambled non-targeting shRNA (shVec) was employed as negative control. The shRNAs sequences were: for shSNHG1-1, 5′-CACCGCTCCTGTCTGCAGGATTTACTTCAAGA GAGTAAATCCTGCAGACAGGAGCTTTTTTG-3′ (forward) and 5′-GATCCAAAAAAGCTCCTGTCTGCAGGA TTTACTCTCTTGAAGTAAATCCTGCAGACAGGAGC-3′ (reverse); for shSNHG1-2, 5′-CACCGGTTTGCTGTGTATCACATTTTT CAAGAGAAAATGTGATACACAGCAAACCTTTTTTG-3′ (forward) and 5′-GATCCAAAAAAGGTTTGCTGTGTATCACATTTTCTCTTGAAAAATGTGATACACAGCAAACC-3′ (reverse); for shVec, 5′-CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG-3′ (forward) and 5′-GATCCAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGACGTGACACGTTCGGAGAAC-3′ (reverse). SNHG1 containing miR-3127-5p binding site was PCR-amplified using TransTaq® DNA Polymerase High Fidelity and the primers 5′-CTAGCTAGCACATAGCATAGCACGCACT-3′ (forward) and 5′-GCGTCGACCAGCCTGGGTAACAAAAC-3′ (reverse) and cloned into Nhe I and Sal I sites of pmirGLO (Promega) to synthesize luciferase reporter pmirGLO-SNHG1. miR-3127-5p binding site mutated SNHG1 luciferase reporter pmirGLO-SNHG1-mut was synthesized by GenScript. FZD4 3′-UTR containing miR-3127-5p binding sites was PCR-amplified using TransTaq® DNA Polymerase High Fidelity and the primers 5′-CTAGCTAGCTTATTGA GTGCCGACTGTAG-3′ (forward) and 5′-GCGTCGACAGCAAGGGG TTTTCTTTGTAC-3′ (reverse) and cloned into Nhe I and Sal I sites of pmirGLO (Promega) to synthesize luciferase reporter pmirGLO-FZD4. miR-3127-5p binding site mutated FZD4 3′-UTR luciferase reporter pmirGLO-FZD4-mut was synthesized by GenScript. miR-3127 mimics and inhibitors were obtained from GenePharma. Transfection of vectors and miRNAs was carried out using Lipofectamine 3000 (Invitrogen).
Stable cell lines construction
pcDNA3.1, pcDNA3.1-SNHG1, or pcDNA3.1-SNHG1-mut was transfected into HeLa cells. pcDNA3.1 or pcDNA3.1-SNHG1 was transfected into SiHa cells. shVec, shSNHG1-1, and shSNHG1-2 were transfected into HeLa cells; 48 h later, the cells were treated with 500 μg/mL neomycin for four weeks to select HeLa and SiHa cells stably overexpressing SNHG1 and HeLa cells stably silencing SNHG1.
Cell viability, apoptosis, and migration assays
Cell viability was evaluated by Glo cell viability and ethynyl deoxyuridine (EdU) incorporation assays. Glo cell viability and EdU incorporation assays were undertaken as previously described.12 Cell apoptosis was assessed by TdT-mediated dUTP nick end labeling (TUNEL) assay as previously described.12 Cell migration was assessed using transwell migration assay as described in our previous report.11
Xenograft assay
Cells (3.0 × 106) were subcutaneously inoculated into six-weeks-old female BALB/c nude mice. After subcutaneous injection, the mice were intraperitoneally injected with 0.1 mL DMSO (0.25%) or baicalein (10 mg/kg/day). Subcutaneous xenografts volume was measured by a caliper and calculated as 0.5 × L × W2 (L, length; W, width). At the 28th day after injection, the subcutaneous xenograft was resected and weighed. The resected xenograft was subjected to Ki67 immunohistochemistry (IHC) staining and TUNEL assay as previously described.11 The xenograft assay was approved by the Ethical Committee of West China Second University Hospital, Sichuan University.
Isolation of cytoplasmic and nuclear RNA
Cytoplasmic and nuclear RNA was extracted with the Cytoplasmic & Nuclear RNA Purification Kit (Norgen) as per the provided manual. The extracted cytoplasmic and nuclear RNA were measured using qRT-PCR.
Dual luciferase reporter assays
pmirGLO-SNHG1 was cotransfected with 1 pmol, 5 pmol, or 10 pmol miR-3127-5p mimics into HeLa cells in 48-well plates per well. pmirGLO-SNHG1 was cotransfected with 5 pmol miR-3127-5p inhibitors into HeLa cells in 48-well plates per well. pmirGLO-FZD4 was cotransfected with pcDNA3.1, pcDNA3.1-SNHG1, pcDNA3.1-SNHG1-mut, shVec, shSNHG1-1, or shSNHG1-2 into HeLa cells. The β-catenin reporter TOPFlash and control reporter FOPFlash were purchased from Addgene. TOPFlash or FOPFlash was cotransfected with pRL-TK (Promega) into indicated cervical cancer cells. pRL-TK expressing Renilla luciferase, served as endogenous control. The luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (Promega) 48 h after transfection.
RNA pull-down assay
Wild type SNHG1 and miR-3127-5p binding site mutated SNHG1 were transcribed from pSPT19-SNHG1 and pSPT19-SNHG1-mut, and concurrently biotinylated by Biotin RNA Labeling Mix (Roche) and Sp6 RNA polymerase (Roche). After treatment with DNase I and purification using RNeasy Mini Kit (Qiagen), 3 µg of purified biotinylated wild type SNHG1 or mutated SNHG1 was mixed with 1 mg of HeLa lysates at 25°C for 1 h. Next, the complexes were enriched using streptavidin agarose bead (Invitrogen) and the enriched RNA was detected using qRT-PCR. Furthermore, the binding between SNHG1 and miR-3127-5p was detected using the EZ-Magna ChIRP RNA Interactome Kit (Millipore) and SNHG1 antisense biotinylated probes. The sequences of SNHG1 antisense probes were: 1, 5ʹ-aagaccacgaagccacttac-3ʹ; 2, 5ʹ-tgctacttttaagcctcgag-3ʹ; 3, 5ʹ-cctcagagttatttatcctc-3ʹ; 4, 5ʹ-taaatcctgcagacaggagc-3ʹ; 5, 5ʹ-cgctggctttgcataaagat-3ʹ; 6, 5ʹ-ctgctctgtaagatagaggc-3ʹ; 7, 5ʹ-aggttgtccaagaacttgga-3ʹ; 8, 5ʹ-ctcaactgctgtttcattgg-3ʹ; 9, 5ʹ-ctatacagtgcctgagtttg-3ʹ; 10, 5ʹ-ggaacagaaggtggctgtat-3ʹ; 11, 5ʹ-cctcaagggaaccttttttg-3ʹ.
Western blot
Western blot was undertaken as previously described with FZD4 (Abcam) and β-actin (Proteintech) primary antibodies.11 β-actin serves as loading control.
Statistical analysis
GraphPad Prism 6.0 software was used to carry out all statistical analyses. Two-tailed t-test was carried out to compare the differences among two groups. One-way ANOVA followed by Dunnett's multiple comparisons test or Kruskal–Wallis test followed by Dunn's multiple comparisons test was carried out to compare the differences among more than two groups. P < 0.05 was considered significant.
Results
Ectopic expression of SNHG1 repressed the tumor-suppressive roles of baicalein
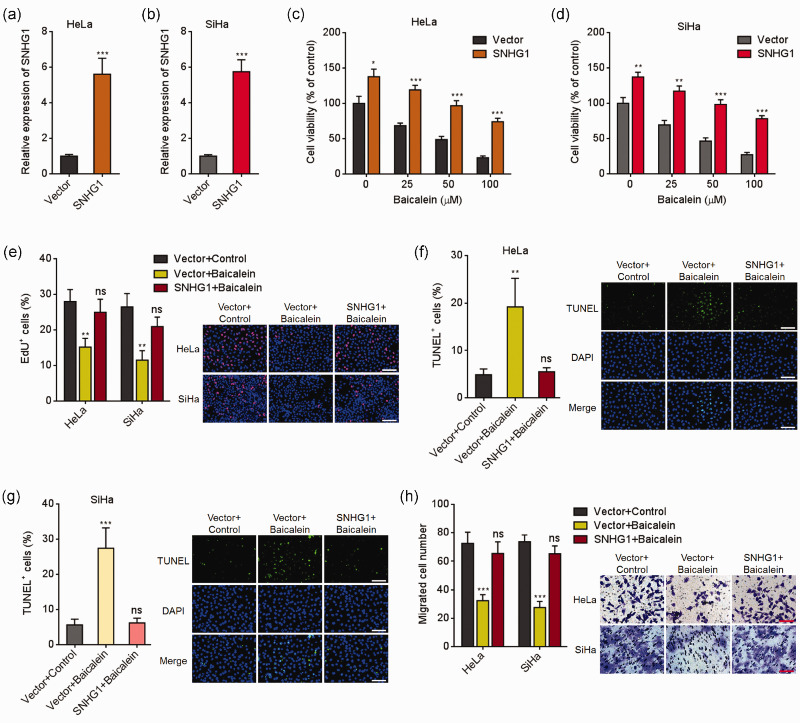
To assess the potential influences of SNHG1 in the tumor-suppressive roles of baicalein, we overexpressed SNHG1 in HeLa and SiHa cells (Figure 1(a) and (b)). HeLa and SiHa cells overexpressing SNHG1 or control were treated with indicated doses of baicalein for 24 h. Glo cell viability assay presented that baicalein repressed CC cell viabilities in dose-dependent manners (Figure 1(c) and (d)). Overexpression of SNHG1 remarkably increased cell viabilities, which were repressed by baicalein (Figure 1(c) and (d)). EdU incorporation assay also presented that baicalein repressed cell viabilities of both cells (Figure 1(e)). Ectopic expression of SNHG1 remarkably attenuated the repression of cell viabilities induced by baicalein (Figure 1(e)). TUNEL assay presented that baicalein promoted apoptosis of both cells (Figure 1(f) and (g)). Ectopic expression of SNHG1 remarkably reversed the cell apoptosis caused by baicalein (Figure 1(f) and (g)). In addition, transwell migration assays presented that baicalein repressed cell migration of both cells (Figure 1(h)). Overexpression of SNHG1 remarkably attenuated the repression of cell migration induced by baicalein (Figure 1(h)). Therefore, these data suggested that ectopic expression of SNHG1 reversed the antitumorigenic roles of baicalein in CC cell viability and migration.
Figure 1.
Ectopic expression of SNHG1 reversed baicalein-caused cell viability repression, apoptosis induction, and migration repression in cervical cancer cells. (a) SNHG1 expression in HeLa cells overexpressing SNHG1 or control was assessed using qRT-PCR. (b) SNHG1expression in SiHa cells overexpressing SNHG1 or control was assessed using qRT-PCR. (c) HeLa cells overexpressing SNHG1 or control were treated with different doses of baicalein (0, 25, 50, 100 µM) for 24 h. Then, cell viability was assessed with Glo cell viability assay. (d) SiHa cells overexpressing SNHG1 or control were treated with different doses of Baicalein (0, 25, 50, 100 µM) for 24 h, followed by Glo cell viability assay to measure cell viability. (e) HeLa and SiHa cells overexpressing SNHG1 or control were treated with 100 µM baicalein for 24 h, followed by EdU incorporation assay to detect cell viability. (f) HeLa cells overexpressing SNHG1 or control were treated with 100 µM baicalein for 24 h, followed by TUNEL assay to detect cell apoptosis. (g) SiHa cells overexpressing SNHG1 or control were treated with 100 µM baicalein for 24 h, followed by TUNEL assay to detect cell apoptosis. (h) Cell migration of HeLa and SiHa cells overexpressing SNHG1 or control and simultaneous treatment with 50 µM baicalein for 48 h was detected with transwell migration assay. Results are presented as mean ± SD. Scale bars, 100 µm. *P < 0.05, **P < 0.01, ***P < 0.001, ns: not significant. (A color version of this figure is available in the online journal.)
SNHG1 silencing enhanced the antitumorigenic roles of baicalein
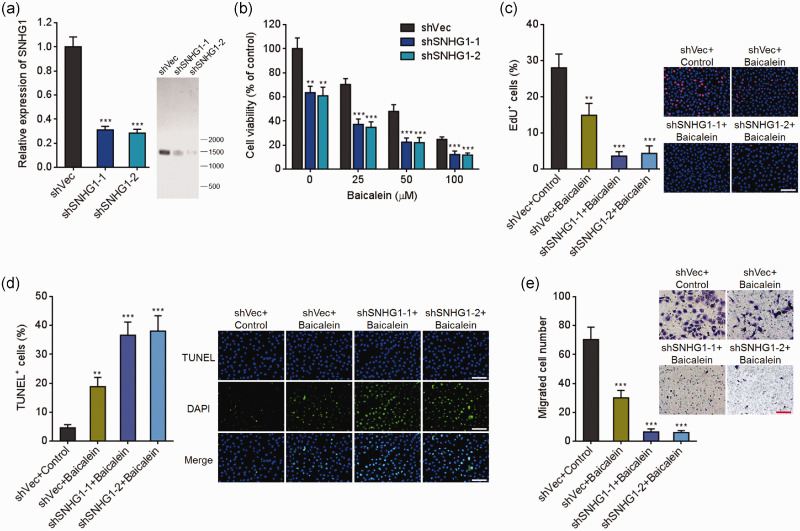
HeLa cells stably silencing SNHG1 or control were constructed (Figure 2(a)). HeLa cells silencing SNHG1 or control were treated with indicated doses of baicalein for 24 h. Next, cell viabilities were assessed using Glo cell viability assays. As presented in Figure 2(b), the repressive roles of baicalein in CC cell viabilities were remarkably enhanced by SNHG1 silencing. EdU incorporation assays also presented that SNHG1 silencing remarkably enhanced the repressive effects of baicalein in CC cell viabilities (Figure 2(c)). TUNEL assays presented that SNHG1 silencing remarkably enhanced the effects of baicalein in promoting cell apoptosis (Figure 2(d)). Additionally, transwell migration assays presented that SNHG1 silencing remarkably enhanced the repressive roles of baicalein in cell migration (Figure 2(e)). Therefore, these observations demonstrated that SNHG1 silencing strengthened the tumor-suppressive roles of baicalein.
Figure 2.
SNHG1 silencing enhanced baicalein-caused cell viability repression, apoptosis induction, and migration repression in cervical cancer cells. (a) SNHG1 expression in HeLa cells silencing SNHG1 or control was assessed using qRT-PCR and northern blot. (b) HeLa cells silencing SNHG1 or control were treated with different doses of baicalein (0, 25, 50, 100 µM) for 24 h, followed by Glo cell viability assay to detect cell viability. (c) HeLa cells silencing SNHG1 or control were treated with 100 µM baicalein for 24 h, followed by EdU incorporation assay to detect cell viability. (d) HeLa cells silencing SNHG1 or control were treated with 100 µM baicalein for 24 h, followed by TUNEL assay to detect cell apoptosis. (e) Cell migration of HeLa cells silencing SNHG1 or control and simultaneous treatment with 50 µM baicalein for 48 h was detected by transwell migration assay. Results are shown as mean ± SD. Scale bars, 100 µm. **P < 0.01, ***P < 0.001. (A color version of this figure is available in the online journal.)
SNHG1 repressed the antitumorigenic roles of baicalein in CC growth in vivo
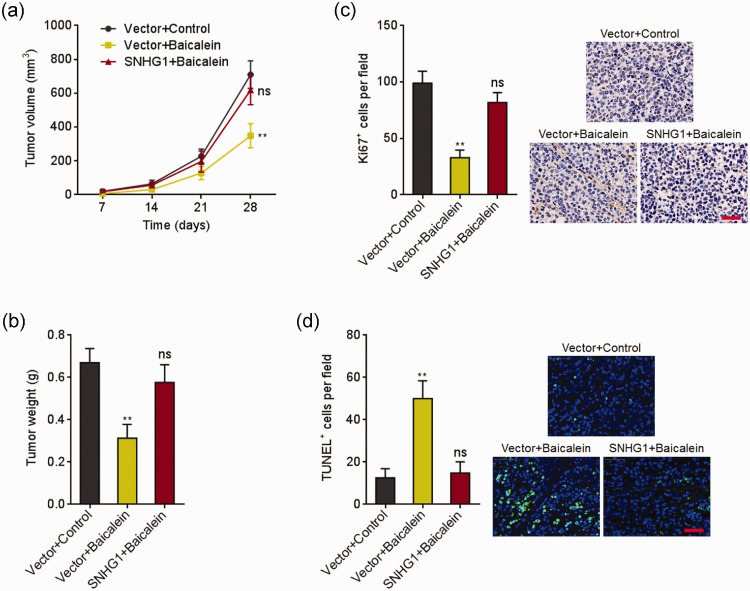
Next, the effects of SNHG1 in the in vivo tumor-suppressive processes of baicalein were explored. HeLa cells overexpressing SNHG1 or control were subcutaneously inoculated into nude mice. Next, the mice were intraperitoneally administrated with 0.1 mL 0.25% DMSO (carrier) or baicalein (10 mg/kg/day). The growth of subcutaneous xenografts was recorded. As presented in Figure 3(a) and (b), baicalein significantly repressed subcutaneous xenografts growth, which was reversed by ectopic expression of SNHG1. Ki67 IHC staining of the xenografts presented that baicalein repressed cell growth in vivo, which was reversed by ectopic expression of SNHG1 (Figure 3(c)). TUNEL staining of the xenografts presented that baicalein induced cell apoptosis in vivo, which was attenuated by ectopic expression of SNHG1 (Figure 3(d)). Therefore, these observations showed that SNHG1 repressed the tumor-suppressive roles of baicalein in vivo.
Figure 3.
SNHG1 reversed baicalein-induced cervical cancer growth inhibition in vivo. (a) HeLa cells overexpressing SNHG1 or control were subcutaneously inoculated into nude mice. The mice were intraperitoneally injected with 0.1 mL DMSO (0.25%) or baicalein (10 mg/kg/day). Subcutaneous xenografts volume was measured every seven days. (b) Xenografts were excised and weighed at 28th day after inoculation. (c) Ki67 IHC staining of xenografts from (b). (d) TUNEL staining of xenografts from (b). Data are presented as mean ± SD based on five mice in each group. Scale bars, 50 µm. **P < 0.01, ns: not significant. (A color version of this figure is available in the online journal.)
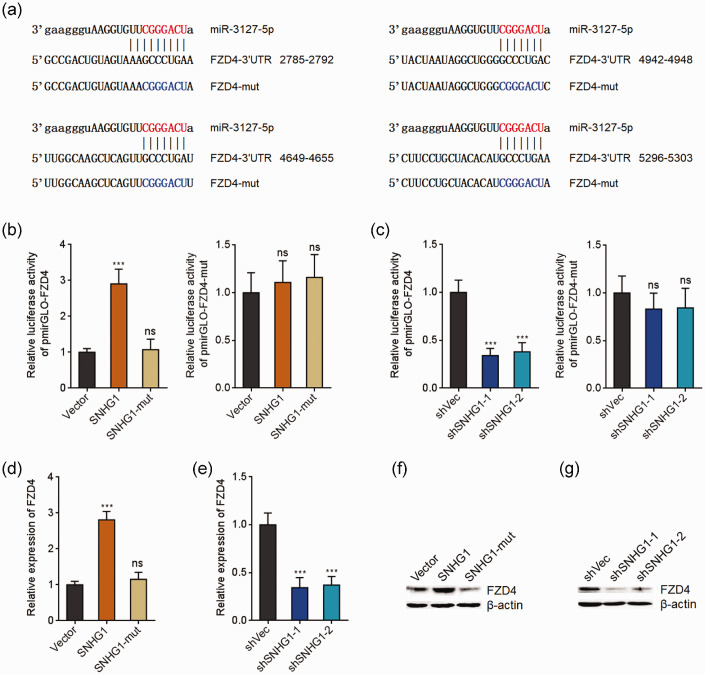
SNHG1 directly bound miR-3127-5p
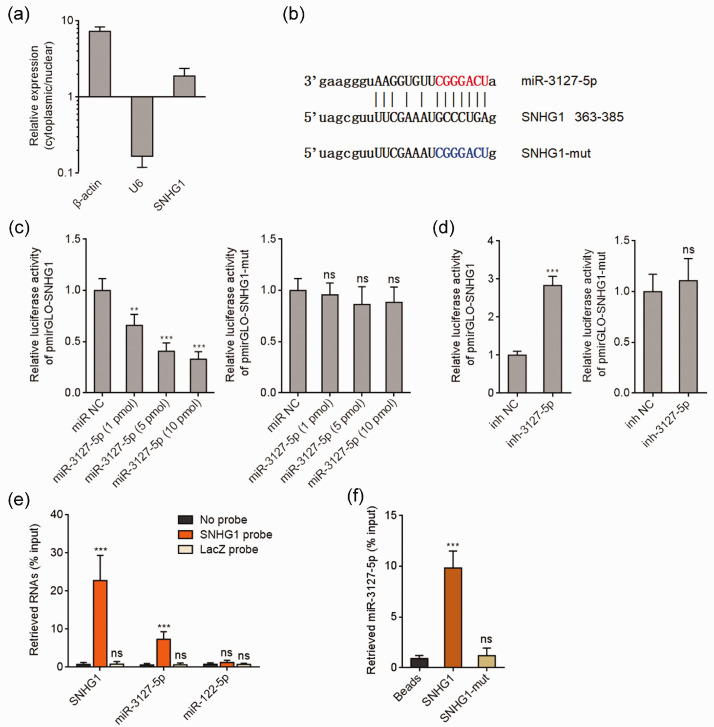
We further elucidated the molecular mechanisms mediating the effects of SNHG1 in the tumor-suppressive processes of baicalein in CC. Bioinformatics analysis indicated that SNHG1 was mainly located in cytoplasm (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/).40 Cytoplasmic and nuclear RNA purification further verified the cytoplasmic location of SNHG1 (Figure 4(a)). A variety of cytoplasmic lncRNAs was demonstrated to agglutinate microRNAs and modulate the biological roles of the interacted microRNAs.41 Analyzing by the TargetScan prediction algorithm (http://www.targetscan.org/) and ENCORI (http://starbase.sysu.edu.cn/),42 we noted a miR-3127-5p binding site on SNHG1 (Figure 4(b)). To explore whether miR-3127-5p binds to SNHG1, dual luciferase reporter assay was conducted. As presented in Figure 4(c), miR-3127-5p mimics remarkably reduced the luciferase activity of the reporter containing wild-type SNHG1, but not miR-3127-5p binding site mutated SNHG1 in a dose-dependent manner. Conversely, miR-3127-5p inhibitors remarkably increased the luciferase activity of the reporter containing wild-type SNHG1, but not miR-3127-5p binding site mutated SNHG1 (Figure 4(d)). To detect the binding of SNHG1 to miR-3127-5p, SNHG1 antisense biotinylated probes were used to pull-down endogenous SNHG1 and the bound miRNAs. As presented in Figure 4(e), both SNHG1 and miR-3127-5p were remarkably enriched in SNHG1 antisense probes group, which supports the interaction between SNHG1 and miR-3127-5p. miR-122-5p was used as a negative control miRNA. Furthermore, affinity pull-down of endogenous miR-3127-5p with in vitro transcribed biotinylated SNHG1 verified the binding of miR-3127-5p by SNHG1 (Figure 4(f)). Therefore, these observations showed that SNHG1 directly bound miR-3127-5p in cervical cancer.
Figure 4.
SNHG1 bound to miR-3127-5p. (a) Subcellular location of SNHG1 was assessed using cytoplasmic and nuclear RNA purification, followed by qRT-PCR. (b) The predicted binding site of miR-3127-5p on SNHG1. (c) Luciferase activity in HeLa cells cotransfected with indicated doses of miR-3127-5p mimics and luciferase reporter containing wild-type or miR-3127-5p binding site mutated SNHG1. Results are presented as the relative ratio of firefly to renilla luciferase activity. (d) Luciferase activity in HeLa cells cotransfected with miR-3127-5p inhibitors and luciferase reporter containing wild-type or miR-3127-5p binding site mutated SNHG1. Data are presented as the relative ratio of firefly to renilla luciferase activity. (e) HeLa cell lysates were mixed with SNHG1 antisense biotinylated probes; after pull-down, RNA was enriched and assessed with qRT-PCR. (f) HeLa cell lysates were mixed with biotinylated SNHG1; after pull-down, RNA was enriched and measured with qRT-PCR. Results are presented as mean ± SD. ***P < 0.001, ns: not significant. (A color version of this figure is available in the online journal.)
SNHG1 up-regulated FZD4 expression via sponging miR-3127-5p
FZD4 was previously revealed as a target of miR-3127-5p and modulate Wnt/β-catenin signaling in several malignancies.43 Next, we investigated whether SNHG1 modulates FZD4 via sponging miR-3127-5p in cervical cancer. Four miR-3127-5p binding sites were predicted in FZD4 3ʹ-untranslated region (3ʹ-UTR) (Figure 5(a)). Ectopic expression of SNHG1 remarkably upregulated the luciferase activity of the reporter containing wild-type FZD4 3ʹ-UTR, but not miR-3127-5p binding site mutated FZD4 3ʹ-UTR (Figure 5(b)). Furthermore, the mutation of miR-3127-5p binding site on SNHG1 reversed the upregulation of the luciferase activity (Figure 5(b)). Conversely, SNHG1 silencing remarkably deceased the luciferase activity of the reporter containing wild-type FZD4 3ʹ-UTR, but not miR-3127-5p binding site mutated FZD4 3ʹ-UTR (Figure 5(c)). Additionally, ectopic expression of SNHG1 remarkably increased FZD4 mRNA and protein levels (Figure 5(d) and (f)). The mutation of miR-3127-5p binding site on SNHG1 reversed the increasing (Figure 5(d) and (f)). Conversely, SNHG1 silencing markedly reduced FZD4 mRNA and protein levels (Figure 5(e) and (g)). Therefore, these observations showed that SNHG1 increased FZD4 expression via sponging miR-3127-5p.
Figure 5.
SNHG1 up-regulated FZD4 via sponging miR-3127-5p. (a) The predicted miR-3127-5p binding sites on FZD4 3ʹ-UTR. (b) Luciferase activity in HeLa cells cotransfected with wild-type or miR-3127-5p binding site mutated SNHG1 expression plasmids and luciferase reporter containing wild-type or miR-3127-5p binding site mutated FZD4 3ʹ-UTR. Result is shown as the relative ratio of firefly to renilla luciferase activity. (c) Luciferase activity in HeLa cells cotransfected with SNHG1 specific shRNAs and luciferase reporter containing wild-type or miR-3127-5p binding site mutated FZD4 3ʹ-UTR. Result is shown as the relative ratio of firefly to renilla luciferase activity. (d) FZD4 mRNA expression level in HeLa cells overexpressing wild-type or miR-3127-5p binding site mutated SNHG1 was assessed using qRT-PCR. (e) FZD4 mRNA expression level in HeLa cells silencing SNHG1 or control was assessed using qRT-PCR. (f) FZD4 protein expression level in HeLa cells overexpressing wild-type or miR-3127-5p binding site mutated SNHG1 was detected using Western blot. (g) FZD4 protein expression level in HeLa cells silencing SNHG1 or control was assessed using Western blot. Results are presented as mean ± SD. ***P < 0.001, ns: not significant. (A color version of this figure is available in the online journal.)
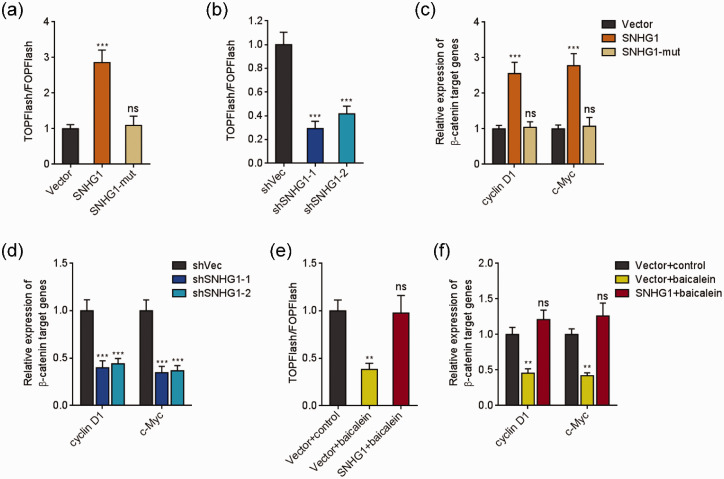
SNHG1 activated Wnt/β-catenin signaling via modulating miR-3127-5p/FZD4
Next, the roles of SNHG1 in Wnt/β-catenin signaling were investigated. TOPFlash/FOPFlash luciferase activities and Wnt/β-catenin targets expressions were detected to evaluate Wnt/β-catenin signaling in SNHG1 stably overexpressed and silenced HeLa cells. Overexpression of SNHG1 remarkably up-regulated the ratio of TOPFlash/FOPFlash (Figure 6(a)). The increasing of the ratio was reversed by the mutation of miR-3127-5p binding site (Figure 6(a)). Conversely, SNHG1 silencing remarkably decreased the ratio of TOPFlash/FOPFlash (Figure 6(b)). Ectopic expression of SNHG1 remarkably increased Wnt/β-catenin targets expression (Figure 6(c)). The mutation of miR-3127-5p binding site on SNHG1 reversed the increasing (Figure 6(c)). Conversely, SNHG1 silencing remarkably decreased Wnt/β-catenin targets expressions (Figure 6(d)). Then, TOPFlash/FOPFlash luciferase activities and Wnt/β-catenin targets expressions were measured in HeLa cells stably overexpressing SNHG1 or control and simultaneous treatment with 50 μM baicalein for 48 h. As presented in Figure 6(e) and (f), baicalein repressed the ratio of TOPFlash/FOPFlash and Wnt/β-catenin target genes expression. Ectopic expression of SNHG1 reversed the decreasing of Wnt/β-catenin signaling activity caused by baicalein (Figure 6(e) and (f)). Therefore, the results demonstrated that baicalein repressed Wnt/β-catenin signaling, which was reversed by SNHG1 via modulating miR-3127-5p/FZD4.
Figure 6.
SNHG1 activated Wnt/β-catenin signaling via modulating miR-3127-5p/FZD4. (a) After transient transfection of TOPFlash or FOPFlash plasmids into HeLa cells overexpressing wild-type or miR-3127-5p binding site mutated SNHG1, luciferase activity was assessed and presented as the ratio of TOPFlash/FOPFlash. (b) After transient transfection of TOPFlash or FOPFlash plasmids into HeLa cells silencing SNHG1 or control, luciferase activity was assessed and presented as the ratio of TOPFlash/FOPFlash. (c) Wnt/β-catenin targets expression level in HeLa cells overexpressing wild-type or miR-3127-5p binding site mutated SNHG1 was measured using qRT-PCR. (d) Wnt/β-catenin targets expression level in HeLa cells silencing SNHG1 or control was measured using qRT-PCR. (e) Luciferase activity in HeLa cells overexpressing SNHG1 or control and simultaneous treatment with 50 µM baicalein after transient transfection of TOPFlash or FOPFlash plasmids was measured and presented as the ratio of TOPFlash/FOPFlash. (f) Wnt/β-catenin targets expression level in HeLa cells overexpressing SNHG1 or control and simultaneous treatment with 50 µM baicalein for 48 h was measured using qRT-PCR. Data are presented as mean ± SD. **P < 0.01, ***P < 0.001, ns: not significant. (A color version of this figure is available in the online journal.)
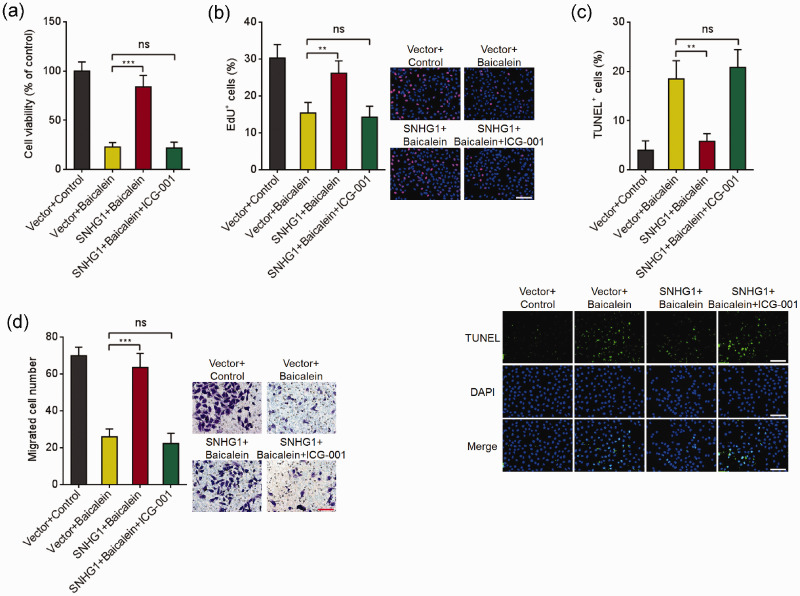
Blockage of Wnt/β-catenin signaling reversed the effects of SNHG1 in modulating the anti-cancer roles of baicalein
To explore whether SNHG1 represses the tumor-suppressive roles of baicalein through modulating Wnt/β-catenin signaling, HeLa cells overexpressing SNHG1 or control were treated with baicalein and Wnt/β-catenin signaling inhibitor ICG-001. Glo cell viability and EdU incorporation assays presented that ICG-001 blocked the effects of SNHG1 in reversing the decreased cell viability caused by baicalein (Figure 7(a) and (b)). TUNEL assays presented that ICG-001 blocked the effects of SNHG1 in repressing cell apoptosis induced by baicalein (Figure 7(c)). Transwell migration assays presented that ICG-001 blocked the roles of SNHG1 in reversing the decreased cell migration caused by baicalein (Figure 7(d)). Therefore, these observations presented that SNHG1 repressed the tumor-suppressive roles of baicalein through activating Wnt/β-catenin signaling.
Figure 7.
Repression of Wnt/β-catenin signaling blocked the effects of SNHG1 in modulating the tumor-suppressive roles of baicalein. (a) HeLa cells overexpressing SNHG1 were treated with 100 μM baicalein and 25 μM ICG-001 for 24 h, followed by Glo cell viability assay to detect cell viability. (b) HeLa cells overexpressing SNHG1 were treated with 100 μM baicalein and 25 μM ICG-001 for 24 h, followed by EdU incorporation assay to detect cell viability. (c) SNHG1 stably overexpressed HeLa cells were treated with 100 μM baicalein and 25 μM ICG-001 for 24 h, followed by TUNEL assay to measure cell apoptosis. (d) Cell migration of HeLa cells overexpressing SNHG1 and simultaneous treatment with 50 μM baicalein and 12.5 μM ICG-001 for 48 h was detected by transwell migration assay. Results are shown as mean ± SD. Scale bars, 100 µm. **P < 0.01, ***P < 0.001, ns: not significant. (A color version of this figure is available in the online journal.)
Discussion
Increasing cellular and animal experiments have shown that baicalein has attractive anti-cancer effects,6 whereas the low bioavailability of baicalein restricts the clinical utilization.5 Developing novel combined therapies to enhance the tumor-suppressive roles of baicalein may facilitate the broader application of baicalein.
Here, we identified lncRNA SNHG1 as a crucial regulator of the antitumorigenic roles of baicalein in CC. Functional experiments demonstrated that SNHG1 attenuates the effects of baicalein in inhibiting cell viability, promoting apoptosis, and suppressing migration. Oppositely, SNHG1 silencing enhances the roles of baicalein in inhibiting cell viability, promoting apoptosis, and suppressing migration. In addition, in vivo xenograft assays showed that SNHG1 blocks the effects of baicalein in repressing xenograft growth and cell proliferation and promoting cell apoptosis in vivo. Collectively, these findings identified SNHG1 as a functional antagonist of baicalein, and suggested that therapeutic intervention of SNHG1 represents a potential treatment method to enhance the antitumorigenic roles of baicalein in CC.
SNHG1, also known as small nucleolar RNA host gene 1, is a classical lncRNA.33 The gene SNHG1 locates at chromosome 11q12.3. SNHG1 encodes several different transcripts. SNHG1 transcript variant 2 (NR_152575) is the main transcript in cervical cancer, which has 10 exons and 1535 nucleotides in length. Therefore, we focused on this transcript. Previous reports showed that SNHG1 were dysregulated in several malignancies, including cervical cancer.34–36,44 Other reports revealed that SNHG1 facilitates cancer cell proliferation, migration, invasion, and drug-resistance in colorectal cancer, laryngeal cancer, breast cancer, and lung cancer via various mechanisms.35–39 Here, we revealed that SNHG1 regulates the tumor-suppressive roles of baicalein in CC. Due to the wide tumor-suppressive effects of baicalein in several different malignancies, the influences of SNHG1 on the antitumorigenic roles of baicalein in other cancers need further investigation.
lncRNAs function in multitude levels. Many cytoplasmic lncRNAs are presented to agglutinate common microRNAs and relieve the suppressive effects of microRNAs on their targets.41 Here, combining bioinformatics analyses and experimental verification, we observed that SNHG1 directly bound miR-3127-5p and upregulated the expression of miR-3127-5p target FZD4. Via up-regulating FZD4, SNHG1 activated Wnt/β-catenin signaling. Previous reports have presented the repressive roles of baicalein in Wnt/β-catenin signaling in osteosarcoma and breast cancer.15,45 In this study, we further observed that baicalein repressed Wnt/β-catenin signaling in CC. SNHG1 attenuated the repressive influences of baicalein on Wnt/β-catenin signaling in CC. The effects of SNHG1 during the tumor-suppressive process of baicalein were abolished by Wnt/β-catenin signaling inhibitor. Mutation of miR-3127-5p binding site on SNHG1 abolished the effects of SNHG1 on miR-3127-5p/FZD4/Wnt/β-catenin axis. Therefore, miR-3127-5p binding site on SNHG1 is the critical region mediating the roles of SNHG1. To fully understand the effects of SNHG1 on its downstream targets, transcriptomic analysis of differential genes regulated by SNHG1 could provide more information, which needs further investigation.
In conclusion, these observations showed that SNHG1 represses the antitumorigenic roles of baicalein in CC through modulating miR-3127-5p/FZD4/Wnt/β-catenin axis. These findings suggested that targeting SNHG1 represents a potential strategy to enhance the antitumorigenic roles of baicalein in cervical cancer.
Footnotes
Authors’ contributions: XY and ZL designed this study; XY, JX, YC, LT, and XT conducted the experiments; XY and ZL draft this manuscript. All authors approved this manuscript.
Declaration OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors received grants from Administration of Traditional Chinese Medicine of Sichuan Province [2018JC029]; Southwest Medical University-Luzhou Traditional Chinese Medicine Hospital [2017-LH020]; and Liangshan Science and Technology Bureau [18YYJS0041]; Luzhou-Southwest Medical University joint project (No. 2019LZXNYDJ53).
ORCID iD: Zhengyu Li https://orcid.org/0000-0002-9957-2687
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 2.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, Rao A, Schultz A, Li X, Sumazin P, Williams C, Mestdagh P, Gunaratne PH, Yau C, Bowlby R, Robertson AG, Tiezzi DG, Wang C, Cherniack AD, Godwin AK, Kuderer NM, Rader JS, Zuna RE, Sood AK, Lazar AJ, Ojesina AI, Adebamowo C, Adebamowo SN, Baggerly KA, Chen TW, Chiu HS, Lefever S, Liu L, MacKenzie K, Orsulic S, Roszik J, Shelley CS, Song Q, Vellano CP, Wentzensen N, Cancer Genome Atlas Research N, Weinstein JN, Mills GB, Levine DA, Akbani R. A comprehensive Pan-Cancer molecular study of gynecologic and breast cancers. Cancer Cell 2018; 33:690–705.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, Ogunbiyi OJ, Azevedo ESG, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP, Group CW. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018; 391:1023–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Z, Liu B, Sun J, Lu L, Liu L, Qiu J, Li Q, Yan C, Jiang S, Mohammadtursun N, Ma W, Li M, Dong J, Gong W. Scutellaria flavonoids effectively inhibit the malignant phenotypes of non-small cell lung cancer in an Id1-dependent manner. Int J Biol Sci 2019; 15:1500–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bie B, Sun J, Guo Y, Li J, Jiang W, Yang J, Huang C, Li Z. Baicalein: a review of its anti-cancer effects and mechanisms in hepatocellular carcinoma. Biomed Pharmacother 2017; 93:1285–91 [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Dong Y, Gao Y, Du Z, Wang Y, Cheng P, Chen A, Huang H. The fascinating effects of baicalein on cancer: a review. Int J Mol Sci 2016; 17:1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang WT, Li J, Vanden Hoek MS, Zhu X, Li CQ, Huang HH, Hsu CW, Zhong Q, Li J, Chen SJ, Vanden Hoek TL, Shao ZH. Baicalein preconditioning protects cardiomyocytes from ischemia-reperfusion injury via mitochondrial oxidant signaling. Am J Chin Med 2013; 41:315–31 [DOI] [PubMed] [Google Scholar]
- 8.Chang WT, Li J, Haung HH, Liu H, Han M, Ramachandran S, Li CQ, Sharp WW, Hamann KJ, Yuan CS, Hoek TL, Shao ZH. Baicalein protects against doxorubicin-induced cardiotoxicity by attenuation of mitochondrial oxidant injury and JNK activation. J Cell Biochem 2011; 112:2873–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Zhang C, Jiang J, Li Y. Baicalein retards proliferation and collagen deposition by activating p38MAPK-JNK via microRNA-29. J Cell Biochem 2019; 120:15625–34 [DOI] [PubMed] [Google Scholar]
- 10.Yu X, Yang Y, Li Y, Cao Y, Tang L, Chen F, Xia J. Baicalein inhibits cervical cancer progression via downregulating long noncoding RNA BDLNR and its downstream PI3K/akt pathway. Int J Biochem Cell Biol 2018; 94:107–18 [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Tang W, Yang Y, Tang L, Dai R, Pu B, Feng C, Xia J. Long noncoding RNA NKILA enhances the anti-cancer effects of baicalein in hepatocellular carcinoma via the regulation of NF-kappaB signaling. Chem Biol Interact 2018; 285:48–58 [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Cao Y, Tang L, Yang Y, Chen F, Xia J. Baicalein inhibits breast cancer growth via activating a novel isoform of the long noncoding RNA PAX8-AS1-N. J Cell Biochem 2018; 119:6842–56 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Chang WT, Li CQ, Lee C, Huang HH, Hsu CW, Chen WJ, Zhu X, Wang CZ, Vanden Hoek TL, Shao ZH. Baicalein preventive treatment confers optimal cardioprotection by PTEN/akt/NO activation. Am J Chin Med 2017; 45:987–1001 [DOI] [PubMed] [Google Scholar]
- 14.Tang Q, Ji F, Sun W, Wang J, Guo J, Guo L, Li Y, Bao Y. Combination of baicalein and 10-hydroxy camptothecin exerts remarkable synergetic anti-cancer effects. Phytomedicine 2016; 23:1778–86 [DOI] [PubMed] [Google Scholar]
- 15.He N, Zhang Z. Baicalein suppresses the viability of MG-63 osteosarcoma cells through inhibiting c-MYC expression via Wnt signaling pathway. Mol Cell Biochem 2015; 405:187–96 [DOI] [PubMed] [Google Scholar]
- 16.Lu C, Wang H, Chen S, Yang R, Li H, Zhang G. Baicalein inhibits cell growth and increases cisplatin sensitivity of A549 and H460 cells via miR-424-3p and targeting PTEN/PI3K/akt pathway. J Cell Mol Med 2018; 22:2478–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung H, Choi HS, Seo EK, Kang DH, Oh ES. Baicalin and baicalein inhibit transforming growth factor-beta1-mediated epithelial-mesenchymal transition in human breast epithelial cells. Biochem Biophys Res Commun 2015; 458:707–13 [DOI] [PubMed] [Google Scholar]
- 18.Yu M, Qi B, Xiaoxiang W, Xu J, Liu X. Baicalein increases cisplatin sensitivity of A549 lung adenocarcinoma cells via PI3K/akt/NF-kappaB pathway. Biomed Pharmacother 2017; 90:677–85 [DOI] [PubMed] [Google Scholar]
- 19.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015; 47:199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu WL, Jin L, Xu A, Wang YF, Thorne RF, Zhang XD, Wu M. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol 2018; 20:492–502 [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Yuan J, Hu H, Chen W, Liu M, Zhang J, Sun S, Guo Z. Long non-coding RNA CHCHD4P4 promotes epithelial-mesenchymal transition and inhibits cell proliferation in calcium oxalate-induced kidney damage. Braz J Med Biol Res 2018; 51:e6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esposito R, Bosch N, Lanzos A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell 2019; 35:545–57 [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, Jia L, Li S, Cancer Genome Atlas Research N, Xie W, Yang D. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes Cell-Cycle progression in cancer. Cancer Cell 2018; 33:e9 706–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondal T, Juvvuna PK, Kirkeby A, Mitra S, Kosalai ST, Traxler L, Hertwig F, Wernig-Zorc S, Miranda C, Deland L, Volland R, Bartenhagen C, Bartsch D, Bandaru S, Engesser A, Subhash S, Martinsson T, Caren H, Akyurek LM, Kurian L, Kanduri M, Huarte M, Kogner P, Fischer M, Kanduri C. Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell 2018; 33:417–34.e7 [DOI] [PubMed] [Google Scholar]
- 25.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol 2017; 19:820–32 [DOI] [PubMed] [Google Scholar]
- 26.Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B, Wu DS, Fang ZY, Bao Y, Jiang MM, Yuan JH, Qu L, Wang LH. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer 2017; 16:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J 2016; 283:3739–54 [DOI] [PubMed] [Google Scholar]
- 28.Samir N, Matboli M, El-Tayeb H, El-Tawdi A, Hassan MK, Waly A, El-Akkad HAE, Ramadan MG, Al-Belkini TN, El-Khamisy S, El-Asmar F. Competing endogenous RNA network crosstalk reveals novel molecular markers in colorectal cancer. J Cell Biochem 2018; 119:6869–81 [DOI] [PubMed] [Google Scholar]
- 29.Rui X, Xu Y, Jiang X, Ye W, Huang Y, Jiang J. Long non-coding RNA C5orf66-AS1 promotes cell proliferation in cervical cancer by targeting miR-637/RING1 axis. Cell Death Dis 2018; 9:1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao LM, Zhang FH, Yao GJ, Ai SF, Zheng M, Huang L. Role of long noncoding RNA 799 in the metastasis of cervical cancer through upregulation of TBL1XR1 expression. Mol Ther Nucleic Acids 2018; 13:580–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys 2018; 637:1–8 [DOI] [PubMed] [Google Scholar]
- 32.Zhang JJ, Fan LP. Long non-coding RNA CRNDE enhances cervical cancer progression by suppressing PUMA expression. Biomed Pharmacother 2019; 117:108726. [DOI] [PubMed] [Google Scholar]
- 33.Thin KZ, Tu JC, Raveendran S. Long non-coding SNHG1 in cancer. Clin Chim Acta 2019; 494:38–47 [DOI] [PubMed] [Google Scholar]
- 34.Bao XL, Zhang L, Song WP. LncRNA SNHG1 overexpression regulates the proliferation of acute myeloid leukemia cells through miR-488-5p/NUP205 axis. Eur Rev Med Pharmacol Sci 2019; 23:5896–903 [DOI] [PubMed] [Google Scholar]
- 35.Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, Xu X, Xu T, Hu X, Sun L, He B, Pan Y, Sun H, Wang S. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer 2018; 17:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu T, Zuo JJ, Li F, Xu YC, Zheng AY, Tao ZZ. LncRNA SNHG1 promotes cell proliferation in laryngeal cancer via Notch1 signaling pathway. Eur Rev Med Pharmacol Sci 2019; 23:6562–9 [DOI] [PubMed] [Google Scholar]
- 37.Zheng S, Li M, Miao K, Xu H. SNHG1 contributes to proliferation and invasion by regulating miR-382 in breast cancer. Cancer Manag Res 2019; 11:5589–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W, Dong X, He C, Tan G, Li Z, Zhai B, Feng J, Jiang X, Liu C, Jiang H, Sun X. LncRNA SNHG1 contributes to sorafenib resistance by activating the akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res 2019; 38:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J 2018; 32:3957–67 [DOI] [PubMed] [Google Scholar]
- 40.Cao Z, Pan X, Yang Y, Huang Y, Shen HB. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 2018; 34:2185–94 [DOI] [PubMed] [Google Scholar]
- 41.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis Cascade in hepatocellular carcinoma. Cancer Cell 2014; 25:666–81 [DOI] [PubMed] [Google Scholar]
- 42.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014; 42:D92–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Sun Y, Wu Y, Tang D, Ding X, Xu W, Su B, Gao W. Downregulation of miR-3127-5p promotes epithelial-mesenchymal transition via FZD4 regulation of wnt/beta-catenin signaling in non-small-cell lung cancer. Mol Carcinog 2018; 57:842–53 [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G, Song H. LncRNA SNHG1 enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol 2018; 96:38–43 [DOI] [PubMed] [Google Scholar]
- 45.Ma X, Yan W, Dai Z, Gao X, Ma Y, Xu Q, Jiang J, Zhang S. Baicalein suppresses metastasis of breast cancer cells by inhibiting EMT via downregulation of SATB1 and wnt/beta-catenin pathway. Drug Des Devel Ther 2016; 10:1419–41 [DOI] [PMC free article] [PubMed] [Google Scholar]