Abstract
Background
Amid the enduring pandemic, there is an urgent need for expanded access to rapid, sensitive, and inexpensive coronavirus disease 2019 (COVID-19) testing worldwide without specialized equipment. We developed a simple test that uses colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) to detect severe acute resrpiratory syndrome coronavirus 2 (SARS-CoV-2) in 40 minutes from sample collection to result.
Methods
We tested 135 nasopharyngeal specimens from patients evaluated for COVID-19 infection at Massachusetts General Hospital. Specimens were either added directly to RT-LAMP reactions, inactivated by a combined chemical and heat treatment step, or inactivated then purified with a silica particle–based concentration method. Amplification was performed with 2 SARS-CoV-2-specific primer sets and an internal specimen control; the resulting color change was visually interpreted.
Results
Direct RT-LAMP testing of unprocessed specimens could only reliably detect samples with abundant SARS-CoV-2 (>3 000 000 copies/mL), with sensitivities of 50% (95% CI, 28%–72%) and 59% (95% CI, 43%–73%) in samples collected in universal transport medium and saline, respectively, compared with quantitative polymerase chain reaction (qPCR). Adding an upfront RNase inactivation step markedly improved the limit of detection to at least 25 000 copies/mL, with 87.5% (95% CI, 72%–95%) sensitivity and 100% specificity (95% CI, 87%–100%). Using both inactivation and purification increased the assay sensitivity by 10-fold, achieving a limit of detection comparable to commercial real-time PCR-based diagnostics.
Conclusions
By incorporating a fast and inexpensive sample preparation step, RT-LAMP accurately detects SARS-CoV-2 with limited equipment for about US$6 per sample, making this a potentially ideal assay to increase testing capacity, especially in resource-limited settings.
Keywords: COVID-19, diagnostics, isothermal amplification, LAMP, nucleic acid technology, rapid tests, SARS-CoV-2
The worldwide spread of coronavirus disease 2019 (COVID-19) has led to an unprecedented need for rapid, accurate, affordable, and readily available severe acute resrpiratory syndrome coronavirus 2 (SARS-CoV-2) tests. Hundreds of molecular assays for the detection of SARS-CoV-2 RNA have received regulatory approval in the United States, Europe, and Asia to date, but they have not met the need for widespread testing demand due to several critical factors, including a high cost per reportable result (in the US$15–40 range) and costly upfront capital equipment such as proprietary testing platforms, real-time amplification and detection platforms, and automated RNA extraction equipment and consumables, which are in limited supply [1, 2]. In general, these tests must be performed by highly trained molecular laboratory professionals in well-resourced laboratories. The development of more simple, rapid, and low-cost diagnostics that do not rely on the same supply chains, reagents, or consumables as other COVID-19 tests could help rapidly and substantially expand testing capabilities, especially in resource-limited settings.
Alternative rapid tests to detect SARS-CoV-2 rely on detection of viral antigen using lateral-flow immunoassays (LFAs). While extremely convenient, respiratory viral LFAs tend to be less sensitive than nucleic acid amplification methods, with an average sensitivity of 61%–75% [3, 4]. As an alternative to antigen detection methods and resource-intensive real-time PCR tests, isothermal amplification methods such as loop-mediated isothermal amplification (LAMP) [5] and recombinase polymerase amplification (RPA) [6, 7] enable sensitive detection of nucleic acids with just the use of a stable heat source in as little at 15 minutes. Colorimetric RT-LAMP expands on the basic LAMP technology with a 1-pot reaction that contains both reverse transcriptase and DNA polymerase with visual detection of nucleic acid amplification due to a pH indicator dye within the master mix, obviating the need for additional detection equipment [8]. LAMP has been used to detect many pathogens including SARS [9], Zika virus [10], Mycobacterium tuberculosis [11], malaria [12], and human leishmaniasis [13]. RT-LAMP has also been performed for SARS-CoV-2 on extracted RNA with a colorimetric read-out [14–18] and for subsequent CRISPR-Cas12-based detection [19].
To develop a truly accessible sample-to-answer nucleic acid–based diagnostic test, one must couple a simple detection method with an equally simple sample preparation method. The simplest sample preparation method is to directly add sample to the amplification reaction [18], but this can be problematic for several reasons. Endogenous RNases present in body fluids can degrade target RNA, and infectious virus contained in the sample may increase the risk of laboratory-acquired infection among technologists who handle the specimens. While heat inactivation alone can partially reduce RNase activity and inactivate virions [20], we and others have shown that RNAses can be fully inactivated by combining heat inactivation with chemical inactivation using the shelf-stable reducing agent Tris(2-carboxyethyl)phosphine (TCEP) and the divalent cation chelator ethylenediaminetetraacetic acid (EDTA) [21, 22]. An additional challenge specific to colorimetric RT-LAMP is that the buffer and phenol red in viral transport media (VTM) may interfere with the pH-mediated color change. To circumvent these issues, most SARS-CoV-2 molecular tests use extracted RNA as input, but RNA extraction is expensive, time-consuming, laborious, and extraction kits are in short supply.
As part of an ongoing quality improvement initiative, we tested 135 clinical nasopharyngeal (NP) samples collected from Massachusetts General Hospital patients who were admitted or evaluated in the emergency department during the COVID-19 pandemic to determine the testing characteristics of 3 diagnostic strategies using colorimetric RT-LAMP (Figure 1). The first was the direct-from-sample approach, including samples collected in either universal transport media or sterile physiologic saline. The second incorporated an upfront 5-minute chemical and heat inactivation step to inhibit RNases and lyse virions. The third strategy uses the aforementioned inactivation step with an additional nucleic acid purification step using a solution of silica particles (“glass milk”) to increase the effective sample input volume into the RT-LAMP reaction [21].
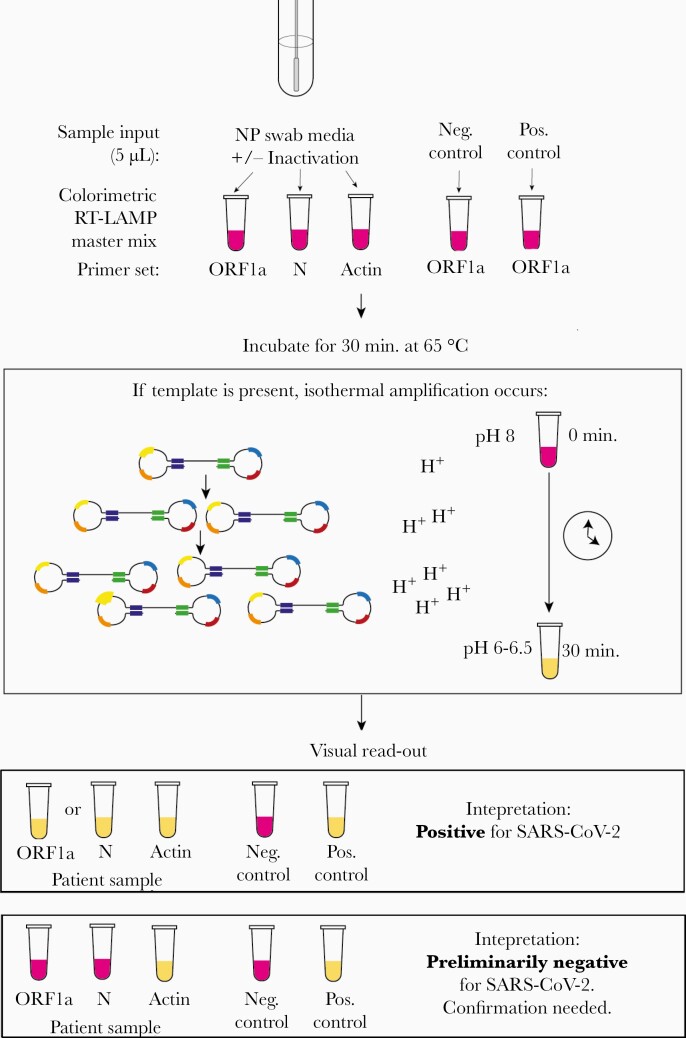
Figure 1.
Schematic for the use of RT-LAMP directly from NP specimen using 2 SARS-CoV-2 specific primers, which target the ORF1a and N genes, and 1 internal specimen control targeting the human actin gene. Before sample addition to the RT-LAMP reaction, the NP specimen can undergo a 5-minute heat and chemical inactivation step to destroy endogenous RNases and lyse viral particles and human cells. The RT-LAMP reaction occurs at 65°C for 30 minutes, during which the amplification of SARS-CoV-2 RNA generates protons that decrease the pH of the reaction mix and result in a color change due to the media’s colorimetric pH indicator. Samples are removed from the heat block, immersed in ice to enhance the color brightness, and color change is visually determined. If the controls are valid, a yellow color change with the ORF1a and/or N gene primers indicates the presence of SARS-CoV-2 RNA in the sample. Abbreviations: NP, nasopharyngeal; RT-LAMP, reverse transcription loop-mediated isothermal amplification; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
METHODS
Clinical Sample Collection, qRT-PCR, and Storage
Nasopharyngeal samples were collected in 1 mL of sterile physiologic saline from the inpatient units and the emergency department (ED) of Massachusetts General Hospital (MGH) between March and April 2020. The inpatient samples were a prospectively collected convenience set obtained from patients whose COVID-19 status was known (20 qPCR positive, 17 qPCR negative). The ED samples were collected prospectively as part of a laboratory quality improvement initiative from patients who presented within a 24-hour period and required clinical COVID-19 testing (22 qPCR positive, 45 qPCR negative). In addition, the nasopharyngeal samples collected in 3 mL of VTM were obtained from excess material collected for routine clinical care (16 qPCR positive, 15 qPCR negative).
Upon receipt in the laboratory, samples were tested with a Food and Drug Administration (FDA) Emergency Use Authorization (EUA)–approved quantitative real-time PCR method: a lab-developed test (LDT) based on the US Centers for Disease Control and Prevention (CDC) assay, the cobas SARS-CoV-2 test for the 6800 system (Roche Diagnostics, Bazel, Switzerland), or the Xpert Xpress SARS-CoV-2 test (Cepheid, Sunnyvale, CA, USA). Of the 42 qPCR-positive saline samples tested using RT-LAMP, 30 had Ct values obtained using the cobas instrument, 11 with the LDT, and 1 with the Xpert. The cobas 6800 system’s cycle threshold tends to be within 2 cycles of the LDT's. If the Ct of a saline specimen was not available, the Ct from the paired VTM specimen that was collected simultaneously was used as a proxy. Though these qPCR assays are not truly quantitative, approximate conversions between cycle thresholds and viral copies/µL were calculated with a standard curve generated on the LDT by spiking 0, 101, 102, 103, and 104 copies/µL of SARS-CoV-2 N gene RNA into SARS-CoV-2-negative nasopharyngeal specimens. Samples were aliquoted and promptly frozen at –20°C for additional testing to avoid RNA degradation. RT-LAMP assay performance was not affected after samples underwent a freeze-thaw cycle.
Patient Consent Statement
This study was approved by the Partners Human Research Committee at the Massachusetts General Hospital with a waiver of written informed consent.
RT-LAMP Primers
The SARS-CoV-2 ORF1a gene (HMS Assay 1e) [21], SARS-CoV-2 N gene (NEB N-A) [23], and human actin B gene (generously provided by New England Biolabs) primer sequences are listed in Supplementary Table 1. The ORF1a primers were combined into a 10X primer stock using 16 μM of Forward Inner Primer (FIP), 16 μM of Backward Inner Primer (BIP), 2 μM of F3, 2 μM of B3, 4 μM of Forward Loop (LF), and 4 μM of Backward Loop (LB). The N gene and human actin primer stocks consisted of the same primer proportions at a 25X concentration.
RT-LAMP Assay
RT-LAMP testing was performed in biosafety level 2, Clinical Laboratory Improvement Amendments (CLIA)–certified clinical laboratory space. Regardless of the sample preparation method, each sample was amplified with 2 SARS-CoV-2-specific primer sets for the ORF1a and N genes. An additional primer set for the human actin B gene also served as an internal specimen control to detect the presence of inhibitory substances. A negative and positive control were tested with every set of clinical samples. Each 25-µL RT-LAMP reaction was performed as described by the manufacturer’s protocols with WarmStart Colorimetric RT-LAMP 2X Master Mix (New England Biolabs, Ipswich, MA, USA) using a 1-µL sample input for samples collected in VTM and a 5-µL input for samples collected in normal saline. After the 30-minute heating step, the results were visually interpreted (Figure 1, Table 1). The interpretative criteria are listed in Table 1. After interpretation, reaction tubes were discarded or stored in sealed bags without re-opening to prevent postamplification contamination of workspaces.
Table 1.
Interpretation Matrix
| RT-LAMP Primer Set Result | |||
|---|---|---|---|
| N | ORF1a | Actin | Action |
| + | + | ± | Report “Positive for SARS-CoV-2 (COVID-19).” |
| - | + | ± | Report “Positive for SARS-CoV-2 (COVID-19).” |
| + | - | ± | Report “Positive for SARS-CoV-2 (COVID-19).” |
| - | - | + | Reflex to qPCR or glass milk purification. |
| - | - | - | Invalid result. Report: “Specimen re-collection and submission is recommended.” |
| Ambiguous color change | + | Indeterminant result. Repeat test. | |
Abbreviations: COVID-19, coronavirus disease 2019; qPCR, quantitative polymerase chain reaction; RT-LAMP, reverse transcription loop-mediated isothermal amplification; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Inactivation and Purification Procedures
The 100X inactivation reagent and purification reagents were prepared as described elsewhere [21]. The inactivation reagent was comprised of 0.25 M of Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl; MilliporeSigma, Burlington, MA, USA), 0.1 M of RNase-free EDTA (ThermoFisher Scientific, Waltham, MA, USA), and 1.1 N of NaOH, diluted in UltraPure water (ThermoFisher Scientific, 10977015). The saline NP sample was mixed with 1/100th volume of the 100X TCEP/EDTA-based inactivation reagent and heated at 95°C for 5 minutes. The sample was then cooled on ice and directly added to the RT-LAMP reaction or used for purification. Purification was performed using glass milk, a predecessor of today’s silica-based column purification methods, which is comprised of a suspension of clean silicon dioxide particles in an equal volume of water [21]. When purification was performed, 250–500 µL of the inactivated sample was mixed with 5 µL of glass milk in a 1.5-mL tube, thoroughly resuspended, and mixed with half the initial sample volume of binding reagent. The binding reagent was comprised of 6 M of NaI (MilliporeSigma), 10 mM of HCl (Millipore Sigma), and 2% Triton X-100 (MilliporeSigma). The sample was then incubated at room temperature for 10 minutes with manual inversions approximately every 2 minutes to resuspend the silica. The samples were briefly spun in a mini benchtop centrifuge for several seconds, and the supernatant was poured off. The pellet was washed with 700 µL of 80% ethanol and briefly spun. The supernatant was poured off again and briefly respun. Any visibly remaining ethanol was removed with a P20 pipette, and the pellet was air-dried on a heat block at 65°C for 5 minutes or until the pellet was visibly dry. Twenty-five microliters of colorimetric RT-LAMP reaction mix was added to the pellet, resuspended, and transferred to a 0.2-mL tube for incubation at 65°C for 30 minutes, briefly placed in ice to enhance the color change, and visually inspected.
Limit of Detection
An initial limit of detection (LoD) study was performed for each SARS-CoV-2 primer by spiking in serially diluted synthetic SARS-CoV-2 RNA (Twist Bioscience, San Francisco, CA, USA) into an inactivated, COVID-negative nasopharyngeal saline media. Five microliters of sample was tested in triplicate at final concentrations of 104, 103, 100, 50, 25, 10, 5, and 0 copies per µL. An additional 20 replicates were performed at the concentration predicted to be the LoD, as defined by the US FDA at the lowest concentration at which 19/20 replicates are positive. The purification dilution experiments were performed by making serial dilutions of a SARS-CoV-2-positive sample in SARS-CoV-2-negative nasopharyngeal specimens.
Cross-Reactivity
Cross-reactivity of the N gene primer set with SARS-CoV-1 and Middle East respiratory syndrome (MERS) was assessed with plasmid controls (Integrated DNA Technologies, Coralville, IA, USA). Plasmids containing the ORF1a region of SARS-CoV-1 and MERS were not available; thus in vitro testing of ORF1a primers with SARS-CoV-1 and MERS was not performed. In addition, to assess for primer cross-reactivity with common respiratory pathogens, the assay was performed on 10 clinical samples collected before the outbreak of SARS-CoV-2 (February through April 2019) that were known to contain a respiratory virus confirmed using clinical multiplexed PCR testing (Film Array Respiratory Panel 2, BioFire Diagnostics, Salt Lake City, UT, USA). As the samples were collected in VTM, RNA was first extracted from 140 µL of nasopharyngeal swab VTM using a Viral RNA Mini Kit (Qiagen, Hilden, Germany) and eluted in 60 µL, and 5 µL of extracted RNA was added to each RT-LAMP reaction.
Statistical Analyses
Confidence intervals were calculated using the Wilson/Brown method in GraphPad Prism (version 7.0a).
RESULTS
Comparison of VTM vs Saline Transport Media
With the goal of directly adding unprocessed sample into the RT-LAMP reaction (Figure 2A), we first optimized the transport media input volume. We added increasing amounts of transport media to a standardized reaction containing 1000 copies of SARS-CoV-2 control RNA. VTM interfered with the colorimetric readout, with complete inhibition of the pH-mediated color change with 3 µL of input, while saline had little effect (Figure 2B). Subsequent experiments were conservatively performed with 1 µL of VTM and 5 µL of saline sample input to facilitate robust assay performance in the setting of intrinsic clinical sample variability.
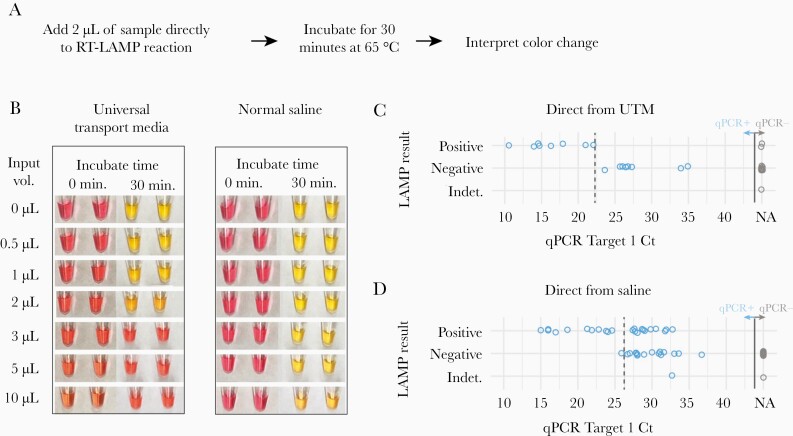
Figure 2.
Detection of SARS-CoV-2 directly from nasopharyngeal samples collected in VTM or 0.9% normal saline. A, Schematic for the 35-minute protocol of direct-from-sample testing. B, Determination of the optimal sample input volume for VTM and saline using a standardized 1000-copy/µL synthetic SARS-CoV-2 input. Samples are pictured before and after the 30-minute amplification step. C, Comparison of the sensitivity of qPCR with RT-LAMP with direct addition of 1 µL of clinical NP specimen collected in universal transport media (16 qPCR-positive samples, 15 qPCR-negative samples) and testing using the N gene primers alone. D, Comparison of the sensitivity of qPCR to RT-LAMP with direct addition of 5 µL of clinical NP samples collected in saline (40 qPCR-positive samples, 45 qPCR-negative samples), using both the N gene and ORF1a primer sets. One invalid result occurred from a sample that had a negative human actin control and was noted to be bloody. The approximate clinical limit of detection is shown with a dotted line. Abbreviations: NA, no amplification; NP, nasopharyngeal; qPCR, quantitative polymerase chain reaction; RT-LAMP, reverse transcription loop-mediated isothermal amplification; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VTM, viral transport media.
Direct-From-Sample Detection
We next asked whether SARS-CoV-2 could be consistently detected from unprocessed clinical VTM and saline samples. We tested 16 qPCR-positive and 15 qPCR-negative NP specimens collected in VTM by adding 1 µL of VTM directly to the RT-LAMP reactions. When compared with qPCR on an FDA EUA–approved platform, the sensitivity of RT-LAMP performed with the SARS-CoV-2 N gene and human actin gene primer sets and direct addition of a VTM specimen was only 50% (95% CI, 28%–72%) (Figure 2C). RT-LAMP could only detect VTM samples with a cycle threshold <23, corresponding to ~3 000 000 copies/mL in internal validation studies. There were 2 false-positive results, possibly related to interpretation difficulties due to a limited dynamic color range and higher background of the N gene primer set.
We next tested NP specimens directly inoculated into saline transport media using both the N and ORF1a primer sets and the interpretation criteria listed (Table 1). Of 40 qPCR-positive samples tested with direct saline addition, RT-LAMP consistently detected samples with cycle thresholds <25 and as high as 32, yet the assay sensitivity was only 59% (95% CI, 43%–73%) (Figure 2D). Among 45 qPCR-negative saline samples tested, the color changes were crisper and easier to interpret compared with VTM, and there were no false positives. This was consistent with in silico and in vitro analyses that did not demonstrate cross-reactivity between the ORF1a and N gene primer sets and other coronaviruses or respiratory viruses (Supplementary Table 2). Overall, normal saline appeared to be a more amenable sample collection media compared with VTM, but direct sample addition to the RT-LAMP reaction remained too insensitive for routine clinical use.
Assay Performance With Sample Inactivation
We next tested whether the assay sensitivity would improve with a simple inactivation step consisting of TCEP/EDTA addition to neutralize endogenous RNase activity and heat to release the viral RNA contained within virions and human cells (Figure 3A) [20-22]. The inactivation step appeared highly effective in nasopharyngeal specimens spiked with serially diluted SARS-CoV-2 control RNA (Supplementary Figure 1) and enabled performance of limit of detection (LoD) studies. As little as 25 copies/µL (25 000 copies/mL) of control SARS-CoV-2 RNA could be detected in all 20 replicates using the ORF1a primers, and the N gene primers appeared slightly less sensitive (Figure 3B).
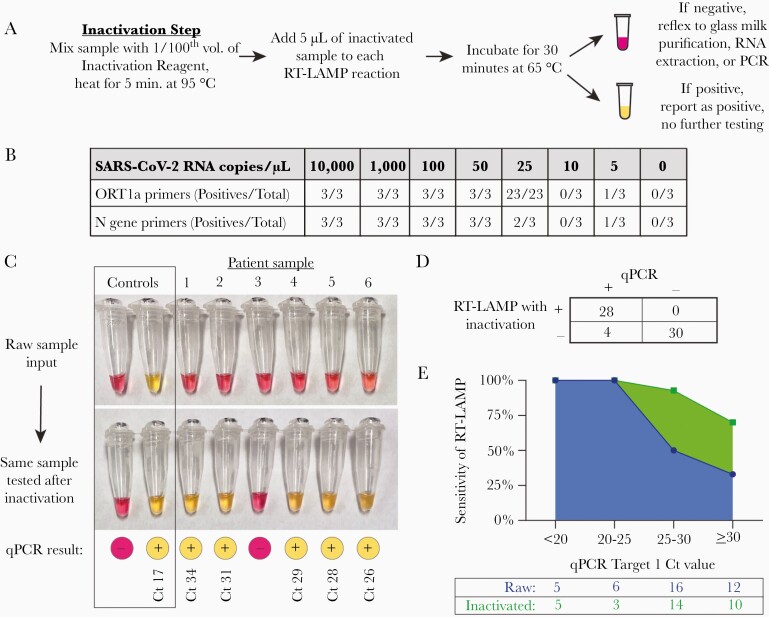
Figure 3.
Detection of SARS-CoV-2 after inactivation of RNases and viral lysis from clinical nasopharyngeal swabs collected in saline. A, Schematic for a 40-minute rule-in protocol where samples are first treated with an inactivation reagent (TCEP/EDTA), and heated to 95°C for 5 minutes before sample addition to the RT-LAMP reaction. B, Determination of the analytic sensitivity of the RT-LAMP assay with each primer set, as determined by synthetic SARS-CoV-2 RNA spiked into inactivated SARS-CoV-2-negative nasopharyngeal samples collected in saline. C, Representative clinical samples illustrating the improvement of RT-LAMP sensitivity after inactivation, with corresponding qPCR results and Ct values. Amplification reactions using the ORF1a primer set are shown. D, Overall performance of RT-LAMP in 62 inactivated clinical samples. RT-LAMP results were categorized as positive or negative using the criteria outlined in Table 1. E, Sensitivity of RT-LAMP with or without inactivation as a function of the input SARS-CoV-2 RNA concentration, as determined by qPCR. The total number of samples tested within each Ct range is shown in the table. Abbreviations: Ct, cycle threshold; qPCR, quantitative polymerase chain reaction; RT-LAMP, reverse transcription loop-mediated isothermal amplification; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TCEP, tris(2-carboxyethyl)phosphine.
We then incorporated the inactivation step into the testing of clinical samples and observed a substantial improvement in the assay sensitivity. COVID-positive samples that were previously falsely negative with unprocessed sample addition were subsequently positive with both SARS-CoV-2 primer sets after inactivation (Figure 3C). Importantly, COVID-negative samples remained negative after inactivation (Figure 3C, Sample 3). To systematically test the efficacy of inactivation, we repeated the assay using inactivation with the available 32 qPCR-positive and 30 qPCR-negative samples that had originally been tested by direct sample addition. We found 100% specificity (95% CI, 87%–100%) and 87.5% sensitivity (95% CI, 72%–95%) with this sample set (Figure 3D). In addition, we found that inactivation enabled the detection of >95% of samples with a cycle threshold <30, corresponding to about 40 viral copies/µL (40 000 copies/mL), and could detect SARS-CoV-2 in samples with cycle thresholds as high as 33.5 (Figure 3E). Thus, the combination of a very simple inactivation step followed by RT-LAMP provided a robust rule-in test for SARS-CoV-2 with a sample-to-result time of ~40 minutes and minimal labor.
Assay Performance After Sample Purification
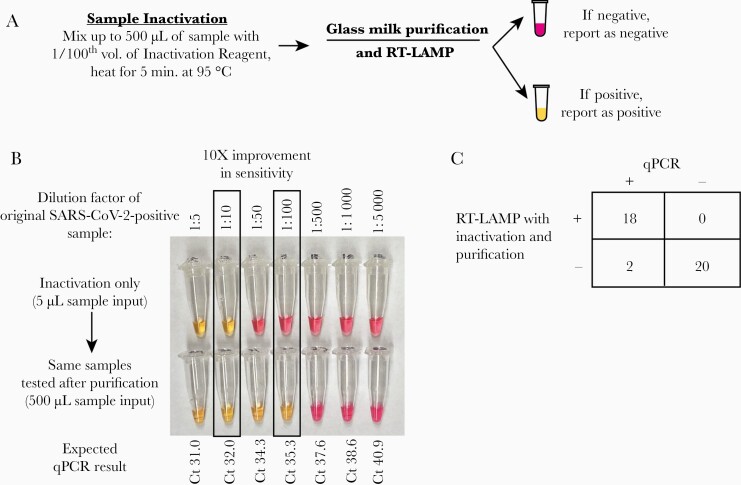
Finally, we asked whether increasing the effective sample input volume using a concentration and purification step could enable detection of samples with very low levels of virus that would otherwise not be detected (falsely negative) with inactivation alone (Figure 4A). We used the glass milk protocol to concentrate up to 500 µL of heat and TCEP/EDTA-inactivated sample into a single RT-LAMP reaction [21]. Using serial dilutions of qPCR-positive samples, we demonstrated that glass milk purification improved the LoD by 10-fold (Figure 4B). We then retested 20 qPCR-positive and 20 qPCR-negative samples using glass milk inactivation. Unsurprisingly, qPCR-positive samples that were previously positive using inactivation alone were also positive after purification. Two of the 4 clinical samples that were falsely negative by inactivation alone were positive after the addition of a purification step, demonstrating the improved limit of detection using glass milk purification. The 2 remaining specimens that tested negative after the purification step may have been truly qPCR-negative due to undergoing ~3 freeze-thaw cycles, but insufficient material remained for repeat testing. Thus, we may be underestimating the sensitivity of inactivation with purification. All 20 qPCR-negative samples tested negative after purification (100% specificity) (Figure 4C). Overall, purification improved the assay sensitivity by increasing the effective sample input volume.
Figure 4.
Further improvement in sensitivity by reflexing RT-LAMP-negative inactivated samples to a glass milk purification procedure. A, Schematic for combining inactivation with glass milk purification. B, Demonstration that purification improves the assay detection limit by ~10-fold using a serially diluted SARS-CoV-2-positive patient’s nasopharyngeal specimen. Using the ORF1a primer set, the lowest dilution detected from the inactivated sample was 1:10. After purifying 500 µL of each dilution, the 1:100 dilution could be detected by the same primer set. C, Overall performance of RT-LAMP in 40 clinical samples that were inactivated and then purified, including 4 specimens that were falsely negative with inactivation alone. Abbreviations: RT-LAMP, reverse transcription loop-mediated isothermal amplification.
DISCUSSION
Here we have demonstrated a simple and inexpensive loop-mediated isothermal amplification assay for the detection SARS-CoV-2 that achieves 87.5% overall sensitivity and 100% specificity compared with qPCR when performed directly from a clinical sample with the inclusion of an upfront, 5-minute sample inactivation step. Performing an additional glass milk purification step resulted in increased assay sensitivity that was comparable to qPCR with an additional assay cost of only US$0.07 per sample. Laboratorians can determine whether the increased sensitivity afforded by glass milk purification justifies the additional labor and 20-minute longer assay turnaround time based on considerations such as their regional disease prevalence (Supplementary Table 3). This assay can be performed in any clinical laboratory or even ad hoc settings, like a mobile laboratory, as it does not require any specialized equipment or highly trained laboratory personnel. As the required reagents are easily manufactured by multiple manufacturers, access to this test does not rely on traditional commercial diagnostic supply chains that have hindered the broad distribution of SARS-CoV-2 testing. The manufacturer of colorimetric RT-LAMP master mix has large-scale production in place, with millions of reactions’ worth of product available. We estimate an overall per-sample cost ~US$6, though personnel and overhead costs will also contribute and vary greatly depending upon the setting. We believe this 40-minute sample-to-answer assay addresses a pressing need for COVID-19 diagnostics worldwide.
Sample preparation is often a time-consuming and expensive step of the testing process. We have demonstrated that RT-LAMP can be performed directly from a nasopharyngeal sample but that the assay sensitivity increases by 30% with chemical RNase inactivation using TCEP/EDTA and heat-mediated lysis. In addition to improving assay performance, the inactivation step described here likely reduces the infectivity of the sample as well [24], reducing the risk of exposure for laboratory personnel. The assay’s sensitivity is further improved by glass milk purification, which is both extremely inexpensive compared to commercial RNA extraction kits (~US$5 per sample) and can be performed without a microcentrifuge, enabling its use in low-resource laboratory environments.
We foresee this assay being used in 3 ways. The first is a 40-minute rule-in test that uses inactivation followed by RT-LAMP. If the sample is positive and controls are valid, the test is reported as positive for SARS-CoV-2 to enable effective infection control practices and clinical management. If a rule-out test is desired, a sample that tests negative using inactivation alone can then be reflexed to a glass milk purification or a qPCR-based test. The third method tests pooled specimens using the inactivation and purification protocols for the rapid screening of large groups, that is, in a school or employment setting, assuming adequate assay performance in this setting is demonstrated with additional clinical validation.
While this clinical validation is focused on nasopharyngeal swabs, which are recommended by the World Health Organization and US CDC as the most sensitive specimen type for SARS-CoV-2 detection [25, 26], the same methods can be applied to other sample types as well, possibly including saliva [21]. Oropharyngeal specimens are likely to be compatible given their similar composition to nasopharyngeal specimens. Sputum is generally a challenging sample type due to high viscosity and heterogeneity, but we expect the TCEP/EDTA chemical inactivation step to mimic the current recommendation to pretreat sputum with dithiothreitol, an alternative reducing agent to TCEP [27]. Further clinical studies to assess the range of compatible sample types are underway.
There are several limitations of this assay. The assay is qualitative and does not provide a semiquantitative cycle threshold number. Additionally, the visual interpretation affords substantial flexibility, but it can also be prone to user errors. Objective color measurements can be performed by measuring the absorbance at 432 and 560 nm, or potentially with a smartphone application [28]. It is critical to use the positive and negative controls as interpretative aids to avoid misinterpreting an orange intermediate color change as positive. The N gene primer set is more likely to give subtle background color changes, and additional primer sets will be tested in the future. Like any nucleic acid amplification test, systems must be in place to avoid environmental and sample contamination with postamplification products. One such precaution is to refrain from opening the reaction vessel after amplification; reactions should be discarded or transferred to a sealed container for later reference, as the color change remains stable for days to weeks. While the assay generally requires very little infrastructure, the operator must abide by laboratory biosafety guidelines, and the procedure is most safely performed within a class II biosafety cabinet, although we recognize that in many settings this may not be possible [29]. Additionally, the RT-LAMP master mix currently requires storage at –20°C, which is not ideal for low-resource or remote settings, but this may be ameliorated by lyophilization.
In summary, we present the implementation of a simple RT-LAMP assay for the detection of SARS-CoV-2 that achieves a high sensitivity and specificity in a challenging clinical sample set obtained during the peak of the spring 2020 COVID-19 pandemic. Future work includes validating additional sample types, validating a specimen pooling approach, eliminating the cold chain requirement through reagent lyophilization, and assessing the feasibility and performance of the assay in resource-limited settings.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Maria Vareschi, John Texeira, and MGH ED staff enabled the ED sample collection. Dr. Tyler Miller and MGH nursing staff enabled the inpatient saline sample collection. Dr. Hetal Marble provided insight into the development of the assay into a commercially available kit. Dr. Virginia Pierce and Dr. Sarah Turbett provided feedback and invaluable guidance on the clinical validation process. Last but certainly not least, the incredibly dedicated MGH microbiology technologists performed the qPCR assays as part of their daily excellence in clinical care.
Financial support. New England Biolabs generously provided the N gene and human actin primer sets and initial master mix. Funding was provided to C.L.C. from the Howard Hughes Medical Institute. Neither organization had a role in study design, data collection and interpretation, or the decision to submit the work for publication.
Potential conflicts of interest. M.N.A. is a co-founder, consultant, and equity interest holder in Day Zero Diagnostics, outside of the submitted work. N.A.T. is an employee of New England Biolabs and is on multiple patents related to pH-based amplification detection and DNA polymerases. J.A.B. reports grants from Zeus Scientific, bioMerieux, Lyme Disease Biobank Foundation, and Immunetics, personal fees from T2 Biosystems, DiaSorin, and Roche Diagnostics, and grants from Bay Area Lyme Foundation, outside the submitted work. Harvard University has filed patent applications regarding this technology on behalf of the inventors. Harvard believes strongly that while intellectual property rights serve to incentivize creation of new products, such rights should not become a barrier to addressing urgent and essential health-related needs. To address the global COVID-19 pandemic, Harvard is implementing technology transfer strategies to allow for rapid utilization of available technologies that may be useful for preventing, diagnosing, and treating COVID-19 infection. More information can be found at otd.harvard.edu. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. M.N.A., G.E.G.M, and E.S.R. conceived and executed the clinical assessment and validation studies. B.A.R. and C.L.C. developed the inactivation and purification sample preparation methods and provided reagents for sample preparation. B.A.W., J.K.M.L., and J.A.B. contributed to the clinical study design and execution. N.A.T. contributed expertise in colorimetric RT-LAMP and provided primers. All authors discussed the results and contributed to the final manuscript.
References
- 1. Zhen W, Smith E, Manji R, et al. Clinical evaluation of three sample-to-answer platforms for the detection of SARS-CoV-2. J Clin Microbiol 2020; 58:e00783–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhen W, Manji R, Smith E, Berry GJ. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol 2020; 58:e00743–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bruning AHL, Leeflang MMG, Vos JMBW, et al. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin Infect Dis 2017; 65:1026–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lambert-Niclot S, Cuffel A, Le Pape S, et al. Evaluation of a rapid diagnostic assay for detection of SARS CoV-2 antigen in nasopharyngeal swab. J Clin Microbiol 20202; 58:e00977–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagamine K, Hase T, Notomi T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 2002; 16:223–9. [DOI] [PubMed] [Google Scholar]
- 6. Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol 2006; 4:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behrmann O, Bachmann I, Spiegel M, et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (exo-IQ). Clin Chem 2020; 66:1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tanner NA, Zhang Y, Evans TC Jr. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 2015; 58:59–68. [DOI] [PubMed] [Google Scholar]
- 9. Hong TC, Mai QL, Cuong DV, et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol 2004; 42:1956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calvert AE, Biggerstaff BJ, Tanner NA, et al. Rapid colorimetric detection of Zika virus from serum and urine specimens by reverse transcription loop-mediated isothermal amplification (RT-LAMP). PLoS One 2017; 12:e0185340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. World Health Organization. The use of loop-mediated isothermal amplification (TB-LAMP) for the diagnosis of pulmonary tuberculosis. Policy guidance. 2016. Available at: https://apps.who.int/iris/bitstream/handle/10665/249154/9789241511186-eng.pdf. Accessed 28 January 2021. [PubMed]
- 12. Reboud J, Xu G, Garrett A, et al. Paper-based microfluidics for DNA diagnostics of malaria in low resource underserved rural communities. Proc Natl Acad Sci U S A 2019; 116:4834–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nzelu CO, Kato H, Peters NC. Loop-mediated isothermal amplification (LAMP): an advanced molecular point-of-care technique for the detection of Leishmania infection. PLoS Negl Trop Dis 2019; 13:e0007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 2020; 26:773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baek YH, Um J, Antigua KJC, et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect 2020; 9:998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park GS, Ku K, Baek SH, et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2. J Mol Diagn 2020; 22:729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu L, Wu S, Hao X, et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem 2020; 66:975–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dao Thi VL, Herbst K, Boerner K, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med 2020; 12:eabc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol 2020; 38:870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill 2020; 25:2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci U S A 2020; 117:24450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018; 360:444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Odiwuor N, Xiong J, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv 2020.02.26.20028373 [Preprint]. 29 February 2020. Available at: 10.1101/2020.02.26.20028373. Accessed 28 January 2021. [DOI]
- 24. Pastorino B, Touret F, Gilles M, et al. Evaluation of heating and chemical protocols for inactivating SARS-CoV-2. Viruses 2020; 12:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. World Health Organization. Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: interim guidance. 19 March 2020. Available at: https://apps.who.int/iris/handle/10665/331501. Accessed 28 January 2021.
- 26. Centers for Disease Control and Prevention. Interim guidelines for collecting, handling, and testing clinical specimens from persons for coronavirus disease 2019 (COVID-19). Updated 5 November 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed 2 December 2020.
- 27. Centers for Disease Control and Prevention. Processing of sputum specimens for nucleic acid extraction. Published 2 July 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/downloads/processing-sputum-specimens.pdf. Accessed 2 December 2020.
- 28. Sun F, Ganguli A, Nguyen J, et al. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip 2020; 20:1621–7. [DOI] [PubMed] [Google Scholar]
- 29. Centers for Disease Control and Prevention. Interim laboratory biosafety guidelines for handling and processing specimens associated with coronavirus disease 2019 (COVID-19). Updated 19 September 2020. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html. Accessed 2 December 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.