Abstract
Bacteria interact with their environment including microbes and higher eukaryotes. The ability of bacteria and fungi to affect each other are defined by various chemical, physical and biological factors. During physical association, bacterial cells can directly attach and settle on the hyphae of various fungal species. Such colonization of mycelia was proposed to be dependent on biofilm formation by the bacteria, but the essentiality of the biofilm matrix was not represented before. Here, we demonstrate that secreted biofilm matrix components of the soil-dwelling bacterium, Bacillus subtilis are essential for the establishment of a dense bacterial population on the hyphae of the filamentous black mold fungus, Aspergillus niger and the basidiomycete mushroom, Agaricus bisporus. We further illustrate that these matrix components can be shared among various mutants highlighting the community shaping impact of biofilm formers on bacteria-fungi interactions.
Keywords: Bacillus subtilis, Aspergillus niger, Agaricus bisporus, Fungal hyphae, Biofilm matrix
1. Introduction
Biofilm development by a plethora of bacteria and fungi has been pragmatically studied separately in the laboratory. Biofilm formation is abundant in the environment [1] and these biofilm communities likely comprise both bacteria and fungi in addition to higher eukaryotes grazing or residing these habitats. Interaction among bacteria and fungi extends physical associations, and furthermore includes chemical communications, ranging from antibiosis and metabolic exchange to specific signaling and chemotaxis [2]. These direct or indirect interactions among bacteria and fungi alters the physiology, growth, movement, differentiation, pathogenesis, reproduction and/or survival of either or both partners [2]. During physical interaction between bacteria and fungi, stable association is possibly facilitated by the production of a viscous biofilm matrix. Indeed, the production of a biofilm matrix has been suggested for various bacterial isolates on the hyphae of the ectomycorrhizal fungus, Laccaria bicolor [3], however, the essentiality of the biofilm matrix in long term attachment has not been confirmed before. Several molecular determinants have been described to be important for bacteria-fungi interactions, including motility, quorum sensing, bacterial secretion system, secondary metabolites (see an extended list reviewed in Ref. [4]). Here, we examined the role of biofilm components of Bacillus subtilis during association with filamentous Ascomycota and Basidiomycota. B. subtilis is a soil-derived Gram-positive bacterium and long-studied model for physiology, genetics and differentiation, including motility, sporulation, biofilm formation [5]. Phenotypic diversification of this bacterium is influenced by intertwinement of global regulators, including Spo0A that has been originally described to be involved in production of heat and pressure resistant spores [6]. In addition, Spo0A has been later shown to impact protease secretion, toxin production, and biofilm development, thus involved in regulation of genes related to cell fate decision of the bacterium [7]. Formation of biofilm in B. subtilis requires a complex matrix composed of exopolysaccharide (EPS), TasA amyloid fiber, and a surface localized hydrophobin, BslA [[8], [9], [10], [11], [12], [13]]. The former two matrix components are essential for the establishment of a floating biofilm on the air-medium interface (referred to as pellicle), the creation of complex vein-like structures on agar medium, or attachment to the root surface of plants [[8], [9], [10],14,15]. BslA protein creates a shield around the pellicle and colony biofilm to avoid penetration of hydrophobic fluids and its deletion alters microstructure of colonies [12,13,16], but it is not indispensable for establishment of floating biofilms [17,18]. The ability and degree of biofilm matrix production correlates with attachment to the plant root surface in hypotonic cultures and also with the ability of the bacterial strains to protect tomato plant against wilt disease caused by Ralstonia solanacearum [10,19]. As the matrix components are secreted from the cells, mutants lacking either EPS or TasA are able to complement each other and create a functional biofilm both in vitro and on the plant root [10,11,20].
A previous study demonstrated that attachment of B. subtilis to the mycelia of Aspergillus niger leads to metabolic alterations in both partners [21]. Microarray experiments revealed that genes responsible for single cell motility in B. subtilis are downregulated 3 h after attachment [21], suggesting settlement of the bacterial cells on the fungal hyphae. However, transcriptions of biofilm-related genes were not altered at such an early stage of interaction compared to planktonic cells.
Here, we revisited the interaction of B. subtilis with A. niger dissecting the co-cultures after 24 h of incubation. We demonstrate that long-term attachment depends on the genes required for exopolysaccharide and amyloid fiber production, but neither the synthesis of hydrophobin nor the ability for single cell motility are required for the bacterial settlement on the fungal mycelium. Moreover, the secreted matrix components can complement single gene deletion mutants. Finally, we show that such a matrix-dependent colonization of fungal hyphae is not only restricted to A. niger, but similarly required for biofilm formation on the mycelia of the basidiomycete, Agaricus bisporus.
2. Material and methods
2.1. Strains, media composition and culturing conditions
All the bacterial and fungal strains used in this study are listed in Table 1. Bacterial strains were cultivated in Lysogeny Broth medium (LB-Lennox, Carl Roth, Germany; 10 g l−1 tryptone, 5 g l−1 yeast extract and 5 g l−1 NaCl) supplemented with 1.5% Bacto agar if required. Supplemented LB medium was prepared by adding 1% 1 mol l−1 MgSO4 and 0.1% 0.1 mol l−1 MnCl2 to basic LB medium. A. niger was cultivated on potato dextrose glucose agar (PDA) medium (Carl Roth, Germany; potato infusion 4 g l−1, glucose 20 g l−1, agar 15 g l−1, pH value 5.2 ± 0.2) to harvest spores and in LB medium for formation of macro-colonies. A. bisporus was cultivated on PDA plates or potato dextran glucose broth (PDB) medium (Carl Roth, Germany; potato infusion 4 g l−1, glucose 20 g l−1, pH value 5.2 ± 0.2).
Table 1.
Bacterial and fungal strains used in this study. CmR, SpecR, KmR, and TetR denote chloramphenicol, spectinomycin, kanamycin, and tetracycline resistance cassettes, respectively.
| Bacterial strains | ||
|---|---|---|
| B. subtilis strains | Characteristics | Reference |
| DK1042 | NCBI3610 comIQ12I | [31] |
| TB34 | DK1042 amyE::Phyperspank-gfp, CmR | [32] |
| TB35 | DK1042 amyE::Phyperspank-mKATE2, CmR | [33] |
| TB36 | DK1042 Δhag::KmR; amyE::Phyperspank-gfp, CmR | [34] |
| TB37 | DK1042 Δhag::KmR; amyE::Phyperspank-mKATE2, CmR | [34] |
| TB421 | DK1042 Δspo0A::KmR; amyE::Phyperspank-gfp, CmR | [32] |
| TB524 | DK1042 Δeps::TetR; amyE::Phyperspank-gfp, SpecR | [11] |
| TB525 | DK1042 Δeps::TetR; amyE::Phyperspank- mKATE2, SpecR | [11] |
| NRS2097 | NCIB3610 ΔbslA::CmR | [35] |
| TB526 | DK1042 ΔbslA::CmR; amyE::Phyperspank-gfp, SpecR | This study |
| TB538 | DK1042 ΔtasA::KmR; amyE::Phyperspank-gfp, SpecR | [11] |
| TB539 |
DK1042 ΔtasA::KmR; amyE::Phyperspank- mKATE2, SpecR |
[11] |
| Fungal strains | ||
|
Species name |
Strain and collection numbers |
Reference |
| A. bisporus | H39/CBS 122262 | [36] |
| A. niger | N402/ATCC 64974/CBS 132248 | [37] |
TB526 was obtained using natural competence [22] by transforming genomic DNA extracted from NRS2097 to TB34 and selecting for chloramphenicol resistance, followed by verifying the mutation by PCR. To select for resistant bacterial colonies after transformation, 5 μg l−1 chloramphenicol was used.
2.2. Hyphal colonization assay of A. niger
Spores of A. niger cultures grown at 28 °C for 7–12 days on PDA plates were harvested using 10 ml saline tween solution (8 g l−1 NaCl and 0.05 ml l−1 Tween 80) and filtered through Miracloth (Millipore; Billerica, USA) following the protocol described in Ref. [21]. The spore solution was centrifuged 5000 rpm for 10 min and resuspended in saline tween solution. Spores were stored at 4 °C until use, but no more than 14 days. Around 3*105 spores ml−1 were inoculated into 25 ml of LB medium and shaken at 120 rpm at 28ᵒC for 24 h 4–5 macro-colonies were transferred into a single well of a 24-well plate and culture was supplemented with 0.01 mol l−1 MgSO4 and 0.1 mmol l−1 MnCl2 as indicated for supplemented LB medium. 10 μl of B. subtilis overnight culture grown at 37 °C was inoculated into each well (single or mixed culture). The co-cultures were incubated at 28 °C with shaking at 170 rpm for 24 h. Following incubation, the pellicle biofilm formed at the air-liquid interface was removed from each well. Subsequently, A. niger macro-colonies were washed two times with sterile MilliQ water before imaging.
2.3. Hyphal colonization assay of A. bisporus
Three-week old A. bisporus culture, grown on PDA plates at 25 °C was wetted with 10 ml of physiological saline and scraped thoroughly with a spreader. Afterwards, 1 ml of the hyphal suspension was incubated in 25 ml PDB at 25 °C without agitation for 8–14 days. After eight days, A. bisporus macro-colonies were floating within the media. One macro-colony each was transferred into a well of a 24-well microtiter plate. Remaining PDB was removed and the macro-colonies were washed once with physiological saline. 1 ml of supplemented LB medium was added to each macro-colony and inoculated with 10 μl of B. subtilis overnight culture grown at 37 °C (single or mixed culture). The microtiter plate was incubated at 25 °C with shaking at 170 rpm for 22–24 h. Subsequently, A. bisporus macro-colonies were washed three times with physiological saline.
2.4. Sample preparation and confocal laser scanning microscopy (CLSM)
Washed fungal macro-colonies were transferred to microscope slides and gently sealed with cover slips. Fungal hyphae colonization was analysed with two confocal laser scanning microscopes (LSM 710 (Carl Zeiss) or TCS SP8 (Leica) both equipped with an argon laser and a Plan-Apochromat 63x/1.4 Oil objective). Fluorescent reporter excitation was performed at 488 nm for green fluorescence and at 561 nm for red fluorescence, while the emitted fluorescence was recorded at 540/40 nm and 668/86 nm (Zeiss) or 520/23 nm and 700/90 nm (Leica) for GFP and mKATE, respectively, (wavelength/bandwidth). For generating multi-layer images, Z-stack series with 1 μm steps were acquired and processed with the software ImageJ (National Institutes of Health). Maximum intensity was used to merge five chosen stacks of the green fluorescent, red fluorescent and overlay channel. Merging of the five stacks was done to integrate signals from the bacterial cells at different focus planes. The bright field channel is represented by only one brightness-adjusted image from one of the stacks used to obtain the fluorescent images.
3. Results and discussion
3.1. Colonization of the A. niger hyphae by B. subtilis depends on the global regulator, Spo0A
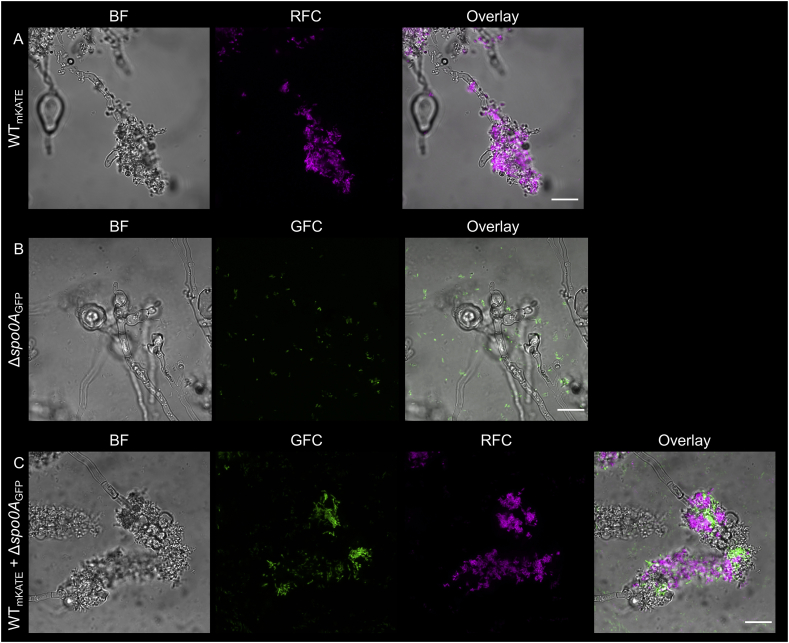
Transcriptome analysis of attaching B. subtilis cells on A. niger mycelia has revealed that expression of genes related to single cell motility is reduced compared to non-attached cells [21] suggesting that the bacterial cells switch from planktonic to sessile state of growth. The global transcriptional regulator, Spo0A influences hundreds of genes in B. subtilis that determine bacterial cell fate, e.g. differentiation into swimming, biofilm forming, or sporulating cell types [7]. Therefore, the impact of spo0A gene deletion in B. subtilis was first assayed during colonization of the hyphae of A. niger. As expected, bacterial cells of the spo0A strain showed reduced hyphal colonization compared to the wild type and only planktonic cells were observed around A. niger (Fig. 1A and B). Driven by the observation that colonization of plant root by B. subtilis requires secreted biofilm matrix components [10,19], we have inspected if addition of wild type strain restores attachment of spo0A mutant cells on the fungal hyphae. Indeed, co-inoculation of wild type and spo0A cells in 1:10 ratio rescued attachment of spo0A strain to the hyphae, suggesting that secreted factors by the wild type strains are sufficient for the establishment of biofilm by the two strains on the fungal mycelia (Fig. 1C). Alternatively, signaling molecules produced by the wild type strain, but absent in the spo0A mutant could induce the molecular factors responsible for hyphal biofilms.
Fig. 1.
Colonization of the A. niger hyphae by B. subtilis (A) wild type, (B) spo0A mutant and (C) the co-culture of 10:1 GFP-labeled spo0A mutant and mKATE-labeled wild type. GFC: green fluorescent channel; RFC: red fluorescent channel. Scale bars indicate 20 μm. The images presented are representative examples selected from independent samples repeated on different days. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Hyphal colonization depends on B. subtilis biofilm matrix components, EPS and TasA
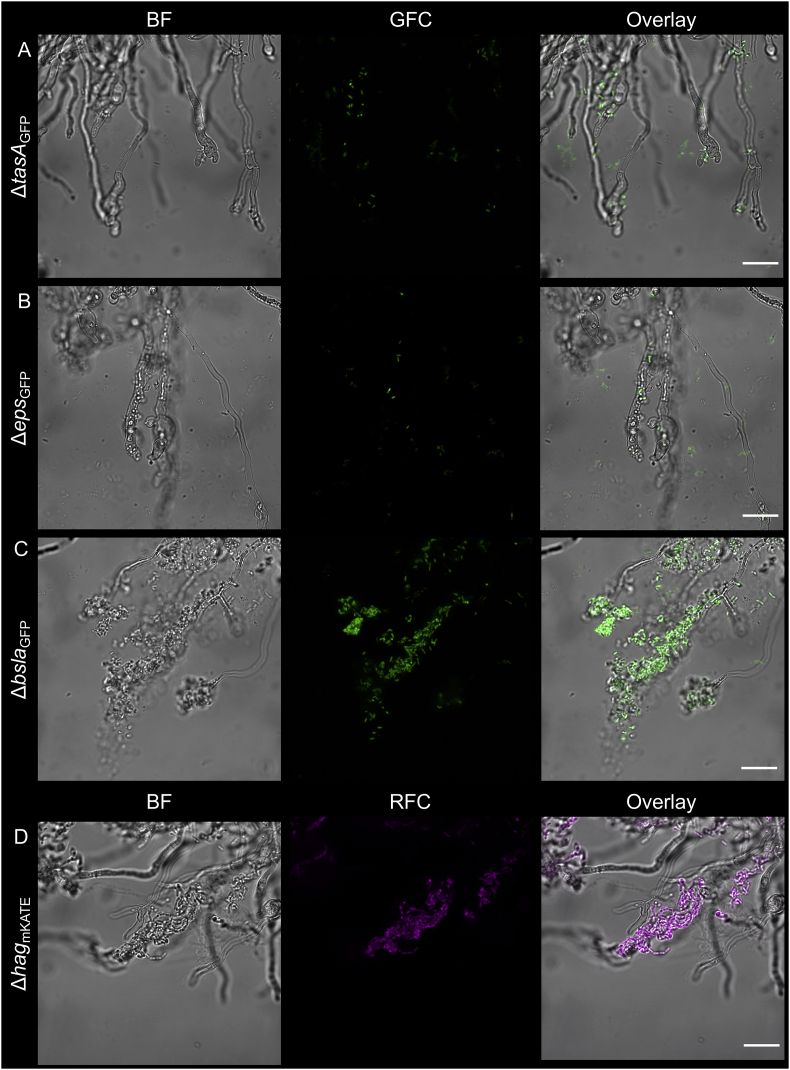
Biofilm formation of B. subtilis depends on various secreted components, including EPS, TasA and BslA. The transcription of these operons involved in the production of matrix is indirectly dependent on Spo0A. Therefore, we have tested how single deletion of eps operon, tasA or bslA genes impacts fungal colonization by B. subtilis. Unlike the bslA mutant, removal of either EPS or TasA hindered the bacterial biofilm development on the hyphae of A. niger (Fig. 2A–C). This suggests that both core components of the matrix, EPS and TasA contribute to establishment of stable biofilm on the mycelia, while the hydrophobin BslA is not necessary for stable attachment. In accordance, previous studies found that BslA is not essential for the establishment of floating pellicle biofilm of B. subtilis, but is required for its repellency and the microstructure of colonies [12,13,17,18]. Finally, gene deletion in the flagellin coding gene was assayed for biofilm establishment during mycelia colonization. Although motility is not essential for generation of biofilms, it has been described to be critical for fast formation of B. subtilis biofilm [23]. Cells lacking single cell motility could colonize the hyphae of A. niger (Fig. 2D), similar to the wild type strain (Fig. 1A). The importance of bacterial flagella and type 4 pili was previously described during co-migration of Burkholderia terrae with fungal hyphae through soil [24], suggesting that essentiality of bacterial motility for fungal interaction could depend on the environment, bacterial physiology or specific properties of the fungal and bacterial cell surfaces. In addition, other examples highlight that fungal hyphae could facilitate bacterial swimming along the hyphae and therefore niche colonization [25,26]. In our simple laboratory system, the bacterium-fungus co-cultures are mildly agitated, which allows firm contact of the bacterial cells and the mycelia. Under these conditions, motility is plausibly not essential.
Fig. 2.
B. subtilis lacking biofilm components, (A) TasA or (B) EPS are unable to grow on the A. niger hyphae, while strains lacking (C) the production of hydrophobin, BslA or (D) motility established stable colonization on the mycelia. GFC: green fluorescent channel; RFC: red fluorescent channel. Scale bars indicate 20 μm. The images presented are representative examples selected from independent samples repeated on different days. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Secreted biofilm matrix components are shared among mutants
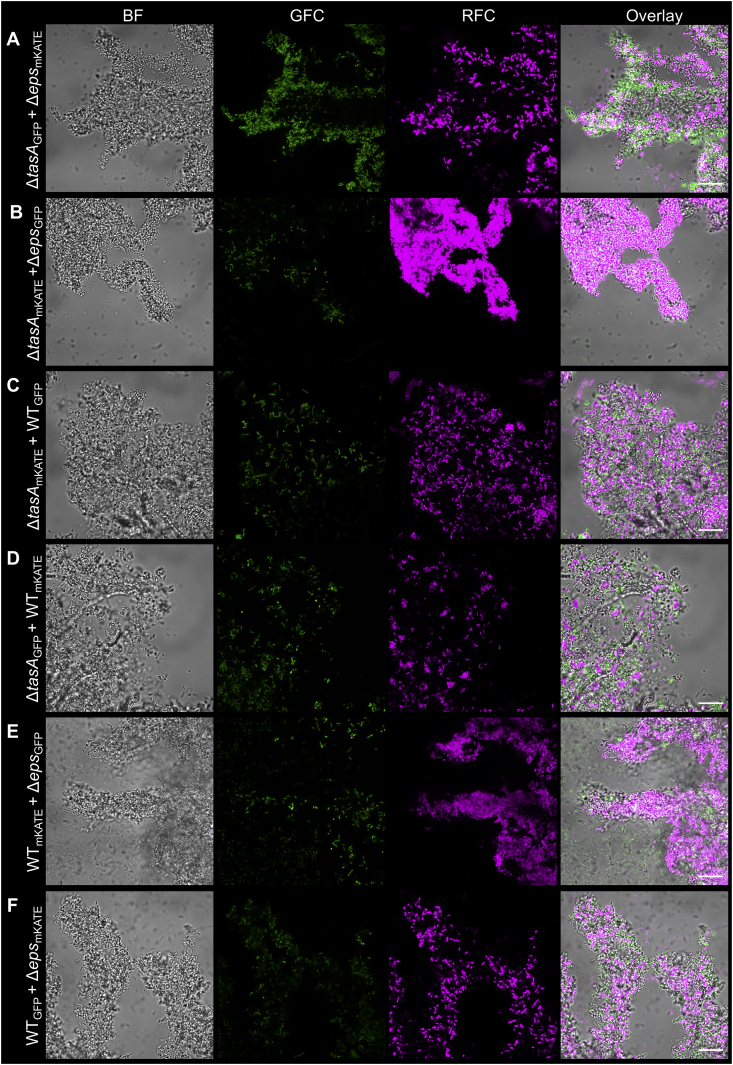
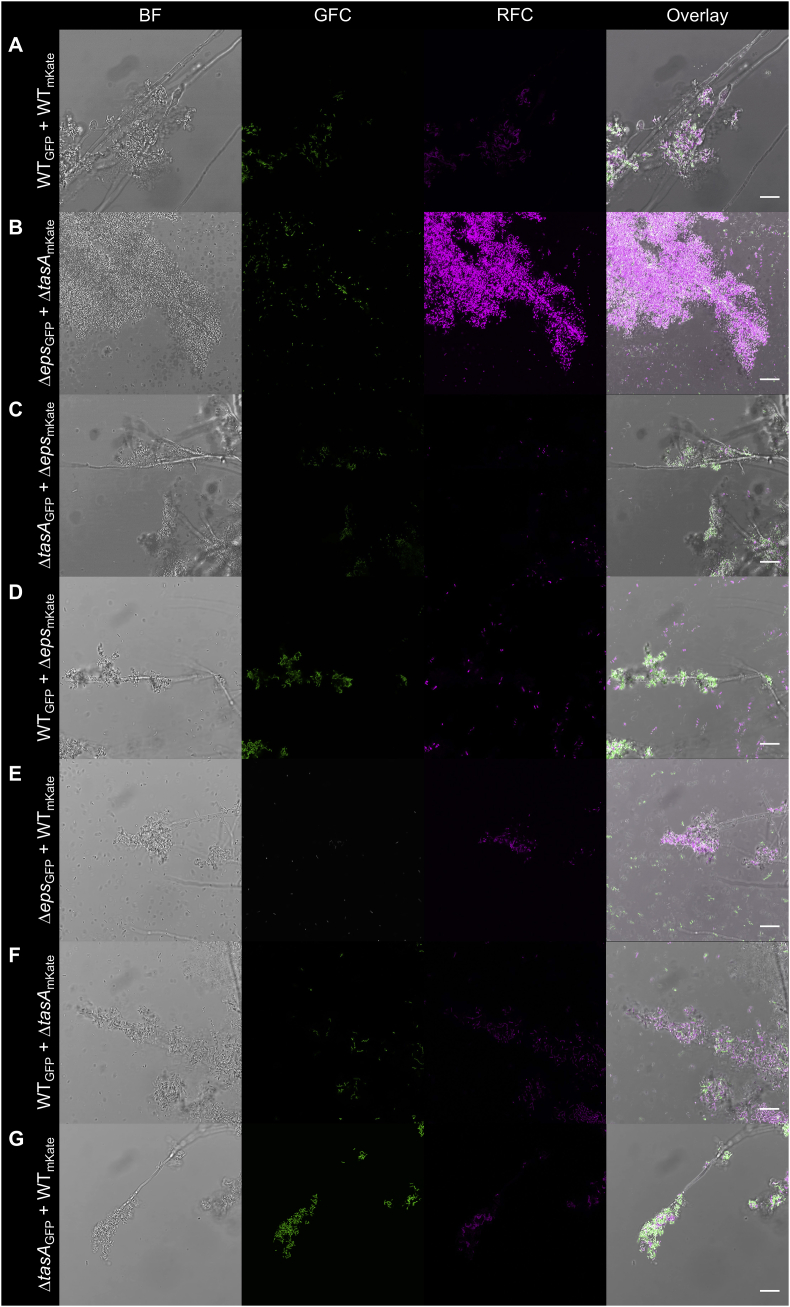
To complement the mutations and lack of biofilm formation ability of the bacterial strains on the hyphae of A. niger, various co-cultivations were tested. Deficiency in EPS or TasA production was complemented either by co-cultivation of mutant strains together or by addition of the wild type strain to biofilm matrix mutant B. subtilis, irrespective of fluorescent labeling (Fig. 3). Previous studies demonstrated that secreted biofilm matrix components complement pellicle biofilm formation [20,27] as well as plant colonization [10,11]. Notably, mixing eps and tasA mutant B. subtilis strains results in genetic division of labor, thus increased population productivity both during pellicle biofilm formation at the air-liquid interface and during plant root colonization [11,28].
Fig. 3.
Mutations in tasA and eps genes were complemented by the co-inoculation of the mutants (A and B), and by the wild type strain (C–F) including a fluorescent marker swap, resulting in fungal colonization by the mutants. GFC: green fluorescent channel; RFC: red fluorescent channel. Scale bars indicate 20 μm. The images presented are representative examples selected from independent samples repeated on different days. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Biofilm formation of B. subtilis on the basidiomycete mycelia
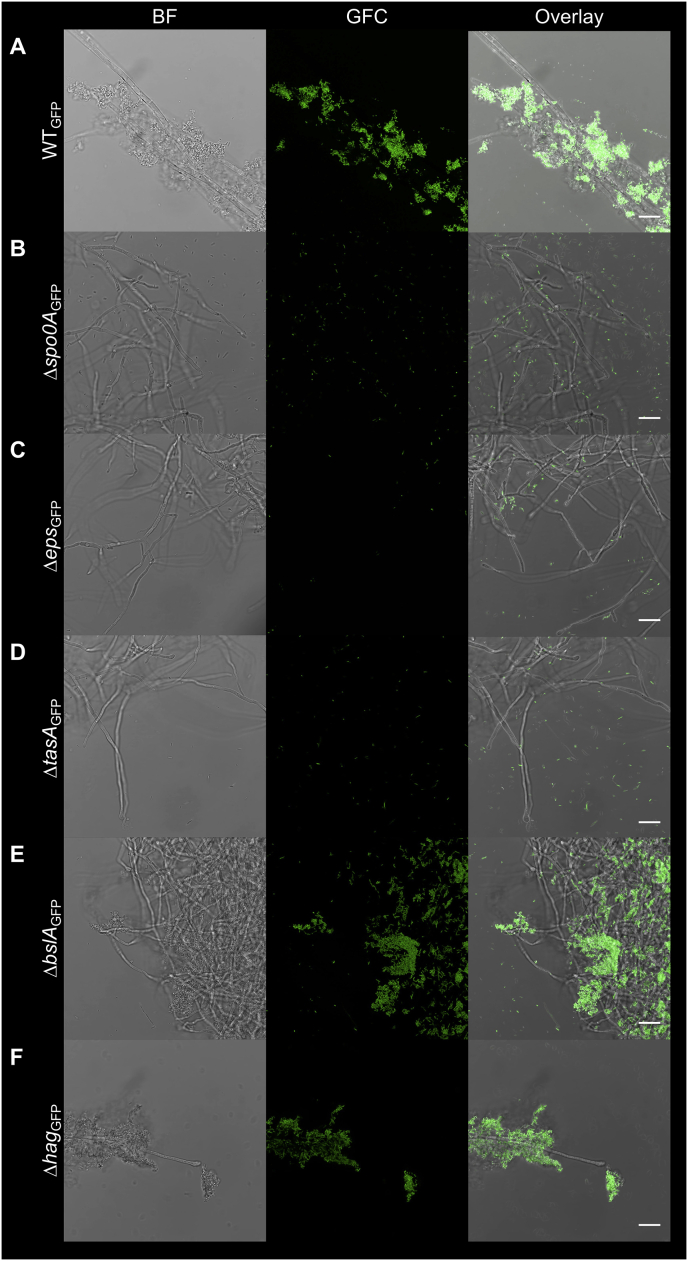
The above described simple co-culture system comprises a bacterium, B. subtilis and an Ascomycota fungus, A. niger that can be both isolated from soil, however, their co-occurrence in nature has never been reported according to our knowledge. Therefore, we examined fungus-attached biofilm formation by B. subtilis on a more ecologically relevant host. In vitro laboratory experiments suggest that B. subtilis cells can colonize the hyphae of the ectomycorrhizal fungus, L. bicolor [3] and the basidiomycete Coprinopsis cinerea [29]. In addition, B. subtilis has been isolated from the soil directly underneath a troop of growing Paxillus involutus mushrooms [30] and below an Agaricus sp. fruiting body (Kiesewalter and Kovács, unpublished observation). Therefore, we set out to examine the importance of biofilm matrix components during mycelia colonization of button mushroom, A. bisporus. In agreement with the above observations on A. niger from this study, hyphal biofilm formation by B. subtilis was diminished by deletion of spo0A, epsA-O, or tasA genes, while removal of bslA or hag genes did not impact colonization properties (Fig. 4).
Fig. 4.
Attachment of B. subtilis on A. bisporus hyphae. B. subtilis wild type (A) and its corresponding mutants were co-cultivated with A. bisporus macro-colonies. Fungal macro-colonies were subsequently washed and visualized with CLSM. The images were recorded at the edge of the fungal macro-colonies. GFC: green fluorescent channel. Scale bars represent 20 μm. The images presented are representative examples selected from different positions of the fungal macro-colonies on independent samples repeated on different days. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Further, complementation of the matrix mutants, eps and tasA strains was performed using a co-culture of the mutants or by addition of wild type to the mutants (Fig. 5). The experiments with the basidiomycete, A. bisporus strengthens the observation that fungal hyphae colonization and biofilm formation by B. subtilis depends on secreted biofilm components. This suggest that production of a matrix by biofilm-proficient bacterial species in nature could potentially facilitate the establishment of multi-species biofilms on fungal hyphae. Indeed, the presence of biofilm matrix and extracellular DNA has been demonstrated for a numerous bacterial species during colonization of ectomycorrhizal fungi, including L. bicolor [3]. Our genetic approach further supports the importance of bacterial biofilm matrix production during long term colonization of fungal mycelia.
Fig. 5.
Attachment of mixed-culture B. subtilis strains to A. bisporus hyphae. Overnight cultures of different B. subtilis strains were mixed and co-cultivated with A. bisporus macro-colonies. Fungal macro-colonies were subsequently washed and visualized with CLSM; pictures were taken approximately near the edge of the macro-colonies. GFC: green fluorescent channel; RFC: red fluorescent channel. Scale bars represent 20 μm. The images presented are representative examples selected from different positions on independent samples. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusion
Bacterial biofilms in the laboratory have been mostly studied using inert substrates, however, during infections or in the environment, bacteria also interact with eukaryotes, including their hosts (from humans and animals to plants) or the co-habitants of their milieu. Here, we demonstrate that biofilm matrix components of B. subtilis are essential for colonization of the hyphae of A. niger and A. bisporus. In addition, the secretion of these matrix components is sufficient to rescue biofilm formation of matrix deficient strains suggesting that social interaction likely shapes the co-evolution of fungi and bacteria in the environment. This leads to the appearance of specific interactions, including primary metabolite cross-feeding, molecular recognition, and potential induction of secondary metabolite production.
Funding information
This work was supported by the Danish National Research Foundation (DNRF137) within the Center for Microbial Secondary Metabolites.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgement
We thank Nicola Stanley-Wall for the comments on our bioRxiv submission.
References
- 1.Flemming H.C., Wuertz S. Bacteria and archaea on Earth and their abundance in biofilms. Nat Rev Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- 2.Frey-Klett P., Burlinson P., Deveau A., Barret M., Tarkka M., Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75:583–609. doi: 10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guennoc C.M., Rose C., Labbé J., Deveau A. Bacterial biofilm formation on the hyphae of ectomycorrhizal fungi: a widespread ability under controls? FEMS Microbiol Ecol. 2018;94:fiy093. doi: 10.1093/femsec/fiy093. [DOI] [PubMed] [Google Scholar]
- 4.Deveau A., Bonito G., Uehling J., Paoletti M., Becker M., Bindschedler S. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–352. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 5.Kovács Á. Bacillus subtilis. Trends Microbiol. 2019;27:724–725. doi: 10.1016/j.tim.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Trowsdale J., Chen S., Hoch J. Evidence that spo0A mutations are recessive in spo0A-/spo0A+ merodiploid strains of Bacillus subtilis. J Bacteriol. 1978;135:99–113. doi: 10.1128/jb.135.1.99-113.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López D., Vlamakis H., Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 8.Branda S.S., González-Pastor J.E., Ben-Yehuda S., Losick R., Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beauregard P.B., Chai Y., Vlamakis H., Losick R., Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A. 2013;110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragoš A., Kiesewalter H., Martin M., Hsu C.-Y., Hartmann R., Wechsler T. Division of labor during biofilm matrix production. Curr Biol. 2018;28:1903–1913. doi: 10.1016/j.cub.2018.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi K., Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012;85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 13.Hobley L., Ostrowski A., Rao F.V., Bromley K.M., Porter M., Prescott A.R. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A. 2013;110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branda S.S., González-Pastor J.E., Dervyn E., Ehrlich S.D., Losick R., Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos-Monterrosa R, Mhatre E, Kovács Á. Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology. 2016;162:1922–1932. doi: 10.1099/mic.0.000371. [DOI] [PubMed] [Google Scholar]
- 16.Kovács Á.T., van Gestel J., Kuipers O.P. The protective layer of biofilm: a repellent function for a new class of amphiphilic proteins. Mol Microbiol. 2012;85:8–11. doi: 10.1111/j.1365-2958.2012.08101.x. [DOI] [PubMed] [Google Scholar]
- 17.Ostrowski A., Mehert A., Prescott A., Kiley T.B., Stanley-Wall N.R. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol. 2011;193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovács Á.T., Kuipers O.P. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J Bacteriol. 2011;193:998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Yan F., Chai Y., Liu H., Kolter R., Losick R. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol. 2013;15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Branda S.S., Chu F., Kearns D.B., Losick R., Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 21.Benoit I., van den Esker M.H., Patyshakuliyeva A., Mattern D.J., Blei F., Zhou M. Bacillus subtilis attachment to Aspergillus niger hyphae results in mutually altered metabolism. Environ Microbiol. 2015;17:2099–2113. doi: 10.1111/1462-2920.12564. [DOI] [PubMed] [Google Scholar]
- 22.Anagnostopoulos C., Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hölscher T., Bartels B., Lin Y.-C., Gallegos-Monterrosa R., Price-Whelan A., Kolter R. Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J Mol Biol. 2015;427:3695–3708. doi: 10.1016/j.jmb.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang P., Zhang M., Van Elsas J.D. Role of flagella and type four pili in the co-migration of Burkholderia terrae BS001 with fungal hyphae through soil. Sci Rep. 2017;7:2997. doi: 10.1038/s41598-017-02959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingham C.J., Kalisman O., Finkelshtein A., Ben-Jacob E. Mutually facilitated dispersal between the nonmotile fungus Aspergillus fumigatus and the swarming bacterium Paenibacillus vortex. Proc Natl Acad Sci U S A. 2011;108:19731–19736. doi: 10.1073/pnas.1102097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlmeier S., Smits T.H.M., Ford R.M., Keel C., Harms H., Wick L.Y. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–4646. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 27.Martin M., Dragoš A., Hölscher T., Maróti G., Bálint B., Westermann M. De novo evolved interference competition promotes the spread of biofilm defectors. Nat Commun. 2017;8:15127. doi: 10.1038/ncomms15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovács Á, Dragoš A. Evolved Biofilm: review on the experimental evolution studies of Bacillus subtilis pellicles. J Mol Biol. 2019 doi: 10.1016/j.jmb.2019.02.005. in press. [DOI] [PubMed] [Google Scholar]
- 29.Stanley C.E., Stöckli M., Van Swaay D., Sabotič J., Kallio P.T., Künzler M. Probing bacterial-fungal interactions at the single cell level. Integr Biol. 2014;6:935–945. doi: 10.1039/c4ib00154k. [DOI] [PubMed] [Google Scholar]
- 30.Tauber JP, Gallegos-Monterrosa R, Kovács Á, Shelest E, Hoffmeister D. Dissimilar pigment regulation in Serpula lacrymans and Paxillus involutus during inter-kingdom interactions. Microbiology. 2018;164:65–77. doi: 10.1099/mic.0.000582. [DOI] [PubMed] [Google Scholar]
- 31.Konkol M.A., Blair K.M., Kearns D.B. Plasmid-encoded comI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol. 2013;195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seccareccia I, Kovács Á, Gallegos-Monterrosa R, Nett M. Unraveling the predator-prey relationship of Cupriavidus necator and Bacillus subtilis. Microbiol Res. 2016;192:231–238. doi: 10.1016/j.micres.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Hölscher T., Dragoš A., Gallegos-Monterrosa R., Martin M., Mhatre E., Richter A. Monitoring spatial segregation in surface colonizing microbial populations. J Vis Exp. 2016;2016 doi: 10.3791/54752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter A., Hölscher T., Pausch P., Sehrt T., Brockhaus F., Bange G. Hampered motility promotes the evolution of wrinkly phenotype in Bacillus subtilis. BMC Evol Biol. 2018;18:155. doi: 10.1186/s12862-018-1266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verhamme D.T., Murray E.J., Stanley-Wall N.R. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J Bacteriol. 2009;191:100–108. doi: 10.1128/JB.01236-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenberg A.S.M., Gao W., Lavrijssen B., Hendrickx P., Sedaghat-Tellgerd N., Foulongne-Oriol M. A detailed analysis of the recombination landscape of the button mushroom Agaricus bisporus var. bisporus. Fungal Genet Biol. 2016;93:35–45. doi: 10.1016/j.fgb.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Bos C.J., Debets A.J.M., Swart K., Huybers A., Kobus G., Slakhorst S.M. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr Genet. 1988;14:437–443. doi: 10.1007/BF00521266. [DOI] [PubMed] [Google Scholar]