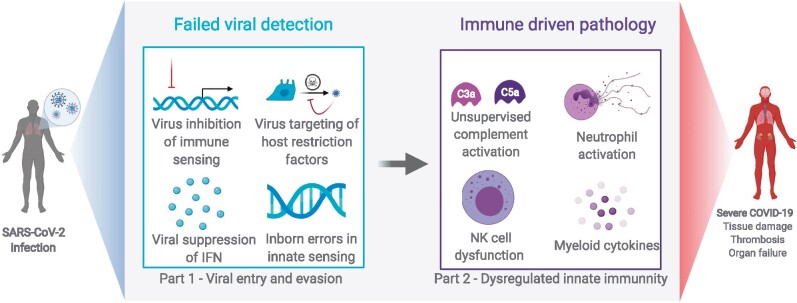
Abstract
The coronavirus infectious disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remains a world health concern and can cause severe disease and high mortality in susceptible groups. While vaccines offer a chance to treat disease, prophylactic and anti-viral treatments are still of vital importance, especially in context of the mutative ability of this group of viruses. Therefore, it is essential to elucidate the molecular mechanisms of viral entry, innate sensing and immune evasion of SARS-CoV-2, which control the triggers of the subsequent excessive inflammatory response. Viral evasion strategies directly target anti-viral immunity, counteracting host restriction factors and hijacking signalling pathways to interfere with interferon production. In Part I of this review, we examine SARS-CoV-2 viral entry and the described immune evasion mechanisms to provide a perspective on how the failure in initial viral sensing by infected cells can lead to immune dysregulation causing fatal COVID-19, discussed in Part II.
Keywords: SARS-CoV-2, COVID-19, viral entry, viral evasion, host response, interferon
Graphical Abstract
Graphical Abstract.
Box 1: Why does your reviewed topic matter in the pandemic?
The Coronavirus disease 2019 (COVID-19) pandemic is caused by a novel virus termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Viral entry into host cells is the first stage of infection and a promising drug target. After viral entry, viral sensing by the host cell is essential for the innate immune response to be triggered. However, SARS-CoV-2 can evade the host immune response by targeting cellular host factors. Therefore, it is pivotal to elucidate the mechanisms of viral entry, innate sensing and immune evasion of SARS-CoV-2, which can provide a basis for therapeutic development.
Box 2: What is the consensus?
To initiate infection, the spike (S) protein of SARS-CoV-2 engages the host cell receptor, angiotensin-converting enzyme II (ACE2), which is highly expressed in lung tissues. Viral materials released into the cytosol can be sensed by the host cell viral recognition machinery which subsequently activates the anti-viral interferon (IFN) pathway. However, SARS-CoV-2 encodes for proteins that counteract these viral recognition pathways or function as IFN antagonists, leading to reduced IFN signalling. In addition, subsets of patients present with genetic mutations in the TLR3 and IRF7 signalling pathways, which lead to defective IFN responses and a worse outcome. This impaired sensing of virus may allow viral replication, which leads to cell damage and systematic immune dysregulation.
INTRODUCTION
The coronavirus infectious disease 2019 (COVID-19) pandemic has had devastating global impacts on human health and the economy. Caused by a novel virus termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), COVID-19 is heterogeneous in clinical presentation and outcome: it is estimated that 20% of patients are asymptomatic, most progress with mild infection and a minority experience acute respiratory distress syndrome [1] that can be fatal. While long-term health implications of SARS-CoV-2 infection are unknown, acute symptoms can cause respiratory, intestinal, kidney and neurological complications as well as loss of smell and taste [2–5]. Disease severity is correlated with age, ethnicity and underlying health conditions including diabetes, obesity and heart disease. It is unclear why some patients develop severe disease, though lack of proper viral recognition and a dysregulated immune response are involved in pathogenicity. After entry into the host cell, viral pathogens have evolved strategies to prevent triggering of anti-viral immune responses by counteracting host anti-viral machinery and pathogen recognition. Innate immune cells are the primary responders to viral infections and key to elicit specific adaptive and memory immune responses through cytokine secretion and antigen presentation [6]. In severe COVID-19, a major dysregulation of innate responses with excessive cytokine production triggers widespread systemic inflammation exacerbating disease [6]. In this review (Part I), we examine recent research elucidating the mechanisms of SARS-CoV-2 cell entry, viral sensing and immune evasion. In Part II, we will discuss the responses of innate immune cells including natural killer (NK) cells, macrophages, monocytes and neutrophils to SARS-CoV-2 infection, including their role in mediating antiviral responses and their contribution to disease pathogenesis. Finally, we highlight promising targets for therapeutic interventions.
Viral entry
Viral entry into a host cell usually requires proteins in the viral envelope engaging with cell surface receptors. In COVID-19, SARS-CoV-2 spike (S) glycoprotein binds to the angiotensin-converting enzyme II (ACE2) [7] receptor with high affinity [8, 9], mediating viral entry. ACE2 is a carboxypeptidase and an important component of the renin–angiotensin system (RAS), a system of regulatory mediators of blood pressure and organ homeostasis involved in the regulation of inflammatory pathways in the lung [10]. ACE2 is highly expressed in endothelial and epithelial cells of the heart, kidney, cornea, liver, gut and airways [11]. In the airways, multi-ciliated respiratory tract cells play an important role as a first line of defence through mucociliary pathogen clearance. Ciliated epithelia cells express high levels of ACE2 mRNA [11, 12] and SARS-CoV-2 infection results in loss of motile cilia, impacting airway immunity [13]. ACE2 expression varies by age and ethnicity and has been associated with comorbidities and severe COVID-19 [14–16]. This can be explained by the protective role of ACE2 in RAS of tissues severely affected by COVID-19 [17]. Additionally, factors that maintain ACE2 expression are important, such as HMGB1, which was demonstrated to be crucial for viral entry [18]. Potential prophylactic treatments to block viral entry by impairing ACE2-S interaction are listed in Table 1.
Table 1:
Potential treatments
| Therapy | Target | Mechanism | References |
|---|---|---|---|
| Monoclonal antibody | ACE2 | Block the interaction between SARS-CoV-2 S protein and ACE2. | Chen et al. [19] |
| Angiotensive enzyme (ACE) inhibitor and angiotensin receptor blocker (ARB) drugs | ACE, ACE2, angiotensin receptor | Inhibition of ACE activity or blockage of the angiotensin receptor activity. | Hippisley-Cox et al. [20] |
| Type I IFN supplementation | IFN-β, IFN-α2b, IFN-α1b | Severe COVID-19 patients have shown reduced type I IFN responses. Type I IFN supplementation reduced the duration of inflammatory markers in mild disease and prevented COVID-19 infection in highly exposed individuals. |
Hung et al. [21] Zhou et al. [22] Meng et al. [23] (prevented infection in highly exposed individuals) |
| Paricalcitol | ADAM17/ACE2 | Regulates ACE2 shedding through ADAM17. | Riera et al. [24] |
| Camosat mesyalte | TMPRSS2 | Inhibitor of protease | Hoffmann et al. [7] |
| Decanoyl-RVKR-chloromethylketone, Naphthofluorescein | Furin | Inhibitor of protease | Cheng et al. [25] |
| Chloroquine, hydroxychloroquine, chlorpromazine | Endocytic pathway | Block the fusion of viral membrane and the endosomal/lysosomal membrane |
Chen et al. [26] Plaze et al. [27] |
| Remdesivir, Enisamium | SARS-CoV-2 RNA polymerase | Inhibition of the RNA-dependent, RNA polymerase. |
Beigel et al. [28] Walker et al. [29] |
Following receptor engagement, fusion of SARS-CoV-2 with the host cell requires transmembrane serine protease 2 (TMPRSS2) to cleave the S protein at the S1/S2 cleavage site [7]. S1 mediates receptor binding, while S2 is required for membrane fusion; both are needed for endocytosis [30] into the host cell via dynamin/clathrin machinery [31]. Additionally, SARS-CoV-2 S protein can be cleaved and primed by the protease furin between its S1/S2 domains [32–34]. The dominant mutant D614G, with a single amino acid mutation in the SARS-CoV-2 S protein, has been linked to increased infectivity, possibly via reduced shedding of the S1 domain [35, 36]. S1 also requires binding of Neuropilin-1 (NRP1) for the viral entry process and blocking this interaction reduced SARS-CoV-2 cell entry [37]. Alternatively, SARS-CoV-2 can enter through the late endosome/lysosome pathway, wherein the cellular cathepsin L proteinase cleaves the S protein and initiates fusion of viral and endosomal/lysosomal membranes [30]. An additional mediator of SARS-CoV-2 infection is A Disintegrin And Metalloproteinase 17 (ADAM17), which cleaves ACE2, thus shedding the receptor and reducing viral uptake in cells [38–40]. Drugs impairing endocytic routes, targeting these proteases and the ADAM17/ACE2 axis are listed in Table 1.
Viral tropism determines the target cell of the virus. Therefore, it is important to note other putative receptors for SARS-CoV-2 entry including kidney injury molecule-1 (KIM-1—highly expressed in kidney cells) [41], CD147 [42], CD4 [43] and CD26 [44]. More work is needed to confirm these, although it is already known that anti-KIM-1 IgG and a KIM-1 inhibitor (TW-37) impaired SARS-CoV-2 endocytosis in human alveolar basal epithelial cells [41, 45]. It is still debated whether SARS-CoV-2 replicates in innate immune cells such as monocytes and macrophages which express ACE2, TMPRSS2 and ADAM17 [46] but SARS-CoV-2 infection did not induce a cytopathic effect, suggesting abortive infection in these cells [47].
Innate sensing
Following virus entry, the viral genome is exposed, and viral proteins are synthesized by the host cell machinery for assembly of new virions. However, host cells can recognize this viral material (pathogen-associated molecular patterns—PAMPs) via pattern recognition receptors (PRRs). PRRs initiate signalling cascades culminating in the production of pro-inflammatory cytokines and IFNs, which upregulate IFN-stimulated genes (ISGs) that further direct innate and adaptive immunity.
PRRs include cytosolic RNA sensors such as retinoic acid-inducible Gene I (RIG-I) and melanoma differentiation gene 5 (MDA-5), while cytosolic DNA triggers the cyclic GMP–AMP synthase/stimulator of IFN genes (cGAS-STING) pathway. Indeed, in vitro infection with SARS-CoV-2 upregulated pathways for RIG-I signalling in Huh7 cells (liver cell line) [48] and activated MDA-5 in primary human epithelia and cell lines, inducing a robust type I and type III IFN response, though this induction failed to control viral replication [49]. SARS-CoV-2 also induces a cGAS-STING mediated NF-kB activation of inflammatory immune responses [50] and polymorphisms in STING have been suggested to produce a delayed over-secretion of IFN-β in SARS-CoV-2 infection [51].
Endosomal toll-like receptor 7 (TLR7) recognizes single-stranded viral RNA (ssRNA) and TLR3 binds double-stranded RNA (dsRNA) generated during viral replication. Therefore, inborn genetic errors affecting the TLR3 signalling pathway result in a defective type I IFN response and are associated with life-threatening COVID-19 [52]. Sex differences in innate immune sensing can also explain epidemiology underlying COVID-19 severity, as TLR genes are encoded on the X chromosome, causing higher expression and stronger innate immune activation in women [53]. Sex differences in COVID-19 have been evaluated extensively by Takashaki et al. who found that females produced more IFNα2 and that elevated innate cytokines correlated with disease progression, but only in females [54].
Fine tuning of IFN responses appears to be key for COVID-19 outcome, as shown by dysregulated responses attributed to auto-antibodies against IFN found in 10% of severe patients [55]. While IFNs are usually the primary anti-viral cytokines produced by cells sensing respiratory viruses, SARS-CoV-2 infection has been shown to elicit a dampened IFN-I and IFN-III response in human alveolar cells (A549) and ferrets [56]. The IFN response may not be absent but rather delayed, as shown in in vitro infection of lung Calu-3 cells [56], especially when compared to the respiratory Sendai virus [57]. Similarly, in infected human lung tissues, SARS-CoV-2 did not significantly induce types I, II or III IFNs [58]. These lower levels of IFN can be explained by effective viral immune evasion mechanisms. Therefore, early enhancement of IFN signalling may offer therapeutic benefit, and trials of IFN supplementation are listed in Table 1.
Other innate activation pathways involved in SARS-CoV-2 sensing include NOD-like receptor (NLR) activation and tumor necrosis factor (TNF)-α production [48]. NLR family pyrin domain-containing 3 (NLPR3) inflammasome activation by SARS-CoV-2 ORF3a was predicted based on SARS-CoV studies [59, 60] and has now been demonstrated via both ASC-dependent and ASC-independent pathways [61].
Immune evasion
Viruses have evolved mechanisms to evade the activation of host innate immune responses [62]. For example, SARS-CoV-2 displays a range of molecules directly targeting the type I IFN pathway: ORF6 protein has been reported to inhibit both Type I IFN production and downstream signalling, the C-terminus region being critical for this antagonistic effect [57]. ORF6 has been shown to localize in the nuclear pore complex and block nuclear translocation for pSTAT1 [63] and IFN responsive factor (IRF) 3 [64], impairing IFN signalling. The IFN response was also found to be attenuated and linked to viral suppression of STAT1 phosphorylation in monocyte-derived macrophages and dendritic cells [65]. Studies using Sendai virus to mimic IFN response to SARS-CoV-2 revealed that together with ORF6, ORF8 and Nucleocapsid (N) contribute to the inhibition of the type I IFN response [66] and subsequently the NF-κB-responsive promoter via IFN-stimulated response element (ISRE) [67]. SARS-CoV-2 ORF3b is truncated and suppressed IFN induction more than the SARS-CoV variant when using Sendai virus [68]. Furthermore, SARS-CoV-2 PLpro cleaves ISG15 from IRF3, dampening the IFN response [69]. Non-structural proteins (NSP) also perform as IFN antagonists: SARS-CoV-2 NSP6 suppressed IRF3 phosphorylation through binding TANK binding kinase (TBK1), while NSP13 blocked TBK1 phosphorylation [64]. Screening SARS-CoV-2 proteins, Lei et al. found that NSP1, NSP3, NSP12, NSP13, NSP14, but also ORF3, ORF6 and structural M protein could inhibit the activation of the IFN-ß promoter after infection with Sendai virus [57]. SARS-CoV-2 NSP13, NSP14 and NSP15 can also act as IFN antagonists [66] but the mechanisms are still unclear. Interestingly, NSP2 and S protein activate IFN [57]; however, subsequent viral activity perhaps dampens this response. SARS-CoV-2 is also likely to share other evasion mechanisms with SARS and MERS, which have been extensively discussed elsewhere [65, 70–73].
Viral proteins also target cellular intrinsic mechanisms of defence, such as anti-viral host restriction factors: proteins that interfere with the viral life cycle. The C-terminus of SARS-CoV-2 NSP1 obstructs the mRNA entry tunnel of the 40S ribosomal subunit, resulting in translation shutoff of host mRNAs [74]. Other viral proteins including NSP5, NSP8, NSP13, N and envelope protein E interact with host factors involved in epigenetic and RNA regulation, which could interfere with the host response [75]. For instance, NSP16 inhibits pre-mRNA splicing [76], while NSP8 and NSP9 bind to the 7SL RNA component of the signal recognition particle (SRP) complex, interfering with protein trafficking to the cell membrane [76]. Martin-Sancho et al. extensively screened for ISGs acting as host restriction factors in the context of SARS-CoV-2 infection [77]. These ISGs include endosomal factors inhibiting viral entry, nucleic acid binding proteins, inhibitors of viral translation, regulators of membrane lipids and vesicle transport. For example, tetherin (BST2) binds newly synthesized viruses to the plasma membrane impairing viral release. SARS-CoV-2 ORF7a was shown to counteract tetherin to allow viral release [77]. SARS-CoV-2 ORF8 has also been suggested to downregulate surface MHC-I [78] by targeting it for lysosomal degradation, which would impact the function of NK cells and CD8 T-cells.
Conclusion
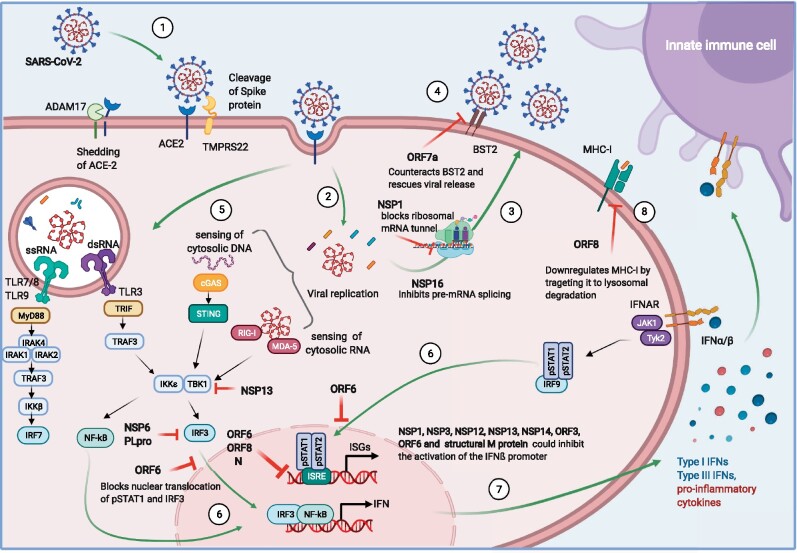
The mechanisms of SARS-CoV-2 immune evasion at the early stages of infection are shown in Figure 1 and this understanding is a key for generating effective treatments against COVID-19, such as those listed in Table 1. The immune dysregulation causing fatal COVID-19 likely starts at the cellular level and identifying pathways involved in defective or altered responses is crucial to dampen the pathogenicity of the infection. As with most viruses, SARS-CoV-2 encodes numerous proteins that counteract host restriction factors or act as IFN antagonists, impairing recognition of the virus which allows for viral replication in the absence of antiviral immunity. SARS-CoV-2 has evolved diverse ways to evade innate mechanisms, which may facilitate high transmissibility and virulence. However, this evasion of the immune system cannot explain why only a small minority of patients progress to severe disease. Thus, other factors such as inborn genetic errors in sensing, existing autoantibodies, immunogenic tolerance and the immune response itself must also influence the outcomes of COVID-19. In Part II of this review, we investigate the consequences of failed immune sensing on the innate immune system, which results in immunopathology.
Figure 1:
Viral entry and immune evasion. Graphical depiction of the reported SARS-CoV-2 mechanisms of immune detection and viral evasion in host cells. (1) Virus engagement to ACE2 and fusion to the cell membrane, (2) viral genetic material release into the cytoplasm, (3) late phase of the viral cycle: viral replication and release, (4) host restriction factors interfering with the viral life cycle: BST2 impeding viral release, (5) host immune sensing of viral components, (6) nuclear translocation of different regulatory factors that trigger transcriptional programs for pro-inflammatory cytokines, ISGs and IFN; and their release (7). (8) MHC-I antigenic presentation; and corresponding viral counteraction potentially affecting induction of immune responses.
ACKNOWLEDGEMENTS
We would like to thank all members of the Oxford-Cardiff COVID19 Literature Consortium for their hard work and commitment during the pandemic. Figures were created using Biorender.com.
AUTHORS’ CONTRIBUTIONS
C.C. and M.T. conceptualized the review, wrote the original draft and reviewed and edited the manuscript. F.L. was involved in conceptualization and wrote the original draft. P.R.S.R., A.A., E.P., V.M.T.B., R.J. and S.M.T. were involved in conceptualization and reviewed and edited the manuscript. F.C.R. and D.O.S. administered the project and reviewed and edited the manuscript. J.R. reviewed and edited the manuscript. L.C.D. was involved in conceptualization, reviewed and edited the manuscript, and administered the project. E.G.-M. conceptualized the review, wrote the original draft, reviewed and edited the manuscript and administered the project.
CONFLICT OF INTEREST STATEMENT
The authors have no conflict of interest to declare.
DATA AVAILABILITY
All data are contained within the manuscript. This review was facilitated by weekly releases of the Oxford-Cardiff COVID19 Literature Consortium journal club—a database of reviewed articles and journals will be made available on request.
REFERENCES
- 1. Buitrago-Garcia D, Egli-Gany D, Counotte MJ et al. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLOS Med 2020;17:e1003346. doi:10.1371/journal.pmed.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carignan A, Valiquette L, Grenier C et al. Anosmia and dysgeusia associated with SARS-CoV-2 infection: an age-matched case-control study. CMAJ 2020;192:E702–e707. doi:10.1503/cmaj.200869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fanelli V, Fiorentino M, Cantaluppi V et al. Acute kidney injury in SARS-CoV-2 infected patients. Crit Care 2020;24:155. doi:10.1186/s13054-020-02872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mao R, Qiu Y, He J-S et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2020;5:667–78. doi:10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang X, Yu Y, Xu J et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020;8:475–81. doi:10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKechnie JL, Blish CA. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe 2020;27:863–9. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoffmann M, Kleine-Weber H, Schroeder S et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80.e8. doi:10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li W, Moore MJ, Vasilieva N et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003;426:450–4. doi:10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li F, Li W, Farzan M et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005;309:1864–8. doi:10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 10. Alfano G, Guaraldi G, Fontana F et al. The role of the renin-angiotensin system in severe acute respiratory syndrome-CoV-2 infection. Blood Purif 2020. doi:10.1159/000507914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sungnak W, Huang N, Bécavin C et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 2020;26:681–7. doi:10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee IT, Nakayama T, Wu C-T et al. PREPRINT: Robust ACE2 protein expression localizes to the motile cilia of the respiratory tract epithelia and is not increased by ACE inhibitors or angiotensin receptor blockers. medRxiv 2020:2020.05.08.20092866. doi:10.1101/2020.05.08.20092866. [Google Scholar]
- 13. Robinot R, Hubert M, de Melo GD et al. PREPRINT: SARS-CoV-2 infection damages airway motile cilia and impairs mucociliary clearance. bioRxiv 2020:2020.10.06.328369. doi:10.1101/2020.10.06.328369. [Google Scholar]
- 14. Baker SA, Kowk S, Berry GJ et al. PREPRINT: Angiotensin-converting enzyme 2 (ACE2) expression increases with age in patients requiring mechanical ventilation. medRxiv 2020:2020.07.05.20140467. doi:10.1101/2020.07.05.20140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swärd P, Edsfeldt A, Reepalu A et al. Age and sex differences in soluble ACE2 may give insights for COVID-19. Crit Care 2020;24:221. doi:10.1186/s13054-020-02942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen J, Jiang Q, Xia X et al. Individual variation of the SARS-CoV-2 receptor ACE2 gene expression and regulation. Aging Cell 2020;19. doi:10.1111/acel.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS-COV-2. Front Cell Infect Microbiol 2020;10. doi:10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei J, Alfajaro MM, DeWeirdt PC et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 2020. doi:10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen X, Li R, Pan Z et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol 2020;17:647–9. doi:10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hippisley-Cox J, Young D, Coupland C et al. Risk of severe COVID-19 disease with ACE inhibitors and angiotensin receptor blockers: cohort study including 8.3 million people. Heart 2020;106:1503–11. doi:10.1136/heartjnl-2020-317393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hung IF-N, Lung K-C, Tso EY-K et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. The Lancet 2020;395:1695–704. doi:10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Q, Wei X-S, Xiang X et al. PREPRINT: Interferon-a2b treatment for COVID-19. medRxiv 2020:2020.04.06.20042580. doi:10.1101/2020.04.06.20042580. [Google Scholar]
- 23. Meng Z, Wang T, Li C et al. PREPRINT: An experimental trial of recombinant human interferon alpha nasal drops to prevent coronavirus disease 2019 in medical staff in an epidemic area. medRxiv 2020:2020.04.11.20061473. doi:10.1101/2020.04.11.20061473. [Google Scholar]
- 24. Riera M, Anguiano L, Clotet S et al. Paricalcitol modulates ACE2 shedding and renal ADAM17 in NOD mice beyond proteinuria. Am J Physiol Renal Physiol 2016;310:F534–46. doi:10.1152/ajprenal.00082.2015. [DOI] [PubMed] [Google Scholar]
- 25. Cheng Y-W, Chao T-L, Li C-L et al. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep 2020:108254. doi:10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Shen T, Zhong L et al. Research progress of chloroquine and hydroxychloroquine on the COVID-19 and their potential risks in clinic use. Front Pharmacol 2020; 11. doi:10.3389/fphar.2020.01167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plaze M, Attali D, Petit AC et al. Repurposing chlorpromazine to treat COVID-19: the reCoVery study. L'Encéphale 2020; 46:169–72. doi:10.1016/j.encep.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beigel JH, Tomashek KM, Dodd LE et al. Remdesivir for the treatment of Covid-19—final report. N Engl J Med 2020. doi:10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walker AP, Fan H, Keown JR et al. PREPRINT: Enisamium is a small molecule inhibitor of the influenza A virus and SARS-CoV-2 RNA polymerases. bioRxiv 2020:2020.04.21.053017. doi:10.1101/2020.04.21.053017. [Google Scholar]
- 30. Ou X, Liu Y, Lei X et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020;11:1620. doi:10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bayati A, Kumar R, Francis V et al. PREPRINT: SARS-CoV-2 uses clathrin-mediated endocytosis to gain access into cells. bioRxiv 2020:2020.07.13.201509. doi:10.1101/2020.07.13.201509. [Google Scholar]
- 32. Shang J, Wan Y, Luo C et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci 2020;117:11727–34. doi:10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 2020;23:101212. doi:10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walls AC, Park Y-J, Tortorici MA et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181:281–92.e6. doi:10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korber B, Fischer WM, Gnanakaran S et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020;182:812–27.e19. doi:10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang L, Jackson CB, Mou H et al. PREPRINT: The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv 2020:2020.06.12.148726. doi:10.1101/2020.06.12.148726. [Google Scholar]
- 37. Daly JL, Simonetti B, Klein K et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020: eabd3072. doi:10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Palau V, Riera M, Soler MJ. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol Dial Transplant 2020;35:1071–2. doi:10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lambert DW, Yarski M, Warner FJ et al. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J Biol Chem 2005;280:30113–9. doi:10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Haga S, Yamamoto N, Nakai-Murakami C et al. Modulation of TNF-α-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-α production and facilitates viral entry. Proc Natl Acad Sci 2008;105:7809–14. doi:10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ichimura T, Mori Y, Aschauer P et al. PREPRINT: KIM-1/TIM-1 is a receptor for SARS-CoV-2 in lung and kidney. medRxiv 2020:2020.09.16.20190694. doi:10.1101/2020.09.16.20190694. [Google Scholar]
- 42. Wang K, Chen W, Zhou Y-S et al. PREPRINT: SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020:2020.03.14.988345. doi:10.1101/2020.03.14.988345. [Google Scholar]
- 43. Davanzo GG, Codo AC, Brunetti NS et al. PREPRINT: SARS-CoV-2 uses CD4 to infect T helper lymphocytes. medRxiv 2020:2020.09.25.20200329. doi:10.1101/2020.09.25.20200329. [Google Scholar]
- 44. Radzikowska U, Ding M, Tan G et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020:75:2829–45. doi:10.1111/all.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mori YA, Ajay AK, Chang J-H et al. KIM-1 mediated tubular fatty acid uptake leads to progressive diabetic kidney disease. SSRN Electron J 2020. doi:10.2139/ssrn.3641944. [Google Scholar]
- 46. Abassi Z, Knaney Y, Karram T et al. The lung macrophage in SARS-CoV-2 infection: a friend or a foe? Front Immunol 2020;11. doi:10.3389/fimmu.2020.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yilla M, Harcourt BH, Hickman CJ et al. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res 2005;107:93–101. doi:10.1016/j.virusres.2004.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Appelberg S, Gupta S, Svensson Akusjärvi S et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect 2020;9:1748–60. doi:10.1080/22221751.2020.1799723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Antoine R, Valadão Ana Luiza C, Marine T et al. PREPRINT: SARS-CoV-2 replication triggers an MDA-5-dependent interferon production which is unable to efficiently control replication. bioRxiv 2020:2020.10.28.358945. doi:10.1101/2020.10.28.358945. [Google Scholar]
- 50. Neufeldt CJ, Cerikan B, Cortese M et al. PREPRINT: SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. bioRxiv 2020:2020.07.21.212639. doi:10.1101/2020.07.21.212639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Berthelot J-M, Lioté F. COVID-19 as a STING disorder with delayed over-secretion of interferon-beta. EBioMedicine 2020; 56:102801. doi:10.1016/j.ebiom.2020.102801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Q, Bastard P, Liu Z et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020:eabd4570. doi:10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y, Jerkic M, Slutsky AS et al. Molecular mechanisms of sex bias differences in COVID-19 mortality. Crit Care 2020;24:405. doi:10.1186/s13054-020-03118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takahashi T, Ellingson MK, Wong P et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020. doi:10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bastard P, Rosen LB, Zhang Q et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020:eabd4585. doi:10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Blanco-Melo D, Nilsson-Payant BE, Liu WC et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181:1036–45.e9. doi:10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lei X, Dong X, Ma R et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 2020;11:3810. doi:10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chu H, Chan JF, Wang Y et al. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin Infect Dis 2020. doi:10.1093/cid/ciaa410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen I-Y, Moriyama M, Chang M-F et al. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front Microbiol 2019;10. doi:10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. van den Berg DF, te Velde AA. Severe COVID-19: NLRP3 inflammasome dysregulated. Front Immunol 2020;11. doi:10.3389/fimmu.2020.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu H, Chitre SA, Akinyemi IA et al. PREPRINT: SARS-CoV-2 viroporin triggers the NLRP3 inflammatory pathway. bioRxiv 2020:2020.10.27.357731. doi:10.1101/2020.10.27.357731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 2008; 8:911–22. doi:10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Miorin L, Kehrer T, Sanchez-Aparicio MT et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci 2020:202016650. doi:10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xia H, Cao Z, Xie X et al. Evasion of type i interferon by SARS-CoV-2. Cell Rep 2020; 33:108234. doi:10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang D, Chu H, Hou Y et al. Attenuated interferon and proinflammatory response in SARS-CoV-2–infected human dendritic cells is associated with viral antagonism of STAT1 phosphorylation. J Infect Dis 2020;222:734–45. doi:10.1093/infdis/jiaa356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yuen CK, Lam JY, Wong WM et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect 2020;9:1418–28. doi:10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li J-Y, Liao C-H, Wang Q et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 2020;286:198074. doi:10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Konno Y, Kimura I, Uriu K et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 2020; 32:108185. doi:10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shin D, Mukherjee R, Grewe D et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020;587:657–662. doi:10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bouayad A. Innate immune evasion by SARS-CoV-2: comparison with SARS-CoV. Rev Med Virol 2020: e2135. doi:10.1002/rmv.2135. [DOI] [PubMed] [Google Scholar]
- 71. Park A, Iwasaki A. Type I and Type III interferons – induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 2020;27:870–8. doi:10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Amor S, Fernández Blanco L, Baker D. Innate immunity during SARS-CoV-2: evasion strategies and activation trigger hypoxia and vascular damage. Clin Exp Immunol 2020;202:193–209. doi:10.1111/cei.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Deng X, Buckley AC, Pillatzki A et al. Inactivating three interferon antagonists attenuates pathogenesis of an enteric coronavirus. J Virol 2020;94:e00565–20. doi:10.1128/jvi.00565-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thoms M, Buschauer R, Ameismeier M et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020;369:1249–55. doi:10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gordon DE, Jang GM, Bouhaddou M et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020;583:459–68. doi:10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Banerjee AK, Blanco MR, Bruce EA et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell 2020;183:P1325–39.E21. doi:10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Martin-Sancho L, Lewinski MK, Pache L et al. PREPRINT: functional landscape of SARS-CoV-2 cellular restriction. bioRxiv 2020:2020.09.29.319566. doi:10.1101/2020.09.29.319566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang Y, Zhang J, Chen Y et al. PREPRINT: The ORF8 Protein of SARS-CoV-2 mediates immune evasion through potently downregulating MHC-I. bioRxiv 2020:2020.05.24.111823. doi:10.1101/2020.05.24.111823. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript. This review was facilitated by weekly releases of the Oxford-Cardiff COVID19 Literature Consortium journal club—a database of reviewed articles and journals will be made available on request.