Abstract
MicroRNAs (miRNAs or miRs) are non-coding small RNAs that target specific messenger RNAs to inhibit protein translation. miR-200a and miR-141 function as tumor suppressors by targeting STAT4. These two miRNAs belong to the same family, and their expression is often decreased in various cancer types, but are located on different chromosomes of the human genome. The present study showed that the expression levels of miR-141 and miR-200a in serum and cells of liver cancer are significantly downregulated. The expression levels of miR-141 and miR-200a are closely associated with clinicopathological features of liver cancer, especially metastasis and invasion. It is first reported that STAT4 is the new common target gene of miR-141 and miR-200a. In the present study, miR-141 and miR-200a were confirmed to inhibit the expression of E-cadherin and vimentin synergistically during epithelial-mesenchymal transition to regulate the proliferation, migration and invasion of liver cancer cells by targeting STAT4. Simultaneous overexpression of miR-200a and miR-141 resulted in stronger effects compared with each miRNA alone. In addition, overexpression of STAT4 significantly reversed the tumor suppressive roles of miR-200a and miR-141 in liver cancer cells. These findings enrich the tumor suppressor mechanisms of the miR-200 family, and may also provide new experimental and theoretical basis for the use of miRNAs for early diagnosis, prognosis and thorough treatment of liver cancer.
Keywords: liver cancer, microRNA-200a, microRNA-141, STAT4
Introduction
Liver cancer is one of the most common malignant tumors of the digestive system. Liver cancer ranks sixth in the global most-common tumors and is the fourth leading cause of cancer-associated mortality (1). The incidence and mortality of liver cancer are increasing year by year. At present, the treatment of liver cancer is mainly surgical resection, supplemented with radiotherapy and chemotherapy (2). Although the rapid development of modern medical technology has significantly improved the efficiency and safety of resection of liver cancer, the rate of recurrence and metastasis in postoperative patients with liver cancer is high, which markedly lowers the patients long-term survival rate (3). Therefore, elucidating the mechanism of the development and progression of liver cancer and finding more effective biomarkers with high sensitivity and high specificity for early diagnosis and prognosis of liver cancer are still some of the hot topics of cancer research.
MicroRNAs (miRNAs or miRs) are endogenous non-coding small RNAs with 18–22 nucleotides that are found in eukaryotes, and are involved in regulating a variety of complex pathophysiological processes in the human body by complementarily binding to the 3-untranslated region (UTR) of the target gene mRNA (4). Previous studies found that ~50% of miRNAs in chromosomes are localized in tumor-associated fragile sites (5,6). Abnormal expression of miRNAs can promote or inhibit the development of malignancies. Previous studies have reported the roles of a single miRNA in the development of cancer (7–10). Some studies have found that two or more different miRNAs cooperatively participate in the biological process of cancer by targeting the same molecule (4,11,12). miR-200a and miR-141 are two important members of the miR-200 family, which is aberrantly expressed in liver cancer (13). miR-200a and miR-141 have a similar sequence, which suggests that both have similar biological functions (14,15). Although some studies have reported that miR-200a and miR-141 have low expression in liver cancer and play important tumor-suppressing roles (16–19), whether there is a common biological function between miR-200a and miR-141 in liver cancer and their specific molecular mechanism need to be further explored.
The main purpose of the present study was to identify new common molecule STAT4 targeted by both miR-200a and miR-141 in liver cancer, in order to provide a solid theoretical foundation for accelerating clinical biomedical transformation and early implementation of miRNA-based biomarkers for early screening, diagnosis and prognosis monitoring of liver cancer.
Materials and methods
Human specimens
Blood samples from 30 (22 male and 8 female) patients with liver cancer aged from 25 to 70 (average, 51.2±12.85) years old and 30 (22 male and 8 female) normal subjects aged from 22 to 66 (average, 50.37±11.24) years old were collected from the Affiliated Huashan Hospital of Fudan University (Shanghai, China) between January 2015 and January 2016, with the written consent of the patients and approval by the Ethics Committee of Huashan Hospital (Shanghai, China). Serum was isolated from whole blood by centrifugation at 4°C and 800 × g for 10 min, and then stored at −80°C.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). RT-qPCR assay was performed to quantify miRNA expression levels using miScript II RT kit (Qiagen GmbH) and QuantiTect SYBR Green PCR Master Mix (Qiagen GmbH) according to the manufacturers instructions. Cel-miR-39 was used as a control, and the primers for each miRNA were purchased from Qiagen GmbH. To determine the mRNA levels of STAT4, E-Cadherin, vimentin, GAPDH and U6 small nuclear RNA (RNU6), total RNA was reversely transcribed using PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.). Reverse transcription reaction was performed using SYBR Premix Ex Taq (Takara Bio, Inc.) at 37°C for 15 min and 85°C for 5 sec. qPCR was performed to detect mRNA expression levels using SYBR Premix Ex Taq (Takara Bio, Inc.) on an ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR thermocycling conditions were as follows: 95°C for 30 sec followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. The primers used in this study were: STAT4 forward, 5′-AGCCATCTCGGAGGAATA-3′ and reverse, 5′-CAGACAACCGGCCTTTAT-3′; E-cadherin forward, 5′-TCCATTTCTTGGTCTACGCC-3′ and reverse, 5′-CACCTTCAGCCAACCTGTTT-3′; vimentin forward, 5′-CGGTTGAGACCAGAGATGGA-3′ and reverse, 5′-TGCTGGTACTGCACTGTTGC-3′; RNU6 forward, 5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse, 5′-GGAACGCTTCACGAATTTG-3′; and human GAPDH forward, 5′-CCACCCATGGCAAATTCCATGGCA-3′ and reverse, 5′-TCTAGACGGCAGGTCAGGTCCACC-3′. The expression levels of RNU6 or GAPDH were used as internal controls. The expression levels of mRNAs in each group were calculated by relative quantification using the 2−ΔΔCq method (20).
Cell culture and lentivirus infection
Human liver cancer cell lines (HepG2, Huh7 and MHCC97H), human normal hepatocyte cell (THLE-2) and 293T cell were obtained from the American Type Culture Collection, and cultured in DMEM/high-glucose medium (HyClone; Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (EMD Millipore), at 37°C and 5% CO2. The cell lines were characterized by Genetic Testing Biotechnology Corporation using short tandem repeat profile analysis. GV309 (Shanghai GeneChem Co., Ltd.) was used as an expression plasmid for lentiviral packaging. In this construct, the hU6 promoter is used for miRNA translation, and the ubiquitin promoter is used for the GFP tag, which is not fused. pHelper 1.0 vector plasmid and pHelper 2.0 vector plasmid was used as lentiviral auxiliary packaging plasmid (Shanghai GeneChem Co., Ltd.). The miR-200a or miR-141 sequence was chemically synthesized from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and cloned into the lentiviral expression vector GV309, and the recombinant plasmid containing the target fragment was obtained. GV309-GFP served as negative control (NC). A total of 20 µg recombinant plasmid (GV309-miR-200a-GFP, GV309-miR-141-GFP or GV309-GFP), 15 µg pHelper 1.0 vector plasmid and 10 µg pHelper 2.0 vector plasmid were co-transfected into HEK293T cells used by Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) to produce lentivirus. Virus soups were used to infect liver cancer cells (HepG2). Infected cells were screened with puromycin (Shanghai GeneChem Co., Ltd.). Stable cell lines expressing miR-NC, miR-200a, miR-141 and miR-200a+miR-141 were established and were used for subsequent functional experiments 72 h after lentiviral transfection.
Cell proliferation assay
A total of 5×103 miR-NC, miR-200a, miR-141 or miR-200a+miR-141-transfected cells per well were plated in 96-well plates, and cultured at 37°C in an incubator with 5% CO2. Cell proliferation was measured using a Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc.) according to the manufacturers instructions.
Cell migration and invasion assays
Cell migration and invasion were evaluated using a Transwell assay (Corning Inc.). miR-NC, miR-200a, miR-141 and miR-200a+miR-141-transfected HepG2 cells (1×105 cells/100 µl serum-free medium) were placed into the upper chamber. High-glucose DMEM supplemented with 20% FBS (600 µl) was placed into the lower chamber. Chambers were assembled and incubated at 37°C in the presence of 5% CO2 for 24 h for the migration assay or 48 h for the invasion assay. After the incubation, cells from the upper surface of the membranes were removed, and the migrated cells on the lower surface of the membranes were fixed with methanol for 10 min at 37°C and stained with crystal violet at room temperature for 10 min, and counted in three randomly selected fields using an inverted microscope (magnification, ×100; Nikon TE2000). Representative photographs were selected. For the invasion assay, the upper chambers were pre-incubated with Matrigel for 4 h to form a layer of Matrigel, and the migrated cells in the lower chamber were counted 48 h after plating.
Western blot analysis
Cells were lysed by RIPA lysis buffer containing protease inhibitors (Beyotime Institute of Biotechnology, Inc.). Protein concentration was determined using bicinchoninic acid assay kit. Proteins (30 µg per lane) were separated by PAGE using 10% Bis-Tris gels (Beyotime Institute of Biotechnology, Inc.) and transferred onto a polyvinylidene fluoride membrane. Immunoblotting was performed with diluted antibodies against STAT4 (1:1,000; cat. no. 2653; Cell Signaling Technology, Inc.), E-cadherin (1:1,000; cat. no. 3195; Cell Signaling Technology, Inc.), and vimentin (1:1,000; cat. no. 5741; Cell Signaling Technology, Inc.), and an internal reference antibody against β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.) overnight at 4°C. The membrane was washed with 0.1% Tween-20 in TBST three times and then incubated with goat anti-rabbit IgG (1:5,000; cat. no. HAF008; R&D Systems, Inc.) for 1 h at room temperature. Specific complexes were visualized with an enhanced chemiluminescence detection system (Thermo Fisher Scientific, Inc.).
Luciferase activity assays
psiCHECK-2 (Promega Corporation) was used as a luciferase reporter plasmid. The multiple cloning site region was located behind the T7 promoter in this vector, and contained the Luc marker of gene expression. The target fragment was inserted into the XhoI and NotI sites, and the expression was regulated by the SV40 promoter. Luciferase reporter plasmids containing the wild-type (WT) or mutant (MUT) sequence of the STAT4-3′-UTR region were inserted into the psiCHECK-2 luciferase vector and into a plasmid used as an internal expression control, and then transfected into 293T/miR-NC, 293T/miR-200a or 293T/miR-141 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h after transfection, luciferase assays were performed using a Dual-luciferase reporter assay kit (Promega Corporation). The Renilla luciferase activity was used as internal reference.
Statistical analysis
Values were obtained from at least three independent experiments and presented as the mean ± SD. The receiver operating characteristic (ROC) curve of miR-200a and miR-141 was plotted. When data conforms to normal distribution, comparison between two groups was conducted using the Students t-test while comparisons between multiple groups of data were performed using ANOVA. The Least Significant Difference post hoc test (three groups) and Tukeys test (more than three groups) were used for pairwise comparison between multiple groups. Non-parametric tests (Mann-Whitney U test and Kruskal-Wallis H test) were performed when making comparisons in datasets that are not normally distributed. All statistical analyses were performed using SPSS 19.0 software (SPSS, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of miR-200a and miR-141 expression in serum of patients with liver cancer is closely associated with clinicopathological factors
Circulating miR-200a and miR-141 levels were detected in samples from 30 patients with liver cancer and 30 normal subjects. The expression levels of circulating miR-200a and miR-141 were markedly decreased in the preoperative serum of patients with liver cancer compared with those found in healthy individuals. Besides, the expression level of miR-200a and miR-141 in the postoperative serum of the same patients increased significantly compared with the levels in preoperative serum (Fig. 1A). These results indicate that the expression level of miR-200a or miR-141 can be detected in blood samples, and may be used as a potential biomarker.
Figure 1.
miR-200a and miR-141 are downregulated and closely associated with metastasis and invasion of liver cancer. (A) miR-200a (left panel) and miR-141 (right panel) expression levels in the serum of healthy individuals, preoperative serum of liver cancer patients and postoperative serum of patients with liver cancer were detected by qPCR, and the endogenous nematode cel-miR-39 was used as the internal reference. (B) ROC curve analysis of miR-200a, miR-141 and their combination for evaluating metastasis (left panel) and invasion (right panel) of liver cancer. (C) miR-200a (left panel) and miR-141 (right panel) in human normal hepatocyte cell (THLE-2) and liver cancer cells (HepG2, Huh7 and MHCC97H) were detected by qPCR and U6 was used as the internal reference. *P<0.05; **P<0.01; ***P<0.001. miR, microRNA; qPCR, quantitative PCR; ROC, receiver operating characteristic.
The present study further evaluated the association between the preoperative serum level of miR-200a/miR-141 and clinicopathological factors, which is summarized in Table I. The expression of miR-200a was significantly associated with tumor size (P=0.001), Barcelona Clinic Liver Cancer (BCLC) stage (P=0.002), differentiation (P=0.011), metastasis (P=0.026), invasion (P=0.024) and recurrence (P=0.029), while the expression of miR-200a was not significantly associated with sex, age, α-fetoprotein (AFP), virus infection or cirrhosis (P>0.05). The expression of miR-141 was associated with the status of liver cancer metastasis and invasion (P<0.01, P<0.05), but not with sex, age, AFP, virus infection, cirrhosis, tumor size, BCLC staging, differentiation or recurrence. These results show that miR-200a and miR-141 are both associated with metastasis and invasion in patients with liver cancer.
Table I.
Associations between the expression levels of miR-200a/miR-141 in preoperative serum, and clinicopathological factors of patients with hepatocellular carcinoma.
| miR-200a | miR-141 | ||||
|---|---|---|---|---|---|
| Clinicopathological parameters | N | Median | P-value | Median | P-value |
| Sex | 0.476 | ||||
| Male | 22 | 0.370 | 0.781 | 0.435 | |
| Female | 8 | 0.425 | 0.535 | ||
| Age (years) | 0.950 | ||||
| ≤55 | 12 | 0.415 | 0.755 | 0.400 | |
| >55 | 18 | 0.370 | 0.490 | ||
| AFP (ng/ml) | 0.471 | ||||
| ≤20 | 11 | 0.370 | 0.767 | 0.490 | |
| >20 | 19 | 0.370 | 0.380 | ||
| Viral infection | 0.389 | ||||
| With | 15 | 0.370 | 0.806 | 0.420 | |
| Without | 15 | 0.370 | 0.490 | ||
| Cirrhosis | 0.432 | ||||
| With | 17 | 0.370 | 0.483 | 0.310 | |
| Without | 13 | 0.370 | 0.530 | ||
| Tumor size (cm) | 0.746 | ||||
| ≤5 | 19 | 0.530 | 0.001b | 0.420 | |
| >5 | 11 | 0.190 | 0.450 | ||
| BCLC stage | 0.9510 | ||||
| A | 14 | 0.650 | 0.002b | 0.435 | |
| B + C | 16 | 0.210 | 0.435 | ||
| Differentiation | 0.6500 | ||||
| Middle or high | 13 | 0.650 | 0.011a | 0.380 | |
| Low | 17 | 0.280 | 0.450 | ||
| Migration | 0.003b | ||||
| With | 15 | 0.28 | 0.026a | 0.31 | |
| Without | 15 | 0.48 | 0.69 | ||
| Invasion | 0.018a | ||||
| With | 8 | 0.170 | 0.024a | 0.315 | |
| Without | 22 | 0.380 | 0.635 | ||
| Recrudescence | 0.781 | ||||
| With | 6 | 0.115 | 0.029a | 0.435 | |
| Without | 24 | 0.380 | 0.455 | ||
P<0.05
P<0.01. miR, microRNA; AFP, α-fetoprotein; BCLC, Barcelona Clinic Liver Cancer.
Furthermore, ROC curve was used to analyze the sensitivity and specificity of miR-200a and miR-141 (individually and combined) for evaluating metastasis and invasion in liver cancer (Fig. 1B). The area under the curve (AUC) was also calculated. As shown in Table II, the AUC values of miR-200a, miR-141 and their combination for comparing metastasis of patients with liver cancer were 0.696 (P>0.05), 0.783 (P<0.01) and 0.795 (P<0.01), respectively. There was no significant difference between the three AUC values. The sensitivity and specificity of miR-200a, miR-141 and their combined detection for diagnosing metastasis of liver cancer were 50.0/50.0, 78.6/68.8 and 78.6/75.0%, respectively (Table II). Although there was no significant difference between the three AUC values, the AUC values in the combined group were higher than those using miR-200a or miR-141 alone. Moreover, the sensitivity and specificity of the combined group were higher compared with those of miR-200a or miR-141 alone, which suggests that the combination of miR-200a and miR-141 can help to improve the AUC values, specificity and sensitivity of diagnosing metastasis in patients with liver cancer, and have potential use in early clinical diagnosis.
Table II.
AUC value of miR-200a, miR-141 and their combination for evaluating the metastasis of hepatocellular carcinoma.
| Index | AUC (x±s) | P-value | 95% CI | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| miR-200a | 0.696±0.097 | 0.067 | 0.506–0.887 | 50.0 | 50.0 |
| miR-141 | 0.783±0.087 | 0.008 | 0.613–0.954 | 78.6 | 68.8 |
| miR-200a+miR-141 | 0.795±0.083 | 0.006 | 0.632–0.957 | 78.6 | 75.0 |
miR, microRNA; AUC, area under the curve.
As shown in Table III, the AUC values of miR-200a, miR-141 and their combination for comparing tumor invasion in patients with liver cancer were 0.773 (P<0.05), 0.781 (P<0.05) and 0.892 (P<0.01), respectively. There was no significant difference between the three AUC values. The sensitivity and specificity of miR-200a, miR-141 and their combined detection for diagnosing tumor invasion in liver cancer were 62.5/77.3, 62.5/72.7 and 75.0/86.4%, respectively (Table III). Although there were no significant differences between the three AUC values, the AUC values in the combined group were higher compared with those using miR-200a or miR-141 alone. Moreover, the sensitivity and specificity of the combined group were higher than those of miR-200a or miR-141 alone, which suggests that the combination of miR-200a and miR-141 can help to improve the AUC values, specificity and sensitivity of diagnosing tumor invasion in patients with liver cancer, and have a potential use in early clinical diagnosis.
Table III.
Area under the curve value of miR-200a, miR-141 and their combination for evaluating the invasion of hepatocellular carcinoma.
| Index | AUC (x±s) | P-value | 95% CI | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| miR-200a | 0.773±0.099 | 0.024 | 0.579–0.967 | 62.5 | 77.3 |
| miR-141 | 0.781±0.085 | 0.020 | 0.616–0.947 | 62.5 | 72.7 |
| miR-200a+miR-141 | 0.892±0.065 | 0.001 | 0.764–1.000 | 75.0 | 86.4 |
miR, microRNA; AUC, area under the curve.
The results of ROC curve showed that the combination of miR-200a and miR-141 can improve the sensitivity and specificity of diagnosis for patients with liver cancer exhibiting metastasis or tumor invasion. Therefore, miR-200a and miR-141 may be involved in the development of liver cancer, especially the processes of metastasis and invasion.
Overexpression of miR-200a and miR-141 has a combined effect on inhibiting the proliferation, migration and invasion of liver cancer cells
The results of serology revealed that miR-200a and miR-141 are downregulated in the preoperative serum of patients with liver cancer and can be used as potential non-invasive markers. To further explore the expression of miR-200a and miR-141 in hepatocarcinoma cell lines, qPCR was used to analyze the expression levels of miR-200a and miR-141 in a human normal hepatocyte cell line (THLE-2) and four hepatoma cell lines (HepG2, Huh7 and MHCC97H). The results showed that the expression levels of miR-200a and miR-141 in these four hepatocarcinoma cell lines were downregulated to different degrees compared with those in THLE-2 (Fig. 1C). As HepG2 cells are easy to be cultured and are commonly used for co-transfection, HepG2 cells were selected for subsequent experiments.
To evaluate the function of miR-200a and miR-141 in liver cancer cells and to explore whether there is a combined effect of miR-200a and miR-141, GV309 (whose map is shown in Fig. 2A) was used as an expression plasmid for lentiviral packaging. Stable cell lines expressing NC (HepG2/miR-NC), miR-200a (HepG2/miR-200a), miR-141 (HepG2-miR/141) and miR-200a+miR-141 (HepG2/miR-200a+miR-141) were established by lentiviral transduction. The expression levels of miR-200a and miR-141 in each stable strain were analyzed by qPCR. The results showed that the levels of miR-200a in stable HepG2/miR-200a and HepG2/miR-200a+miR-141 cells were significantly higher compared with those in the control group (P<0.001 both), which indicates that stable cell lines expressing miR-200a were successfully constructed (Fig. 2A). Similarly, the expression levels of miR-141 in the stable strains HepG2/miR-141 and HepG2/miR-200a+miR-141 were significantly higher compared with those in the control group (P<0.001 both), which showed that stable cell lines expressing miR-141 were successfully constructed (Fig. 2A).
Figure 2.
miR-200a and miR-141 synergistically inhibit the proliferation, migration and invasion of liver cancer cells. (A) The GV309 vector map is shown (left panel) and the levels of miR-200a (middle panel) and miR-141 (right panel) were detected using reverse transcription-quantitative PCR in stable cell lines overexpressing miR-200a, miR-141 or miR-negative control. (B) The morphological changes of HepG2/miR-NC, HepG2/miR-200a, HepG2/miR-141 and HepG2/miR-200a+miR-141 were observed by microscope (magnification, ×100). (C) The effects of miR-200a, miR-141 and miR-200a+miR-141 overexpression on cell proliferation were analyzed using a Cell Counting Kit-8 assay. (D) Transwell assay was used to measure cell migration in each stable cell line. (E) Transwell assay was used to measure cell invasion in each stable cell line (magnification, ×100; Nikon TE2000). *P<0.05; **P<0.01, ***P<0.001. miR, microRNA.
In the present study, cellular morphology was monitored. There were some differences between miR-200a- or miR-141-overexpressing cells and control cells. HepG2/miR-NC cells were loosely adhered to each other, slender and looked spindle-like, showing the appearance of mesenchymal cells. HepG2/miR-200a, HepG2/miR-141 or HepG2/miR-200a+miR-141 cells were closely connected to each other and polygonal arranged in a cobblestone-like arrangement, which is a typical epithelial cell morphology (Fig. 2B).
Meanwhile, the effects of miR-200a and miR-141 on cell proliferation, migration and invasion were also assessed. Overexpression of miR-200a or miR-141 in liver cancer cells attenuated cell proliferation (Fig. 2C). Overexpression of miR-200a or miR-141 in liver cancer cells decreased migration and invasion compared with the effects observed in miR-NC-transfected cells (Fig. 2D and E). It is noteworthy that the inhibition of cell function was most obvious in the stable cell line overexpressing simultaneously miR-200a and miR-141.
The results of the above biological function experiments showed that miR-200a and miR-141 exert synergistic inhibitory effects on the proliferation, migration and invasion of hepatocarcinoma cells.
Downregulation of miR-200a and miR-141 enhances the epithelial-mesenchymal transition (EMT) through targeting STAT4
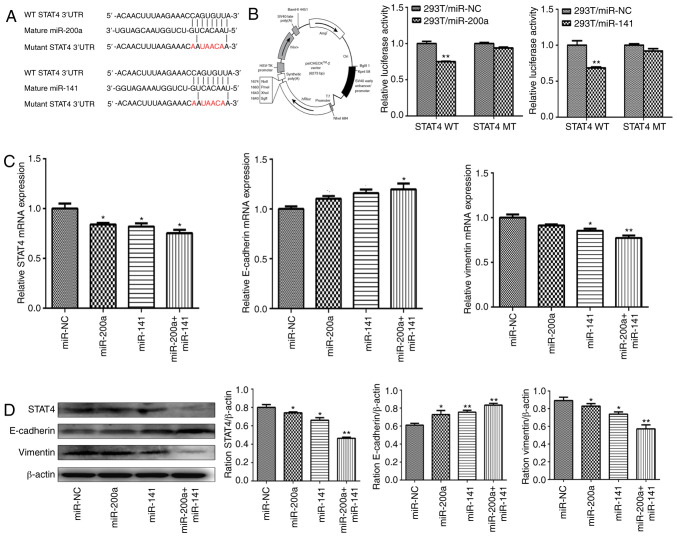
To further understand the molecular mechanism of miR-200a and miR-141 in synergistically inhibiting tumorigenesis, potential common targets of miR-200a and miR-141 were searched using dedicated software. First, published literature on the miR-200 family was searched through PubMed (http://www.ncbi.nlm.nih.gov/pubmed), and the miRBase (http://www.mirbase.org/index.shtml) online tool was used to obtain the base sequence, chromosome location and target prediction of miR-200a and miR-141. Next, TargetScan (www.targetscan.org), picTar (https://pictar.mdc-berlin.de/) and miRDB (www.mirdb.org) were applied to predict the common targets of miR-200a and miR-141, and their intersection was considered as the target gene set for further analysis. The three calculation methods predicted that STAT4 is a potential novel direct target of miR-200a and miR-141 with the binding site at its 3′-UTR region. Reporter luciferase vectors containing the WT or MUT miR-200a/miR-141 binding site of STAT4 3′-UTR were constructed (Fig. 3A). psiCHECK-2, whose map is shown in Fig. 3B, was used as a luciferase reporter plasmid. Luciferase activity assay showed that overexpression of miR-200a and miR-141 markedly decreased the activity of the WT reporter to 24.89 and 31.58%, respectively (P<0.01 both), but not of the MUT reporter (Fig. 3B), suggesting that miR-200a and miR-141 both inhibited the 3-UTR function of STAT4, and the point mutation of this target sequence abolished the effect of miR-200a or miR-141. The present study found that overexpression of miR-200a or miR-141 significantly downregulated the mRNA and protein levels of STAT4 in liver cancer cell lines (Fig. 3C and D).
Figure 3.
miR-200a and miR-141 regulate epithelial-mesenchymal transition progression in liver cancer through targeting STAT4. (A) Schematic diagram of the putative miR-200a/miR-141 binding site in the 3′-UTR of the human STAT4 gene. The mutated nucleotides of the STAT4 3′-UTR are labeled in red. (B) The psiCHECK-2 vector map is shown (left) and 293T cells were co-transfected with miR-NC, miR-200a or miR-141 mimics along with a wild-type or mutant STAT4 luciferase reporter (middle and right). Luciferase activities were measured 48 h after transfection using the dual-luciferase reporter assay system. (C) The mRNA expression levels of STAT4 (left), E-cadherin (middle) and vimentin (right) were measured by reverse transcription-quantitative PCR in HepG2 cells stably overexpressing miR-NC, miR-200a, miR-141 and miR-200a+miR-141, and were normalized to the level of GAPDH. (D) The protein expression levels of STAT4, E-cadherin and vimentin were determined by western blotting in HepG2 stably overexpressing miR-NC, miR-200a, miR-141 and miR-200a+miR-141. β-actin was used as an internal control. *P<0.05; **P<0.01. UTR, untranslated region; STAT4, signal transducer and activator of transcription 4; NC, negative control; miR, microRNA.
EMT is an important way for epithelial cells to obtain migration and invasion abilities, which plays an important role in tumor metastasis and invasion. The change in E-cadherin and vimentin protein is an important molecular characteristic of the EMT process. Consistent with the decrease in STAT4, overexpression of miR-200a and miR-141 in HepG2 cells also inhibited EMT progress (Fig. 3C and D). Furthermore, this inhibition was most pronounced in the combined group.
Forced expression of STAT4 restores miR-200a/miR-141-inhibited EMT progress
Some studies have shown that abnormal expression of STAT4 causes changes in EMT-associated molecules. Therefore, the present study further explored whether miR-200a and miR-141 regulate the downstream EMT process via targeting STAT4. Stable cell lines expressing miR-200a, miR-141 and miR-200a+miR-141 were transfected with lentivirus carrying STAT4. The proteins of each group were extracted, and the expression of E-cadherin, vimentin and STAT4 was detected by western blotting. The results showed that restoring the expression level of STAT4 partially reverses the inhibitory effect produced by miR-200a and miR-141 on the EMT process (Fig. 4).
Figure 4.
Overexpression of STAT4 reverses the inhibitory effects of miR-200a and miR-141 on epithelial-mesenchymal transition progression. (A) HepG2 cells stably expressing miR-negative control, miR-200a, miR-141 or miR-200a+miR-141 were transduced with lentivirus carrying STAT4. Western blotting was used to detect the expression level of STAT4, E-cadherin and vimentin in different treatment groups. β-actin was used as the internal reference. (B) Histogram of each protein expression levels in different treatment groups. *P<0.05; **P<0.01. STAT4, signal transducer and activator of transcription 4; miR, microRNA.
Discussion
The major finding of the present study is that miR-200a and miR-141 synergistically inhibit the expression of the same target gene, STAT4, thereby restraining the EMT process of liver cancer. Several studies have demonstrated that miR-141 retards liver cancer cell growth or enhances chemical sensitivity by targeting sperm associated antigen 9 (SPAG9) (21), zinc finger E-box binding (ZEB1) (22), kelch like ECH associated protein 1 (Keap1) (23) or hepatocyte nuclear factor-3β (HNF-3β) (24), whereas miR-200a does so by targeting MET transcriptional regulator MACC1 (17), Grb2-associated binding protein 1 (GAB1) (16) and dual-specific phosphatase 6 (DUSP6) (25), in liver cancer. The association between miRNA and target can be one-to-many or many-to-one; that is, one miRNA can inhibit many proteins, and one protein can be regulated by many miRNAs. Previous studies have found that the miR-200 family can directly target the E-box-binding transcription regulators ZEB1 and ZEB2, causing abnormal expression of E-cadherin and vimentin in cancer cell lines, thereby inhibiting the tumor EMT process (26,27). Although the present study showed that the common target genes ZEB1 and ZEB2 of the miR-200 family play a crucial role in the EMT process, they did not clearly clarify the specific synergy. The present study provides a better insight into the synergetic role of miR-141 and miR-200a by co-targeting STAT4 in liver cancer cells. To the best of our knowledge, this is the first study to demonstrate the cooperative effect of miR-200a and miR-141 on inhibiting the EMT of liver cancer by targeting a new gene (STAT4) directly.
miR-200a and miR-141 belong to the same miRNA family, miR-200, which is one of the miRNAs families found to be expressed abnormally in numerous tumors (28–30). In the present study, the expression levels of miR-200a and miR-141 were first detected in serum. It was found that, compared with those in the serum of healthy subjects and in the postoperative serum of patients with liver cancer, the expression levels of miR-200a and miR-141 were downregulated in the preoperative serum of patients. The present study further analyzed the association between the expression levels of miR-200a and miR-141 in the preoperative serum of patients and clinicopathological features. The results showed that the relative expression levels of miR-200a and miR-141 were significantly associated with metastasis and invasion in patients with liver cancer. The combination of miR-200a and miR-141 can improve the sensitivity and specificity of the diagnosis of liver cancer with metastasis and invasion. These results suggest that miR-200a and miR-141 may play important roles in mediating the genesis and development of liver cancer. The combined detection of miR-200a and miR-141 in serum has potential clinical application for early screening, diagnosis and prognosis of liver cancer.
Previous studies have shown that miRNAs from the same cluster can play a synergistic role in cancer (31,32). Although miR-200a and miR-141 are located on chromosomes 1p36.33 and12p13.31, respectively, they share the same target sequence (AACACUG in 5 to 3 orientation) (33,34). The present and other studies have reported that miR-200a and miR-141 have low expression in liver cancer and play important roles in suppressing tumors. Thus, the present study aimed to investigate whether there is a common biological function in the process of hepatocellular growth, metastasis and invasion.
To further clarify these questions, a series of stable cell lines of liver cancer overexpressing miR-200a, overexpressing miR-141, and simultaneously overexpressing miR-200a and miR-141, were successively constructed. The biological function and common mechanism of miR-200a and miR-141 were studied at the cellular level, respectively. The results showed that overexpression of miR-200a or miR-141 in hepatocarcinoma cell lines significantly inhibited cell proliferation, migration, invasion and EMT. In vitro inhibition was most pronounced in cell lines overexpressing miR-200a and miR-141 simultaneously, indicating that miR-200a and miR-141 have synergistic effects on inhibiting the biological function of liver cancer. Next, the potential molecular mechanism of this phenomenon was further studied. STAT4, a member of the STAT family, is an important transcription factor that regulates the expression of various molecules in vivo, and mediates cell proliferation, apoptosis, metastasis and other processes (35–37). In recent years, numerous studies have shown that single nucleotide polymorphisms of STAT4 are closely associated with the occurrence and development of liver cancer (38,39). The present study used TargetScan, PicTar, microRNA and other biological information software to predict that STAT4 may be a common target gene of miR-200a and miR-141. Although it has been shown that miR-141 can inhibit the growth of gastric cancer by targeting STAT4 (40), whether miR-200a can target STAT4 and whether miR-141 can also mediate the expression of STAT4 in liver cancer are still not reported. The present study used bioinformatics software, luciferase reporter gene assay, qPCR and western blotting to predict and confirm that STAT4 is a direct and common target gene of miR-200a and miR-141. As an important transcription factor, STAT4 is upregulated in multiple tumors, and promotes tumor metastasis and invasion through various mechanisms, especially regulation of EMT. Zhao et al (41) found that activated STAT4 was overexpressed in epithelial cells of ovarian cancer, and mediated the metastasis and invasion of ovarian cancer via the EMT process. EMT is a process by which epithelial cells transform into interstitial cells in a dynamic way. This change causes cells to lose polarity; that the adhesion molecules on the cell surface are expressed abnormally; and that cells acquire some characteristics of mesenchymal cells. This process is also an important way to make cells involved in metastasis and invasion. The abnormal expression of E-cadherin and vimentin is one of the most important features of EMT progression. Thus, the present study aimed to clarify whether miR-200a and miR-141 affect the expression levels of two downstream EMT-associated important molecules such as E-cadherin and vimentin to participate in the process of EMT by targeting STAT4, which promotes the progression of tumors. For that purpose, stable cell lines overexpressing miR-200a, miR-141 and miR-200a+miR-141 were cultured with lentivirus carrying the STAT4 gene, which restored the expression of STAT4 inhibited by miR-200a or miR-141 in stable strains. The changes in expression of E-cadherin and vimentin in the stable strains were detected by western blotting. The results showed that restoring the expression levels of STAT4 could partly reverse the expression changes of downstream EMT-related characteristic molecules.
In summary, this study conducted investigations at a clinical and cellular level, and showed that the expression levels of miR-200a and miR-141 in the preoperative serum of patients with liver cancer were downregulated. The expression levels of miR-200a and miR-141 were reported to be closely associated with clinicopathological features of liver cancer, especially metastasis and invasion. This study is the first to report that STAT4 is the new common target gene of miR-200a and miR-141. miR-200a and miR-141 were confirmed to inhibit the expression of E-cadherin and vimentin in EMT synergistically to regulate the proliferation, migration and invasion of hepatocarcinoma cells by targeting STAT4 at a cellular level. These results enrich the knowledge on the tumor suppressor mechanism of the miR-200 family, and also provide a new experimental and theoretical basis for the use of miRNAs for the early diagnosis, prognosis and thorough treatment of liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Key Research and Development Plan of China (grant no. 2018YFC2000200), the National Natural Science Foundation of China (grant no. 81772673), the Shanghai Sailing Program (grant no. 19YF1405500) and the Initial Scientific Research Fund of the Huashan Hospital Affiliated to Fudan University (grant no. 2019QD003).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors contributions
YM, WC, CL and HW collected the case material and collected the data. SC, JZ, JC and XC performed the experiments and analyzed the data. SC, YM, WC, CL and HW drafted the manuscript and revised the manuscript. QC, YuL and YoL conceived and designed the study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by The Ethics Committee of Huashan Hospital Affiliated to Fudan University (approval no. 557, 2019). All the healthy individuals and patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380:1450–1462. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American association for the study of liver diseases. Hepatology. 2018;68:723–750. doi: 10.1002/hep.29913. [DOI] [PubMed] [Google Scholar]
- 3.Ye J, Wu S, Pan S, Huang J, Ge L. Risk scoring based on expression of long non-coding RNAs can effectively predict survival in hepatocellular carcinoma patients with or without fibrosis. Oncol Rep. 2020;43:1451–1466. doi: 10.3892/or.2020.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heusschen R, van Gink M, Griffioen AW, Thijssen VL. MicroRNAs in the tumor endothelium: Novel controls on the angioregulatory switchboard. Biochim Biophys Acta. 2010;1805:87–96. doi: 10.1016/j.bbcan.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Xu WP, Liu JP, Feng JF, Zhu CP, Yang Y, Zhou WP, Ding J, Huang CK, Cui YL, Ding CH, et al. MiR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut. 2020;69:1309–1321. doi: 10.1136/gutjnl-2019-318830. [DOI] [PubMed] [Google Scholar]
- 8.Lin J, Shen J, Yue H, Cao Z. MiRNA-183-5p.1 promotes the migration and invasion of gastric cancer AGS cells by targeting TPM1. Oncol Rep. 2019;42:2371–2381. doi: 10.3892/or.2019.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Xu S, Xu J, Li Y, Zhang J, Zhang J, Lu X. MiR-767-5p inhibits glioma proliferation and metastasis by targeting SUZ12. Oncol Rep. 2019;42:55–66. doi: 10.3892/or.2019.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velazquez-Torres G, Shoshan E, Ivan C, Huang L, Fuentes-Mattei E, Paret H, Kim SJ, Rodriguez-Aguayo C, Xie V, Brooks D, et al. A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat Commun. 2018;9:461. doi: 10.1038/s41467-018-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khella HW, Bakhet M, Allo G, Jewett MA, Girgis AH, Latif A, Girgis H, Von Both I, Bjarnason GA, Yousef GM. MiR-192, miR-194 and miR-215: A convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis. 2013;34:2231–2239. doi: 10.1093/carcin/bgt184. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W, Qian P, Zhang X, Zhang M, Wang H, Wu M, Kong X, Tan S, Ding K, Perry JK. Autocrine/paracrine human growth hormone-stimulated MicroRNA 96-182-183 cluster promotes epithelial-mesenchymal transition and invasion in breast cancer. J Biol Chem. 2015;290:13812–13829. doi: 10.1074/jbc.M115.653261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng X, Wang Z, Fillmore R, Xi Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014;344:166–173. doi: 10.1016/j.canlet.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belgardt BF, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, von Meyenn F, Villena FN, Herrmanns K, Bosco D, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 15.Kim YK, Wee G, Park J, Kim J, Baek D, Kim JS, Kim VN. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol. 2013;20:1458–1464. doi: 10.1038/nsmb.2701. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Song W, Shen W, Yang X, Sun W, Qu S, Shang R, Ma B, Pu M, Tao K, et al. MicroRNA-200a suppresses cell invasion and migration by directly targeting GAB1 in hepatocellular carcinoma. Oncol Res. 2017;25:1–10. doi: 10.3727/096504016X14685034103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng J, Wang J, Chen M, Chen G, Wu Z, Ying L, Zhuo Q, Zhang J, Wang W. MiR-200a suppresses cell growth and migration by targeting MACC1 and predicts prognosis in hepatocellular carcinoma. Oncol Rep. 2015;33:713–720. doi: 10.3892/or.2014.3642. [DOI] [PubMed] [Google Scholar]
- 18.Hou X, Yang L, Jiang X, Liu Z, Li X, Xie S, Li G, Liu J. Role of microRNA-141-3p in the progression and metastasis of hepatocellular carcinoma cell. Int J Biol Macromol. 2019;128:331–339. doi: 10.1016/j.ijbiomac.2019.01.144. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Xu Z, Zhou J, Yang H. MiR-141 inhibits proliferation, migration and invasion in human hepatocellular carcinoma cells by directly downregulating TGFβR1. Oncol Rep. 2019;42:1656–1666. doi: 10.3892/or.2019.7325. [DOI] [PubMed] [Google Scholar]
- 20.Mansini AP, Pisarello MJ, Thelen KM, Cruz-Reyes M, Peixoto E, Jin S, Howard BN, Trussoni CE, Gajdos GB, LaRusso NF, et al. MicroRNA (miR)-433 and miR-22 dysregulations induce histone-deacetylase-6 overexpression and ciliary loss in cholangiocarcinoma. Hepatology. 2018;68:561–573. doi: 10.1002/hep.29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lou G, Dong X, Xia C, Ye B, Yan Q, Wu S, Yu Y, Liu F, Zheng M, Chen Z, Liu Y. Direct targeting sperm-associated antigen 9 by miR-141 influences hepatocellular carcinoma cell growth and metastasis via JNK pathway. J Exp Clin Cancer Res. 2016;35:14. doi: 10.1186/s13046-016-0289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng L, Xu M, Xu J, Wu K, Fang Q, Liang Y, Zhou S, Cen D, Ji L, Han W, Cai X. ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis. 2018;9:387. doi: 10.1038/s41419-018-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Pan C, Wei X, Shi Y, Zheng J, Lin X, Shi L. lncRNA KRAL reverses 5-fluorouracil resistance in hepatocellular carcinoma cells by acting as a ceRNA against miR-141. Cell Commun Signal. 2018;16:47. doi: 10.1186/s12964-018-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin L, Liang H, Wang Y, Yin X, Hu Y, Huang J, Ren T, Xu H, Zheng L, Chen X. MicroRNA-141 inhibits cell proliferation and invasion and promotes apoptosis by targeting hepatocyte nuclear factor-3β in hepatocellular carcinoma cells. BMC Cancer. 2014;14:879. doi: 10.1186/1471-2407-14-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, Kim C, Kang H, Tak H, Ahn S, Yoon SK, Kuh HJ, Kim W, Lee EK. MicroRNA-200a-3p increases 5-fluorouracil resistance by regulating dual specificity phosphatase 6 expression. Exp Mol Med. 2017;49:e327. doi: 10.1038/emm.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 27.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15:180–191. doi: 10.1593/neo.121828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang GL, Sun J, Lu Y, Liu Y, Cao H, Zhang H, Calin GA. MiR-200 family and cancer: From a meta-analysis view. Mol Aspects Med. 2019;70:57–71. doi: 10.1016/j.mam.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M. MiR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villadsen SB, Bramsen JB, Ostenfeld MS, Wiklund ED, Fristrup N, Gao S, Hansen TB, Jensen TI, Borre M, Ørntoft TF, et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br J Cancer. 2012;106:366–374. doi: 10.1038/bjc.2011.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michael MZ, OConnor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 35.Miklossy G, Hilliard TS, Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat Rev Drug Discov. 2013;12:611–629. doi: 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 37.Zhang YS, Xin DE, Wang Z, Song X, Sun Y, Zou QC, Yue J, Zhang C, Zhang JM, Liu Z, et al. STAT4 activation by leukemia inhibitory factor confers a therapeutic effect on intestinal inflammation. EMBO J. 2019;15:e99595. doi: 10.15252/embj.201899595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Xu K, Liu C, Chen J. Meta-analysis reveals an association between signal transducer and activator of transcription-4 polymorphism and hepatocellular carcinoma risk. Hepatol Res. 2017;47:303–311. doi: 10.1111/hepr.12733. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Liang L, Liu Y, Luo Y, Liang X, Luo D, Feng Z, Dang Y, Yang L, Chen G. Clinicopathological significance of STAT4 in hepatocellular carcinoma and its effect on cell growth and apoptosis. Onco Targets Ther. 2016;9:1721–1734. doi: 10.2147/OTT.S100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Y, Zhong JH, Gong FS, Xiao J. MiR-141-3p suppresses gastric cancer induced transition of normal fibroblast and BMSC to cancer-associated fibroblasts via targeting STAT4. Exp Mol Pathol. 2019;107:85–94. doi: 10.1016/j.yexmp.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L, Ji G, Le X, Luo Z, Wang C, Feng M, Xu L, Zhang Y, Lau WB, Lau B, et al. An integrated analysis identifies STAT4 as a key regulator of ovarian cancer metastasis. Oncogene. 2017;36:3384–3396. doi: 10.1038/onc.2016.487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.