Abstract
Procaryotes starve and face myriad stresses. The bulk population actively resists the stress, but a small population weathers the stress by entering a resting stage known as persistence. No mutations occur, and so persisters behave like wild-type cells upon removal of the stress and regrowth; hence, persisters are phenotypic variants. In contrast, resistant bacteria have mutations that allow cells to grow in the presence of antibiotics, and tolerant cells survive antibiotics better than actively-growing cells due to their slow growth (such as that of the stationary phase). In this review, we focus on the latest developments in studies related to the formation and resuscitation of persister cells and propose the guanosine pentaphosphate/tetraphosphate (henceforth ppGpp) ribosome dimerization persister (PRDP) model for entering and exiting the persister state.
Keywords: Persistence, Antimicrobial agents, Tolerance
Review
Persister cells: dormant and ubiquitous
Hobby et al. [1] first noted that penicillin does not kill about 1% of Staphylococcus aureus cells and also showed non-dividing (dormant cells) were penicillin resistant; hence, convincing evidence was provided immediately that persister cells are metabolically inactive. Two years later, Bigger [2] named these same surviving S. aureus cells “persisters” and corroborated the 1942 findings of Hobby et al. by showing with three experiments that penicillin does not kill non-dividing cells. After 62 years, a ribosomal reporter was used to show persister cells have low metabolic activity [3].
Although the original literature strongly suggested persisters are dormant, there has been some controversy and confusion about their metabolic state. For example, the Brynildsen group [4] claimed, most likely by incorrectly interpreting FACS data [5], that persisters were not necessarily dormant, then three years later concluded that persisters were dormant [6]. Critically, with Salmonella enterica, it was shown again recently that persisters stem primarily from slow growth [7].
Although they are not formed in the exponential phase, persisters are found in the stationary phase and in biofilms [8,9] since cells are stressed. All bacterial cells tested to date form persisters [10] and this includes pathogens; when these dormant cells revive, they likely reconstitute infections [11]. Since all bacterial cells starve [12], persistence is a general phenotype [13]. We have found, based on antibiotic tolerance, morphology, resuscitation rates, and lack of metabolic activity, that the cells capable of resuscitation that are part of the “viable but non-culturable” population generated from starvation [14] are persister cells [13]. Therefore, due to its ubiquity, persistence is arguably one of the most important metabolic states, and insights on eradicating persisters is important for disease (e.g., tuberculosis, cystic fibrosis, ear infections) and agriculture [15].
Studying persister cells
Since persister cells are found naturally at very low concentrations, from 0.0001 to 1% [10], some groups have studied slowly-growing cells rather than persister cells, such as those created by a nutrient shift [[16], [17], [18], [19]] and those of the stationary stage [20]. This results in incorrectly attributing the characteristics of slowly-growing (tolerant) cells to persister cells. Another difficulty is that persister cells can respond to nutrients immediately, so adding fresh medium (e.g., during cell washing steps) causes the persister cells to resuscitate [21].
Since persister cells are dormant and cells are 50% protein, we reasoned inactivating ribosomes should increase persistence. Hence, we [22] devised methods to convert nearly all of the exponentially-growing cells into persisters by pretreating with (i) rifampicin to stop transcription, (ii) with tetracycline to stop translation, or (iii) with carbonyl cyanide m-chlorophenylhydrazone (CCCP) to stop translation by eliminating ATP production. These pretreatment methods may be used to convert nearly all the exponentially-growing cells into a population of persister cells, resulting in a 10,000-fold increase in persistence [22]. By increasing the population of persister cells dramatically, mechanistic insights into their formation and resuscitation may be made. Furthermore, these methods produce bona fide persister cells as they have been verified eight ways: multi-drug tolerance, immediate change from persister to non-persister in the presence of nutrients, dormancy via flow cytometry with the metabolic dye redox sensor green, dormancy via a lack of resuscitation on gel pads that lack nutrients (some exponential cells divide under these conditions), no change in MIC compared to exponential cells, no resistance phenotype, similar morphology to ampicillin-induced persisters, and similar resuscitation as ampicillin-induced persisters [21]. Critically, our methods have been validated for both Pseudomonas aeruginosa and S. aureus [23] and have been verified to date by six independent groups [[23], [24], [25], [26], [27], [28]].
Similar to pretreating with rifampicin, tetracycline, and CCCP, fluoroquinolones may be used to increase persistence by inducing the SOS response via DNA strand breaks [29]. Also, pre-treatment with oxidative stress or acid stress increases persistence 12,000-fold [30] and increasing the toxicity of a toxin such as MqsR of the MqsR/MqsA TA system increases persistence dramatically [30]. By using these methods to increase the concentration of persister cells, it was determined that bacterial persistence increases when the cells are less fit to mount an active fight against an environmental stress [30]. Also, these methods led to the insight that indole decreases persistence in Escherichia coli [31,32]. From this realization, halogenated indoles such as 5-iodoindole, 4-fluoroindole, 7-chloroindole, and 7-bromoindole were identified that effectively kill E. coli and S. aureus persister cells [33].
Persister cells form by inactivating ribosomes via ppGpp
With few exceptions, the alarmone ppGpp is central for the mechanism of how cells become persistent. Originally, ppGpp had an important role in the study of the first mutation related to persistence, hipA7. In E. coli, the gain-of-function set of mutations in toxin gene hipA known as hipA7 (causing substitutions G22S and D291A in HipA) of the HipA/HipB TA system increased persistence 1,000 fold [34] due to reduced antitoxin HipB binding [35]. These substitutions render the HipA7 variant not toxic [36], so the link of TA systems to persistence from the use of this variant is problematic. In addition, ppGpp has been linked to the MazF/MazE TA system since ppGpp is required to activate MazF toxicity [37].
However, there is a trend to distance persistence from intracellular TA systems since a credible mechanism relating ppGpp to the activation of TA systems has not been discerned. Specifically, the model in which Lon protease is activated by polyphosphate (which builds up as ppGpp is activated) and then degrades antitoxins to activate toxins has been retracted [38]. Also, two other manuscripts relating persistence to the deletion of 10 type II TA systems [39] and relating persistence to HipA [40] have been retracted. Basically, data from other laboratories disputed these findings [41] including (i) polyphosphate inactivates Lon protease in vitro, rather than activates Lon activity [42], (ii) there is little connection between Lon and persistence [43,44], (iii) degradation of YefM antitoxin of the YoeB/YefM TA system is independent of ppGpp and polyphosphate [45], and (iv) degradation of antitoxins RelB and MazE is independent of ppGpp [41]. Also, the TA system deletion strain had 79 coding mutations beyond those related to the 10 TA systems [46], and the strain was contaminated with phage [47]. Note, in contrast to intracellular TA systems, Lon protease and ppGpp are implemented for creating persister cells via contact-dependent growth inhibition [48].
Further evidence of the absence of a link between TA systems and persistence is that of another E. coli strain with 10 type II TA systems deleted (but with a large genomic inversion) which showed TA systems are not related to persistence [49]. Also, deletion of 12 TA systems in S. enterica had no effect on persistence [7]. Furthermore, using single cells and a verified E. coli strain with 10 type II TA systems deleted, ATP concentrations and TA systems did not influence persistence [50], and ppGpp was confirmed as important for persister formation [50]. However, the authors used a RpoS-mCherry proxy for ppGpp that may not be accurate [49]. Of course, high concentrations of persister cells may be formed by producing nearly any toxic protein in E. coli [51], so production of toxin MqsR of the MqsR/MqsA TA system [[52], [53], [54], [55]] has been used to increase persistence by 14,000-fold for E. coli [30]. However, use of these non-physiological levels of toxins does not mean cells use toxins of TA systems for persistence. Instead, the primary physiological roles of TA systems (to date) seem more likely to be [56] (i) inhibiting phage [57], (ii) maintaining genetic elements such as plasmids [58], (iii) reducing metabolism as a response to stress [54,59], and (iv) forming biofilms [60,61].
Since the link between ppGpp and TA systems is problematic, we reasoned that it may be possible to directly relate ppGpp to persistence based on its dominant role in the stress response; we also utilized the realization that reducing ribosome activity is the critical step for persistence [22]. Upon experiencing myriad stresses (e.g., nutrient, antibiotic, or oxidative stress), most cells mount an active response against the stress by altering replication, transcription, and translation via ppGpp [62]. ppGpp slows replication by inhibiting DNA primase [62], and ppGpp reduces translation by decreasing ribosome synthesis [63]. ppGpp also directly controls the activity of several enzymes; for example, ppGpp inhibits GTPases [62]. ppGpp changes transcription by activating RpoS (sigmaS, the stress response sigma factor for the stationary phase) and RpoE (sigmaE, the stress response sigma factor for misfolded proteins in the periplasm) [64]. Notably, the physiological roles of ppGpp continue to expand, such as inhibition of the synthesis of purine nucleotides [65], regulation of purine homeostasis through activation of nucleosidase PpnN [66], inhibition of the ribosome-associated GTPase Era that is important for assembling 30S ribosome subunits [67], and binding and likely inactivating the GTPase HflX that activates 100S ribosomes [68].
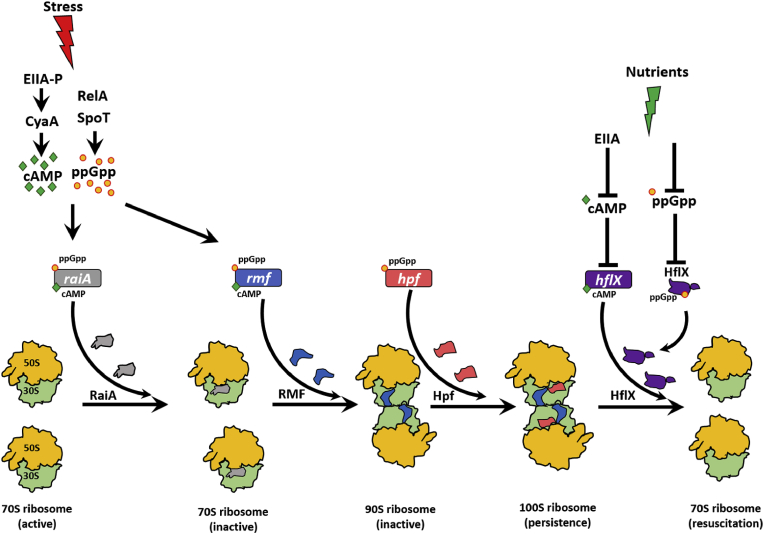
In contrast to the vast majority of cells that mount an active response to stress, the sub-population of cells that become persisters may be created when ppGpp turns off an excessive number of ribosomes by activating the expression of the small (55 aa) ribosome modulation factor (RMF) [69], which converts active 70S ribosomes into inactive 100S ribosomes [70] via an inactive 90S dimer complex [71]. Also, ppGpp induces the hibernation promoting factor the hibernation promoting factor (Hpf) (95 aa) which converts 90S ribosomes into 100S ribosomes and is highly conserved as it is found in most bacteria and some plants [72]. This dimerization is essential for survival in the stationary phase [73]. Hence, we reasoned that ppGpp directly modulates persistence through its direct inactivation of ribosomes. In agreement with this hypothesis, we found overproduction of RMF and Hpf increase persistence as well as reduce single-cell resuscitation while ppGpp has no effect on single-cell resuscitation [74]. We have termed this mechanism the ppGpp ribosome dimerization persister (PRDP) model (Fig. 1) in which persister cells form not through TA systems but by the direct control of ribosome activity by ppGpp. In support of this model, E. coli cells have 100 times less persistence in the absence of ppGpp [44], and higher ppGpp levels increases the persistence of single cells by 1,000-fold [50].
Fig. 1.
Schematic of the ppGpp ribosome dimerization persister (PRDP) model. Stress such as nutrient limitation, osmotic stress, and acid stress induces the stringent response which results in ppGpp formation by RelA/SpoT and generation of cAMP (e.g., upon glucose depletion via the phosphorylated glucose phosphotransfer enzyme, EIIA-P) in E. coli. ppGpp induces hpf, and both ppGpp and cAMP together induce raiA and rmf. RaiA inactivates 70S ribosomes, RMF converts 70S ribosomes into inactive 90S ribosomes, and Hpf converts inactive 90S ribosomes into inactive 100S ribosomes. Moreover, ppGpp binds HflX to likely inactivate it, and cAMP represses hflX. Upon addition of nutrients and removal of the stress, cAMP decreases (due to unphosphorylated EIIA) which stimulates HflX production; HflX dissociates inactive 100S ribosomes into active 70S ribosomes and growth resumes. Since persister cells form in the absence of ppGpp (although at much reduced levels), cAMP by itself and perhaps other mechanisms activate RMF and Hpf as well.
Another model for persister cell formation is based on reducing ATP [46,[75], [76], [77], [78]] or uncoupling the membrane potential with compounds like CCCP to reduce ATP [22]. However, conflicting results have been obtained from other labs suggesting ATP does not control persistence [7,50].
Persister cell resuscitation is heterogeneous and is based on ribosome content
Single cell analysis has revealed that most persister cells do not resuscitate spontaneously, but instead, wake upon sensing fresh nutrients [79]. This is in opposition to the ‘Scout Model’ [80] in which it was proposed (without data) that persister cells wake spontaneously and periodically to see if it is safe to grow.
Upon waking, persister resuscitation is heterogeneous and includes five phenotypes: (i) immediate division, (ii) immediate elongation followed by division, (iii) immediate elongation but no division, (iv) delayed elongation/division, and (v) no growth [21]. Critically, this heterogeneity in resuscitation is due to the different levels of active ribosomes: by using a validated green fluorescence protein reporter of ribosome levels, we observed that higher ribosome levels result in faster waking [21]. Hence, cells with low levels of ribosomes have to increase their ribosome levels to a threshold value, then they begin to divide or elongate [21]. The varying ribosome levels result from cell-to-cell differences in the numbers of ribosomes in exponentially-growing cells upon ceasing their metabolic activity [21].
This heterogeneity in persister cell resuscitation [21] has been corroborated by several groups. For example, using E. coli and time-lapse microscopy, Şimşek and Kim [81] followed 12,800 of what appears to be persister cells (or stationary cells, it is not clear which they used since it is not clear if the stationary-phase cells that they used were pre-treated with ampicillin to eliminate non-persisters) to find that most cells wake immediately and that their rejuvenation probability decays exponentially, resulting in lag time before resuscitation from 0 to over 1000 min. Pu et al. [28], also found that single persister cells wake heterogeneously (from less than 12 h–40 h), but their explanation based on the degree of protein refolding required for their resuscitation is unlikely as it would require an enzyme that could re-nature a thousand different proteins [82]. In addition, Goormaghtigh and Van Melderen [83] found persister cells wake heterogeneously and frequently include elongation.
In addition, when persisters resuscitate and begin dividing, their growth rate is the same as exponential cells; hence, once the formerly-dormant cell commences division, it is fully recovered [21]. Also, it is important to note that residual stress may be detrimental to sustained resuscitation in that cells with residual ampicillin antibiotic fail to resuscitate [21].
Persister cells resuscitate by activating dormant ribosomes
It has been proposed that toxins are inactivated and in this way persister cells may revive. However, as with the model that relies on TA systems for persister cell formation, there is little data that support this. For example, it was suggested that the peptidyl-tRNA hydrolase Pth counteracts toxin TacT in S. Typhimurium, “explaining how bacterial persisters can resume growth”; however, there were no resuscitation data to validate this claim [84]. Similarly, it was claimed that deactivation of polymerized (active) HokB toxin by monomerization controls persister waking; however, single-cell observations were not made but instead delays in resuscitation were estimated from growth data [85] for this type I system. Also, the key features for the HokB system have been discerned from non-physiological levels of HokB (i.e., from overproduction of HokB) [85,86], and there is no persister phenotype for deleting hokB [87]. Critically, for type II TA systems, there is little data for the paradigm that antitoxins are degraded from TA complexes and cells revive; reactivation of toxins bound to antitoxins is unlikely given that the antitoxin is bound tightly to the toxin in most cases (for type II TA systems).
Since there is little evidence of toxins of TA systems being inactivated to revive cells, the simplest model for persister cell resuscitation is re-activation of ribosomes. Based on single-cell observations [74,79], we propose the PRDP model (Fig. 1) holds for persister resuscitation: after ppGpp generates persister cells directly by inactivating ribosomes via increased production of RaiA (inactivates 70S ribosomes) [72], production of the ribosome modulation factor (RMF), and production of Hpf. Once the stress dissipates and the cells have fresh nutrients, inactive 100S ribosomes reactivate by the ribosome resuscitation factor HflX [88] which will no longer be inactivated by ppGpp [68] and repressed by cAMP [89]; HflX also rescues ribosomes from mRNA [90]. In support of this, we found RaiA, RMF, and Hpf increase persistence and reduce single-cell persister resuscitation and that RMF increases 100S ribosome formation in persister cells [74]. In addition, we found producing HflX increases single-cell waking [79]. Furthermore, the resuscitating cells sense the fresh nutrients by chemotaxis and phosphotransferase membrane proteins [79], and transport of nutrients reduces the level of secondary messenger cAMP through enzyme IIA [79]; this reduction in cAMP levels stimulates ribosome resuscitation and rescue [79]. The waking persister cells also immediately commence chemotaxis toward nutrients [79]. Therefore, persister cells wake by sensing nutrients via membrane receptors, and this external waking signal is conveyed via the secondary messenger cAMP to inactive ribosomes which are used by the persisters to wake and begin chemotaxis to acquire nutrients [79].
Corroborating the ribosome model for persister cell resuscitation [74], a 10,000-member library of druglike compounds was screened to identify compounds that increase persister cell resuscitation and 2-[[2-(4-bromophenyl)-2-oxoethyl]thio]-3-ethyl-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one (BPOET) was identified that stimulates persister cell resuscitation by activating ribosomes via pseudouridine modification of 23S rRNA by pseudouridine synthase RluD [91]; hence, ribosomes are modified to facilitate persister resuscitation. Also, 100S ribosome recovery is extremely rapid and occurs in less than 1 min [72], which matches our data indicating some persister cells wake immediately [21].
Perspectives
Implications of the PRDP model include that persister cell formation is an elegantly-regulated response to stress rather than a bet-hedging (“stochastic”) process. Experimental data support this idea in that spontaneous persisters are rare [92] but external stress (e.g., antibiotics, hydrogen peroxide, acid) converts nearly the whole population into persister cells [22,29,30]. In addition, the PRDP model suggests resuscitation is also an elegant response to improved environmental conditions rather than spontaneous. Since all cells starve and need deep resting states, it is logical that cells utilize elegant pathways for both persister cell formation and resuscitation.
Also, clearly there is something more than just stress that makes cells become persistent. For example, all stationary-phase cells are starving but only a small percentage become persisters. The PRDP model suggests that a small population of stressed cells has a larger number of their ribosomes inactivated so this heterogeneity in ribosome dimerization is perhaps the “switch” that makes cells persistent.
Remaining questions include whether the PRDP model is general; i.e., holds for other species, and whether it is relevant for clinical isolates. The PRDP model is likely applicable to myriad stresses since RMF dramatically increases persistence with for stress from ampicillin [74], ciprofloxacin [74], netilmicin [93], gentamicin [94], acid [95], osmotic stress [96] and nutrient limitation in the stationary phase [97,98]. Furthermore, since RMF is conserved in bacteria [72] and Hpf is even more widely distributed [99], there is the possibility that the PRDP model may be applicable for the formation of the persister state of many strains. For example, like E. coli, the persistence of P. aeruginosa also requires ppGpp [100] and so it would be interesting to test further whether ribosome dimerization also controls persister cell formation and resuscitation in this strain. It is already established that both Hpf and ppGpp (but not RMF) are necessary for the long term survival of P. aeruginosa during starvation by preserving ribosomes [101] and that ppGpp plays a role in hpf expression [99]. However, a preliminary report indicates that after treating with ciprofloxacin, high persister variants are present in cystic fibrosis patents, but mutations related to ppGpp were not found [102].
Since persister cells are formed in the absence of ppGpp but at much lower levels [44], there must be other means to activate Rmf and Hpf without ppGpp, which leads to the formation of inactive and dimerized ribosomes. For example, cAMP, which is generated upon starvation (e.g., glucose depletion), induces rmf [63] and raiA [72], and plays a role in the formation of persister cells [32] (Fig. 1). In addition, cAMP represses hflX [89]. Hence, cAMP is as likely to be as important as ppGpp for persistence. Critically, there may be several mechanisms for forming persister cells, but the net result is that ribosomes are inactivated.
New compounds will indubitably be identified for killing persister cells since it now possible to create large libraries of bona fide persister cells in a facile manner [21,22]. For example, by screening a 10,000-member library of druglike compounds, we identified that the indole derivative, 5-nitro-3-phenyl-1H-indol-2-yl-methylamine hydrochloride (NPIMA) [103], kills E. coli persister cells better than 5-iodoindole [33] and better than the DNA crosslinking agent cisplatin [104,105]. NPIMA also eliminated P. aeruginosa persister cells and had activity on the representative Gram positive pathogen, S. aureus [103]. The mode of action of NPIMA with both Gram-positive and Gram-negative bacteria was determined to be via membrane damage which leads to lysis. Critically, understanding the steps for persister formation and resuscitation (Fig. 1) will provide additional targets for fighting persistence.
There are also parallels being discovered between bacterial persisters and cancer cells that are recalcitrant to known treatments [106]. Dormancy for cancer cells occurs when the growth of the cells is temporarily stopped before initiating a new primary tumor that may be even more aggressive the original tumor. Hence, insights gleaned from bacterial persisters may be relevant for treating cancer.
Acknowledgements
This work was supported by funds derived from the Biotechnology Endowed Professorship at the Pennsylvania State University. The authors appreciate their discussions over the years with Prof. Michael Benedik.
References
- 1.Hobby G.L., Meyer K., Chaffee E. Observations on the mechanism of action of penicillin. Exp Biol Med. 1942;50:281–285. [Google Scholar]
- 2.Bigger J.W. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet. 1944;244:497–500. [Google Scholar]
- 3.Shah D., Zhang Z., Khodursky A., Kaldalu N., Kurg K., Lewis K. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orman M.A., Brynildsen M.P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother. 2013;57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood T.K., Knabel S.J., Kwan B.W. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henry T.C., Brynildsen M.P. Development of Persister-FACSeq: a method to massively parallelize quantification of persister physiology and its heterogeneity. Sci Rep. 2016;6:25100. doi: 10.1038/srep25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pontes M.H., Groisman E.A. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal. 2019;12 doi: 10.1126/scisignal.aax3938. eaax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis K. Persister cells, dormancy, and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 9.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Van den Bergh B., Fauvart M., Michiels J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol Rev. 2017;41:219–251. doi: 10.1093/femsre/fux001. [DOI] [PubMed] [Google Scholar]
- 11.Defrain V., Fauvart M., Michiels J. Fighting bacterial persistence: current and emerging anti-persister strategies and therapeutics. Drug Resist Updates. 2018;38:12–26. doi: 10.1016/j.drup.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt T.M. Bacteria battling for survival. In: Kolter R., S.M, editors. Microbes and evolution. American Society of Microbiology; Washington, D.C.: 2012. pp. 59–64. [Google Scholar]
- 13.Kim J.-S., Chowdhury N., Yamasaki R., Wood T.K. Viable but non-culturable and persistence describe the same bacterial stress state. Environ Microbiol. 2018;20:2038–2048. doi: 10.1111/1462-2920.14075. [DOI] [PubMed] [Google Scholar]
- 14.Xu H.S., Roberts N., Singleton F.L., Attwell R.W., Grimes D.J., Colwell R.R. Survival and viability of nonculturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 15.Martins P.M.M., Merfa M.V., Takita M.A., De Souza A.A. Persistence in phytopathogenic bacteria: do we know enough? Front Microbiol. 2018;9:1099. doi: 10.3389/fmicb.2018.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amato Stephanie M., Orman Mehmet A., Brynildsen Mark P. Metabolic control of PersisteMol cellr formation in Escherichia coli. Mol Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Amato S.M., Brynildsen M.P. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amato Stephanie M., Brynildsen Mark P. Persister heterogeneity arising from a single metabolic stress. Curr Biol. 2015;25:2090–2098. doi: 10.1016/j.cub.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 19.Radzikowski J.L., Vedelaar S., Siegel D., Ortega Á.D., Schmidt A., Heinemann M. Bacterial persistence is an active σS stress response to metabolic flux limitation. Mol Syst Biol. 2016;12:882. doi: 10.15252/msb.20166998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orman M.A., Brynildsen M.P. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nat Commun. 2015;6:7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J.-S., Yamasaki R., Song S., Zhang W., Wood T.K. Single cell observations show persister cells wake based on ribosome content. Environ Microbiol. 2018;20:2085–2098. doi: 10.1111/1462-2920.14093. [DOI] [PubMed] [Google Scholar]
- 22.Kwan B.W., Valenta J.A., Benedik M.J., Wood T.K. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother. 2013;57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grassi L., Di Luca M., Maisetta G., Rinaldi A.C., Esin S., Trampuz A., Batoni G. Generation of persister cells of Pseudomonas aeruginosa and Staphylococcus aureus by chemical treatment and evaluation of their susceptibility to membrane-targeting agents. Front Microbiol. 2017;8:1917. doi: 10.3389/fmicb.2017.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui P., Niu H., Shi W., Zhang S., Zhang W., Zhang Y. Identification of genes involved in bacteriostatic antibiotic-induced persister formation. Front Microbiol. 2018;9:413. doi: 10.3389/fmicb.2018.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayanaswamy V.P., Keagy L.L., Duris K., Wiesmann W., Loughran A.J., Townsend S.M., Baker S. Novel glycopolymer eradicates antibiotic- and CCCP-induced persister cells in Pseudomonas aeruginosa. Front Microbiol. 2018;9:1724. doi: 10.3389/fmicb.2018.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulaiman J.E., Hao C., Lam H. Specific enrichment and proteomics analysis of Escherichia coli persisters from rifampin pretreatment. J Proteome Res. 2018;17:3984–3996. doi: 10.1021/acs.jproteome.8b00625. [DOI] [PubMed] [Google Scholar]
- 27.Tkhilaishvili T., Lombardi L., Klatt A.-B., Trampuz A., Di Luca M. Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int J Antimicrob Agents. 2018;52:842–853. doi: 10.1016/j.ijantimicag.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Pu Y., Li Y., Jin X., Tian T., Ma Q., Zhao Z. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell. 2019;73:143–156. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Dörr T., Lewis K., Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000760. e1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong S.H., Wang X., O’Connor H.F., Benedik M.J., Wood T.K. Bacterial persistence increases as environmental fitness decreases. Microbial Biotechnol. 2012;5:509–522. doi: 10.1111/j.1751-7915.2011.00327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y., Kwan B.W., Osbourne D.O., Benedik M.J., Wood T.K. Toxin YafQ increases persister cell formation by reducing indole signalling. Environ Microbiol. 2015;17:1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- 32.Kwan B.W., Osbourne D.O., Hu Y., Benedik M.J., Wood T.K. Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol Bioeng. 2015;112:588–600. doi: 10.1002/bit.25456. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.-H., Kim Y.-G., Gwon G., Wood T.K., Lee J. Halogenated indoles eradicate bacterial persister cells and biofilms. Amb Express. 2016;6:123. doi: 10.1186/s13568-016-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyed H.S., Bertrand K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher M.A., Balani P., Min J., Chinnam N.B., Hansen S., Vulic M. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature. 2015;524:59–64. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korch S.B., Henderson T.A., Hill T.M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 37.Aizenman E., Engelberg-Kulka H., Glaser G. An Escherichia coli chromosomal "addiction module" regulated by guanosine 3’,5’-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maisonneuve E., Castro-Camargo M., Gerdes K. Retraction notice to: ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2018;172:1135. doi: 10.1016/j.cell.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 39.Retraction for Maisonneuve Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci. 2018;115 doi: 10.1073/pnas.1803278115. E2901-E2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Retraction for Germain Stochastic induction of persister cells by HipA through (p)ppGpp-mediated activation of mRNA endonucleases. Proc Natl Acad Sci. 2019;116 doi: 10.1073/pnas.1906160116. 11077-11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Melderen L., Wood T.K. Commentary: what is the link between stringent response, endoribonuclease encoding type II toxin-antitoxin systems and persistence? Front Microbiol. 2017;8:191. doi: 10.3389/fmicb.2017.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osbourne D.O., Soo V.W.C., Konieczny I., Wood T.K. Polyphosphate, cyclic AMP, guanosine tetraphosphate, and c-di-GMP reduce in vitro Lon activity. Bioengineered. 2014;5:264–268. doi: 10.4161/bioe.29261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan Y., Lazinski D., Rowe S., Camilli A., Lewis K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio. 2015;6 doi: 10.1128/mBio.00078-15. e00078-00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury N., Kwan B.W., Wood T.K. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci Rep. 2016;6:20519. doi: 10.1038/srep20519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramisetty B.C.M., Ghosh D., Roy Chowdhury M., Santhosh R.S. What is the link between stringent response, endoribonuclease encoding type II toxin–antitoxin systems and persistence? Front Microbiol. 2016;7:1882. doi: 10.3389/fmicb.2016.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan Y., Brown Gandt A., Rowe S.E., Deisinger J.P., Conlon B.P., Lewis K. ATP-dependent persister formation in Escherichia coli. mBio. 2017;8:e02267. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harms A., Fino C., Sørensen M.A., Semsey S., Gerdes K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio. 2017;8 doi: 10.1128/mBio.01964-17. e01964-01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghosh A., Baltekin Ö., Wäneskog M., Elkhalifa D., Hammarlöf D.L., Elf J., Koskiniemi S. Contact-dependent growth inhibition induces high levels of antibiotic-tolerant persister cells in clonal bacterial populations. EMBO J. 2018;37 doi: 10.15252/embj.201798026. e98026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goormaghtigh F., Fraikin N., Putrinš M., Hallaert T., Hauryliuk V., Garcia-Pino A. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio. 2018;9 doi: 10.1128/mBio.00640-18. e00640-00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svenningsen M.S., Veress A., Harms A., Mitarai N., Semsey S. Birth and resuscitation of (p)ppGpp induced antibiotic tolerant persister cells. Sci Rep. 2019;9:6056. doi: 10.1038/s41598-019-42403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chowdhury N., Kwan B.W., McGibbon L.C., Babitzke P., Wood Thomas K. Toxin MqsR cleaves single-stranded mRNA with various 5’ ends. MicrobiologyOpen. 2016;5:370–377. doi: 10.1002/mbo3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren D., Bedzyk L.A., Thomas S.M., Ye R.W., Wood T.K. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64:515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 53.Brown B.L., Wood T.K., Peti W., Page R. Structure of the Escherichia coli antitoxin MqsA (YgiT/b3021) bound to its gene promoter reveals extensive domain rearrangements and the specificity of transcriptional regulation. J Biol Chem. 2011;286:2285–2296. doi: 10.1074/jbc.M110.172643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Kim Y., Hong S.H., Ma Q., Brown B.L., Pu M. Antitoxin MqsA helps mediate the bacterial general stress response. Nat Chem Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X., Lord D.M., Hong S.H., Peti W., Benedik M.J., Page R., Wood T.K. Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ Microbiol. 2013;15:1734–1744. doi: 10.1111/1462-2920.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song S., Wood T.K. Post-segregational killing and phage inhibition are not mediated by cell death through toxin/antitoxin systems. Front Microbiol. 2018;9:814. doi: 10.3389/fmicb.2018.00814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pecota D.C., Wood T.K. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J Bacteriol. 1996;178:2044–2050. doi: 10.1128/jb.178.7.2044-2050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.González Barrios A.F., Zuo R., Hashimoto Y., Yang L., Bentley W.E., Wood T.K. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y., Wang X., Ma Q., Zhang X.-S., Wood T.K. Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaca A.O., Colomer-Winter C., Lemos J.A. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimada T., Yoshida H., Ishihama A. Involvement of cyclic AMP receptor protein in regulation of the rmf gene encoding the ribosome modulation factor in Escherichia coli. J Bacteriol. 2013;195:2212–2219. doi: 10.1128/JB.02279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalebroux Z.D., Swanson M.S. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 65.Wang B., Dai P., Ding D., Del Rosario A., Grant R.A., Pentelute B.L., Laub M.T. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol. 2019;15:141–150. doi: 10.1038/s41589-018-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y.E., Bærentsen R.L., Fuhrer T., Sauer U., Gerdes K., Brodersen D.E. ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Mol Cell. 2019;74:1239–1249. doi: 10.1016/j.molcel.2019.03.035. e1234. [DOI] [PubMed] [Google Scholar]
- 67.Wood A., Irving S.E., Bennison D.J., Corrigan R.M. The (p)ppGpp-binding GTPase Era promotes rRNA processing and cold adaptation in Staphylococcus aureus. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008346. e1008346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y., Zborníková E., Rejman D., Gerdes K. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio. 2018;9 doi: 10.1128/mBio.02188-17. e02188-02117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Izutsu K., Wada A., Wada C. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells. 2001;6:665–676. doi: 10.1046/j.1365-2443.2001.00457.x. [DOI] [PubMed] [Google Scholar]
- 70.Wada A., Igarashi K., Yoshimura S., Aimoto S., Ishihama A. Ribosome modulation factor: stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214:410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 71.Ueta M., Yoshida H., Wada C., Baba T., Mori H., Wada A. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10:1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 72.Prossliner T., Winther K.S., Sørensen M.A., Gerdes K. Ribosome hibernation. Annu Rev Genet. 2018;52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 73.Yoshida H., Shimada T., Ishihama A. Coordinated hibernation of transcriptional and translational apparatus during growth transition of Escherichia coli to stationary phase. mSystems. 2018;3 doi: 10.1128/mSystems.00057-18. e00057-00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song S., Wood T.K. ppGpp ribosome dimerization model for bacterial persister formation and resuscitation. bioRXiv. 2019:663658. doi: 10.1016/j.bbrc.2020.01.102. [DOI] [PubMed] [Google Scholar]
- 75.Dörr T., Vulić M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000317. e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng H.-Y., Soo V.W.C., Islam S., McAnulty M.J., Benedik M.J., Wood T.K. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ Microbiol. 2014;16:1741–1754. doi: 10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 77.Conlon B.P., Rowe S.E., Gandt A.B., Nuxoll A.S., Donegan N.P., Zalis E.A. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 2016;1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 78.Cameron D.R., Shan Y., Zalis E.A., Isabella V., Lewis K. A genetic determinant of persister cell formation in bacterial pathogens. J Bacteriol. 2018;200 doi: 10.1128/JB.00303-18. e00303-00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamasaki R., Song S., Benedik M.J., Wood T.K. Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience. 2020;23:100792. doi: 10.1016/j.isci.2019.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Epstein S.S. Microbial awakenings. Nature. 2009;457:1083. doi: 10.1038/4571083a. [DOI] [PubMed] [Google Scholar]
- 81.Şimşek E., Kim M. Power-law tail in lag time distribution underlies bacterial persistence. Proc Natl Acad Sci. 2019;116:17635–17640. doi: 10.1073/pnas.1903836116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wood T.K., Song S., Yamasaki R. Ribosome dependence of persister cell formation and resuscitation. J Microbiol. 2019;57:213–219. doi: 10.1007/s12275-019-8629-2. [DOI] [PubMed] [Google Scholar]
- 83.Goormaghtigh F., Van Melderen L. Single-cell imaging and characterization of Escherichia coli persister cells to ofloxacin in exponential cultures. Sci Adv. 2019;5 doi: 10.1126/sciadv.aav9462. eaav9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheverton Angela M., Gollan B., Przydacz M., Wong Chi T., Mylona A., Hare Stephen A., Helaine S. A Salmonella toxin promotes persister formation through acetylation of tRNA. Mol Cell. 2016;63:86–96. doi: 10.1016/j.molcel.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilmaerts D., Dewachter L., De Loose P.-J., Bollen C., Verstraeten N., Michiels J. HokB monomerization and membrane repolarization control persister awakening. Mol Cell. 2019;75:1031–1042. doi: 10.1016/j.molcel.2019.06.015. e1034. [DOI] [PubMed] [Google Scholar]
- 86.Wilmaerts D., Bayoumi M., Dewachter L., Knapen W., Mika J.T., Hofkens J. The persistence-inducing toxin HokB forms dynamic pores that cause ATP leakage. mBio. 2018;9 doi: 10.1128/mBio.00744-18. e00744-00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Verstraeten N., Knapen Wouter J., Kint Cyrielle I., Liebens V., Van den Bergh B., Dewachter L. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell. 2015;59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 88.Gohara D.W., Yap M.-N.F. Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management. Curr Genet. 2018;64:753–760. doi: 10.1007/s00294-017-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin H.H., Hsu C.C., Yang C.D., Ju Y.W., Chen Y.P., Tseng C.P. Negative effect of glucose on ompA mRNA stability: a potential role of cyclic AMP in the repression of hfq in Escherichia coli. J Bacteriol. 2011;193:5833–5840. doi: 10.1128/JB.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y., Mandava C.S., Cao W., Li X., Zhang D., Li N. HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat Struct Mol Biol. 2015;22:906. doi: 10.1038/nsmb.3103. [DOI] [PubMed] [Google Scholar]
- 91.Song S., Wood T.K. Persister cells resuscitate via ribosome modification by 23S rRNA pseudouridine synthase RluD. Environ Microbiol. 2020 doi: 10.1111/1462-2920.14828. in press. [DOI] [PubMed] [Google Scholar]
- 92.Balaban N.Q., Merrin J., Chait R., Kowalik L., Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 93.Tkachenko A.G., Kashevarova N.M., Tyuleneva E.A., Shumkov M.S. Stationary-phase genes upregulated by polyamines are responsible for the formation of Escherichia coli persister cells tolerant to netilmicin. FEMS Microbiol Lett. 2017;364 doi: 10.1093/femsle/fnx084. fnx084-fnx084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKay S.L., Portnoy D.A. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother. 2015;59:6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.El-Sharoud W.M., Niven G.W. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology. 2007;153:247–253. doi: 10.1099/mic.0.2006/001552-0. [DOI] [PubMed] [Google Scholar]
- 96.Shcherbakova K., Nakayama H., Shimamoto N. Role of 100S ribosomes in bacterial decay period. Genes Cells. 2015;20:789–801. doi: 10.1111/gtc.12273. [DOI] [PubMed] [Google Scholar]
- 97.Yamagishi M., M.H, Wada A., Sakagami M., Fujita N., Ishihama A. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: growth phase-and growth rate dependent control. EMBO J. 1993;12:625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bubunenko M., Baker T., Court D.L. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J Bacteriol. 2007;189:2844–2853. doi: 10.1128/JB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Akiyama T., Williamson K.S., Franklin M.J. Expression and regulation of the Pseudomonas aeruginosa hibernation promoting factor. Mol Microbiol. 2018;110:161–175. doi: 10.1111/mmi.14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen D., Joshi-Datar A., Lepine F., Bauerle E., Olakanmi O., Beer K. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Akiyama T., Williamson K.S., Schaefer R., Pratt S., Chang C.B., Franklin M.J. Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci. 2017;114:3204–3209. doi: 10.1073/pnas.1700695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mojsoska B., Cameron D.R., Bartell J.A., Haagensen J.A., Sommer L.M., Lewis K. The high persister phenotype of Pseudomonas aeruginosa is associated with increased fitness and persistence in cystic fibrosis airways. 2019. [DOI]
- 103.Song S., Gong T., Yamasaki R., Kim J.-S., Wood T.K. Identification of a potent indigoid persister antimicrobial by screening dormant cells. Biotechnol Bioeng. 2019;116:2263–2274. doi: 10.1002/bit.27078. [DOI] [PubMed] [Google Scholar]
- 104.Chowdhury N., Wood T.L., Martínez-Vázquez M., García-Contreras R., Wood T.K. DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol Bioeng. 2016;113:1984–1992. doi: 10.1002/bit.25963. [DOI] [PubMed] [Google Scholar]
- 105.Cruz-Muñiz M.Y., López-Jacome L.E., Hernández-Durán M., Franco-Cendejas R., Licona-Limón P., Ramos-Balderas J.L. Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. Int J Antimicrob Agents. 2016;49:88–92. doi: 10.1016/j.ijantimicag.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 106.Vallette F.M., Olivier C., Lézot F., Oliver L., Cochonneau D., Lalier L. Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer. Biochem Pharmacol. 2019;162:169–176. doi: 10.1016/j.bcp.2018.11.004. [DOI] [PubMed] [Google Scholar]