Abstract
The effects of microRNAs (miRNAs/miRs) on glioblastoma have attracted the attention of researchers in the last 7 years. However, the role of miR-640 and its targeted gene, Slit guidance ligand 1 (SLIT1), in the development of glioblastoma are not yet fully understood. The present study aimed to investigate the role of miR-640 in the proliferation and adhesion of glioblastoma. Reverse transcription-quantitative PCR analysis was performed to detect miR-640 and SLIT1 expression in glioblastoma tissues and cells. In addition, the Dual-luciferase reporter and RNA-pull down assays were performed to assess the association between miR-640 and SLIT1. The Cell Counting Kit-8, BrdU ELISA, cell adhesion and caspase-3 activity assays were also performed to assess cell viability, proliferation, adhesion and apoptosis of glioblastoma cells, respectively. The results demonstrated that miR-640 expression was upregulated in glioblastoma tissues and cells. In addition, miR-640 promoted the cell viability, proliferation and adhesion of glioblastoma cells, while inhibiting cell apoptosis. SLIT1, a direct downstream target of miR-640, was demonstrated to be downregulated in glioblastoma tissues and cells. Furthermore, overexpression of SLIT1 attenuated the promotive effect of miR-640 on glioblastoma cells. Taken together, these results suggest that miR-640 accelerates the proliferation and adhesion of glioblastoma cell lines by targeting and suppressing SLIT1.
Keywords: glioblastoma, microRNA-640, Slit guidance ligand 1
Introduction
Glioblastoma refers to an astrocytic tumor that is typically observed in the nervous system (1). Glioblastoma has a 97% mortality rate during advanced age (>60 years) in Finland (2), not only because the brain and spinal cord are complex and difficult-to-understand, but also because the existing surgical techniques have limited benefits (3). Additionally, glioblastoma has a poor prognosis, thereby contributing to the low 10.1% 3-year survival rate and the high recurrence rate of patients with this cancer (4,5). Recently, the survival rate of patients with glioblastoma has considerably improved with the use of standard surgical treatments, conventional radiotherapy and medication such as temozolomide (6–8). Increasing research on immunotherapy has also provided novel insights into glioblastoma treatments (9). While considerable effort and notable progress have been made to improve the outcomes of patients with glioblastoma, researchers are yet to determine the underlying molecular mechanisms that trigger the development of glioblastoma.
Regarded as small non-coding single-stranded RNAs encoded by an endogenous gene, microRNAs (miRNAs/miRs) were discovered to play crucial regulatory roles in the development of cancer by regulating mRNA degradation or translational inhibition of downstream target genes (10–12). Previous studies have reported that miRNAs act as tumor suppressors or promoters in the abnormal biological process of glioblastoma (13–15). Despite the discovery of several miRNAs associated with the development and progression of glioma (16–18), researchers are yet to investigate the effects of other miRNAs on glioblastoma cells (19–21). For example, miR-640 inhibits the formation of capillaries (22); however, its role in cancer development has not yet been investigated. In their research, Li et al (23) and Zhai et al (24) both demonstrated that miR-640 exerts tumor-suppressive effects in ovarian cancer and hepatocellular carcinoma. In another study, miR-640 was reported to enhance the development of prostate cancer (25). However, the role of miR-640 in glioblastoma remains unclear.
Slit guidance ligand 1 (SLIT1) is located on chromosome 10q24.1 and consists of 37 exons (26). SLIT1 was first identified as a member of the axon guidance molecule SLITs family, which is predominantly located in the central nervous system, and mediates axon branching, elongation and metastatic repulsion of nerve cells (27,28). Previous studies have demonstrated that SLITs regulate the signaling pathways of different types of cancer, including breast cancer and colorectal cancer (29,30). It has also been reported that low levels of SLIT1, caused by methylation, can further deteriorate colorectal cancer (31). SLITs exhibit different expression levels in different subtypes of gastric cancer (32). Although Amodeo et al (33) demonstrated the involvement of SLIT1 in glioblastoma cell migration, the influence of SLIT1 on glioblastoma is not yet fully understood.
The present study aimed to investigate the underlying molecular mechanism of glioblastoma by assessing the regulatory impact of miR-640 and its target genes on the progression of glioblastoma cells. The results demonstrated that miR-640 exerted a cancer-promoting effect on glioblastoma cells by targeting SLIT1. Taken together, these results provide novel insight to help decrease the mortality rate of patients with glioblastoma.
Materials and methods
Microarray analysis
A total of two mRNA microarray datasets [GSE90886 (34) and GSE104291 (35)] were obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The data were subjected to microarray analysis using limma 3.26.8 (36), with P<0.05. Similarly, the GSE103229 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103229) was downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds), and microarray analysis of the miRNAs was performed using limma 3.26.8, with P<0.05 and log-fold change (logFC)>0. The data were subsequently uploaded onto Metascape (https://metascape.org/gp/index.html#/main/step1), UALCAN (http://ualcan.path.uab.edu/index.html) for The Cancer Genome Atlas analysis, ENCORI Pan-Cancer Analysis Platform, ENCORI Pan-Cancer Analysis Platform (http://starbase.sysu.edu.cn/panCancer.php), miRDB (http://mirdb.org) and Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/) for gene enrichment and gene analyses.
Tissue acquisition and cell culture
The present study was approved by the Ethics Committee of Puai Hospital (Wuhan, China) and written informed consent was provided by all patients prior to the study start. A total of 32 pairs of glioblastoma tissues and adjacent normal tissues were collected from Puai Hospital between June 2017 and January 2020. The clinical characteristics of the patients with glioblastoma are presented in Table I.
Table I.
Clinicopathological characteristics of patients with glioblastoma (n=32).
| Characteristics | Number of patients, n | Percentage, % |
|---|---|---|
| Age, years | ||
| ≤50 | 17 | 53.1 |
| >50 | 15 | 46.9 |
| Sex | ||
| Male | 19 | 59.4 |
| Female | 13 | 40.6 |
| Tumor origin | ||
| Primary | 21 | 65.6 |
| Secondary | 11 | 34.4 |
| KPS value | ||
| ≤70 | 20 | 62.5 |
| >70 | 12 | 37.5 |
| Location | ||
| Frontal | 5 | 15.6 |
| Temporal | 11 | 34.4 |
| Parietal | 9 | 28.1 |
| Other | 7 | 21.9 |
KPS, Karnofsky performance score.
The glioblastoma cell lines (A172, U251, H4 and SHG44) and normal astrocyte cell line (HA) were purchased from the BeNa Culture Collection. H4, A172, SHG44 and HA cells were maintained in DMEM (cat. no. E600004; Sangon Biotech Co., Ltd.) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml streptomycin (both Gibco; Thermo Fisher Scientific, Inc.), at 37°C in 5% CO2. U251 cells were maintained in MEM-EBSS medium (cat. no. E600020; Sangon Biotech Co., Ltd.) supplemented with 10% FBS and 100 U/ml streptomycin, at 37°C in 5% CO2.
Cell transfection
miR-640 inhibitor, miR-640 mimic and their corresponding negative controls (mimic-NC and inhibitor-NC) were purchased from Guangzhou RiboBio Co., Ltd. SLIT1 overexpression plasmid and empty vector were purchased from GeneCopoeia, Inc. All sequences are listed in Table SI. A172 and U251 cells were transfected with 50 nM inhibitor-NC, 50 nM miR-640 inhibitor, 50 nM mimic-NC, 50 nM miR-640 mimic and 2.5 µg SLIT1 overexpression plasmid or empty vector using Lipofectamine® 2000 reagent (cat. no. 11668019; Thermo Fisher Scientific, Inc.) at room temperature, according to the manufacturer's instructions. Following incubation for 2 days at 37°C, transfection efficiency was assessed via reverse transcription-quantitative (RT-q)PCR analysis. Due to similar transfection efficiencies of inhibitor-NC, mimic-NC or empty vector (Fig. S1), cells in the NC group were co-transfected with inhibitor-NC and mimic-NC. Subsequent experiments were performed 48 h after transfection.
RT-qPCR
Total RNA from cells and tissues was extracted using TRIzol® reagent (cat. no. 15596026; Thermo Fisher Scientific, Inc.) and reverse transcribed into cDNA. miRNA was reverse-transcribed using the mirVana™ qRT-PCR miRNA Detection kit (cat. no. AM1558, Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The reaction condition was as follows: 37°C for 30 min and 95°C for 10 min. mRNA was reverse-transcribed using SuperScript™ III First-Strand Synthesis SuperMix (cat. no. 11752050; Thermo Fisher Scientific, Inc.). The reaction condition was as follows: 25°C for 10 min, 50°C for 30 min and 85°C for 5 min. qPCR was subsequently performed using the SYBR Premix Ex Taq (cat. no. RR420A; Takara Biotechnology Co., Ltd.) using the StepOnePlus™ Real-Time PCR System (cat. no. 4376600; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The reaction condition was as follows: 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. The primer sequences used for qPCR are listed in Table II. Relative miRNA and mRNA expression levels were calculated using the 2−ΔΔCq method (37). U6 and β-actin were used as the internal controls for miRNA and mRNA, respectively.
Table II.
Primer sequences used for quantitative PCR.
| Gene | Primer sequences (5′-3′) |
|---|---|
| miR-640 | Forward: GTGACCCTGGGCAAGTTCCT |
| Reverse: CCCCAAGGCAACCGTAGAGG | |
| SLIT1 | Forward: CTGCTCCCCGGATATGAACC |
| Reverse: TAGCATGCACTCACACCTGG | |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| β-actin | Forward: TCGTGCGTGACATTAAGGAG |
| Reverse: GTCAGGCAGCTCGTAGCTCT |
miR, microRNA; SLIT1, Slit guidance ligand 1.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to assess cell viability. A172 and U251 cell suspensions (100 µl) in a logarithmic growth phase were seeded into 96-well plates at a density of 2,000 cells/well. Following incubation overnight at 37°C, cells were transfected with miR-640 mimic, miR-640 inhibitor and corresponding NCs, or SLIT1 overexpression plasmid and corresponding empty vector for 0, 24, 48 and 72 h at 37°C. CCK-8 solution (10 µl; cat. no. E606335; Sangon Biotech Co., Ltd.) was added to each well, according to the manufacturer's instructions. Following incubation for 2 h, cell viability was analyzed at a wavelength of 450 nm, using a microplate reader.
BrdU ELISA assay
The BrdU ELISA assay was performed to assess cell proliferation, using the BrdU Cell Proliferation ELISA kit (cat. no. ab126556; Abcam). Transfected cells were seeded into 96-well plates at a density of 3,000 cells/well and incubated for 2 days in DMEM for A172 cells and MEM-EBSS medium for U251 cells at 37°C. Subsequently, cells were incubated with 100 µl 1X BrdU reagent (included in the kit) for 2 h at 37°C. The medium was discarded and 200 µl/well fixing solution (included in the kit) was added to the cells, which were incubated for 30 min at 25°C. Cells were washed three times with PBS, prior to incubation with 100 µl/well anti-BrdU monoclonal detector antibody (1:1,000; included in the kit) for 1 h at 25°C. Subsequently, 100 µl/well goat anti-mouse IgG conjugate (1:2,000; included in the kit) was added and cells were incubated for 30 min at 25°C. Cells were subsequently incubated with 100 µl/well TMB peroxidase substrate for 30 min in the dark at 25°C and analyzed at a wavelength of 450 nm, using a microplate reader.
Cell adhesion assay
For the cell adhesion assay, a 96-well plate was pre-coated with 10 µg/ml collagen I (cat. no. C7661; Sigma-Aldrich; Merck KGaA) and blocked with 1% BSA (Beijing Solarbio Science & Technology Co., Ltd.) overnight at 4°C. The transfected cells were suspended in serum-free medium and seeded into the pre-coated 96-well plate at a density of 1×105 cells/well. After 1 h, the adherent cells were fixed with 4% paraformaldehyde for 15 min at 25°C, while the non-adherent cells were removed using PBS. The fixed cells were subsequently stained with 2.5% crystal violet for 20 min at 25°C in methanol and measured at a wavelength of 450 nm, using a microplate reader.
Caspase-3 activity assay
Caspase-3 activity was measured to assess cell apoptosis, as it is highly expressed during cell apoptosis (38). The Caspase-3 Assay kit (cat. no. ab39401; Abcam) was used to measure caspase-3 activity. Following cell transfection for 48 h, 1×106 cells were collected and incubated with 50 µl cell lysis buffer for 15 min at 25°C. Protein concentration was measured and adjusted to 50–200 µg protein per 50 µl reaction buffer. Cell lysates were subsequently incubated with 50 µl reaction buffer supplemented with 10 mM DTT and 5 µl 4 mM DEVD-p-NA substrate at 25°C for 30 min. Caspase-3 activity was measured at a wavelength of 405 nm, using a microplate reader.
Dual-luciferase reporter assay
A Dual-luciferase reporter system was used to assess the luciferase activity of cells. miRDB predicted two binding sites of SLIT1 3′-untranslated region (UTR) or the area to which miR-640 could bind. These two binding sites (2442–2448 and 2945–2952) of SLIT1 3′-UTR were mutated using the QuikChange XL Site-Directed Mutagenesis kit (cat. no. 200522; Agilent Technologies, Inc.). The wild-type SLIT1 3′-UTR and mutant SLIT1 3′-UTR were inserted into the pEZX-MT05 reporter plasmids (GeneCopoeia, Inc.) and subsequently co-transfected into A172 and U251 cells with miR-640 mimic using Lipofectamine® 2000 reagent (cat. no. 11668019; Thermo Fisher Scientific, Inc.). Following incubation for 48 h at 37°C, luciferase activity was detected using the Secrete-Pair Dual Luminescence Assay kit (cat. no. LF031; GeneCopoeia, Inc.). The activity of Gaussia Luciferase was normalized to the activity of secreted alkaline phosphatase.
RNA pull-down assay
The RNA pull-down assay was performed to verify the association between SLIT1 and miR-640. Briefly, 0.5 ml of 25 mM Tris-HCl, 0.05% NP-40, 70 mM KCl, 2.5 mM EDTA, 80 U/ml RNase inhibitor and 1X protease inhibitor cocktail (cat. no. 539136; Sigma-Aldrich; Merck KGaA) were used to incubate 3×106 A172 and U251 cells for 20 min on ice. The mixture was centrifuged at 1,400 × g at 4°C for 20 min and the supernatant lysate was collected. Subsequently, 10 nM biotin-coupled miR-22-3p (bio-miR-22-3p) or biotin-coupled NC (bio-NC) (Guangzhou RiboBio Co., Ltd.) was added to the supernatant lysate and incubated for 2 h at 25°C. Following incubation, 100 µl streptavidin magnetic beads (cat. no. 88816; Thermo Fisher Scientific, Inc.) were added to the supernatant lysate. After 4 h, the beads were washed with PBS and the expression of pull-down RNA was assessed via RT-qPCR analysis.
Western blotting
Total protein was extracted from cells (A172, H4, U251, SHG44 and HA) and tissues (normal and glioblastoma tissues) using RIPA buffer (Keygene N.V.), according to the manufacturer's instructions. Total protein was separated via SDS-PAGE, transferred onto cellulose nitrate membranes (Bio-Rad Laboratories, Inc.) and blocked with 5% non-fat milk in TBST buffer (TBS containing 0.1% Tween-20) for 1 h at room temperature. The membranes were incubated with primary antibodies against GAPDH (cat. no. ab8245; 1:2,500; Abcam) and SLIT1 (cat. no. ab151724; 1:2,000; Abcam) overnight at 4°C. Membranes were washed three times with TBST buffer. Following the primary incubation, membranes were incubated with goat anti-rabbit IgG (cat. no. ab97051) and goat anti-mouse IgG (cat. no. ab175783) secondary antibodies labeled with HRP (both 1:10,000; Abcam) at room temperature for 1 h. Protein blots were subsequently washed with TBST buffer and visualized using ECL-Plus reagent (cat. no. 345818; EMD Millipore).
Statistical analysis
All experiments were performed in triplicate and data are presented as the mean ± standard deviation. Data were analyzed using SPSS 19.0 software (IBM Corp.). Log-rank test was performed for prognosis analysis. A paired or unpaired two-tailed Student's t-test was used to compare differences between two groups of paired tissues or unpaired cells, respectively, while one-way ANOVA followed by Dunnett's or Tukey's post-hoc test was used to compare differences among multiple groups. Spearman's correlation analysis was used for analyzing the correlation between SLIT1 and miR-640 expression. P<0.05 was considered to indicate a statistically significant difference.
Results
SLIT1 and miR-640 are key regulators of glioblastoma
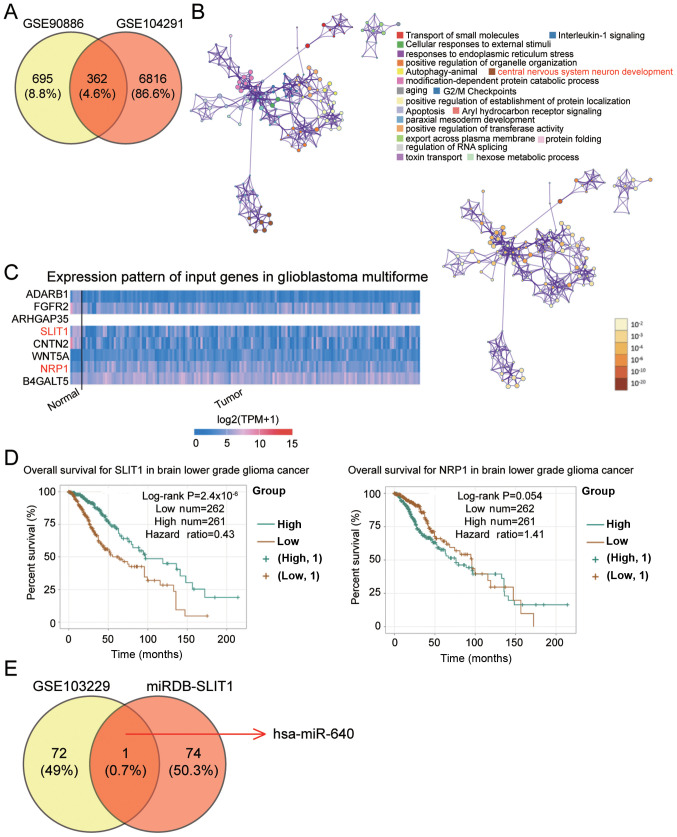
The GSE90886 and GSE104291 datasets were downloaded from the GEO database to identify the key gene in glioblastoma. Venny 2.1.0 identified 362 common differentially expressed genes (DEGs) that overlapped between the two mRNAs microarray (Fig. 1A). The 362 DEGs were subsequently uploaded onto Metascape for biological processes analysis. The central nervous system neuron development, which involves eight DEGs, was selected as the key biological process as glioblastoma is associated with central nervous system neuron development (39) (Fig. 1B). Based on The Cancer Genome Atlas data (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) on glioblastoma multiforme, low SLIT1 expression and high NRP1 expression were observed in glioblastoma multiforme (Fig. 1C). Due to its association with poor prognosis, SLIT1 was investigated in the present study (Fig. 1D). The GSE103229 dataset was used to identify the differentially expressed miRNAs, while miRDB was used to predict the miRNAs binding to SLIT1. hsa-miR-640 was identified as the key miRNA in glioblastoma (Fig. 1E), so its role was further investigated in the present study.
Figure 1.
SLIT1 and miR-640 are associated with glioblastoma. (A) A total of 362 DEGs were identified to overlap between the GSE90886 and GSE104291 datasets using Venny 2.1.0. (B) The central nervous system neuron development involving eight DEGs (ADARB1, FGFR2, ARHGAP35, SLIT1, CNTN2, WNT5A, NRP1 and B4GALT5) was selected as the key biological process using Metascape. (C) SLIT1 and NRP1 were abnormally expressed in glioblastoma multiforme based on The Cancer Genome Atlas analysis. (D) Low SLIT1 expression was significantly associated with poor prognosis of brain lower grade glioma cancer using the ENCORI Pan-Cancer Analysis platform. (E) miR-640 was identified as the key miRNA that could bind to the SLIT1 3′-UTR, and was associated with glioblastoma. The GSE103229 dataset was the miRNA expression profile. miRDB was used to predict the miRNAs binding to mRNA 3′-UTR. SLIT1, Slit guidance ligand 1; miR, microRNA; UTR, untranslated region.
miR-640 promotes the malignant phenotype of glioblastoma cells
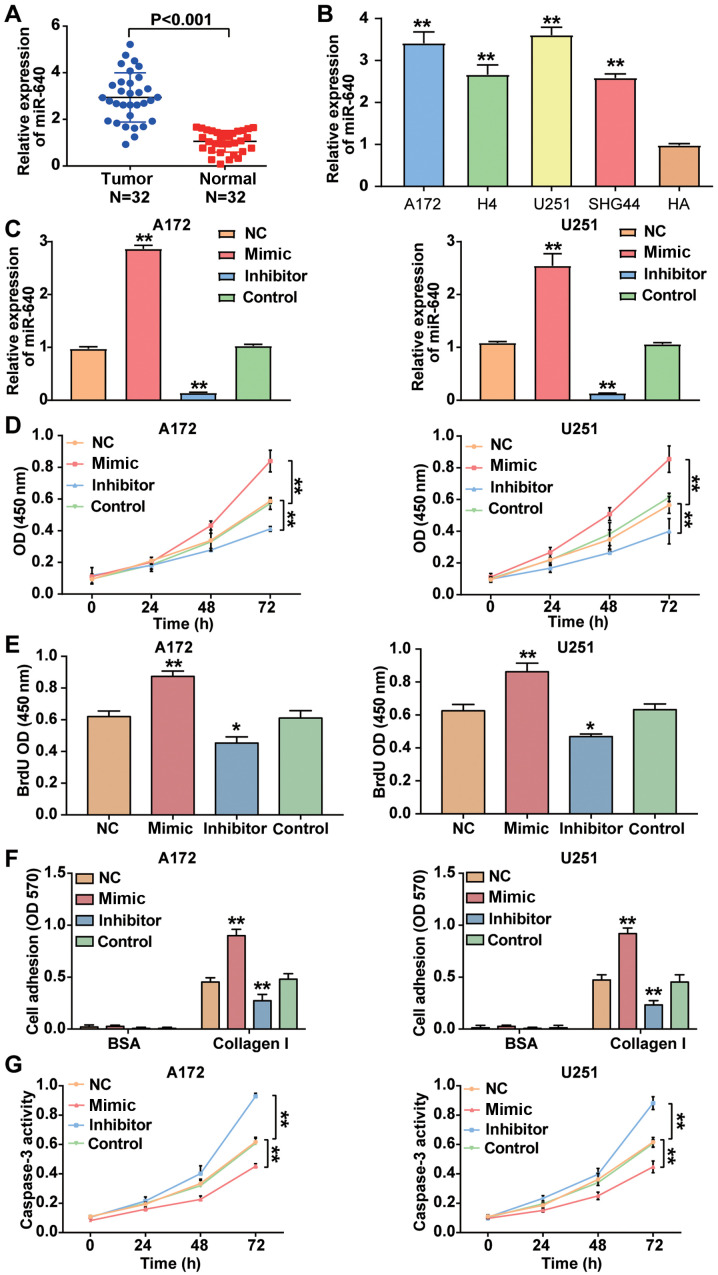
RT-qPCR analysis was performed to detect miR-640 expression in glioblastoma tissues and cells. The result demonstrated that miRNA expression was higher in glioblastoma tissues by 2.78-fold compared with normal tissues (P<0.001; Fig. 2A). Similarly, miR-640 expression was significantly higher in the glioblastoma cell lines (A172, H4, U251 and SHG44) compared with the normal astrocyte HA cell line (P<0.001; Fig. 2B). Given that miR-640 expression was higher in A172 and U251 cells compared with the other glioblastoma cell lines, the A172 and U251 cell lines were selected for subsequent cytological experiments. Following transfection of A172 and U251 cells with miR-640 mimic, miR-640 inhibitor and plasmids, the results demonstrated that miR-640 expression increased by ~2.5-fold in A172 and U251 cells transfected with miR-640 mimic, while miR-640 expression decreased by ~85% in A172 and U251 cells transfected with miR-640 inhibitor (P<0.001; Fig. 2C). The results of the CCK-8 assay demonstrated that overexpression of miR-640 enhanced cell viability, the effects of which were reversed following downregulation of miR-640 (P<0.001; Fig. 2D).
Figure 2.
miR-640 promotes cell viability, proliferation and adhesion but inhibits apoptosis in glioblastoma cells. (A) RT-qPCR analysis was performed to detect miR-640 expression in glioblastoma tissues and normal tissues. (B) RT-qPCR analysis was performed to detect miR-640 expression in the glioblastoma cell lines (A172, H4, U251 and SHG44) and the normal astrocyte cell line, HA. (C) Transfection efficiency of miR-640 mimic and miR-640 inhibitor were verified via RT-qPCR analysis. (D) The Cell Counting Kit-8 assay was performed to assess the viability of A172 and U251 cells. (E) The BrdU ELISA assay was performed to assess the proliferation of A172 and U251 cells. (F) The cell adhesion assay was performed to assess the adhesive ability of transfected A172 and U251 cells. (G) The caspase-3 activity assay was performed to assess apoptosis of transfected A172 and U251 cells. Data are presented as the mean ± standard deviation (n=3). *P<0.05 vs. control group; **P<0.001 vs. HA, control group or NC. miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; OD, optical density.
The results of the BrdU ELISA assay demonstrated that overexpression of miR-640 promoted A172 and U251 cell proliferation, the effects of which were reversed following downregulation of miR-640 (Fig. 2E). The cell adhesion assay demonstrated that the cell adhesive ability was enhanced by almost 2-fold in A172 and U251 cells transfected with miR-640 mimic, while the cell adhesive ability was impaired by ~50% following transfection with miR-640 inhibitor (P<0.001; Fig. 2F). Caspase-3 activity assay was measured to assess cell apoptosis, as caspase-3 activity is enhanced during cell apoptosis (37). The results demonstrated that caspase-3 activity was decreased in glioblastoma cells transfected with miR-640 mimic, while it was elevated in glioblastoma cells transfected with miR-640 inhibitor compared with the control group (P<0.001; Fig. 2G). Taken together, these results suggest that miR-640 acts as an oncogene in glioblastoma cells.
Association between miR-640 and SLIT1
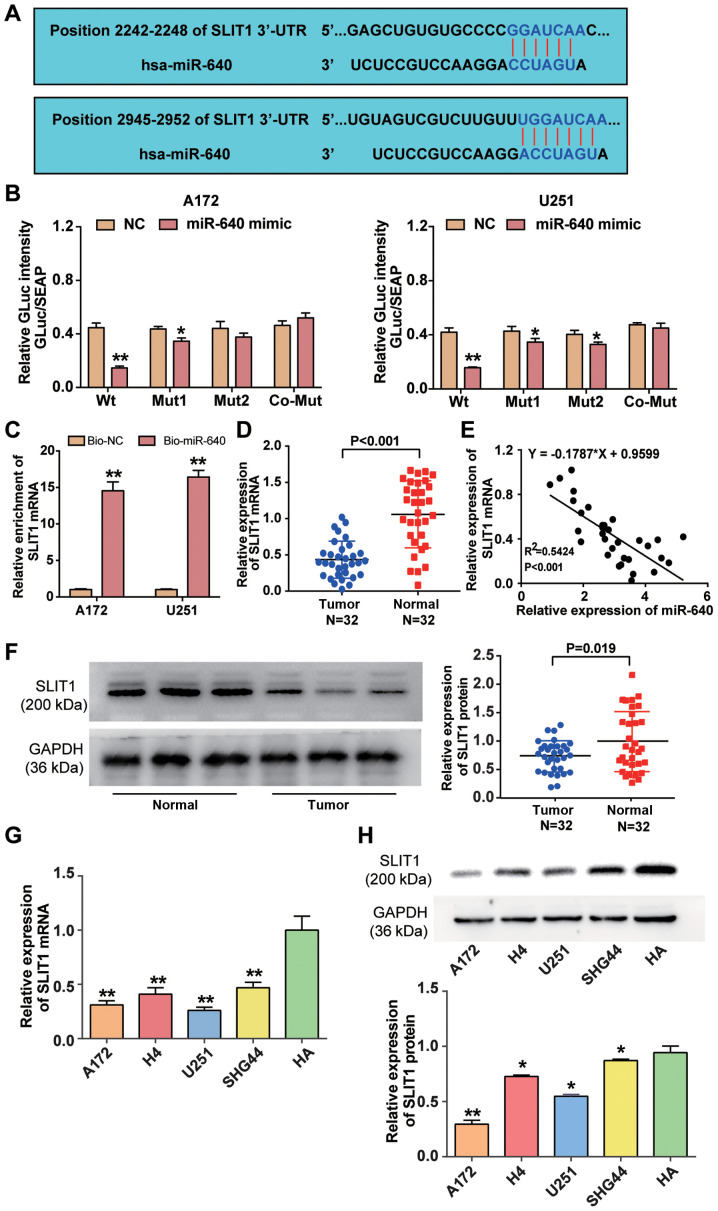
miRDB analysis predicted that SLIT1 3′-UTR contained two binding sites to which miR-640 could bind (Fig. 3A). The two binding sites in SLIT1 3′-UTR were co-mutated and subsequently inserted into the luciferase reporter plasmids. The results of the dual-luciferase reporter assay demonstrated that fluorescence intensity decreased >50% in A172 and U251 cells co-transfected with wild-type SLIT1 3′-UTR and miR-640 mimic (P<0.001), whereas the targeting effect of miR-640 declined when the two binding sites of SLIT1 3′-UTR were mutated (Fig. 3B). The results of the RNA pull-down assay demonstrated that SLIT1 mRNA expression was enriched in A172 and U251 cells transfected with biotin-coupled miR-640 (P<0.001; Fig. 3C). Furthermore, RT-qPCR analysis revealed that SLIT1 mRNA expression was downregulated by 55% in glioblastoma tissue samples (P<0.001; Fig. 3D), and SLIT1 expression was negatively correlated with miR-640 expression in glioblastoma tissues (Fig. 3E). Western blot analysis demonstrated that SLIT1 expression was downregulated in glioblastoma tissues compared with normal tissues (P<0.05; Fig. 3F). Similarly, RT-qPCR (P<0.001) and western blot (P<0.05) analyses indicated that SILT1 expression decreased in the glioblastoma cell lines (A172, H4, U251 and SHG44) compared with the normal astrocyte cell line (HA) (Fig. 3G and H). Collectively, these results suggest that SLIT1 is directly suppressed by miR-640 as a target downstream in glioblastoma.
Figure 3.
miR-640 directly binds to SLIT1 3′-UTR. (A) The potential binding site between miR-640 and SLIT1 was predicted using miRDB. (B) The potential binding site between miR-640 and 3′-UTR of SLIT1 was determined via the Dual-luciferase reporter assay. (C) The RNA pull-down assay was performed to detect SLIT1 enrichment in A172 and U251 cells. (D) RT-qPCR analysis was performed to detect SLIT1 expression in glioblastoma tissues and normal tissues. (E) Spearman's correlation analysis was performed to determine the association between miR-640 and SLIT1 expression levels. (F) Western blot analysis was performed to detect SLIT1 protein expression in glioblastoma tissues and normal tissues. (G) RT-qPCR analysis was performed to detect SLIT1 mRNA expression in the glioblastoma cell lines (A172, H4, U251 and SHG44) and normal astrocyte HA cells. **P<0.001 vs. HA. (H) The expression level of SLIT1 protein in glioblastoma cell lines (A172, H4, U251 and SHG44) and normal astrocyte cell line HA was measured using a western blot system. Data are presented as the mean ± standard deviation (n=3). *P<0.05 vs. co-transfection with Wt and miR-640 mimic or HA; **P<0.001 vs. co-transfection with Wt and miR-640 mimic, Bio-NC or HA. miR, microRNA; SLIT1, Slit guidance ligand 1; UTR, untranslated region; Wt, wild-type; Mut, mutant; Bio-NC, biotin-coupled negative control; RT-qPCR, reverse transcription-quantitative PCR.
miR-640 contributes to the development of glioblastoma by inhibiting SLIT1
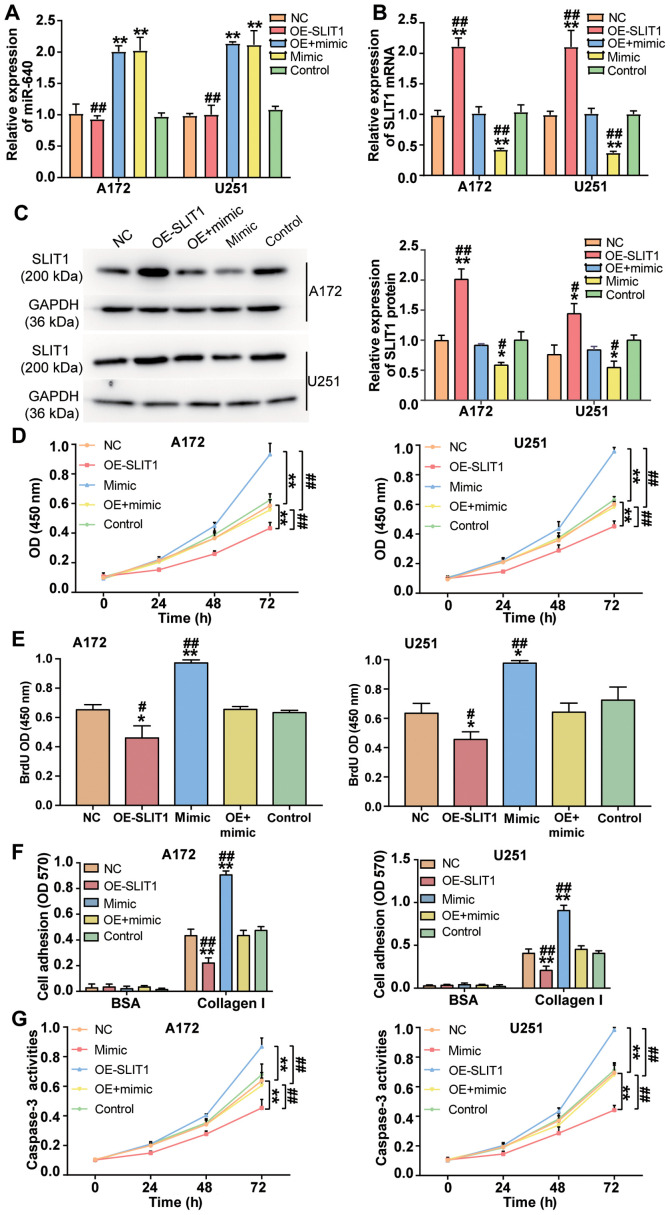
To investigate the effect of miR-640 on glioblastoma cells, miR-640 mimic and SLIT1 overexpression plasmid were transfected or co-transfected into A172 and U251 cells. RT-qPCR analysis demonstrated that transfection with SLIT1 overexpression plasmid did not affect miR-640 expression (P<0.001; Fig. 4A). Conversely, SLIT1 overexpression plasmid enhanced SLIT1 expression by 2.1-fold, while transfection with miR-640 mimic decreased SLIT1 expression by ~60% in both glioblastoma cell lines (P<0.001; Fig. 4B). Western blot analysis demonstrated that transfection with SLIT1 overexpression plasmid increased SLIT1 protein expression, while overexpression of miR-640 decreased SLIT1 protein expression in A172 and U251 cells (P<0.05; Fig. 4C). These results indicate that A172 and U251 cells were successfully transfected or co-transfected with miR-640 mimic and SLIT1 overexpression plasmid.
Figure 4.
miR-640 exerts a promoting effect on glioblastoma cells by inhibiting SLIT1 expression. (A) miR-640 and (B) SLIT1 expression was detected in the transfected A172 and U251 cells via reverse transcription-quantitative PCR analysis. (C) Western blot analysis was performed to detect SLIT1 protein expression in A172 and U251 cells transfected with miR-640 mimic or OE-SLIT1. (D) The Cell Counting Kit-8 assay was performed to assess the viability of transfected A172 and U251 cells. (E) The BrdU ELISA assay was performed to assess the proliferation of transfected A172 and U251 cells. (F) The cell adhesion assay was performed to assess the adhesive ability of transfected A172 and U251 cells. (G) The caspase-3 activity assay was performed to assess apoptosis of transfected A172 and U251 cells. Data are presented as the mean ± standard deviation (n=3). *P<0.05, **P<0.001 vs. the control group; #P<0.05, ##P<0.001 vs. control co-transfection with mimic and the OE-SLIT1 group. miR, microRNA; SLIT1, Slit guidance ligand 1; OE, overexpression vector; NC, negative control; OD, optical density.
Several cytological experiments were performed in transfected A172 and U251 cells. The results of the CCK-8 assay demonstrated that the promotive effect of miR-640 mimic on cell viability was attenuated following transfection with SLIT1 overexpression plasmid in A172 and U251 cells (P<0.001; Fig. 4D). Similarly, the results of the BrdU ELISA assay demonstrated that the promotive effect of miR-640 mimic on cell proliferation was relieved following transfection with SLIT1 overexpression plasmid (P<0.05; Fig. 4E). The cell adhesive ability in the co-transfection group was impaired by ~50% compared with the miR-640 mimic group (P<0.001; Fig. 4F). Notably, the inhibitory effect of miR-640 mimic on caspase-3 activity was attenuated in both A172 and U251 cells following transfection with SLIT1 overexpression plasmid (P<0.001; Fig. 4G). Taken together, these results suggest that the enhanced function of miR-640 in the progression of glioblastoma cells is repressed following overexpression of SLIT1.
Discussion
The results of the present study demonstrated high miR-640 expression and the low SLIT1 expression in glioblastoma tissues and cells. Furthermore, miR-640 was demonstrated to enhance cell proliferation and adhesion, but impaired apoptosis in glioblastoma cells. In addition, SLIT1 exhibited a suppressive role in glioblastoma cells, and miR-640 was demonstrated to bind to SLIT1 3′-UTR, thereby suppressing SLIT1 expression. Overexpression of SLIT1 attenuated the promoting effect of miR-640 on glioblastoma cells.
The effect of miR-640 on different types of human cancer has been extensively studied. For example, by performing miRNA microarray analysis, Li et al (23) demonstrated that miR-640 expression is decreased in paclitaxel-resistant ovarian carcinoma samples, and that high miR-640 expression yields a more favorable prognosis (23). Furthermore, miR-640 expression has been reported to be downregulated in hepatocellular carcinoma (HCC) tissues, which contributes to the development of HCC cells (24). Lodes et al (25) used a pan-human miRNA microarray to assess miRNA expression in five forms of human cancers, including prostate carcinoma, colon carcinoma, ovarian carcinoma, breast carcinoma and lung carcinoma. The results demonstrated that miR-640 is overexpressed in serum from patients with prostate cancer. Taken together, these findings suggest that miR-640 plays different roles in different types of cancer. In glioblastoma, miR-640 was upregulated using microarray analysis (40). Consistent with this finding, the results of the microarray and RT-qPCR analyses performed in the present study demonstrated that miR-640 expression was upregulated in glioblastoma. In addition, the CCK-8, BrdU (41,42) and cell adhesion (43) assays were performed to determine the molecular mechanism of miR-640 in glioblastoma cells. The results of these experiments demonstrated that overexpression of miR-640 promoted the viability, proliferation and adhesion of glioblastoma cells, while interference with miR-640 inhibited the proliferation and adhesion of these cells. Caspases, which are key regulators of mammalian apoptosis, are activated following cell injury and are responsible for regulating apoptosis (44). Caspase-3 is the main mediator of apoptosis, and it is responsible for the proteolysis of several downstream key proteins associated with apoptosis (45). To determine the molecular mechanism of miR-640-induced apoptosis in glioblastoma multiforme cells, caspase-3 activity was analyzed. The results demonstrated that by interfering with A172 and U251 cells, miR-640 could promote the activity of caspase-3; however, these effects were reversed following overexpression of miR-640. Therefore, it was concluded that miR-640 contributed to cell proliferation and adhesion, and impaired apoptosis in glioblastoma cells.
Previous studies have demonstrated that SLIT1 participates in intracellular metabolism and nerve cell elongation, branching and functional regeneration through its receptor, Robo (30,46,47). Kim et al (32) reported that SLIT1 is methylated in different types of human cancer, and in 11 gastric cancer cell lines. In addition, SLIT1 methylation occurs in patients with early gastric cancer and advanced gastric cancer (32). Similar to gastric cancer, the SLIT1 promoter region is methylated in glioma tumor cells (48). Another study on glioblastoma demonstrated that SLIT1 improves the As2O3 resistance of glioblastoma cells (31). Based on previous findings (32,33), it was speculated that SLIT3 may not only be downregulated but also be a tumor suppressor in glioblastoma. In the present study, microarray and RT-qPCR analyses proved that SLIT1 expression was downregulated in glioblastoma. Based on the results of the cytological experiments, overexpression of SLIT1 inhibited cell viability, proliferation and adhesion, and promoted apoptosis of glioblastoma cells. Additionally, SLIT1 was identified as a target gene of miR-640, suggesting that overexpression of SLIT1 may attenuate the promotive effect of miR-640 on glioblastoma. Thus, the present study revealed that miR-640 may act as a tumor promoter in glioblastoma cells by targeting SLIT1.
The present study is not without limitations as it fails to explain the potential signaling pathways affected by SLIT1 in the development of glioblastoma. Thus, prospective studies will focus on investigating the effect of miR-640 on the malignant behavior of tumors in animals.
In conclusion, the results of the present study demonstrated that miR-640 contributed to the progression of glioblastoma cells by directly targeting SLIT1. Taken together, these results suggest that the miR-640/SLIT1 axis may have the potential to provide novel targets for the treatment and diagnosis of glioblastoma.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CL performed the experiments and the data analysis. ZL and SD conceived and designed the study. YC and XC contributed to the acquisition of the data. NL, ZL and JC contributed to the analysis and interpretation of data. JC and SD wrote the manuscript. NL, XC and YC reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Puai Hospital (approval no. KY-2019-04702; Wuhan, China) and written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Korja M, Raj R, Seppä K, Luostarinen T, Malila N, Seppälä M, Mäenpää H, Pitkäniemi J. Glioblastoma survival is improving despite increasing incidence rates: A nationwide study between 2000 and 2013 in Finland. Neuro Oncol. 2019;21:370–379. doi: 10.1093/neuonc/noy164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander BM, Cloughesy TF. Adult glioblastoma. J Clin Oncol. 2017;35:2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 4.Kim H, Leiby BE, Shi W. Too little, too soon: Short-course radiotherapy in elderly patients with glioblastoma. J Clin Oncol. 2016;34:2191–2192. doi: 10.1200/JCO.2015.66.3195. [DOI] [PubMed] [Google Scholar]
- 5.Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, Wang AC, Ellingson BM, Rytlewski JA, Sanders CM, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sukumar UK, Bose RJC, Malhotra M, Babikir HA, Afjei R, Robinson E, Zeng Y, Chang E, Habte F, Sinclair R, et al. Intranasal delivery of targeted polyfunctional gold-iron oxide nanoparticles loaded with therapeutic microRNAs for combined theranostic multimodality imaging and presensitization of glioblastoma to temozolomide. Biomaterials. 2019;218:119342. doi: 10.1016/j.biomaterials.2019.119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho KH, Cheng CH, Chou CM, Chen PH, Liu AJ, Lin CW, Shih CM, Chen KC. miR-140 targeting CTSB signaling suppresses the mesenchymal transition and enhances temozolomide cytotoxicity in glioblastoma multiforme. Pharmacol Res. 2019;147:104390. doi: 10.1016/j.phrs.2019.104390. [DOI] [PubMed] [Google Scholar]
- 8.Cloughesy T, Finocchiaro G, Belda-Iniesta C, Recht L, Brandes AA, Pineda E, Mikkelsen T, Chinot OL, Balana C, Macdonald DR, et al. Randomized, double-blind, placebo-controlled, multicenter phase II study of onartuzumab plus bevacizumab versus placebo plus bevacizumab in patients with recurrent glioblastoma: Efficacy, safety, and hepatocyte growth factor and O(6)-Methylguanine-DNA methyltransferase biomarker analyses. J Clin Oncol. 2017;35:343–351. doi: 10.1200/JCO.2015.64.7685. [DOI] [PubMed] [Google Scholar]
- 9.Goswami S, Walle T, Cornish AE, Basu S, Anandhan S, Fernandez I, Vence L, Blando J, Zhao H, Yadav SS, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med. 2020;26:39–46. doi: 10.1038/s41591-019-0694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 11.Citron F, Segatto I, Vinciguerra GLR, Musco L, Russo F, Mungo G, D'Andrea S, Mattevi MC, Perin T, Schiappacassi M, et al. Downregulation of miR-223 expression is an early event during mammary transformation and confers resistance to CDK4/6 inhibitors in luminal breast cancer. Cancer Res. 2020;80:1064–1077. doi: 10.1158/0008-5472.CAN-19-1793. [DOI] [PubMed] [Google Scholar]
- 12.Carotenuto P, Hedayat S, Fassan M, Cardinale V, Lampis A, Guzzardo V, Vicentini C, Scarpa A, Cascione L, Costantini D, et al. Modulation of biliary cancer chemo-resistance through microRNA-mediated rewiring of the expansion of CD133+ cells. Hepatology. 2020;72:982–996. doi: 10.1002/hep.31094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang T, Wan X, Alvarez AA, James CD, Song X, Yang Y, Sastry N, Nakano I, Sulman EP, Hu B, Cheng SY. MIR93 (microRNA-93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15:1100–1111. doi: 10.1080/15548627.2019.1569947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han M, Wang S, Fritah S, Wang X, Zhou W, Yang N, Ni S, Huang B, Chen A, Li G, et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain. 2020;143:512–530. doi: 10.1093/brain/awz406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang CH, Wang Y, Sims M, Cai C, Pfeffer LM. MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer Lett. 2019;465:59–67. doi: 10.1016/j.canlet.2019.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Jiang J, Wang X, Lu J. PWRN1 suppressed cancer cell proliferation and migration in glioblastoma by inversely regulating hsa-miR-21-5p. Cancer Manag Res. 2020;12:5313–5322. doi: 10.2147/CMAR.S250166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X, Yan P, Feng J, Zhang F. Expression of microRNA-210 and the prognosis in glioma patients: A meta-analysis. Biomark Med. 2020;14:795–805. doi: 10.2217/bmm-2019-0448. [DOI] [PubMed] [Google Scholar]
- 18.Xia W, Zhu J, Tang Y, Wang X, Wei X, Zheng X, Hou M, Li S. PD-L1 inhibitor regulates the miR-33a-5p/PTEN signaling pathway and can be targeted to sensitize glioblastomas to radiation. Front Oncol. 2020;10:821. doi: 10.3389/fonc.2020.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Q, Gao J, Zhao Y, Hou R. Long non-coding RNA LBX2-AS1 enhances glioma proliferation through downregulating microRNA-491-5p. Cancer Cell Int. 2020;20:411. doi: 10.1186/s12935-020-01433-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Pan CM, Chan KH, Chen CH, Jan CI, Liu MC, Lin CM, Cho DY, Tsai WC, Chu YT, Cheng CH, et al. MicroRNA-7 targets T-Box2 to inhibit epithelial-mesenchymal transition and invasiveness in glioblastoma multiforme. Cancer Lett. 2020;493:133–142. doi: 10.1016/j.canlet.2020.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Lin H, Zuo D, He J, Ji T, Wang J, Jiang T. Long noncoding RNA WEE2-AS1 plays an oncogenic role in glioblastoma by functioning as a molecular sponge for microRNA-520f-3p. Oncol Res. 2020 Aug 24; doi: 10.3727/096504020X15982623243955. doi: 10.3727/096504020X15982623243955 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harel S, Sanchez-Gonzalez V, Echavarria R, Mayaki D, Hussain SN. Roles of miR-640 and Zinc finger protein 91 (ZFP91) in angiopoietin-1-induced in vitro angiogenesis. Cells. 2020;9:1602. doi: 10.3390/cells9071602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Lu Y, Chen Y, Lu W, Xie X. MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma based on formalin-fixed paraffin-embedded samples. BMC Cancer. 2013;13:216. doi: 10.1186/1471-2407-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai Z, Fu Q, Liu C, Zhang X, Jia P, Xia P, Liu P, Liao S, Qin T, Zhang H. Emerging roles of hsa-circ-0046600 targeting The miR-640/HIF-1α signalling pathway in the progression of HCC. Onco Targets Ther. 2019;12:9291–9302. doi: 10.2147/OTT.S229514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tong M, Jun T, Nie Y, Hao J, Fan D. The role of the Slit/Robo signaling pathway. J Cancer. 2019;10:2694–2705. doi: 10.7150/jca.31877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blockus H, Chedotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- 28.Ma L, Tessier-Lavigne M. Dual branch-promoting and branch-repelling actions of Slit/Robo signaling on peripheral and central branches of developing sensory axons. J Neurosci. 2007;27:6843–6851. doi: 10.1523/JNEUROSCI.1479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marlow R, Strickland P, Lee JS, Wu X, Pebenito M, Binnewies M, Le EK, Moran A, Macias H, Cardiff RD, et al. SLITs suppress tumor growth in vivo by silencing Sdf1/Cxcr4 within breast epithelium. Cancer Res. 2008;68:7819–7827. doi: 10.1158/0008-5472.CAN-08-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaworski A, Tom I, Tong RK, Gildea HK, Koch AW, Gonzalez LC, Tessier-Lavigne M. Operational redundancy in axon guidance through the multifunctional receptor Robo3 and its ligand NELL2. Science. 2015;350:961–965. doi: 10.1126/science.aad2615. [DOI] [PubMed] [Google Scholar]
- 31.Shuai W, Wu J, Chen S, Liu R, Ye Z, Kuang C, Fu X, Wang G, Li Y, Peng Q, et al. SUV39H2 promotes colorectal cancer proliferation and metastasis via tri-methylation of the SLIT1 promoter. Cancer Lett. 2018;422:56–69. doi: 10.1016/j.canlet.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Kim M, Kim JH, Baek SJ, Kim SY, Kim YS. Specific expression and methylation of SLIT1, SLIT2, SLIT3, and miR-218 in gastric cancer subtypes. Int J Oncol. 2016;48:2497–2507. doi: 10.3892/ijo.2016.3473. [DOI] [PubMed] [Google Scholar]
- 33.Amodeo V, A D, Betts J, Bartesaghi S, Zhang Y, Richard-Londt A, Ellis M, Roshani R, Vouri M, Galavotti S, et al. A PML/Slit axis controls physiological cell migration and cancer invasion in the CNS. Cell Rep. 2017;20:411–426. doi: 10.1016/j.celrep.2017.06.047. [DOI] [PubMed] [Google Scholar]
- 34.Long H, Liang C, Zhang X, Fang L, Wang G, Qi S, Huo H, Song Y. Prediction and analysis of key genes in glioblastoma based on bioinformatics. Biomed Res Int. 2017;2017:7653101. doi: 10.1155/2017/7653101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sciuscio D, Diserens AC, van Dommelen K, Martinet D, Jones G, Janzer RC, Pollo C, Hamou MF, Kaina B, Stupp R, et al. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clin Cancer Res. 2011;17:255–266. doi: 10.1158/1078-0432.CCR-10-1931. [DOI] [PubMed] [Google Scholar]
- 36.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 39.Pomeroy SL. Neural development and the ontogeny of central nervous system tumors. Neuron Glia Biol. 2004;1:127–133. doi: 10.1017/S1740925X04000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nawaz Z, Patil V, Thinagararjan S, Rao SA, Hegde AS, Arivazhagan A, Santosh V, Somasundaram K. Impact of somatic copy number alterations on the glioblastoma miRNome: miR-4484 is a genomically deleted tumour suppressor. Mol Oncol. 2017;11:927–944. doi: 10.1002/1878-0261.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piao XY, Li W, Li Z, Zhang N, Fang H, Zahid D, Qu Q. Forced FoxO1:S249V expression suppressed glioma cell proliferation through G2/M cell cycle arrests and increased apoptosis. Neurol Res. 2019;41:189–198. doi: 10.1080/01616412.2018.1548724. [DOI] [PubMed] [Google Scholar]
- 42.Liang HX, Sun LB, Liu NJ. Neferine inhibits proliferation, migration and invasion of U251 glioma cells by down-regulation of miR-10b. Biomed Pharmacother. 2019;109:1032–1040. doi: 10.1016/j.biopha.2018.10.122. [DOI] [PubMed] [Google Scholar]
- 43.Russo MA, Paolillo M, Sanchez-Hernandez Y, Curti D, Ciusani E, Serra M, Colombo L, Schinelli S. A small-molecule RGD-integrin antagonist inhibits cell adhesion, cell migration and induces anoikis in glioblastoma cells. Int J Oncol. 2013;42:83–92. doi: 10.3892/ijo.2012.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Cai T, Zhang W, Zhu W, Lv S. Effects of Saikosaponin D on apoptosis in human U87 glioblastoma cells. Mol Med Rep. 2017;16:1459–1464. doi: 10.3892/mmr.2017.6765. [DOI] [PubMed] [Google Scholar]
- 45.Mazumder S, Plesca D, Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol Biol. 2008;414:13–21. doi: 10.1007/978-1-59745-339-4_2. [DOI] [PubMed] [Google Scholar]
- 46.Leyva-Diaz E, del Toro D, Menal MJ, Cambray S, Susín R, Tessier-Lavigne M, Klein R, Egea J, López-Bendito G. FLRT3 is a Robo1-interacting protein that determines Netrin-1 attraction in developing axons. Curr Biol. 2014;24:494–508. doi: 10.1016/j.cub.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 47.Kaneko N, Herranz-Pérez V, Otsuka T, Sano H, Ohno N, Omata T, Nguyen HB, Thai TQ, Nambu A, Kawaguchi Y, et al. New neurons use Slit-Robo signaling to migrate through the glial meshwork and approach a lesion for functional regeneration. Sci Adv. 2018;4:eaav0618. doi: 10.1126/sciadv.aav0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickinson RE, Dallol A, Bieche I, Krex D, Morton D, Maher ER, Latif F. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Cancer. 2004;91:2071–2078. doi: 10.1038/sj.bjc.6602222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.