Abstract
Batrachochytrium dendrobatidis is a globally distributed generalist pathogen that has driven many amphibian populations to extinction. The life cycle of B. dendrobatidis has two main cell types, motile zoospores, and sessile reproductive sporangia. When grown in a nutrient-rich liquid medium, B. dendrobatidis forms aggregates of sporangia that transition into monolayers on surfaces and at the air-liquid interface. Pathogenic microorganisms use biofilms as mechanisms of group interactions to survive under harsh conditions in the absence of a suitable host. We used fluorescent and electron microscopy, crystal violet, transcriptomic, and gas chromatographic analyses to understand the characteristics of B. dendrobatidis monolayers. The cell-free monolayer fraction showed the presence of extracellular ribose, mannose, xylose, galactose, and glucose. Transcriptome analysis showed that 27%, 26%, and 4% of the genes were differentially expressed between sporangia/zoospores, monolayer/zoospores, and sporangia/monolayer pairs respectively. In pond water studies, zoospores developed into sporangia and formed floating aggregates at the air-water interface and attached film on the bottom of growth flasks. We propose that B. dendrobatidis can form surface-attached monolayers in nutrient-rich environments and aggregates of sporangia in nutrient-poor aquatic systems. These monolayers and aggregates may facilitate dispersal and survival of the fungus in the absence of a host. We provide evidence for using a combination of plant-based chemicals, allicin, gingerol, and curcumin as potential anti-chytrid drugs to mitigate chytridiomycosis.
Keywords: Biofilm, Transcriptomics, Gene expression, Phytochemicals, Anti-chytrid drugs
Graphical abstract
Introduction
Batrachochytrium dendrobatidis is an aquatic non-filamentous fungus that belongs to the phylum Chytridiomycota [1]. It causes chytridiomycosis, an emerging fungal disease that has driven the amphibian populations to the brink of extinction worldwide [[2], [3], [4], [5], [6], [7]]. The life cycle of B. dendrobatidis consists of two distinct cell types, free-swimming zoospores and sessile sporangia [8]. Zoospores are unicellular and wall-less and can swim approximately 2 cm using a posteriorly oriented single flagellum before encysting and enlarging into chitin-containing walled thalli. Thalli mature into sporangia (>10 μm), on a suitable host or substrate within 24 h [9]. Sporangia serve as reproductive structures and are anchored to substrates by rhizoids [8]. The fungus infects the keratinized mouthparts of tadpoles and the skin of the adult amphibians. Infected animals suffer from hyperkeratosis, where the epidermal layer of the animal thickens and may result in abnormal sloughing [8,10]. As a result, it is believed that infected frogs die due to the interference with normal skin functions, especially sodium and potassium ion transport which lead to cardiac arrest [11,12].
To date, most of the B. dendrobatidis research has focused on host-pathogen interactions with little emphasis on the pathogen’s survivability in the environment in the absence of animal hosts. Several studies have shown the presence of B. dendrobatidis DNA in pond water [13,14] and its ability to live in alternative hosts such as crayfish, zebrafish, waterfowls, and nematodes [[15], [16], [17], [18]]. The ability of the fungus to form monolayers in liquids is incompletely understood. When grown in nutrient-rich artificial media, B. dendrobatidis grows into a monolayer attached to the liquid-contacting polystyrene surfaces of the tissue culture-treated flask after four to five days of incubation. Additionally, a mat-like film forms on the surface and at the air-liquid interface. Recently, Brutyn et al. described the identification of biofilm-associated proteins in the supernatant of B. dendrobatidis zoospores suspension [19]. In this study, we investigated the monolayer and monolayer-associated cells that is a poorly studied life form of B. dendrobatidis. We hypothesize that B. dendrobatidis can survive in the environment in the absence of biotic hosts by forming a monolayer of aggregated sporangia. The major goals were to test the ability of B. dendrobatidis to form monolayers in nutrient rich and nutrient-limited conditions, characterize the architecture of monolayers, understand gene expression differences of the monolayers from zoospores and sporangial forms, and determine the resistance profiles of monolayers to different abiotic factors and chemicals, including the anti-chytrid activities of plant-based chemicals. We characterized the monolayers formed by B. dendrobatidis using microscopic, biochemical, and molecular analyses. Gene expression differences among zoospores, sporangia, and monolayer-associated cells were compared using next-generation sequencing and RNA-seq analysis. The resistance profiles of zoospores, sporangia and monolayer-associated cells to current anti-chytrid chemicals and those derived from plants including allicin, 6-gingerol, and curcumin as well as abiotic stressors were also investigated and provide a potential treatment additive to infected animals.
Materials and methods
Cultivation of the chytrid
Batrachochytrium dendrobatidis VM1 isolate was kindly provided by Louise Rollins-Smith (Vanderbilt University), which was obtained from a diseased Western Chorus Frog (Pseudacris triseriata) by Verma Miera and Elizabeth Davidson (Arizona State University). The fungus was grown and maintained on TGhL agar (1.6% tryptone, 0.2% gelatin hydrolysate, 0.4% lactose, and 0.8% agar), H-agar (1% tryptone, 0.32% glucose, 0.8% agar) or H-broth (1% tryptone, 0.32% glucose) at 23 °C in the dark. Five to six-day-old cultures grown in the media described above were used throughout the experiments described in this work.
Synchronization of zoospores
B. dendrobatidis zoospores were synchronized to be mostly at the same size and age and harvested as described previously by Moss et al. [20]. Six-day-old cultures of B. dendrobatidis grown on TGhL or H-agar plates were flooded with 2 ml sterile water and left undisturbed for 5 min. The remaining liquid was removed and discarded. H-broth (2 ml) was added and allowed to incubate for 30 min. The zoospores released after 30 min were determined to be synchronous by microscopy (≥95%).
Crystal violet assay
Three 24-well culture plates (polystyrene, VWR) containing 1 ml H-broth were inoculated with separate synchronized zoospore cultures of B. dendrobatidis from five-day old plates and incubated in the dark at 23 °C. The increase in biomass was quantified using crystal violet (CV) at different incubation periods using the method developed by Djordjevic and colleagues [21] with slight modifications. After six days of incubation, the contents of the plates were emptied and left to dry in a circulating air hood for approximately 20 min. One milliliter of 0.2% CV solution (W/V) in distilled water was added to each well and allowed to stain for 45 min in the dark. The plates were then emptied and gently washed three to five times with water to remove any non-attached cells and excess CV from the wells. One milliliter of 95% (v/v) ethanol was added and incubated for 30 min to decolorize the cells in the dark. Absorbance values of the liquid fractions were recorded at a wavelength of 595 nm. The magnitude of the absorbance value is indicative of the culture attached to the surface.
Growth of B. dendrobatidis in natural pond and lake water, Milli-Q water, tap water, and distilled water
To test the ability of B. dendrobatidis to form monolayers in low nutrient environments, the zoospores were grown in water collected from different natural ponds or lakes around Texas. Additionally, the growth of zoospores in Milli-Q water, tap water, and distilled water were tested. Zoospores (3 x 106 in 10 μl) suspended in sterile distilled water from five to six-day old H-agar plates were inoculated into 2 ml of sterilized pond water, Milli-Q water, tap water, or distilled water in 24-well culture plates and incubated for six-days in the dark at 23 °C. The CV assay was performed as previously described to assess the formation of monolayers of surface-attached aggregates. Following the formation of visible aggregates on the surface of the water (approximately 6-days), 50 μl of culture were transferred to fresh pond water containing flasks and growth was observed after 10 days.
Water in exposed systems are constantly subjected to movement due to wind, rain, surface runoff and other disturbances. This is particularly evident in shallow areas. To simulate this environment, the monolayers formed in pond water in 24-well plates were washed three times every day with sterile distilled water over a period of five days to test the robust adherence of monolayer-associated cells to surfaces. On the sixth day, the CV assay was performed to assess the biomass of the monolayers.
Microscopy
Scanning electron microscopy (SEM) was carried out to determine the development of adherent B. dendrobatidis monolayer on 22 mm poly-l-lysine coated glass coverslips (Neuvitro Corporation) in 1 ml H-broth at 23 °C in the dark. A coverslip for each day for six days was inoculated. After the desired incubation period, the coverslips were transferred into a 6-well polystyrene plate and washed very carefully to remove non-adherent cells and zoospores with phosphate buffered saline (PBS) (pH 8.0, calcium and magnesium-free, Invitrogen). The adhered material was fixed in the primary fixing solution (2.5% glutaraldehyde, 2.0% paraformaldehyde solution in 0.05 M sodium cacodylate buffer pH 7.4) overnight at 4 °C. Coverslips were then washed in 0.05 M sodium cacodylate buffer three times for 10 min each. Secondary fixation was carried out in 1% osmium tetroxide in 0.5 M sodium cacodylate buffer for 30 min. Following three 10-min washes in 0.05 M sodium cacodylate buffer, the material was dehydrated in an ethanol series (25%, 50%, 75%, 95% for 15 min each and four times in 100% for 15 min each). After dehydration, the samples were dried at the critical point (31.1 °C, 1073 psi). Samples were mounted on SEM stubs and coated with gold in a sputter coater for 2.5 min before viewing with a Hitachi S–3400 N scanning electron microscope (Hitachi High-Technologies America, Inc., Pleasanton, CA, USA).
Glycosyl composition analyses
Extracellular polysaccharides were isolated as previously described with modifications [22,23]. Monolayers were harvested from four to six-day-old B. dendrobatidis cultures grown in 10 ml of H-broth at 23 °C. Attached and floating films were harvested as monolayer-associated cells to distinguish them from free-swimming zoospores. The monolayer-associated cells were re-suspended in 10 ml of sterile Milli-Q water and vortexed for 1 min, sonicated for 3 min (20 kHz), and then vortexed again for 2 min to separate cells. This preparation was centrifuged at 370 x g for 20 min. The resulting supernatant (matrix proteins, DNA, and polysaccharides) was recovered and filtered using 0.45 μm filters to remove any cellular debris. Extracellular polysaccharides were precipitated by adding chilled ethanol (95%) at a 1:4 ratio to the cell-free supernatants. Dried samples were used for the glycosyl composition analysis. Aliquots from cultures were inoculated on H-agar before and after sonication to confirm that the cells were viable. After confirming the viability of the sonicated cells, the activity of glucose-6 phosphate dehydrogenase (SIGMA), a marker protein for cell lysis, was determined according to the manufacturer’s instructions.
Glycosyl composition was analyzed by the Complex Carbohydrate Research Center at The University of Georgia. The analysis was performed by combined gas chromatography/mass spectrometry (GC/MS) of the per-O-trimethylsilyl (TMS) derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis as described previously [24]. After preliminary hydrolysis with 2 M, trifluoroacetic acid (TFA) at 120 °C for 1 h and complete removal of TFA by evaporation under an N2-stream, the samples (600 μg with inositol 20 μg) were heated with methanolic HCl in a sealed screw-top glass test tube for 18 h at 80 °C. After cooling and removal of the solvent under a stream of nitrogen, the samples were treated with a mixture of methanol, pyridine, and acetic anhydride for 30 min. The solvents were evaporated, and the samples were derivatized with Tri-Sil® (Pierce) at 80 °C for 30 min. GC/MS analysis of the TMS methyl glycosides was performed on an Agilent 7890A GC interfaced to a 5975C MSD, using a Supelco Equity-1 fused silica capillary column (30 m x 0.25 mm ID).
Staining of extracellular matrix-like material
Monolayers were grown on glass coverslips in 24-well polystyrene plates with 1 ml of H-broth for six days at 23 °C. They were washed twice with PBS and fixed with 3.8% formaldehyde in PBS for 30 min and again washed twice with PBS. The material was stained with 1 μl of 4′,6-diamidino-2-phenylindole (DAPI, 100 μg/ml in water) and 8 μl of Texas Red-conjugated concanavalin A (1 mg/ml) in 200 μl of PBS. While DAPI stains nucleic acids, concanavalin A (Con A) binds to glucosyl and mannosyl residues of the polysaccharides in the ECM. Stained films were imaged using an Olympus BX41 Fluorescence Microscope in the College of Arts and Sciences Imaging Center at Texas Tech University.
Total RNA extraction and sequencing
B. dendrobatidis was cultivated in H-broth and H-agar media separately at 23 °C in the dark and on the third day, monolayer-associated cells and sporangia were collected. Monolayer-associated cells and sporangia were washed three times with sterile water to remove any zoospores and unattached cells. Cells attached on the bottom of tissue culture-treated polystyrene flasks (VWR) and on H-agar plates were scraped, separately collected and filtered through sterile coffee filters (cone coffee filters #4) to further remove any zoospores. Synchronized zoospores were collected from 6-day-old H-agar plates as described above. Cells were ground to a fine powder in liquid nitrogen using a pre-chilled mortar and pestle. The ground cells were then transferred into microcentrifuge tubes containing lysis buffer and the total RNA extraction was carried out following the instructions in Spectrum™ Plant Total RNA Kit (SIGMA-ALDRICH). The quantity and the quality of the RNA were determined by Nanodrop and Hi-sensitivity DIK TapeStation (Agilent 2200 TapeStation).
The whole transcriptome sequencing library of B. dendrobatidis was generated according to the manufacturer’s instructions in the TruSeq RNA Sample Preparation low-throughput (LT) protocol of Illumina. The prepared libraries were sequenced using the Illumina MiSeq® system. These data have been deposited with links to BioProject accession number PRJNA563776 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
RNA-seq data analysis
Reads from three technical replicates in each experimental group were quality-filtered and mapped to B. dendrobatidis JAM81 v1.0 reference transcriptome (https://mycocosm.jgi.doe.gov/Batde5/Batde5.home.html). Mapping was done using the pseudo-alignment-based tool, Kallisto (pachterlab.github.io/kallisto/) [25], with 100 bootstraps per sample for quantification of transcript sequences. Kallisto output files were imported to R software (Version 3.2.3) [26] and Sleuth (https://github.com/pachterlab/sleuth) [27] was used to normalize transcript counts and test for differential gene expression. To test for differential expression, the Wald test (WT) was implemented within the Sleuth package. The Q-value was calculated after adjusting p-value for multiple testing by means of the false discovery rate, FDR using the Benjamini-Hochberg procedure [28]. Differentially expressed genes were determined if their corresponding corrected p-value (Q-value) was ≤0.001.
ClustVis was used to generate the PCA and heatmap plots after unit variate scaling and centering of the genes expression (https://biit.cs.ut.ee/clustvis/) [29]. Protein functions of the B. dendrobatidis transcriptome were annotated using the relevant description acquired from JGI (https://mycocosm.jgi.doe.gov/Batde5/Batde5.home.html). This was further confirmed with PANNZER2 (Protein ANNotation with Z-scoRE) annotation, which provides prediction of the functional annotation and estimated predictive value (the closer the value to one, the stronger the prediction) (Supplementary Table S1) [30].
Determining the minimum inhibitory concentrations of plant-based chemicals
The inhibition of the growth of B. dendrobatidis zoospores by the phytochemicals curcumin, allicin, and 6-gingerol was determined using microdilution in 96-well plate (non-treated, polystyrene, Cole-Parmer), as previously described [31], with minor modifications. Briefly, synchronized zoospores were cultured in at least three replicates in 200 μl of H-broth for six days with or without the addition of different concentrations of curcumin (3, 6, 10, 20, 30, 50 μg/ml) (Sigma), 6-gingerol (20, 40, 60, 80, 100, 200 μg/ml) (Sigma) and allicin (3.375, 6.75, 13.5, 27 μg/ml) (Allimax) in H-broth. Curcumin and 6-gingerol were insoluble in water and were therefore solubilized in 100% dimethyl sulfoxide (DMSO) and 100% methanol respectively and filter sterilized. Allicin was resuspended in H-broth and filter sterilized freshly, before every assay. Each assay plate contained a positive control with zoospores in 200 μl H-broth and media control of 200 μl H-broth with no cells. The effect of vehicle controls, DMSO and methanol, were tested at their respective solvent concentrations in a similar set-up as above. There was no change in the pH of the media containing any of the chemicals tested in these experiments. At the beginning of each assay and at 24 h intervals, absorbance was measured at 492 nm with a BioTek PowerWave microplate reader (BioTek® Instruments, Inc., Winooski, VT), and 10 μl of samples were spotted on H-agar plates after 24 h to check cell viability. The change in absorbance reflected the amount of fungal growth during the incubation period. Data are presented as the percentage growth of the H-broth control. The minimum inhibitory concentration (MIC) was defined as the lowest drug concentration in which there was no growth visible on H-agar plates. These selected concentrations of plant-based chemicals were tested for their fungicidal activities on all three cell types: zoospores, sporangia, and monolayer-associated cells at 24 h post-inoculation. Assays were repeated three times with at least three replicates for trial.
Fungal growth inhibition by antifungal drugs and physical conditions
Fungicidal effects of different physical conditions: high temperature and pH, antifungal drugs: sodium chloride, amphotericin B, curcumin, 6-gingerol, and allicin were tested on B. dendrobatidis zoospores, sporangia, and monolayer-associated cells in vitro. All the assays were set up similarly as described below. The effects of all these treatments were analyzed at 24 h post-inoculation. Tissue culture-treated polystyrene plates were used for the growth of monolayers while non-treated plates were used for the growth of zoospores and sporangia to minimize cell attachment. Synchronized zoospores at a concentration of 3 x 106 per well were used throughout the experiments. Sporangia (3 x 106) were harvested by incubating zoospores for 24 h in liquid media. Six-day-old monolayers were washed three times with sterile distilled water and used for the assays. After 24 h of incubation at different conditions, cell viability was measured by the 2, 3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) assay (cell proliferation kit II-XTT, Sigma). Twenty-five microliters of XTT labeling mixtures were added to each well and incubated at 23 °C for 19 h in the dark for color development. Absorbance was measured at 475 nm. The conversion of the yellow XTT dye to an orange color formazan, takes place in the presence of metabolically active viable cells. The mitochondrial succinoxidase, cytochrome P450 systems, and flavoprotein oxidases are involved in this XTT-to-formazan conversion [32].
Cell viability was assessed as a percentage of the no-treatment control following calculation of specific absorbance (A) using the equation, ((A475nm (test) – A475nm (control)) x 100/(A475nm (no-treatment control) – A475nm (control)). Heat-treated cells (>60 °C for 10 min) were used as the negative control. Means were corrected using the negative control. The treatments that had no significantly different absorbance from the negative control of heat-treated cells (>60 °C for 20 min was used for zoospores and sporangia while 100% ethanol for 10 min was used to kill monolayer-associated cells). These heat-treated cells were considered non-living and used to confirm the MIC obtained from the spot plate technique. The base color change that was observed in the presence of XTT may be due to the released enzymes such as dehydrogenases and succinoxidase from dead cells. The absorbance values for the living cells were corrected by subtracting the absorbance values for the negative controls with heat-treated or ethanol-treated cells. All the assays were repeated three times with a minimum of three replicates at each time.
Temperature
Plates were incubated at 23 °C, 28 °C, and 34 °C in water baths to assess the impact of temperature on cell viability. After 24 h, cell viability was measured using the XTT assay.
pH
The cells were grown in H-broth adjusted to different pH levels: 3, 5, 7, and 9. After 24 h incubation at these levels, the XTT assay was performed to measure the cell viability.
Antifungal drugs
All three cell types were grown for 24 h in the presence of a concentration gradient of antifungals: sodium chloride (5–20 mg/ml) and amphotericin B (0.5–4 μg/ml). Cell viability was measured by the XTT assay. Minimum inhibitory concentrations of curcumin (6 μg/ml, pH 6.96), 6-gingerol (200 μg/ml, pH 6.96), and allicin (3.375 μg/ml, pH 6.83) were tested against the different cells in H-broth for 24 h. The combinatorial effect of all three plant-based antifungal drugs at MIC, MIC/2, and MIC/4 was tested and cell viability was measured by the XTT reduction assay after 24 h.
Statistical analysis
All data were analyzed using GraphPad Prism 6 (https://www.graphpad.com/) statistical software. All the assays conducted for testing the growth of the fungus in H-broth and different water samples were analyzed using ordinary one-way ANOVA with Dunnett’s multiple comparison test. The viability assays were analyzed using the percentage mean values of three independent experiments. We corrected the absorbance readings by subtracting the absorbance of heat-treated cells from the live cells of each type. The two-way ANOVA with Dunnett’s multiple comparison tests were performed to determine the statistical significance of non-treated H-broth control.
Results
Kinetics of B. dendrobatidis monolayer formation suggests increased biomass over time
When grown in nutrient-rich H-broth media in dark at 23 °C, B. dendrobatidis attached to tissue culture-treated polystyrene surfaces 24 h post-inoculation, and in three to six days, a monolayer was formed covering the whole surface of the flask in contact with the liquid. A mat-like film which mostly consisted of empty sporangia and some intact sporangia floated off the solid surface to the air-liquid interface after 4–5 days while the monolayer remained attached on the surface. The standard CV based assay was performed on the tissue culture-treated polystyrene wells where B. dendrobatidis was grown to a surface-attached monolayer over a period of eight-days in H-broth (Fig. S1a). The biomass of the surface attached monolayer increased over eight days with significant increments between days four and eight (p < 0.0001), four and six (p < 0.0001), and six and eight (p < 0.05).
Growth of B. dendrobatidis in pond water, Milli-Q water, tap water, and distilled water
Growth of B. dendrobatidis was monitored in natural pond and lake water samples collected from different locations in Texas. A noticeable monolayer was observed in all samples tested in this study by crystal violet assay at the 6-day time point (Fig. S1b). After six days, further growth at the air-liquid interface was observed as large aggregates when grown in flasks. After six-days of incubation in Milli-Q water, tap water, and distilled water, zoospores had attached to the surfaces of the wells as dispersed aggregates and had not produced monolayers (Fig. S2a). Monolayers formed from growth in pond or lake water samples, however, were stable even after washing every day between days two and six (Fig. S2b).
The B. dendrobatidis monolayer has a heterogeneous structure
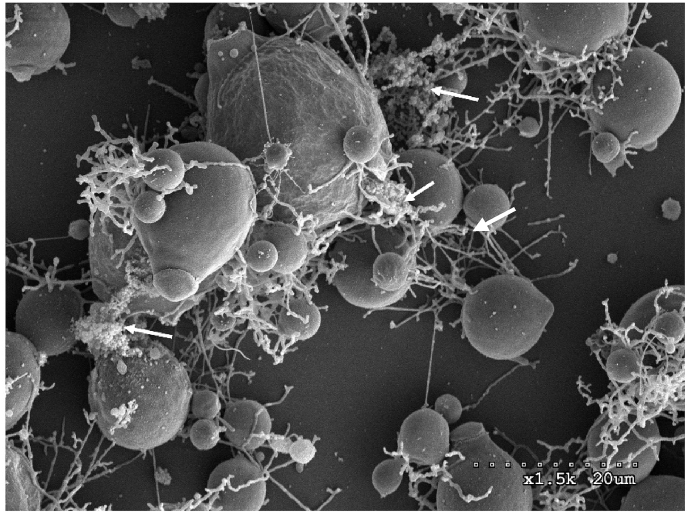
The architecture of the mature B. dendrobatidis monolayer was observed using SEM and fluorescence microscopy. We chose these microscopic methods because the fixation and dehydration in specimen preparation for SEM could distort the architecture of the monolayer by shrinking the extracellular matrix material. All images showed a heterogeneous architecture of the monolayer consisting mainly of different developmental stages of sporangia and zoospores (Fig. 1, Fig. 2). The development of microcolony-containing film from initial attachment through maturation is shown in Fig. 1 a and b. Scanning electron microscopic images showed the initial attachment of zoospores to the surface and transition to young sporangia after 24 h of growth (Fig. 1 a and b -Day 1). Microcolony formation was observed after 48 h (Fig. 1 a and b -Day 2). A very thin extracellular matrix-like material connecting microcolonies was visible in some areas on SEM images of mature monolayers (Fig. 2 arrows). Because of the dehydration effect inherent in sample preparation for SEM, matrix materials took on a stringy cobweb-like appearance. This effect has similarly been noted in other biofilm images visualized by SEM [33].
Fig. 1.
Scanning electron microscopic images showing the time course of the B. dendrobatidis monolayer development with (a) low power and (b) high power. Monolayer development was studied every day up to 6-days under the SEM. Arrows (white) indicate microcolonies formed after 48 h of incubation. Images indicate the transition of zoospores to sporangia in 24 h then to a mature monolayer in 6-days.
Fig. 2.
Scanning electron microscopic image of a 6-day old mature monolayer film. The monolayer was grown on tissue-culture treated glass coverslips in H-broth media. Mature monolayers mainly consisted of sporangia at different development stages. Thread-like rhizoids and extracellular matrix materials hold sporangia together, making the monolayer. Mature sporangia with discharge papilla and plug are clearly visible. Arrows indicate the extracellular matrix-like material.
Texas Red-conjugated concanavalin A and DAPI, stained the extracellular matrix-like material in red, and DNA containing nuclei in blue, respectively. Fig. 3 shows sporangia and mature monolayers stained with Texas Red-conjugated concanavalin A and DAPI. Staining around sporangia isolated from H-agar plates was notably reduced (Fig. 3a). Staining of cell walls of sporangia and rhizoids by Texas Red-conjugated concanavalin A was observed (Fig. 3a). Interestingly, extracellular matrix-like material was visible in monolayers stained with Texas Red-conjugated concanavalin A and high around microcolonies (Fig. 3b).
Fig. 3.
Epifluorescence images of sporangia and mature B. dendrobatidis monolayers. (a) sporangia (b) mature monolayer. Mature monolayers and sporangia were stained with Texas Red conjugated concanavalin A which binds to glucose and mannose residues and DAPI which intercalated into DNA. Blue indicates DAPI stained nucleic acids while the red color indicates concanavalin A stained thin extracellular matrix-like materials. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Glycosyl composition analysis confirms the presence of the extracellular matrix
The glycosyl composition of extracellular matrix material from B. dendrobatidis monolayers was analyzed using gas chromatography coupled with mass spectrometry. This analysis revealed a total carbohydrate content of 6.17% (wt/wt) with five relatively abundant monosaccharides, ribose, xylose, mannose, galactose and glucose, which accounted for 30.3%, 10.1%, 19.1%, 12%, and 23.4% of the total carbohydrate pool respectively (Table 1).
Table 1.
Composition of the extracellular matrix with respect to available monosaccharides.
| Glycosyl Residue | Mass (μg) | Mol %a |
|---|---|---|
| Ribose (Rib) | 10.0 | 30.3 |
| Xylose (Xyl) | 3.3 | 10.1 |
| Mannose (Man) | 7.6 | 19.1 |
| Galactose (Gal) | 4.7 | 12.0 |
| Glucose (Glc) | 9.3 | 23.4 |
| OMe-Hexose | 2.2 | 5.1 |
Values are expressed as mole percent of total carbohydrate which was 6.17% (weight, %). The total percentage may not add to exactly 100% due to rounding.
RNA-seq analysis showed differential gene expression between zoospores, sporangia, and monolayer-associated cells
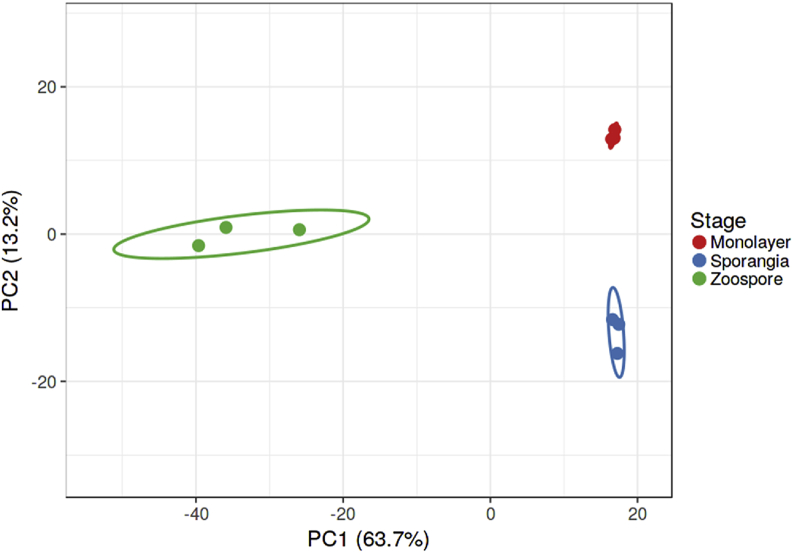
We used multivariate analyses to explore the transcriptome variation in an unbiased manner between the different life stages of B. dendrobatidis. The two principal components, PC1, and PC2, of principal component analysis (PCA) accounted for 76.9% of the total variance between samples (Fig. 4). The PCA showed clustering of the replicates of each life stage separately. Furthermore, PC1 explained 63.7% of the variance in gene expression among the samples; a higher percentage indicates that a high number of genes have been differentially expressed between samples. The zoospore transcriptome was separated along the PC1 axis from sporangia and monolayer transcriptomes. On the other hand, sporangia and monolayer transcriptomes were separated along the PC2 axis, which accounted for only 13.2% of the variance (Fig. 4). We complemented this PCA with hierarchical clustering analysis (HCA), which corroborated the PCA results. This analysis is shown with a heatmap based on the scaled and centered expression values of genes, with purple indicating lower levels and orange indicating higher levels (Fig. 5). From the heatmap, we observed relatively close clustering of genes expressed in the sporangia samples with those of the monolayer samples. Genes expressed in the zoospore samples were notably different from the other two life stage samples. The dendrograms showed the close clustering of the sporangia and monolayer samples in columns, while differentially expressed genes were clustered in rows.
Fig. 4.
Transcriptomic multivariate analysis for B. dendrobatidis at different life stages. Principal component analysis (PCA) score plot showing that the transcriptome of B. dendrobatidis in different life stages. Colored circles represent life cycle stages; red – monolayer-associated cells, blue – sporangia, and green – zoospores. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Heatmap of the differentially expressed genes among three life stages of B. dendrobatidis. Based on the scaled and centered expression values of the genes. The hierarchical clustering of the samples is in columns while genes are clustered in rows. Different colors indicate differences in the gene expression levels. Purple indicates the lower relative abundance of mRNA while orange indicates the higher relative abundance. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The differential gene expression patterns of sporangia vs zoospores (S/Z), monolayer-associated cells vs zoospores (M/Z) and sporangia vs monolayer-associated cells (S/M) were compared. Expression was considered significantly different if the p-value (after multiple-testing correction) was ≤0.001 among the different cell types. The complete set of genes differentially expressed in all three comparisons are shown in supporting information (Supplementary Table S1). Among a total of 2408 genes differentially expressed in S/Z comparison (Supplementary Table S1), relative abundance of mRNA of 1193 genes was higher in zoospores while 1215 genes were higher in sporangia. Among 2266 total differentially expressed genes in the M/Z comparison, 1358 genes expressed more in zoospores and 908 genes showed higher expression in monolayer-associated cells (Supplementary Table S1). Lastly, among 350 genes which were expressed differently in the S/M comparison, the relative abundance of 288 genes was higher in sporangia while 62 genes were more highly expressed in monolayer-associated cells (Supplementary Table S1). As a percentage of the whole transcriptome (8732 genes), differentially expressed genes represented a change in 27%, 26%, and 4% of the transcriptome in S/Z, M/Z, and S/M respectively. Many of these differentially expressed genes encoding proteins are involved in signal transduction, cell adhesion, proteolysis, sugar hydrolysis, flagellar structure and function, and lipid biosynthesis (Table 2).
Table 2.
Selected number of differentially expressed genes of different growth forms of B. dendrobatidis.
| Sporangia versus zoospores | |||
|---|---|---|---|
| Functional classification | Number of transcripts | Transcript Ids | Average log2(S/Z) |
| Aspartyl protease | 12 | 22086; 28984; 22179; 92592; 25223; 20107; 12639; 16209; 90861; 27037; 4465; 35816 | −0.64 |
| Cystathionine gamma-synthase/cystathionine beta-lyase | 1 | 84873 | −2.38 |
| Ergosterol biosynthetic proteins | 2 | 12887; 10216 | −2.67 |
| Extracellular metalloproteinase | 6 | 5304; 34483; 36120; 1480; 35259; 36196; 1501 | −2.10 |
| Fasciclin and related glycoproteins for cell adhesion | 3 | 91430; 85835; 35182 | −1.24 |
| Flagellar proteins (structural; dynein, radial spoke, regulatory; transport) | 28 | 26475; 13321; 34804; 86893; 36206; 11418; 26315; 22880; 28204; 21337; 27665; 35252; 85955; 22301; 88754; 26624; 91375; 92813; 26721; 87960; 36288; 34404; 91203; 90742; 25191; 34914; 11890; 35750 | −3.52 |
| N-terminal nucleophile aminohydrolase | 5 | 17403; 19045; 88011; 92136; 10771 | −2.18 |
| Putative bifunctional chitinase/lysozyme | 1 | 36170 | −4.33 |
| Serine protease | 2 | 92713; 20557 | −1.94 |
| Signal transduction serine/threonine kinase | 7 | 84593; 88514; 24743; 86769; 21324; 8822; 34594 | −1.36 |
| Sugar hydrolase | 1 | 14954 | −5.56 |
| Zn-dependent exopeptidase |
1 |
19340 |
−1.41 |
| Monolayers versus zoospores | |||
| Functional classification |
Number of transcripts |
Transcript Ids |
Average log2 (M/Z) |
| Alkyl hydroperoxide reductase | 3 | 18846; 86762; 21125 | 1.35 |
| Cyclopropane-fatty-acyl phospholipid synthase (Lipid biosynthesis) | 3 | 86144; 9371; 18849 | 2.88 |
| Cystathionine gamma-synthase/cystathionine beta-lyase | 1 | 84873 | 2.79 |
| Enoyl-CoA hydratase | 2 | 14474; 85657 | 2.18 |
| Ergosterol biosynthetic proteins |
4 |
33694; 35090; 10216; 12887 |
1.83 |
| Monolayers versus sporangia | |||
| Functional classification |
Number of transcripts |
Transcript Ids |
Average log2 (M/S) |
| Alkyl hydroperoxide reductase | 1 | 19546 | 0.82 |
| Cyclopropane-fatty-acyl phospholipid synthase (Lipid biosynthesis) | 1 | 86144 | 1.08 |
| Cystathionine gamma-synthase/cystathionine beta-lyase | 1 | 84903 | 1.51 |
| Enoyl-CoA hydratase | 1 | 85657 | 0.70 |
| Extracellular metalloproteinase | 5 | 34483; 1483; 28410; 16271; 36120; 36196 | −1.72 |
| Nucleotide-diphospho-sugar transferase | 3 | 20856; 33853; 16693 | −0.70 |
| Serine protease | 3 | 36738; 12494; 1094 | −1.50 |
| Zn-dependent exopeptidase | 2 | 19977; 33390 | −1.49 |
Relative sensitivity of B. dendrobatidis cell stages to high temperature and variations in pH
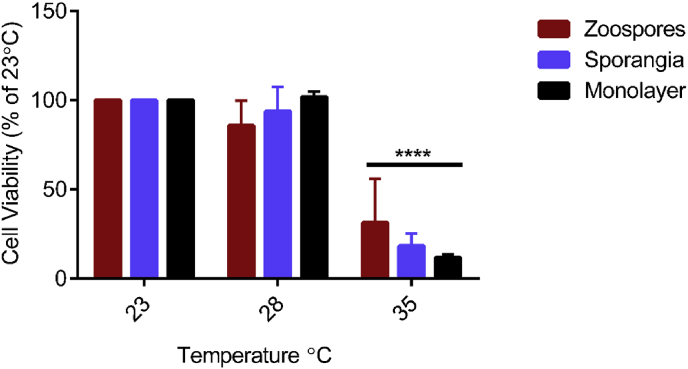
Fig. 6 shows the effect of temperature on the viability of zoospores, sporangia and monolayer-associated cells as measured by the XTT reduction assay. Values represent the percentage cell viability of the no treatment control (23 °C). All three cell types were viable at 28 °C while all three cell types were equally affected by 35 °C (p < 0.0001).
Fig. 6.
Effect of temperature on cell viability. After exposure to different temperatures for 24 h, cell viability was measured by XTT reduction. Bars represent the percentage mean ± SEM of three independent experiments analyzed using two-way ANOVA with Dunnett’s multiple comparison test. Statistical significance of differences is indicated by asterisks. (**** p < 0.0001, F = 49.85).
The effect of acidic and alkaline conditions on the viability of B. dendrobatidis cell types was assessed using XTT reduction (Fig. 7). All the cell types were dead at pH 3 implying higher sensitivity to strong acidic conditions (p < 0.0001). Metabolic activities at pH 5 and pH 7 were not statistically significantly different except sporangia which showed mild sensitivity (p = 0.01). Overall, cells had significantly lower viability at pH 9 compared to pH 7 (p < 0.05). Monolayer-associated cells showed higher survivability at pH 9 compared to sporangia and zoospores (p < 0.0001).
Fig. 7.
Effect of pH on B. dendrobatidis cell types. Bars represent the percentage mean ± SEM of three independent experiments analyzed using two-way ANOVA with Dunnett’s multiple comparison tests. Statistical significance of differences is indicated by asterisks. **** p < 0.0001, * p < 0.05, F = 43.76.
Sensitivity of zoospores, sporangia and monolayer-associated cells to antifungals
Amphotericin B and sodium chloride are antifungal agents available for treating chytridiomycosis [34,35]. Sodium chloride has been recommended for use in captive breeding habitats as salt refuges [36]. We tested the effect of these agents on B. dendrobatidis cell types at 24 h (Fig. 8). All three cell types showed similar susceptibilities to sodium chloride at all concentrations tested (Fig. 8a). Sporangia showed resistance to amphotericin B at 0.5 μg/ml and 1 μg/ml (Fig. 8b).
Fig. 8.
Cytotoxicity of amphotericin B and sodium chloride to zoospores, sporangia, and monolayer-associated cells in vitro. (a) the effects of different concentrations of sodium chloride and (b) amphotericin B on B. dendrobatidis survival were tested against all three cell types by the XTT reduction. Cells were grown in H-broth alone or in H-broth with different concentrations of the chemicals. The percentage means of three independent experiments were analyzed using two-way ANOVA with Dunnett’s multiple comparison tests comparing the viabilities to H-broth media. Asterisks on bars indicate the statistical significance. **** p < 0.0001, ** p < 0.01, F(a) = 22.49, F(b) = 112.4.
Previous studies in our laboratory have shown that water and/or methanol extracts of turmeric, garlic, and ginger were inhibitory to B. dendrobatidis growth (unpublished). We examined sensitivty of B. dendrobatidis cell types to assess the effect of active ingredients from each of these plant products, curcumin (turmeric), allicin (garlic) and 6-gingerol (ginger) respectively. The minimum inhibitory concentrations against zoospores were determined by measuring the absorbance at 492 nm as an indirect measurement of growth and spotting on H-agar plates to further test the viability of the cells. Fig. 9 shows the effect of these chemicals on B. dendrobatidis zoospores. All three plant chemicals were effective against zoospores, killing at least 50% of the zoospore population within 24 h.
Fig. 9.
Dose-dependent effects of plant-based chemicals on the growth of zoospores in vitro. The effects of different concentrations of (a) curcumin (b) allicin (c) 6-gingerol were tested against zoospores for 6-days at 23 °C by measuring the absorbance (492 nm) at 24 h intervals. Zoospores were grown in H-broth alone or in H-broth with various concentrations of plant-based chemicals. Each data point represents the mean ± SEM of three independent experiments. Data were analyzed using repeated measure two-way ANOVA with Dunnett’s multiple comparison tests. **** p < 0.0001, ** p < 0.01, * p < 0.05, F(a) = 24.58, F(b) = 26.03, F(c) = 17.27.
Minimum inhibitory concentrations of curcumin and allicin against zoospores were 6 μg/ml and 3.375 μg/ml respectively. Zoospores showed growth at 200 μg/ml of 6-gingerol, the highest concentration tested without an effect of the solvent, methanol.
Fig. 10 shows the relative sensitivity of all three cell types at the respective MICs of zoospores to each of the three chemical extracts (curcumin (6 μg/ml), gingerol (200 μg/ml) and allicin (3.375 μg/ml)). Sporangia and monolayer-associated cells showed significant resistance to at least one of the chemicals. Sporangia showed resistance to curcumin, 6-gingerol, and allicin at the concentrations tested while monolayer-associated cells were most resistant to allicin.
Fig. 10.
Cytotoxicity of plant-based chemicals on B. dendrobatidis cell types. The MICs of curcumin (6 μg/ml), allicin (3.375 μg/ml), and 6-gingerol (200 μg/ml) for zoospores were tested for survivability of sporangia and monolayer-associated cells by XTT reduction. Cells were grown in H-broth only or in H-broth with chemicals for 24 h at 23 °C. Bars represent the percentage mean ± SEM of three independent experiments analyzed by two-way ANOVA with Dunnett’s multiple comparison test. Statistically significant differences from H-broth are indicated by asterisks. *** p < 0.001, ** p < 0.01, F = 14.89.
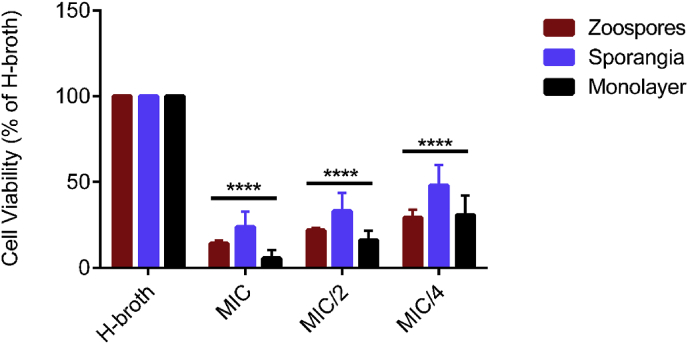
The combinatorial effects of plant chemicals at their MIC, MIC/2, and MIC/4 on B. dendrobatidis cell types are shown in Fig. 11. The mixtures of chemicals at their MIC, MIC/2, and MIC/4 similarly affected survivability of all three cell types.
Fig. 11.
Combinatorial treatment of plant-based chemicals. Mixtures of the chemicals were tested in full, half and quarter strengths of MICs. Cells were grown in H-broth or in H-broth with chemicals for 24 h at 23 °C. The viability of the cells was measured by XTT reduction. Bars represent the percentage mean ± SEM of three independent experiments analyzed by two-way ANOVA with Dunnett’s multiple comparison test. Asterisks on bars indicate the statistical significance of cell viability compared to H-broth. **** p < 0.0001, F = 100.1.
Discussion
The purpose of this study was to assess the characteristics of in vitro formation of a monolayer by the chytrid B. dendrobatidis on solid surfaces and at the air-liquid interface in nutrient-rich media and the ability of the fungus to grow in nutrient-poor environments. We addressed the development, chemical nature and architecture of the monolayer and evaluated zoospores, sporangia, and monolayer-associated cells for their gene expression profiles and resistance to physical conditions and antifungal chemicals.
Scanning electron microscopic images suggest that monolayer formation of B. dendrobatidis goes through a distinct sequence of events, including initial adhesion, microcolony formation, and maturation. This sequence has been observed in other fungi and bacteria that form biofilms [[37], [38], [39]]. After 24 h of incubation, B. dendrobatidis zoospores enlarged and adhered to the surface of poly-l-lysine-coated glass coverslips by their rhizoids. The formation of microcolonies was clear after 48 h. Thereafter, the density of the monolayer increased and matured over 6 days. When grown in liquid cultures, the matured outermost layer of the monolayer detached from the lower surface and floated like a mat at the air-liquid interface. This could represent a dispersal mechanism in ponds, lakes and other water systems where the chytrid may be found. Scanning electron microscopic images revealed that mature B. dendrobatidis monolayers were mainly composed of a mixture of different development stages of sporangia and zoospores. Similar observations have been made in biofilm-forming fungi Candida sp. and Trichosporon asahii [37,40].
Mature B. dendrobatidis monolayers showed a thin extracellular matrix-like material around cells in fluorescence microscopic images stained with Texas Red-conjugated concanavalin A, a lectin which specifically binds to glucose and mannose residues in sugars, suggesting their presence in this extracellular matrix-like material (Fig. 3). The thickness of this layer was high around the microcolonies. Sporangia that were not associated with microcolonies showed limited staining, possibly due to the presence of glucans associated with the cell wall. The presence of an extensive extracellular matrix was not observed in SEM images. This may be due to the intense dehydration process in preparing materials for SEM imaging. Further confirmation of the presence of extracellular matrix-like material was made by gas chromatography/mass spectrometry. The total carbohydrate pool was found to be composed mainly of ribose, xylose, mannose, galactose, and glucose strongly resembling the matrix composition of other fungal biofilms [[41], [42], [43], [44]]. Lack of enzyme activity observed for glucose 6-phosphate dehydrogenase, a cytoplasmic marker enzyme, confirmed the lack of cell lysis during the processing of the matrix material. The most abundant sugar in the chytrid extracellular matrix was ribose which accounted for the 30% of the total carbohydrate pool. This may have resulted from the hydrolysis of nucleic acids which are released from dead cells by strong DNA-degrading enzymes (Maiti and San Francisco, unpublished) or from extracellular DNA within the matrix produced by B. dendrobatidis.
In addition to the phenotypic characterization of monolayers formed by B. dendrobatidis, we provide the gene expression profiles of B. dendrobatidis zoospores, sporangia, and monolayer-associated cells. Our results indicate that monolayers are a distinct entity in the life cycle of B. dendrobatidis. We focused our analysis on the differential gene expression observed through pairwise comparisons. The zoospore gene expression profile was significantly different from sporangia and monolayer-associated cells. We found in zoospores, higher abundance of mRNA for genes related to signal transduction, cell adhesion, aspartyl and serine proteases and metalloprotein families, hydrolases, flagellar and structural proteins. Higher mRNA abundance for genes encode proteins similar to adhesins may play a role in initial attachment of zoospores to surfaces. Higher mRNA abundance was observed for radial spoke-like proteins that have been involved in flagella movement of Chlamydomonas reinhardtii [45]. In addition to spoke-like proteins, relatively more mRNA of genes encoding proteins involved in flagella movement were found in zoospores.
Even though the monolayers are mainly composed of different development stages of sporangia, the gene expression profiles were different in a pairwise comparison of the two cell types. Pairwise comparison of sporangia and monolayer-associated cells showed a 4% difference in their gene expression. We found high mRNA abundance of genes encoding proteins in signal transduction, serine, metallo-, and aspartyl protease families, chitin synthase, enzymes in lipid metabolism, and multidrug resistance-related ABC transporters. Interestingly, we found higher relative abundance of mRNA for genes encoding alkyl hydroperoxide reductase [46,47], cyclopropane-fatty-acyl phospholipid synthase [48,49], cystathionine gamma-synthase/cystathionine beta-lyase [50], and enoyl-CoA hydratase [[51], [52], [53]]. These proteins have been observed to be important in biofilm formation in other microorganisms. Enoyl-CoA hydratase that catalyzes the formation of cis-2-decenoic acid [52,53], a small lipid molecule which acts as an autoinducer has been proposed to play a role in biofilm dispersal in P. aeruginosa and C. albicans [51]. This suggests and supports our hypothesis of monolayer dispersion in the environment.
Biofilms facilitate survival of microorganisms in the presence of extreme abiotic factors or antimicrobial agents. We compared monolayer-associated cells with sporangia and zoospores for their resistance profiles to known antifungal stressors and environments. We found that monolayer-associated cells are more resistant to alkaline pH and allicin (extracted from garlic), compared to zoospores and sporangia. Alkaline conditions damage proteins, lipids, and nucleic acids [54] while allicin binds to free sulfhydryl (-SH) groups of cysteine residues [55]. The relatively higher abundance of mRNA for genes encoding Hsp70, cyclopropane-fatty-acyl phospholipid synthase, alkyl hydroperoxide reductase, and glutathione transferase that provide oxidative stress resistance [[56], [57], [58], [59], [60]] supports this observation of monolayer-cell resistance.
Sporangia showed resistance to curcumin, 6-gingerol, and amphotericin B at concentrations that negatively impacted survival of zoospores and monolayer-associated cells. Although the mode of action of 6-gingerol is not known, it has been established that curcumin and amphotericin B exert their antifungal activities by interfering with ergosterol in fungal membranes by forming reactive oxygen species [61,62]. Expression of genes encoding oligopeptide transporter activity (OPT) and those involved in fatty acid biosynthesis as we observed in sporangia have been observed in amphotericin resistance in Candida auris and could explain their observed resistance to the drug in our experiments [60]. Our data also show that amphotericin B can effectively inhibit the growth of zoospores within 24 h at 0.5 μg/ml, a concentration much lower than what is currently prescribed [31,34], and may reduce the possible toxic side effects to amphibians. Sodium chloride concentrations affected all cell types similarly. Cells were comparatively less sensitive up to 10 mg/ml concentration of sodium chloride. The expression of genes linked to salt tolerance, such as glycerol-3-phosphate dehydrogenase [63], may be involved in showing such salt tolerance in all three cell types. The concentrations we tested were higher than those proposed as treatment measures for chytridiomycosis creating salt refuges and may be useful in decontamination of equipment in the field. The phytochemical extracts used in this study have not been tested yet to determine their safety and efficacy in treating infected amphibians. The specific role of genes involved in monolayer formation and environmental resistance is beyond the scope of this work as there is no facile system currently available to generate mutants in B. dendrobatidis. Therefore, biological function assessment of these genes is presently speculative.
Conclusions
Our study characterizes B. dendrobatidis monolayers that can form in the absence of the host and highlights their importance in resistance to environmental stressors. We show that monolayers resemble biofilms and could play a role in adherence and dispersal of the fungus in aquatic environments. Our findings provide support for additional methods to detect chytrid eDNA not only in water but also on surfaces around pond and lake environments. This suggestion is supported by a study by Bosch et al., that showed the successful use of Virkon S to disinfect surfaces around the ponds to prevent chytrid infections of treated frogs [64]. Finally, in this study, we introduce plant-based chemicals, curcumin, 6-gingerol, and allicin as promising anti-chytrid drugs alternatives to toxic azole drugs currently in use to treat infected animals. It is important, however, to determine the toxicity levels of these chemicals on amphibians before employing them for treating chytrid infections.
Author contribution
SS designed and carried out experiments to assess the solid surface and air-liquid interface associated monolayer cells; performed data analysis, visualization, discussion, wrote and revised the manuscript.
LM performed the initial assessment studies of the solid surface and air-liquid interface associated monolayer cells.
MME performed RNA sequencing data analysis, visualization and revised the manuscript.
MJS initiated the project and oversaw all aspects of the work; provided financial support and editing of the manuscript.
Acknowledgments
This work was supported by the Texas Tech University Biological Sciences and grants from the Association of Biologists of Texas Tech University to SS. We thank Louise Rollins-Smith for kindly providing B. dendrobatidis VM1 strain for this study. Authors would also like to thank Sydney Gassiott for collecting pond water samples from around the state of Texas, Nancy Carty for her early work on the B. dendrobatidis matrix formation and Haleigh Gerold for her work on pond water studies. We thank Angela Moss for reviewing this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2019.100009.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Longcore J.E., Pessier A.P., Nichols D.K. Batrachochytrium dendrobatidis gen. et sp. nov., a chytrid pathogenic to amphibians. Mycologia. 1999;91:219. doi: 10.2307/3761366. [DOI] [Google Scholar]
- 2.Yu Y.-H., Zhang S.-C., Zhao Y.-J., Shi J., Yu L.-H., Lu Q.-F. Chytridiomycosis and amphibian population declines. CHINESE J Zool. 2006;41:118–122. [Google Scholar]
- 3.Borovsky D., Smith E.E., Whelan W.J. Purification and properties of potato 1,4-alpha-D-glucan:1,4-alpha-D-glucan 6-alpha-(1,4-alpha-glucano)-transferase. Evidence against a dual catalytic function in amylose-branching enzyme. Eur J Biochem. 1975;59:615–625. doi: 10.1007/s10393-007-0093-5. [DOI] [PubMed] [Google Scholar]
- 4.Wake D.B., Vredenburg V.T. Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc Natl Acad Sci. 2008;105:11466–11473. doi: 10.1073/pnas.0801921105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher M.C., Garner T.W.J., Walker S.F. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- 6.Hyatt A., Speare R., Cunningham A., Carey C. Amphibian chytridiomycosis. Dis Aquat Organ. 2010;92:89–91. doi: 10.3354/dao02308. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez C.A., Dosta M.D., Partida A.H., Alberto J., Torres R., Mejia J.C. Amphibian chytridiomycosis: a threat to global biodiversity. Int J Aquat Sci. 2014;5:94–109. [Google Scholar]
- 8.Berger L., Hyatt A.D., Speare R., Longcore J.E. Life cycle stages of the amphibian chytrid Batrachochytrium dendrobatidis. Dis Aquat Organ. 2005;68:51–63. doi: 10.3354/dao068051. [DOI] [PubMed] [Google Scholar]
- 9.Piotrowski J.S., Annis S.L., Longcore J.E. Physiology of Batrachochytrium dendrobatidis , a chytrid pathogen of amphibians. Mycologia. 2004;96:9–15. doi: 10.1080/15572536.2005.11832990. [DOI] [PubMed] [Google Scholar]
- 10.Ohmer M.E.B., Cramp R.L., Russo C.J.M., White C.R., Franklin C.E. Skin sloughing in susceptible and resistant amphibians regulates infection with a fungal pathogen. Sci Rep. 2017 doi: 10.1038/s41598-017-03605-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voyles J., Young S., Berger L., Campbell C., Voyles W.F., Dinudom A. Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science. 2009;326:582–585. doi: 10.1126/science.1176765. [DOI] [PubMed] [Google Scholar]
- 12.Wu N.C., McKercher C., Cramp R.L., Franklin C.E. Mechanistic basis for the loss of water balance in green tree frogs infected with a fungal pathogen. Am J Physiol Integr Comp Physiol. 2019 doi: 10.1152/ajpregu.00355.2018. [DOI] [PubMed] [Google Scholar]
- 13.Kirshtein J.D., Anderson C.W., Wood J.S., Longcore J.E., Voytek M.A. Quantitative PCR detection of Batrachochytrium dendrobatidis DNA from sediments and water. Dis Aquat Organ. 2007;77:11–15. doi: 10.3354/dao01831. [DOI] [PubMed] [Google Scholar]
- 14.Chestnut T., Anderson C., Popa R., Blaustein A.R., Voytek M., Olson D.H. Heterogeneous occupancy and density estimates of the pathogenic fungus Batrachochytrium dendrobatidis in waters of North America. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garmyn A., Van Rooij P., Pasmans F., Hellebuyck T., Van Den Broeck W., Haesebrouck F. Waterfowl: potential environmental reservoirs of the chytrid fungus Batrachochytrium dendrobatidis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon T a, Brannelly L a, Chatfield M.W.H., Johnson P.T.J., Joseph M.B., McKenzie V.J. Chytrid fungus Batrachochytrium dendrobatidis has nonamphibian hosts and releases chemicals that cause pathology in the absence of infection. Proc Natl Acad Sci. 2013;110:210–215. doi: 10.1073/pnas.1200592110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liew N., Mazon Moya M.J., Wierzbicki C.J., Hollinshead M., Dillon M.J., Thornton C.R. Chytrid fungus infection in zebrafish demonstrates that the pathogen can parasitize non-amphibian vertebrate hosts. Nat Commun. 2017;8:15048. doi: 10.1038/ncomms15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapard E.J., Moss A.S., San Francisco M.J. Batrachochytrium dendrobatidis can infect and cause mortality in the nematode Caenorhabditis elegans. Mycopathologia. 2012;173:121–126. doi: 10.1007/s11046-011-9470-2. [DOI] [PubMed] [Google Scholar]
- 19.Brutyn M., D’Herde K., Dhaenens M., Van Rooij P., Verbrugghe E., Hyatt A.D. Batrachochytrium dendrobatidis zoospore secretions rapidly disturb intercellular junctions in frog skin. Fungal Genet Biol. 2012;49:830–837. doi: 10.1016/j.fgb.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Moss A.S., Reddy N.S., Dortaj I.M., San Francisco M.J. Chemotaxis of the amphibian pathogen Batrachochytrium dendrobatidis and its response to a variety of attractants. Mycologia. 2008;100:1–5. doi: 10.3852/mycologia.100.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Djordjevic D., Wiedmann M., McLandsborough L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl Environ Microbiol. 2002;68:2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azeredo J., Henriques M., Sillankorva S., Oliveira R. Extraction of exopolymers from biofilms: the protective effect of glutaraldehyde. Water Sci Technol. 2003;47:175–179. [PubMed] [Google Scholar]
- 23.Wu S., Baum M.M., Kerwin J., Guerrero D., Webster S., Schaudinn C. Biofilm-specific extracellular matrix proteins of nontypeable Haemophilus influenzae. Pathog Dis. 2014;72:143–160. doi: 10.1111/2049-632X.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santander J., Martin T., Loh A., Pohlenz C., Gatlin D.M., Curtiss R. Mechanisms of intrinsic resistance to antimicrobial peptides of Edwardsiella ictaluri and its influence on fish gut inflammation and virulence. Microbiology. 2013;159:1471–1486. doi: 10.1099/mic.0.066639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 26.Kunze H., Bohn E., Bahrke G. Effects of psychotropic drugs on prostaglandin biosynthesis in vitro. J Pharm Pharmacol. 1975;27:880–881. doi: 10.2514/2.2417. [DOI] [PubMed] [Google Scholar]
- 27.Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995 doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 29.Metsalu T., ClustVis Vilo J. A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Törönen P., Medlar A., Holm L. PANNZER2: a rapid functional annotation web server. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martel A., Van Rooij P., Vercauteren G., Baert K., Van Waeyenberghe L., Debacker P. Developing a safe antifungal treatment protocol to eliminate Batrachochytrium dendrobatidis from amphibians. Med Mycol. 2011;49:143–149. doi: 10.3109/13693786.2010.508185. [DOI] [PubMed] [Google Scholar]
- 32.Altman F.P. Tetrazolium salts and formazans. Prog Histochem Cytochem. 1976;9:1–56. doi: 10.1016/S0079-6336(76)80015-0. [DOI] [PubMed] [Google Scholar]
- 33.Dohnalkova A.C., Marshall M.J., Arey B.W., Williams K.H., Buck E.C., Fredrickson J.K. Imaging hydrated microbial extracellular polymers: comparative analysis by electron microscopy. Appl Environ Microbiol. 2011;77:1254–1262. doi: 10.1128/AEM.02001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holden W.M., Ebert A.R., Canning P.F., Rollins-Smith L.A. Evaluation of amphotericin B and chloramphenicol as alternative drugs for treatment of chytridiomycosis and their impacts on innate skin defenses. Appl Environ Microbiol. 2014;80:4034–4041. doi: 10.1128/AEM.04171-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stockwell M.P., Storrie L.J., Pollard C.J., Clulow J., Mahony M.J. Effects of pond salinization on survival rate of amphibian hosts infected with the chytrid fungus. Conserv Biol. 2015;29:391–399. doi: 10.1111/cobi.12402. [DOI] [PubMed] [Google Scholar]
- 36.Stockwell M.P., Clulow J., Mahony M.J. Evidence of a salt refuge: chytrid infection loads are suppressed in hosts exposed to salt. Oecologia. 2015;177:901–910. doi: 10.1007/s00442-014-3157-6. [DOI] [PubMed] [Google Scholar]
- 37.Di Bonaventura G., Pompilio A., Picciani C., Iezzi M., D’Antonio D., Piccolomini R. Biofilm formation by the emerging fungal pathogen Trichosporon asahii: development, architecture, and antifungal resistance. Antimicrob Agents Chemother. 2006;50:3269–3276. doi: 10.1128/AAC.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaur S., Singh S. Biofilm formation by Aspergillus fumigatus. Med Mycol. 2014;52:2–9. doi: 10.3109/13693786.2013.819592. [DOI] [PubMed] [Google Scholar]
- 39.Martinez L.R., Casadevall A. Biofilm Formation by Cryptococcus neoformans. Microbiol Spectr. 2015;3:1–11. doi: 10.1128/microbiolspec.MB-0006-2014. [DOI] [PubMed] [Google Scholar]
- 40.Chandra J., Kuhn D.M., Mukherjee P.K., Hoyer L.L., McCormick T., Ghannoum M.A. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez L.R., Casadevall A. Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Appl Environ Microbiol. 2007;73:4592–4601. doi: 10.1128/AEM.02506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beauvais A., Loussert C., Prevost M.C., Verstrepen K., Latgé J.P. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 2009;9:411–419. doi: 10.1111/j.1567-1364.2009.00482.x. [DOI] [PubMed] [Google Scholar]
- 43.Zarnowski R., Westler W.M., Lacmbouh G.A., Marita J.M., Bothe J.R., Bernhardt J. Novel entries in a fungal biofilm matrix encyclopedia. mBio. 2014;5 doi: 10.1128/mBio.01333-14. e01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faria-Oliveira F., Carvalho J., Belmiro C.L.R., Ramalho G., Pavão M., Lucas C. Elemental biochemical analysis of the polysaccharides in the extracellular matrix of the yeast Saccharomyces cerevisiae. J Basic Microbiol. 2015;55:685–694. doi: 10.1002/jobm.201400781. [DOI] [PubMed] [Google Scholar]
- 45.Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci. 2006 doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh E., Jeon B. Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang I.-A., Kim J., Park W. Endogenous hydrogen peroxide increases biofilm formation by inducing exopolysaccharide production in Acinetobacter oleivorans DR1. Sci Rep. 2016;6:21121. doi: 10.1038/srep21121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benamara H., Rihouey C., Jouenne T., Alexandre S. Impact of the biofilm mode of growth on the inner membrane phospholipid composition and lipid domains in Pseudomonas aeruginosa. Biochim Biophys Acta. 2011;1808:98–105. doi: 10.1016/j.bbamem.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Dubois-Brissonnet F., Trotier E., Briandet R. The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front Microbiol. 2016;7:1673. doi: 10.3389/fmicb.2016.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D.-D., Wang Y., Dai B.-D., Li X.-X., Zhao L.-X., Cao Y.-B. ECM17-dependent methionine/cysteine biosynthesis contributes to biofilm formation in Candida albicans. Fungal Genet Biol. 2013;51:50–59. doi: 10.1016/j.fgb.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Davies D.G., Marques C.N.H. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Amari D.T., Marques C.N.H., Davies D.G. The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J Bacteriol. 2013;195:4600–4610. doi: 10.1128/JB.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marques C.N.H., Davies D.G., Sauer K. Control of biofilms with the fatty acid signaling molecule cis-2-Decenoic acid. Pharmaceuticals. 2015;8:816–835. doi: 10.3390/ph8040816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr Biol. 2014 doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller A., Eller J., Albrecht F., Prochnow P., Kuhlmann K., Bandow J.E. Allicin induces thiol stress in bacteria through S-allylmercapto modification of protein cysteines. J Biol Chem. 2016;291:11477–11490. doi: 10.1074/jbc.M115.702308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giotis E.S., Muthaiyan A., Blair I.S., Wilkinson B.J., McDowell D.A. Genomic and proteomic analysis of the alkali-tolerance response (AlTR) in Listeria monocytogenes 10403S. BMC Microbiol. 2008 doi: 10.1186/1471-2180-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giotis E.S., Muthaiyan A., Natesan S., Wilkinson B.J., Blair I.S., McDowell D.A. Transcriptome analysis of alkali shock and alkali adaptation in Listeria monocytogenes 10403S. Foodb Pathog Dis. 2010 doi: 10.1089/fpd.2009.0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panmanee W., Hassett D.J. Differential roles of OxyR-controlled antioxidant enzymes alkyl hydroperoxide reductase (AhpCF) and catalase (KatB) in the protection of Pseudomonas aeruginosa against hydrogen peroxide in biofilm vs. planktonic culture. FEMS Microbiol Lett. 2009;295:238–244. doi: 10.1111/j.1574-6968.2009.01605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X., Zhao X. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother. 2009;53:1395–1402. doi: 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muñoz J.F., Gade L., Chow N.A., Loparev V.N., Juieng P., Berkow E.L. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018 doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finkelstein A., Holz R. Aqueous pores created in thin lipid membranes by the polyene antibiotics nystatin and amphotericin B. Membranes. 1973 [PubMed] [Google Scholar]
- 62.Mesa-Arango A.C., Trevijano-Contador N., Román E., Sánchez-Fresneda R., Casas C., Herrero E. The production of reactive oxygen species is a universal action mechanism of amphotericin B against pathogenic yeasts and contributes to the fungicidal effect of this drug. Antimicrob Agents Chemother. 2014 doi: 10.1128/AAC.03570-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu K.H., Ding X.W., Narsing Rao M.P., Zhang B., Zhang Y.G., Liu F.H. Morphological and transcriptomic analysis reveals the osmoadaptive response of endophytic fungus Aspergillus montevidensis ZYD4 to high salt stress. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosch J., Sanchez-Tomé E., Fernández-Loras A., Oliver J.A., Fisher M.C., Garner T.W.J. Successful elimination of a lethal wildlife infectious disease in nature. Biol Lett. 2015;11:20150874. doi: 10.1098/rsbl.2015.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.