Abstract
The ubiquitin system is complex, multifaceted, and is crucial for the modulation of a vast number of cellular processes. Ubiquitination is tightly regulated at different levels by a range of enzymes including E1s, E2s, and E3s, and an array of DUBs. The UPS directs protein degradation through the proteasome, and regulates a wide array of cellular processes including transcription and epigenetic factors as well as key oncoproteins. Ubiquitination is key to the dynamic regulation of programmed cell death. Notably, the TNF signaling pathway is controlled by competing ubiquitin conjugation and deubiquitination, which governs both proteasomal degradation and signaling complex formation. In the inflammatory response, ubiquitination is capable of both activating and dampening inflammasome activation through the control of either protein stability, complex formation, or, in some cases, directly affecting receptor activity. In this review, we discuss the enzymes and targets in the ubiquitin system that regulate fundamental cellular processes regulating cell death, and inflammation, as well as disease consequences resulting from their dysregulation. Finally, we highlight several pre-clinical and clinical compounds that regulate ubiquitin system enzymes, with the aim of restoring homeostasis and ameliorating diseases.
Subject terms: Ubiquitins, Tumour-suppressor proteins, Inflammasome
Facts
Signaling pathways activated by TNF are intricately controlled by opposing ubiquitination and deubiquitination thus allowing precise spacial and temporal activation of signaling complexes.
Inflammatory responses utilize ubiquitination to carefully regulate protein activity and stability for the optimal inflammasome activation.
Ubiquitin signaling can be genetically dysregulated in human disease and during infection by pathogens. Targeting such dysregulation is of therapeutic value to combat these diseases.
An increasing number of targeted therapeutics are being developed that are enabling safer and more efficacious treatment regimens, particularly when dosed in combination.
Open questions
Why are many signaling proteins ubiquitinated without a clear functional relevance for the signaling outcome(s)?
How do immunoregulatory drugs elicit their anti-inflammatory effect through CRBN-dependent and -independent mechanisms?
An efficacious frontline therapeutic regimen for patients with multiple myeloma includes treatment with IMiDs, which achieve therapeutic benefit by promoting the proteasomal degradation of critical cellular targets, combined with proteasome inhibitors. Mechanistically, how do these seemingly opposing treatment strategies achieve therapeutic benefit?
Introduction—the ubiquitin system
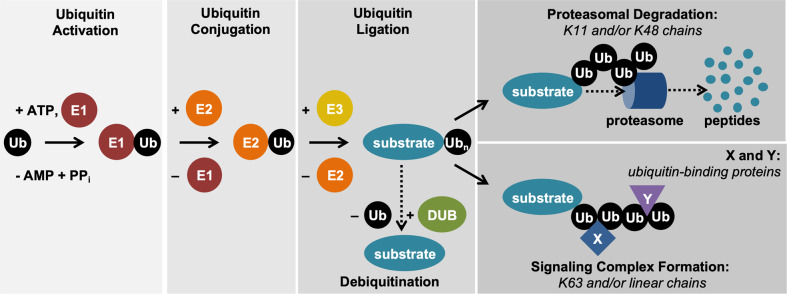
Ubiquitination is an essential posttranslational modification that covalently links the 76-amino acid ubiquitin protein to a target protein. The reaction is a multistep process mediated by three classes of enzymes: ubiquitin activating enzymes (E1), ubiquitin conjugating enzymes (E2), and ubiquitin ligases (E3) [1] (Fig. 1). E1s activate and transfer ubiquitin to E2s, while E3s recruit the substrate proteins for the transfer of ubiquitin moieties. E1 and E2 families are relatively small with 2 and 42 members, respectively, whereas several hundred ubiquitin ligases have been identified [2, 3]. Single ubiquitin molecules can be conjugated to the target (monoubiquitination) or ubiquitin chains can be formed by linking individual ubiquitin molecules by seven internal lysines (K6, K11, K27, K29, K33, K48, K63) or amino-terminal methionine to form linear ubiquitin chains (polyubiquitination) [4]. Different ubiquitin chains are recognized by specific ubiquitin-binding domains [5]. This variety of ubiquitin modifications and ubiquitin-binding proteins is the foundation of selectivity of the ubiquitin system and allows the transmission of defined signals in a precise spatial and temporal manner. The various types of chains play a critical role in distinct cellular processes with K48, K63, and linear ubiquitin chains being the most extensively studied. K48-linked ubiquitin chains and mixed K11/K48-linked ubiquitin chains [6] generally induce degradation of the modified protein via the proteasome, a multisubunit complex that recruits and proteolyzes ubiquitin-modified substrates [5] (Fig. 1). This process is used during signaling events and in transcriptional regulation, but also plays a critical role in protein homeostasis [1]. K63-linked, linear chains or branched/mixed chains can form the scaffolding platform for further protein recruitment and signaling [7]. Cellular events mediated by ubiquitination are counteracted by ubiquitin hydrolases/deubiquitinating enzymes (DUBs), which can cleave specific ubiquitin linkages and/or remove ubiquitin modifications more generally [8] (Fig. 1). DUBs play a critical role in maintaining the ubiquitin system at an equilibrium, but also in restricting signaling to avoid pathway hyperactivation [9]. Given the importance of ubiquitination for a vast number of cellular processes and for overall organismal homeostasis, it is not surprising that this instrumental posttranslational modification has to be tightly controlled and regulated at many levels. In this review, we discuss the enzymes and targets in the ubiquitin system that regulate the fundamental processes of inflammation and cell death, as well as the pathophysiology resulting from their dysregulation. Finally, we highlight the role of targeted therapeutics in the inhibition of the proteasome in the inflammatory response, the interplay of immunomodulatory drugs (iMiDs) in the ubiquitin proteasome system (UPS), and the DUB-like enzyme PLpro whose activity plays a role in the pathogenesis of the topical SARS-CoV-2 pandemic.
Fig. 1. Ubiquitin proteasome system.
Ubiquitination is a multistep process that involves ubiquitin activation by E1 enzymes, ubiquitin conjugation to E2 enzymes, and ubiquitin ligation to the substrate protein via E3 enzymes. Ubiquitination can result in proteasomal degradation of the substrate or in recruitment of the substrate to multiprotein complexes, depending on the topology of the polyubiquitin chain linkages. X and Y indicate ubiquitin chain-binding proteins.
Ubiquitination in the regulation of inflammasomes and cell death
Ubiquitination in TNF-mediated signaling
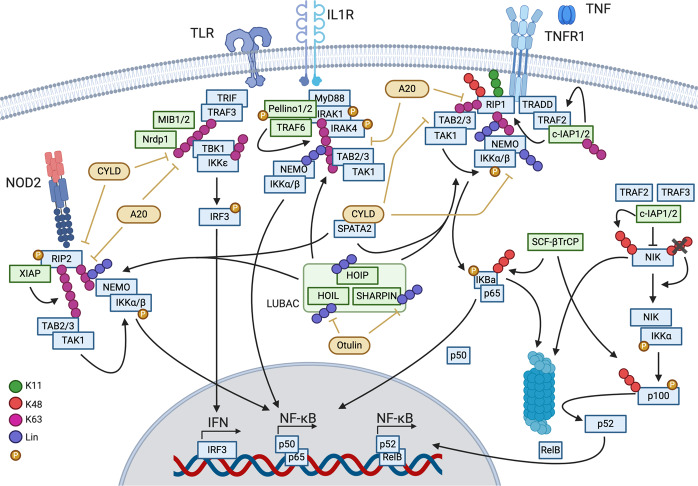
Ubiquitination plays a critical role during TNF (tumor necrosis factor)-mediated inflammatory signaling and cell death regulation. TNF is the most-studied member of the TNF family of inflammatory cytokines [10], and induces NF-κB (nuclear factor κ-light-chain-enhancer of activated B cells) and MAPK (mitogen-activated protein kinase) mediated gene expression, but also apoptotic and necroptotic cell death. Ubiquitination and deubiquitination of various components of the TNF-induced signaling complexes regulate the stability of these complexes and directly influence cellular fate. TNF binds to its cell surface receptor TNFR1 (TNF receptor 1) to trigger protein complex assembly, which is initially ubiquitin independent. TRADD (TNFRSF1A associated via death domain) and RIP1 (receptor interacting protein kinase 1; RIPK1) are recruited to the TNFR1-associated complex via homotypic death domain (DD) interactions [11, 12]. The N-terminal domain of TRADD allows the recruitment of TRAF2 (TNF receptor–associated factor 2) via its TRAF domain [13]. TRAF2 brings along c-IAP1/2 (cellular inhibitor of apoptosis) via the interaction of c-IAP BIR1 (baculovirus IAP repeat) domain and the c-IAP1/2 interacting motif of TRAF2 [14–17].
c-IAP1/2 are RING (really interesting new gene) domain-containing ubiquitin ligases that mediate K11-, K48-, and K63-linked ubiquitination of several components of the TNFR1 complex, including RIP1 and themselves [18–21] (Fig. 2). Ubiquitination of RIP1 on lysine 377 in human (376 in mice) was shown to be critical for NF-kB activation [22, 23] as well as embryonal development in mice [24–26]. Mutagenesis of K376 to arginine resulted in reduced NF-κB activation, reduced complex I formation, and increased cell death [24–26]. Besides the stabilization of RIP1 within the complex, c-IAP1 can promote RIP1 degradation by adding K48-linked ubiquitin chains resulting in reduced cell death potential by RIP1 [27]. Loss of either c-IAP1 or c-IAP2 does not result in developmental defects, but the combined loss is embryonic lethal and partially rescued by the loss of TNFR1 [28].
Fig. 2. Ubiquitination in inflammatory signaling.
Signaling mediated by TNFR1, Il-1R, TLR3/4, or NOD2 relies on complex ubiquitination involving multiple ubiquitin chains to activate inflammatory gene expression. Green indicates ubiquitin ligases and yellow deubiquitinase. Ubiquitin linkage types are indicated in the figure.
Another ubiquitin ligase, LUBAC (linear ubiquitin chain assembly complex) [29], is recruited to the complex by binding c-IAP1/2-generated K63-linked ubiquitin chains [30] (Fig. 2). LUBAC consists of SHARPIN (SHANK-associated RH domain interactor) [31–33], HOIP (HOIL-1-interacting protein) [29], and HOIL-1 (heme-oxidized iron-responsive element binding protein 2 ubiquitin ligase-1) [29], and it mediates the linear ubiquitination of proteins in signaling complexes. HOIP is the E3 responsible for the assembly of linear ubiquitin chains mediated by its RBR domain (RING in-between RING–RING) and the LDD (linear ubiquitin chain determining domain) regions [32, 34–36]. HOIP deficiency induces apoptosis resulting in embryonic lethality at E10.5, which can be rescued by the loss of TNFR1 to E17.5 [37]. HOIL-1 has an RBR domain as well, but its activity is not critical for the linear ubiquitination of LUBAC substrates [38]. A recent publication, however, showed that HOIL-1 E3 activity mediates monoubiquitination of LUBAC components thus forming a platform for the extension of autoinhibitory ubiquitin chains by HOIP [39]. Knockout of HOIL-1 in mice is embryonic lethal, but can be rescued by combined loss of caspase-8 and MLKL, demonstrating the clear interplay of cell death and ubiquitination [40]. The third component of LUBAC, SHARPIN, has no catalytic activity but it is critical for proper signaling and complex stability [33]. Mice with mutations in the SHARPIN gene (cpdm-mice) develop chronic proliferative dermatitis [41, 42], which can be prevented by the deletion of TNF, thus proving the critical role of functional LUBAC for TNF-mediated signaling [32]. Once LUBAC is recruited to the TNFR1 complex, it ubiquitinates several complex components including NEMO, RIP1, TRADD, and TNFR1 [32, 43, 44].
The key effectors of TNF-mediated prosurvival signaling are the kinase complexes IKK (IκB kinase) and TAK1/TAB2/3 (transforming growth factor β-activated kinase 1, TAK1-binding proteins 2/3), which are recruited in a ubiquitin dependent fashion. TAB2 and 3 bind to K63 ubiquitin chains via their carboxy-terminal zinc-finger (ZnF) domain and recruit TAK1 to the signaling complexes that activate NF-κB and MAPK pathways [45, 46]. The IKK complex consists of kinases IKKα/β and the adapter protein NEMO, which recruits the complex through binding of its LZ (leucine zipper) domain to K11 and K63 chains [23, 47] and via high affinity binding of its UBAN (ubiquitin binding in ABIN and NEMO proteins) domain to linear ubiquitin chains [48]. Linear ubiquitination further stabilizes signaling complexes and facilitates MAPK and NF-κB activation [44, 48]. The signaling platform created by K63-linked and linear ubiquitin chains brings kinase complexes into close proximity and enables the activation of IKKβ by TAK1 [49] leading to the phosphorylation of IκBα and NF-κB1. Phosphorylated IκBα undergoes degradative ubiquitination by SCFβ-TrCP (SKP1-cullin-F-box - Beta-transducin repeats-containing proteins), while NF-κB1 is partially degraded, also through the E3 activity of SCFβ-TrCP [50]. This results in free p65 and the NF-κB1 processing product p50, which then can translocate to the nucleus (reviewed by Zhang et al. [51]).
Besides playing a critical role during TNF-mediated signaling, cellular IAPs also regulate noncanonical NF-κB signaling. Together with TRAF2 and TRAF3, c-IAP1/2 form a complex that controls the stability of the kinase NIK (NF-κB–inducing kinase) [52, 53]. Without any stimulus, NIK is constantly ubiquitinated by the E3 activity of c-IAPs from this complex leading to its proteasomal degradation [54] (Fig. 2). Inflammatory pathway activation, such as stimulation of TRAF3-binding and some TRAF2-binding TNF family receptors (e.g., CD40, Fn14), leads to disruption of the TRAF2/TRAF3/c-IAP1/2 complex by inducing degradation and/or translocation of the complex components to insoluble cellular fraction [55–57]. This allows the accumulation of NIK leading to IKKα activation and phosphorylation of p100 [58]. p100 is then ubiquitinated and processed to p52 [59] resulting in noncanonical NF-κB signaling.
Deubiquitination in TNF signaling
The activity and stability of TNF-mediated prosurvival signaling can be restricted by ubiquitin hydrolases or deubiquitinases (DUBs). So far, three DUBs have been described with critical function within these signaling complexes: A20/TNFAIP3, CYLD (cylindromatosis), and Otulin (OTU deubiquitinase with linear linkage specificity) (Fig. 2). Their molecular details as well as phenotypes associated with altered protein function will be discussed below. Initially identified as a TNF/NF-κB target gene [60], A20 is recruited to the TNF receptor complex via ZnF motifs: ZnF4 binds K63-linked, while ZnF7 has a preference for linear ubiquitin chains [61–63]. A20 is also a ubiquitin-editing enzyme that can mediate ubiquitination (using its ZnF4) or deubiquitination (through its OTU domain) of signaling complex components (e.g., RIP1) [64]. Whole body deficiency of A20 results in a severe multiorgan inflammation, which is not surprising given its crucial role in many signaling processes [65]. In addition, extensive studies with conditional knockouts of A20 in mice further underline the importance of A20 during infections and in maintenance of homeostasis (summarized by Martens and van Loo [66]). Inactivation of A20 by mutagenesis of the DUB active site (C103A) or disruption of ZnF4 function (C609A, C612A) resulted in viable animals with no overt phenotype [67, 68]. In both cases, mutagenesis sensitized mice in inflammatory disease models, such as DSS-induced colitis [68], EAE (experimental autoimmune encephalomyelitis), or TNF-induced shock (SIRS) [67]. In contrast, mutagenesis of the ZnF7 domain (C764A, C767A) resulted in inflammatory arthritis [69, 70], and combined inactivation of ZnF4 and ZnF7 domains phenocopied A20 KO mice [69]. Polykratis et al. additionally showed that ZnF7 mutagenesis resulted in less linear chains in complex I [70] providing further evidence that A20 protects linear chains from degradation via its recruitment to modified RIP1 as described before by Draber et al. [43].
The other TNF signaling–associated DUB, CYLD, is a USP domain–containing DUB with specificity for K63-linked and linear ubiquitin chains [71, 72]. CYLD is recruited to the TNFR complex in a LUBAC-dependent fashion through interaction bridged by the adapter protein SPATA2 (Spermatogenesis Associated 2) [73–75]. CYLD can limit TNF-induced prosurvival signaling and promote cell death [76–78]. Accordingly, absence of SPATA2 provides protection against TNF-induced cell death [79]. Genetic disruption of CYLD function can lead to embryonic lethality, increased cancer formation or developmental defects, suggesting a complex role of CYLD in regulating multiple signaling pathways [80].
Otulin (FAM105B) is a DUB with high specificity for hydrolyzing linear ubiquitin chains [81]. Like CYLD, Otulin interacts with LUBAC [43, 82]. However, Otulin is not recruited to the TNFR1 complex; instead, it regulates TNF signaling indirectly by acting on LUBAC [43]. Indeed, studies with catalytic inactive Otulin (Otulin C129A) showed that Otulin plays a critical role in deubiquitinating LUBAC components thus regulating their stability [83]. Furthermore, Otulin inactivation reduces TNFR1 complex formation and increases cell death [83]. Loss of Otulin in the hematopoietic compartment resulted in a TNF-dependent inflammatory phenotype [84]. Otulin C129A/C129A embryos died at E10.5 with extensive cell death in placenta and yolk sac, which was rescued partially by inactivating TNF cell death effectors caspase-8 and RIPK3 [83]. These findings suggest that dysregulated linear ubiquitination triggers cell death and implicate LUBAC, Otulin, and associated proteins in the regulation of cell death activation and inhibition.
Ubiquitination during TNF-induced cell death
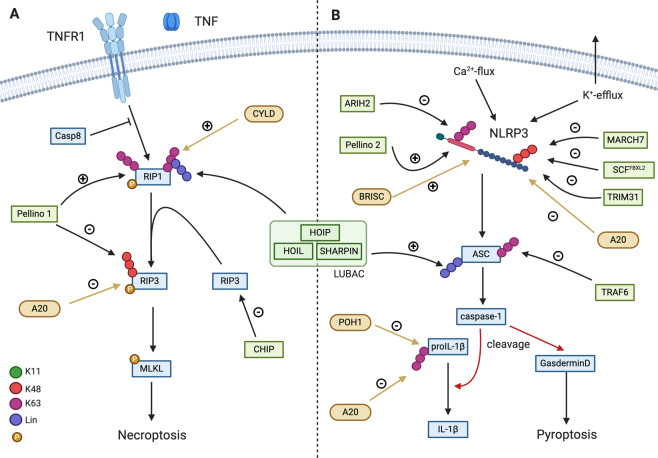
Apart from mediating TNFR1-associated complex I assembly and MAPK and NF-κB activation, ubiquitination also plays a critical role in TNF-induced cell death complexes. These complexes include the cytosolic complex II and the necrosome, as well as the recently described bridging complex [85], which mediates the transition of RIP1 from complex I to the cytosol. This bridging complex consists of heavily ubiquitinated RIP1 and, based on caspase-8 activity, it can either induce RIP1 kinase–dependent apoptosis or necroptosis [85]. During necroptotic cell death, RIP1 undergoes ubiquitination with K63 and linear ubiquitin chains [86] (Fig. 3), with K115 as a prominent site of ubiquitination [87]. Ripk1K115R/K115R mice did not show a developmental phenotype, but were more sensitive to TNF-induced systemic shock [26]. Although ubiquitin ligase(s) that modify RIP1 during necroptotic signaling are not well known, a study found that the E3 PELI1 (Pellino1) can promote RIP1 ubiquitination during necroptosis [88]. In addition to RIP1, PELI1 can also modulate RIP3 protein levels. Phosphorylation of RIP3 at T182 recruits PELI1 to promote K48-linked RIP3 ubiquitination on Lys363 leading to its proteasomal degradation thus restricting necroptosis [89]. Similarly, ubiquitin ligase CHIP (carboxyl terminus of Hsp70-interacting protein) can induce RIP3 lysosomal degradation and restrict necroptosis in a phosphorylation independent manner [89] (Fig. 3). Loss of RIP3 in vivo can partially rescue the phenotype observed in CHIP KO mice, suggesting that the absence of CHIP chaperone function can lead to cell death [90].
Fig. 3. Ubiquitination in the regulation of cell death and inflammasome.
Ubiquitination of the key components of TNF-stimulated cell death and NLRP3 inflammasome–mediated signaling regulate cell survival and inflammation. Green indicates ubiquitin ligases and yellow deubiquitinase. Ubiquitin linkage types are indicated in the figure.
In addition to its role in proliferative signaling, A20 can directly influence necroptosis by deubiquitinating RIP3 at Lysine5, thus leading to reduced cell death [91]. Deubiquitination of RIP1 by CYLD in the necrosome was also suggested to facilitate necroptosis [92]. While it is clear that ubiquitination of complex II components take place during TNF-mediated cell death signaling, there is further research needed to fully understand its significance in regulating cell death and inflammatory responses.
E3 activity of IAPs in direct regulation of cell death signaling
IAPs have been studied initially due to their ability to inhibit caspases [93]. However, the inhibitory activity of IAPs can be counteracted by the mitochondrial protein SMAC [94, 95]. Once released from the mitochondria, SMAC binds BIR domains of IAPs thus blocking their inhibitory ability [93, 94, 96]. This finding has led to the development of a class of small molecules called IAP antagonists or SMAC mimetics [97]. Treatment with IAP antagonists causes a conformational change that opens the c-IAP structure and allows c-IAP RING domain dimerization and consequent activation of c-IAP1/2 E3 ligase activity leading to autoubiquitination and subsequent proteasomal degradation [98, 99]. The prompt activation of c-IAP E3 activity triggers initial RIP1 ubiquitination and NF-κB and MAPK activation [20]. However, proteasomal degradation of c-IAPs allows NIK stabilization and stimulation of the noncanonical NF-κB pathway [55, 100]. NF-κB and MAPK activation cause upregulation of TNF and trigger TNFR1 mediated signaling, which cannot promote RIP1 ubiquitination and prosurvival signaling in the absence of c-IAPs [18, 59, 100]. Instead, RIP1 will form the apoptotic complex with FADD/caspase-8 or complex with RIP3 to promote apoptotic or necroptotic cell death, respectively [20]. This is a great example of the link between exacerbated ubiquitination and cell death induction, and it represents a mechanistic platform for development of IAP antagonists for treatment of various human diseases: from cancer to HIV latency [96, 97, 101].
Pattern recognition receptor signaling and ubiquitination
The innate immune system must operate in an efficient and safe manner to maintain organismal homeostasis by recognizing invading pathogens and initiating the host defense mechanisms [102]. Important components of this defense mechanism are proteins that recognize microbe-associated molecular patterns (MAMPs), such as nucleotide-binding oligomerization domain (NOD)-like receptor (NLRs) family [103]. Among the NLR group, NOD1 and NOD2 are crucial for innate immune responses to some bacterial infections as they sense bacterial peptidoglycans and stimulate NF-κB and MAPK signaling resulting in the expression of proinflammatory cytokines and chemokines [103]. NOD2 is particularly relevant for human pathologies as mutations in the NOD2 locus are linked with inflammatory genetic diseases such as Crohn’s disease, early onset sarcoidosis, and Blau syndrome [104].
NOD2-mediated signaling relies on RIP2 (kinase receptor-interacting protein 2; RIPK2) [105], which recruits a number of signaling regulators to the NOD2-associated protein complex including several ubiquitin ligases and DUBs [106]. Several RIP2-modifying E3s have been reported including c-IAP1/2, XIAP (X-linked inhibitor of apoptosis), Pellino3, ITCH (Itchy E3 Ubiquitin Protein Ligase), TRAF6, and TRIM27 (Tripartite Motif Containing 27), but the critical E3 in NOD/RIP2 pathways is XIAP [106]. XIAP binds the kinase domain of RIP2 with its BIR2 domain to promote K63-linked ubiquitination of RIP2 [107] (Fig. 2). This enables LUBAC recruitment and linear ubiquitination of RIP2 resulting in the activation of NF-κB and MAPK to promote production of inflammatory cytokines and chemokines [108]. Interestingly, the kinase domain of RIP2 does not have enzymatic role in NOD2 signaling, only scaffolding, as it serves as a docking module that enables XIAP to bind and ubiquitinate RIP2 [109, 110]. Disruption of RIP2-XIAP binding blocks RIP2 ubiquitination and inhibits NOD2 signaling, demonstrating that XIAP-mediated RIP2 ubiquitination is critical in this inflammatory pathway [109]. Comparable to TNF signaling, the same trio of DUBs (A20, CYLD, and Otulin) removes K63-linked and linear ubiquitin chains from RIP2 thus restricting NOD2 proinflammatory signaling [108].
Inflammasomes
While some pattern recognition receptors (PRRs) recognize MAMPs, others are stimulated by damage-associated molecular patterns (DAMPs) leading to inflammasome activation and an inflammatory response (Fig. 3) [111]. Activated inflammasomes lead to catalytically active capase-1, which process the proinflammatory cytokines IL-1β and IL-18 [112, 113]. The most studied PRR is NLRP3 (nucleotide-binding and leucine-rich repeat-containing protein 3), which can be stimulated by a variety of DAMPs or MAMPs in a two-step process that involves priming (step 1) and subsequent activation (step 2) [114, 115]. Besides a cytokine response, inflammasome activation can lead to pyroptotic cell death. Pyroptosis is induced by caspase-1/11-mediated cleavage of GSDMD (gasdermin D), which separates the inhibitory and the lytic domains of GSDMD. The cell death inducing N-terminal domain forms membrane pores and allows spillage of cellular contents and ion flux [116, 117].
NLRP3 activity is regulated by various mechanisms. Its levels can be induced by the priming step [118] and ubiquitination can regulate its stability as well as its activity [119]. Several E3 ligases (TRIM31, SCFFBXL2, and MARCH7) can induce degradation of NLRP3 by K48-linked ubiquitination and subsequent proteasomal or autophagosomal degradation [120–122] (Fig. 3). The E3 ARIH2 (Ariadne homolog 2) inactivates NLRP3 by K63-linked ubiquitination without induction of degradation [123]. NLRP3 ubiquitination can also be stimulatory, as is the case with E3 Pellino2, which promotes K63-linked NLRP3 modification [124]. Ubiquitination of other inflammasome-associated proteins such as ASC (apoptosis-associated speck-like protein containing a CARD) can modulate their activity as well. TRAF6 directly or indirectly induces ASC degradation by K63-linked ubiquitin chains [125], while LUBAC can contribute to inflammasome activation by linear ubiquitination [126, 127]. In an analogous fashion to other signaling pathways, deubiquitination can also regulate inflammasome activity. The BRISC DUB complex components BRCC3 (BRCA1/BRCA2-containing complex subunit 3) and ABRO (Abraxas brother 1) are critical in mediating NLRP3 inflammasome activation by deubiquitination [128, 129]. In contrast, A20 activity dampens inflammasome activation although it likely does not act directly on NLRP3 but rather on multiple proteins whose activity and/or stability play important role in inflammasome activity (RIP1, RIP2, RIP3, IL1-β) [130, 131].
IL-1β and TLR signaling
IL-1β is one of the two major proinflammatory cytokines released during pyroptosis. It is activated by cleavage and released from the cell in a passive manner. Upon binding of IL-1β to IL-1 receptor, the adapter protein MyD88 (myeloid differentiation primary response 88) is recruited via TIR (Toll/interleukin-1 receptor) domain interaction [132, 133]. Through a series of homotypic DD–DD interactions, MyD88, IRAK1/2 (Interleukin-1 receptor–associated kinase), and subsequently IRAK4 are recruited to the complex. IRAK4 transactivates itself by autophosphorylation and also phosphorylates and activates IRAK1 [134]. E3s Pellino1/2 and TRAF6 can bind IRAK1, which activates Pellino1/2 by phosphorylation [135, 136] (Fig. 2). These ubiquitin ligases mediate K63-linked ubiquitination of the complex [137, 138], resulting in LUBAC recruitment as well as the activation of MAPK and NF-κB signaling via TAB2/3-TAK1 and the IKK complex (as described above). Signaling by LPS-binding receptor TLR4 (toll-like receptor 4) shares the same pathway components such as IL-1R signaling complex, and both TLR/IL-1R-mediated signaling events can be restricted by DUBs A20 and CYLD68, [139] (Fig. 2). In addition, IL-1β is also deubiquitinated by POH1 inhibiting its cleavage by caspase-1 [140] (Fig. 3). TLRs can form a different kind of intercellular complex. The TIR domain of TLR4 can recruit RHIM domain–containing adapter TRIF, which allows the recruitment of a different subsets of signaling mediators leading to MyD88-independent NF-κB signaling via RIP1 and TRAF6 [141, 142]. During TLR4-mediated necroptotic cell death, TRIF can recruit RIP3, and with it MLKL, therefore enabling RIP3 and MLKL ubiquitination [143]. The pathway is further activated by E3 ligases such as TRIM56, TRIM62, and Pellino, or it can be restricted by TRIM38 (summarized by Ullah et al. [144]). TRIF may also activate the TBK1 (tank-binding kinase 1) and IKKε to activate the transcription factor IRF3, resulting in the induction of Type 1 interferon response [145, 146]. K63-linked polyubiquitination of TBK1 is required for the induction of Type 1 interferons that is mediated by the E3s MIB1 (Mind Bomb 1), MIB2, and Nrdp1, whereas the DUBs CYLD and A20 reverse TBK1 ubiquitination and dampen Type 1 interferon signaling [147] (Fig. 2).
Pathologies associated with inflammatory pathway components
Mutations in E3s and DUBs participating in inflammatory pathways have been associated with various disease. Besides causing autoimmune and autoinflammatory diseases, mutations in these E3s/DUBs can be found in various types of cancers due to the prosurvival signaling function of the pathways. CYLD was first identified as a gene mutated in cylindromatosis, a hereditary form of skin cancer [148], due to dysregulation of NF-κB signaling [76]. A20 seems to have a dual role in cancer. In some cancers, A20 loss of function mutations were identified, suggesting a tumor suppressive role, while high expression of A20 was associated with poor survival in other types (summarized by Martens and van Loo [66]). Copy number gains of c-IAP1/2 have been described in diffuse large B cell lymphoma [149] and carcinomas [150], indicating a role as mediators of TNF-mediated NF-κB signaling. Moreover, deletions of c-IAP1/2 or their adapters TRAF2/3 were described in patients with multiple myeloma that lead to increased noncanonical NF-κB signaling [151, 152]. The IAP family member XIAP, and inherited loss of XIAP, has been implication in X-linked lymphoproliferative type 2 disorder (XLP2), which manifests with lymphohystiocytosis, hypogammaglobulinemia, and lymphomas [153].
Besides their roles in oncogenesis, several TNF pathway components have been associated with autoinflammatory diseases in patients. Haploinsufficiency of A20 was identified in multiple studies resulting in inflammatory diseases marked by hyperactivated NF-κB signaling [154] (for detailed summary [66]). Mutations in Otulin were identified in three patients suffering from autoinflammatory disease (named ORAS or OTULIN-related autoinflammatory syndrome) manifested by panniculitis and recurrent fever [84, 155]. Analysis of one mutation showed reduced protein stability leading to dysregulation of TNF signaling and sensitization to TNF-induced cell death [155]. Treatment of patients with ORAS with anti-TNF antibodies resulted in a decrease of measured inflammatory parameters such as CRP or neutrophil counts [84]. Mutations in the LUBAC components HOIP and HOIL-1 have also been associated with inflammatory phenotypes in human patients [156–158]. Fatal HOIL-1 mutations were identified in three patients of two families, resulting in systemic autoinflammation and susceptibility to infections [157]. Mutations in HOIP leading to loss of expression were associated with multiorgan autoinflammation and immunodeficiency [156, 158]. All described HOIL-1 or HOIP mutations lower the expression of all LUBAC components, which speaks to the critical role of all components for the complex stability. Furthermore, it is increasingly apparent that dysregulation of linear ubiquitination affects many critical signaling events and leads to inflammatory diseases.
Therapeutic modulation of the ubiquitin system
In addition to the IAP antagonists described above, we highlight proteasome inhibitors, immunomodulatory compounds, and SARS-CoV2 protease inhibitors as emerging strategies to target the ubiquitin/proteasome enzymes and pathways featured in this review.
Proteasome inhibition and inflammation response
The proteasome is an important mediator of proliferative response in tumor and immune cells by regulating NF-κB signaling. More specifically, the proteasome participates in the activation of the canonical NF-κB pathway by degrading the negative regulator IκBα to release the transcriptionally active p50 and p65 heterodimer, and in the non-canonical NF-κB pathway, by proteolytically processing the active p52 NF-κB subunit from the longer precursor p100 (Fig. 2) [51]. The clinically approved inhibitors of the b5 proteasomal protease are bortezomib, carfilzomib, and ixazomib, which have multifaceted antiproliferative effects, including inhibition of NF-κB signaling, and have shown the most clinical benefit in multiple myeloma [159].
The clinical benefit of proteasome inhibitors in multiple myeloma was originally proposed to be a consequence of the genetic aberrations that activate NF-κB signaling in multiple myeloma cells, for example as mentioned above [151, 152], that impose NF-κB-dependence [160]. That said, bortezomib was also shown to paradoxically activate NF-κB signaling in multiple myeloma cell lines, primary multiple myeloma cells, and in xenografts, indicating that bortezomib-induced cytotoxicity in multiple myeloma cells is not fully attributable to inhibition of NF-κB signaling [161, 162]. However, the more selective proteasome inhibitors carfilzomib and ixazomib have demonstrated NF-κB repression in multiple myeloma cells [163, 164]. This inhibitory activity reduces the expression of NF-κB–induced cytokines and growth factors that regulate multiple myeloma cells and also pre-osteoclasts, resulting in less bone destruction [165–167].
Proteasome inhibition also exhibits multiple effects on the host immune cells. At clinical doses (≤20 nM), bortezomib treatment increases the levels of immunostimulatory cytokines (IL-2, IL-12, IL-15) in lymphocytes, stabilizes their receptors, and increases their effector function against tumor cells [168]. Bortezomib was also shown to decrease expression of HLA class 1 molecules in multiple myeloma cells and sensitize them to NK cell–mediated lysis [169]. In addition, bortezomib treatment upregulates immunostimulatory tumor antigens, priming the cancer cells for NK cell toxicity. [170]. These studies have shown that bortezomib treatment mediates a dual antitumor effect, by inhibiting tumor cell proliferation and by increasing sensitivity to cytotoxic immune cells. Therefore, immunotherapeutic strategies with antitumor T cell and/or NK cell could provide a combinatorial benefit with bortezomib treatment. In agreement, bortezomib is reported to synergize with the apoptotic potential of cytokines in tumors [171].
Immunomodulatory drugs and the ubiquitin proteasome system
The iMiDs, such as thalidomide, are a class of compounds that display potent anti-inflammatory, antiangiogenic, antiproliferative, and immunomodulatory effects. One of the defining characteristics of the iMiDs is their ability to inhibit the LPS-induced production of the proinflammatory cytokine TNF-α [172], and iMiDs are capable of rescuing mice from endotoxic shock [173]. Similarly, iMiDs reduce the production of IL12 [174], but IL6 is unaffected [172]. In the production of cytokines induced by the MYD88-independent TLR4 pathway, translocation of the transcription factor IRF3 (interferon regulatory factor 3) to the nucleus following its phosphorylation is critical in the regulation of the IFN-1 response [145]. It has been shown that iMiDs prevent this translocation, leading to a reduction in the IFN-1 response [175]. While it is known that IRF3 is tightly regulated by polyubiquitination and subsequent proteasomal degradation [145], how this process is directly or indirectly affected by iMiD treatment is unknown.
Thalidomide, and the structurally related iMiD analogs lenalidomide and pomalidomide, have been shown to harness ubiquitin machinery through recruitment of neo-substrates to the ubiquitin ligase complex CRL4CRBN (CUL4-RBX1-DDB1-CRBN) for ubiquitination and subsequent proteasomal degradation. The clinical efficacy of lenalidomide and pomalidomide in treating multiple myeloma is linked to the degradation of the transcription factors IKZF1/3 (Ikaros zinc finger proteins 1 and 3), as well as the accompanied IL2 upregulation [176–179]. In addition, lenalidomide uniquely induces the degradation of CK1α (casein kinsase 1α) and leads to synthetic lethality in del(5q) myelodysplastic syndrome [180]. A more recent study revealed that the “molecular glue” feature of iMiDs induces ternary complex formation between cereblon (CRBN) and a number of Cys2-His2 (C2H2) ZnF proteins, promoting their subsequent ubiquitination and degradation [181]. This structural plasticity of CRBN has inspired the development of next-generation iMiDs (known as CRBN modulators) and heterobifunctional protein degraders [182, 183]. It remains to be explored whether the degradation of non-IKZF1/3 targets may contribute to the therapeutic activities of established and novel iMiDs.
While the antitumor effects of iMiDs are dependent on CRBN [184], CRBN-independent activity accounts for 30–40% TNF-α inhibition following TLR4 activation [172, 175]. IRF3 translocation and resultant IFN-1 signaling has been shown to proceed in the absence of CRBN in murine studies, which at least partly accounts for the described CRBN-independent activity [175]. The remainder of the anti-inflammatory activity displayed by iMiDs may therefore be CRBN-dependent. It is possible that iMiDs may similarly recruit key proteins that regulate inflammation to the CRL4CRBN complex to promote their proteasomal degradation.
Modulation of the immune response by the SARS-CoV-2 protease PLpro
SARS-CoV-2 is the human coronavirus responsible for the COVID-19 global pandemic, and a defining factor of SARS-CoV-2 pathogenesis is the inhibition of innate immune sensing and reduced interferon response [185, 186]. CoV-2 PLpro, the papain-like protease of SARS-CoV-2, is required for viral polyprotein cleavage and is thus essential for viral replication [187]. In addition, CoV-2 PLpro cleaves both ubiquitin and the ubiquitin-like protein ISG-15 from host proteins, thereby displaying DUB and de-ISGylase activity [188]. One notable target is IRF3, which is a critical component in the type I interferon pathway activated by TLR4 as previously discussed in the context of iMiDs [146]. ISGylation of IRF3 typically provides an activating role by shielding IRF3 from ubiquitylation and subsequent degradation, allowing its phosphorylation and nuclear translocation [145]. Heterologous expression of PLpro within mammalian cells, or by infection with SARS-CoV-2, is reported to decrease phosphorylation of both IRF3 and its activator; TBK1. TBK1 phosphorylation is also an activator of the NF-κB pathway, causing upregulation of inflammatory signaling [189]. Concordantly, INF-β and NF-κB expression levels following poly(I:C) and TNF-α treatments, respectively, were also significantly decreased by infection or PLpro expression [190]. This shows a clear role for the DUB/deISGylation activity of PLpro in the suppression of both IFN and NF-κB signaling pathways, and may contribute to the pathogenesis of the virus.
The effects of CoV-2 PLpro on the IFN and NF-κB signaling pathways, which facilitate establishment of viral infection, as well as being essential for viral replication, make it a promising target for therapeutics. A number of so-called naphthalene-based inhibitors, originally developed against PLpro enzyme of SARS-CoV responsible for the 2003 SARS outbreak [190–193], have shown biochemical and cellular activity against CoV-2 PLPro, owing to the two proteins’ high homology (82% sequence identity). Two related scaffolds in this class are exemplified by GRL-0617 [190] and 3k [194], which both inhibit purified CoV-2 PLpro and prevent viral replication in cells. Interestingly, inhibitors in this series occupy the S4/S3 binding pockets, which are located away from the catalytic triad but block the entry of the ISG-15 C-terminus to cause inhibition. Two newly developed peptide-based inhibitors of CoV-2 PLpro, VIR250 and VIR251, mimic the sequence of PLPro cleavage sites and irreversibly bind to the catalytic Cys111 [195]. While these compounds have not been investigated for antiviral activity in cells, VIR251 showed a clear inhibition of the DUB/deISGylation activity of CoV-2 PLpro.
Conclusions
The ubiquitin system is complex, multifaceted, and is crucial for the modulation of a vast number of cellular processes. Ubiquitination is tightly regulated by a number of enzymes including E1s, E2s, E3s, and DUBs, and dynamically regulates inflammation and a number of programmed cell death mechanisms. Notably, the TNF signaling pathway is controlled by competing ubiquitin conjugation and deubiquitination that governs both proteasomal degradation and signaling complex formation. Aberrant ubiquitin regulation of inflammatory pathways is reported in a number of pathologies including cancers and autoinflammatory diseases. Ubiquitination is capable of both activating and dampening inflammasome activation through the control of either protein stability, complex formation, and, in some cases, directly affecting receptor activity.
Targeting dysfunctional aspects of the ubiquitin system is a promising approach in the treatment of inflammatory disease, cancers, and infectious disease. Proteasome inhibitors decrease tumor cell proliferation, reduce osteoblast activity, and promote sensitivity to immune cells. Although the antiproliferative effects of proteasome inhibitors are complex and not fully understood, inhibition of the NF-κB signaling pathway appears to play a key role. iMiDs co-opt the UPS by recruiting neosubstrates to the ubiquitin ligase complex CRL4CRBN, thus directing their degradation, and have been designed to show promising specificity. The mechanisms underlying the therapeutic benefits of IMiDs are also complex, as their anti-inflammatory activity is partially CRBN-independent, and their regulation of IRF3 signaling remains to be fully elucidated. Finally, the novel coronavirus SARS-CoV-2 modifies the TLR4-induced production of host cytokines through the DUB/deISGylating activity of its papain-like protease PLPro. Pre-clinical inhibitors of PLpro reverse this effect and attenuate viral replication, thus PLPro may be a therapeutic target to combat SARS-CoV-2 morbidity and mortality, and potentially future coronaviruses that utilize similar mechanisms. Looking forward, future work that deepens our understanding of the complex cellular role of the ubiquitin system, including its role in disease processes, is vital. Such work could lead to the identification of novel targets and efficacious therapeutics to treat the wide array of diseases in which the ubiquitin system is implicated.
Acknowledgements
We thank Anna Shemorry, Simon Vidal, and all members of Wertz and Vucic labs for stimulating discussions and helpful comments. Figures 2 and 3 were created with BioRender.com.
Compliance with ethical standards
Conflict of interest
All authors are employees of Genentech/Roche.
Footnotes
Edited by G. Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peter E. Cockram, Matthias Kist
Contributor Information
Ingrid E. Wertz, Email: ingrid@gene.com
Domagoj Vucic, Email: domagoj@gene.com.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Clague MJ, Heride C, Urbé S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–26. doi: 10.1016/j.tcb.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 4.Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J Cell Sci. 2016;129:875–80. doi: 10.1242/jcs.183954. [DOI] [PubMed] [Google Scholar]
- 5.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 6.Haakonsen DL, Rape M. Branching out: improved signaling by heterotypic ubiquitin chains. Trends Cell Biol. 2019;29:704–16. doi: 10.1016/j.tcb.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Emmerich CH, Bakshi S, Kelsall IR, Ortiz-Guerrero J, Shpiro N, Cohen P. Lys63/Met1-hybrid ubiquitin chains are commonly formed during the activation of innate immune signalling. Biochem Biophys Res Commun. 2016;474:452–61. doi: 10.1016/j.bbrc.2016.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta. 2014;1843:114–28. doi: 10.1016/j.bbamcr.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 10.Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119:651–65. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 12.Hsu H, Huang J, Shu H-B, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–96. doi: 10.1016/S1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 13.Hsu H, Shu H-B, Pan M-G, Goeddel DV. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/S0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 14.Shu H-B, Takeuchi M, Goeddel DV. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA. 1996;93:13973–8. doi: 10.1073/pnas.93.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothe M, Pan M-G, Henzel WJ, Ayres TM, V Goeddel D. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–52. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 16.Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC, et al. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (TNF) to efficiently activate NF-κB and to prevent TNF-induced apoptosis. J Biol Chem. 2009;284:35906–15. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varfolomeev E, Wayson SM, Dixit VM, Fairbrother WJ, Vucic D. The inhibitor of apoptosis protein fusion c-IAP2·MALT1 stimulates NF-κB activation independently of TRAF1 AND TRAF2. J Biol Chem. 2006;281:29022–9. doi: 10.1074/jbc.M605116200. [DOI] [PubMed] [Google Scholar]
- 18.Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, et al. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor α (TNFα)-induced NF-κB activation. J Biol Chem. 2008;283:24295–9. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertrand MJM, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, et al. Both cIAP1 and cIAP2 regulate TNFα-mediated NF-κB activation. Proc Natl Acad Sci USA. 2008;105:11778–83. doi: 10.1073/pnas.0711122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor α-induced NF-κB activation. J Biol Chem. 2006;281:13636–43. doi: 10.1074/jbc.M600620200. [DOI] [PubMed] [Google Scholar]
- 23.Ea C-K, Deng L, Xia Z-P, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–57. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Tang Y, Tu H, Zhang J, Zhao X, Wang Y, Qin J, et al. K63-linked ubiquitination regulates RIPK1 kinase activity to prevent cell death during embryogenesis and inflammation. Nat Commun. 2019;10:4157. doi: 10.1038/s41467-019-12033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Zhang H, Xu C, Li X, Li M, Wu X, et al. Ubiquitination of RIPK1 suppresses programmed cell death by regulating RIPK1 kinase activation during embryogenesis. Nat Commun. 2019;10:4158. doi: 10.1038/s41467-019-11839-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kist M, Kőműves LG, Goncharov T, Dugger DL, Yu C, Roose-Girma M, et al. Impaired RIPK1 ubiquitination sensitizes mice to TNF toxicity and inflammatory cell death. Cell Death Differ. 2020. https://www.nature.com/articles/s41418-020-00629-3. [DOI] [PMC free article] [PubMed]
- 27.Annibaldi A, Wicky John S, Vanden Berghe T, Swatek KN, Ruan J, Liccardi G, et al. Ubiquitin-mediated regulation of RIPK1 kinase activity independent of IKK and MK2. Mol Cell. 2018;69:566–e5. doi: 10.1016/j.molcel.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moulin M, Anderton H, Voss AK, Thomas T, Wong WW-L, Bankovacki A, et al. IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J. 2012;31:1679–91. doi: 10.1038/emboj.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–87. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–44. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda F, Deribe YL, Skånland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–41. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–6. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 33.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–6. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 34.Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31:3833–44. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stieglitz B, Rana RR, Koliopoulos MG, Morris-Davies AC, Schaeffer V, Christodoulou E, et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–6. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stieglitz B, Morris‐Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–6. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peltzer N, Rieser E, Taraborrelli L, Draber P, Darding M, Pernaute B, et al. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 2014;9:153–65. doi: 10.1016/j.celrep.2014.08.066. [DOI] [PubMed] [Google Scholar]
- 38.Smit JJ, van Dijk WJ, El Atmioui D, Merkx R, Ovaa H, Sixma TK. Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J Biol Chem. 2013;288:31728–37. doi: 10.1074/jbc.M113.495846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuseya Y, Fujita H, Kim M, Ohtake F, Nishide A, Sasaki K, et al. The HOIL-1L ligase modulates immune signalling and cell death via monoubiquitination of LUBAC. Nat Cell Biol. 2020;22:663–73. doi: 10.1038/s41556-020-0517-9. [DOI] [PubMed] [Google Scholar]
- 40.Peltzer N, Darding M, Montinaro A, Draber P, Draberova H, Kupka S, et al. LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature. 2018;557:112–7. doi: 10.1038/s41586-018-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.HogenEsch H, Gijbels MJ, Offerman E, van Hooft J, van Bekkum DW, Zurcher C. A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am J Pathol. 1993;143:972–82. [PMC free article] [PubMed] [Google Scholar]
- 42.Seymour RE, Hasham MG, Cox GA, Shultz LD, HogenEsch H, Roopenian DC, et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8:416–21. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 43.Draber P, Kupka S, Reichert M, Draberova H, Lafont E, de Miguel D, et al. LUBAC-recruited CYLD and A20 regulate gene activation and cell death by exerting opposing effects on linear ubiquitin in signaling complexes. Cell Rep. 2015;13:2258–72. doi: 10.1016/j.celrep.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 45.Kanayama A, Seth RB, Sun L, Ea C-K, Hong M, Shaito A, et al. TAB2 and TAB3 Activate the NF-κB Pathway through Binding to Polyubiquitin Chains. Mol Cell. 2004;15:535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Shim J-H. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 2005;19:2668–81. doi: 10.1101/gad.1360605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu C-J, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 48.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Clark K, Lawrence T, Peggie MW, Cohen P. An unexpected twist to the activation of IKKβ: TAK1 primes IKKβ for activation by autophosphorylation. Biochem J. 2014;461:531–7. doi: 10.1042/BJ20140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heissmeyer V, Krappmann D, Hatada EN, Scheidereit C. Shared pathways of IκB kinase-induced SCFβTrCP-mediated ubiquitination and degradation for the NF-κB precursor p105 and IκBα. Mol Cell Biol. 2001;21:1024–35. doi: 10.1128/MCB.21.4.1024-1035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-κB: a blossoming of relevance to human pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng P-H, Keats JJ, Wang H, et al. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-κB signaling. Nat Immunol. 2008;9:1364–70. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, et al. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol. 2008;9:1371–8. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao G, Zhang M, Harhaj EW, Sun S-C. Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem. 2004;279:26243–50. doi: 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- 55.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 2007;131:669–81. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 56.Varfolomeev E, Goncharov T, Maecker H, Zobel K, Komuves LG, Deshayes K, et al. Cellular inhibitors of apoptosis are global regulators of NF-κB and MAPK activation by members of the TNF family of receptors. Sci Signal. 2012;5:ra22. doi: 10.1126/scisignal.2001878. [DOI] [PubMed] [Google Scholar]
- 57.Vince JE, Chau D, Callus B, Wong WW-L, Hawkins CJ, Schneider P, et al. TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1–TRAF2 complex to sensitize tumor cells to TNFα. J Cell Biol. 2008;182:171–84. doi: 10.1083/jcb.200801010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Senftleben U. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–9. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 59.Amir RE, Haecker H, Karin M, Ciechanover A. Mechanism of processing of the NF-κB2 p100 precursor: identification of the specific polyubiquitin chain-anchoring lysine residue and analysis of the role of NEDD8-modification on the SCFβ-TrCP ubiquitin ligase. Oncogene. 2004;23:2540–7. doi: 10.1038/sj.onc.1207366. [DOI] [PubMed] [Google Scholar]
- 60.Dixit VM, Green S, Sarma V, Holzman LB, Wolf FW, O’Rourke K, et al. Tumor necrosis factor-alpha induction of novel gene products in human endothelial cells including a macrophage-specific chemotaxin. J Biol Chem. 1990;265:2973–8. doi: 10.1016/S0021-9258(19)39896-5. [DOI] [PubMed] [Google Scholar]
- 61.Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–70. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, et al. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–55. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol Cell. 2010;40:548–57. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 64.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 65.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–4. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martens A, van Loo G. A20 at the crossroads of cell death, inflammation, and autoimmunity. Cold Spring Harb Perspect Biol. 2020;12:a036418. doi: 10.1101/cshperspect.a036418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wertz IE, Newton K, Seshasayee D, Kusam S, Lam C, Zhang J, et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature. 2015;528:370–5. doi: 10.1038/nature16165. [DOI] [PubMed] [Google Scholar]
- 68.Lu TT, Onizawa M, Hammer GE, Turer EE, Yin Q, Damko E, et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Razani B, Whang MI, Kim FS, Nakamura MC, Sun X, Advincula R, et al. Non-catalytic ubiquitin binding by A20 prevents psoriatic arthritis–like disease and inflammation. Nat Immunol. 2020;21:422–33. doi: 10.1038/s41590-020-0634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Polykratis A, Martens A, Eren RO, Shirasaki Y, Yamagishi M, Yamaguchi Y, et al. A20 prevents inflammasome-dependent arthritis by inhibiting macrophage necroptosis through its ZnF7 ubiquitin-binding domain. Nat Cell Biol. 2019;21:731–42. doi: 10.1038/s41556-019-0324-3. [DOI] [PubMed] [Google Scholar]
- 71.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–64. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 72.Komander D, Reyes‐Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63‐linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–73. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elliott PR, Leske D, Hrdinka M, Bagola K, Fiil BK, McLaughlin SH, et al. SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol Cell. 2016;63:990–1005. doi: 10.1016/j.molcel.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kupka S, De Miguel D, Draber P, Martino L, Surinova S, Rittinger K, et al. SPATA2-mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep. 2016;16:2271–80. doi: 10.1016/j.celrep.2016.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schlicher L, Wissler M, Preiss F, Brauns‐Schubert P, Jakob C, Dumit V, et al. SPATA 2 promotes CYLD activity and regulates TNF‐induced NF‐κB signaling and cell death. EMBO Rep. 2016;17:1485–97. doi: 10.15252/embr.201642592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brummelkamp TR, Nijman SMB, Dirac AMG, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 77.Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 2003;424:793–6. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 78.Kovalenko A, Chable-Bessia C, Cantarella G, Israël A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 79.Wei R, Xu LW, Liu J, Li Y, Zhang P, Shan B, et al. SPATA2 regulates the activation of RIPK1 by modulating linear ubiquitination. Genes Dev. 2017;31:1162–76. doi: 10.1101/gad.299776.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–83. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–26. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elliott PR, Nielsen SV, Marco-Casanova P, Fiil BK, Keusekotten K, Mailand N, et al. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol Cell. 2014;54:335–48. doi: 10.1016/j.molcel.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heger K, Wickliffe KE, Ndoja A, Zhang J, Murthy A, Dugger DL, et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature. 2018;559:120–4. doi: 10.1038/s41586-018-0256-2. [DOI] [PubMed] [Google Scholar]
- 84.Damgaard RB, Walker JA, Marco-Casanova P, Morgan NV, Titheradge HL, Elliott PR, et al. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell. 2016;166:1215–.e20. doi: 10.1016/j.cell.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Amin P, Florez M, Najafov A, Pan H, Geng J, Ofengeim D, et al. Regulation of a distinct activated RIPK1 intermediate bridging complex I and complex II in TNFα-mediated apoptosis. Proc Natl Acad Sci USA. 2018;115:E5944–53. doi: 10.1073/pnas.1806973115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Almagro MC, Goncharov T, Newton K, Vucic D. Cellular IAP proteins and LUBAC differentially regulate necrosome-associated RIP1 ubiquitination. Cell Death Dis. 2015;6:e1800. doi: 10.1038/cddis.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Almagro MC, Goncharov T, Izrael-Tomasevic A, Duttler S, Kist M, Varfolomeev E, et al. Coordinated ubiquitination and phosphorylation of RIP1 regulates necroptotic cell death. Cell Death Differ. 2017;24:26–37. doi: 10.1038/cdd.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, Meng H, Li X, Zhu K, Dong K, Mookhtiar AK, et al. PELI1 functions as a dual modulator of necroptosis and apoptosis by regulating ubiquitination of RIPK1 and mRNA levels of c-FLIP. Proc Natl Acad Sci USA. 2017;114:11944–9. doi: 10.1073/pnas.1715742114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi S-W, Park H-H, Kim S, Chung JM, Noh H-J, Kim SK, et al. PELI1 selectively targets kinase-active RIP3 for ubiquitylation-dependent proteasomal degradation. Mol Cell. 2018;70:920–.e7. doi: 10.1016/j.molcel.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 90.Seo J, Lee E-W, Sung H, Seong D, Dondelinger Y, Shin J, et al. CHIP controls necroptosis through ubiquitylation- and lysosome-dependent degradation of RIPK3. Nat Cell Biol. 2016;18:291–302. doi: 10.1038/ncb3314. [DOI] [PubMed] [Google Scholar]
- 91.Onizawa M, Oshima S, Schulze-Topphoff U, Oses-Prieto JA, Lu T, Tavares R, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16:618–27. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moquin DM, McQuade T, Chan FK-M. CYLD deubiquitinates RIP1 in the TNFα-induced necrosome to facilitate kinase activation and programmed necrosis. Harhaj EW, editor. PLoS One. 2013;8:e76841. doi: 10.1371/journal.pone.0076841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salvesen GS, Abrams JM. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–84. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 94.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c–dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/S0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 95.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/S0092-8674(00)00009-X. [DOI] [PubMed] [Google Scholar]
- 96.Morrish E, Brumatti G, Silke J. Future therapeutic directions for Smac-mimetics. Cells. 2020;9:406. doi: 10.3390/cells9020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Disco. 2012;11:109–24. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 98.Dueber EC, Schoeffler AJ, Lingel A, Elliott JM, Fedorova AV, Giannetti AM, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376–80. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- 99.Feltham R, Bettjeman B, Budhidarmo R, Mace PD, Shirley S, Condon SM, et al. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem. 2011;286:17015–28. doi: 10.1074/jbc.M111.222919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vince JE, Wong WW-L, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 101.Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, et al. Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature. 2020;578:160–5. doi: 10.1038/s41586-020-1951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen G, Shaw MH, Kim Y-G, Nuñez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol Mech Dis. 2009;4:365–98. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 103.Caruso R, Warner N, Inohara N, Núñez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Caso F, Galozzi P, Costa L, Sfriso P, Cantarini L, Punzi L. Autoinflammatory granulomatous diseases: from Blau syndrome and early-onset sarcoidosis to NOD2-mediated disease and Crohn’s disease. RMD Open. 2015;1:e000097. doi: 10.1136/rmdopen-2015-000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kobayashi K, Inohara N, Hernandez LD, Galán JE, Núñez G, Janeway CA, et al. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–9. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 106.Witt A, Vucic D. Diverse ubiquitin linkages regulate RIP kinases-mediated inflammatory and cell death signaling. Cell Death Differ. 2017;24:1160–71. doi: 10.1038/cdd.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Damgaard RB, Nachbur U, Yabal M, Wong WW-L, Fiil BK, Kastirr M, et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell. 2012;46:746–58. doi: 10.1016/j.molcel.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 108.Topal Y, Gyrd-Hansen M. RIPK2 NODs to XIAP and IBD. Semin Cell Dev Biol. 2020. [DOI] [PubMed]
- 109.Goncharov T, Hedayati S, Mulvihill MM, Izrael-Tomasevic A, Zobel K, Jeet S, et al. Disruption of XIAP-RIP2 association blocks NOD2-mediated inflammatory signaling. Mol Cell. 2018;69:551–.e7. doi: 10.1016/j.molcel.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 110.Hrdinka M, Schlicher L, Dai B, Pinkas DM, Bufton JC, Picaud S, et al. Small molecule inhibitors reveal an indispensable scaffolding role of RIPK 2 in NOD 2 signaling. EMBO J. 2018;37:e99372. doi: 10.15252/embj.201899372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 112.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, et al. A novel heterodimeric cysteine protease is required for interleukin-1βprocessing in monocytes. Nature. 1992;356:768–74. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 113.Gu Y, Kuida K, Tsutsui H, Ku G, Hsiao K, Fleming MA, et al. Activation of interferon-γ inducing factor mediated by interleukin-1β converting enzyme. Science. 1997;275:206–9. doi: 10.1126/science.275.5297.206. [DOI] [PubMed] [Google Scholar]
- 114.Swanson KV, Deng M, Ting JP-Y. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–89. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chauhan D, Vande Walle L, Lamkanfi M. Therapeutic modulation of inflammasome pathways. Immunol Rev. 2020;297:123–38. doi: 10.1111/imr.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–71. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 117.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 118.Groslambert M, Py B. Spotlight on the NLRP3 inflammasome pathway. J Inflamm Res. 2018;ume 11:359–74. doi: 10.2147/JIR.S141220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lopez‐Castejon G. Control of the inflammasome by the ubiquitin system. FEBS J. 2020;287:11–26. doi: 10.1111/febs.15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J, et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun. 2016;7:13727. doi: 10.1038/ncomms13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z, et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell. 2015;160:62–73. doi: 10.1016/j.cell.2014.11.047. [DOI] [PubMed] [Google Scholar]
- 122.Han S, Lear TB, Jerome JA, Rajbhandari S, Snavely CA, Gulick DL, et al. Lipopolysaccharide primes the NALP3 inflammasome by inhibiting its ubiquitination and degradation mediated by the SCF FBXL2 E3 ligase. J Biol Chem. 2015;290:18124–33. doi: 10.1074/jbc.M115.645549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kawashima A, Karasawa T, Tago K, Kimura H, Kamata R, Usui-Kawanishi F, et al. ARIH2 ubiquitinates NLRP3 and negatively regulates NLRP3 inflammasome activation in macrophages. J Immunol. 2017;199:3614–22. doi: 10.4049/jimmunol.1700184. [DOI] [PubMed] [Google Scholar]
- 124.Humphries F, Bergin R, Jackson R, Delagic N, Wang B, Yang S, et al. The E3 ubiquitin ligase Pellino2 mediates priming of the NLRP3 inflammasome. Nat Commun. 2018;9:1560. doi: 10.1038/s41467-018-03669-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chiu H-W, Chen C-H, Chang J-N, Chen C-H, Hsu Y-H. Far-infrared promotes burn wound healing by suppressing NLRP3 inflammasome caused by enhanced autophagy. J Mol Med. 2016;94:809–19. doi: 10.1007/s00109-016-1389-0. [DOI] [PubMed] [Google Scholar]
- 126.Guan K, Wei C, Zheng Z, Song T, Wu F, Zhang Y, et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J Immunol. 2015;194:4880–90. doi: 10.4049/jimmunol.1402851. [DOI] [PubMed] [Google Scholar]
- 127.Rodgers MA, Bowman JW, Fujita H, Orazio N, Shi M, Liang Q, et al. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. J Exp Med. 2014;211:1333–47. doi: 10.1084/jem.20132486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Py BF, Kim M-S, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49:331–8. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 129.Ren G, Zhang X, Xiao Y, Zhang W, Wang Y, Ma W, et al. ABRO1 promotes NLRP3 inflammasome activation through regulation of NLRP3 deubiquitination. EMBO J. 2019;38:e100376. doi: 10.15252/embj.2018100376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duong BH, Onizawa M, Oses-Prieto JA, Advincula R, Burlingame A, Malynn BA, et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity. 2015;42:55–67. doi: 10.1016/j.immuni.2014.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vande Walle L, Van Opdenbosch N, Jacques P, Fossoul A, Verheugen E, Vogel P, et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–5. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 133.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–47. doi: 10.1016/S1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 134.Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, et al. Sequential control of Toll-like receptor–dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–91. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 135.Smith H, Peggie M, Campbell DG, Vandermoere F, Carrick E, Cohen P. Identification of the phosphorylation sites on the E3 ubiquitin ligase Pellino that are critical for activation by IRAK1 and IRAK4. Proc Natl Acad Sci USA. 2009;106:4584–90. doi: 10.1073/pnas.0900774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baud V, Liu Z-G, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ordureau A, Smith H, Windheim M, Peggie M, Carrick E, Morrice N, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 139.Shembade N, Ma A, Harhaj EW. Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–9. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang L, Liu Y, Wang B, Xu G, Yang Z, Tang M, et al. POH1 deubiquitinates pro-interleukin-1β and restricts inflammasome activity. Nat Commun. 2018;9:4225. doi: 10.1038/s41467-018-06455-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, et al. RIP1 is an essential mediator of Toll-like receptor 3–induced NF-κB activation. Nat Immunol. 2004;5:503–7. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 142.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the toll-like receptor signaling. J Immunol. 2003;171:4304–10. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 143.Lawlor KE, Khan N, Mildenhall A, Gerlic M, Croker BA, D’Cruz AA, et al. RIPK3 promotes cell death and NLRP3 inflammasome activation in the absence of MLKL. Nat Commun. 2015;6:6282. doi: 10.1038/ncomms7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ullah MO, Sweet MJ, Mansell A, Kellie S, Kobe B. TRIF-dependent TLR signaling, its functions in host defense and inflammation, and its potential as a therapeutic target. J Leukoc Biol. 2016;100:27–45. doi: 10.1189/jlb.2RI1115-531R. [DOI] [PubMed] [Google Scholar]
- 145.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–58. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 146.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–81. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 147.Hu H, Sun S-C. Ubiquitin signaling in immune responses. Cell Res. 2016;26:457–83. doi: 10.1038/cr.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Biggs PJ, Wooster R, Ford D, Chapman P, Mangion J, Quirk Y, et al. Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12–q13: evidence for its role as a tumour suppressor gene. Nat Genet. 1995;11:441–3. doi: 10.1038/ng1295-441. [DOI] [PubMed] [Google Scholar]
- 149.Yang Y, Kelly P, Shaffer AL, Schmitz R, Yoo HM, Liu X, et al. Targeting non-proteolytic protein ubiquitination for the treatment of diffuse large B cell lymphoma. Cancer Cell. 2016;29:494–507. doi: 10.1016/j.ccell.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–67. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng W-J, et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell. 2007;12:131–44. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rigaud S, Fondanèche M-C, Lambert N, Pasquier B, Mateo V, Soulas P, et al. XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature. 2006;444:110–4. doi: 10.1038/nature05257. [DOI] [PubMed] [Google Scholar]
- 154.Zhou Q, Wang H, Schwartz DM, Stoffels M, Park YH, Zhang Y, et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat Genet. 2016;48:67–73. doi: 10.1038/ng.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Damgaard RB, Elliott PR, Swatek KN, Maher ER, Stepensky P, Elpeleg O, et al. OTULIN deficiency in ORAS causes cell type‐specific LUBAC degradation, dysregulated TNF signalling and cell death. EMBO Mol Med. 2019;11:e9324. doi: 10.15252/emmm.201809324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Boisson B, Laplantine E, Dobbs K, Cobat A, Tarantino N, Hazen M, et al. Human HOIP and LUBAC deficiency underlies autoinflammation, immunodeficiency, amylopectinosis, and lymphangiectasia. J Exp Med. 2015;212:939–51. doi: 10.1084/jem.20141130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boisson B, Laplantine E, Prando C, Giliani S, Israelsson E, Xu Z, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol. 2012;13:1178–86. doi: 10.1038/ni.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Oda H, Beck U, Kuehn HS, Sampaio Moura N, Hoffmann P, Ibarra M, et al. Second case of HOIP deficiency expands clinical features and defines inflammatory transcriptome regulated by LUBAC. Front Immunol. 2019;10:479. doi: 10.3389/fimmu.2019.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wertz IE, Wang X. From discovery to bedside: targeting the ubiquitin system. Cell Chem Biol. 2019;26:156–77. doi: 10.1016/j.chembiol.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 160.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitinproteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–85. doi: 10.1016/S0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 161.Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K, et al. Bortezomib induces canonical nuclear factor-κB activation in multiple myeloma cells. Blood. 2009;114:1046–52. doi: 10.1182/blood-2009-01-199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li C, Chen S, Yue P, Deng X, Lonial S, Khuri FR, et al. Proteasome inhibitor PS-341 (bortezomib) induces calpain-dependent IκBα degradation. J Biol Chem. 2010;285:16096–104. doi: 10.1074/jbc.M109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Allegra A, Speciale A, Molonia MS, Guglielmo L, Musolino C, Ferlazzo G, et al. Curcumin ameliorates the in vitro efficacy of carfilzomib in human multiple myeloma U266 cells targeting p53 and NF-κB pathways. Toxicol Vitr. 2018;47:186–94. doi: 10.1016/j.tiv.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 164.Chauhan D, Tian Z, Zhou B, Kuhn D, Orlowski R, Raje N, et al. In vitro and in vivo selective antitumor activity of a novel orally bioavailable proteasome inhibitor MLN9708 against multiple myeloma cells. Clin Cancer Res. 2011;17:5311–21. doi: 10.1158/1078-0432.CCR-11-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ang E, Pavlos NJ, Rea SL, Qi M, Chai T, Walsh JP, et al. Proteasome inhibitors impair RANKL-induced NF-κB activity in osteoclast-like cells via disruption of p62, TRAF6, CYLD, and IκBα signaling cascades. J Cell Physiol. 2009;220:450–9. doi: 10.1002/jcp.21787. [DOI] [PubMed] [Google Scholar]
- 166.Hongming H, Jian H. Bortezomib inhibits maturation and function of osteoclasts from PBMCs of patients with multiple myeloma by downregulating TRAF6. Leuk Res. 2009;33:115–22. doi: 10.1016/j.leukres.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 167.Garcia-Gomez A, Quwaider D, Canavese M, Ocio EM, Tian Z, Blanco JF, et al. Preclinical activity of the oral proteasome inhibitor MLN9708 in myeloma bone disease. Clin Cancer Res. 2014;20:1542–54. doi: 10.1158/1078-0432.CCR-13-1657. [DOI] [PubMed] [Google Scholar]
- 168.Pellom ST, Dudimah DF, Thounaojam MC, Uzhachenko RV, Singhal A, Richmond A, et al. Bortezomib augments lymphocyte stimulatory cytokine signaling in the tumor microenvironment to sustain CD8+T cell antitumor function. Oncotarget. 2017;8:8604–21. doi: 10.18632/oncotarget.14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Shi J, Tricot GJ, Garg TK, Malaviarachchi PA, Szmania SM, Kellum RE, et al. Bortezomib down-regulates the cell-surface expression of HLA class I and enhances natural killer cell–mediated lysis of myeloma. Blood. 2008;111:1309–17. doi: 10.1182/blood-2007-03-078535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, et al. ATM-ATR–dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–11. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- 171.Khan T, Stauffer JK, Williams R, Hixon JA, Salcedo R, Lincoln E, et al. Proteasome inhibition to maximize the apoptotic potential of cytokine therapy for murine neuroblastoma tumors. J Immunol. 2006;176:6302–12. doi: 10.4049/jimmunol.176.10.6302. [DOI] [PubMed] [Google Scholar]
- 172.Sampaio EP, Sarno EN, Galilly R, Cohn ZA, Kaplan G. Thalidomide selectively inhibits tumor necrosis factor alpha production by stimulated human monocytes. J Exp Med. 1991;173:699–703. doi: 10.1084/jem.173.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Moreira AL, Wang J, Sarno EN, Kaplan G. Thalidomide protects mice against LPS-induced shock. Braz J Med Biol Res. 1997;30:1199–207. doi: 10.1590/S0100-879X1997001000010. [DOI] [PubMed] [Google Scholar]
- 174.Moller DR, Wysocka M, Greenlee BM, Ma X, Wahl L, Flockhart DA, et al. Inhibition of IL-12 production by thalidomide. J Immunol. 1997;159:5157–61. [PubMed] [Google Scholar]
- 175.Millrine D, Miyata H, Tei M, Dubey P, Nyati K, Nakahama T, et al. Immunomodulatory drugs inhibit TLR4-induced type-1 interferon production independently of Cereblon via suppression of the TRIF/IRF3 pathway. Int Immunol. 2016;28:307–15. doi: 10.1093/intimm/dxw005. [DOI] [PubMed] [Google Scholar]
- 176.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–5. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–9. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reid G, Hübner MR, Métivier R, Brand H, Denger S, Manu D, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/S1097-2765(03)00090-X. [DOI] [PubMed] [Google Scholar]
- 179.Gandhi AK, Kang J, Havens CG, Conklin T, Ning Y, Wu L, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4 CRBN. Br J Haematol. 2014;164:811–21. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Krönke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, et al. Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature. 2015;523:183–8. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sievers QL, Petzold G, Bunker RD, Renneville A, Słabicki M, Liddicoat BJ, et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362:eaat0572. [DOI] [PMC free article] [PubMed]
- 182.Hansen JD, Correa M, Nagy MA, Alexander M, Plantevin V, Grant V, et al. Discovery of CRBN E3 ligase modulator CC-92480 for the treatment of relapsed and refractory multiple myeloma. J Med Chem. 2020;63:6648–76. doi: 10.1021/acs.jmedchem.9b01928. [DOI] [PubMed] [Google Scholar]
- 183.Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol. 2018;14:706–14. doi: 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ito T, Handa H. Cereblon and its downstream substrates as molecular targets of immunomodulatory drugs. Int J Hematol. 2016;104:293–9. doi: 10.1007/s12185-016-2073-4. [DOI] [PubMed] [Google Scholar]
- 185.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–41. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600–12. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ratia K, Kilianski A, Baez-Santos YM, Baker SC, Mesecar A. Structural basis for the ubiquitin-linkage specificity and deISGylating activity of SARS-CoV papain-like protease. Rey FA, editor. PLoS Pathog. 2014;10:e1004113. doi: 10.1371/journal.ppat.1004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–62. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ghosh AK, Takayama J, Aubin Y, Ratia K, Chaudhuri R, Baez Y, et al. Structure-based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome−coronavirus papain-like protease. J Med Chem. 2009;52:5228–40. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ghosh AK, Takayama J, Rao KV, Ratia K, Chaudhuri R, Mulhearn DC, et al. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein−ligand X-ray structure and biological evaluation. J Med Chem. 2010;53:4968–79. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Báez-Santos YM, Barraza SJ, Wilson MW, Agius MP, Mielech AM, Davis NM, et al. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem. 2014;57:2393–412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP, et al. Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2. EMBO J. 2020;39:e106275. doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rut W, Lv Z, Zmudzinski M, Patchett S, Nayak D, Snipas SJ, et al. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci Adv. 2020;6:eabd4596. doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]