Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected millions of people globally. Virus infection requires the receptor-binding domain (RBD) of the spike protein. Although studies have demonstrated anti-spike and -RBD antibodies to be protective in animal models, and convalescent plasma as a promising therapeutic option, little is known about immunoglobulin isotypes capable of blocking infection.
Methods
We studied spike- and RBD-specific immunoglobulin isotypes in convalescent and acute plasma/serum samples using a multiplex bead assay. We also determined virus neutralization activities in plasma and serum samples, and purified immunoglobulin fractions using a vesicular stomatitis pseudovirus assay.
Results
Spike- and RBD-specific immunoglobulin (Ig) M, IgG1, and IgA1 were produced by all or nearly all subjects at variable levels and detected early after infection. All samples displayed neutralizing activity. Regression analyses revealed that IgM and IgG1 contributed most to neutralization, consistent with IgM and IgG fractions’ neutralization potency. IgA also exhibited neutralizing activity, but with lower potency.
Conclusion
IgG, IgM, and IgA are critical components of convalescent plasma used for treatment of coronavirus disease 2019 (COVID-19).
Keywords: SARS-CoV-2, COVID-19, antibody isotypes, neutralization, convalescent plasma
Immunoglobulin (Ig) M, IgG1, and IgA1 antibodies against severe acute respiratory syndrome coronavirus 2 spike glycoprotein and its receptor-binding domain are present in plasma from patients with convalescent coronavirus disease 2019 (COVID-19). IgG, IgM, and IgA contribute to virus neutralization, providing the basis for optimal selection of convalescent plasma for COVID-19 treatment.
Since the first patients with coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), were identified in Wuhan, China [1], the epidemic has spread worldwide, infecting millions of people. Effective therapeutics and vaccines are urgently needed. Convalescent plasma transfusions have shown promising results in patients with severe COVID-19 [2–4]. Clinical trials to evaluate the efficacy of this treatment in ambulatory and hospitalized patients are underway [5–7] and FDA Emergency Usage Authorization has been issued [8]. To this end, information is needed about immunoglobulin isotypes in convalescent plasma that have antiviral activities. The data would likewise inform vaccine development [9]. Most vaccines are based on the SARS-CoV-2 spike protein [9, 10], a membrane-anchored protein present on the virus envelope along with 2 others (membrane and envelope proteins) and it contains the receptor-binding domain (RBD) for binding and entry into cells [11–13]. The vaccines aim to protect by inducing neutralizing antibodies (Abs) that block viral infection.
SARS-CoV-2 spike-, RBD-, and nucleocapsid-specific serum and plasma Abs of immunoglobulin (Ig) M, IgG, and IgA isotypes are found in most patients with COVID-19 [14–19], with neutralizing activities developing within 2 weeks of infection and declining over time [16, 17, 20, 21]. However, the neutralizing titers vary greatly [16, 17, 20, 21] and are correlated with Ab binding levels against RBD, spike, and/or nucleocapsid, and with age, symptom duration, and symptom severity [16, 17]. Several RBD-specific monoclonal IgG Abs with neutralizing activity have been generated, and these confer protection in animal models [16, 20, 22, 23]. A monoclonal Ab of IgA isotype recognizing both SARS-CoV-1 and SARS-CoV-2 spike proteins and blocking angiotensin-converting enzyme 2 receptor binding was recently described [24]. However, no direct evidence is available regarding the neutralizing capacity of plasma IgM and IgA from patients with COVID-19.
Studies on other respiratory viruses such as influenza show that, in addition to IgG, IgA could also mediate virus neutralization, and their relative contributions depend on the physiologic compartment in which they are found, with IgA contributing to the protection of mostly the upper respiratory tract, while IgG protects the lower respiratory tract [25, 26]. An anti-hemagglutinin monoclonal polymeric IgA has been demonstrated to mediate more potent anti-influenza activities than monoclonal IgG against the same epitope [27]. An IgM monoclonal Ab with neutralizing activity against influenza B has also been described [28]. In addition, respiratory syncytial virus–specific mucosal IgA is a better correlate of protection than serum IgG counterparts [29]. In the case of SARS-CoV-1, high titers of IgA in the lungs are correlated with reduced pathology in animal models [30]. Whether IgA in the blood and the respiratory tract mucosa offer protection against SARS-CoV-2 remains an open question. Moreover, scant data are available regarding the IgM contribution to neutralization and protection against viruses, including SARS-CoV-2. Of note, in terminally ill patients, systemic SARS-CoV-2 infection affects multiple organs [31]. Thus, the capacity of plasma immunoglobulin to suppress virus spread is critical for effective therapy against severe COVID-19.
Our group recently described a multiplex bead Ab-binding assay using Luminex technology to detect total immunoglobulin against spike and RBD [32]. In this study we characterized the immunoglobulin isotype profiles using the Luminex assay that detects spike- and RBD-specific IgM, IgG1-4, and IgA1-2. Using a pseudovirus assay [33], our group also measured plasma or serum neutralization and determined the neutralizing capacity of IgM, IgA, and IgG fractions. The data indicate a high prevalence of spike- and RBD-specific IgM and IgA, similar to that of IgG1, in plasma and serum samples from patients with COVID-19, and their contributions to virus neutralization. In addition, by testing purified IgG, IgM, and IgA fractions from convalescent plasma samples, the current study presents the first direct evidence that plasma IgG, IgM, and IgA all contribute to SARS-CoV-2 neutralization.
METHODS
Recombinant Proteins
SARS-CoV-2 spike and RBD proteins were produced as described elsewhere [34, 35].
Human Samples
All COVID-19–positive and COVID-19–negative samples tested in this study are tabulated in Supplementary Table 1. Twenty-five citrated COVID-19–convalescent plasma samples destined for transfusion to SARS-CoV-2-infected individuals (TF1–25, collected between 26 March and 7 April 2020) and 10 contemporary COVID-19–negative specimens (N4–13) were obtained from the Division of Transfusion Medicine of the Department of Pathology, Molecular and Cell-Based Medicine (Mount Sinai Hospital System, institutional review board [IRB] no. 20-03574). The convalescent specimens TF1–25 were from donors prescreened to have serum IgG reciprocal titer ≥320 in the Mount Sinai Hospital enzyme-linked immunosorbent assay anti-IgG COVID-19 assay. Four serum samples from deidentified individuals with COVID-19 (P5–8) were provided by the Clinical Pathology Division of the Department of Pathology, Molecular and Cell-Based Medicine at the Icahn School of Medicine at Mount Sinai.
The following samples were obtained from volunteers enrolled in IRB-approved protocols at the Icahn School of Medicine at Mount Sinai (IRB nos. 16-00772, 1600791 and 17-01243) and the James J. Peter Veterans Affairs Medical Center (IRB BAN-1604): serum samples from 7 participants with documented SARS-CoV-2 infection (P1 on days 8, 11, and 15 after symptom onset, P2 on days 7 and 10 after symptom onset, and RP1–5 after convalescence), and prepandemic serum samples from 12 healthy donors (N1–3 and N14–22). All study participants provided written consent. All samples were heat inactivated before use.
Immunoglobulin Fractionation
IgA was isolated first from plasma using peptide M agarose beads (InvivoGen; GEL-PDM). The pass-through plasma was enriched sequentially for IgG using protein G agarose beads (InvivoGen; GEL-AGG) and for IgM using a HiTrap IgM column (GE Healthcare; no. 17-5110-01). An additional step was performed using Protein A Plus mini-spin columns to separate IgG from IgM. Protein concentrations were determined with a NanoDrop spectrophotometer (Thermo Scientific).
Multiplex Bead Ab-Binding Assay
SARS-CoV-2 spike and RBD antigens were coupled to beads and experiments performed as described elsewhere [32], except for the use of different secondary Abs designated in the figure legends.
COV2pp Production and Titration
SARS-CoV-2 pseudoviruses (COV2pp) with wild-type (WT) or D614G-mutated spike proteins were produced as described elsewhere [33]. Pseudoviruses were titrated on 20 000 Vero-CCL81 cells seeded 24 hours before infection. At 18–22 hours after infection, the infected cells were washed, and Renilla luciferase activity was measured with the Renilla-Glo Luciferase Assay System (Promega no. E2720) on a Cytation3 (BioTek) instrument.
COV2pp Neutralization
Virus was preincubated with diluted plasma or serum samples for 30 minutes. The virus-sample mix was then added to Vero-CCL81 cells seeded 24 hours earlier and spinoculated. Infection was measured after 18–22 hours by luciferase activity. The percentage of neutralization was calculated as follows: 100 - 100[(sample RLU − cell control RLU)/virus control RLU], where RLU represents relative light units. Inhibitory concentration (IC50) and 90% inhibitory concentration (IC90) titers were calculated as the reciprocal sample dilution or purified immunoglobulin fraction concentration achieving 50% and 90% neutralization, respectively.
Statistical Analysis
Two-tailed Mann-Whitney test, Spearman rank-order correlation test, and simple linear regressions were performed as described in the figure legends, using GraphPad Prism 8 software.
RESULTS
Variable Levels of Immunoglobulin Isotypes Against SARS-CoV-2 Spike and RBD in Convalescent Individuals
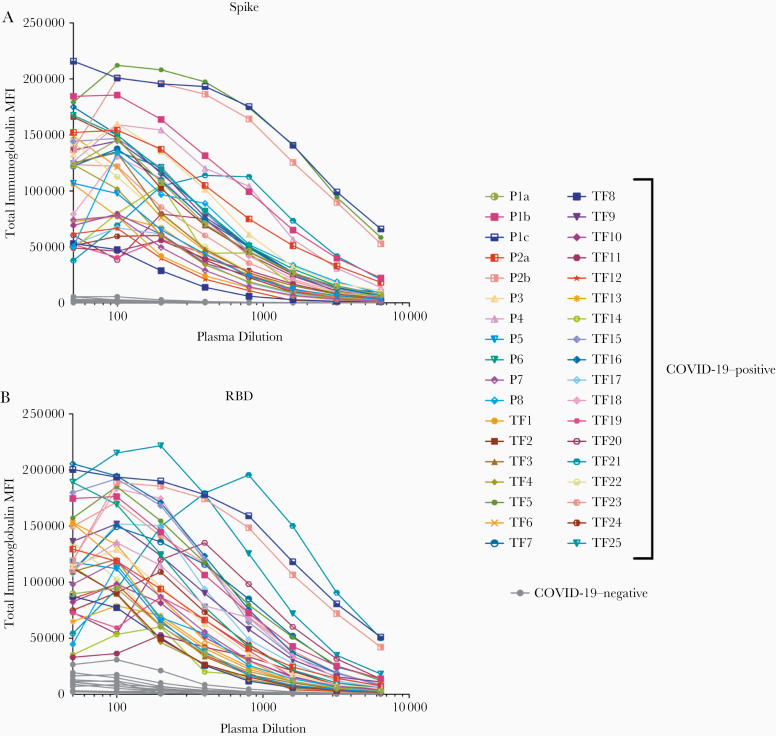
A total of 29 serum (P5–8) and plasma (TF1–25) specimens from COVID-19–convalescent individuals was tested. TF1–25 were collected about 4–8 weeks after the initial outbreak in North American, and used for transfusion into hospitalized patients with COVID-19 [2]. Ten plasma samples from COVID-negative contemporaneous blood bank donors (N4–13) were included for comparison. Serum or plasma samples from 12 uninfected individuals banked before the COVID-19 outbreak (N1–3 and N14–22) were used to establish background values. The specimens were initially titrated for total immunoglobulin against spike and RBD (Figure 1). All 29 COVID-19–positive specimens exhibited titration curves of total immunoglobulin Abs against spike, while none of the negative controls displayed reactivity.
Figure 1.
Titration of severe acute respiratory syndrome coronavirus 2 spike and receptor-binding domain (RBD) total immunoglobulin in plasma or serum samples from individuals with convalescent coronavirus disease 2019 (COVID-19). Titration of spike-specific (A) or RBD-specific (B) total immunoglobulin from 29 individuals with convalescent COVID-19, 2 patients with acute COVID-19 with longitudinal samples, and 13 COVID-19–negative individuals. Specimens were diluted at 2-fold dilutions from 1:50 to 1:6400. Abbreviation: MFI, mean fluorescence intensity.
Similar results were observed with RBD, except that 1 contemporaneous COVID-19–negative sample had a low level of RBD-specific immunoglobulin (N10). Overall, the background mean fluorescence intensity (MFI) values were higher for RBD than spike. To assess the reproducibility of the assay, the samples were tested in ≥2 separate experiments run on different days, and a strong correlation was observed between the MFI values from these independent experiments (Supplementary Figure 1). The areas under the curve highly correlated with the MFI values from specimens diluted 1:200 (P < .001; Supplementary Figure 2); consequently, all samples were tested for isotyping at this dilution. At the 1:200 dilution we were able to discern a diverse range of immunoglobulin isotype levels among individual samples (Figure 2). To evaluate for the presence of spike-specific and RBD-specific total immunoglobulin, IgM, IgG1, IgG2, IgG3, IgG4, IgA1, and IgA2, the specificity and strength of the secondary Abs used to detect the different isotypes were first validated with Luminex beads coated with myeloma proteins of known immunoglobulin isotypes (IgG1, IgG2, IgG3, IgG4, IgA1, IgA2, and IgM). All 8 secondary Abs were able to detect their specific immunoglobulin isotypes with MFI values reaching >60 000 (Supplementary Figure 3).
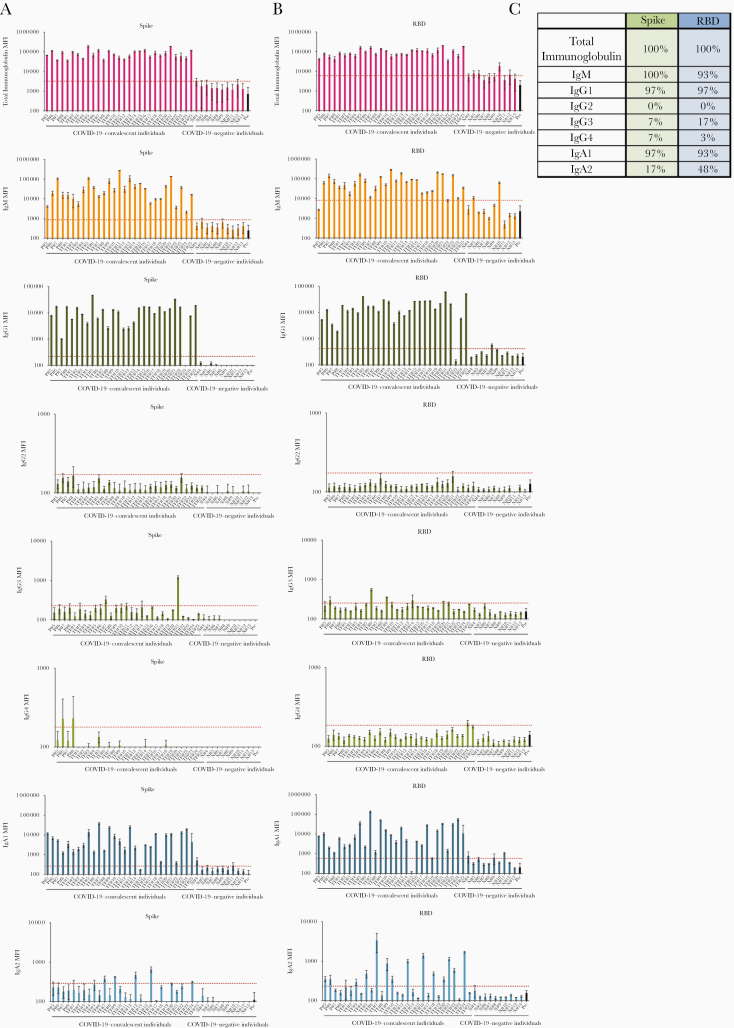
Figure 2.
Levels of immunoglobulin isotypes against the severe acute respiratory syndrome coronavirus 2 spike and receptor-binding domain (RBD) vary in plasma or serum samples from individuals with convalescent coronavirus disease 2019 (COVID-19). A, B, Total immunoglobulin and immunoglobulin (Ig) M, IgG1, IgG2, IgG3, IgG4, IgA1, and IgA2 against spike (A) and RBD (B) in specimens from 29 COVID-19–convalescent individuals, 13 COVID-19–negative contemporaneous samples, and prepandemic controls were detected using the following secondary antibodies: rabbit biotinylated-anti-human total immunoglobulin (Abcam; catalog no. ab97158) at 2 μg/mL, mouse biotinylated-anti-human IgG1 Fc (Invitrogen; no. MH1515) at 4 μg/mL, mouse biotinylated-anti-human IgG2 Fc (Southern Biotech; no. 9060-08) at 1 μg/mL, mouse biotinylated-anti-human IgG3 hinge (Southern Biotech; no. 9210-08) at 3 μg/mL, mouse biotinylated-anti-human IgG4 Fc (Southern Biotech; no. 9200–08) at 4 μg/mL, mouse biotinylated-anti-human IgA1 Fc (Southern Biotech; no. 9130-08) at 4 μg/mL, mouse biotinylated-anti-human IgA2 Fc (Southern Biotech; no. 9140-08) at 4 μg/mL, or goat biotinylated-anti-human IgM (Southern Biotech; no. 2020-08) at 3 μg/mL. The samples were tested at a dilution of 1:200 and data are shown as mean fluorescence intensity (MFI) (+ standard deviation [SD]) of duplicate measurements from ≥2 independent experiments. The prepandemic controls are shown as MFI (+ SD) of 12 samples (Pre; black bar). Horizontal red dotted lines represent cutoff values, determined as mean + 3 SDs of 12 prepandemic samples for each of the isotypes. C, Percentages of responders above the cutoff for each spike- or RBD-specific immunoglobulin isotype.
All 29 convalescent individuals had anti-spike and anti-RBD total immunoglobulin (Figure 2), but the immunoglobulin levels were highly variable, with MFI values ranging from 36 083 to 190 150. In addition, all 29 convalescent individuals also displayed IgM Abs against spike at varying levels, and 93% were positive for anti-RBD IgM when evaluated using cutoff values calculated as mean + 3 standard deviations of the 12 prepandemic samples (Figures 2B and 2C). In contrast, IgG2, IgG3, and IgG4 Abs against spike and RBD were detected in only a small fraction of the subjects, and the levels were very low (MFI < 1300) (Figure 2).
Surprisingly, almost all individuals produced IgA1 Abs against spike (97%) and RBD (93%), while 17% exhibited IgA2 against spike, and 48% exhibited IgA2 against RBD (Figure 2). Low levels, slightly above cutoff, of spike- and RBD-binding total immunoglobulin, IgM, IgG1, and IgA1 were detected sporadically in contemporaneous COVID-19 samples, such as N8, N10, and N11. The responses against spike and RBD were highly correlated for every isotype (Supplementary Figure 4). Overall, these data demonstrate that IgM, IgG1, and IgA1 Abs were induced against spike and RBD in all or almost all COVID-19–convalescent individuals (Figure 2). The levels, however, were highly variable among individuals. No significant difference was observed between female and male individuals (Supplementary Figure 5).
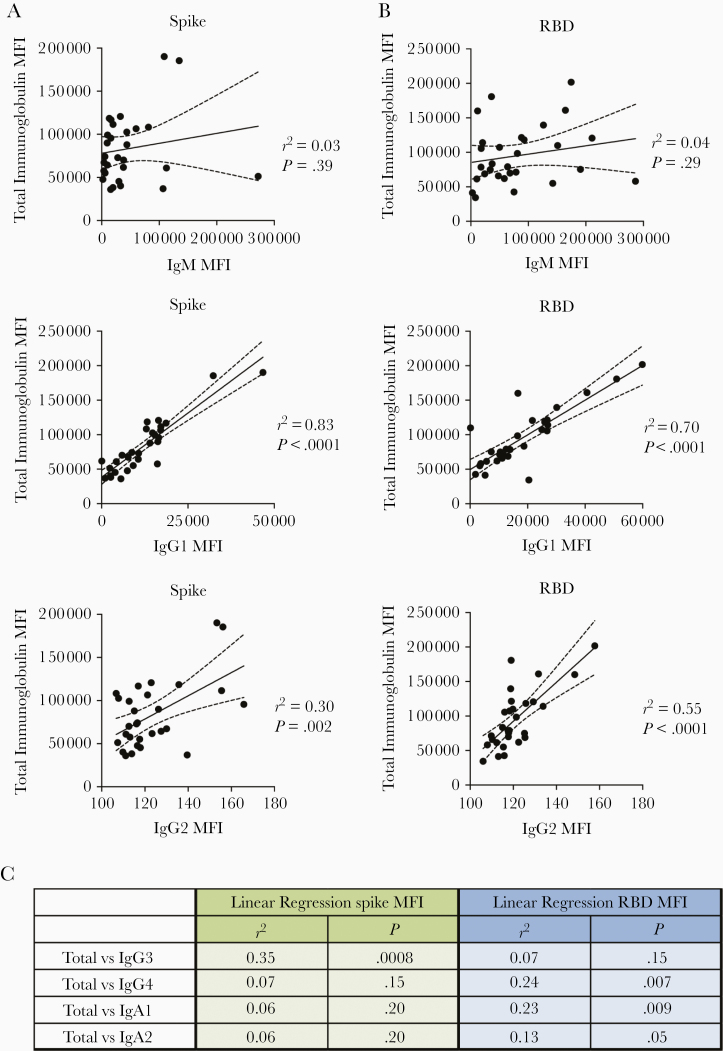
In Figure 3, regression analyses to assess the impact of individual isotypes on the total immunoglobulin binding showed that IgG1 had the highest r2 values (0.83 and 0.70 for spike-binding and RBD-binding IgG1, respectively; P < .001), indicating that IgG1 is the major isotype induced by SARS-CoV-2 infection against spike and RBD (Figures 3A and 3B). IgG2 Abs against RBD had an r2 value of 0.55 (P < .001), but IgG2 levels were very low. For all other isotypes, including IgM, the r2 values were <0.40 (Figure 3C). Thus, despite the presence of many isotypes in serum and plasma samples, as expected, the major isotype of spike and RBD-specific Abs is IgG1.
Figure 3.
Immunoglobulin (Ig) G1 is the dominant isotype induced in individuals with convalescent coronavirus disease 2019 (COVID-19). Results are shown for samples from the 29 COVID-19–convalescent individuals from Figure 1. A, B, Simple linear regression of spike-specific (A) or receptor-binding domain (RBD)–specific (B) total immunoglobulin levels versus IgM, IgG1, and IgG2 levels. C, Linear regression of total immunoglobulin versus spike- and RBD-specific IgG3, IgG4, IgA1, and IgA2 levels. Dashed dash lines represent 95% confidence intervals.
Specimens from 2 patients (P1 and P2) were obtained during the acute phase of the infection. Serial specimens from these patients were tested to determine the isotypes of Abs present early in infection. The earliest samples from both patients, drawn at 7 or 8 days after symptom onset were already positive for total immunoglobulin, IgG1, IgA1, and IgM Abs against spike and RBD (Supplementary Figure 6), and these levels increased over the following 3–7 days. On the contrary, IgA2 Ab levels were near or below background on days 7–8 and remained unchanged over the 2 weeks after onset. IgG4 Abs also remained low or near background, whereas IgG2 and IgG3 Abs increased slightly to above background after 10–15 days.
Neutralizing Activity in Specimens From All Individuals With Convalescent COVID-19
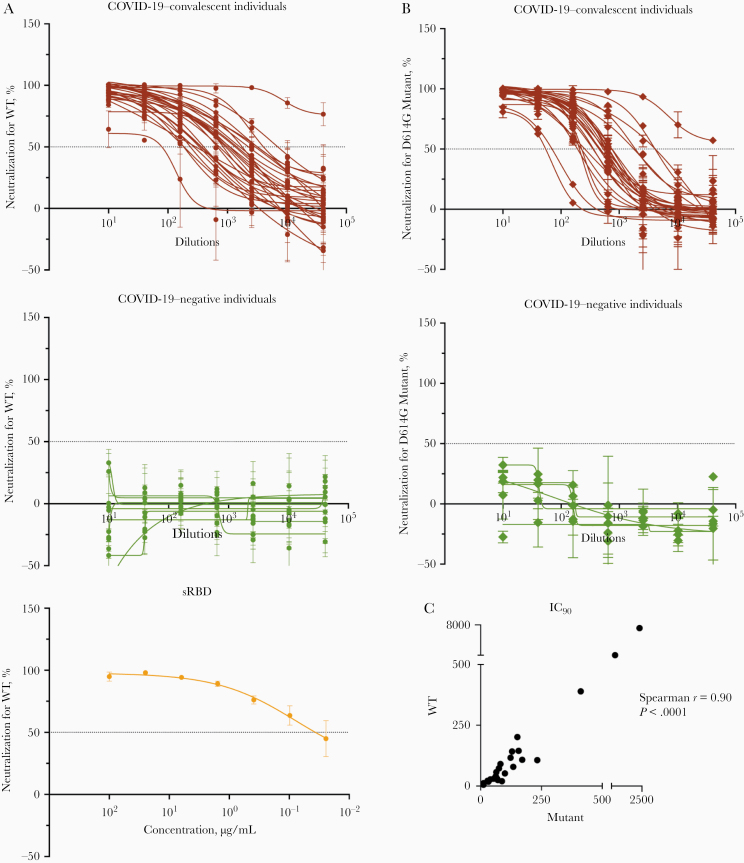
We subsequently tested the ability of samples from convalescent subjects to neutralize a VSV∆G pseudovirus bearing the SARS-CoV-2 spike protein (COV2pp). This pseudovirus assay demonstrated a strong positive correlation with neutralization of the authentic SARS-CoV-2 virus [33]. The titration of neutralizing activity against the WT COV2pp is shown in Figure 4A for specimens from 28 COVID-19–convalescent individuals and 11 uninfected individuals, tested over a range of 7 serial 4-fold dilutions. A soluble recombinant RBD (sRBD) protein capable of blocking virus infection was tested in parallel as a positive control.
Figure 4.
Neutralization activities are detected in all individuals with convalescent coronavirus disease 2019 (COVID-19). A, B, Neutralization of COV2pp with wild-type (WT) (A) or D614G mutated (B) spike proteins by samples from 28 COVID-19–convalescent and 11 COVID-19–negative individuals. The neutralizing activity of recombinant soluble receptor-binding domain (sRBD) is shown as a positive control. At 24 hours before infection, 20 000 Vero-CCL81 cells per well were seeded. Virus (82.5 µL per well) was preincubated with serially diluted samples (27.5 µL per well; 4-fold from 1:10 to 1:40 960) for 30 minutes at room temperature. The virus-sample mix was then added to the cells and spinoculated by centrifugation (1250 rpm for 1 hour at room temperature). Six virus-only and 6 medium-only wells were kept for each plate. After 18–22 hours at 37°C, infection was measured by luciferase activity. sRBD was tested as a positive control at 4-fold dilutions from 100 to 0.02 µg/mL. Data are shown as mean percentage of neutralization (plus standard deviation) of triplicates. Extrapolated titration curves were generated using a nonlinear regression model in GraphPad Prism software (inhibitor vs response – variable slope [4 parameters]; least squares regression). Dotted horizontal lines denote 50% neutralization. C, Spearman correlation between 90% inhibitory concentration (IC90) titers against COV2pp WT versus D614G.
All specimens from COVID-19–convalescent individuals were able to neutralize the virus at levels above 50% (Figure 4A). For 26 of 28 specimens, neutralization reached >90% (Figure 4A). The sample with the lowest titer (reciprocal IC50 titer, 37) reached a neutralization plateau of only about 60%. Of note, 1 sample (TF11) demonstrated highly potent neutralization with a reciprocal IC50 titer > 40 960, and neutralization was still 75% at the highest dilution tested. None of the samples from uninfected individuals reached 50% neutralization (Figure 4A), while the sRBD positive control demonstrated potent neutralization with an IC50 of 0.06 µg/mL (Figure 4A), similar to that recently reported [33].
The samples were also tested for neutralization against a COV2pp bearing the spike with a D614 mutation (D614G mutant), as the D614G variant has become the most prevalent circulating strain in the global pandemic [36]. Similar to the WT COV2pp, all COVID-19–convalescent samples had neutralizing activity reaching >50%, while none of the negative samples did (Figure 4B). The IC90 titers against WT and D614 mutant differed on average by only 1.7-fold and correlated strongly with each other (P < .0001, Figure 4C).
Contributions of IgM and IgG1 to SARS-CoV-2 Neutralization
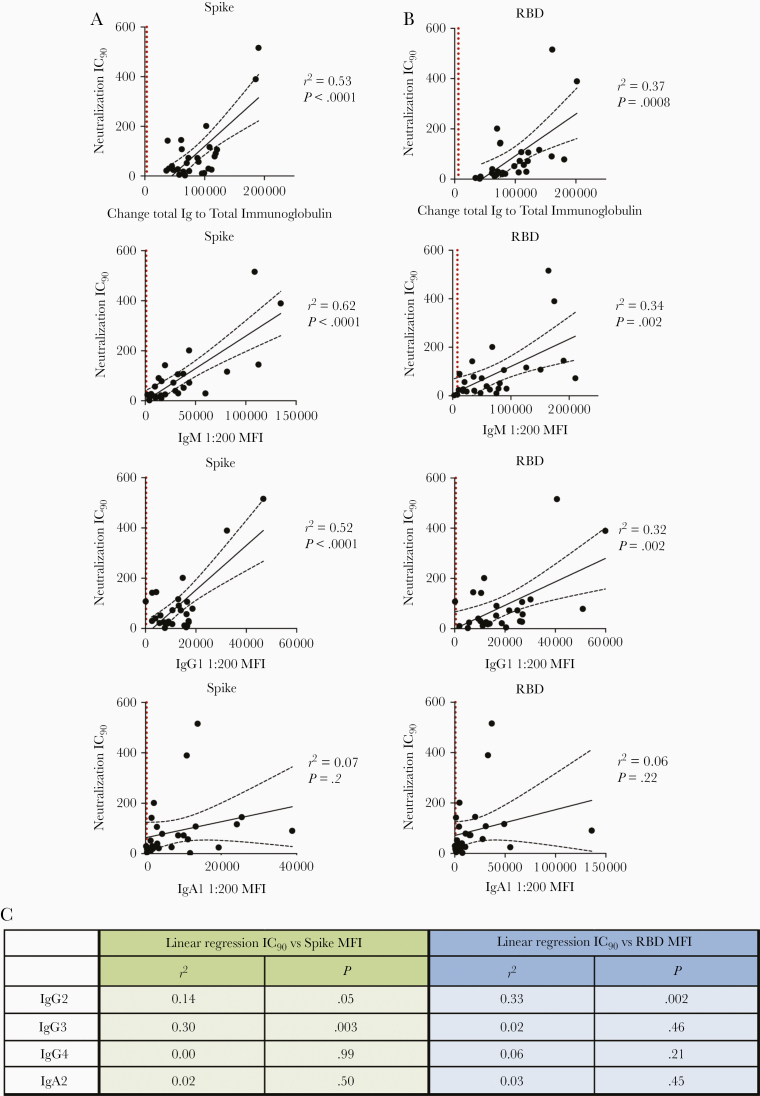
Given our observation that Ab isotype levels and neutralization titers varied tremendously among individuals with convalescent COVID-19 (Figures 2 and 5), we investigated the relative contribution of each Ab isotype to the neutralizing activities. Regression analyses were performed on 27 COVID-19–convalescent samples (TF11 was excluded due to its outlier neutralization titer). As expected, relatively high r2 values (0.32–0.62) and significant P values were observed with total immunoglobulin, IgM and IgG1; in each case, r2 values were higher for spike than for RBD (Figure 6A). The highest r2 value was achieved in the analysis of IC90 neutralizing titers and IgM binding to spike (r2 = 0.62). For other isotypes, significant P values were sporadically achieved, but r2 values were weak (Figures 6A and 6B).
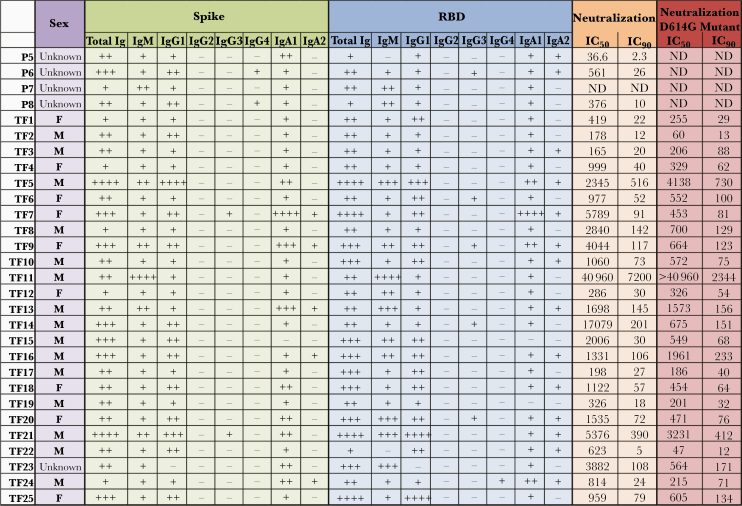
Figure 5.
Summary of relative immunoglobulin (Ig) isotype levels and neutralization titers. Table shows sex, relative levels of spike-specific and receptor-binding domain (RBD)–specific immunoglobulin isotypes (+ indicates bottom quartile; ++, second quartile; +++, third quartile; ++++, top quartile; and −, nonresponder), and reciprocal median inhibitory concentration (IC50) and 90% inhibitory concentration (IC90) neutralization titers against wild-type (WT) and D614G pseudoviruses in 29 plasma samples from individuals with convalescent coronavirus disease 2019. Abbreviations: F, female; M, male; ND, not done.
Figure 6.
Immunoglobulin (Ig) M and IgG1 contribute most to severe acute respiratory syndrome coronavirus 2 neutralization. A, B, Simple linear regression of reciprocal 90% inhibitory concentration (IC90) neutralization titers of 27 individuals with convalescent coronavirus disease 2019 (COVID-19) versus spike-specific (A) or receptor-binding domain (RBD)–specific (B) total immunoglobulin, IgM, IgG1, and IgA1 antibody levels. Black dashed lines represent the 95% confidence intervals; dotted vertical red lines, cutoff (mean of 12 prepandemic samples + 3 standard deviations) for each isotype from Figure 1. C, Statistical results of simple linear regression analyses of reciprocal IC90 neutralization titers of 27 COVID-19–convalescent individuals versus spike-specific or RBD-specific antibody levels for IgG2-4 and IgA2. Abbreviation: MFI, mean fluorescence intensity.
Mediation of Neutralizing Activities by Plasma IgM, IgG, and IgA Fractions
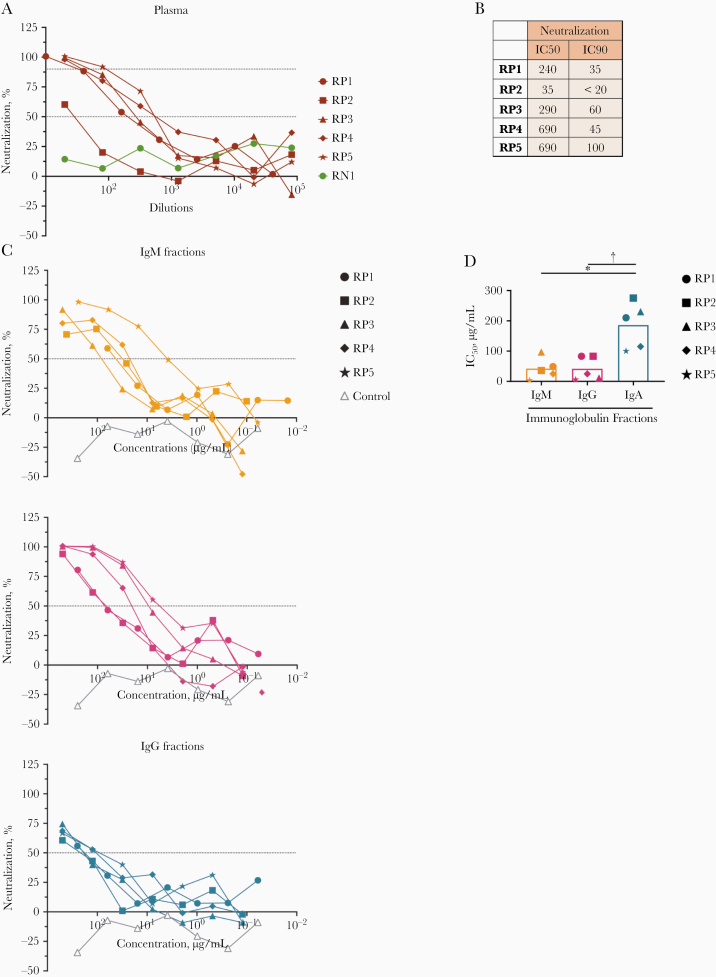
To assess directly the capacity of different isotypes to mediate neutralization, we evaluated the neutralization activities of IgM, IgG, and IgA fractions purified from the plasma of 5 COVID-19–convalescent individuals (RP1–5). The enrichment of IgM, IgG1, and IgA1 Abs reactive with spike and RBD was validated using the isotyping method used above (Supplementary Figure 7 and data not shown). These IgM, IgG, and IgA fractions were then evaluated for neutralizing activity along with the original plasma (Figure 7). The RP1–5 plasma neutralizing reciprocal IC50 titers ranged from 35 to 690 (Figures 7A and 7B). Purified IgM and IgG fractions from RP1–5 all mediated neutralization reaching >50%. Unexpectedly, plasma IgA fractions also displayed neutralizing activity, although not with the same potency as IgM and IgG (Figures 7C, D). In contrast, IgM, IgG, and IgA fractions from the negative control (RN1) showed no neutralization (Figures 7C, D).
Figure 7.
Purified immunoglobulin (Ig) M, IgG, and IgA fractions display neutralizing activities against severe acute respiratory syndrome coronavirus 2. A, Neutralization of COV2pp in plasma samples from 5 individuals infected with coronavirus disease 2019 (COVID-19) (RP1–5) compared to a specimen from a COVID-19–uninfected individual (RN1; green circles). Plasma samples were tested at 4-fold dilutions from 1:10 to 1:40 960 or 1:20 to 1:81 920. Data are shown as the mean percentage of neutralization; dotted horizontal lines highlight 50% and 90% neutralization. B, Reciprocal median inhibitory concentration (IC50) and 90% inhibitory concentration (IC90) neutralization titers of RP1–5 plasma samples. C, Neutralization of COV2pp by purified IgM, IgG, and IgA fractions from 5 COVID-19–infected individuals (RP1–5) compared with a control immunoglobulin fraction (open gray triangles). IgA was isolated first from plasma samples by mixing 1:2 diluted plasma with peptide M agarose beads (600 µL/28 mL plasma; InvivoGen GEL-PDM) for 1.5 hours at room temperature. After washing of beads, IgA was eluted with a pH 2.8 buffer (Thermo Scientific no. 21004) and neutralized with pH 9 Tris buffer. The pass-through plasma sample was collected for IgG enrichment using protein G agarose beads (InvivoGen GEL-AGG) and subsequently for IgM isolation using a HiTrap IgM column (GE Healthcare; no. 17-5110-01). An additional purification step was performed using Protein A Plus mini-spin columns to separate IgG from IgM. The fractions were tested at 4-fold dilutions from 500 to 0.02 µg/mL. Data are shown as the mean percentage of neutralization; dotted horizontal lines highlight 50% neutralization. D, IC50 of purified IgM, IgG, and IgA fractions from RP1–5. Statistical significance was determined with a 2-tailed Mann-Whitney test. *P < .05; †P < .01.
DISCUSSION
Our study demonstrates that IgG1, IgA1, and IgM Abs against SARS-CoV-2 spike and RBD were prevalent in plasma of patients with convalescent COVID-19 approximately 1–2 months after infection. These isotypes were present within 7–8 days after the onset of symptoms. Importantly, all 3 isotypes showed the capacity to mediate virus neutralization. While regression analyses demonstrated the strongest contributions of IgM and IgG1 to neutralizing activity, direct testing of purified isotype fractions showed that IgA also was able to neutralize, indicating the protective potential of all 3 major immunoglobulin isotypes. These data carry important implications for the use of convalescent plasma and hyperimmunoglobulin as COVID-19 therapeutics, suggesting that their selection would optimally be based on the presence of all of these immunoglobulin isotypes.
While all COVID-19–convalescent individuals exhibited neutralization activities reaching >50% and 26 of 28 specimens attained 90% neutralization, neutralization levels were highly variable with IC50 and IC90 titers ranging over 3 orders of magnitude. The titers were comparable against the initial Wuhan strain and the currently prevalent D614G strain of SARS-CoV-2. Similarly, the levels of spike-binding and RBD-binding total immunoglobulin and immunoglobulin isotypes varied greatly.
A trend toward higher levels of total immunoglobulin and each immunoglobulin isotype was seen in female compared with male subjects, as reported in another study [37]. Moreover, except for TF11 (a male elite neutralizer), the median neutralizing IC90 titer was higher in female than in male subjects, although the difference did not reach significance (data not shown). Sex differences in Ab induction have been observed following influenza vaccination in humans and mice and were shown to result from the impact of sex-related steroids [38]. Whether and to what extent this contributes to the sex differences seen in clinical outcomes of COVID-19 remains to be investigated. Other studies have shown that Ab levels were associated with multiple factors, including time from disease onset [39] and disease severity [15]. However, other than sex, clinical data are not available for the subjects studied here, limiting our analysis only to neutralization and immunoglobulin isotypes.
One remarkable finding from our study is that although neutralization titers correlated with binding levels of IgM and IgG1 and not with those of IgA1 or IgA2, purified IgA fractions from patients with convalescent COVID-19 exhibited significant neutralizing activities. The importance of this finding is underscored by the data showing that IgA1 was the prominent isotype in some samples such as TF7 and TF24 and that IgA1 could be detected early after symptom onset. Data from other studies also support the significance of IgA in that purified IgA fractions exhibited more, or as potent neutralizing activities as purified IgG, and that RBD-binding IgA correlated as strongly as IgG with microneutralization titers [40].
IgA was also detected in saliva and bronchoalveolar lavage from patients with COVID-19 [41]. Nonetheless, Wang et al [42] reported that plasma IgA monomers were less potent than the plasma IgG and secretory IgA counterparts. In our study, neutralization activities detected in the IgA fractions were mediated mainly by IgA1, the predominant IgA isotype in plasma, and the IC50 potency of the IgA fraction was approximately 4-fold lower than the potency of IgM and IgG1 fractions. This difference cannot be explained entirely by lower amounts of spike-specific IgA1 in the tested fractions, as estimations using spike-specific monoclonal IgA and IgM Abs yielded similar IgA and IgM concentrations in the respective purified fractions (median, 2 and 2.5 µg/mL, respectively). Fine epitope specificities and affinities may differ for IgA, IgM, and IgG to affect neutralization potency but have yet to be evaluated.
In addition to neutralization, nonneutralizing Ab activities have been implicated in protection from various virus infections through potent Fc-mediated functions such as Ab-dependent cellular cytotoxicity, Ab-dependent cellular phagocytosis, and complement-mediated lysis; this is reported for human immunodeficiency, influenza, Marburg, and Ebola viruses [26, 43–45]. The Fc activities were not evaluated in our study, and their contribution to protection against SARS-CoV-2 is yet unclear [46, 47].
A recent study demonstrated enrichment of spike-specific IgM and IgA1 Abs and spike-specific phagocytic and Ab-dependent complement deposition (ADCD) activity in plasma of individuals who recovered from SARS-CoV-2 infection, while nucleocapsid-specific IgM and IgA2 responses and nucleocapsid-specific ADCD activity were features enriched in deceased patients [48]. DNA vaccines expressing full-length and truncated spike proteins could curtail SARS-CoV-2 infection in the respiratory tract by varying degrees in rhesus macaques. This virus reduction correlated with levels of neutralization and also with Fc-mediated effector functions, such as ADCD [45]. Interestingly, these DNA vaccines elicited spike- and RBD-specific IgG1, IgG2, IgG3, IgA, and IgM Abs, and similar to our findings, neutralization correlated most strongly with IgM.
Adenovirus serotype 26 vaccine vectors encoding 7 SARS-CoV-2 spike variants also showed varying protection levels, and virus reduction correlated best with neutralizing titers together with IgM-binding levels, FcγRII-binding, and ADCD responses [49]. Defining the full functional potential of Abs against SARS-CoV-2—including neutralizing, nonneutralizing, and enhancing activities—are vital for determining the optimal Ab treatment modalities against COVID-19 and the potential efficacy of COVID-19 vaccine candidates.
When we examined plasma specimens collected within 7–8 days after COVID-19 symptom onset, we detected IgG and IgA against spike and RBD, as well as IgM. This is consistent with published reports showing that 100% of COVID-19–infected individuals developed IgG within 19 days after symptom onset and that IgG and IgM seroconversion could occur simultaneously [15]. IgA was also found early after infection (4–6 days after symptom onset) and increased over time [14, 19, 41]. These studies suggest that measuring total immunoglobulin, rather than IgG only, could contribute to improved outcomes for early disease diagnosis. We found no correlation between the levels of different isotypes in the specimens examined in our study (data not shown). Of note, IgA presence early during acute infection may suggest the potential contribution of natural IgA, which, similar to natural IgM, arises spontaneously from innate B1 cells to provide the initial humoral responses before the induction of adaptive classic B cells [50].
In summary, the current study demonstrates that spike- and RBD-specific IgM, IgG1, and IgA1 are produced by all or almost all analyzed COVID-19–convalescent subjects and can be detected at early stages of infection. The plasma samples of convalescent individuals also display neutralization activities mediated by IgM, IgG, and IgA1, although neutralization titers correlated more strongly with IgM and IgG levels. The contribution of IgM, IgG, and IgA to SARS-CoV-2–neutralizing activities demonstrates their importance in the efficacy of passively transferred Abs for SARS-CoV-2 treatment.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Florian Krammer, Viviana Simon, and Rebecca Powell for donation of samples and reagents, and all the donors for their contribution to the research.
Author contributions. J. K., S. W., G. E. A., S. Z. P., and C.E.H. wrote and edited the manuscript. J. K., S. W., S. Z. P., and C. E. H. designed the experiments. J. K., S. W., V. I., and X. L. performed the experiments and collected the data. J. K., A. N., S. Z. P., and C. E. H. analyzed the data. K. Y. O., C. S., S. I., C. T. H., F. A., and B. L. provided protocols, antigens, cells, and pseudovirus stocks. G. E. A., I. B., S. A., J. C. B., E. M. K., J. S., D. J., M. B. G., and S. L. provided specimens. All authors read and approved the final manuscript.
Financial support . This work was supported by the Microbiology Laboratory Clinical Services at the Mount Sinai Health System and the Mount Sinai Health System Translational Science Hub; the Department of Medicine of the Icahn School of Medicine at Mount Sinai Department of Medicine (S. Z. P. and C. E. H.); the Department of Microbiology and the Ward-Coleman estate, endowing the Ward-Coleman chairs at the Icahn School of Medicine at Mount Sinai (B. L.); the Department of Veterans Affairs (Merit Review grant I01BX003860 to C.E.H. and Research Career Scientists award 1IK6BX004607 to C. E. H.); the National Institutes of Health (grants AI139290 to C. E. H., R01 AI123449 and R21 AI1498033 to B. L., U54TR001433, Viral-Host Pathogenesis Training grant T32AI07647 to K. Y. O. and C. S., and grant F31 AI154739 to K. Y. O.). Clinical Hematology Oncology Treatment - Study Group (CHOT-SG) Fukuoka University, Japan, postdoctoral fellowship to S.I.; and the Ministry of Science and Technology (MOST), Taiwan, postdoctoral fellowship to C.T.H.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu STH, Lin HM, Baine I, et al. Convalescent plasma treatment of severe COVID-19: a propensity score-matched control study. Nat Med 2020; 26:1708–13. [DOI] [PubMed] [Google Scholar]
- 3. Zeng H, Wang D, Nie J, et al. The efficacy assessment of convalescent plasma therapy for COVID-19 patients: a multi-center case series. Signal Transduct Target Ther 2020; 5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ibrahim D, Dulipsingh L, Zapatka L, et al. Factors associated with good patient outcomes following convalescent plasma in COVID-19: a prospective phase II clinical trial. Infect Dis Ther 2020; 8:913–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Janssen M, Schäkel U, Fokou CD, et al. A randomized open label phase-II clinical trial with or without infusion of plasma from subjects after convalescence of SARS-CoV-2 infection in high-risk patients with confirmed severe SARS-CoV-2 disease (RECOVER): a structured summary of a study protocol for a randomised controlled trial. Trials 2020; 21:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. ClinicalTrials.gov. Convalescent plasma to limit SARS-CoV-2 associated complications (CSSC-004).https://clinicaltrials.gov/ct2/show/NCT04373460. Accessed 2 November 2020.
- 7. ClinicalTrials.gov. Convalescent plasma in outpatients with COVID-19 (C3PO). https://clinicaltrials.gov/ct2/show/NCT04355767. Accessed 2 November 2020.
- 8. FDA Issues Emergency Use Authorization for Convalescent Plasma as Potential Promising COVID–19 Treatment, Another Achievement in Administration’s Fight Against Pandemic. https://www.fda.gov/news-events/press-announcements/fda-issues-emergency-use-authorization-convalescent-plasma-potential-promising-covid-19-treatment
- 9. Thanh Le T, Andreadakis Z, Kumar A, et al. The COVID-19 vaccine development landscape. Nat Rev Drug Discov 2020; 19:305–6. [DOI] [PubMed] [Google Scholar]
- 10. Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity 2020; 52:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020; 367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181:271–80.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020; 181:281–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma H, Zeng W, He H, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol 2020; 17:773–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 16. Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 2020; 584:437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 2020; 26:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 2020; 71:778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Padoan A, Sciacovelli L, Basso D, et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin Chim Acta 2020; 507:164–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020; 584:115–9. [DOI] [PubMed] [Google Scholar]
- 21. Prévost J, Gasser R, Beaudoin-Bussières G, et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep Med 2020; 1:100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020; 369:956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020; 584:443–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ejemel M, Li Q, Hou S, et al. IgA MAb blocks SARS-CoV-2 spike-ACE2 interaction providing mucosal immunity. BioRxiv [Preprint: not peer reviewed]. 15 May 2020. Available from: https://www.biorxiv.org/content/10.1101/2020.05.15.096719v1. [Google Scholar]
- 25. Renegar KB, Small PA, Boykins LG, Wright PF. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J Immunol Baltim Md 1950 2004; 173:1978–86. [DOI] [PubMed] [Google Scholar]
- 26. Krammer F. The human antibody response to influenza A virus infection and vaccination. Nat Rev Immunol 2019; 19:383–97. [DOI] [PubMed] [Google Scholar]
- 27. Muramatsu M, Yoshida R, Yokoyama A, et al. Comparison of antiviral activity between IgA and IgG specific to influenza virus hemagglutinin: increased potential of IgA for heterosubtypic immunity. PLoS One 2014; 9:e85582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shen C, Zhang M, Chen Y, et al. An IgM antibody targeting the receptor binding site of influenza B blocks viral infection with great breadth and potency. Theranostics 2019; 9:210–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Habibi MS, Jozwik A, Makris S, et al. ; Mechanisms of Severe Acute Influenza Consortium Investigators . Impaired antibody-mediated protection and defective IgA B-cell memory in experimental infection of adults with respiratory syncytial virus. Am J Respir Crit Care Med 2015; 191:1040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat Rev Microbiol 2009; 7:226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe 2020; 1:e290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiss S, Klingler J, Hioe C, et al. A high-throughput assay for circulating antibodies directed against the S protein of severe acute respiratory syndrome coronavirus 2. J Infect Dis 2020; 222:1629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oguntuyo KY, Stevens CS, Hung CT, et al. Quantifying absolute neutralization titers against SARS-CoV-2 by a standardized virus neutralization assay allows for cross-cohort comparisons of COVID-19 sera. mBio 2021. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 2020; 26:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stadlbauer D, Amanat F, Chromikova V, et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 2020; 57:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020; 182:812–27.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zeng F, Dai C, Cai P, et al. A comparison study of SARS-CoV-2 IgG antibody between male and female COVID-19 patients: a possible reason underlying different outcome between gender. J Med Virol 2020; 92:2050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Potluri T, Fink AL, Sylvia KE, et al. Age-associated changes in the impact of sex steroids on influenza vaccine responses in males and females. NPJ Vaccines 2019; 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020; 26:1200–4. [DOI] [PubMed] [Google Scholar]
- 40. Mazzini L, Martinuzzi D, Hyseni I, et al. Comparative analyses of SARS-CoV-2 binding (IgG, IgM, IgA) and neutralizing antibodies from human serum samples. J Immunol Methods 2021; 489:112937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Isho B, Abe KT, Zuo M, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol 2020; 5:eabe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Z, Lorenzi JCC, Muecksch F, et al. Enhanced SARS-CoV-2 neutralization by secretory IgA in vitro. bioRxiv [Preprint: not peer reviewed]. 9 September 2020. Available from: https://www.biorxiv.org/content/10.1101/2020.09.09.288555v1. [Google Scholar]
- 43. Horwitz, Bar-On Y, Lu CL, et al. Non-neutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 2017; 170:637–48.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ilinykh PA, Huang K, Santos RI, et al. Non-neutralizing antibodies from a Marburg infection survivor mediate protection by Fc-effector functions and by enhancing efficacy of other antibodies. Cell Host Microbe 2020; 27:976–91.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gunn BM, Yu WH, Karim MM, et al. A role for Fc function in therapeutic monoclonal antibody-mediated protection against Ebola virus. Cell Host Microbe 2018; 24:221–33.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020; 369:806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zohar T, Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat Rev Immunol 2020; 20:392–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atyeo C, Fischinger S, Zohar T, et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 2020; 53:524–32.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mercado NB, Zahn R, Wegmann F, et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 2020; 586:583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meyer-Bahlburg A. B-1 cells as a source of IgA. Ann N Y Acad Sci 2015; 1362:122–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.