Abstract
Interspecies interactions in bacterial biofilms have important impacts on the composition and function of communities in natural and engineered systems. To investigate these interactions, synthetic communities provide experimentally tractable systems. Biofilms grown on agar-surfaces have been used for investigating the eco-evolutionary and biophysical forces that determine community composition and spatial distribution of bacteria. Prior studies have used genetically identical bacterial strains and strains with specific mutations, that express different fluorescent proteins, to investigate intraspecies interactions. Here, we investigated interspecies interactions and, specifically, determined the community composition and spatial distribution in synthetic communities of Pseudomonas aeruginosa, Pseudomonas protegens and Klebsiella pneumoniae. Using quantitative microscopic imaging, we found that interspecies interactions in multispecies colonies were influenced by type IV pilus mediated motility, extracellular matrix secretion, environmental parameters, and these effects were also influenced by the specific partner in the dual species combinations. These results indicate that the patterns observable in mixed species colonies can be used to understand the mechanisms that drive interspecies interactions, which are dependent on the interplay between specific species’ physiology and environmental conditions.
Keywords: Pseudomonas, Klebsiella, Biofilm, Colony, Interspecies interactions, Model community
Highlights
-
•
Spatial patterns in bacterial colonies are species and interaction dependent.
-
•
Surface motility and extracellar matrix production affect interspecies interactions.
-
•
Agar surface colonies show how bacteria interact in biofilms.
Introduction
It is now widely accepted that bacteria form biofilms as an adaptive strategy that facilitates growth and protects them from environmental stresses [[1], [2], [3]]. The genetic systems, adherence mechanisms and consequences of biofilm formation have also been well studied for a number of model bacterial species [[4], [5], [6]]. While such studies have greatly improved our understanding of biofilms and their function, most studies focus on a single species in isolation, whereas in nature, biofilms exist as communities of different species [7], driving the need to study multi-species biofilms. The spatial structure and dense growth of biofilms facilitates interactions between these different species, which in turn strongly influence the growth and survival of bacteria in natural and engineered communities [8,9]. Interactions between community members vary considerably; some interactions facilitate growth and survival, while others inhibit growth or even result in the death of one species [10]. Both faciliatory and inhibitory interactions can be mediated by secreted products, e.g. metabolite exchange [11] and antibiotic production [12] or by contact-dependent mechanisms, e.g. adhesion [13,14] and Type VI secretion mediated killing [15]. Biofilms also create spatial structure, which can enable the genetic division of labour to improve pellicle formation [16] and influences the evolution [17], formation [18] and outcome [19] of interspecies interactions. Furthermore, spatial structure directly enables metabolite exchange for cross-feeding as well as influencing access to essential nutrients and oxygen [20]. Using a synthetic, three-species community, we have shown that co-culture biofilms exhibit increased growth by particular members and enhanced stress tolerance for the entire community [21]. In this community, we observe close association between Klebsiella pneumoniae and Pseudomonas aeruginosa, while P. protegens is more randomly distributed. However, the genetic dissection of the factors driving the organization and function of this community is limited by the long experimental time frames associated with growing biofilms in flow cells. Therefore, we sought for a higher throughput experimental system to investigate factors hypothesized to be involved in interspecies interactions.
Colonies grown on a nutrient surface are an efficient system for manipulating and visualizing the spatial distribution of different microorganisms [22]. For example, in colonies, various spatiotemporal aspects of microbial interactions have been characterized including the co-localization of mutually auxotrophic strains of Saccharomyces cerevisiae [23,24], the vertical separation of differently sized cells of Escherichia coli [25], the sequential range expansion of nitrate reducing Pseudomonas stutzeri [26], the spatial separation patterns of strains of Vibrio cholerae [27] or Bacillus subtilis [28] that differ in the quantity of extracellular matrix produced, and the proportion of different antibiotic resistant/sensitive strains of P. aeruginosa [29]. All of these studies have focused on different strains or mutants of a single species (intraspecies interactions), differing only in the fluorescent marker they express and targeted deletion of specific genes. However, it is also clear that this approach has considerable potential for investigating interactions between different species and addresses our need for increased throughput.
We previously showed that our model biofilm community comprised of P. aeruginosa PAO1, P. protegens Pf-5 (formerly P. fluorescens [30,31]) and K. pneumoniae KP-1 exhibits properties not observed in biofilms of the individual species or liquid cultures. These include an overall increase in total biofilm biomass, sharing of resistance to sodium dodecyl sulfate allowing the sensitive species, P. protegens, to survive [21], and reduced production of morphotypic variants for all species [32]. We hypothesized that many of the factors that are important for monospecies biofilm formation would play important roles in mixed species community interactions as well. For example, we previously showed that changes in matrix production by P. aeruginosa altered the biofilm community composition in flow cells [33]. In this study, we adapted the colony biofilm approach to investigate the mechanisms that drive positive and negative interactions in dual-species biofilms. We determined whether these interactions were beneficial or detrimental by quantifying the area colonized by each species and qualifying where each species was found. We showed that each pairing of species resulted in different spatial distributions. To determine the cause of these differences, we tested the effects of biofilm related traits, specifically the type IV pilus of P. aeruginosa and matrix production by K. pneumoniae, and showed how they influence interspecies interactions and spatial distribution in an environment-dependent fashion. This work demonstrates that co-culturing in colony biofilms is a useful tool for determining the mechanisms and outcomes of interactions between bacterial community members.
Materials and methods
Strains, media and growth conditions: Colonies were grown on minimal medium (48 mM Na2HPO4; 22 mM KH2PO4; 9 mM NaCl; 19 mM NH4Cl; 2 mM MgSO4; 0.1 mM CaCl2; 0.04 mM FeSO4; 2 mM glucose and 0.4% casamino acids) with either 1.5% or 0.6% agar in 24 well plates or 8 well Ibidi™ slides. Previously described strains (Table 1) [21] of P. aeruginosa PAO1, P. protegens Pf-5 and K. pneumoniae KP-1 constitutively expressing fluorescent protein genes inserted into the chromosome using a Tn7 transposon were recovered from freezer stocks directly onto LB agar plates and grown for 24–48 h at room temperature. Bacteria were inoculated into liquid medium from agar plates (because they are highly mucoid, wild-type K. pneumoniae strains were homogenized by passing repeatedly through a 30 G needle), the inoculum was standardized to OD600 = 0.1 and 1/100 dilutions were used directly or mixed 1:1 and 0.5 μL was spotted at the center of each well. Spots were dried for 30 min in a laminar flow cabinet, plates were sealed with parafilm and incubated at room temperature. To confirm that the inoculum ratio determined by OD was 1:1, colony forming units (CFUs) were determined separately for the inoculum for each species/strain. If CFU counts indicated that the inoculum used would have resulted in an incorrect ratio (≥10:1), the sample was not used. For co-culture colonies, only combinations of CFP or YFP labeled strains with dsRed or mCherry were used, with the exception of KP-1-YFP and Pf-5-CFP, due to issues of overlapping excitation and emission spectra. When multiple combinations were possible (e.g. KP-1-dsRed with KP-1-CFP or KP-1-YFP) both combinations were treated as equivalent i.e. the combination of dsRed with CFP was considered a replicate of combinations of dsRed with YFP.
Table 1.
Species and strains used in this study.
| Species and strain | Genotypic and phenotypic characteristics | Tn7 chromosomal fluorescent protein | Source or reference |
|---|---|---|---|
| P. aeruginosa PAO1 | Wild-type | YFP, mCherry | ATCC BAA-47 [19], this study |
|
P. aeruginosa PAO1 Small colony variant (SCV) |
pilT Thr210 frameshift, twitching defect | YFP | [32] |
| P. protegens Pf-5 | Wild-type | CFP, mCherry | ATCC BAA-477 [19], this study |
| K. pneumoniae KP-1 | Wild-type | dsRed, CFP, YFP | [19] |
| K. pneumoniae KP-1 Non-mucoid variant (NMV) | UDP-glucose lipid carrier transferase Phe123Ile, matrix secretion defect | dsRed | [32] |
Image acquisition: Colonies were imaged on a Zeiss Axio Observer.Z1 inverted (24 well plates) or Imager.M2 upright (8 well slides) microscope using an HXP 120C halogen light source. On the Z1 a 5X/0.16 objective was used and a 10X/0.3 objective on the M2. The filter sets (excitation/emission) used were: Zeiss Set 10 for CFP (450–490/515-565), Set 46 for YFP (490–510/520-550) and Set 31 (550–580/590-660) for dsRed and mCherry on the Z1. Set 43 (520–570/565-630) was used for dsRed and mCherry on the M2. Both filter sets are compatible with acquiring fluorescence images for dsRed and mCherry expressing strains. Exposure times and focus were manually optimized for each sample then colonies were imaged using ‘Tilescan’ mode in Zeiss Zen (Blue edition) with 10% overlap. Twelve-bit images were captured using an AxioCamMRm3. Shading correction was applied using pre-captured profiles of fluorescent slides. Images from both microscopes were treated equivalently in downstream processing.
Image Presentation: Representative images from each experimental combination were selected for presentation. Images were processed for viewing to improve contrast and remove artefacts, but only raw images were used for quantitative analysis. Images were corrected for flat-field illumination using BaSiC [34], stitched together in ImageJ [35,36] and then the brightness was scaled manually for each channel in each image. In cases when there was high background signal, the background was either subtracted manually by segmenting the colony area or by using the background subtraction tool.
Image Analysis: For full details, see the supplementary material. Briefly, tilescan images stitched together using Zeiss Zen were imported into R and processed using the package ‘imager’ [37]. Each channel was normalized to a range of 0 to 1 and any background signal was subtracted. To segment the colony from the background, a mask of the colony was generated by taking the sum of all channels (including the inverted brightfield) then thresholding this image using either “IJDefault” or “triangle” in the R package ‘auto_thresholdr’ [38], as described in the supplementary methods. Fractal dimensions were determined according to the methods of Rudge et al. [39] and Bérubé and Jebrak [40]. The path length ratio (sinuosity) of sector boundaries were determined from a single thresholded channel, for full details see the supplementary methods. To determine the amount of overlap between the two strains within a colony, the ratio between channels was calculated at every pixel by subtracting one channel from the other and dividing them by the sum of both ((c1-c2)/(c1+c2)). Thus, values can range from 1/-1 indicating only the presence of one strain, to 0, indicating an exactly equal mix of the two strains. The proportion of mixed pixels was then calculated by dividing the number of bins in the range −0.25:0.25 by the total number of pixels within the colony as defined by the segmentation. We then attempted to determine the area occupied by each strain within the colony by thresholding each fluorescent channel with several different methods (including Li, Huang and triangle), then manually selecting the most accurate method. Due to the manual intervention required to pick an appropriate threshold, which varied substantially between images, the area of each strain was instead manually determined in ImageJ. For all conditions a minimum of 6 biological replicates were analyzed. The image data is available openly available in the NTU research data repository DR-NTU (Data) at DOI: https://doi.org/10.21979/N9/FBXSXV.
Statistics: Analysis of variance and Tukey’s honest significant difference post-hoc test, which includes multiple-testing p-value adjustment were used to determine if there were significant differences with p ≤ 0.05 as a significance cut-off.
Results
Monospecies colonies
To benchmark the method and as controls, we first compared patterns of interactions for mixtures of two strains of the same species with previously published studies (Fig. 1). This also formed the basis for comparison of how each species behaved on their own to when they were interacting with a second species. Qualitative inspection of the colonies after 24 and.
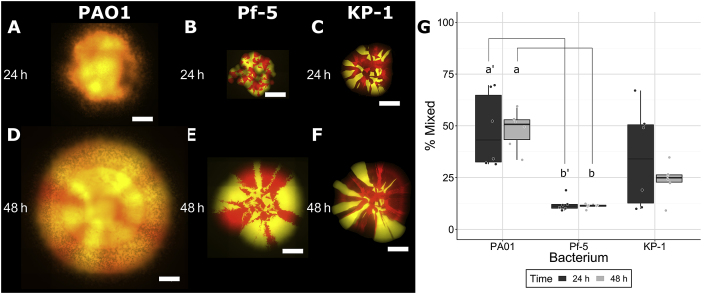
Fig. 1.
Colonies consisting of P. aeruginosa PAO1 (A, D), P. protegens Pf-5 (B, E) or K. pneumoniae KP-1 (C, F). Each species was labeled separately with two different fluorescent protein genes and are presented here as red or yellow (false colouring). Strains were mixed 1:1 (OD600) at the time of inoculation and colonies were imaged after 24 and 48 h of growth. Scale bars indicate 1000 μm. Overlap between strains was calculated from raw images as the percentage of mixed pixels (G). Boxplots are presented in Tukey style. Lines connecting letters indicate statistically significant differences between a and b (p ≤ 0.05) according to ANOVA and Tukey’s honest significant differences test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
48 h of growth indicated that each species differed in how they separate into sectors. P. protegens (Fig. 1B,E) and K. pneumoniae (Fig. 1C,F) showed clear segregation between strains, although the shape of the borders differed. For P. aeruginosa (Fig. 1A,D), separation into sectors was only apparent at 48 h and the borders between strains were less distinct than for the other species. To quantify these differences in sectoring, we calculated the fractal dimensions and path-length ratios of sector boundaries (Supplementary Figure 1, Supplementary Table 1). The fractal dimension and path length ratios for K. pneumoniae were significantly lower than for P. protegens, indicating that the sectors for K. pneumoniae are straighter than P. protegens. This analysis thus supports the qualitative description of the colony sectors. Due to the lack of defined sectors for P. aeruginosa, it was not possible to compare with the other two strains.
To better characterize the differences in colony growth, quantitative analyses were performed by determining the percentage of pixels with similar relative signal within each colony (Fig. 1, G, Supplementary Figure 2, Supplementary Table 2). Around 50% of pixels were mixed in the P. aeruginosa colonies compared to 25% or less in the other two species after 48 h of growth. Even at the leading edge of P. aeruginosa regions that appeared to consist of a single strain, cells of both colours were present and were well mixed (Supplementary Video 1). Movement and mixing of the two strains was visible up to 500 μm inward from the colony edge (Supplementary Video 2). For all three species, there was no change in mixing between 24 and 48 h. These results show that both P. protegens and K. pneumoniae behave similarly to previous studies as they form colonies with clearly defined sectors. In contrast, P. aeruginosa colonies were highly mixed (no discernable sector boundaries), likely due to a specific physiological trait that is absent in the other two species.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.bioflm.2020.100035
The following is/are the supplementary data related to this article:
1
2
Dual species colonies
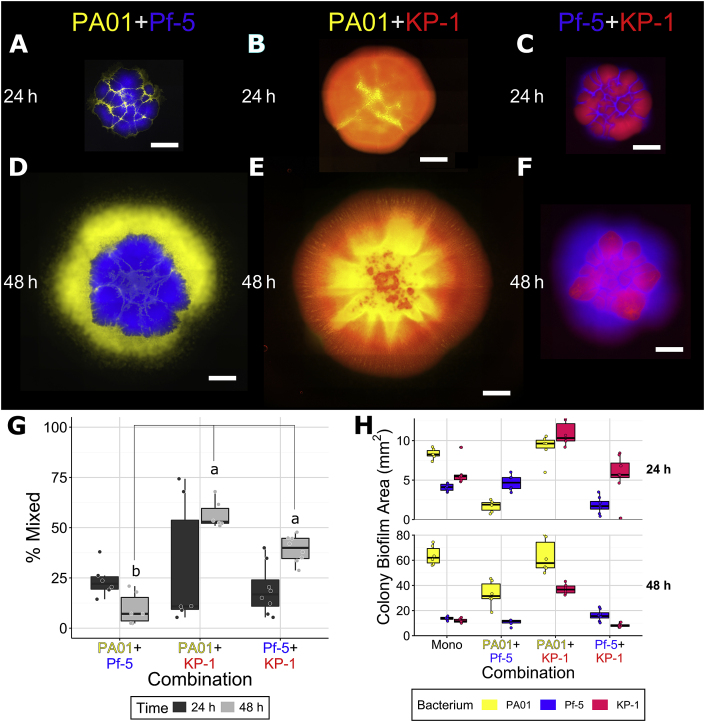
Based on the results from the monospecies experiments, we next wanted to determine the spatial patterning of dual species colonies to understand how interspecies interactions impacted colony formation. It was hypothesized that cooperative interactions would be evidenced by overlap between species while competition would result in spatial exclusion. None of the combinations of species separated into clear radial sectors, as observed for the dual-strain colonies described above, and instead formed distinct patterns (Fig. 2). While P. aeruginosa appeared to be restricted to the interior of the colony when cultured with K. pneumoniae (Fig. 2B,E), closer inspection of brightfield images (Supplementary Figure 3) showed that P. aeruginosa was also present around the outside perimeter of K. pneumoniae, as visible with its distinct lattice morphology. This combination changed from unmixed (~25%, though variable) at 24 h to mixed at 48 h (consistently 50%). P. aeruginosa localized around the perimeter of P. protegens colonies (Fig. 2A,D), especially after 48 h and these species did not mix at either timepoint, despite P. aeruginosa being able to mix with K. pneumoniae. Based on the monospecies results, where both K. pneumoniae and P. protegens separated into sectors, it was predicted that they would also make clear sectors when grown together. However, when grown with K. pneumoniae, P. protegens grew around the outer edge of the colony (Fig. 2C, F) and these species were also partially mixed at 48 h (~40%, significantly higher than P. aeruginosa combined with P. protegens), which was not predicted. Thus, while both Pseudomonas species were able to mix with K. pneumoniae, which did not mix with itself, P. protegens may be able to block P. aeruginosa from occupying its territory.
Fig. 2.
Dual species colonies of P. aeruginosa PAO1 and P. protegens Pf-5 (A, D), P. aeruginosa PAO1 and K. pneumoniae KP-1 (B, E), and P. protegens Pf-5 and K. pneumoniae KP-1 (C, F), overlap between differently labeled strains (G) and the area covered by individual species in colonies (H). Each species was labeled with different fluorescent protein genes and are presented here as yellow (P. aeruginosa PAO1), blue (P. protegens) and red (K. pneumoniae). Strains were mixed 1:1 (OD600) at the time of inoculation and colonies were imaged after 24 and 48 h of growth. Scale bar indicates 1000 μm. Boxplots are presented in Tukey style. Paired letters indicate statistically significant differences between a and b (p ≤ 0.05) according to ANOVA and Tukey’s honest significant differences test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The area covered by each strain in the dual-species colonies was compared to the area covered by single species colonies as a measure of whether the two exhibit positive or negative interactions during co-culture (Table 2, Fig. 2, H). At 24 h, the area covered by P. aeruginosa alone (~8 mm2) was lower than when co-cultured with P. protegens (~2 mm2) and was similarly reduced at 48 h, from ~70 mm2 (monospecies) compared to ~35 mm2 (dual species). In contrast, there was no difference in the area covered by P. protegens when co-cultured with P. aeruginosa, although it was significantly decreased (from ~4 to ~1.5 mm2) at 24 h, but not 48 h, with K. pneumoniae. The area covered by K. pneumoniae increased when co-cultured with P. aeruginosa from ~5 to ~10 mm2 at 24 h and ~12 to ~40 mm2 at 48 h, but was not affected by P. protegens. These results differed substantially from the monospecies colonies as interspecies interactions affected the surface area colonized and mixing within colonies, dependent upon the specific combination of species. As K. pneumoniae appeared to benefit from mixing with P. aeruginosa, but was negatively affected by mixing with P. protegens, we aimed to investigate why. As P. aeruginosa can crawl on agar surfaces using its type IV pili and K. pneumoniae produces a capsular polysaccharide which makes for very thick, sticky colonies, we therefore hypothesized that these physiological differences may be responsible for the observed differences in colony organisation interspecies interactions.
Table 2.
Comparison of dual species interactions for P. aeruginosa, P. aeruginosa small colony variant (SCV), P. protegens, K. pneumoniae and K. pneumoniae non-mucoid variant (NMV).
| Actor | Recipient |
||||
|---|---|---|---|---|---|
| P. aeruginosa | P. aeruginosa SCV | P. protegens | K. pneumoniae | K. pneumoniae NMV | |
| P. aeruginosa | ↑ | ||||
| P. aeruginosa-SCV | ↓ | ND | |||
| P. protegens | ↓ | ↓ | ↓ | ||
| K. pneumoniae | ↓ | ||||
| K. pneumoniae-NMV | ND | ||||
∗ Empty cells indicate no significant difference between single and dual species colonies was observed, ND indicates a combination which was not determined. Interactions that resulted in a significant increase in area are indicated by ↑ and decreases by ↓ compared to the monospecies colony by analysis of variance and Tukey’s honest significant differences post-hoc test. Differences were considered significant if their multiple-testing adjusted p-value was below 0.05 for 48 h comparisons. For all p-values see Supplementary Table 3.
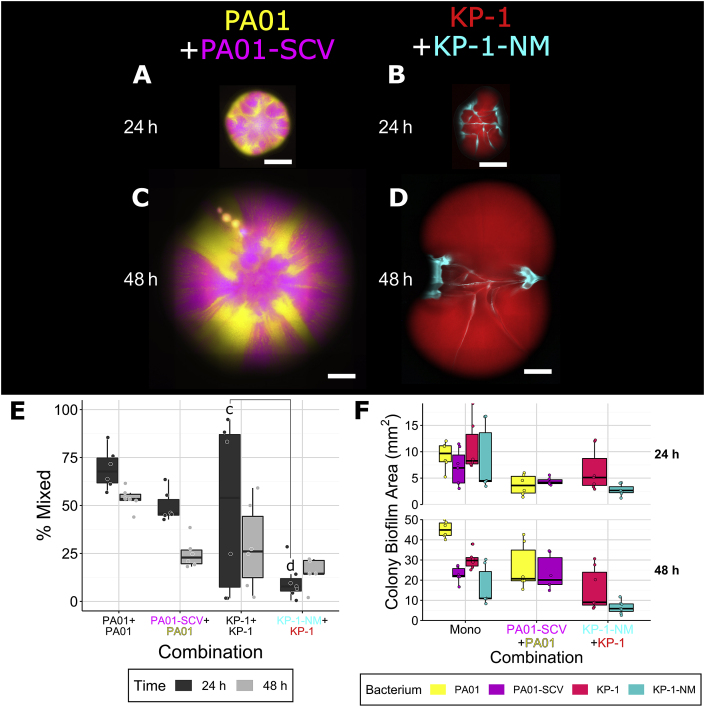
Co-cultures with a P. aeruginosa small colony variant
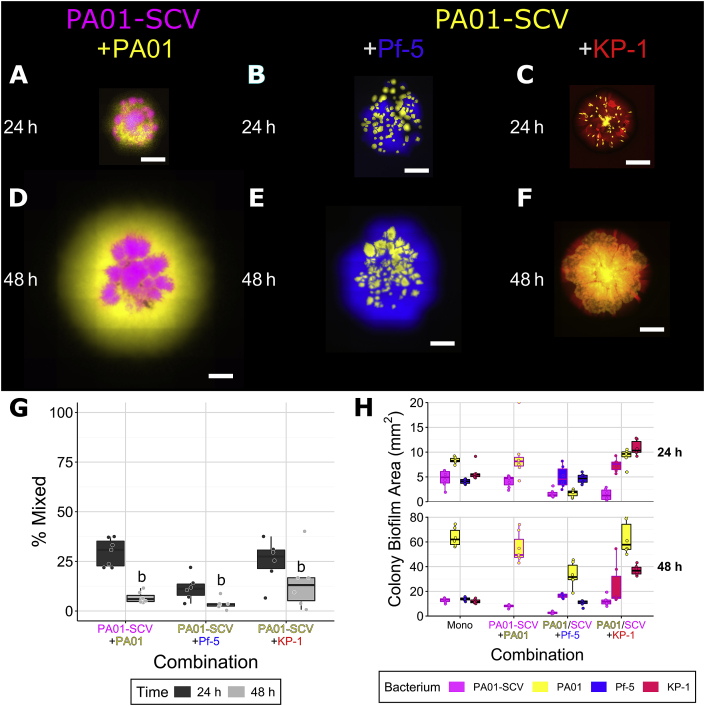
P. aeruginosa uses type IV pili (TFP) for twitching based motility on surfaces (including agar) [41] and TFP have also been shown to be important for structure formation during flow cell biofilm development [42]. Additionally, while common in monospecies biofilms, we observed that small colony variants (SCV) are significantly reduced or absent in mixed species biofilms, suggesting there may be strong selection against these TFP mutants [32]. We therefore hypothesized that TFP may play an important role in mixing and colony development. To investigate the effect of the TFP of P. aeruginosa in colonies, we used a SCV, which we previously determined to have a single mutation in pilT [32], preventing pilus retraction and therefore TFP motility [43,44]. This strain was cultured in dual species colonies with its parental strain, P. protegens and K. pneumoniae. Colonies formed with the SCV differed from those with wild-type P. aeruginosa and the mixing of strains/species also differed (Fig. 3). Both P. aeruginosa (Fig. 3A,D) and P. protegens (Fig. 3B,E) formed a perimeter around the SCV, but there was very little (≤10%) mixing of strains/species. The SCV only mixed slightly more with K. pneumoniae (Fig. 3C,F). The SCV colonized less area than wild-type P. aeruginosa, ~12 mm2 compared to ~70 mm2, respectively, after 48 h (Table 2, Fig. 3, H). In dual species colonies, the area covered by the SCV was reduced to ~2 mm2 at 48 h in the presence of P. protegens. When grown with K. pneumoniae, the SCV covered less area at 24 h but this was not apparent at 48 h. The SCV did not affect the area covered by the other strains and in contrast to when the parental strain of P. aeruginosa was present, the SCV did not increase the area covered by K. pneumoniae. These results suggest that TFP based surface motility of P. aeruginosa may be the main factor responsible for mixing with itself and K. pneumoniae. It also suggests that TFP may enable P. aeruginosa to facilitate the expansion of K. pneumoniae.
Fig. 3.
Co-culture colonies consisting of P. aeruginosa PAO1 and P. aeruginosa PAO1 small colony variant (SCV) (A, D), P. aeruginosa PAO1 SCV and P. protegens Pf-5 (B, E) and P. aeruginosa PAO1 SCV and K. pneumoniae KP-1 (C, F), overlap between differently labeled strains (G), and the area covered by individual species/strains (H). Each species was labeled with different fluorescent protein genes and are presented here as yellow (P. aeruginosa wild-type when paired with the SCV, SCV when paired with the other two species), magenta (P. aeruginosa SCV), blue (P. protegens) and red (K. pneumoniae). Strains were mixed 1:1 (OD600) at the time of inoculation and colonies were imaged after 24 and 48 h of growth. Scale bar indicates 1000 μm. Boxplots are presented in Tukey style. For the area boxplots (H) combinations of P. aeruginosa with P. protegens or K. pneumoniae, boxes outlined in black represent the wild-type (same data as Fig. 2H), outlines in magenta represent combinations with the SCV. Letters indicate statistically significant differences between a and b (p ≤ 0.05) (see previous figures) according to ANOVA and Tukey’s honest significant differences test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Co-cultures with a non-mucoid K. pneumoniae variant
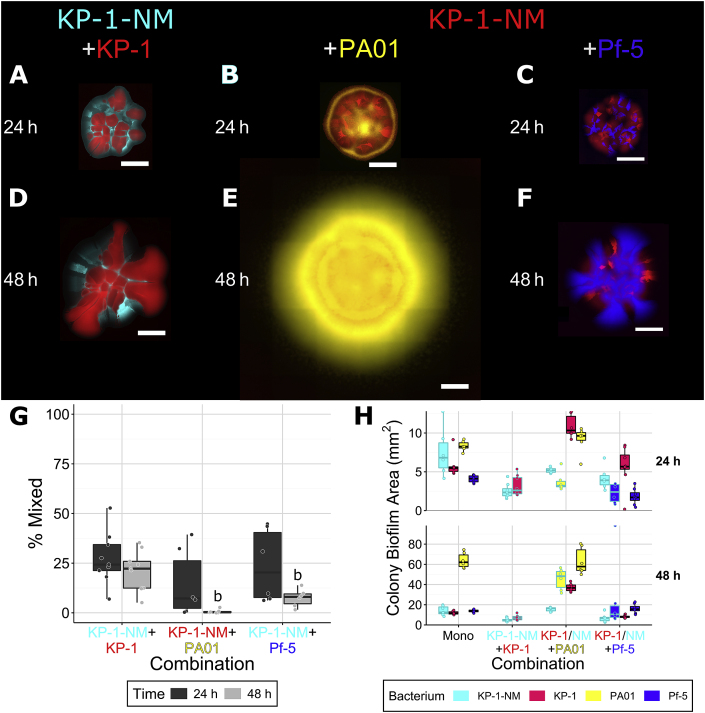
As a hallmark of biofilms, we also expected secreted extracellular matrix (ECM) polymers to influence interactions between species and the resulting spatial distributions. Similarly to the P. aeruginosa SCV, we observed a high proportion (30–40%) of non-mucoid variants (NMV) spontaneously produced by monospecies biofilms of K. pneumoniae, while almost none are observed in mixed species biofilms [32]. To investigate the importance of ECM produced by K. pneumoniae, the NMV strain was cultured in dual strain/species colonies with the parental strain, P. aeruginosa and P. protegens. This previously characterized NMV has a mutation within a predicted colanic acid synthesis gene cluster, preventing it from producing its normal, thick ECM [32]. Single-strain colonies formed by the NMV differed from wild-type K. pneumoniae (Supplementary Figure 4). Boundaries between sectors of the NMV were jagged like those of P. protegens instead of straight as was observed for the wild-type (Supplementary Figure 1). Dual-species colonies with the NMV also differed as it did not mix with wild-type K. pneumoniae or P. protegens (Fig. 4). When grown with P. aeruginosa (Fig. 4B,E), mixing was low at 24 h (10–20%) and was not readily observed at 48 h. Thus, it appears that, in addition to TFP, the capsule of K. pneumoniae is also important in mediating mixing, which was unexpected.
Fig. 4.
Co-culture colonies consisting of K. pneumoniae KP-1 with the K. pneumoniae KP-1 non-mucoid variant (NMV) (A, D), K. pneumoniae KP-1 NMV with P. aeruginosa (B, E), and K. pneumoniae KP-1 NMV with PAO1 P. protegens Pf-5 (C, F), overlap between differently labeled strains (G) and the area covered by individual species/strains (H). Each species was labeled with different fluorescent protein genes and are presented here as yellow (P. aeruginosa), blue (P. protegens), red (K. pneumoniae wild-type when paired with the NMV, K. pneumoniae NMV when paired with the other species) and cyan (K. pneumoniae NMV). Strains were mixed 1:1 (OD600) at the time of inoculation and colonies were imaged after 24 and 48 h of growth. Scale bar indicates 1000 μm. Boxplots are presented in Tukey style. For the area boxplots (H), combinations of K. pneumoniae with P. aeruginosa or P. protegens, boxes outlined in black represent the wild-type (same data as Fig. 2, H), outlines in cyan represent the NMV. Letters indicate statistically significant differences between a and b (p ≤ 0.05) (see previous figures) according to ANOVA Tukey’s honest significant differences test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
When grown as single-strain colonies, wild-type K. pneumoniae and the NMV each covered a similar amount of area, suggesting both strains grow equally well as colonies. When the two were co-cultured together (Table 2, Fig. 4A,D), the NMV covered less area, ~2.5 mm2, compared to when grown alone, ~7 mm2, at 24 h, and was also reduced at 48 h (from ~12 to ~5 mm2). This indicates that the NMV is not as competitive as its parental strain under these conditions. The area covered by the NMV was also decreased when paired with P. protegens (Fig. 4C,F), however, neither of these differences persisted at 48 h. In contrast, the area covered by P. aeruginosa was decreased when grown in the presence of the K. pneumoniae NMV at 24 h but not 48 h. When paired with P. aeruginosa, the area covered by the NMV was not significantly increased as was observed for the parental K. pneumoniae strain when paired with P. aerguinosa. These results indicate that the capsule produced by K. pneumoniae may also play an important role in the surface area expansion of K. pneumoniae when co-cultured with P. aeruginosa, possibly in conjunction with its TFP.
Colonies cultured on 0.6% agar
Given that the P. aeruginosa SCV and K. pneumoniae NMV differed substantially from their respective wild-types in their mixing and colony area covered, we were interested in examining how these traits were influenced when the environment was modified. The percentage of agar influences motility, as lower percentages enable swarming and/or swimming motility and affects the colony growth of highly mucoid strains [27]. Culturing on a lower percentage of agar was thus expected to affect interactions between the wild-type strains of P. aeruginosa and K. pneumoniae and their non-motile (SCV) and non-mucoid (NMV) variants. Colony morphologies consisting of wild-type and mutant strains grown on 0.6% agar differed from those on 1.5% (Fig. 5) as did the areas colonized (Table 3). However, those consisting of two differently labeled wild-type strains did not. Additionally, the agar concentration did not influence the formation of sector boundaries in the wild-type strains (Supplementary Figure 1,5). When the P. aeruginosa SCV was co-cultured with its parental strain (Fig. 5A,C), both strains expanded outwards together, but were less mixed (~22% at 48 h) compared to the two differently coloured wild-type P. aeruginosa strains (~55% at 48 h, Fig. 1G), which was similar to that observed on 1.5% agar (50%, Fig. 1G). When the K. pneumoniae NMV was co-cultured with the wild-type (Fig. 5B,D), the two strains showed little mixing, however by 48 h the wild-type had expanded much more than the NMV (Fig. 5, D). When two wild-type K. pneumoniae strains were grown together, they mixed comparably on both percentages of agar (~25%, Figs. 5E and 1G).
Fig. 5.
Co-culture colonies on 0.6% agar consisting of P. aeruginosa PAO1 with P. aeruginosa small colony variant (SCV) (A, C), and K. pneumoniae with K. pneumoniae KP-1 non-mucoid variant (NMV) (B, D), overlap between differently labeled strains (E) and the area covered by individual species/strains (F). Each species was labeled with different fluorescent protein genes and are presented here as yellow (P. aeruginosa wild-type), magenta (P. aeruginosa SCV), red (K. pneumoniae wild-type) and cyan (K. pneumoniae NMV). Strains were mixed 1:1 (OD600) at the time of inoculation and colonies were imaged after 24 and 48 h of growth. Scale bar indicates 1000 μm. Boxplots are Tukey style. Paired letters indicate statistically significant differences between c and d (but not c and a, see previous figures) (p ≤ 0.05) according to ANOVA and Tukey’s honest significant differences test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Comparison of effects of 0.6% agar on the area colonized in colonies of P. aeruginosa, P. aeruginosa small colony variant (SCV), K. pneumoniae and K. pneumoniae non-mucoid (NM) variant.
| Wild-type compared to mutant |
Monospecies 0.6% agar |
Wild-type and mutant 0.6% agar |
||||||
|---|---|---|---|---|---|---|---|---|
| 1.5% Agar | 0.6% Agar | Effect of decreasing agar concentration | Effect of being in co-culture compared to mono-culture | |||||
| Time | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | 24 h | 48 h |
| PAO1 | SCV ↓ | SCV ↓ | SCV ↓ | ↓ | ↓ | ↓ | ||
| PAO1-SCV | ↑ | |||||||
| KP-1 | NMV ↓ | ↑ | ↓ | |||||
| KP-1-NMV | ||||||||
∗ Results from comparisons between the wild-type and mutant on each percentage of agar, between agar percentages for each strain and the between cultivation alone or in combination are shown. Interactions that resulted in a significant increase in area are indicated by ↑ and decreases by ↓, using analysis of variance and Tukey’s honest significant differences post-hoc test. Differences were considered significant if their multiple-testing adjusted p-value (95% confidence) was below 0.05. Empty cells indicate no significant change. For all p-values see Supplementary Table 3.
Colony area was significantly higher for the P. aeruginosa SCV on 0.6% compared to 1.5% agar at 48 h (~20 vs ~10 mm2, Fig. 5F). This was significantly lower than the wild-type on 0.6% agar (~50 mm2), which was also significantly decreased compared to on 1.5% agar (~70 mm2). When cultured with the SCV on 0.6% agar, wild-type P. aeruginosa area was significantly reduced at both timepoints (from 45 to 22 mm2 at 48 h). The K. pneumoniae wild-type colonized a significantly increased area on 0.6% agar (~27 mm2) compared to the NMV (~10 mm2) and the wild-type on 1.5% agar (~11 mm2). When they were co-cultured, the area colonized by wild-type, but not NMV K. pneumoniae, was significantly lower after 48 h. These results show that as expected, modifying the environment changed the outcomes of interactions between the SCV and NMV and their parental strains. Lowering the percentage of agar did not affect mixing, although it did however, affect expansion by the two bacteria species. Under the lower agar concentration, the SCV benefited (increased colony expansion) while the parental P. aeruginosa did not. In contrast, the parental K. pneumoniae benefited from the lower agar concentration, while the NMV did not.
Discussion
Colonies highlight differences in intraspecies interactions
Our model community of P. aeruginosa, P. protegens and K. pneumoniae has been shown to reproducibly establish biofilms with differential abundances of the three bacteria, it displays shared defence mechanisms and reduced production of natural genetic variants, suggesting strong selection pressure against those variants in the community. To begin to unravel the mechanisms involved in these interactions, here we have adapted a colony biofilm community system as a higher throughput approach to test a matrix of combinations of mutants, species interactions and physiological conditions.
For each of the three species from our model community, intraspecific competition resulted in species-specific patterns of sector formation. Separation of mixed colonies into sectors has been attributed to genetic drift at the leading edge of the colony, as despite having equal fitness, stochastic effects result in new territory being colonized by either one strain or the other, but not both [45]. Here, based on quantitative and qualitative assessment, we show that the shape of the boundaries between sectors depends on the bacterium, which likely reflects their individual traits. The separation of wild-type K. pneumoniae strains into sectors with straight boundaries resembled that of Saccharomyces cerevisiae [24], whereas the jagged boundaries of P. protegens were more similar to those of P. stutzeri [26], simulated rod-shaped cells [46] or E. coli [45]. In the case of E. coli, the formation of fractal boundaries between sectors has been explained by the anisotropic forces of cell division and growth causing unlinked chains of rod-shaped cells to buckle [39]. Thus, similar buckling likely causes the formation of jagged edges between boundaries of the wild-type P. protegens strains. The jagged boundaries visible between sectors within K. pneumoniae NMV colonies (Supplementary Figure 4) indicate that its secreted extracellular matrix causes the straight boundaries between sectors observed in the wild-type. Colonies of P. aeruginosa grown on nutrient rich LB medium were previously shown to be well mixed [47], similar to our observations here using minimal medium. The observation that pilB mutants segregate into clear sectors [47] and the resemblance of videos of the edge of P. aeruginosa colonies (Supplementary Video 1) to those showing TFP motility [48] indicates that this motility likely enabled the mixing of the P. aeruginosa wild-type strains.
Interspecies interactions differ from intraspecies interactions
Previous studies have focused on colonies of either identical, but differently labeled strains of the same species, or strains with specific genetic modifications, whereas here, we investigated interactions between different species. While the patterns for our single strain data resemble previous work, it is clear that patterns of species distribution in mixed species colonies differ markedly. Thus, the data presented here show how the different physiological traits of the three studied bacteria determine community composition and spatial distribution in colonies.
We have assessed the three pairwise interactions tested here using the notation of Momeni et al. [23], where A [~,~] B indicates a neutral interaction and A[↑,↓]B indicates a relationship where A benefits and B is negatively affected (Table 2). The spatial distribution patterns of P. aeruginosa with P. protegens, and P. protegens with K. pneumoniae are similar as one strain expanded outward faster and surrounded the other. Although the area covered by the inner strains was not reduced compared to when they were grown alone, both interactions negatively affect the inner strain as it no longer had equal access to space/nutrients. For the first pair, the area covered by the outer strain, P. aeruginosa, was significantly decreased, indicating that both strains experienced negative outcomes from the interaction, so P. aeruginosa [↓,↓] P. protegens. For the second pair, P. protegens was the outer strain but its area was not reduced so, P. protegens [~,↓] K. pneumoniae. In the third case, when P. aeruginosa was paired with K. pneumoniae, P. aeruginosa did not differ in the amount of area covered but the area of K. pneumoniae was significantly increased, so, P. aeruginosa [~,↑] K. pneumoniae. The results therefore suggest that differences in expansion rate may be a key factor in the development of the patterns observed here. Thus, future experiments that collect images every couple of hours, could enable a detailed quantification of expansion rates for single and mixed species colonies. Consistent with our previous work comparing planktonic and biofilm growth modes, these results differ from our observations in planktonic culture where P. protegens outgrew the other two species by between 10-100 fold and P. aeruginosa and K. pneumoniae equally reduced each other’s growth by ~100 fold [21].
The interactions between K. pneumoniae and the two Pseudomonas species were starkly different. When it was grown with P. aeruginosa it colonized more than twice as much area than it could alone, indicating a strong benefit from being co-cultured. P. fluorescens Pf0-1 has been observed to rapidly evolve a division of labour where two different mutant strains are able to make a faster expanding colony than either the wild-type or each strain individually [49]. In those mixed colonies, one strain produced the force for colony expansion through cell division while the other produced an extracellular polymer which acts as a lubricant at the expanding edge of the colony. In our work, K. pneumoniae may have been taking advantage of the extracellular DNA [41] or rhamnolipid surfactants [50] produced by P. aeruginosa to enable colony expansion. Non-motile E. coli was observed to take advantage of motile Acinetobacter baylyi, forming intricately branched floral patterns [51]. These patterns were qualitatively dissimilar to those we observed, indicating that the mechanism by which K. pneumoniae takes advantage of P. aeruginosa motility is also likely different. This strong effect of P. aeruginosa on K. pneumoniae is consistent with our observations in flow cell biofilms where despite making up only a small (1–5%) proportion of the 3-species community, P. aeruginosa influences the relative proportions of both other bacteria [21]. Conversely, P. protegens did not assist K. pneumoniae but was found around the outside of K. pneumoniae and these two strains were also more mixed at 48 h than monospecies colonies of each species. In strains of E. coli engineered to have equal growth rates but different cell shapes, small cells were found to reside on top of the colonies and larger cells were below, at the agar surface, where they maintained access to nutrients obtained from an agar surface [25]. K. pneumoniae cells are larger than Pseudomonas which may have allowed P. protegens to overgrow it and prevent K. pneumoniae from expanding further. In the third case, P. aeruginosa was also found around the outside of P. protegens, but these species were not mixed. This indicates that P. protegens could not take advantage of P. aeruginosa’s motility and these species mutually exclude each other in space. Based on the observations here, future developments of this experimental system could also explore the three-dimensional distribution of the strains. For example, Liu et al. (2017) showed that species in community biofilms organised themselves spatially to optimise fitness that this spatial organisation was facilitated by the other community members [20].
P. aeruginosa twitching motility is important for interactions
Twitching motility by P. aeruginosa has been best studied in interstitial biofilms where cells are sandwiched between an agar surface and a glass coverslip [41,44,48,52]. The colonies studied here are qualitatively similar, although the expanding front does not form as intricate lattices. Without a functional TFP, the SCV could only colonize about 20% of the area of the wild-type. When paired with wild-type P. aeruginosa or P. protegens, the SCV was outcompeted for space, indicating that TFP motility is a competitive trait. It also appears to be a commensal trait as the area colonized by K. pneumoniae was not increased when paired with the P. aeruginosa SCV, whereas it was increased three-fold when paired with TFP motile, wild-type P. aeruginosa. TFP motility can also be abrogated by deleting the gene for pilin subunits, pilA [52]. It will be interesting to contrast our observations of a hyper-piliated pilT mutant with a non-piliated pilA mutant. In flow-cell biofilms, a pilA mutant of P. aeruginosa was less competitive with Agrobacterium tumefaciens [53], but conversely, outcompeted Staphylococcus aureus [54]. Motility, as a competitive trait, has been suggested to allow strains better access nutrients, to cover other organisms or to disrupt their biofilms [55], which is supported by our results. Even though P. aeruginosa was able to cover K. pneumoniae with and without a functional TFP, only the motile wild-type P. aeruginosa had a commensal relationship with K. pneumoniae, indicating that motility can also be a commensal trait.
K. pneumoniae secreted matrix is important for interactions
Self-secreted extracellular matrix is a hallmark of biofilms that influences the spatial positioning and interactions between cells within a biofilm [22]. Matrix secretion allows producing cells to better access nutrients in colonies of P. fluorescens [56] and for simulated cells under flow conditions [57], by excluding other cells. In flow-cell biofilms, the K. pneumoniae NMV outcompeted its isogenic wild-type strain, but was less fit when grown with P. aeruginosa and P. protegens [32]. In colony biofilms, the K. pneumoniae NMV colonized the same total area as the wild-type strain when alone, but did not mix when the two were co-cultured. Furthermore, the NMV did not colonize more area when paired with P. aeruginosa and was outcompeted by P. protegens, resulting in less area covered compared to the NMV when cultured alone. This indicates that the biofilm matrix normally produced by K. pneumoniae KP-1 improves interspecies, but not [58] intraspecies, competition. The mutual exclusion observed between the wild-type and NMV also indicates that the NMV cannot take advantage of the wild-type, similarly to exclusion by B. subtilis [59] and V. cholerae [60]. When co-cultured, Pantoea agglomerans and B. subtilis form colonies with structural properties not observed in either single species alone, even with a B. subtilis mutant that does not produce EPS, indicating that it could share the exopolysaccharide being produced by P. agglomerans [58]. Our results indicate that the K. pneumoniae NMV can not similarly make use of Pseudomonas matrix components.
Agar concentration changes inter-strain interaction outcomes
It is well understood that agar concentration influences bacterial motility [61]. It also affects the ability of biofilms to extract nutrients as it determines the osmotic pressure of an environment [27]. Here we showed that lowering the agar concentration from 1.5% to 0.6% increased the area colonized by the TFP motility deficient P. aeruginosa SCV, though it was still less than the wild-type. However, in co-culture it colonized as much area as the wild-type and was not encircled by the parental wild-type (Table 2). Conversely, the K. pneumoniae NMV was less effective at colonization compared to the wild-type on 0.6% but not 1.5% agar, but was still able to compete with the wild-type when co-cultured in both situations. For the P. aeruginosa SCV, lowering the agar concentration likely enabled flagella-based motility that could partially compensate for the defect in twitching motility. Motility is important for colonization of roots [62], the gastrointestinal tract of Zebra fish [63] and for the persistence of uropathogenic E. coli [64]. Here, we showed that motility influenced intraspecies competition but the outcome depended on environmental conditions. Previously, we observed that the P. aeruginosa SCV completely outcompeted wild-type P. aeruginosa in flow cell biofilms [32]. The increased attachment provided by hyper-piliation of the SCV [44,65] likely provides this benefit in flow cells, whereas the lack of motility was a detriment in colonies. This highlights the differences in environmental pressures between the culturing methods. Hyper-piliation also leads to aggregation [66], which may explain how the P. aeruginosa wild-type and SCV separated into sectors with straight boundaries on 0.6% agar. This appeared to be similar to the differently tagged strains of K. pneumoniae, indicating that this community morphology may not exclusively be caused by matrix secretion.
For K. pneumoniae (which is non-motile), lowering the agar concentration allowed the wild-type to colonize more area, which was similar to how a rugose strain of V. cholerae that hypersecretes extracellular matrix (ECM)formed larger colonies on lower concentrations of agar [27]. In V. cholerae, it was demonstrated that matrix secretion generates an osmotic pressure gradient between the agar and the colony, allowing the colony to expand by physical swelling and also drawing more nutrients out of the agar. It is likely that this is a general mechanism attributable to the biofilm matrix. In this context, the wild-type K. pneumoniae would be similar to the rugose V. cholerae, producing larger amounts of ECM, while the K. pneumoniae NMV and wild-type V. cholerae are analogous in their relatively lower amount of ECM production. The community morphology of dual strain colonies of ECM secretors and non-secretors differed between the two species. In V. cholerae, the hyper-secretor was encircled by the non-secretor at both 1.5 and 0.6% agar, but colonized far more area at 0.6% as the non-secretor was pushed to the outside of the colony. Conversely, the K. pneumoniae NMV was not displaced and even prevented spreading by wild-type K. pneumoniae (Fig. 5, D). Similar to V. cholerae, the K. pneumoniae NMV did not benefit from the matrix secreted by wild-type K. pneumoniae as its area was not increased in co-culture. In B. subtilis, it was also observed that ECM-producing cells outcompete non-secretors [28], and to a higher degree when the humidity is higher (which is similar to lower agar concentration). Additionally, osmotic pressure generated by the ECM enhances colony spreading in this species [67]. Thus, matrix production appears to be a general strategy of bacteria that increases competitiveness in colonies.
Conclusions
Interactions between bacteria are key for determining the composition and function of communities. Here we used colonies to investigate intra- and inter-species interactions. Using the members of our three species model community, we showed that they differ in how they interact with members of their own species due to their physiological traits: TFP motility in P. aeruginosa allowed populations to mix whereas ECM in K. pneumoniae caused straight borders between population sectors. Using mutants deficient in these traits, we showed that their impact depended on the agar concentration. These physiological traits were also important when interspecies interactions were examined. The motility of P. aeruginosa enabled it to outcompete P. protegens and to facilitate increased colonization area by K. pneumoniae. Importantly, the spatial distribution of species, observed for dual-species colonies did not resemble any patterns previously seen for experimental or simulated monospecies and dual-strain colonies. These experiments show that interspecies interactions differ substantially from intraspecies interactions and that co-culture colonies are a powerful way to investigate how bacterial physiology determines these interactions.
CRediT authorship contribution statement
Sean C. Booth: Conceptualization, Methodology, Software, Formal analysis, Analysis, Investigation, Writing - original draft, Writing, Visualization. Scott A. Rice: Conceptualization, Methodology, Resources, Writing - original draft, Writing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank Sujatha Subramoni for helpful advice regarding the three species community, Talgat Sailov for microscopy assistance and Diane McDougald for reading the manuscript and providing helpful comments. We would also like to thank the Singapore Centre for Environmental Life Sciences Engineering (SCELSE), whose research is supported by the National Research Foundation Singapore, Ministry of Education, Nanyang Technological University and National University of Singapore, under its Research Centre of Excellence Programme.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2020.100035.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mah T.F.C., O’Toole G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 2.Toole G.O., Kaplan H.B., Kolter R. 2000. Biofilm formation as microbial development; pp. 49–79. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the Natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 4.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008 doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlamakis H., Chai Y., Beauregard P. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013 doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fazli M., Almblad H., Rybtke M.L. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ Microbiol. 2014 doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 7.Røder H.L., Sørensen S.J., Burmølle M. Studying bacterial multispecies biofilms: where to start? Trends Microbiol. 2016 doi: 10.1016/j.tim.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Elias S., Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- 9.Rendueles O., Ghigo J.-M. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol Rev. 2012;36:972–989. doi: 10.1111/j.1574-6976.2012.00328.x. [DOI] [PubMed] [Google Scholar]
- 10.Ghoul M., Mitri S. The ecology and evolution of microbial competition. Trends Microbiol. 2016;24:833–845. doi: 10.1016/j.tim.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 11.Morris B.E.L., Henneberger R., Huber H., Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 12.Cornforth D.M., Foster K.R. Antibiotics and the art of bacterial war. Proc Natl Acad Sci U S A. 2015;112 doi: 10.1073/pnas.1513608112. 10827–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A., Inagaki S., Sigurdson W., Kuramitsu H.K. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol. 2005;20:39–42. doi: 10.1111/j.1399-302X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamada M., Ikegami A., Kuramitsu H.K. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett. 2005;250:271–277. doi: 10.1016/j.femsle.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez D., Sabnis A., Foster K.R., Mavridou D.A.I. Costs and benefits of provocation in bacterial warfare. Proc Natl Acad Sci. 2018;115:7593–7598. doi: 10.1073/pnas.1801028115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragoš A., Kiesewalter H., Martin M. Division of labor during biofilm matrix production. Curr Biol. 2018;28:1903–1913. doi: 10.1016/J.CUB.2018.04.046. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen S.K., Rainey P.B., Haagensen J.A.J., Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 18.Haagensen J.A.J., Hansen S.K., Christensen B.B. Development of spatial distribution patterns by biofilm cells. Appl Environ Microbiol. 2015;81:6120–6128. doi: 10.1128/AEM.01614-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen B.B., Haagensen J.A.J., Heydorn A., Molin S. Metabolic commensalism and competition in a two-species microbial consortium. Appl Environ Microbiol. 2002;68:2495–2502. doi: 10.1128/aem.68.5.2495-2502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W., Russel J., Røder H.L. Low-abundant species facilitates specific spatial organization that promotes multispecies biofilm formation. Environ Microbiol. 2017;19:2893–2905. doi: 10.1111/1462-2920.13816. [DOI] [PubMed] [Google Scholar]
- 21.Lee K.W.K., Periasamy S., Mukherjee M. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadell C.D., Drescher K., Foster K.R. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 23.Momeni B., Shou W., Brileya K.A., Fields M.W. Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife. 2013;2013 doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller M.J.I., Neugeboren B.I., Nelson D.R., Murray A.W. Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci U S A. 2014;111:1037–1042. doi: 10.1073/pnas.1313285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith W.P.J., Davit Y., Osborne J.M. Cell morphology drives spatial patterning in microbial communities. Proc Natl Acad Sci U S A. 2017;114:E280–E286. doi: 10.1073/pnas.1613007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldschmidt F., Regoes R.R., Johnson D.R. Successive range expansion promotes diversity and accelerates evolution in spatially structured microbial populations. ISME J. 2017;11:2112–2123. doi: 10.1038/ismej.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J., Nadell C.D., Stone H.A. Extracellular-matrix-mediated osmotic pressure drives Vibrio cholerae biofilm expansion and cheater exclusion. Nat Commun. 2017;8:327. doi: 10.1038/s41467-017-00401-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Gestel J., Weissing F.J., Kuipers O.P., Kovács Á.T. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 2014;8:2069–2079. doi: 10.1038/ismej.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frost I., Smith W.P.J., Mitri S. Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 2018;12:1582–1593. doi: 10.1038/s41396-018-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramette A., Frapolli M., Saux M.F., Le Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol. 2011;34:180–188. doi: 10.1016/j.syapm.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Lim C.K., Hassan K.A., Penesyan A. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ Microbiol. 2013;15:702–715. doi: 10.1111/j.1462-2920.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- 32.Kelvin Lee K.W., Hoong Yam J.K., Mukherjee M. Interspecific diversity reduces and functionally substitutes for intraspecific variation in biofilm communities. ISME J. 2016;10:846–857. doi: 10.1038/ismej.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Periasamy S., Nair H.A.S., Lee K.W.K. Pseudomonas aeruginosa PAO1 exopolysaccharides are important for mixed species biofilm community development and stress tolerance. Front Microbiol. 2015;6:851. doi: 10.3389/fmicb.2015.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng T., Thorn K., Schroeder T. A BaSiC tool for background and shading correction of optical microscopy images. Nat Commun. 2017;8:14836. doi: 10.1038/ncomms14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preibisch S., Saalfeld S., Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barthelme S. 2018. Imager: image processing library based on “CImg”. [Google Scholar]
- 38.Landini G., Randell D.A., Fouad S., Galton A. Automatic thresholding from the gradients of region boundaries. J Microsc. 2017;265:185–195. doi: 10.1111/jmi.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudge T.J., Federici F., Steiner P.J. Cell polarity-driven instability generates self-organized, fractal patterning of cell layers. ACS Synth Biol. 2013;2:705–714. doi: 10.1021/sb400030p. [DOI] [PubMed] [Google Scholar]
- 40.Bérubé D., Jébrak M. High precision boundary fractal analysis for shape characterization. Comput Geosci. 1999 doi: 10.1016/S0098-3004(99)00067-9. [DOI] [Google Scholar]
- 41.Turnbull L., Toyofuku M., Hynen A.L. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun. 2016;7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klausen M., Heydorn A., Ragas P. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 43.Whitchurch C.B., Hobbs M., Livingston S.P. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-V. [DOI] [PubMed] [Google Scholar]
- 44.Whitchurch C.B., Mattick J.S. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol Microbiol. 1994;13:1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 45.Hallatschek O., Hersen P., Ramanathan S., Nelson D.R. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci U S A. 2007;104 doi: 10.1073/pnas.0710150104. 19926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blanchard A.E., Lu T. Bacterial social interactions drive the emergence of differential spatial colony structures. BMC Syst Biol. 2015;9:59. doi: 10.1186/s12918-015-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitri S., Clarke E., Foster K.R. Resource limitation drives spatial organization in microbial groups. ISME J. 2016;10:1471–1482. doi: 10.1038/ismej.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gloag E.S., Turnbull L., Huang A. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 2013;110:11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim W., Levy S.B., Foster K.R. Rapid radiation in bacteria leads to a division of labour. Nat Commun. 2016;7:10508. doi: 10.1038/ncomms10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murray T.S., Kazmierczak B.I. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol. 2008;190:2700–2708. doi: 10.1128/JB.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiong L., Cao Y., Cooper R. Flower-like patterns in multi-species bacterial colonies. Elife. 2020 doi: 10.7554/eLife.48885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semmler A.B.T., Whitchurch C.B., Mattick J.S. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 53.An D., Danhorn T., Fuqua C., Parsek M.R. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci. 2006;103:3828–3833. doi: 10.1073/pnas.0511323103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang L., Liu Y., Markussen T. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol. 2011;62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- 55.Rendueles O., Ghigo J.-M.J.-M. Mechanisms of competition in biofilm communities. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0009-2014. [DOI] [PubMed] [Google Scholar]
- 56.Kim W., Racimo F., Schluter J. Importance of positioning for microbial evolution. Proc Natl Acad Sci U S A. 2014;111:E1639–E1647. doi: 10.1073/pnas.1323632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jayathilake P.G., Jana S., Rushton S. Extracellular polymeric substance production and aggregated bacteria colonization influence the competition of microbes in biofilms. Front Microbiol. 2017;8:1865. doi: 10.3389/fmicb.2017.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yannarell S.M., Grandchamp G.M., Chen S.Y. A dual-species biofilm with emergent mechanical and protective properties. J Bacteriol. 2019 doi: 10.1128/JB.00670-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M., Dragoš A., Hölscher T. De novo evolved interference competition promotes the spread of biofilm defectors. Nat Commun. 2017;8:15127. doi: 10.1038/ncomms15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nadell C.D., Drescher K., Wingreen N.S., Bassler B.L. Extracellular matrix structure governs invasion resistance in bacterial biofilms. ISME J. 2015;9:1700–1709. doi: 10.1038/ismej.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greenberg E.P., Canale-Parola E. Motility of flagellated bacteria in viscous environments. J Bacteriol. 1977;132:356–358. doi: 10.1128/jb.132.1.356-358.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pliego C., de Weert S., Lamers G. Two similar enhanced root-colonizing Pseudomonas strains differ largely in their colonization strategies of avocado roots and Rosellinia necatrix hyphae. Environ Microbiol. 2008;10:3295–3304. doi: 10.1111/j.1462-2920.2008.01721.x. [DOI] [PubMed] [Google Scholar]
- 63.Stephens W.Z., Wiles T.J., Martinez E.S. Identification of population bottlenecks and colonization factors during assembly of bacterial communities within the zebrafish intestine. mBio. 2015;6 doi: 10.1128/mBio.01163-15. e01163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright K.J., Seed P.C., Hultgren S.J. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun. 2005;73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiang P., Burrows L.L. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J Bacteriol. 2003;185:2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Déziel E., Comeau Y., Villemur R. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol. 2001;183:1195–1204. doi: 10.1128/JB.183.4.1195-1204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seminara A., Angelini T.E., Wilking J.N. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci. 2012 doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2