Abstract
Background
The epidemiological features and outcomes of hospitalized adults with coronavirus disease 2019 (COVID-19) have been described; however, the temporal progression and medical complications of disease among hospitalized patients require further study. Detailed descriptions of the natural history of COVID-19 among hospitalized patients are paramount to optimize health care resource utilization, and the detection of different clinical phenotypes may allow tailored clinical management strategies.
Methods
This was a retrospective cohort study of 305 adult patients hospitalized with COVID-19 in 8 academic and community hospitals. Patient characteristics included demographics, comorbidities, medication use, medical complications, intensive care utilization, and longitudinal vital sign and laboratory test values. We examined laboratory and vital sign trends by mortality status and length of stay. To identify clinical phenotypes, we calculated Gower’s dissimilarity matrix between each patient’s clinical characteristics and clustered similar patients using the partitioning around medoids algorithm.
Results
One phenotype of 6 identified was characterized by high mortality (49%), older age, male sex, elevated inflammatory markers, high prevalence of cardiovascular disease, and shock. Patients with this severe phenotype had significantly elevated peak C-reactive protein creatinine, D-dimer, and white blood cell count and lower minimum lymphocyte count compared with other phenotypes (P < .01, all comparisons).
Conclusions
Among a cohort of hospitalized adults, we identified a severe phenotype of COVID-19 based on the characteristics of its clinical course and poor prognosis. These findings need to be validated in other cohorts, as improved understanding of clinical phenotypes and risk factors for their development could help inform prognosis and tailored clinical management for COVID-19.
Keywords: clinical phenotype, COVID-19, medical complications, mortality, multisystem inflammatory syndrome
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused more than 10 million infections and 239 590 deaths in the United States [1]. As the number of cases increases, the pandemic continues to place the US health care system under pressure. Detailed descriptions of the natural history of coronavirus disease 19 (COVID-19) among hospitalized patients are paramount to optimize health care resource utilization, and the detection of different clinical presentations may allow more tailored clinical management strategies. Clinicians have recognized that in severely ill patients, COVID-19 may present with nonpulmonary manifestations rarely seen with other respiratory viruses [2–5]; direct cytopathic viral effects in organs, such as the brain and kidneys, and an exaggerated proinflammatory response have been proposed as possible mechanisms to explain this difference [6].
While advanced age, comorbid conditions, and inflammatory markers have been identified as independent risk factors for severe disease [7–15], how these risk factors interact and their relationship to severe pulmonary and extrapulmonary complications remains poorly understood. Recognizing patterns of clinical progression based on host characteristics, physiologic and laboratory measurements, complications, and outcomes will help clinicians better understand the spectrum and natural history of COVID-19 and can influence decisions about clinical management. Methods to identify heterogeneous phenotypes have been used for other broad disease entities, such as sepsis and asthma [16–18].
We collected longitudinal clinical data on 305 hospitalized patients with laboratory-confirmed SARS-CoV-2 infection in the US state of Georgia. Our primary objective was to identify and describe COVID-19 clinical phenotypes based on demographics, comorbidities, presenting signs and symptoms, laboratory values, complications, and outcomes. Our secondary objectives were to (1) differentiate clinical syndromes at hospital presentation, (2) characterize temporal trends of vital signs and laboratory parameters by disease severity, and (3) describe medical complications and outcomes among hospitalized patients.
METHODS
Study Design and Participants
The US Centers for Disease Control and Prevention (CDC), the Georgia Department of Public Health (DPH), and 3 hospital networks within Georgia partnered to review patient medical records at 8 Georgia hospitals. Seven hospitals were in metropolitan Atlanta, and 1 was in southern Georgia; they comprised academic medical centers, a public teaching hospital, and community hospitals; all provided tertiary care.
Hospitals provided lists of patients with a positive SARS-CoV-2 reverse transcription polymerase chain reaction test who were admitted during March 1–March 30, 2020 (n = 698). Clinicians reviewed electronic medical records on a convenience sample of sequentially identified adults aged ≥18 years from each hospital’s patient list [19]. We conducted as many initial chart abstractions as possible through April 20 (n = 305). Transfers between participating hospitals and multiple admissions of the same patient (10 patients had a single readmission, and 1 had 2 readmissions) were analyzed as a single hospitalization.
Because some patients were still hospitalized at the time of initial data abstraction, all patient records were re-examined on May 8 to assess outcomes. Study data were collected and managed using Research Electronic Data Capture (REDCap) tools hosted at the CDC [20, 21]. Medical records were examined for clinical, laboratory, and radiologic data, and audits were performed on all chart abstractions to correct data missingness and identify implausible values. This investigation was determined by the CDC and DPH to be public health surveillance [22].
Analyses
Clinical Phenotypes
To identify patterns in the clinical course, we grouped patients into clusters based upon demographic characteristics (age, sex); comorbidities (diabetes mellitus, cardiovascular disease, hypertension, chronic lung disease, extreme obesity [body mass index {BMI} ≥40 kg/m2], neurologic disorders); presenting signs and symptoms (fever [defined as documentation of fever in the medical record or recorded temperature >38.0°C on presentation to the hospital], cough, dyspnea, diarrhea, chills, altered mental status); days from symptom onset to admission; maximum values of C-reactive protein (CRP), lactate dehydrogenase (LDH), ferritin, creatine phosphokinase (CPK), D-dimer, partial thromboplastin time (PTT), aspartate aminotransferase (AST), alanine aminotransferase (ALT), and creatinine; minimum values of white blood cell count (WBC) and absolute lymphocyte count; and complications as diagnosed by the treating physician (shock, acute respiratory distress syndrome [ARDS], venous thromboembolic events [VTE], acute kidney injury [AKI], acute liver injury, and bacterial coinfection). Comorbidities, presenting symptoms, and complications were selected based on known or suspected risk factors, frequency observed in prior studies, and associations with severe COVID-19 [8, 23, 24]. Laboratory values were selected to represent markers of inflammation, hypercoagulability, and end organ damage, and we used peak or nadir values to capture the highest severity of illness. Race was not included as a factor in the clustering analysis because of high prevalence of black race (83.2%). Eighteen patients initially admitted for reasons other than COVID-19 (eg, trauma, postsurgical complication, device-related issues) were excluded from the analysis. The median proportion of imputed data per patient was 11.8%, and all imputed values were removed after clustering for subsequent analysis and visualization (Supplementary Methods).
We computed Gower’s dissimilarity matrix [25] between observations, which allows a similarity metric to be calculated from continuous and categorical variables. Because we wanted to emphasize clinical over demographic characteristics in the phenotypes, age and sex were downweighted by 50%. We used the partitioning around medoids algorithm [26] to cluster observations. For visualization, we calculated standard deviations for categorical variables and median absolute deviations for numeric variables.
Vital Signs and Laboratory Values
Admission vital signs and laboratory values were abstracted for all patients. For serial vital signs and laboratory data, patients were stratified by phenotype, and we plotted the daily average among patients by stratum, inversely weighting observations by the number of observations per patient per day. Wilcoxon rank-sum tests were used to evaluate differences in distributions.
Analyses and visualizations were conducted using R software, version 3.6.2 (Vienna, Austria).
RESULTS
Patient Characteristics
Baseline characteristics of patients have been previously described. Among 305 patients, the median age (interquartile range [IQR]) was 60 (46–69) years, 51% were female, and 83% were non-Hispanic Black. One hundred nineteen patients (39%) were admitted to the intensive care unit (ICU), 92 (30%) received invasive mechanical ventilation (IMV), and 51 (17%) died. Two hundred eighty-seven (94%) were initially admitted for confirmed or suspected COVID-19. Among 18 patients not originally admitted with suspected COVID-19, SARS-CoV-2 was detected by RT-PCR at median hospital day 2; 7 were diagnosed ≥5 days after admission (range, 5–38 days).
Initial Presentation
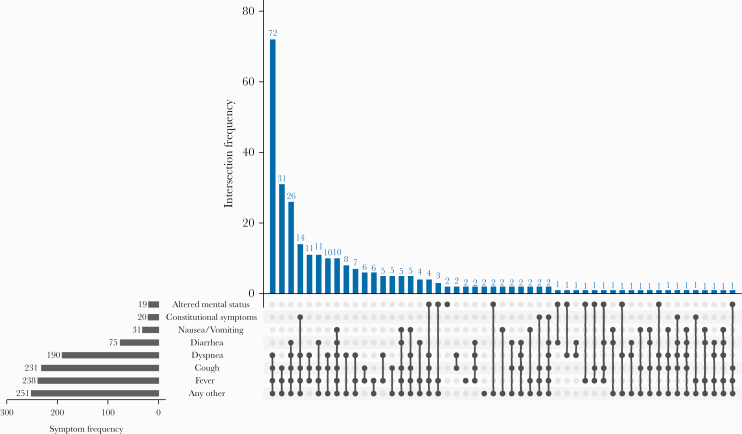
Among the subset of 287 patients whose initial reason for admission was COVID-19, the median time from symptom onset to admission (IQR) was 7 (4.0–9.0) days. While most presented with fever or respiratory symptoms (Figure 1), there were 10 (3%) patients who did not present with fever, cough, or shortness of breath. Of these 10, 7 developed a fever within 2 days of admission, and 5, all of whom were >70 years old, presented with new or worsened altered mental status. After febrile respiratory syndromes, syndromes involving myalgia, fatigue, or gastrointestinal symptoms were the most common. Diarrhea occurred in 75 (26%) patients, usually in combination with fever or respiratory symptoms. Nineteen (7%) patients presented with altered mental status (median age [range], 79 [62–95] years).
Figure 1.
Presenting signs and symptoms among adults hospitalized for COVID-19 whose initial reason for admission was COVID-19 symptoms (n = 287)—Georgia, March 2020. The blue bars show the number of patients with each mutually exclusive symptom combination, indicated by the connected black dots. The black bars show the frequency of each symptom. “Constitutional Symptoms” include myalgia and fatigue. “Any Other” includes sore throat, chills, headaches, anosmia, chest pain, dizziness, rash, or other symptoms. Abbreviation: COVID-19, coronavirus disease 2019.
Hospital Course and Clinical Management
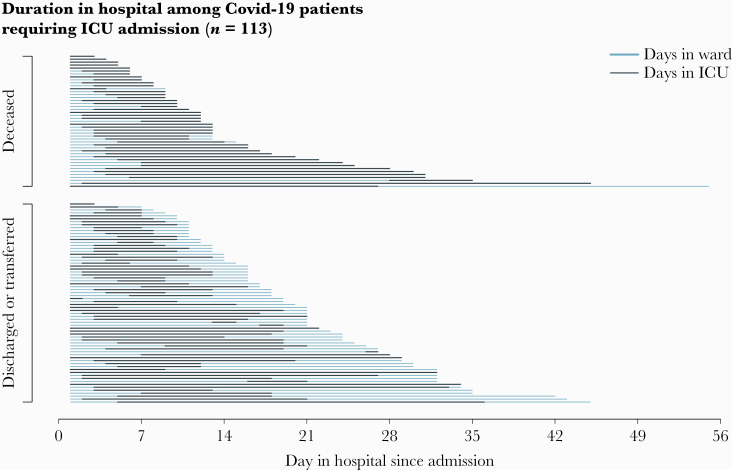
Median length of stay (LOS) (IQR) was 7.5 (4.0–14.0) days for patients discharged alive and 11.0 (7.5–18.0) days for patients who died. Among ICU patients (n = 119), the median duration of ICU stay (IQR) was 8.0 (5.0–12.50) days (Figure 2). ICU admission occurred at a median (IQR) of hospital day 3.0 (2.0–4.0), with IMV initiation at a median (IQR) of hospital day 3.5 (2.0–5.0) and vasopressor initiation at median (IQR) hospital day 5.0 (3.0–8.0). The median duration of IMV (IQR) was 9.0 (5.0–12.0) days. Among 119 ICU patients, 13 (11%) underwent prone positioning while ventilated, 6 (5%) received a tracheostomy, and 2 (2%) received extracorporeal membrane oxygenation.
Figure 2.
Hospitalization status by day and final outcome among patients requiring ICU admission (n = 113)—Georgia, United States, March 2020. Abbreviation: ICU, intensive care unit.
In addition to supportive therapy, hydroxychloroquine was administered to 117 (38%) patients, of whom 37 did not require intensive care. Corticosteroids were initiated in 17% of patients. Eight patients received lopinavir/ritonavir, and none received tocilizumab. No patients received immunomodulatory therapies (eg, interleukin [IL]-1 inhibitors and IL-6 blockers). Antibiotics covering common community-acquired pathogens were administered to 95% within 48 hours after admission (99% of ICU patients); 51% received antibiotics after 48 hours, mostly as an escalation of empiric antibiotics. Data on anticoagulation use were not available for analysis.
Complications
AKI was diagnosed by the treating clinician in 124 (41%) patients; 22 (7%) required renal replacement therapy (RRT) during hospitalization (Table 1). VTEs were documented in 17 (6%) patients, including 11% of ICU patients; arterial thrombotic events (stroke, peripheral artery thrombosis, or acute coronary syndrome) were seen in 7 (2%) patients. Among patients admitted to the ICU, 69 (58%) had a diagnosis of shock; of these, 58 (84%) died.
Table 1.
Clinical Complications Among Adults Hospitalized With COVID-19 (n = 305)—Georgia, United States, March 2020
| Complications | Total (n = 305), No. (%) | Non-ICU (n = 186), No. (%) | ICU (n = 119), No. (%) |
|---|---|---|---|
| Acute kidney injury | 124 (40.7) | 45 (24.2) | 79 (66.4) |
| Shock | 69 (22.6) | 0 (0) | 69 (58.0) |
| Acute respiratory distress syndrome | 62 (20.3) | 0 (0) | 62 (52.1) |
| Liver dysfunction | 40 (13.1) | 12 (6.5) | 28 (23.5) |
| Rhabdomyolysis | 14 (4.6) | 5 (2.7) | 9 (7.6) |
| Venous thrombotic events | 17 (5.6) | 4 (2.2) | 13 (10.9) |
| Deep vein thrombosis | 14 (4.6) | 2 (1.1) | 12 (10.1) |
| Pulmonary embolism | 4 (1.3) | 2 (1.1) | 2 (1.7) |
| Arterial thrombotic events | 7 (2.3) | 0 (0) | 7 (5.9) |
| Peripheral arterial thrombosis | 1 (0.3) | 0 (0) | 1 (0.8) |
| Acute coronary syndrome | 4 (1.3) | 0 (0) | 4 (3.4) |
| Stroke | 2 (0.7) | 0 (0) | 2 (1.7) |
| Other cardiovascular complications | 47 (15.4) | 10 (5.4) | 37 (31.1) |
| Cardiac arrhythmia | 37 (12.1) | 8 (4) | 29 (24.4) |
| Congestive heart failure | 11 (3.6) | 3 (1.6) | 8 (6.7) |
| New-onset cardiomyopathy | 5 (1.6) | 0 (0) | 5 (4.2) |
| Myocarditis | 1 (0.3) | 0 (0) | 1 (0.8) |
| Hospital-acquired pneumonia | 3 (1.0) | 0 (0) | 3 (2.5) |
| Ventilator-associated pneumonia | 4 (1.3) | 0 (0) | 4 (3.4) |
Abbreviations: COVID-19, coronavirus 2019; ICU, intensive care unit.
Viral coinfections were uncommon (n = 10); the most common viral coinfection was with influenza (n = 8 [3%], 275/305 were tested). Bacterial coinfections, defined as positive culture with corresponding antibacterial treatment, were identified in 32 (11%) patients. Among 74 patients tested for either pathogen, no urine Legionella or Streptococcus pneumoniae antigen tests were positive. Seven patients had documented hospital-acquired or ventilator-associated pneumonia. Eight (3%) patients had bacteremia; the most common pathogens were coagulase-negative Staphylococcus species (Supplementary Table 1).
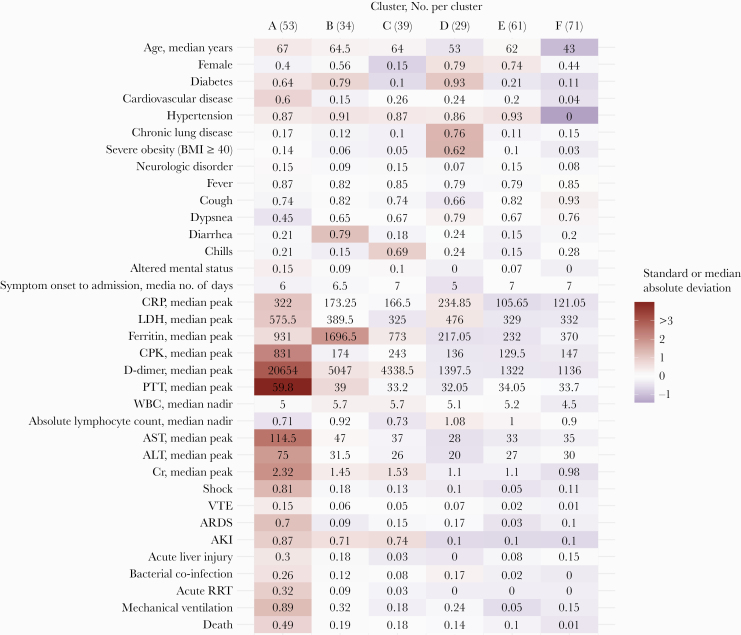
Clustering of Patient Characteristics
The partitioning around medoids analysis grouped patients into 6 patterns (clusters A–F, Figure 3). Mortality was highest in cluster A (49%, n = 53) compared with other clusters (<20%). Proportions of IMV (89%), shock (81%), and AKI (87%) were also highest in cluster A. This cluster had the highest median age (67 years), the highest prevalence of cardiovascular disease (64%), and was predominantly male (60%). Cluster A patients had elevated median peak CRP, LDH, CPK, D-dimer, PTT, AST, ALT, and creatinine compared with the cohort (Supplementary Figure 1). The median nadir absolute lymphocyte count for this cluster was relatively low (0.72 cells/mm3).
Figure 3.
Clinical phenotypes among adults hospitalized with COVID-19—Georgia, United States, March 2020. Cell labels show median cluster values for age, symptom onset to admission days, and laboratory values (CRP peak through Cr peak) and cluster proportions for female sex, comorbidities (diabetes through neurologic disorder), presenting symptoms (fever through altered mental status), complications (shock through bacterial coinfection), and outcomes (acute RRT, mechanical ventilation, death). Cell color indicates the median absolute deviation from the median or standard deviation from the mean. Analysis excludes 18 patients who were initially admitted for reasons other than COVID-19. Abbreviations: AKI, acute kidney injury; ALT, alanine aminotransferase; ARDS, acute respiratory distress syndrome; AST, aspartate aminotransferase; BMI, body mass index; COVID-19, coronavirus disease 2019; CPK, creatine phosphokinase; Cr, creatinine; CRP, C-reactive protein; LDH, lactate dehydrogenase; PTT, partial thromboplastin time; RRT, renal replacement therapy; VTE, venous thromboembolism; WBC, white blood cell count.
Clusters B (n = 34) and C (n = 39) had a high prevalence of AKI (>70%) and similar mortality rates (19% and 18%, respectively). Cluster B was distinguished by high prevalence of diabetes (79%) and diarrhea as a presenting symptom (79%), and cluster C patients had a high prevalence of chills (69%). There were 2 clusters (D, n = 29, and E, n = 61) that were predominantly female. These clusters differed by comorbidities: Cluster D had higher prevalence of diabetes, chronic lung disease, and severe obesity (BMI ≥40). Cluster E had the lowest prevalence of mechanical ventilation (5%).
Cluster F (n = 71) patients were the youngest (median age, 43 years) and had few comorbidities. There were few complications reported in these patients relative to the cohort. While 15% of these patients required mechanical ventilation, this cluster had the lowest mortality rate (1%).
Treatment and co-infections by cluster are shown in Supplementary Table 2. Antibiotic use overall was >90% among all clusters, although continued antibiotic use >48 hours after admission was higher among cluster A patients (87%) than others (range, 33%–41%). Cluster A had the highest proportions of hydroxycloroquine (69.8%) and azithromycin (83.0%) use. The proportions of patients receiving corticosteroids and remdesivir were not distinctively different in cluster A. The highest proportions of bacterial co-infections were seen in clusters A (26.4%) and D (17.2%); all patients with ventilator-associated pneumonia were in cluster A, accounting for 5.7% of this group.
Serial Vital Signs and Laboratory Values
Serial vital signs and laboratory data were available for 240 (81%) patients admitted to 5 of the hospitals. The value distribution and number of observations per patient stratified by LOS are shown in Supplementary Table 3. Differences in laboratory values and vital signs were observed in patients with the severe cluster A phenotype compared with the rest of the cohort (Figure 4). Patients in cluster A had a higher heart rate, respiratory rate, and temperature and lower mean arterial pressure (MAP) compared with patients in other clusters. While CRP was most elevated at approximately hospital day 7 in all patients, those in cluster A had higher mean CRP values throughout the hospital course compared with those in other clusters (207 vs 116 mg/L; P < .001). Mean creatinine and BUN were also more elevated in cluster A throughout the hospital course (creatinine 2.3 vs 1.5 mg/dL; P < .001). D-dimer was higher among cluster A patients in the first days of hospitalization. While mean WBC count was higher in cluster A patients (10.8 vs 6.9 × 109/L; P < .001), median nadir absolute lymphocyte count was lower compared with patients in other phenotype clusters (0.71 vs .95 × 109/L; P = .009). AST and ALT were elevated in cluster A patients, particularly at the beginning of the hospital course and during the second week of hospitalization, and there was a 2:1 ratio in average values of AST to ALT. Alkaline phosphatase was similar between cluster A and other clusters until the second week of hospitalization, when cluster A patients had more elevated values compared with the rest of the cohort.
Figure 4.
Key laboratory values among adults hospitalized with COVID-19 by phenotype (n = 240)—Georgia, United States, March 2020. The average daily value among patients in cluster A vs other clusters is shown for 4 key laboratory values. For patients with multiple observations per day, the average is weighted inversely by the number of observations per patient per day. Abbreviations: AST, aspartate aminotransferase; COVID-19, coronavirus disease 2019.
Discharge, Readmissions, and Deaths
Final outcomes were known for 297 (97%) patients. Eleven patients were readmitted to the same hospital system during the data collection period; 6 were readmitted with worsening COVID-19 symptoms, 3 with complaints likely unrelated to COVID-19, and in 2 patients cause of readmission was not available. Among 51 patients (17%) with a recorded cause of death, the primary event leading to death was respiratory failure in 26 (51%) and multiorgan system failure or shock in 13 (26%). Among patients who received IMV, mortality was 44%.
Among 246 patients discharged alive, 205 (83%) were discharged to their prehospital level of care, and 38 (15%) were discharged to a higher level of care than their prehospital baseline (eg, someone who was previously independent at home being discharged home with new services or to a rehabilitation facility) (Supplementary Table 4). There were 37 (12%) patients with new oxygen requirements upon discharge. Three patients were discharged for prolonged mechanical ventilation weaning to long-term acute care facilities, 4 were discharged with a tracheostomy, and 6 were discharged with new RRT needs.
DISCUSSION
In a cohort of 305 hospitalized patients with COVID-19 in Georgia, we conducted a cluster analysis to identify clinical phenotypes. Patient characteristics clustered into patterns based on host factors, symptoms, laboratory findings, and complications. One cluster was characterized by high mortality, elevated inflammatory markers, laboratory evidence of end organ damage, shock, and VTE. These features share similarities to the multisystem inflammatory syndrome in children (MIS-C) with COVID-19 in Europe and the United States [27, 28], and they also share similarities to cytokine release syndrome due to acute infection. Patients with this phenotype were more often older, male, and had a high prevalence of cardiovascular disease compared with other clusters. The phenotype identified in this study may overlap with case reports in adults of an MIS-A phenotype characterized by inflammatory markers and extrapulmonary involvement [29–31].
Acute kidney injury was the most common nonrespiratory complication among this cohort, and some patients required new renal replacement therapy upon discharge. This finding is consistent with those from other reports [3, 10, 13], which suggest that SARS-CoV-2 may have a specific tropism for renal tissue [2]. However, the degree to which acute kidney injury can be attributed to effects of SARS-CoV-2 in our cohort is unclear given the high proportion of shock in some patients. VTE was common, particularly among patients with the severe phenotype. Other studies have described higher rates of VTE among critically ill COVID-19 patients compared with other patients [32]; it is possible that thromboembolic events were underdetected in this cohort if diagnostic imaging was limited for SARS-CoV-2 infection control reasons. Microbiologic evidence of bacterial coinfection was uncommon (~10%), similar to other studies [33], although the widespread use of empiric antibiotics may limit the ability to detect bacterial infections in these patients.
This study has several limitations. Although it included a cohort enrolled from academic, community, and public hospitals, it was limited by the relatively small sample size. Therefore, the phenotypes observed in our cohort may not be generalizable to other patient populations, and our findings need replication. Diagnostic and treatment interventions were performed as needed for routine clinical care and not as part of a protocol; thus, serial laboratory values are likely to be biased toward patients with more severe disease and may not represent patients with milder illness. The interpretation of laboratory values and vital signs over time was affected by censoring. Those with more normal values are likely to be discharged, and those with severely abnormal values may die, which may result in the appearance of misleading trends. Some information, particularly presenting symptoms, may not have been systematically recorded in the medical record. Data were collected on hospitalization early in the course of the US pandemic in Georgia (March and April), and clinical practice and public health guidance have rapidly evolved since then. Decisions about which patients received SARS-CoV-2 testing were not standardized across sites and were likely affected by test availability, and they may have led to increased ascertainment of patients who had fever and respiratory symptoms due to the need to prioritize patients for diagnostic testing. This analysis aimed at exploring clinical phenotypes; prognostic factors identified in this cohort have been carefully described elsewhere [23].
Despite our small cohort size, our analysis has several strengths. It presents a novel strategy to identify clinical phenotypes of COVID and aid in the understanding of therapeutic options. The clustering analysis probes complex interactions between multiple features of the patient and clinical course; this methodology should be considered in other cohorts, at different time points during the pandemic, to verify the consistency of the findings [34]. A high level of data completeness and quality allow for confidence in the ascertainment of complications and final outcomes.
This study illustrates the spectrum of the COVID-19 hospital course from presentation to outcome of patients admitted to the hospital during March 2020. We show heterogeneity in how COVID-19 manifests in adults. These findings improve our understanding of the prognosis of COVID-19 based on comorbidities and early clinical data. Additional research to identify early predictors of a severe phenotype may help focus clinical management strategies in those most at risk. Our data suggest that a phenotype of inflammation and multisystem involvement exist in adults; this may represent severe acute COVID-19 but may also overlap with MIS [27, 28]. The impact of different clinical management strategies for severe COVID-19 should be further explored as treatment guidelines evolve, and additional investigation into the pathophysiology of severe disease may inform optimal treatment strategies for different clinical phenotypes.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Chad M. Heilig, Mary E. Evans, Mohleen Kang, Nathan W. Furukawa, Deblina Datta, CDC COVID-19 Response Clinical Team; William A. Bornstein, Debra E. Houry, Kymmi L. Cooley, Glenn Hilburn, Jonathan J. Perkins, Chad Robichaux, Tripura Vadlamani, Keneisha Williams, informatics and information technology staff members at collaborating hospitals; health care personnel on the front lines of COVID-19 care in Georgia.
Financial support. This study was supported by the Centers for Disease Control and Prevention.
Potential conflicts of interest. Dr. Blum reported being a consultant for Clew Medical. No other disclosures were reported. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This investigation was determined by the CDC and DPH to be public health surveillance and thus did not require patient consent, as no personal identifiable data were used. The design of the work was approved by CDC ethics committees.
References
- 1. Centers for Disease Control and Prevention COVID data tracker. Available at: https://www.cdc.gov/covid-data-tracker/index.html#cases. Accessed 27 July 2020.
- 2. Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 2020; 383:590–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirsch JS, Ng JH, Ross DW, et al. ; Northwell COVID-19 Research Consortium; Northwell Nephrology COVID-19 Research Consortium Acute kidney injury in patients hospitalized with COVID-19. Kidney Int 2020; 98:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klok FA, Kruip M, van der Meer NJM, et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oxley TJ, Mocco J, Majidi S, et al. Large-vessel stroke as a presenting feature of Covid-19 in the young. N Engl J Med 2020; 382:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li H, Liu L, Zhang D, et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. Lancet; 395:1517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382:1708–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395:P1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med; 382:2372–4. [DOI] [PMC free article] [PubMed]
- 11. Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019—COVID-NET, 14 States, March 1–30, 2020. MMWR Morb Mortal Wkly Rep 2020; 69;458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020; 323:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richardson S, Hirsch JS, Narasimhan M, et al. ; the Northwell COVID-19 Research Consortium Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City Area. JAMA 2020; 323:2052–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Covid-19 in critically Ill patients in the Seattle region—case series. N Engl J Med 2020; 382:2012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cummings MJ, Baldwin MR, Abrams D, et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet; 395:1763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seymour CW, Kennedy JN, Wang S, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA 2019; 321:2003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gårdlund B, Dmitrieva NO, Pieper CF, et al. Six subphenotypes in septic shock: latent class analysis of the PROWESS Shock study. J Crit Care 2018; 47:70–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore WC, Hastie AT, Li X, et al. ; National Heart, Lung, and Blood Institute’s Severe Asthma Research Program Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J Allergy Clin Immunol 2014; 133:1557–63.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jackson BR, Natarajan P, Rossow J, et al. Predictors on admission of mechanical ventilation and death in an observational cohort of adults hospitalized with COVID-19. Clin Infect Dis. 2020:ciaa1459. doi: 10.1093/cid/ciaa1459. [DOI] [PMC free article] [PubMed]
- 20. Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium The REDCap Consortium: building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harris PA, Taylor R, Thielke R, et al. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gold JAW, Wong KK, Szablewski CM, et al. Characteristics and clinical outcomes of adult patients hospitalized with COVID-19 - Georgia, March 2020. MMWR Morb Mortal Wkly Rep 2020; 69:545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang K, Zuo P, Liu Y, et al. Clinical and laboratory predictors of in-hospital mortality in patients with COVID-19: a cohort study in Wuhan, China. Clin Infect Dis. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Y, Lu X, Li Y, et al. Clinical course and outcomes of 344 intensive care patients with COVID-19. Am J Respir Crit Care Med 2020; 201:1430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gower JC. A general coefficient of similarity and some of its properties. Biometrics 1971; 27:857–71. [Google Scholar]
- 26. Kaufman L, Rousseeuw P. Finding Groups in Data: An Introduction to Cluster Analysis. New York: Wiley, 1990.
- 27. Dufort EM, Koumans EH, Chow EJ, et al. ; New York State and Centers for Disease Control and Prevention Multisystem Inflammatory Syndrome in Children Investigation Team Multisystem inflammatory syndrome in children in New York State. N Engl J Med 2020; 383:347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feldstein LR, Rose EB, Horwitz SM, et al. ; Overcoming COVID-19 Investigators; CDC COVID-19 Response Team Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med 2020; 383:334–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shaigany S, Gnirke M, Guttmann A, et al. An adult with Kawasaki-like multisystem inflammatory syndrome associated with COVID-19. Lancet 2020; 396:e8–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Newton-Cheh C, Zlotoff DA, Hung J, et al. Case 24–2020: a 44-year-old woman with chest pain, dyspnea, and shock. N Engl J Med 2020; 383:475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morris SB, Schwartz NG, Patel P, et al. Case series of multisystem inflammatory syndrome in adults associated with SARS-CoV-2 infection—United Kingdom and United States, March-August 2020. MMWR Morb Mortal Wkly Rep 2020; 69:1450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poissy J, Goutay J, Caplan M, et al. Pulmonary embolism in COVID-19 patients: awareness of an increased prevalence. Circulation 2020; 142(2):184–6. [DOI] [PubMed]
- 33. Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect 2020; 81:266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basile AO, Ritchie MD. Informatics and machine learning to define the phenotype. Expert Rev Mol Diagn 2018; 18:219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.