Abstract
Studies of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–infected patients and experimentally infected animals indicate a critical role for augmented expression of proinflammatory chemokines and cytokines in severe disease. Here, we demonstrate that SARS-CoV-2 infection of human monocyte-derived macrophages (MDMs) and monocyte-derived dendritic cells was abortive, but induced the production of multiple antiviral and proinflammatory cytokines (interferon-α, interferon-β, tumor necrosis factor, and interleukins 1β, 6, and 10) and a chemokine (CXCL10). Despite the lack of efficient replication in MDMs, SARS-CoV-2 induced profound interferon-mediated cell death of host cells. Macrophage activation and death were not enhanced by exposure to low levels of convalescent plasma, suggesting that antibody-dependent enhancement of infection does not contribute to cell death. Together, these results indicate that infection of macrophages and dendritic cells potentially plays a major role in coronavirus disease 2019 pathogenesis, even in the absence of productive infection.
Keywords: COVID-19, SARS-CoV-2, macrophage, IFN, cell death
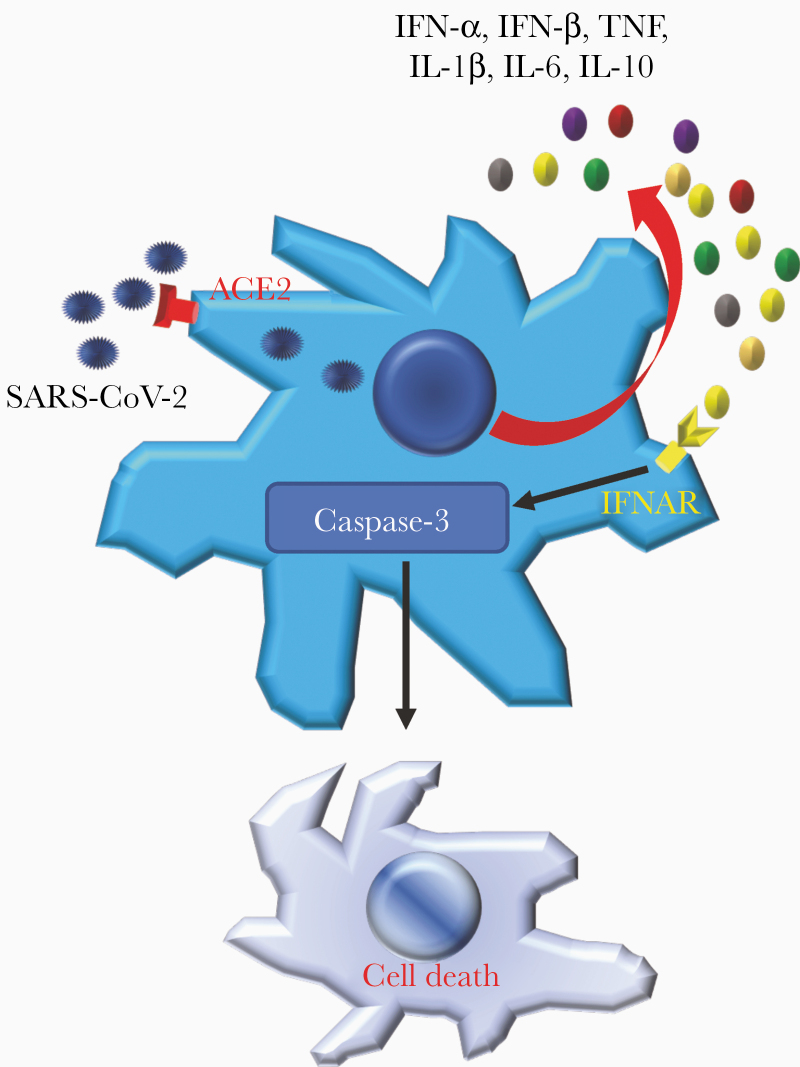
Severe acute respiratory syndrome coronavirus 2 exhibited angiotensin converting enzyme 2 dependent abortive infection of human myeloid cells. The infection induced expression of antiviral and proinflammatory cytokines and chemokines, which ultimately resulted in type I interferon-mediated cell death of human macrophages.
Graphical Abstract
The global pandemic of coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is characterized by severe pneumonia and multiple systemic complications (eg, multiorgan damage including endothelialitis, thrombosis, and myocardial damage) [1]. Although respiratory tract epithelial cells are the primary target cells of SARS-CoV-2 [2], other respiratory tract–resident cells, such as macrophages in alveoli and pulmonary hilum lymphoid tissue, also contain SARS-CoV-2 antigen [3]. Also, circulating monocytes migrate to multiple tissues and, in the context of other infections, have been implicated in the spread of virus to distant organs and tissues [4].
In addition, myeloid cells also orchestrate host immune responses to virus infections. Alveolar macrophages sense and respond to microbial threats by producing bioactive molecules to eliminate pathogens and promote tissue repair [5], while the infiltration of circulating myeloid cells, especially pathogenic inflammatory monocyte-macrophages, may result in elevated lung cytokine/chemokine levels, vascular leakage, and even impaired virus-specific T-cell responses [6]. A dysregulated macrophage response may promote excessive neutrophil infiltration and tissue damage, leading to macrophage activation syndrome and increased morbidity and mortality [7, 8]. The beneficial or deleterious role of lung-resident or infiltrating macrophages is determined largely by their secretion of cytokines and chemokines [9, 10].
Previous studies of the severe acute respiratory syndrome and Middle East respiratory syndrome coronaviruses [11] (SARS-CoV and MERS-CoV, respectively) showed that human macrophages and dendritic cells are potent sources of proinflammatory inflammatory mediators [4, 6, 12]. SARS-CoV abortively infects these myeloid cells [13], but whether MERS-CoV infection is abortive or productive is controversial [12, 14]. SARS-CoV-2–infected macrophages or dendritic cells could potentially produce multiple proinflammatory cytokines and chemokines, such as interleukin (IL) 6, IL-8, tumor necrosis factor (TNF), and CXCL-10, contributing to local tissue injury and pathogenic systemic inflammatory responses (cytokine storm) [15–17]. Proinflammatory mediator induction (IL-6, IL-8, and type I and III interferons [IFNs], among others) enhances NF-kB–mediated [18] activation and, possibly, immune dysfunction and results in multiple organ failure in severe COVID-19 cases [19–21].
In this study, using human circulating monocyte-derived macrophages (MDMs) and monocyte-derived dendritic cells (MDDCs), we showed that SARS-CoV-2 abortively infected both types of cells in an angiotensin-converting enzyme 2 (ACE2)–dependent manner. Abortive infection induced expression of higher levels of cytokines and chemokines (interferon [IFN]–α, TNF, IL-6, IL-8, IL-10, and CXCL-10), leading to type I IFN–mediated cell death. In comparison to SARS-CoV, SARS-CoV-2 induced high levels of most cytokines and chemokines and more cell death.
MATERIALS AND METHODS
Human Subjects Approval
Written informed consent was obtained from subjects to obtain plasma for this study. The study was approved by the Institutional Review Board (IRB) of the University of Iowa (IRB numbers 202003554 and 201402735).
Cells and Virus
Human peripheral blood samples were obtained from anonymous donors at the DeGowin Blood Center at the University of Iowa. Consent forms were approved by the University of Iowa’s IRB. To obtain monocytes, peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation through Ficoll-Paque PLUS density gradient media (Cytiva) and cultured at a seeding density of 1 × 106 cells/mL in RP-10 media (RPMI-1640 medium [Invitrogen] supplemented with 10% fetal bovine serum [FBS; Atlanta Biologicals]), 2 mM L-glutamine) and 5 ng/mL macrophage colony-stimulating factor (eBioscience) (macrophages) or 100 ng/mL granulocyte macrophage colony-stimulating factor (PeproTech) + 50 ng/mL IL-4 (PeproTech) (MDDCs) at 37°C with 5% carbon dioxide. After 96 hours, the plates were washed with Hanks’ balanced salt solution devoid of divalent cations (Invitrogen) to remove nonadherent cells. Adherent cells were then trypsinized, pelleted, and cultured for 10 days.
The 2019n-CoV/USA-WA1/2019 strain of SARS-CoV-2 (accession number: MT123290) used in these studies was passaged on Calu-3 2B4 cells. SARS-CoV was obtained from Dr Kanta Subbarao (National Institutes of Health) and passaged on Vero E6 cells. Calu-3 2B4 cells were grown in Minimal Essential Media (Gibco, Grand Island, New York) supplemented with 20% FBS. Vero E6 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% FBS.
Virus Infection and Titration
Human PBMC, MDMs, or MDDCs were infected with SARS-CoV-2 or SARS-CoV at a multiplicity of infection (MOI) of 2 in DMEM at 37°C. One hour later, the culture medium was replaced with 10% FBS-DMEM with or without washing with phosphate-buffered saline (PBS). In some experiments, virus was inactivated by ultraviolet (UV) irradiation. Inactivation was confirmed by plaque assay as described below.
Virus titers in culture medium were determined using Vero E6 cells (grown in 10% FBS-DMEM). For plaque assays, cells were fixed with 10% formaldehyde 3 days after infection and stained with crystal violet.
Confocal Microscopy
Cultured MDMs were plated in Chamber Slide (Thermo Fisher) before infection. At the indicated time points postinfection, supernatants were removed, and cells were fixed with 4% paraformaldehyde. For staining, sections were blocked with goat serum for 2 hours in humidity chambers at room temperature. Sections were then treated with primary antibodies (rabbit anti–SARS-CoV-2 nucleoprotein [N protein] [Sino Biological] and mouse antihuman caspase-3 [Abcam]) at 4°C overnight. After washing with PBS, samples were treated with secondary antibodies (Alexa 488 goat antirabbit immunoglobulin G [IgG], Alexa 562 goat antimouse IgG, Abcam) for 30 minutes at room temperature. Finally, slides were overlaid with 4′,6-diamidino-2-phenylindole (Vector) and examined using a confocal microscope (Zeiss 710).
Transmission Electron Microscopy
After fixation with 2.5% glutaraldehyde in 0.1 M sodium cacodylate, the cell monolayers were treated with 1% osmium tetroxide containing 1.5% potassium ferrocyanide followed by 2.5% uranyl acetate. Samples were dehydrated using increasing concentrations of ethanol, and embedded in Eponate 12 resin (Ted Pella, Redding, California). Resin was polymerized in a 65°C oven overnight. Ultrathin sections (70–90 nm) were cut with a Leica UC6 ultramicrotome (Leica Microsystems). Sections were collected on copper grids and sequentially stained with 5% uranyl acetate and Reynold lead citrate. The sections were viewed using a Jeol 1230 transmission electron microscope (Jeol USA, Peabody, Massachusetts) equipped with a Gatan Ultrascan 2k × 2k CCD camera.
Flow Cytometry Assays
The following monoclonal antibodies were used: antigen-presenting cell (APC) eFluor780-conjugated mouse antihuman CD8 (SK1, eBioscience), fluorescein isothiocyanate–conjugated mouse antihuman CD3 (HIT3a), APC-conjugated mouse antihuman CD19 (4G7), phycoerythrin-conjugated mouse antihuman CD14 (63D3), Pacific Blue–conjugated mouse antihuman CD56 (5.1H11), PerCP Cy5.5-conjugated mouse antihuman CD4 (RPA-T4, Biolegend), rabbit anti–SARS-CoV-2 N protein (Sino Biological), and Alexa 488 goat antirabbit IgG (Abcam). For surface staining, 1 × 106 cells were stained with the indicated antibodies at 4°C. For SARS-CoV-2 N protein staining, cells were fixed and permeabilized using Cytofix/Cytoperm Solution (BD Biosciences) after washing and stained with the indicated antibodies. A LIVE/DEAD cell viability assay kit (Invitrogen) was used for gating live cells. Flow cytometric data were acquired using a FACSVerse and were analyzed using FlowJo software (Tree Star).
Western Blot Assays
Cell lysates were collected from cells lysed in Nonidet P-40 cell lysis buffer [22] supplemented with a protease inhibitor cocktail [22] and a phosphatase inhibitor cocktail [22]. Protein samples were run on 4%–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to activated polyvinylidene fluoride membranes. Membranes were incubated overnight with isotype control IgG antibody or SARS-CoV-2 nucleocapsid antibody (Sino Biological; 1:1000 in intercept blocking buffer [IBB], Li-Cor). Membranes were then incubated with IRDye 680RD goat antirabbit IgG secondary antibody (Li-Cor) at 1:15 000 for 1 hour in IBB. Membrane was imaged using a Li-Cor Odyssey Imaging System.
Human Plasma Treatment
High-titer COVID-19 convalescent plasma (half maximal inhibitory concentration = 1:1480, determined by a neutralizing titer assay using a luciferase-expressing SARS-CoV-2 S protein pseudovirus system as previously described [23]) and control plasma were collected (IRB-approved protocol numbers 202003554 and 201402735). The plasma was diluted at 1:5 (final concentration: 10%) or 1:50 (final concentration: 1%) in DMEM and mixed 1:1 with SARS-CoV-2 for 1 hour at 37°C. The virus-antibody mixtures were then adsorbed to MDMs for 1 hour at 37°C.
Blocking Assay
To block viral entry, MDMs were incubated with α-ACE2 monoclonal antibody (AF933, R&D Systems) or isotype control IgG at a final concentration of 10 µg/mL for 1 hour prior to SARS-CoV-2 infection.
To block type I IFN signaling, IFN-α receptor (α-IFNAR) monoclonal antibody (clone TAB-722, Creative Biolabs) or isotype control IgG was added to cells at a final concentration of 10 µg/mL at 12 hours after SARS-CoV-2 infection.
Real-Time Polymerase Chain Reaction Assays
Total RNA was extracted from MDMs using Trizol (Invitrogen) according to the manufacturer’s protocol. Following DNase treatment, 200 ng of total RNA was used as a template for first-strand complementary DNA (cDNA). The resulting cDNA was subjected to amplification of selected genes by real-time quantitative polymerase chain reaction (PCR) using Power SYBR Green PCR Master Mix (Applied Biosystems). Average values from duplicate samples were used to calculate the relative abundance of transcripts normalized to hypoxanthine-guanine phosphoribosyltransferase and presented as 2-ΔCT. The following primers were used for cytokine and chemokine detection: IFN-α: 5′-AGA AGG CTC CAG CCA TCT CTG T-3′, 5′-TGC TGG TAG AGT TCG GTG CAG A-3′; IFN-β: 5′-CTT GGA TTC CTA CAA AGA AGC AGC-3′, 5′-TCC TCC TTC TGG AAC TGC TGC A-3′; TNF: 5′-CTC TTC TGC CTG CTG CAC TTT G-3′, 5′-ATG GGC TAC AGG CTT GTC ACT C-3′; IL-1β: 5′-CCA CAG ACC TTC CAG GAG AAT G-3′, 5′-GTG CAG TTC AGT GAT CGT ACA GG-3′; IL-6: 5′-AGA CAG CCA CTC ACC TCT TCA G-3′, 5′-TTC TGC CAG TGC CTC TTT GCT G-3′; IL-8: 5′-GAG AGT GAT TGA GAG TGG ACC AC-3′, 5′-CAC AAC CCT CTG CAC CCA GTT T-3′; IL-10: 5′-TCT CCG AGA TGC CTT CAG CAG A-3′, 5′-TCA GAC AAG GCT TGG CAA CCC A-3′; CCL2: 5′-CAG GTG ACA GAG ACT CTT GGG A-3′, 5′-GGC AAT CCT ACA GCC AAG AGC T-3′; CXCL10: 5′-GGT GAG AAG AGA TGT CTG AAT CC-3′, 5′-GTC CAT CCT TGG AAG CAC TGC A-3′. For detection of viral genomes, the following primers were used (ORF1ab): 5′-CCC TGT GGG TTT TAC ACT TAA-3′, 5′-ACG ATT GTG CAT CAG CTG A-3′, 5′-CCG TCT GCG GTA TGT GGA AAG GTT ATG G-3′ and subgenomic RNA (N protein RNA): SARS-CoV-2: 5′-GAC CCC AAA ATC AGC GAA AT-3′, 5′-TCT GGT TAC TGC CAG TTG AAT CTG-3′; SARS-CoV: 5′-CAG AAC GCT GTA GCT TCA AAA ATC T-3′, 5′-CAG AAC CCT GTG ATG AAT CAA CAG-3′.
Statistical Analysis
Student t test was used to analyze differences in mean values between groups. All results are expressed as mean ± standard error of the mean. P values < .05 were considered statistically significant.
RESULTS
Abortive Infection of Human MDMs and MDDCs by SARS-CoV-2
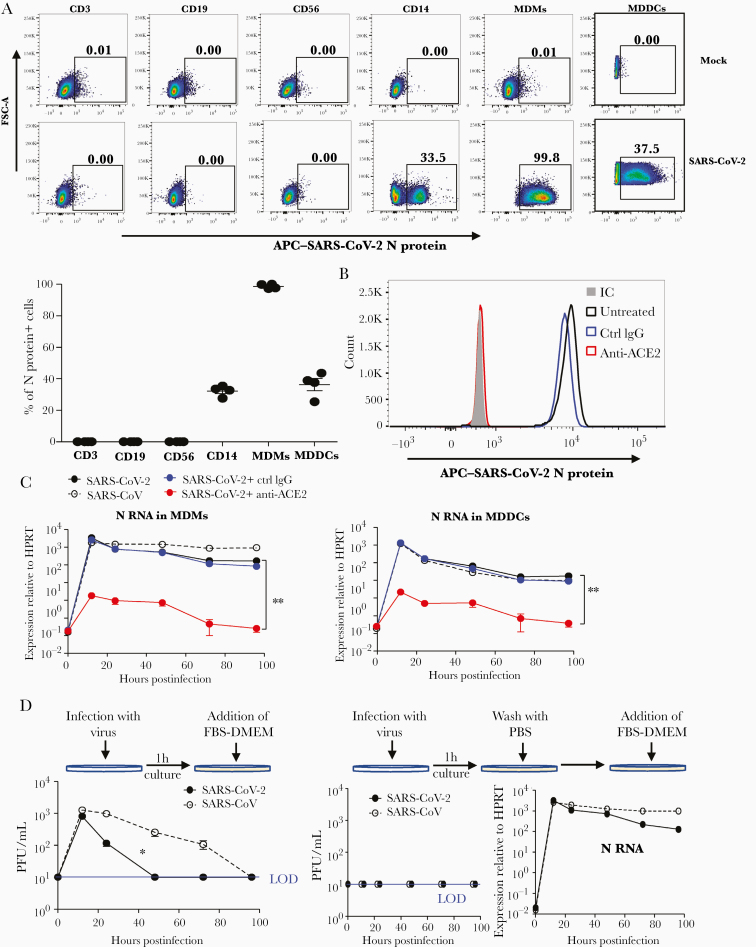
To determine SARS-CoV-2 infectivity of human immune cells, PBMCs and in vitro–differentiated myeloid cells (MDMs or MDDCs) from healthy donors were infected with SARS-CoV-2 at an MOI of 2. Twenty-four hours later, the expression of SARS-CoV-2 N protein in immune cell subsets was determined by flow cytometry. As shown in Figure 1A and 1B, SARS-CoV-2 N protein could be detected in CD14+ monocytes, MDMs, and MDDCs, but not CD3+ T cells, CD19+ B cells, or CD56+ natural killer (NK) cells (gating strategy is shown in Supplementary Figure 1). MDM and MDDC infection required entry through ACE2, the SARS-CoV-2 receptor, since ACE2 blockade prevented virus entry (Figure 1C–E).
Figure 1.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) abortive infection of human monocyte-derived macrophages (MDMs) and monocyte-derived dendritic cells (MDDCs). A and B, Human peripheral blood mononuclear cells (PBMCs), MDMs, and MDDCs were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 2. Infected immune cell subsets were identified by flow cytometry using an anti-SARS-CoV-2 N protein antibody. C, SARS-CoV-2 N protein expression in SARS-CoV-2–infected MDMs treated with an anti–human angiotensin-converting enzyme 2 antibody or its isotype control. D–G, MDMs (D, F, and G) or MDDCs (E) were infected with SARS-CoV-2 or severe acute respiratory syndrome coronavirus at MOI of 2, followed by washing with phosphate-buffered saline (G) or not (D–F), before addition of 10% fetal bovine serum–Dulbecco’s modified Eagle’s medium. At indicated time points, culture media were collected and cells were treated with Trizol for RNA extraction. SARS-CoV-2 N RNA expression was determined by quantitative polymerase chain reaction (D, E, and G). Infectious viral particles in culture medium were detected by plaque assays (F and G). B and D–G, Data are shown as mean ± standard error of the mean and are representative of 3 independent experiments (n = 4–8). *P < .05; **P < .01. See also Supplementary Figures 1 and 2. Abbreviations: ACE2, angiotensin-converting enzyme 2; APC, antigen-presenting cell; Ctrl, control; FBS-DMEM, fetal bovine serum–Dulbecco’s modified Eagle’s medium; FSC-A, forward scatter area; HPRT, hypoxanthine-guanine phosphoribosyltransferase; IC, isotype control antibody; IgG, immunoglobulin G; LOD, limit of detection; MDDC, monocyte-derived dendritic cell; MDM, monocyte-derived macrophage; PBS, phosphate-buffered saline; PFU, plaque-forming units; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
To further examine replication in human MDMs and MDDCs, RNA was harvested from infected cells at the indicated time points postinfection and evaluated for viral RNA by quantitative PCR. Viral N protein RNA could be detected in both MDMs and MDDCs at 12 hours postinfection (hpi) and decreased 10-fold (MDMs) and 100-fold (MDDCs) over the following 96 hours (Figure 1D and 1E). Notably, viral genomic RNA (ORF1ab, gRNA) levels were much lower in MDMs and MDDCs compared to both Vero E6 and Calu-3 (Supplementary Figure 2A and 2B). Expression levels of viral N protein were lower in MDMs compared to Vero E6 cells as determined by Western blot assay (Supplementary Figure 2C). In addition, while virus could be detected in culture medium of MDMs within 24 hpi (Figure 1F), it represented residual virus inoculum because washing cells with PBS after virus adsorption reduced levels in the culture medium to nil, but did not affect N protein RNA expression in infected cells (Figure 1G). Taken together, these data showed that human MDMs and MDDCs could be infected by SARS-CoV-2 but failed to support the production of infectious virus. Similarly and consistent with previous studies [13, 24], SARS-CoV also resulted in abortive infection of MDMs and MDDCs.
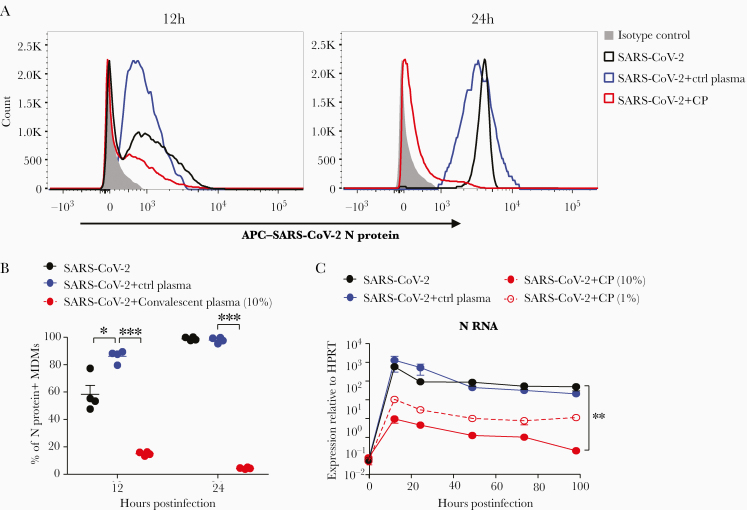
Figure 2.
Coronavirus disease 2019 convalescent plasma (CP) blocks the entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in human monocyte-derived macrophages (MDMs). SARS-CoV-2 was incubated with CP or control plasma as described in the Materials and Methods before infecting human MDMs. A and B, N protein expression in SARS-CoV-2–infected MDMs treated with CP or control plasma was determined by flow cytometry at indicated time points. C, SARS-CoV-2 N RNA expression in MDMs was detected by quantitative polymerase chain reaction. B and C, Data are shown as mean ± standard error of the mean and are representative of 3 independent experiments (n = 4–6). *P < .05; **P < .01; ***P < .001. Abbreviations: APC, antigen-presenting cell; CP, convalescent plasma; Ctrl, control; HPRT, hypoxanthine-guanine phosphoribosyltransferse; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Convalescent Plasma Blocked SARS-CoV-2 Infection of MDMs
To further distinguish ACE2-mediated entry from nonspecific virus uptake by MDMs, virus was incubated with media containing 10% or 1% human COVID-19 convalescent plasma (CP, original titer: 1:1480) or control plasma from a healthy donor before infection. CP treatment decreased the percentage of SARS-CoV-2 N protein+ MDMs (Figure 2A and 2B) and viral RNA expression (Figure 2C). In addition, we found no evidence of enhanced macrophage infection when cells were treated with a low dose of CP (final concentration: 1%, neutralizing titer 1:14.8) (Figure 2C).
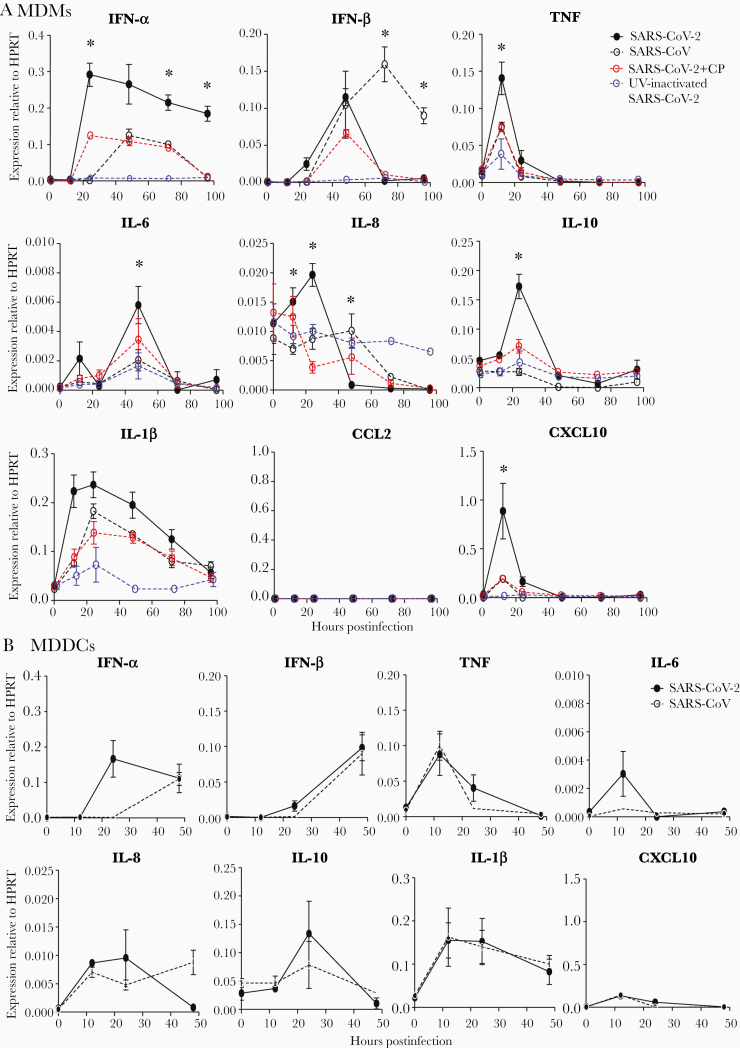
SARS-CoV-2 Induces Cytokine and Chemokine Production by MDMs and MDDCs
Since MDMs-derived cytokines and chemokines play critical roles in limiting viral infection and in pathogenic processes, we evaluated cytokine and chemokine messenger RNA production by virus-infected MDMs (Figure 3A) and MDDCs (Figure 3B) at several time points after infection. To provide a framework for interpreting the results, we infected parallel cultures with SARS-CoV and UV-inactivated SARS-CoV-2. Infectious SARS-CoV-2, but not UV-inactivated SARS-CoV-2, induced higher expression of IFN-α, TNF, IL-6, IL-8, IL-10, and CXCL-10 and similar levels of IL-1β compared to SARS-CoV. SARS-CoV preferentially induced IFN-β whereas SARS-CoV-2 induced higher and prolonged expression of IFN-α. In addition, cytokine and chemokine production was dynamic after infection with expression of some molecules peaking at 24 hours after SARS-CoV-2 infection (eg, CXCL10, IL-10) or earlier (TNF) while maximal levels of IL-6 were reached at 48 hpi. As expected, treatment with low amounts of CP (final concentration: 1%) decreased the expression of these cytokines and chemokines, again inconsistent with antibody disease enhancement. Compared to those of MDMs, the expression levels of cytokines and chemokines were lower in SARS-CoV-2–treated MDDCs, implying distinct roles of these 2 cell types in infection.
Figure 3.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–infected human monocyte-derived macrophages (MDMs) and monocyte-derived dendritic cells (MDDCs) produce multiple cytokines/chemokines. MDMs (A) or MDDCs (B) were exposed to severe acute respiratory syndrome coronavirus (SARS-CoV) or SARS-CoV-2 (with or without convalescent plasma treatment), or ultraviolet-inactivated SARS-CoV-2. RNA was extracted at the indicated time points postinfection. Cytokine/chemokine levels were determined by quantitative polymerase chain reaction as described in the Materials and Methods. Data are shown as mean ± standard error of the mean and are representative of 3 independent experiments (n = 4–6). *Difference between the SARS-CoV-2 and SARS-CoV groups (P < .05). Abbreviations: CP, convalescent plasma; HPRT, hypoxanthine-guanine phosphoribosyltransferase; IFN, interferon; IL, interleukin; MDDC, monocyte-derived dendritic cell; MDM, monocyte-derived macrophage; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TNF, tumor necrosis factor; UV, ultraviolet.
SARS-CoV-2 Infection Results in Type I IFN–Mediated Cell Death of MDMs
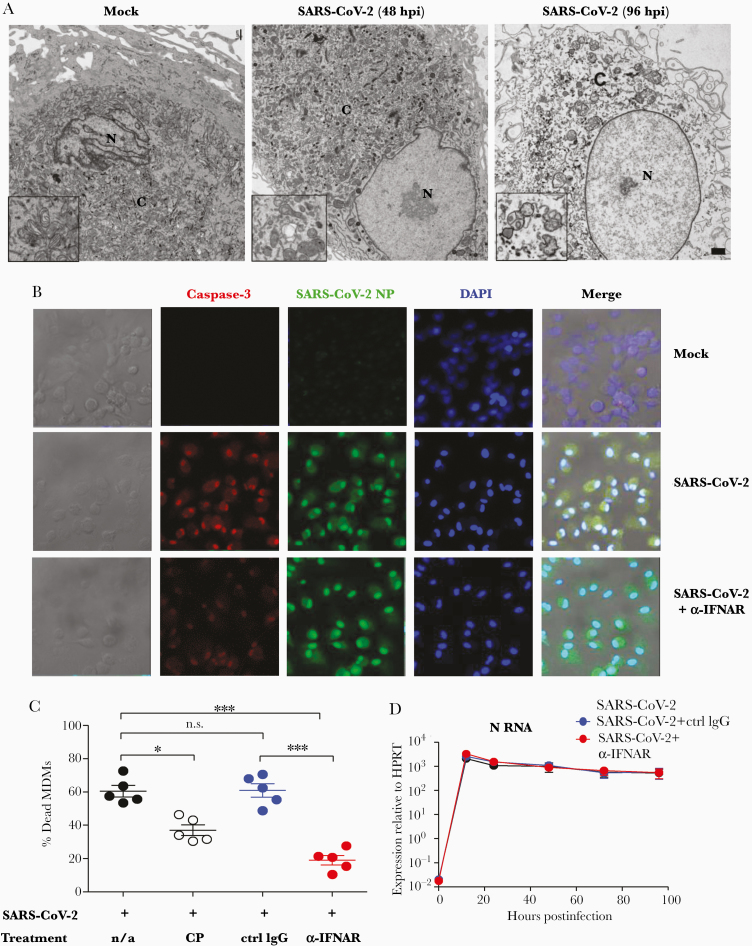
To determine whether abortive infection in combination with elevated expression of proinflammatory cytokines and chemokines resulted in additional phenotype changes such as cell death, MDMs were infected and examined for ultrastructural changes by transmission electron microscopy [18]. The nucleus and cytoplasm of infected macrophages showed progressive electron lucency with dilation of the cyto-cavitary network. At 48 hpi, cells exhibited early signs of death, which progressed to frank cell death by 96 hpi. Consistent with these changes, mitochondria, which were normal in appearance prior to infection (Figure 4A, left panel, inset), were dysmorphic by 48 hpi (Figure 4A, center panel, inset). By 96 hpi, mitochondria exhibited extensively blebbing (right panel, inset), indicative of cell death. Of note, we found no virus particles or virus replication structures such as double-membrane vesicles (DMVs) in these infected cells. To gain further insight into the mechanism of cell death, we analyzed MDMs for the coexpression of SARS-CoV-2 N protein and caspase-3 (a marker of cell apoptosis) by confocal microscopy at 96 hpi (Figure 4B). Nearly all of the cells expressed caspase-3, consistent with extensive apoptosis. To determine whether IFN-α/β contributed to the development of cell death, we treated infected cells with IFN-α/β receptor (IFNAR) blocking antibody. As shown in Figure 4B and 4C and Supplementary Figure 3A, blocking IFNAR at 12 hpi decreased the percentage of dead MDMs, determined using a LIVE/DEAD cell viability assay kit, when cells were examined at 96 hpi. Treatment with IFNAR blocking antibody did not change the percentage of infected cells or levels of virus RNA (Figure 4D and Supplementary Figure 3B), suggesting that type I IFNs contributed to SARS-CoV-2–induced cell death. Moreover, treatment with low concentrations (1%) of CP did not increase cell death (Figure 4C). In contrast to results obtained with MDMs, a smaller fraction of MDDCs underwent death after infection with SARS-CoV-2 (Supplementary Figure 3C).
Figure 4.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of human monocyte-derived macrophages (MDMs) results in interferon (IFN)–dependent cell death. A, Representative transmission electron micrographs of SARS-CoV-2–infected human macrophages from mock and SARS-CoV-2 groups. N: nucleus, C: cytosol; bar = 1 µm. Images shown are representative of 3 independent donors. Insets show progressive loss of mitochondrial integrity as infection proceeds. B, SARS-CoV-2–infected MDMs were fixed at 96 hours postinfection (hpi) and prepared for confocal microscopy as described in the Materials and Methods. Some wells were treated with anti-alpha/beta interferon antibody (α-IFNAR) antibody at 12 hpi. The expression and co-localization of SARS-CoV-2 N protein and caspase-3 are shown. C and D, Virus was incubated with convalescent plasma (final concentration: 1%) or cultures were treated with α-IFNAR antibody at 12 hpi. Isotype matched antibodies were included as controls. Dead MDMs were identified at 96 hpi using a LIVE/DEAD cell viability assay kit with flow cytometry as shown in Supplementary Figure 3A (C). SARS-CoV-2 N protein RNA expression in MDMs was detected by quantitative polymerase chain reaction at the indicated time points postinfection (D). Data are shown as mean ± standard error of the mean and are representative of 3 independent experiments (n = 5). *P < .05; ***P < .001. See also Supplementary Figure 3. Abbreviations: α-IFNAR, anti alpha/beta interferon receptor antibody; CP, convalescent plasma; Ctrl, control; DAPI, 4′,6-diamidino-2-phenylindole; hpi, hours postinfection; HPRT, hypoxanthine-guanine phosphotransferase; IgG, immunoglobulin G; MDM, monocyte-derived macrophage; n/a, not applicable; NP, nucleocapsid protein; n.s., not significant; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
DISCUSSION
Here, we demonstrate that human MDMs are abortively infected by SARS-CoV-2, in agreement with a previous report [24]. We additionally show that MDDCs do not support productive SARS-CoV-2 infection. Levels of intracellular viral RNA and protein were very low in both cell types and we detected no virus or viral replicative structures by TEM. We also show that entry is ACE2 and S protein dependent, since anti-ACE2 antibody (Figure 1C–E) and CP (Figure 2) from a patient with COVID-19 blocked the entry of SARS-CoV-2 into MDMs and MDDCs. Furthermore, a lower dose of CP (final concentration: 1%) did not enhance viral entry, cytokine expression, or cell death (Figures 2C, 3A, and 4C), suggesting that antibody-dependent enhancement of macrophage infection did not occur. Although we observed no antibody-dependent enhancement, it will still be important to carefully evaluate patients for disease enhancement in clinical trials of vaccination and antibody treatment and in patients who show evidence of reinfection [25]. Of note, in disagreement with a recent report [26], we found no evidence that T cells, B cells, or NK cells (Figure 1A) were infected by the virus.
Remarkably, given the low expression level of viral RNA and protein, SARS-CoV-2 infection invoked potent cytokine and chemokine production by human MDMs and less by MDDCs (Figure 3). Chemokine and cytokine expression were not induced solely by virus binding to the ACE2 receptor because they were not induced by UV-inactivated virus (Figure 3A). Whether viral RNA or protein expressed during abortive replication signal through Toll-like receptors or retinoic acid–inducible gene I–like receptors to activate IFN- and NF-kB–dependent pathways will require further investigation. Several SARS-CoV-2 proteins have been reported to activate or counter the innate immune response [18, 27], but at this point, it is not known whether these proteins, double-stranded viral RNA, or the intrinsic process of virus replication (eg, membrane distortion to form sites of RNA replication [28]) are most important in this process. Furthermore, we assessed cytokine/chemokine RNA expression longitudinally and found that infection of MDMs induced higher expression of IFN-α, TNF, IL-6, IL-8, IL-10, and CXCL-10 and similar expression of IFN-β and IL-1β when compared to SARS-CoV (Figure 3A). Of note, neither virus was as potent an inducer of proinflammatory molecule expression as H5N1, H1N1pdm, or MERS-CoV [24]. Since mortality in COVID-19 patients is associated in some cases with dysregulated and elevated proinflammatory molecule expression [1, 3], this observation suggests that other cells, such as lung epithelial cells or uninfected bystander myeloid cells, are major contributors to expression of these molecules. Nonetheless, since macrophage-derived cytokines and chemokines contribute to both host defense and tissue damage, these results suggest that even abortive infection of myeloid cells could contribute to a hyperinflammatory milieu. For example, CXCL10 plasma levels are highly associated with COVID-19 severity and may predict disease progression [29, 30]. TNF levels were found to correlate with the abundance of proinflammatory CXCL10+CCL2+ macrophages in severe COVID-19 [31], which might exacerbate inflammation in a positive feedback loop.
IL-6 and IL-1β expressions were also shown to correlate with disease severity [16, 17, 30]. Although blocking IL-6 therapy was postulated to improve clinical outcome [32–34], recent controlled trials did not support its efficacy in treatment of COVID-19 patients [35]. IL-1β signaling blockade using anakinra was also found to provide benefit in some COVID-19 cases [36–39]. However, confirmation of IL-1β blockade efficacy will require larger controlled trials.
We also demonstrate that human MDM-derived IFN-α/β participated in cell death in an autocrine or paracrine manner after SARS-CoV-2 infection (Figure 4C). Similarly, IFN-α, -β, and -γ treatment of human alveolospheres [2] resulted in cell death [40]. Induction of caspase-3 suggests that MDMs underwent apoptosis. However, this conclusion needs further validation because caspase-3 is also associated with necrotic/pyroptotic cell death in a few settings [41]. These results are in contrast to studies of apoptosis in MDMs infected with a common cold human coronavirus (HCoV-229E), which is dependent on viral replication, and not TNF-related apoptosis-inducing ligand, FasL, TNF, or caspase activity. HCoV-229E infection of PBMC-derived myeloid cells shows preferential killing of dendritic cells but not macrophages [42]. The lower expression of proinflammatory cytokines in SARS-CoV-2–infected MDDCs, compared to that of MDMs, might contribute to their lower rates of death (Supplementary Figure 4C), suggesting that the role of IFN signaling in different myeloid cell types is variable. In addition to effects on expression of proinflammatory mediators, enhanced myeloid cell death may contribute to adaptive immune responses. Despite the lack of viral particles and DMV structures in infected cells (Figure 4A), dying macrophages might contain sufficient viral antigen so that they can be phagocytosed by APCs to induce T-cell– or B-cell–mediated immune responses.
In summary, these results illustrate some of the complexities of the pathogenesis of infections caused by SARS-CoV-2 and other human respiratory coronaviruses. Even when the infection is abortive, the consequence may be expression of proinflammatory mediators that contribute to pathogenic outcomes. These results reinforce the need for antiviral agents, as infectious but not UV-treated virus causes macrophage activation and cell death, and for agents that dampen the host immune response, diminishing the exuberant inflammatory response to which dying macrophages contribute.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Dr Thomas Gallagher for critical review and discussion of the manuscript.
Financial support. This project was supported by grants from the National Institutes of Health (grant numbers RO1 AI129269 and PO1 AI060699 to S. P.).
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed..
References
- 1. Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of Covid-19 in New York city. N Engl J Med 2020; 382:2372–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sungnak W, Huang N, Bécavin C, et al. ; HCA Lung Biological Network . SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 2020; 26:681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang C, Xie J, Zhao L, et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 2020; 57:102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci 2020; 257:118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hashimoto D, Chow A, Noizat C, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 2013; 38:792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016; 19:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McGonagle D, Sharif K, O’Regan A, Bridgewood C. The role of cytokines including interleukin-6 in COVID-19 induced pneumonia and macrophage activation syndrome-like disease. Autoimmun Rev 2020; 19:102537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schurink B, Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe 2020; 1:e290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Channappanavar R, Perlman S. Evaluation of activation and inflammatory activity of myeloid cells during pathogenic human coronavirus infection. Methods Mol Biol 2020; 2099:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol 2020; 20:355–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shubina M, Tummers B, Boyd DF, et al. Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J Exp Med 2020; 217:e20191259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chu H, Zhou J, Wong BH, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology 2014; 454–455:197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung CY, Poon LL, Ng IH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol 2005; 79:7819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scheuplein VA, Seifried J, Malczyk AH, et al. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol 2015; 89:3859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020; 26:1200–4. [DOI] [PubMed] [Google Scholar]
- 16. Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020; 26:1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang X, Tan Y, Ling Y, et al. Viral and host factors related to the clinical outcome of COVID-19. Nature 2020; 583:437–40. [DOI] [PubMed] [Google Scholar]
- 18. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020; 583:459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sallenave JM, Guillot L. Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS-CoV-2 in Covid-19: key therapeutic targets? Front Immunol 2020; 11:1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020; 130:2620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020; 369:718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng J, Wong LR, Li K, et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice [manuscript published online ahead of print 9 November 2020]. Nature 2020. doi:10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hui KPY, Cheung MC, Perera RAPM, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med 2020; 8:687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arvin AM, Fink K, Schmid MA, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 2020; 584:353–63. [DOI] [PubMed] [Google Scholar]
- 26. Pontelli MC, Castro IA, Martins RB, et al. Infection of human lymphomononuclear cells by SARS-CoV-2 [published online ahead of print 2020]. bioRxiv. DOI: 10.1101/2020.07.28.255912. [DOI] [Google Scholar]
- 27. Miorin L, Kehrer T, Sanchez-Aparicio MT, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A 2020; 117:28344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020; 369:1395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Y, Shen C, Li J, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol 2020; 146:119–27.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Laing AG, Lorenc A, Del Molino Del Barrio I, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med 2020; 26:1623–35. [DOI] [PubMed] [Google Scholar]
- 31. Zhang F, Mears JR, Shakib L, et al. IFN- gamma and TNF- alpha drive a CXCL10+CCL2+ macrophage phenotype expanded in severe COVID-19 and other diseases with tissue inflammation. bioRxiv [Preprint]. Posted online 5 August 2020. 10.1101/2020.08.05.238360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guillén L, Padilla S, Fernández M, et al. Preemptive interleukin-6 blockade in patients with COVID-19. Sci Rep 2020; 10:16826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guaraldi G, Meschiari M, Cozzi-Lepri A, et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol 2020; 2:e474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biran N, Ip A, Ahn J, et al. Tocilizumab among patients with COVID-19 in the intensive care unit: a multicentre observational study. Lancet Rheumatol 2020; 2:e603–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. King A, Vail A, O’Leary C, et al. Anakinra in COVID-19: important considerations for clinical trials. Lancet Rheumatol 2020; 2:e379–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dimopoulos G, de Mast Q, Markou N, et al. Favorable anakinra responses in severe Covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell Host Microbe 2020; 28:117–23.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cavalli G, De Luca G, Campochiaro C, et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol 2020; 2:e325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huet T, Beaussier H, Voisin O, et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol 2020; 2:e393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wampler Muskardin TL. Intravenous anakinra for macrophage activation syndrome may hold lessons for treatment of cytokine storm in the setting of coronavirus disease 2019. ACR Open Rheumatol 2020; 2:283–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Katsura H, Sontake V, Tata A, et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 2020; 27:890–904.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun 2017; 8:14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mesel-Lemoine M, Millet J, Vidalain PO, et al. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J Virol 2012; 86:7577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.