Abstract
The contribution of the biofilm extracellular polymeric substance (EPS) matrix to reduced antimicrobial susceptibility in biofilms is widely recognised. As such, the direct targeting of the EPS matrix is a promising biofilm control strategy that allows for the disruption of the matrix, thereby allowing a subsequent increase in susceptibility to antimicrobial agents. To this end, surface-functionalized nanoparticles (NPs) have received considerable attention. However, the fundamental understanding of the interactions occurring between engineered NPs and the biofilm EPS matrix has not yet been fully elucidated. An insight into the underlying mechanisms involved when a NP interacts with the EPS matrix will aid in the design of more efficient NPs for biofilm control. Here we demonstrate the use of highly specific fluorescent probes in confocal laser scanning microscopy (CLSM) to illustrate the distribution of EPS macromolecules within the biofilm. Thereafter, a three-dimensional (3D) colocalization analysis was used to assess the affinity of differently functionalized silica NPs (SiNPs) and EPS macromolecules from Pseudomonas fluorescens biofilms. Results show that both the charge and surface functional groups of SiNPs dramatically affected the extent to which SiNPs interacted and localized with EPS macromolecules, including proteins, polysaccharides and DNA. Hypotheses are also presented about the possible physicochemical interactions which may be dominant in EPS matrix-NP interactions. This research not only develops an innovative CLSM-based methodology for elucidating biofilm-nanoparticle interactions but also provides a platform on which to build more efficient NP systems for biofilm control.
Keywords: Bacterial biofilm, Nanoparticles, Pseudomonas fluorescens, EPS matrix, Physicochemical interactions, Colocalization
Highlights
-
•
Study of NP-EPS interaction via novel combinations of specific probes and CLSM.
-
•
Colocalization analysis revealed surface dependent high affinity NP-EPS interactions.
-
•
NP surface functionalization strongly affects their interaction with the EPS matrix.
-
•
NPs show differential binding to proteins, polysaccharides and eDNA in the EPS.
Introduction
Bacterial biofilms are highly-structured multicellular accumulations of microbes which are enclosed in a self-produced extracellular polymeric substance (EPS) matrix composed of polysaccharides, proteins/enzymes, lipids and exogenous DNA (eDNA), as well as lysed cell debris [1,2]. The EPS matrix allows for the colonization of both biotic and abiotic surfaces and also contributes to the persistence of bacteria at various interfaces by playing a crucial role in the development and survival of the bacterial population [3]. It does this by serving as a scaffold for growth, by aiding in the protection of cells from environmental stresses, by serving as a nutrient reservoir and also by affecting the diffusion of antibacterial agents throughout the biofilm [4]. In addition, biofilms typically grow heterogeneously within complex environments which further contributes to their unique and intricate three-dimensional (3D) architecture and extensive physicochemical gradients [5].
Biofilm control and eradication is of great importance in the processing industries as their accumulation and persistence can lead to significant economic losses stemming from the induction of corrosion in piping [6], contamination of raw material leading to a decrease in product quality [7] or increasing energy usage [8]. In addition to this, bacterial biofilms also have a significant impact in human health and wellbeing [9]. In the food, agricultural and medical sectors, bacterial contamination in the form of biofilms calls for the development of more intensive and novel infection treatment and disinfection methods to control disease outbreaks and reduce food wastage and contamination [10].
Traditionally, biofilm removal strategies have focused on directly targeting the bacterial cells themselves. However, the protection offered by the EPS matrix is believed to lead to a decrease in the effectiveness of antibiofilm agents [11]. Although many bacterial cell-targeting strategies have been employed for biofilm removal, attention is now shifting from targeting individual bacterial cells within the biofilm, to methods for the degradation of the EPS matrix itself [12]. Therefore, the development of novel approaches for the control of unwanted biofilms must include the analysis of the composition of the EPS matrix in relation to the interactions which occur between the EPS matrix and external antibiofilm agents.
Engineered nanoparticles (NPs) have shown great promise in combating and eradicating bacterial biofilms [4]; however, there is currently a lack of fundamental understanding of the mechanisms associated with and also the interactions between the EPS matrix and engineered NPs. The biological and physicochemical interactions occurring between NPs and bacterial biofilms are highly complex and dependent on a myriad of factors including the inherent characteristics of the NPs (e.g., size, charge, surface functionalization), EPS composition and abundance (e.g., biomolecules present, maturity/age, density, bacterial species), and the environmental conditions (e.g., temperature, pH, flow conditions, ionic strength) [4]. A fundamental understanding of the interactions occurring in biofilm-NP systems is of great importance in the use of the nanomaterials for future applications in either the study of, or eradication of persistent biofilms. Currently, few studies have evaluated the fundamental interactions which occur in the EPS matrix when bacterial biofilms are exposed to differentially engineered NPs.
Thus far, the in-situ analysis of the localization and possible high-affinity interactions between NPs and EPS matrix macromolecules with NPs has remained a challenge. Colocalization CLSM studies have frequently been used to determine whether two molecules associated with the same structure or with each other [13]. For example, determination of the association of a protein with a specific cellular organelle [14], or whether two different proteins associate with the same structure [15]. The repeated observation of the association and nanometer proximity of two fluorescent probes increases the certainty that there is a biological or physicochemical interaction occurring between them. To this end, many fluorescent probes have been used for colocalization analysis [[16], [17], [18]]. Lectins are proteins which are highly specific for carbohydrates binding without altering their structure. Their use in high resolution imaging represents an opportunity for non-destructive in-situ analyses and thus have been widely used for glycoconjugate biofilm investigations when used in combination with other fluorescent probes [[19], [20], [21]]. Therefore, the combination of a fluorescence lectin-binding analysis [22] and colocalization analysis in CLSM could aid in evaluating the localization and affinity of differently functionalized NPs with fluorescently labelled EPS matrix macromolecules.
In this work we describe a highly sensitive confocal laser scanning microscopy (CLSM) imaging system developed to quantitatively identify the colocalization of various engineered fluorescent silica NPs (SiNPs) with specific macromolecules within the EPS matrix of P. fluorescens WCS365 biofilms. Using this system, we were able to accurately assess the accumulation, localization and binding affinities of these SiNPs based on variations in their surface charge and surface functionalization as well as hypothesize the possible physicochemical interactions which may be dominant in EPS matrix-NP interactions.
Results
Nanoparticle characterization
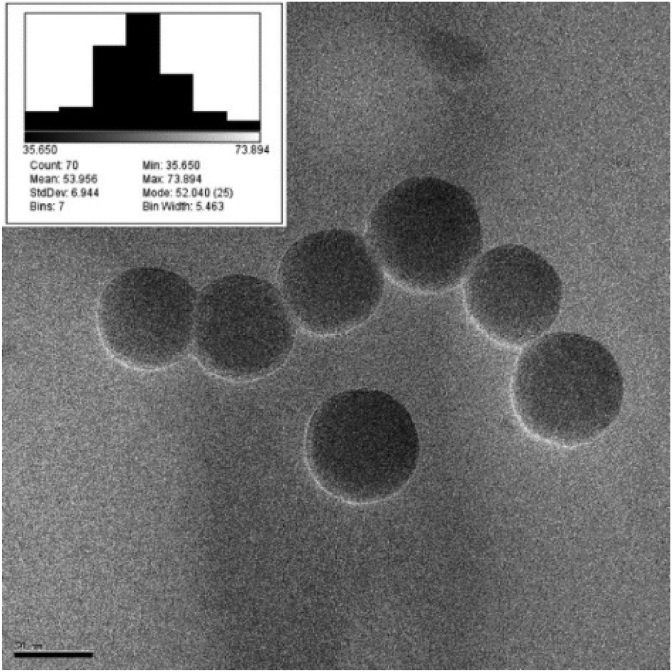
Characterization was carried out on four groups of fluorescent-labelled SiNPs (Table 1), that were synthesized and subsequently functionalized through alkyl-silane grafting. The as-synthesized SiNPs have a diameter of 54.0 ± 6.9 nm, as determined by TEM (Fig. 1). In Table 1, the size distributions and surface charge of SiNPs are reported, determined by TEM and DLS analysis and Z-potential measurements, respectively. The functionalization of SiNPs does not affect the size of the NPs core, as shown by the size distribution extrapolated from TEM images of the functionalized SiNPs (Fig. S1). However, the effect of silane grafting can be observed in both hydrodynamic ratio and surface charge (Table 1). All the functionalized NPs show an increase in size, determined by DLS, consistent with silane grafting onto the surface. Furthermore, all the NPs present a low polydispersity index (PDI), which indicates high stability of the aqueous dispersion. Bare-SiNPs show the characteristic negative charge of SiNPs, whereas the positive charge measured for Amine-SiNPs is due to the presence of the –NH2 group. Epoxy-SiNPs exhibit a negative Z-potential, but higher compared to the non-functionalized NPs; this can be ascribed to the hydrophobic character of the epoxy-group. On the other hand, Hydroxy-SiNPs show a higher negative surface charge, consistent with a greater stability of the aqueous dispersion given by the 3-(2,3-dihydroxypropoxy) propyl]-trimethoxysilane ligand.
Table 1.
Nanoparticle characterisation. Size determined by TEM and DLS, and surface charge measured by Zeta potential.
| Sample | Size by TEM (nm) | Size by DLS (nm) | PDI | Zeta potential (mV) | |
|---|---|---|---|---|---|
| Bare |  |
54.0 (±6.9) | 79.7 (±0.7) | 0.025 (±0.02) | −37.3 (±2.4) |
| Amine | 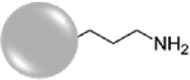 |
57.3 (±4.3) | 105.5 (±4.4) | 0.169 (±0.04) | +29.3 (±0.4) |
| Epoxy | 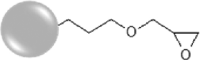 |
58.7 (±5.9) | 83.6 (±3.7) | 0.071 (±0.03) | −27.7 (±0.6) |
| Hydroxy |  |
57.8 (±7.4) | 85.1 (±3.7) | 0.073 (±0.03) | −38.9 (±3.3) |
Fig. 1.
Representative TEM micrograph for as-synthesized Bare-SiNPs, with particle size distribution (nm) reported in the inset. Scale bar is 50 nm.
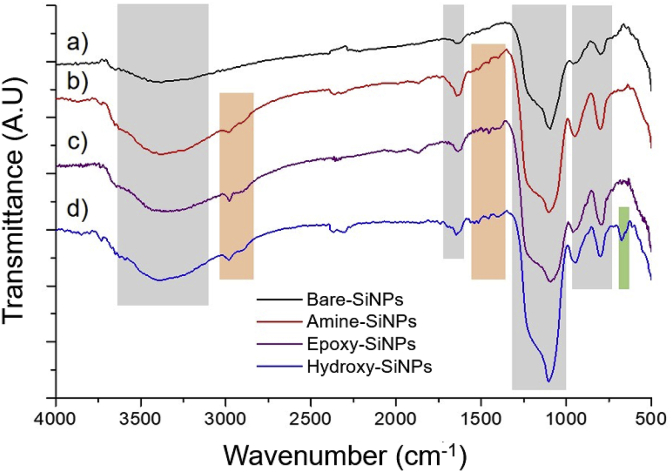
FT-IR spectra were collected on non-dye loaded SiNPs, in order to eliminate the contribution from the internal fluorescent label (Fig. 2). All the spectra show strong characteristic stretching bands for –OH (3300 - 3700 cm−1), Si–O–Si (~1100 cm−1) and Si–OH (~850 cm−1), as well as Si–OH bending (~950 cm−1); the peak at 1634 cm−1 is attributed to vibrations of water molecules adsorbed on the NPs surface [23] For the Amine-, Epoxy- and Hydroxy-functionalized SiNPs we can observe diagnostic bands for C–H stretching (2800 - 2900 cm−1) and bending (1350 - 1500 cm−1), ascribable to the alkyl chains of grafted silanes [24] It must be pointed out that for the Amine-SiNPs a contribution can be considered of the –NH2 groups vibrations to the band at ~1500 cm−1. On the Hydroxy-functionalized SiNPs we can observe a peak at ~670 cm−1, which can be assigned to C–OH vibrations [25] The same band is not present in the Epoxy-SiNPs, and this is consistent with a successful epoxy-ring opening reaction.
Fig. 2.
FT-IR spectra for, respectively, as-synthesized Bare-SiNPs (a), black) and Amine- (b), red), Epoxy- (c), purple) and Hydroxy- (d), blue) functionalized SiNPs. Coloured highlights refer to silica-related bands (grey), alkyl-silane bands (orange) and Hydroxy-SiNPs diagnostic bands (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Localization and distribution of SiNPs within the EPS matrix
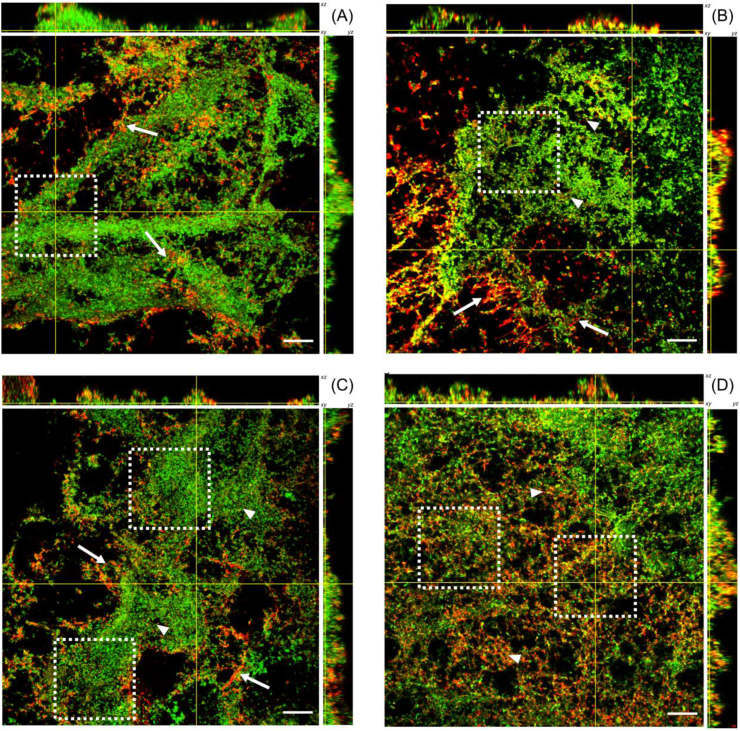
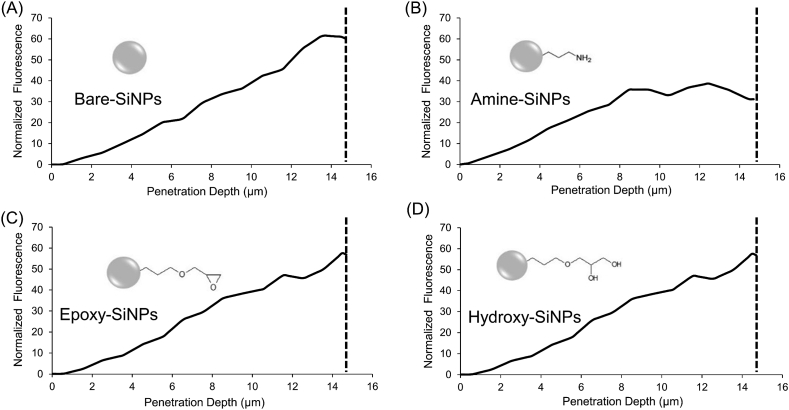
The patterns of localization and distribution of each NPs within the biofilm are unique and vary dramatically with changes in charge and surface-functionalization [26]. Bare-SiNPs are shown to be heterogeneously distributed throughout the biofilm microcolonies (Fig. 3A) and found either in small clustered aggregates on the periphery or within biofilm microcolonies (Fig. 3A, white arrows). Penetration profiles reveal the ability of the Bare-SiNPs to permeate the entire depth of the biofilm (Fig. 4A). The positively charged Amine-SiNPs show an enhanced binding to string-like projections on the periphery of the biofilm microcolonies (Fig. 3B, white arrows), as well as an accumulation on the exterior of the microcolonies (Fig. 3B, white arrowheads). Penetration profiles (Fig. 4B) show two distinct peaks at 8.3 and 12 μm respectively, with a decrease in signal as it approaches the bottom layer. This indicates both the full penetration of the biofilm but also the aggregation towards to bottom periphery of the biofilm. The Epoxy-SiNPs fully penetrate into the bottom layer of the biofilm microcolonies (Fig. 4C) and are also well-distributed throughout the biofilm but accumulate both in heterogenous clusters within the microcolonies (Fig. 3C, white arrowheads) and aggregates at the biofilm periphery with a string-like organization (Fig. 3C, white arrows). The Hydroxy-SiNPs show a more even distribution and penetration of the biofilm (Fig. 4D) and also show preference to the interior of the biofilm microcolonies (Fig. 3D, white arrowheads) with no significant accumulation on the microcolony periphery. For clarity, images with separate channels are included in the supporting information (Fig. S2-S5). The biofilm was shown to entrap between 20 and 25% of the SiNPs from solution (Fig. S6) meaning that the relative percentages of each NPs inside the multiple biofilms could be comparable.
Fig. 3.
CLSM images of 72h P. fluorescens WCS365 biofilms after exposure to (A) Bare-SiNPs, (B) Amine-SiNPs, (C) Epoxy-SiNPs and (D) Hydroxy-SiNPs. Biofilm cells were stained with Syto9 (green) and SiNPs were labelled with RITC (red). The central images show the horizontal (xy) section 0.5 μm above the substrate surface. Upper and side panels represent z-stack images of the xz and yz planes, respectively. The yellow lines indicate the position of xz and yz planes on the xy section images. White arrows and arrowheads indicate the distribution patterns of SiNPs and white dashed squares represents the centre of the microcolonies. Representative images from three independent experiments are displayed. Scale bar represents 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
Penetration profile of P. fluorescens WCS365 biofilms after exposure to (A) Bare-SiNPs, (B) Amine-SiNPs, (C) Epoxy-SiNPs and (D) Hydroxy-SiNPs. The x-axis shows the depth of penetration of biofilms, where 0 μm represents the top layer of biofilm and the dashed vertical line (at approximate average thickness of the 14.7 μm) represents the bottom layer of biofilm. The y-axis is the normalized fluorescence intensity of red channel.
EPS matrix macromolecule spatial organization within the biofilm
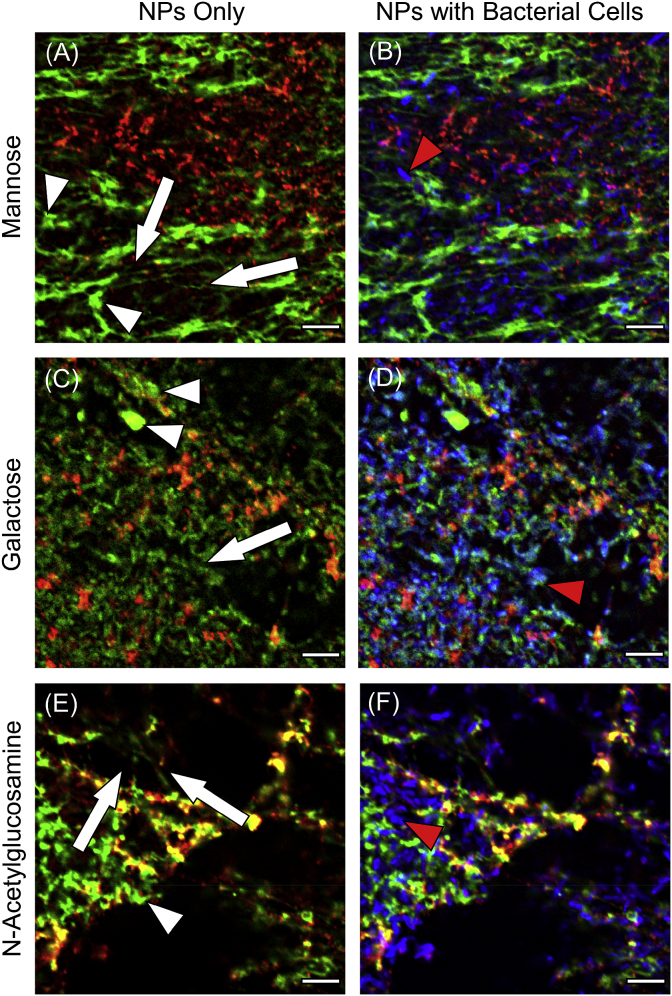
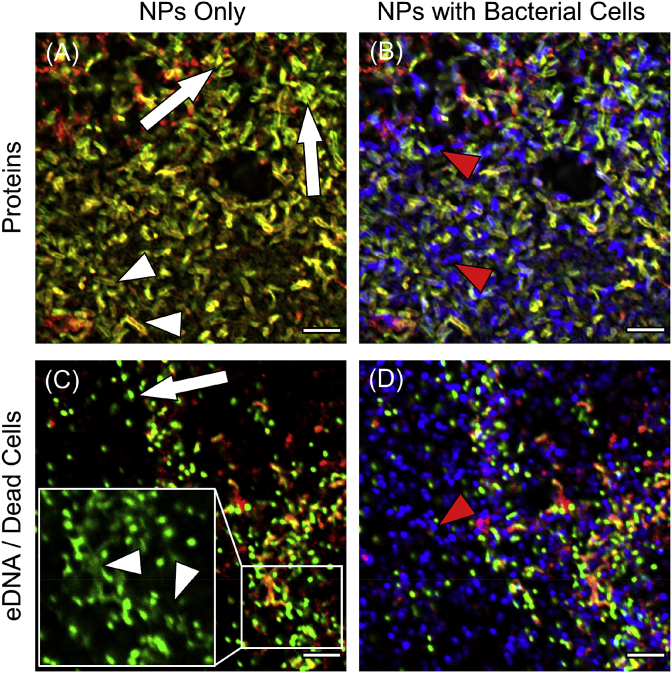
Differential fluorescent staining and CLSM were used to assess the distribution and architecture of EPS macromolecules within the biofilm in relation to bacterial cells and Bare-SiNPs; representative images are shown in Fig. 5, Fig. 6. Mannose-containing polysaccharides stained by the ConA lectin predominantly form a network of string-like projections (Fig. 5A, white arrows) which are more distributed through the entire biofilm and independent of the microcolonies and bacterial cells. Mannose is also found in small clusters at the base of the string-like projections (Fig. 5A, white arrowheads). Galactose-containing polysaccharides stained by the RCA lectin are distributed both uniformly (Fig. 5C, white arrows) and in aggregates (Fig. 5C, white arrowheads) and are also found in close proximity to bacterial cells within the microcolonies (Fig. 5D, red arrowhead). N-Acetylglucosamine (NAG)-containing polysaccharides stained by the WGA lectin are distributed in both string-like projections protruding from the microcolonies (Fig. 5E, white arrows) as well as in clusters between bacterial cells (Fig. 5F, red arrowhead). Proteins within the EPS matrix which are stained by Sypro® Orange show a variety of morphologies including rounded clusters (Fig. 6A, white arrows) and surrounding the membranes or dead or dying cells (Fig. 6, white arrowheads). Sypro® Orange was unable to stain proteins on the surface of live cells and thus did not produce any significant fluorescent signal on the cell surface (Fig. 6B, red arrowhead). Sytox™ Green staining of the biofilm shows both the presence of both eDNA (Fig. 6C, white arrowheads) and genomic DNA within dead/dying cells (Fig. 6C, white arrows) which are distributed into the spaces between live cells (Fig. 6D). SiNPs within the biofilms are shown as cloud-like distributions or in smaller aggregates and can be found in close proximity to polysaccharides, proteins or eDNA but could possibly also be found attached to another unstained component of the biofilm.
Fig. 5.
Representative CLSM images showing the localization and distribution of EPS macromolecules stained with lectins (green) specific to (A) mannose, (C) galactose and (E) NAG, relative to both bacterial cells stained with DAPI (blue), and SiNPs (red). Image channels showing live cells corresponding to (A), (C) and (E) are shown in (B), (D) and (F) respectively. White arrows and arrowheads indicate distribution patterns of polysaccharides while red arrowheads show the distribution of bacterial cells. Each image shows the horizontal (xy) section taken between 1.5 and 2.0 μm above the substrate surface within the centre of a microcolony. Scale bar represents 5 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
Representative CLSM images showing the localization and distribution of EPS macromolecules (green) stained with Sypro® Orange for proteins (A) and Sytox™ Green for eDNA (C), relative to both bacterial cells stained with DAPI (blue), and SiNPs (red). Image channels showing live cells corresponding to (A) and (C) are shown in (B) and (D) respectively. White arrows and arrowheads indicate distribution patterns of proteins and eDNA while red arrowheads show the distribution of bacterial cells. Inset in (C): DNA found as either eDNA (white arrowheads) or genomic DNA within dead/dying cells. Each image shows the horizontal (xy) section taken between 1.5 and 2.0 μm above the substrate surface within the centre of a microcolony. Scale bar represents 5 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Colocalization of SiNPs with EPS matrix macromolecules
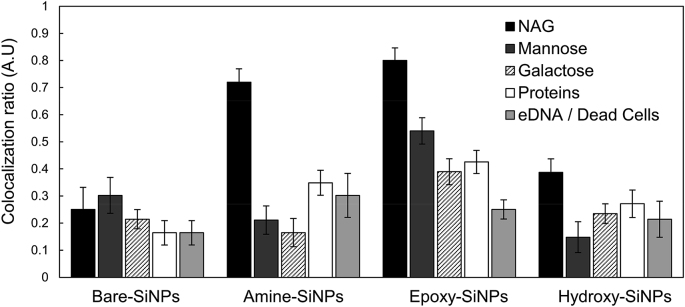
Under semi-quantitative and qualitative instances, the colocalization of two probes in separate fluorescent channels (green and red) can be subjectively (and incorrectly) identified by the appearance of a yellow colour when images are superimposed or “merged” [27]. Therefore a systematic colocalization analysis using image processing that compares the relative position and intensity of each pixel would give the most reliable and reproducible results [13]. The terminology for describing colocalization data considers weak colocalization ranging from 0 to 0.49, moderate colocalization as 0.5–0.79, and strong colocalization as 0.8–1.0 [28]. As shown in Fig. 7, Epoxy-SINPs display the highest overall colocalization with EPS macromolecules compared to the other SiNPs. The highest colocalization ratio was found in interactions between NAG-containing polysaccharides with Epoxy-SiNPs; the only binding pair to show a strong colocalization ratio. Moderate colocalization ratios were found between both Mannose-containing polysaccharides with Epoxy-SiNPs as well as between NAG-containing polysaccharides with Amine-SiNPs. All other SiNPs and probe combinations shows weak colocalization. Colocalization ratios are particularly low for proteins and polysaccharides interacting with Bare-SiNPs, Galactose-containing polysaccharides with Amine-SiNPs and Mannose-containing polysaccharides with Hydroxy-SiNPs.[28]
Fig. 7.
Colocalization analysis of functionalized SiNPs and differentially-labelled EPS components within the biofilm matrix. Fiji’s JaCoP colocalization analysis (Mander’s correlation coefficients) was conducted to examine pixel to pixel correlation in separate channels over space in several z-stack CLSM images. Data is representative of the mean ± standard deviation of three separate experiments, each of which contained at least 3 z-stack CLSM images. A one-way ANOVA, Tukey’s pairwise comparison, (P < 0.05) was used to assess statistical differences.
Protein binding affinity (in-vitro analysis)
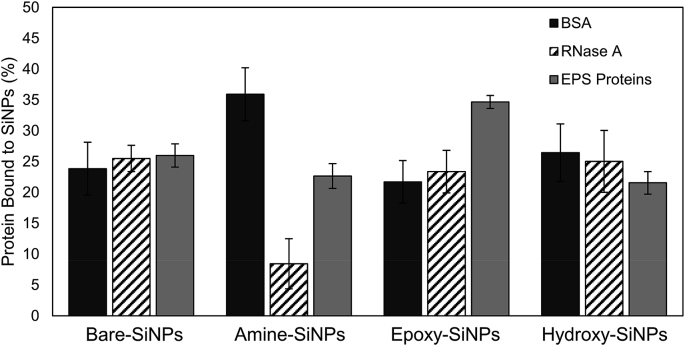
The protein corona formed when biofilms are exposed to extracted EPS proteins in-vitro (Fig. S7) showed that changes in the SiNPs surface chemistry can alter the amount and type of protein which associates with the SiNP surface. To evaluate the level of binding of various proteins to differently functionalized SiNPs and how protein charge and pI affects this, the SiNPs were exposed to either an EPS protein extract or two model proteins, BSA and RNase, each with different surface chemistry and charge (Fig. 8). The in-silico mapping of the electrostatic surface potentials (at physiological pH) of the model proteins demonstrated an overall negative charge for BSA while RNase showed an overall positive charge (Fig. S8). Bare-SiNPs and Hydroxy-SiNPs showed no significant difference in their protein binding affinity ranging between 20 and 25% of the total protein in solution. In terms of positively charged Amine-SiNPs, there was a significant increase in the binding affinity of BSA while RNase showed a decreased binding affinity and the EPS-extract showed an affinity intermediate to the model proteins. Epoxy-SiNPs showed an overall significant increase in the amount of protein bound as compared to the other SiNPs.
Fig. 8.
Effect of surface functional groups on the binding of proteins in solution to SiNPs. Data is representative of the mean ± standard deviation of three separate experiments (n = 6). A one-way ANOVA, Tukey’s pairwise comparison, (P < 0.05) was used to assess statistical differences.
Carbohydrate binding affinity (in-vitro analysis)
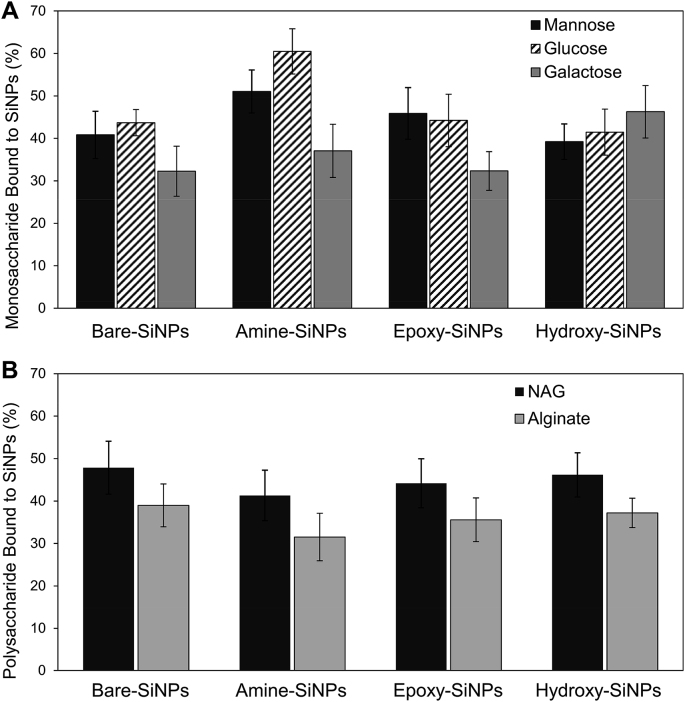
To identify the contribution of specific carbohydrates to the level of colocalization shown, SiNPs were exposed to each mono- or polysaccharide independent of the biofilm. Amine-SiNPs were shown to have the highest overall percentage of monosaccharide molecules bound to their surface while glucose was shown to be the monosaccharide which bound to all SiNPs with the highest proportion and galactose showing the lowest overall affinity (Fig. 9A). Both NAG and alginate polysaccharides showed no significant difference (P < 0.05) in binding preference towards each SiNPs but NAG did have a higher binding affinity as compared to alginate (Fig. 9B).
Fig. 9.
Effect of surface functional groups on the binding of carbohydrates in solution to SiNPs. Graphs show the percentage of A) monosaccharides and B) polysaccharides bound to the surface of the SiNPs after exposure. Data is representative of the mean ± standard deviation of three separate experiments, each of which contained nine replicates. A one-way ANOVA, Tukey’s pairwise comparison, (P < 0.05) was used to assess statistical differences.
Discussion
Distribution of SiNPs within the EPS matrix
Changes in the NP surface chemistry have been shown to affect how they interact with the biofilm as a whole. For instance, Li et al. demonstrated that the control of NP distribution and penetration into bacterial biofilms is determined by variation of surface properties. This research showed the penetration of only positively charged functionalized CdSe–ZnS core–shell NPs (24 nm diameter) into Escherichia coli biofilms [29], with the changes in the distribution of the NPs being governed by their charge and hydrophobicity. In the current study, even though the entrapment of SiNPs by the biofilm did not vary significantly, the distribution patterns and binding affinity of the SiNPs in CLSM imaging varied greatly, further highlighting the need for in-depth high-resolution analyses. Positively charged Amine-SiNPs showed the highest overall entrapment and accumulation in the biofilm (Fig. S6) but all functionalized SiNPs were nevertheless able to fully penetrate the biofilms microcolonies regardless of their overall charge or surface groups present (Fig. 4). Changes in the localization and distribution patterns of SiNPs in the EPS matrix based on their surface chemistry displays a contribution of these groups to the affinity of SiNPs to different macromolecules in the EPS matrix and not their propensity to permeate the biofilm.
Identification and spatial organization of EPS matrix macromolecules
The EPS matrix must be assessed both in terms of its inherent 3D architecture and the role of each molecule within the structure, albeit only qualitatively. If a particular molecule is to be targeted for degradation and subsequent biofilms removal, then molecules involved in the structural rigidity and integrity of the biofilm should be identified and the surface of “smart” NPs should be tailored in order to direct the NP towards its target. To this end, the structure-to-function relationship of many biofilm-associated macromolecules have previously been elucidated [30]. For example, polysaccharides like PEL [31] and PSL [32] have been shown to cross-link and form complexes with other macromolecules for the maintenance and development of the microcolony structure while eDNA has been shown to provide a structural scaffold for biofilm development [33]. Few studies have used differential staining to elucidate the composition of bacterial biofilms. Super-resolution microscopy of Vibrio cholerae biofilms using an in vivo labelling strategy allowed for the visualization of the molecular architecture and composition of these biofilms [34]. In the case of P. fluorescens biofilm, no studies have previously used the highly specific nature of fluorescent probes to assess the affinity of functionalized NPs with various EPS macromolecules. For the current P. fluorescens biofilms, the string-like topography of both NAG- and Mannose-containing polysaccharide EPS molecules suggests that they may be useful for stabilizing the 3D structure of the biofilm [35] and can also provide a scaffold on which the biofilm can develop in its early stages of attachment [36]. Galactose-containing polysaccharides can be produced as part of the differentiation in maturation of the biofilm [37] while the variation of morphology and topography of proteins within the EPS suggests the increased likelihood of proteins forming complexes with other EPS biomolecules with the biofilm [38]. The eDNA in biofilms is either present in the EPS matrix after being released as genomic DNA from lysed cells or as eDNA secreted by metabolically active cells [39]. DNA in the EPS matrix has been shown to be essential in intercellular adhesion and furthering biofilm stability [40]. The binding of fluorescently-labelled lectins to multiple targets within biofilms or bioaggregates has been previously demonstrated [41], especially in the binding of lectins to glycoconjugates [42] and proteins [43] both on the bacterial cell surface as well as within the EPS matrix. However, in the current study, the signal produced by EPS molecule-specific stains was shown to occur independently to that of the signal from live bacterial cells (Fig. 5, Fig. 6), thus mitigating the possible effects of cell-surface binding and therefore erroneous colocalization results.
In-vitro binding affinity analyses
The use of simplified in-vitro models in biofilm research have been instrumental in addressing some fundamental questions with regard to interaction studies but cannot always be compared to in-situ models which are closer resemble a natural environment [44] The interactions involved within biofilms are complex; which makes them difficult to study under in-vitro conditions. Both methodologies are important to include so as to increase the ease of transference to clinical or industrial settings [45]. The protein concentration per mass of P. fluorescens biofilm EPS extracts have been shown to be at least ten times higher than the carbohydrate concentration per mass [46]. Proteins within the biofilm are often closely associated with (or form complexes) with other proteins, polysaccharides and eDNA and this can result in structurally unique niches with multiple exposed surface functional groups for SiNPs to interact with [47]. Due to the increased concentration of proteins in these biofilms, an interaction of SiNPs with a protein component (or more likely protein complex) of the EPS matrix may therefore have a higher probability of occurring. The affinity of proteins for SiNPs was studied using a protein extract precipitated from the biofilm EPS matrix as well as BSA and RNase as model proteins with varying charge and pI. As compared to the other SiNPs, Amine-SiNPs show a unique protein binding pattern. While BSA has a high affinity for this positively charged surface, a significantly lower amount of RNase molecules were bound to the Amine-SiNPs surface. As this phenomenon is only observed in positively charged SiNPs, it suggests a dominant electrostatic contribution. To further demonstrate the implication of surface charge and functionalization in the affinity of proteins for the SiNPs, the exposure of different SiNPs to an EPS protein extract showed a significant variation in the protein corona formed (Fig. S7). On the other hand, the higher binding affinity of EPS proteins for Epoxy-SiNPs when compared to the other SiNPs was again possibly due to the reactivity of the functional groups at the Epoxy-SiNPs surface [24]. The affinity of the carbohydrate component of EPS matrix to SiNPs was also further analyzed through in-vitro studies, using individual monosaccharide and polysaccharide solutions. The contribution of SiNP surface charge became more evident for the binding of sugar molecules to the surface of SiNPs with the highest overall binding shown to be on the surface of Amine-SiNPs.
Colocalization of SiNPs with EPS matrix macromolecules
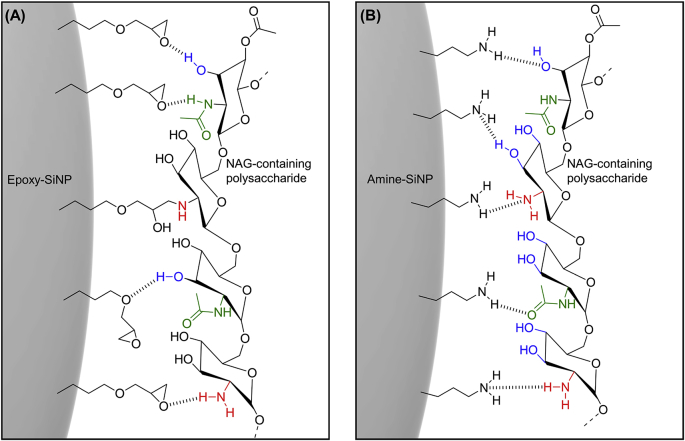
Differences in the colocalization of SiNPs which have relatively similar surface chemistry (Epoxy- and Hydroxy-SiNPs) or SiNPs with a similar charge (Bare-, Epoxy- and Hydroxy-SiNPs) with each respective EPS macromolecule further highlights the extent to which a comparatively small change in the surface chemistry of the SiNPs can dramatically affect the SiNPs binding affinity. Given the complex and heterogeneous nature of the biofilm matrix, complex physicochemical interactions are expected to govern the affinity of SiNPs to EPS macromolecules; however we can hypothesize that certain distinct characteristics of both the SiNPs and EPS macromolecules in question may be dominant in changing their affinity and therefore stabilizing their interactions. For example, higher levels of colocalization between Epoxy-SiNPs and NAG-containing polysaccharides may be due to the electrostatic contribution of the epoxy functional groups [24] interacting with the amino functional groups on the partially deacylated NAG polysaccharides, with possible covalent and hydrogen bonding interactions occurring (Fig. 10A). The increased colocalization between Epoxy-SiNPs and Mannose-containing polysaccharides may also be stabilized by hydrogen bonding between functional groups. Similarly, the electrostatic contributions of the amino, hydroxyl and carbonyl groups of NAG-containing polysaccharides with the amino groups Amine-SiNPs may be stabilized under the same intermolecular forces in these conditions (Fig. 10B).
Fig. 10.
Hypothesized interactions between SiNPs surface functional and polysaccharides in the EPS matrix. Interactions between functional groups on NAG-containing polysaccharides and functional groups on the surface of (A) Epoxy-SiNPs or (B) Amine-SiNPs. These interactions may occur through possible hydrogen bonding or covalent interactions.
While the electrostatic contributions of macromolecules and SiNPs may stabilize and increase the likelihood of physicochemical interactions, these same contributions may lead to repulsion forces between similarly charged entities. For instance, the negative charge on the surface of Bare-SiNPs and the overall negative charge of proteins and eDNA within the matrix may lead to lower colocalization between these binding pairs (Fig. 5). In addition, weak colocalization ratios are not necessarily indicative of a lack of interaction but rather, the discernment of these specific interactions may require additional fluorescent labelling. Void or black areas in CLSM images are not always representative of empty space but rather as areas which have not been fluorescently labelled and may contain other macromolecules. Furthermore, the transport and accumulation of NPs within the EPS matrix may, to a certain extent, also occur through electric double layer effects, hydrophobic, steric and bridging interactions as well as EPS matrix pore clogging, sieving and the effect of biofilm topography [[48], [49], [50]].
Conclusion
The complexity of interactions already occurring between different macromolecules within natural biofilms have been elucidated previously [51], while investigations of inherently complex interactions between functionalized NPs and the biofilm matrix is still in its infancy. In order to optimize and contribute to the development of novel approaches for biofilm eradication and control methods, specifically with the aid of engineered NPs, the understanding of the complex interaction which occur in these systems is vital. This study has demonstrated the successful use of a combination of highly specific fluorescent probes and 3D-CLSM imaging to identify the distribution and topography of various macromolecules (including proteins, polysaccharides and DNA) within P. fluorescens WCS365 biofilms. Building on these results, colocalization and binding analyses revealed various high affinity interactions which may be occurring between functionalized SiNPs and these EPS matrix macromolecules. Although highly specific, the design of unique NP systems which have high selectivity for certain EPS macromolecules have previously been based on a more traditional “lock-and key” or ligand-protein interactions [52], and often require a great deal of optimization. Alternatively, the use of a colocalization analysis approach applied to “smart” NP design may lead to better assessment of NP-biofilm macromolecule interactions and more effective biofilm control measures. For example, if NAG is to be targeted for enzyme degradation, a NP-enzyme conjugate with an overall positive charge may provide more efficient targeted removal. The contribution of this work can add to the fundamental understanding of NP–biofilm matrix interactions, to enable more versatile and efficient NP design and application and provide new avenues for the development of advanced approaches to the prevention, control and eradication of unwanted biofilms.
Methodology
Materials
The following reagents were all purchased from Merck, Ireland, and used as supplied: Acetic Acid, N-Acetylglucosamine (GlcNAc), Ammonium Chloride (NH4Cl), Ammonium Hydroxide (NH4OH), (3-Aminopropyl) Triethoxysilane (APTES), Bovine Serum Albumin (BSA) Calcium Chloride (CaCl2), Cyclohexane, Dowex® cation exchange resin, Ethanol (EtOH), D-Galactose(Gal), 3-Glycidoxypropyltrimethoxysilane (GPTMS), D-Glucose (Glc), Glycerol, King B Agar, Magnesium Sulfate (MgSO4), D-Mannose (Man), Mowiol 4–88, Ribonuclease I (RNase), Rhodamine B Isothiocyanate (RITC) and Tetraethyl Orthosilicate (TEOS). Potassium Phosphate Dibasic was purchased from Honeywell, Fluka™, (Ireland). Pierce™ silver staining kit, Sypro® Orange Protein Gel Stain and Sytox™ Green were purchased from ThermoFischer Scientific (Illinois, USA). Fluorescein (FITC)-labelled Lectin Kit I was purchased from Vector Laboratories (California, USA). Grade 1 pure water (18.2 MΩ cm−1), was obtained from an Elga Process Water System (Biopure 15 and Pureflex 2, Veolia, Ireland).
Nanoparticle synthesis and characterization
RITC-labelled SiNPs (Bare-SiNPs) were synthesized using a microemulsion method with alkaline buffer, according to Quan et al. [53]. Briefly, 40 mL of 0.01 M aqueous solution of NH4Cl were poured in an Erlenmeyer flask and the pH adjusted to 9 using NH4OH. In a separate container, 10 mL of cyclohexane were mixed with 5 mL of TEOS and 1 mL of RITC-APTES dye-conjugate (10 mg dye, 50 μL APTES, 5 mL ethanol); the latter was added to the reaction mixture to achieve fluorescent labelling of the nanoparticles. This mixture was then added to the buffer, and the reaction carried out at 60 °C for 20 h, under constant stirring and in the darkness. At the reaction’s completion, the aqueous phase (containing the SiNPs) was collected and the particles were precipitated through centrifugation (12000 RPM for 15 min) and washed three times with water to remove excess of unreacted reagents.
For the synthesis of positively-charged, amine-functionalized NPs (Amine-SiNPs), the surface was functionalized with (3-aminopropyl)triethoxysilane (APTES), according to Hristov et al. [54]. Briefly, aliquots of 25 mg of NPs (dispersed in water) were mixed with a 10% v/v APTES solution in ethanol. The final concentration of NPs in the reaction mixture was 2.5 mg mL−1. The reaction was initially carried out for 1 h at room temperature under continuous shaking, and then continued for a further hour at 90 °C (always under shaking). Finally, the NPs were precipitated by centrifugation (12000 RPM for 15 min) and washed three times with water.
For the synthesis of epoxy-functionalized NPs (Epoxy-SiNPs), surface modification was carried out using 3-glycidoxypropyltrimethoxysilane (GPTMS), following a procedure adapted from Ojea-Jiménez et al. [23]. To an aqueous solution of as-synthesized NPs (160 mL, 2 mg mL−1 concentration), 134 μL of NaOH 1 mol L−1 were added to adjust the pH to 10, followed by the addition of 167 μL of GPTMS. The reaction was carried out overnight (20 h) at room temperature and under continuous stirring, and afterwards the particles were washed three times with water, through centrifugation (12000 RPM for 20 min). A part of the prepared epoxy-SiNPs was further modified through a ring-opening reaction to yield hydroxy-functionalized NPs (Hydroxy-SiNPs). The reaction, adapted from Worthen et al. [55], consisted in the addition of 300 μL of 0.2 mol L−1 HCl to 20 mL of Epoxy-SiNPs dispersion (10 mg mL−1), under continuous stirring for 30 min. All the fluorescent dye-labelled SiNPs suspensions were stored in 50 mg mL−1 aqueous stock solutions, and afterwards diluted to the desired working concentrations for each specific experiment.
The NPs were characterized, and the surface functionalization monitored by Dynamic Light Scattering (DLS) and Z-Potential measurements (Zetasizer Nano ZS, Malvern Instruments), FT-IR spectroscopy (Vertex 70, Bruker) and transmission electron microscopy (TEM) analysis (FEI Technai G2). DLS and Z-potential analysis were carried out in triplicate, with 10 scans for each run. FT-IR spectra were collected for dried NPs (overnight at 60 °C) from 4000 to 400 cm−1, with a resolution of 1 cm−1. TEM images were acquired after drop casting on carbon-coated grids of SiNPs dispersions in ethanol (0.5 mg mL−1); average size and distributions were obtained using ImageJ [56], by measuring the size of populations of at least 50 particles.
Bacterial culture and maintenance
The bacterial strain used in this investigation was P. fluorescens WCS365. Bacterial cultures were stored at −80 °C in 25% (w/v) glycerol. For cultivation, thawed aliquots were streaked onto King B agar plates and incubated for 24 h at 30 °C. A single bacterial colony was used to inoculate 50 mL of sterile King B medium in a 250 mL Erlenmeyer flask and incubated at 30 °C with shaking at 200 RPM overnight (16–18 h) to an approximate optical density (OD) at 600 nm of between 2.3 and 2.6. The overnight cultures were then diluted to a final OD600 of 1.0 using fresh sterile King B medium.
Biofilm growth
Biofilms were prepared as per Safari et al. (2014) with minor modifications [57]. A 5 mL volume of the OD600 of 1 diluted culture, supplemented with CaCl2 to a final concentration of 1.5 mmol L−1, was added to a sterile 50 mL centrifuge tube containing a glass coverslip (24 mm × 50 mm) and plugged with sterile cotton wool. Tubes were then incubated for 72 h at 30 °C with shaking at 100 RPM.
EPS extraction and protein precipitation
EPS was extracted from 72 h old biofilms grown on glass coverslips using cation-exchange resin (CER) extraction as per Jachlewski et al. with minor modifications [58]. Briefly, the biofilms were gently washed with PBS buffer and then resuspended in 0.5 mL of a NaCl 0.9% (w/v) solution. After sonication for 2 min, the cation exchange Dowex® resin was added (1%, w/v) and the EPS extracts were shaken at 300 RPM for 2 h at 4 °C in the dark. An EPS solution was obtained after centrifugation (20000 g for 20 min at 4 °C) and filtration (Millipore® 0.22 μm pore membrane) of the supernatant. EPS extracts were maintained at −80 °C until their use.
Proteins from an EPS extract were precipitated based on an ammonium sulfate salting-out method with minor modifications [59]. Briefly, to 1 mL of EPS extract, (NH4)2SO4 was added dropwise over a period of 1 h. An increased turbidity was observed after adding 0.49 g of the salt. The EPS extract was then left at 4 °C overnight for complete precipitation and then centrifuged at 20000 g for 20 min at 4 °C. The supernatant was discarded, and the protein pellet was dispersed in the same volume of H2O. The EPS protein solution was then kept at 4 °C until its use.
Sample preparation for CLSM
Various fluorescent probes (Table 2) were used for the identification and subsequent colocalization analysis of polysaccharides, proteins and eDNA within the biofilms. The polysaccharides present in P. fluorescens WCS365 biofilms have not yet been identified. Polysaccharides produced by other Pseudomonads include alginate [60], PEL (a partially deacetylated N-acetyl-d-glucosamine (NAG) and N-acetyl-d-galactosamine polysaccharide (NAC)) and PSL (a Mannose, Galactose and l-rhamnose containing polysaccharide) [61]. Therefore, the specific lectins used in the current study were chosen based on their prominence, highly specific binding and also their overall abundance within the biofilms from the results of a fluorescence lectin-binding analysis [22] (data not shown). To this end, fluorescently-labelled (FITC) lectins and various other probes were used to label the individual components within the biofilm independent of other molecules.
Table 2.
Fluorescently labelled probes for polysaccharide, proteins or eDNA-binding specificities employed for specific staining of P. fluorescens biofilms.
| Probe | Abbreviation | Main Specificity | λEx/λEm (nm) |
|---|---|---|---|
| Concanavalin A (Canavalia ensiformis) | Con A | Mannose, Glucose | 495/519 |
| Wheat Germ Agglutinin (Triticum vulgaris) | WGA | N-acetylglucosamine (NAG) | 495/519 |
| Ricinus communis Agglutinin 120 | RCA | Galactose | 495/519 |
| Sypro® Orange | SO | Proteins | 490/570 |
| Sytox™ Green | SG | eDNA and Dead bacterial cells | 504/523 |
| Syto 9 | SY9 | All cells | 483/503 |
| DAPI | DA | Nucleic acids | 385/451 |
Staining: Lectin stock solutions were diluted in H2O to a final concentration of 20 μg mL −1. A 1000X dilution of Sypro® Orange was used for the identification of proteins within the biofilm, while eDNA was stained with a 5000X dilution of Sytox™ Green. Each biofilm-coated glass coverslip was carefully removed from the centrifuge tubes, and gently rinsed three times in H2O. The coverslip was then placed horizontally on a sample holder and 150 μL of each respective fluorescent probe solution was added directly to the biofilm, followed by incubation in the dark for 15 min. After incubation, the biofilms were gently rinsed three times in H2O to remove all unbound probes.
Exposure to SiNPs: Suspensions of SiNPs were prepared by diluting stock solutions to a working concentration of 0.5 mg mL−1 using H2O. The working solutions were sonicated for 1 h prior to use. Subsequent to fluorescent probe staining of each biofilm component, 150 μL of each respective SiNP solution was placed on top of the biofilm as described above. The biofilms were the rinsed three times in H2O to remove all unbound SiNPs. Each stained biofilm-coated coverslip was mounted onto a glass microscope slide (25 mm × 75 mm x 1 mm) using Tris-buffered Mowiol 4–88 (pH 8.5) mounting medium as per Fulaz et al. [5] (Fig. S9). The prepared slides were then allowed to dry for at least 1 h at room temperature while protected from light. Horizontal plane z-stack images were acquired with an Olympus FluorView FV1000 CLSM attached to an inverted Olympus IX81 microscope with a 60x/1.35 NA UPL SAPO oil immersion objective (Olympus Optical, Tokyo, Japan). The microscope was equipped with 405 nm, 488 nm, 543 nm and 633 nm laser lines.
Image acquisition, processing and analysis
All images were acquired equally; 1x digital zoom, a scanning speed of 2.0 μs/pixel, with 2x Kalman line averaging and sequential channel acquisition. At least three image stacks, with a z-step of 1 μm, from each of three independent experiments were acquired and used for each analysis. All image files were analyzed in Fiji image processing software [62]. All images were background subtracted and median filtered (0.5 pixel radius) prior to all analysis. To quantify the penetration profiles of the SiNPs, fluorescence signals from z-stack CLSM images were used to determine the location and penetration of SiNPs. All colocalization analysis were performed using the “JaCoP” plugin [27] in Fiji image processing software. Mander’s correlation coefficients were used for reporting colocalization between SiNPs and EPS matrix macromolecules and therefore the colocalization ratios were independent of signal intensity and overall abundance of fluorescence signal.
SiNPs entrapment by the biofilm
A method adapted from by Nevius et al. [63], [71] was used to determine the influence on SiNP surface modification on the entrapment of SiNPs into the biofilm. Briefly, biofilms were cultured in black 96-well plates at 30 °C for 24 h with orbital shaking at 125 RPM. After growth, biofilms were rinsed three times in H2O to remove loosely attached cells. SiNP suspensions (0.5 mg mL−1) were added to each biofilm-containing well as well as control wells and incubated for 15 min. The total relative entrapment of SiNPs by biofilms was calculated by measuring the fluorescent intensity of SiNPs which have not yet attached to biofilms (and therefore present in the supernatant) and comparing this to SiNP controls (wells in which NPs have not been exposed to the biofilm). The overall entrapment of SINPs can be calculated using the difference between the two fluorescence intensity values (n = 6). The SiNPs fluorescence intensity (λex = 550, λem = 585 nm) was measured using a plate reader (SpectraMax iD3, Molecular devices).
In-vitro binding interactions
While in-situ experimental work provides an insight into the bacterial biofilm system, in-vitro analyses may provide an insight into the general mechanisms involved in various other applications. For protein binding affinity analysis, 500 μg mL−1 of a precipitated EPS protein extract, BSA or a RNase solution was exposed to each SiNP [2 mg mL−1] for 15 min, similarly as above. BSA and RNase were chosen as model proteins based on their differing surface chemistry to analyse the effect of overall surface charge on the protein attachment to the NP surface. In order to quantify the protein concentration in the supernatant and therefore the concentration of protein bond to the SiNPs surface, the Lowry method for protein quantification was used [64]. For the analysis of the protein corona formed on each NP after exposure to an EPS protein extract, the total precipitated EPS proteins as well as the protein corona formed on each respective SiNP was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method described by Laemmli [65]. Ten microliters of supernatant samples were mixed with 10 μl sample buffer (125 mmol L−1 Tris pH 6.8; 4% SDS; 2% β-mercaptoethanol; 0.25 mol L−1 EDTA; 20% glycerol; 0.04% bromophenol blue), separated by SDS-PAGE (12% w/v) and visualized by silver staining according to the manufacturer’s instructions (Pierce Silver stain kit, Thermo Fischer Scientific).
For carbohydrate binding affinity analysis, Mannose, Galactose, Glucose, NAG and sodium alginate stock solutions of 500 mmol L−1 were prepared in H2O. Based on serial dilutions in H2O, standard curves ranging from 0.03125 to 10 mmol L−1 for Mannose, Galactose, Glucose and Alginate was generated. For NAG, a standard curve ranging from 3.125 to 250 mmol L−1 was prepared. For binding analysis, 250 μL of Mannose [5 mmol L−1], Galactose [5 mmol L−1], Glucose [5 mmol L−1], Alginate [5 mmol L−1] or NAG [100 mmol L−1] was added to 250 μL of each SiNPs [2 mg mL−1] solution, respectively, and incubated at 1000 RPM at 25 °C for 15 min. To assess the concentration of carbohydrates bound to the SiNPs, the SiNP-carbohydrate solution was centrifuged at 20000 g for 20 min and three 50 μL aliquots were removed from the supernatant for analysis. A phenol-sulphuric acid total carbohydrate assay was performed on each aliquot and compared to each respective standard curve [66].
Simulation and visualization of protein surface charge distribution
The electrostatic surface potentials of BSA and RNase was calculated using the Adaptive Poisson–Boltzmann Solver (APBS) software [67], after assigning protonation states to residues at a neutral pH, using PROPKA [68]. Charge distributions were visualized using 3DMol [69] with the program internal threshold for colouring electrostatic potential set to −2.0 kBTe [2]; blue: +2.0 kBTe [2]. The protein crystal structures of BSA (protein data bank entry ID 3V03) and RNase (protein data bank entry ID 5D97) were taken from the RCSB Protein Data Bank (http://www.rcsb.org/) [70].
Statistical analysis
All data presented is representative of the mean ± standard deviation of at least three separate experiments, each of which contained at least 3 replicates. A one-way ANOVA followed by Tukey’s HSD pairwise comparison (P < 0.05) was used to assess statistical significance using IBM SPSS Statistics for windows IBM SPSS Statistics for Windows version 26.0. (IBM Corp., Armonk, N·Y., USA). Where qualitative images are shown, experiments were performed in triplicate, and a representative image is shown.
Funding sources
This research was supported by Science Foundation Ireland (SFI) under grant number 15/IA/3008.
CRediT authorship contribution statement
Dishon Wayne Hiebner: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing - original draft, Writing - review & editing. Caio Barros: Investigation, Writing - review & editing. Laura Quinn: Writing - review & editing. Stefania Vitale: Formal analysis, Investigation, Validation, Writing - review & editing. Eoin Casey: Conceptualization, Writing - review & editing, Funding acquisition, Resources, Supervision, Project administration.
Declaration of competing interest
All authors declare they have no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2020.100029.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rybtke M., Hultqvist L.D., Givskov M., Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J Mol Biol. 2015;427:3628–3645. doi: 10.1016/j.jmb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland I.W. The biofilm matrix - an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/S0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 3.Miyaue S., Suzuki E., Komiyama Y., Kondo Y., Morikawa M., Maeda S. Bacterial memory of persisters: bacterial persister cells can retain their phenotype for days or weeks after withdrawal from colony-biofilm culture. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.01396. 1396-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fulaz S., Vitale S., Quinn L., Casey E. Nanoparticle-biofilm interactions: the role of the EPS matrix. Trends Microbiol. 2019;27:915–926. doi: 10.1016/j.tim.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Fulaz S., Hiebner D., Barros C.H., Devlin H., Vitale S., Quinn L., Casey E. Ratiometric imaging of the in situ pH distribution of biofilms by use of fluorescent mesoporous silica nanosensors. ACS Appl Mater Interfaces. 2019;11:32679–32688. doi: 10.1021/acsami.9b09978. [DOI] [PubMed] [Google Scholar]
- 6.Vigneron A., Alsop E.B., Chambers B., Lomans B.P., Head I.M., Tsesmetzis N. Complementary microorganisms in highly corrosive biofilms from an offshore oil production facility. Appl Environ Microbiol. 2016;82:2545–2554. doi: 10.1128/Aem.03842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zumsteg A., Urwyler S.K., Glaubitz J. Characterizing bacterial communities in paper production-troublemakers revealed. MicrobiologyOpen. 2017;6 doi: 10.1002/mbo3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho G., Balestrino D., Forestier C., Mathias J.D. How do environment-dependent switching rates between susceptible and persister cells affect the dynamics of biofilms faced with antibiotics? Npj Biofilms and Microbiomes. 2018;4 doi: 10.1038/s41522-018-0049-2. UNSP 610.1038/s41522-018-0049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romling U., Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Intern Med. 2012;272:541–561. doi: 10.1111/joim.12004. [DOI] [PubMed] [Google Scholar]
- 10.Bixler G.D., Bhushan B. Biofouling: lessons from nature. Philos. Trans. R. Soc. A. 2012;370:2381–2417. doi: 10.1098/rsta.2011.0502. [DOI] [PubMed] [Google Scholar]
- 11.Algburi A., Comito N., Kashtanov D., Dicks L.M., Chikindas M.L. Control of biofilm formation: antibiotics and beyond. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.02508-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koo H., Allan R.N., Howlin R.P., Stoodley P., Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babbey C.M., Ahktar N., Wang E., Chen C.C.H., Grant B.D., Dunn K.W. Rab10 regulates membrane transport through early endosomes of polarized Madin-Darby Canine Kidney cells. Mol Biol Cell. 2006;17:3156–3175. doi: 10.1091/mbc.E05-08-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fay F.S., Taneja K.L., Shenoy S., Lifshitz L., Singer R.H. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- 16.French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 17.Cho W.J., Daniel E.E. Colocalization between caveolin isoforms in the intestinal smooth muscle and interstitial cells of Cajal of the Cav1(+/+) and Cav1 (-/-) mouse. Histochem Cell Biol. 2006;126:9–16. doi: 10.1007/s00418-005-0128-3. [DOI] [PubMed] [Google Scholar]
- 18.Honeker L.K., Root R.A., Chorover J., Maier R.M. Resolving colocalization of bacteria and metal(loid)s on plant root surfaces by combining fluorescence in situ hybridization (FISH) with multiple-energy micro-focused X-ray fluorescence (ME muXRF) J Microbiol Methods. 2016;131:23–33. doi: 10.1016/j.mimet.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zippel B., Neu T.R. Characterization of glycoconjugates of extracellular polymeric substances in tufa-associated biofilms by using fluorescence lectin-binding analysis. Appl Environ Microbiol. 2011;77:505–516. doi: 10.1128/Aem.01660-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro L., Zhang R., Muñoz J.A., González F., Blázquez M.L., Sand W., Ballester A. Characterization of exopolymeric substances (EPS) produced by Aeromonas hydrophila under reducing conditions. Biofouling. 2014;30:501–511. doi: 10.1080/08927014.2014.892586. [DOI] [PubMed] [Google Scholar]
- 21.Bennke C.M., Neu T.R., Fuchs B.M., Amann R. Mapping glycoconjugate-mediated interactions of marine Bacteroidetes with diatoms. Syst Appl Microbiol. 2013;36:417–425. doi: 10.1016/j.syapm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Neu T.R., Lawrence J.R. Lectin-binding analysis in biofilm systems. Biofilms. 1999;310:145–152. doi: 10.1016/s0076-6879(99)10012-0. [DOI] [PubMed] [Google Scholar]
- 23.Ojea-Jiménez I., Urbán P., Barahona F., Pedroni M., Capomaccio R., Ceccone G., Kinsner-Ovaskainen A., Rossi F., Gilliland D. Highly flexible platform for tuning surface properties of silica nanoparticles and monitoring their biological interaction. ACS Appl Mater Interfaces. 2016;8:4838–4850. doi: 10.1021/acsami.5b11216. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q., Huang R.F., Guo L.H. One-step and high-density protein immobilization on epoxysilane-modified silica nanoparticles. Chin Sci Bull. 2009;54:2620–2626. doi: 10.1007/s11434-009-0210-7. [DOI] [Google Scholar]
- 25.Kumar S., Singh J., Agrawal V.V., Ahamad M., Malhotra B.D. Biocompatible self-assembled monolayer platform based on (3-glycidoxypropyl)trimethoxysilane for total cholesterol estimation. Anal. Methods. 2011;3:2237–2245. doi: 10.1039/c1ay05231d. [DOI] [Google Scholar]
- 26.Golmohamadi M., Clark R., Veinot J., Wilkinson K. The role of charge on the diffusion of solutes and nanoparticles (silicon nanocrystals, nTiO(2), nAu) in a biofilm. Environ Chem. 2013;10:34–41. doi: 10.1071/EN12106. [DOI] [Google Scholar]
- 27.Bolte S., Cordelieres F.P. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 28.Zinchuk V., Wu Y., Grossenbacher-Zinchuk O. Bridging the gap between qualitative and quantitative colocalization results in fluorescence microscopy studies. Sci Rep. 2013;3:1365. doi: 10.1038/srep01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., Yeh Y.C., Giri K., Mout R., Landis R.F., Prakash Y.S., Rotello V.M. Control of nanoparticle penetration into biofilms through surface design. Chem Commun. 2015;51:282–285. doi: 10.1039/c4cc07737g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Martino P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. Aims Microbiology. 2018;4:274–288. doi: 10.3934/microbiol.2018.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennings L.K., Storek K.M., Ledvina H.E., Coulon C., Marmont L.S., Sadovskaya I., Secor P.R., Tseng B.S., Scian M., Filloux A., Wozniak D.J. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc Natl Acad Sci USA. 2015;112:11353–11358. doi: 10.1073/pnas.1503058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S., Liu X., Liu H., Zhang L., Guo Y., Yu S., Wozniak D.J., Ma L.Z. The exopolysaccharide Psl-eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2015;7:330–340. doi: 10.1111/1758-2229.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huseby M.J., Kruse A.C., Digre J., Kohler P.L., Vocke J.A., Mann E.E., Bayles K.W., Bohach G.A., Schlievert P.M., Ohlendorf D.H., Earhart C.A. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc Natl Acad Sci USA. 2010;107:14407–14412. doi: 10.1073/pnas.0911032107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berk V., Fong J.C., Dempsey G.T., Develioglu O.N., Zhuang X., Liphardt J., Yildiz F.H., Chu S. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reichhardt C., Parsek M.R. Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa biofilm architecture and matrix localization. Front Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00677. 677-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kostakioti M., Hadjifrangiskou M., Hultgren S. J. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013;3 doi: 10.1101/cshperspect.a010306. ARTN a01030610.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng B.S., Majerczyk C.D., da Silva D.P., Chandler J.R., Greenberg E.P., Parsek M.R. Quorum sensing influences Burkholderia thailandensis biofilm development and matrix production. J Bacteriol. 2016;198:2643–2650. doi: 10.1128/jb.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lembre P., Lorentz C., Di P. 2012. The complex World of polysaccharides ch. [Chapter 13] [Google Scholar]
- 39.Raymond B.B.A., Jenkins C., Turnbull L., Whitchurch C.B., Djordjevic S.P. Extracellular DNA release from the genome-reduced pathogen Mycoplasma hyopneumoniae is essential for biofilm formation on abiotic surfaces. Sci Rep. 2018;8:10373. doi: 10.1038/s41598-018-28678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z., Gong X., Xie J., Xu Z., Liu G., Zhang G. Investigation of formation of bacterial biofilm upon dead siblings. Langmuir. 2019;35:7405–7413. doi: 10.1021/acs.langmuir.8b01962. [DOI] [PubMed] [Google Scholar]
- 41.Neu T.R., Swerhone G.D.W., Lawrence J.R. Assessment of lectin-binding analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology-Sgm. 2001;147:299–313. doi: 10.1099/00221287-147-2-299. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence J.R., Swerhone G.D.W., Kuhlicke U., Neu T.R. In situ evidence for metabolic and chemical microdomains in the structured polymer matrix of bacterial microcolonies. FEMS Microbiol Ecol. 2016;92 doi: 10.1093/femsec/fiw183. [DOI] [PubMed] [Google Scholar]
- 43.Zubkov M.V., Fuchs B.M., Eilers H., Burkill P.H., Amann R. Determination of total protein content of bacterial cells by SYPRO staining and flow cytometry. Appl Environ Microbiol. 1999;65:3251–3257. doi: 10.1128/aem.65.7.3251-3257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lebeaux D., Chauhan A., Rendueles O., Beloin C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens (Basel, Switzerland) 2013;2:288–356. doi: 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernández-Barat L., Ben-Aicha S., Motos A., Vila J., Marco F., Rigol M., Muñoz L., Bassi G.L., Ferrer M., Torres A. Assessment of in vivo versus in vitro biofilm formation of clinical methicillin-resistant Staphylococcus aureus isolates from endotracheal tubes. Sci Rep. 2018;8:9. doi: 10.1038/s41598-018-30494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molobela I.P., Cloete T.E., Beukes M. Protease and amylase enzymes for biofilm removal and degradation of extracellular polymeric substances (EPS) produced by Pseudomonas fluorescens bacteria. Afr J Microbiol Res. 2010;4:1515–1524. [Google Scholar]
- 47.Fong J.N.C., Yildiz F.H. Biofilm matrix proteins. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0004-2014. ARTN MB-0004-2014 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Habimana O., Steenkeste K., Fontaine-Aupart M.P., Bellon-Fontaine M.N., Kulakauskas S., Briandet R. Diffusion of nanoparticles in biofilms is altered by bacterial cell wall hydrophobicity. Appl Environ Microbiol. 2011;77:367–368. doi: 10.1128/AEM.02163-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peulen T., Wilkinson K. Diffusion of nanoparticles in a biofilm. Environ Sci Technol. 2011;45:3367–3373. doi: 10.1021/es103450g. [DOI] [PubMed] [Google Scholar]
- 50.Wang X.N., Liu B.S., Pan X.L., Gadd G.M. Transport and retention of biogenic selenium nanoparticles in biofilm-coated quartz sand porous media and consequence for elemental mercury immobilization. Sci Total Environ. 2019;692:1116–1124. doi: 10.1016/j.scitotenv.2019.07.309. [DOI] [PubMed] [Google Scholar]
- 51.Flemming H.C., Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 52.Gupta A., Landis R.F., Rotello V.M. Nanoparticle-based antimicrobials: surface functionality is critical. F1000Research. 2016;5:F1000. doi: 10.12688/f1000research.7595.1. Faculty Rev-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quan B., Lee C., Yoo J.S., Piao Y. Facile scalable synthesis of highly monodisperse small silica nanoparticles using alkaline buffer solution and their application for efficient sentinel lymph node mapping. J Mater Chem B. 2017;5:586–594. doi: 10.1039/C6TB02304E. [DOI] [PubMed] [Google Scholar]
- 54.Hristov D.R., Rocks L., Kelly P.M., Thomas S.S., Pitek A.S., Verderio P., Mahon E., Dawson K.A. Tuning of nanoparticle biological functionality through controlled surface chemistry and characterisation at the bioconjugated nanoparticle surface. Sci Rep. 2015;5 doi: 10.1038/srep17040. 17040-17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worthen A.J., Tran V., Cornell K.A., Truskett T.M., Johnston K.P. Steric stabilization of nanoparticles with grafted low molecular weight ligands in highly concentrated brines including divalent ions. Soft Matter. 2016;12:2025–2039. doi: 10.1039/c5sm02787j. [DOI] [PubMed] [Google Scholar]
- 56.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/Nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Safari A., Habimana O., Allen A., Casey E. The significance of calcium ions on Pseudomonas fluorescens biofilms - a structural and mechanical study. Biofouling. 2014;30:859–869. doi: 10.1080/08927014.2014.938648. [DOI] [PubMed] [Google Scholar]
- 58.Jachlewski S., Jachlewski W.D., Linne U., Bräsen C., Wingender J., Siebers B. Isolation of extracellular polymeric substances from biofilms of the thermoacidophilic archaeon Sulfolobus acidocaldarius. Front. Bioeng. Biotech. 2015;3 doi: 10.3389/fbioe.2015.00123. 123-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wingfield P. Protein precipitation using ammonium sulfate. Current Protocols in Protein Science. 2016;84:A.3F.1–A.3F.9. doi: 10.1002/0471140864.psa03fs84. [DOI] [PubMed] [Google Scholar]
- 60.Maleki S., Maerk M., Hrudikova R., Valla S., Ertesvag H. New insights into Pseudomonas fluorescens alginate biosynthesis relevant for the establishment of an efficient production process for microbial alginates. N Biotech. 2017;37:2–8. doi: 10.1016/j.nbt.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 61.Borlee B.R., Goldman A.D., Murakami K., Samudrala R., Wozniak D.J., Parsek M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nevius B.A., Chen Y.P., Ferry J.L., Decho A.W. Surface-functionalization effects on uptake of fluorescent polystyrene nanoparticles by model biofilms. Ecotoxicology. 2012;21:2205–2213. doi: 10.1007/s10646-012-0975-3. [DOI] [PubMed] [Google Scholar]
- 64.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 65.Laemmli U.K. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227:680. doi: 10.1038/227680a0. + [DOI] [PubMed] [Google Scholar]
- 66.Dubois M., Gilles K., Hamilton J.K., Rebers P.A., Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168 doi: 10.1038/168167a0. 167-167. [DOI] [PubMed] [Google Scholar]
- 67.Baker N., Holst M., Wang F. Adaptive multilevel finite element solution of the Poisson-Boltzmann equation II. Refinement at solvent-accessible surfaces in biomolecular systems. J Comput Chem. 2000;21:1343–1352. doi: 10.1002/1096-987x(20001130)21:15<1343::Aid-jcc2>3.3.Co;2-b. [DOI] [Google Scholar]
- 68.Olsson M.H., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical p K a predictions. J Chem Theor Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 69.Rego N., Koes D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics. 2015;31:1322–1324. doi: 10.1093/bioinformatics/btu829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berman H.M. The protein data bank. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 71.Devlin Henry. Colloids and Surfaces B: Biointerfaces. 2020;193 doi: 10.1016/j.colsurfb.2020.111123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.