Abstract
Exosomes carry functional molecules that can regulate cancer progression. Understanding the function of exosomal markers may provide invaluable insights into the mechanism of metastasis in hepatocellular carcinoma (HCC). The aim of the present study was to identify metastasis-associated microRNAs (miRNAs/miRs) expressed in plasma exosomes. A miRNA microarray and reverse transcription-quantitative PCR were used to analyze the plasma exosome miRNA expression profiles of patients with metastatic or non-metastatic HCC. Receiver operating characteristic (ROC) curve and Kaplan-Meier analyses were used to evaluate the predictive performance and prognostic efficacy of candidate miRNAs identified in the Gene Expression Omnibus database (dataset accession no. GSE67140). Bioinformatics analysis was used to examine the role of exosomal miRNAs in HCC metastasis. A total of 32 miRNAs were differentially expressed in plasma exosomes of patients with metastatic HCC compared with in those of patients with non-metastatic HCC. Additionally, the expression levels of six miRNAs were consistent between plasma exosome samples and matched tissue samples. ROC analysis demonstrated that miR-18a, miR-27a and miR-20b could discriminate metastatic HCC from non-metastatic HCC. Furthermore, the prognostic efficacy of the combination of three miRNAs (miR-18a, miR-20b and miR-221) was superior to that of individual miRNAs. Survival analysis demonstrated that high expression levels of the candidate miRNAs were associated with poor prognosis. Bioinformatics analysis indicated that the potential target genes of these miRNAs were involved in biological processes, molecular functions and cellular components that were associated with metastasis. The present findings suggested that these exosomal miRNAs may serve important roles in HCC lung metastasis and could represent a complementary clinical tool for the assessment of HCC prognosis.
Keywords: microRNA, exosome, hepatocellular carcinoma, biomarker, metastasis
Introduction
Hepatocellular carcinoma (HCC) is one of most prevalent malignancies worldwide with a high mortality (1). According to the American Epidemiological Survey of Liver Cancer, the adjusted incidence of HCC increased at a rate of 4.5% annually between 2000 and 2009, making it the fastest-growing cause of cancer-associated mortality in the United States (2). HCC is also a highly invasive tumor with frequent intrahepatic or pulmonary metastasis, both of which account for the high disease recurrence and poor survival following liver resection (3). HCC diagnosis and metastasis identification are usually based on imaging and occasionally verified using biopsy results (4). Although advances in MRI and computed tomography have improved the imaging of focal hypervascular masses consistent with HCC, these procedures are costly and not suitable for daily practice (5). In addition, pathological biopsies are the gold standard for HCC diagnosis, yet biopsy results are associated with a high false negative rate (6), and the procedure can cause discomfort or pain. The detailed molecular mechanisms that contribute to HCC metastasis are largely unknown. Furthermore, effective targeted therapeutic drugs for HCC remain unavailable. Therefore, the identification of non-invasive diagnostic and predictive biomarkers of HCC metastasis is essential.
Exosomes are microscopic vesicles that serve important roles in cell-cell communication and signal transduction (7). Exosomes transport biologically active molecules (8) to target cells, in order to initiate signaling and mediate intercellular communication. Furthermore, the potential use of exosomes as biomarkers has been demonstrated for other cancer types, such as breast cancer and gastric cancer (9), which demonstrates that the use of exosomes as a tumor marker for clinical diagnosis and prediction is feasible. Li et al (10) have highlighted the significance of exosomes in the development of HCC; however, the role of exosomes in tumor growth and metastasis remains a matter of debate. Therefore, the aim of the present study was to identify exosomal molecules that may be used as metastasis-related biomarkers in patients with HCC.
MicroRNAs (miRNAs/miRs) are a class of non-coding RNAs ~22 nucleotides in length, which can modulate gene expression (11). Tumor cells can secrete miRNAs that mediate intracellular signaling cascades, which in turn can affect various physiological processes, such as angiogenesis, metabolic reprogramming and metastasis (12). A number of studies (13,14) have indicated that secretory miRNAs, particularly exosomal miRNAs, are involved in promoting metastasis (15), highlighting the potential for exosomal miRNAs as promising biomarkers. For example, Lin et al (16) identified an miRNA classifier containing seven differentially expressed miRNAs that allows early detection of HCC. However, the role of metastasis-associated miRNAs in HCC has yet to be determined.
The aim of the present study was to identify a comprehensive circulating exosomal miRNA profile in plasma from patients with metastatic HCC. Furthermore, bioinformatics analysis was used to screen target genes of differentially expressed miRNAs and to evaluate their potential role in HCC metastasis. Furthermore, the prognostic efficiency of candidate miRNAs, and their association with overall survival (OS) were also assessed. The study identified a panel consisting of three miRNAs that could be used to discriminate metastatic HCC cases from non-metastatic HCC cases. Overall, the present findings may provide insights into the mechanisms that lead to metastasis and the role of exosomes in this context. The present study also suggested that exosomal miRNAs may represent potential biomarkers for HCC.
Materials and methods
Plasma and human tissue sample collection
A total of 40 patients with HCC diagnosed by clinical pathology at Nanfang Hospital (Guangzhou, China) between January 2017 and April 2018 were recruited. Among them, 20 patients with HCC (13 men and 7 women) with lung metastasis and 20 sex-matched primary cases were enrolled. The median age (age range, 50–80 years) of the metastatic group was 68 years and that of the non-metastatic group (age range, 50–81 years) was 67.5 years. All patients did not undergo radiotherapy and chemotherapy prior to surgery. All patients were referred for extensive evaluation, including measurement of α-fetoprotein levels and MRI before surgical resection. The tumor was examined by postoperative pathology. A 5-ml sample of peripheral venous blood was collected from all patients into EDTA tubes, and then centrifuged at 2,500 × g for 10 min at room temperature. The supernatant was transferred into RNase-free Eppendorf tubes and stored at −80°C until use. In 10 of the patients with metastatic HCC, lung metastasis biopsies were also collected, and stored in the −80°C ultra-low temperature refrigerator until further use. Informed consent was obtained from each patient on the day of admission.
Isolation of exosomes
Exosomes were purified from the plasma of patients with HCC by ultracentrifugation. The plasma was thawed on ice and centrifuged at 500 × g for 10 min and the supernatant was collected. The following steps were completed continuously and without interruption as soon as possible. The supernatant was ultracentrifuged using a W32Ti rotor (L-80XP; Beckman Coulter, Inc.) at 16,800 × g for 30 min to pellet the exosomes. Subsequently, the pellet was washed in PBS to eliminate contaminating proteins, and centrifuged again at 110,000 × g for 70 min. PBS was subsequently removed and the exosomes were re-suspended in 100 µl PBS. All centrifugation steps were performed at 4°C. All samples were stored in a −80°C ultra-low temperature refrigerator until use (within 1 month).
NTA
Exosome pellets were resuspended in PBS at a concentration of 1×107−109/ml, and then examined using a NanoSight NS300 instrument (Malvern Panalytical) to determine the size and concentration. Particle movement was analyzed and recorded in a real-time video using the NTA software (version 2.3; Malvern Panalytical). Exosome size and concentration were determined using NTA v3.2 software (Malvern Panalytical).
Western blotting
Western blotting was performed as previously described (17) Exosome lysates were prepared using RIPA buffer (Nanjing KeyGen Biotech Co., Ltd.). Protein concentrations were determined using a Bradford protein assay (Nanjing KeyGen Biotech Co., Ltd.). The samples were resolved using SDS-PAGE, transferred to PVDF membranes (EMD Millipore) and incubated with primary antibodies overnight at 4°C, as previously described (17). Primary antibodies specific for 70 kilodalton heat shock protein (HSP70; cat. no. ab194360; 1:1,000), CD63 (cat. no. ab59479; 1:1,000) and GAPDH (cat. no. ab8245; 1:10,000) were purchased from Abcam. Following primary antibody incubation, a HRP-conjugated secondary antibody (cat. nos. FDM007 and FDR007; 1:10,000; Fdbio Science) was added. The Fdbio-Femto ECL western blotting detection reagent (Fdbio Science) was used to visualize protein bands.
RNA processing and miRNA profiling
Exosomes from the plasma of 4 patients with primary HCC and 4 patients with HCC lung metastasis were analyzed using Affymetrix miRNA 4.0 arrays (Thermo Fisher Scientific, Inc.). The RNA processing and miRNA profiling experiments were performed at the laboratory of Gene-Cloud of Biotechnology Information (GCBI; Shanghai, China).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
The initial miRNA microarray results from the plasma exosomes samples were validated using RT-qPCR. Exosome and tissue miRNA purification and RT-qPCR were performed as described previously (18). Briefly, total RNA from stored plasma exosome and tissue samples was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. RT-qPCR was performed using SYBR Green and detected using the Applied Biosystems Real-Time PCR system (Thermo Fisher Scientific, Inc.). Each sample was analyzed in triplicate. The primers used for RT-qPCR are listed in Table I. The U6 small nuclear RNA was used as an internal control. The results were quantified using the 2−ΔΔCq method (19) and normalized to U6 expression.
Table I.
Primer sequences for reverse transcription-quantitative PCR used in exosome and tissue miRNA experiments.
| Primer of miRNA | Sequence (5′-3′) |
|---|---|
| hsa-let-7e | TGAGGTAGGAGGTTGTATAGTT |
| hsa-miR-18a | CCCATCTAGTGCAGATAGAAA |
| hsa-miR-27a | TTCACAGTGGCTAAGTTCCGC |
| hsa-miR-221 | CTACATTGTCTGCTGGGTTTC |
| hsa-miR-185 | GAGAGAAAGGCAGTTCCTGA |
| hsa-miR-361 | TTATCAGAATCTCCAGGGGTA |
| hsa-miR-20b | AGTGCTCATAGTGCAGGTAG |
| hsa-miR-652 | CGCCACTAGGGTTGTGAAAA |
| hsa-miR-4454 | CTGAGCGCTGCCAGTCAAA |
| hsa-miR-4720 | CCCTGGCATATTTGGTATAAC |
| hsa-miR-5189 | CAACCGTCAGAGCCCAGAA |
| U6-forward | GGAACGATACAGAGAAGATTAGC |
| U6-reverse | TGGAACGCTTCACGAATTTGCG |
miR, microRNA.
Gene Expression Omnibus (GEO) miRNA dataset and patient information
Datasets that met the following criteria were selected for study: i) The patients did not undergo chemoradiotherapy before surgery and only datasets containing >20 samples were included; and ii) HCC with lung-metastasis and without metastasis was confirmed by postoperative pathology and the patients had complete follow-up data. The Cancer Genome Atlas (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and GEO database were used to mined the data. A dataset containing 91 human HCC tumor samples without vascular invasion/metastasis and 81 samples with vascular invasion/metastasis was downloaded from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database [dataset accession no. GSE67140 (20)]. Receiver operating characteristic (ROC) analysis was carried out on these data using GraphPad Prism 8.0 software (GraphPad Software, Inc.).
Survival data from the Kaplan-Meier plotter database
The present study used the online Kaplan-Meier plotter database (http://www.kmplot.com/analysis/index.php?p=service&cancer=liver_mirna) tool to evaluate the prognostic values of candidate miRNAs (21). Original data were downloaded and Kaplan-Meier plots were generated to analyze potential prognostic values of candidate miRNAs with the platform of RNA sequencing. Analysis was performed by excluding patients who survived >80 months because there might be unrecognized confounding bias. In addition, the online plotter was used to perform the survival analysis, and hazard ratios with 95% confidence intervals and log rank P-values were calculated. P<0.05 was considered to indicate a statistically significant difference.
Bioinformatics analysis of differentially expressed exosomal miRNAs
The miRanda (22) (release miRanda-aug2010; http://www.microrna.org/) and TargetScan (release 7.0; http://www.targetscan.org/vert_70/) databases were used to predict the target genes of candidate exosomal miRNAs. Gene Ontology (GO; release 2019-12; http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG; release 92; http://www.kegg.jp/) analyses were performed to identify the biological functions and pathways for which P<0.05 and false discovery rate <0.05. R language (version 3.6.0; http://cran.r-project.org/) and ggplot2 (version 3.3.2) were used to generate the image (23,24). The miRNA-mRNA networks were generated using GCBI (https://www.gcbi.com.cn/gclib/html/index) and Cytoscape (version 3.6; http://www.cytoscape.org/).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 8.0; GraphPad Software, Inc.) or SPSS version 24.0 statistical software (SPSS, Inc.). Data are presented as the mean ± standard deviation. Differences between two groups were analyzed using a two-tailed unpaired Student's t-test. Pearson correlation and residual analysis were used to measure and assess the correlation between candidate plasma exosome miRNA levels, and tissue ROC curves were established by plotting sensitivity against 1-specificity. The diagnostic values of candidate miRNAs and the panel were evaluated using the area under the curve (AUC). Comparisons of AUC values were based on Delong's test in SPSS using R plug-in package pROC (R 4.0.3 package/pROC v1.16.2), and P-values were adjusted using the Bonferroni strategy when multiple comparisons were conducted (25). Graphs were generated using GraphPad Prism 5.0. P<0.05 was considered to indicate a statistically significant difference.
Results
Identification and characterization of exosomes
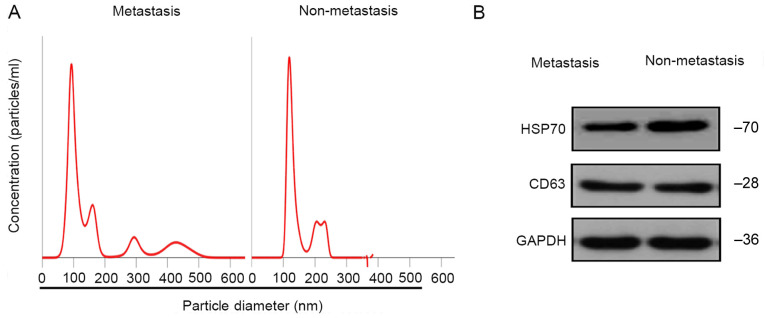
Using NTA, 30–150-nm vesicles were identified, consistent with previously reported features of exosomes (26), suggesting that the isolated particles were likely exosomes (Fig. 1A). Additionally, western blot analysis indicated that the particles were positive for exosome markers CD63 and HSP70 (Fig. 1B). These results demonstrated that the particles isolated from the plasma of patients with HCC were exosomes.
Figure 1.
Identification of exosomes in patients with metastatic and non-metastatic HCC. (A) Nanoparticle tracking analysis profile of exosomes from the plasma of patients with metastatic and non-metastatic HCC. The number of particles/ml (in millions/ml) is indicated on the y-axis. The x-axis indicates the diameter of particles (in nm). (B) Presence of typical exosome-associated proteins (CD63 and HSP70) in exosomes from the plasma of patients with HCC. HCC, hepatocellular carcinoma; HSP70, 70 kilodalton heat shock protein.
Exosomal miRNA profiles are altered between patients with metastatic and non-metastatic HCC
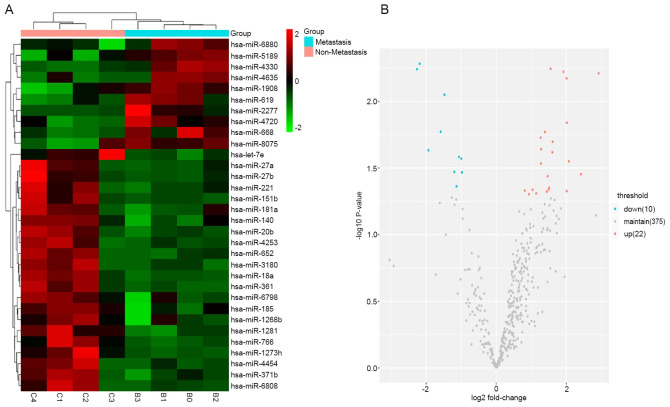
A total of 32 differentially expressed exosomal miRNAs were detected in 4 patients with HCC lung metastasis and 4 sex- and age-matched patients with non-metastatic HCC (Table II). Among these, 18 miRNAs (let-7e, miR-18a, miR-27a, miR-181a, miR-221, miR-27b, miR-185, miR-361, miR-20b, miR-652, miR-140, miR-1280, miR-151b, miR-4253, miR-1268b, miR-4454, miR-6798 and miR-1273h) were significantly upregulated and 5 miRNAs (miR-4330, miR-2277, miR-4720, miR-5189 and miR-8075) were significantly downregulated (fold change >1.2; P<0.05). Group-specific signal intensities of the exosomal miRNA profile and the volcano plot for the differentially expressed miRNAs are shown in Fig. 2. These data indicated that exosomal miRNAs were differentially expressed between patients with HCC with lung metastasis and patients with non-metastatic HCC.
Table II.
Differentially expressed exosomal microRNAs (n=32) in plasma from patients with hepatocellular carcinoma with metastasis and without metastasis.
| MicroRNA | Fold change | diffState | P-value |
|---|---|---|---|
| hsa-let-7e | 2.01425971 | Up | 0.01445168 |
| hsa-miR-18a | 1.91615029 | Up | 0.00600160 |
| hsa-miR-27a | 2.41141531 | Up | 0.03514587 |
| hsa-miR-181a | 1.44064685 | Up | 0.04755760 |
| hsa-miR-221 | 2.00841888 | Up | 0.04713177 |
| hsa-miR-27b | 1.49297559 | Up | 0.04580692 |
| hsa-miR-185 | 2.92150796 | Up | 0.00614653 |
| hsa-miR-361 | 2.00636979 | Up | 0.00671691 |
| hsa-miR-20b | 1.60817127 | Up | 0.02009393 |
| hsa-miR-652 | 1.59785688 | Up | 0.02407410 |
| hsa-miR-140 | 1.38821462 | Up | 0.01697033 |
| hsa-miR-1281 | 1.54732638 | Up | 0.00569455 |
| hsa-miR-1908 | −0.98002981 | Down | 0.03410698 |
| hsa-miR-151b | 1.27543975 | Up | 0.02927815 |
| hsa-miR-4253 | 1.50611955 | Up | 0.04447737 |
| hsa-miR-4330 | −1.47908532 | Down | 0.00890724 |
| hsa-miR-2277 | −1.93607770 | Down | 0.02324236 |
| hsa-miR-3180 | 0.93363816 | Up | 0.04959451 |
| hsa-miR-1268b | 1.46370203 | Up | 0.03639235 |
| hsa-miR-4454 | 2.07258859 | Up | 0.02816307 |
| hsa-miR-4635 | −1.13383056 | Down | 0.04346026 |
| hsa-miR-4720 | −2.26430974 | Down | 0.00572943 |
| hsa-miR-371b | 1.03442024 | Up | 0.04597321 |
| hsa-miR-766 | 0.81375899 | Up | 0.04659530 |
| hsa-miR-619 | −0.99689872 | Down | 0.02691538 |
| hsa-miR-668 | −1.06214814 | Down | 0.02606566 |
| hsa-miR-5189 | −2.18451380 | Down | 0.00521571 |
| hsa-miR-6798 | 1.27908232 | Up | 0.02282279 |
| hsa-miR-6808 | 1.13920150 | Up | 0.04899890 |
| hsa-miR-6880 | −1.19417658 | Down | 0.03378472 |
| hsa-miR-1273h | 1.26636581 | Up | 0.01870905 |
| hsa-miR-8075 | −1.58600573 | Down | 0.01690265 |
diffState, difference State; miR, microRNA.
Figure 2.
Microarray analysis of exosomal miRNAs differentially expressed in plasma samples from patients with metastatic and non-metastatic HCC. (A) Heat map of the differential expression of exosomal miRNAs from patients with metastatic and non-metastatic HCC. Each column represents an individual sample and each row represents a single miRNA. The expression levels of each miRNA in a single sample is depicted according to the color scale. Red represents high expression levels, whereas green represents low expression levels. (B) Volcano plot of the association between the logarithm of the fold change on the x-axis and log of P-value between patients with metastatic and non-metastatic HCC on the y-axis. Red represents miRNAs with expression changes of >1.2 from the remaining miRNAs while blue represents miRNAs with expression changes of <-1.2 fold from the remaining miRNAs. HCC, hepatocellular carcinoma; miRNA/miR, microRNA.
RT-qPCR validation of exosomal miRNA expression profiles
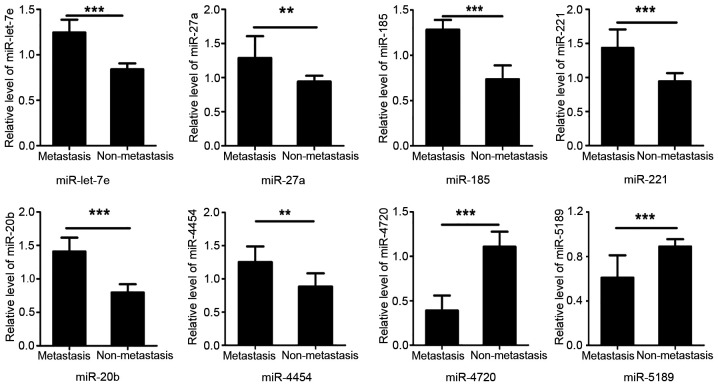
Using RT-qPCR, the eight most dysregulated miRNAs from the miRNA microarray were validated in plasma exosome samples collected from 20 paired patients with HCC with lung metastasis and without metastasis. A total of six exosomal miRNAs (let-7e, miR-27a, miR-221, miR-185, miR-20b and miR-4454) were statistically significantly upregulated in exosome samples from patients with HCC with lung metastasis (P<0.05). The expression levels of the remaining two miRNAs (miR-4720 and miR-5189) were significantly downregulated in samples from patients with metastatic HCC compared with in samples from patients with non-metastatic HCC (P<0.05; Fig. 3). Therefore, the miRNA microarray results were consistent with the results of RT-qPCR.
Figure 3.
Validation of differential exosomal miRNAs in plasma exosomes by reverse transcription-quantitative PCR between patients with metastatic and non-metastatic HCC. Relative expression levels of let-7e, miR-27a, miR-221, miR-185, miR-20b, miR-4454, miR-4720 and miR-5189 in the plasma of 16 patients with lung-metastatic HCC and 16 patients with primary HCC. U6 was used as a reference gene. Data are presented as the mean ± SD of three independent experiments. Student's t-test was used to analyze the data. **P<0.01, ***P<0.001. HCC, hepatocellular carcinoma; miRNA/miR, microRNA.
Correlation analysis of differentially expressed miRNAs in exosomes and matched HCC tissue samples from patients with lung metastasis
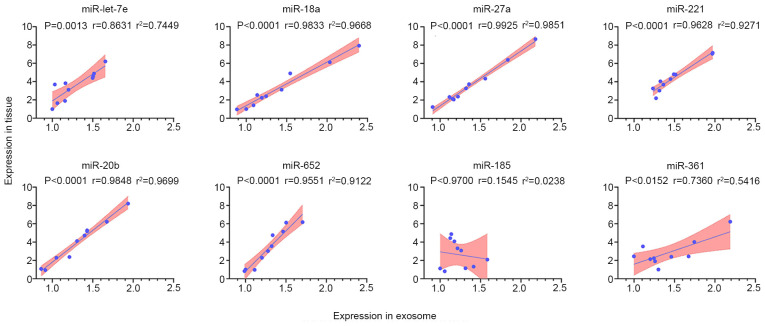
Several significant differentially expressed miRNAs were randomly selected to verify the association between exocrine expression and corresponding tissue expression. The expression levels of the eight highly expressed miRNAs (let-7e, miR-18a, miR-27a, miR-221, miR-185, miR-361, miR-20b and miR-652) in 10 matched plasma exosome samples and tissue samples from patients with HCC with lung metastasis were analyzed by RT-qPCR. The results demonstrated that the expression levels of let-7e, miR-18a, miR-27a, miR-221, miR-20b, miR-361 and miR-652 were positively correlated in exosome and tissue samples (Fig. 4), indicating consistency between plasma and metastatic tissue expression of these miRNAs. However, the levels of miR-185 were not correlated in exosome samples and matched tissue samples. Residual analysis was performed when the correlation was assessed, and these data are shown in Fig. S1. Additionally, a spline curve was used to explore how it may fit the data, and this revealed that the model was suitable (Fig. S2). Therefore, the seven uniformly expressed miRNAs were selected as candidate biomarkers for subsequent experiments.
Figure 4.
Correlation and linear regression analysis of the expression levels of selected miRNAs from plasma exosomes and matched tissue samples in patients with HCC lung metastasis (n=10). The results revealed the correlation of let-7e, miR-18a, miR-27a, miR-221, miR-20b and miR-652 levels between plasma exosomes and lung metastatic tissue samples from 10 cases of HCC lung metastatic patients. The P-value and Pearson's coefficient were displayed from Pearson's test. Linear fit-line, 95% confidence interval band and r2 are shown to indicate the linear correlation between both variables. HCC, hepatocellular carcinoma; miRNA/miR, microRNA.
Candidate exosomal miRNAs and miRNA panels predict HCC metastasis
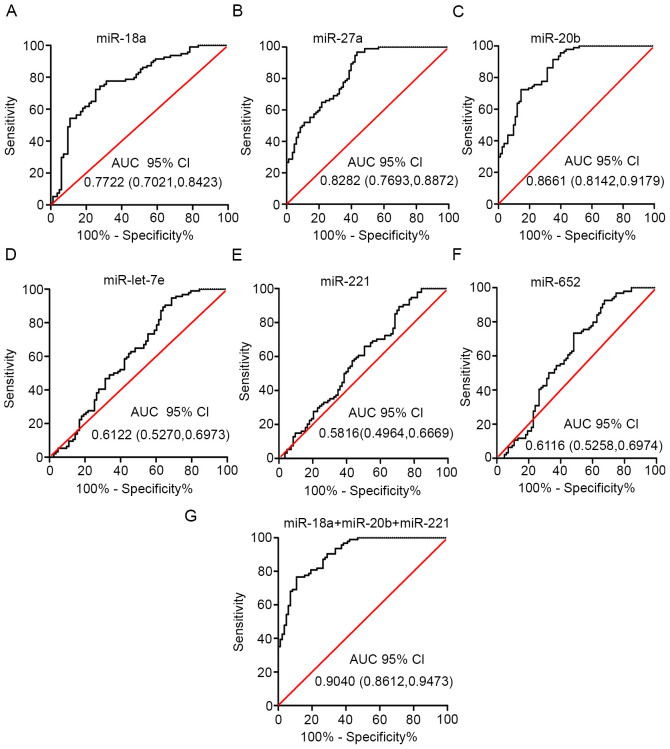
The diagnostic value of each of the six plasma miRNAs (let-7e, miR-18a, miR-27a, miR-221, miR-20b and miR-652) was evaluated using ROC analysis of a GEO dataset. The GEO dataset (accession no. GSE67140) included 172 patients with HCC with or without vascular invasion/metastasis. miR-18a, miR-27a and miR-20b had high AUC values (0.7722, 0.8282 and 0.8661, respectively; Fig. 5A-C). However, the AUC values for the other three miRNAs (let-7e, miR-221 and miR-652) suggested weak classification accuracy (Fig. 5D-F).
Figure 5.
Plasma exosome miRNA score performance. Receiver operating characteristic curves were presented for (A) miR-18a, (B) miR-27a, (C) miR-20b, (D) miR-let-7e, (E) miR-221, (F) miR-652 and (G) for the combination of miR-18a, miR-20b and miR-221. The AUC with 95% CI was used to evaluate the levels of discrimination. 95% CI, 95% confidence interval; AUC, area under the curve; miRNA/miR, microRNA.
Delong's test and Bonferroni correction were performed on various combinations of the miRNA biomarkers. This analysis demonstrated that the combined expression levels of miR-18a, miR-20b and miR-221 exhibited the best diagnostic performance (Fig. 5G). The combination of these three miRNAs increased the AUC to 0.9040 (Tables III and SI). Overall, these findings, combined with the consistent expression levels between plasma exosome samples and tissue samples, demonstrated that the three exosomal miRNAs could be used as a panel for improved detection of vascular invasion/metastasis in patients with HCC.
Table III.
AUC, 95% CI and P-values of the individual candidate microRNAs and the combined panel.
| Exosomal miRNAs | AUC | 95% CI | P-value |
|---|---|---|---|
| let-7e | 0.6122 | 0.5270-0.6973 | 0.0101 |
| miR-18a | 0.7722 | 0.7021-0.8423 | <0.0001 |
| miR-27a | 0.8282 | 0.7963-0.8872 | <0.0001 |
| miR-221 | 0.5816 | 0.4964-0.6669 | 0.0612 |
| miR-20b | 0.8661 | 0.8142-0.9179 | <0.0001 |
| miR-652 | 0.6116 | 0.5258-0.6974 | 0.0105 |
| miR-18a + miR-20b + miR-221 | 0.9040 | 0.8612-0.9473 | <0.0001 |
95% CI, 95% confidence interval; AUC, area under the curve; miR, microRNA.
Candidate exosomal miRNAs are associated with poor prognosis in patients with HCC
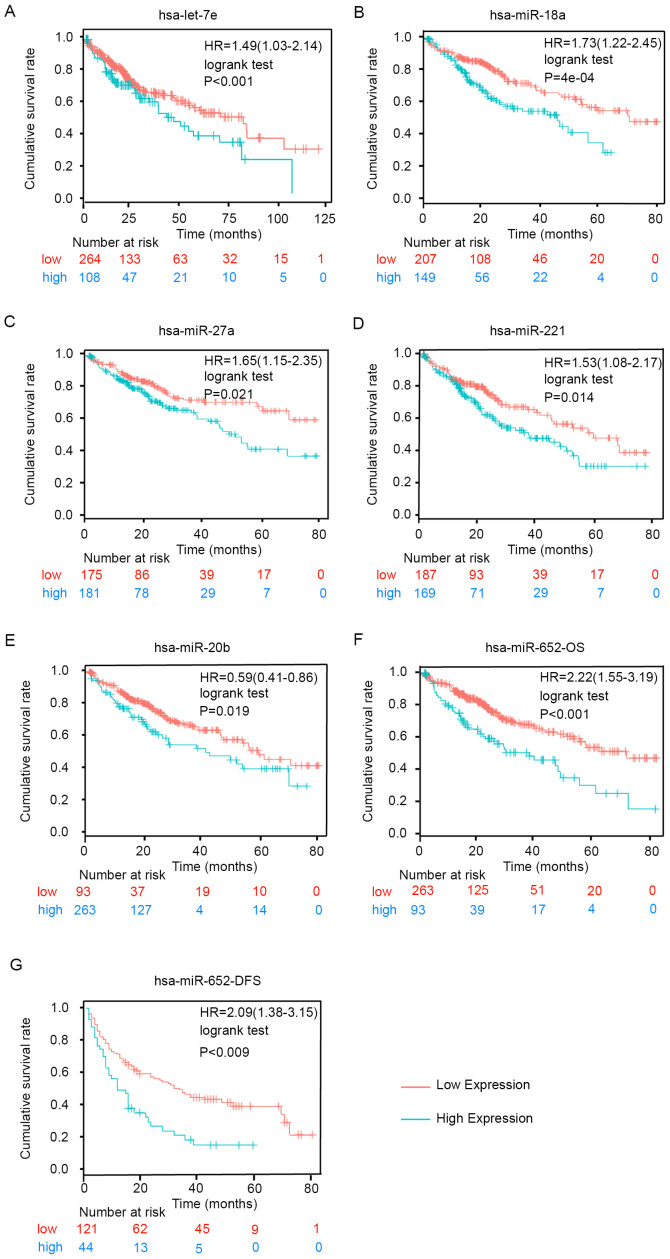
Kaplan-Meier analysis was used to determine whether the candidate miRNAs were associated with OS or disease-free survival (DFS) in patients with HCC using follow-up data collected for 80 months. As shown in Fig. 6, high expression levels of let-7e, miR-18a, miR-27a, miR-221, miR-20b and miR-652 were significantly associated with poor OS (Fig. 6A-F). Furthermore, high expression levels of miR-652 were associated with shorter DFS (Fig. 6G). However, the other miRNAs did not exhibit significant associations with DFS based on the results of the analysis using the Kaplan-Meier plotter tool (data not shown). These results demonstrated that the upregulation of the six exosomal miRNAs was significantly associated with poor survival and increased risk of metastasis in HCC.
Figure 6.
Kaplan-Meier analysis of the prognostic values of the six miRNAs in hepatocellular carcinoma. Kaplan-Meier curve for OS according to the expression levels of (A) let-7e, (B) miR-18a, (C) miR-27a, (D) miR-221 and (E) miR-20b. Kaplan-Meier curve for (F) OS and (G) DFS according to miR-652 expression. HR, hazard ratio (95% confidence interval); miRNA/miR, microRNA; OS, overall survival; DFS, disease-free survival.
Bioinformatics prediction and functional analysis of exosomal miRNAs in HCC metastasis
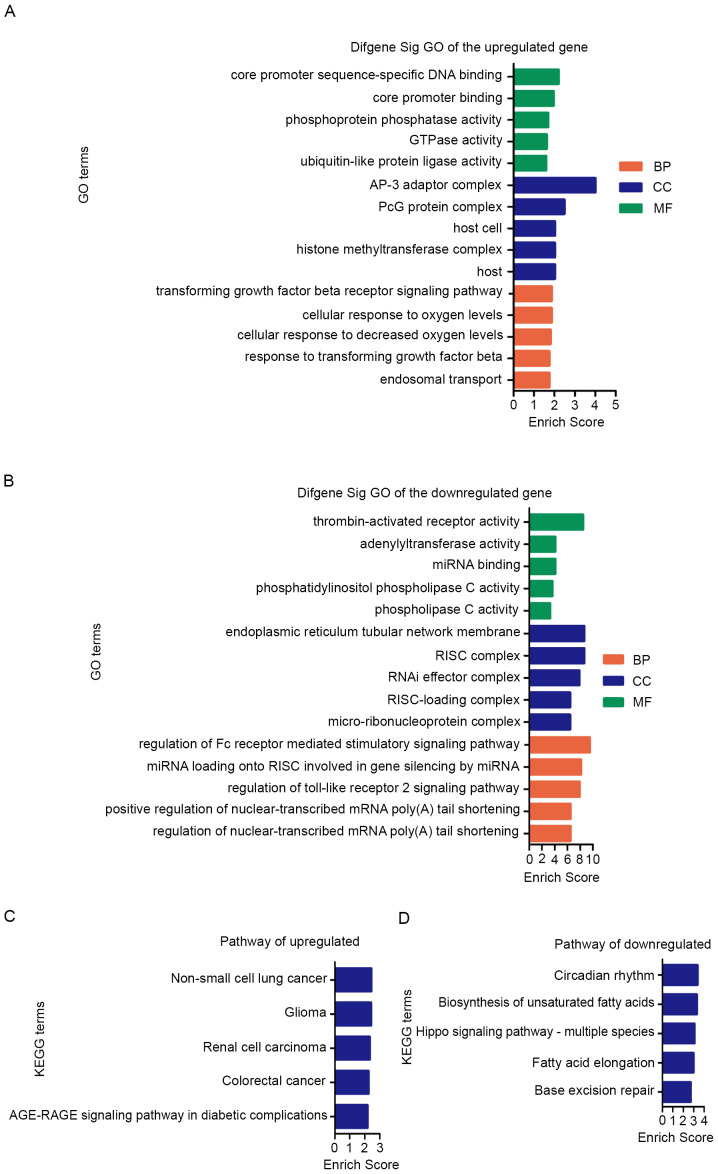
The target genes and signaling pathways associated with the candidate miRNAs were identified using bioinformatics tools. The results were summarized and shown in Tables SII and III). The GO analysis of target gene was mainly classified into three functional groups: Cellular component (CC), molecular function (MF) and biological process (BP). GO enrichment analysis of upregulated miRNAs suggested that most significantly predicted target genes were involved in ‘transforming growth factor beta receptor signaling pathway (BP)’, ‘response to transforming growth factor beta (BP)’, ‘AP-3 adaptor complex (CC)’, ‘PcG protein complex (CC)’ and ‘core promoter sequence-specific DNA binding (MF)’, and these were among the top 5 BP, CC and MF terms (Fig. 7A; Table SII).
Figure 7.
Top 5 enrichment scores in GO enrichment analysis and KEGG pathway enrichment analysis of predicted function of differential exosomal miRNAs. GO terms and signaling pathways terms were sorted by enrichment scores of genes in descending order from top to bottom. In GO analysis, green bars represent MF terms, blue bars represent CC terms and orange red bars represent BP terms of (A) upregulated and (B) downregulated miRNAs. The KEGG analysis was based on potential target genes of (C) upregulated and (D) downregulated miRNAs. miRNA, microRNA; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular component; MF, molecular function.
KEGG pathway analysis of upregulated miRNAs revealed that the top 5 enriched signaling pathways were ‘Non-small cell lung cancer’, ‘Glioma’, ‘Renal cell carcinoma’ and ‘colorectal cancer’ (Fig. 7C; Table SII).
Furthermore, GO enrichment analysis of downregulated miRNAs revealed that most predicted target genes were involved in ‘regulation of the Fc receptor mediated stimulatory signaling pathway’, ‘endoplasmic reticulum tubular network membrane’, ‘RISC complex’, ‘thrombin-activated receptor activity’ and ‘miRNA loading onto RISC involved in gene silencing by miRNA’ (Fig. 7B; Table SIII).
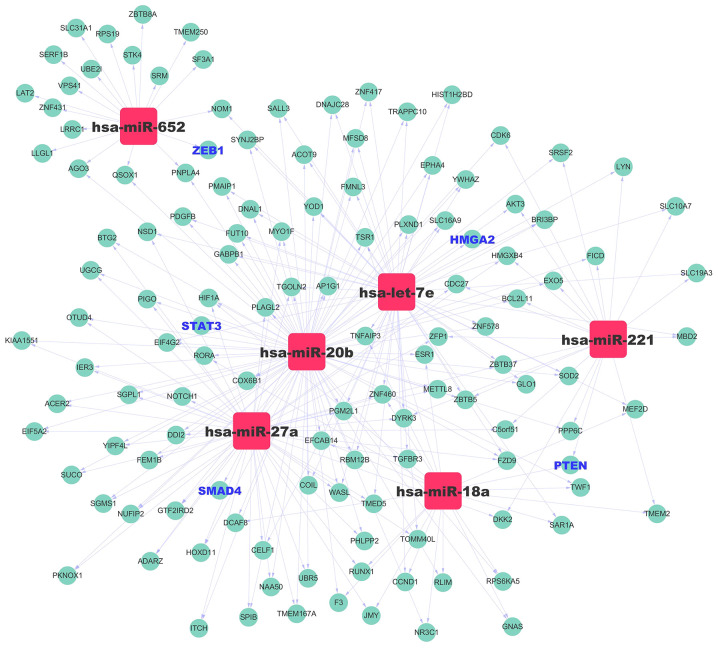
In addition, KEGG pathway analysis of downregulated miRNAs suggested that the top 5 enriched pathways were ‘Circadian rhythm’, ‘Biosynthesis of unsaturated fatty acids’ and ‘Hippo signaling pathway’ (Fig. 7D; Table SIII). To study the regulatory association between the miRNAs and target genes, a miRNA-mRNA network was constructed for the upregulated miRNAs (Fig. 8). Collectively, these findings suggested that the six upregulated exosomal miRNAs were involved in regulating physiological processes associated with metastasis.
Figure 8.
miRNA-gene network. The cyan circles represent genes, whereas the squares represent the miRNAs (let-7e, miR-18a, miR-27a, miR-20b, miR-221 and miR-652). The black-bold genes are upregulated candidate genes, and the blue-bold genes are the target genes that are closely associated with metastasis. The associations between miRNAs and genes are represented by gray lines. miRNA/miR, microRNA.
Discussion
In the present study, plasma exosomal miRNA expression was evaluated in patients with HCC with and without metastasis. A total of 32 exosomal miRNAs were differentially expressed between patients with metastatic and non-metastatic HCC. Among the upregulated miRNAs, the expression levels of six miRNAs were consistent between plasma exosome samples and matched metastatic tissue samples. When comparing the diagnostic value of individual and combined biomarkers, different combination strategies were considered and the combinations were compared with each individual biomarker. Comparisons of AUC values were based on Delong's test and the P-value was adjusted using the Bonferroni strategy for multiple comparisons. The results demonstrated that miR-18a, miR-27a and miR-20b had a high AUC, and the combination of miR-18a, miR-20b and miR-221 exhibited improved performance compared with single miRNA expression. The combined panel had an AUC of 0.9040 in discriminating metastatic cases from non-metastatic cases. Furthermore, high expression levels of let-7e, miR-18a, miR-27a, miR-221, miR-20b and miR-652 were associated with poor OS in patients with HCC.
The aforementioned process was the discovery step. To verify the hypothesis with the help of the public databases, two databases, namely, The Cancer Genome Atlas and GEO, were searched to identify datasets with specific clinical information (patients with HCC with lung-metastasis and without metastasis who did not undergo chemoradiotherapy before surgery). Unfortunately, only the GSE67140 dataset in the GEO database met the inclusion criteria. The other datasets were either too small or scattered to utilize. In the next step, large samples and corresponding follow-up data will be collected to verify the results of OS, DFS and ROC curve results, and to assess the possibility of plasma exosomal miRNA markers for diagnosis or prognosis of patients with HCC. Additionally, clinicopathologic variables, including tumor diameter, number of tumor nodules, histopathologic classification, vein invasion and clinical TNM classification, are also being collected.
Let-7e expression has previously been reported to be downregulated in several human cancer types, and acts as a tumor suppressor that promotes apoptosis and inhibits proliferation, migration and invasion (27,28). By contrast, in the present study, the expression levels of let-7e were increased in plasma from patients with metastatic HCC and high let-7e levels predicted shorter OS. These findings are inconsistent with previous studies (29,30) and require further study. The predicted target genes of let-7e were identified using the miRanda database. High mobility group AT-hook 2 (HMGA2) was among the identified target genes. According to the literature, HMGA2 mediates epithelial-mesenchymal transition (EMT) and regulates transcription factors linking various signaling pathways, such as the TGF-β and MAPK signaling pathways, to regulators of tumor invasiveness and metastasis (31).
miR-18a regulates metastasis in gastric cancer (32), colon cancer (33), breast cancer (34), HCC and nasopharyngeal carcinoma (35,36). In the present study, miR-18a demonstrated a good diagnostic value in discrimination of metastasis. PTEN was among the predicted targets of miR-18a according to the bioinformatics analysis. A previous study has indicated that PTEN-dependent signaling pathways can drive EMT and consequently increase migration, invasion and metastasis (37).
miR-27a influences tumorigenesis, tumor cell proliferation, apoptosis, invasion, migration and angiogenesis by regulating various target genes and could affect clinical therapy, drug sensitivity of patients and patient prognosis (38). Additionally, miR-27a can act as a promising biomarker in serum and is a potential therapeutic target in various tumor types (39). In the present study, miR-27a could accurately predict HCC metastasis. Bioinformatics target prediction demonstrated miR-27a could target SMAD4, a gene involved in the TGF-β signaling pathway. SMAD4/TGF-β signaling is a pivotal regulator of tumor invasion and metastasis following EMT (40). The present study demonstrated that miR-27a served an important role in tumor metastasis.
Dysregulated miR-20b levels are associated with metastasis of breast cancer, colorectal cancer and gastric cancer (41). However, to the best of our knowledge, the potential of miR-20b as an HCC biomarker has not been reported. In the present study, miR-20b could discriminate metastatic HCC from non-metastatic HCC and could be used as a predictive biomarker of HCC metastasis. Target prediction analysis identified STAT3 as a miR-20b target. STAT3 signaling is central to the regulation of metastasis via EMT (42).
miR-652 is defined as an oncomir and can promote metastasis in endometrial cancer, pancreatic cancer, non-small cell lung cancer and prostate cancer cells (43). In the present study, ROC analysis of miR-652 did not demonstrate a high diagnostic accuracy. However, the DFS and OS rates of patients with high miR-652 expression were significantly worse than those of patients with low miR-652 expression.
Finally, bioinformatics tools were used to analyze the possible mechanisms involved in HCC metastasis in the exosomes. The results suggested that the dysregulated miRNAs were involved in various signaling pathways, including ‘GTPase activity’ and ‘cell adhesion molecule binding’, which constitute a complex regulatory network that is relevant to metastasis. An miRNA-mRNA regulatory network was also constructed to examine the associations among the targets of the differentially expressed miRNAs. Nevertheless, bioinformatics prediction can only provide clues for elucidating the role of miRNAs in HCC metastasis and the possible underlying mechanisms. Further experimental validation is still necessary to clarify the detailed regulatory relationship between miRNAs and their target genes in the future.
In conclusion, to the best of our knowledge, the present study was the first to investigate metastasis-associated miRNA profiles in plasma exosomes from patients with HCC. Furthermore, comprehensive analysis of this profile, together with target prediction and survival analysis provided novel insights into the role of exosomes in HCC lung metastasis. A panel consisting of three miRNAs that might serve as an accurate and non-invasive tool for HCC metastasis prediction using plasma exosomes was identified. The present findings suggested that exosomal miRNAs served important roles in HCC metastasis and could represent a complementary clinical tool for the assessment of HCC prognosis.
Supplementary Material
Acknowledgements
The authors would like to thank Professor Zhao Liang and Professor Lin Jie (Southern Medical University, Guangzhou, China) for linguistic advice and experimental assistance.
Funding
The present study was supported by grants from the President Foundation of Nanfang Hospital, Southern Medical University (grant no. 2017B005) and the Natural Science Foundation of Guangdong Province, China (grant no. 2014A030310425).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
BX, CH and ST conceived the study and were responsible for confirming the authenticity of all raw data. CH and ST performed outpatient service, obtained the informed consents, preserved the samples, and performed analysis and interpretation of data. Experiments were performed by BX, ST, CH and XL. BX and CH drafted the manuscript. DS, XL and LL organized the tables and prepared the figure. Data were analyzed by LL, YD and DS. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committee of Southern Medical University Nanfang Hospital (Guangzhou, China) approved the present study. All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, Negri E. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302–309. doi: 10.1016/j.jhep.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–491.e1. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee EC, Kim SH, Park H, Lee SD, Lee SA, Park SJ. Survival analysis after liver resection for hepatocellular carcinoma: A consecutive cohort of 1002 patients. J Gastroenterol Hepatol. 2017;32:1055–1063. doi: 10.1111/jgh.13632. [DOI] [PubMed] [Google Scholar]
- 4.Di Tommaso L, Spadaccini M, Donadon M, Personeni N, Elamin A, Aghemo A, Lleo A. Role of liver biopsy in hepatocellular carcinoma. World J Gastroenterol. 2019;25:6041–6052. doi: 10.3748/wjg.v25.i40.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sersté T, Barrau V, Ozenne V, Vullierme MP, Bedossa P, Farges O, Valla DC, Vilgrain V, Paradis V, Degos F. Accuracy and disagreement of computed tomography and magnetic resonance imaging for the diagnosis of small hepatocellular carcinoma and dysplastic nodules: Role of biopsy. Hepatology. 2012;55:800–806. doi: 10.1002/hep.24746. [DOI] [PubMed] [Google Scholar]
- 6.Russo FP, Imondi A, Lynch EN, Farinati F. When and how should we perform a biopsy for HCC in patients with liver cirrhosis in 2018? A review. Dig Liver Dis. 2018;50:640–646. doi: 10.1016/j.dld.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Mathieu M, Martin-Jaular L, Lavieu G, Thèry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 8.Li I, Nabet BY. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol Cancer. 2019;18:32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Yi M, Dong B, Tan X, Luo S, Wu K. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int J Cancer. 2020 Nov 12; doi: 10.1002/ijc.33386. (Epub ahead of print). doi: 10.1002/ijc.33386, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, Chen D, Li N, Li W. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer. 2020;19:1. doi: 10.1186/s12943-019-1085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: Formation-function-future. Int J Mol Sci. 2028;17:2028. doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kai K, Dittmar RL, Sen S. Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol. 2018;78:22–36. doi: 10.1016/j.semcdb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: Communication from a distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Kahroba H, Hejazi MS, Samadi N. Exosomes: From carcinogenesis and metastasis to diagnosis and treatment of gastric cancer. Cell Mol Life Sci. 2019;76:1747–1758. doi: 10.1007/s00018-019-03035-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, Li SP, Xiong Y, Yuan Y, Min J, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: A multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16:804–815. doi: 10.1016/S1470-2045(15)00048-0. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian XP, Huang WJ, Huang HQ, Liu YH, Wang L, Zhang X, Lin TY, Rao HL, Li M, Liu F, et al. Prognostic and predictive value of a microRNA signature in adults with T-cell lymphoblastic lymphoma. Leukemia. 2019;33:454–2465. doi: 10.1038/s41375-019-0466-0. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Lou W, Chen J, Ding B, Chen D, Zheng H, Jiang D, Xu L, Bao C, Cao G, Fan W. Identification of invasion-metastasis-associated microRNAs in hepatocellular carcinoma based on bioinformatic analysis and experimental validation. J Transl Med. 2018;16:266. doi: 10.1186/s12967-018-1639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan S, Han X, Xue H, Zhang P, Guo X, Li T, Guo X, Yuan G, Deng L, Li G. Let-7f Inhibits glioma cell proliferation, migration, and invasion by targeting periostin. J Cell Biochem. 2015;116:1680–1692. doi: 10.1002/jcb.25128. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen BS, Wu CC. Systems biology as an integrated platform for bioinformatics, systems synthetic biology, and systems metabolic engineering. Cells. 2013;2:635–688. doi: 10.3390/cells2040635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou W, Lai M, Zhang Y, Zheng L, Xing Z, Li T, Zou Z, Song Q, Zhao X, Xia L, et al. Exosome release is regulated by mTORC1. Adv Sci (Weinh) 2018;6:1801313. doi: 10.1002/advs.201801313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding C, Yu H, Shi C, Shi T, Qin H, Cui Y. MiR-let-7e inhibits invasion and magration and regulates HMGB1 expression in papillary thyroid carcinoma. Biomed Pharmacother. 2019;110:528–536. doi: 10.1016/j.biopha.2018.11.057. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Jin S, Liu MD, Pang P, Wu H, Qi ZZ, Liu FY, Sun CF. Hsa-let-7e-5p Inhibits the proliferation and metastasis of head and neck squamous cell carcinoma cells by targeting chemokine receptor 7. J Cancer. 2019;10:1941–1948. doi: 10.7150/jca.29536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Yusheng Z, Qiang C, Zhen W, Boshi S. Methyltransferase-like 1 (METTL1) served as a tumor suppressor in colon cancer by activating 7-methyguanosine (m7G) regulated let-7e miRNA/HMGA2 axis. Life Sci. 2020;249:11748. doi: 10.1016/j.lfs.2020.117480. [DOI] [PubMed] [Google Scholar]
- 30.Bruna C, Laura G, Miyuki U, Isabela W, Fernando A, Glauco B, Edmund C, Katia C. Let-7 miRNA's expression profile and its potential prognostic role in uterine leiomyosarcoma. Cells. 2019;8:1452–1463. doi: 10.3390/cells8111452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou M, Bao X, Luo F, Chen X, Liu L, Wu M. HMGA2 modulates the TGFβ/Smad, TGFβ/ERK and notch signaling pathways in human lens epithelial-mesenchymal transition. Curr Mol Med. 2018;18:71–82. doi: 10.2174/1566524018666180705104844. [DOI] [PubMed] [Google Scholar]
- 32.Yuan J, Tan L, Yin Z, Zhu W, Tao K, Wang G, Shi W, Gao J. MIR17HG-miR-18a/19a axis, regulated by interferon regulatory factor-1, promotes gastric cancer metastasis via Wnt/β-catenin signalling. Cell Death Dis. 2019;10:454. doi: 10.1038/s41419-019-1685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rammer M, Webersinke G, Haitchi-Petnehazy S, Maier E, Hackl H, Charoentong P, Malli T, Steinmair M, Petzer AL, Rumpold H. MicroRNAs and their role for T stage determination and lymph node metastasis in early colon carcinoma. Clin Exp Metastasis. 2017;34:431–440. doi: 10.1007/s10585-017-9863-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang P, Yang Q, Du X, Chen Y, Zhang T. Targeted regulation of Rell2 by microRNA-18a is implicated in the anti-metastatic effect of polyphyllin VI in breast cancer cells. Eur J Pharmacol. 2019;851:161–173. doi: 10.1016/j.ejphar.2019.02.041. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Cai X, Liu E, Tian X, Tian C. MicroRNA-18a promotes proliferation and metastasis in hepatocellular carcinoma via targeting KLF4. Oncotarget. 2017;8:68263–68269. doi: 10.18632/oncotarget.19293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Wang J, Cheng L, Lu MP. miR-18a downregulates DICER1 and promotes proliferation and metastasis of nasopharyngeal carcinoma. Int J Clin Exp Med. 2014;7:847–855. [PMC free article] [PubMed] [Google Scholar]
- 37.Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, Wang S, Liu T, Cai H, Yao K, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun. 2015;6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo W, Zhang D, Ma S, Wang C, Zhang Q, Wang H, He K, Liu Z. miR-27a is highly expressed in H1650 cancer stem cells and regulates proliferation, migration, and invasion. J Cancer Res Ther. 2018;14(Suppl 1):S1004–S1011. doi: 10.4103/0973-1482.199450. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, Xu T, Xu M, Wang S. Circulating exosomal miR-27a and miR-130a act as novel diagnostic and prognostic biomarkers of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:746–754. doi: 10.1158/1055-9965.EPI-18-0067. [DOI] [PubMed] [Google Scholar]
- 40.Siraj AK, Pratheeshkumar P, Divya SP, Parvathareddy SK, Bu R, Masoodi T, Kong Y, Thangavel S, Al-Sanea N, Ashari LH, et al. TGFβ-induced SMAD4-dependent apoptosis proceeded by EMT in CRC. Mol Cancer Ther. 2019;18:1312–1322. doi: 10.1158/1535-7163.MCT-18-1378. [DOI] [PubMed] [Google Scholar]
- 41.Lu P, Gu Y, Li L, Wang F, Yang X, Yang Y. Long noncoding RNA CAMTA1 promotes proliferation and mobility of the human breast cancer cell line MDA-MB-231 via targeting miR-20b. Oncol Res. 2018;26:625–635. doi: 10.3727/096504017X14953948675395. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Wendt MK, Balanis N, Carlin CR, Schiemann WP. STAT3 and epithelial-mesenchymal transitions in carcinomas. JAKSTAT. 2014;3:e28975. doi: 10.4161/jkst.28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W, Zhou C, Luo M, Shi X, Li Y, Sun Z, Zhou F, Chen Z, He J. MiR-652-3p is upregulated in non-small cell lung cancer and promotes proliferation and metastasis by directly targeting Lgl1. Oncotarget. 2016;7:16703–16715. doi: 10.18632/oncotarget.7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.