Abstract
We evaluated the performance of the Abbott BinaxNOW rapid antigen test for coronavirus disease 2019 (Binax-CoV2) to detect virus among persons, regardless of symptoms, at a public plaza site of ongoing community transmission. Titration with cultured severe acute respiratory syndrome coronavirus 2 yielded a human observable threshold between 1.6 × 104-4.3 × 104 viral RNA copies (cycle threshold [Ct], 30.3–28.8). Among 878 subjects tested, 3% (26 of 878) were positive by reverse-transcription polymerase chain reaction, of whom 15 of 26 had a Ct <30, indicating high viral load; of these, 40% (6 of 15) were asymptomatic. Using this Ct threshold (<30) for Binax-CoV2 evaluation, the sensitivity of Binax-CoV2 was 93.3% (95% confidence interval, 68.1%–99.8%) (14 of 15) and the specificity was 99.9% (99.4%–99.9%) (855 of 856).
Keywords: COVID-19, SARS-CoV-2, Rapid Antigen Test, Point of Care testing
This study examines the utility of the Abbott BinaxNOW rapid direct antigen severe acute respiratory syndrome coronavirus 2 in the context of community screening at a public transit hub, in comparison with reverse-transcription polymerase chain reaction.
The global pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has spread at an unprecedented pace [1] fueled by efficient transmission of infection by the respiratory route, including by asymptomatic and presymptomatic persons. Instances of successful control make use of masking, social distancing, and aggressive testing, tracing, and quarantine [2].
To date, the cornerstone of testing has been reverse-transcription polymerase chain reaction (RT-PCR) examination of respiratory secretions, which has excellent sensitivity and specificity but is expensive and requires sophisticated equipment and highly trained personnel [3]. In practice, these features have often generated testing delays compromising their utility [4]. As a result, there is interest in rapid and economical assays that circumvent these limitations [5]. However, methods that do not include an amplification step are inherently less sensitive; their proper deployment will therefore require a rigorous evaluation of performance characteristics in different epidemiologic settings.
Lateral flow antigen detection diagnostics have been deployed for a variety of infectious diseases including malaria, RSV, and influenza. The Abbott BinaxNOW COVID-19 Ag Card (hereafter referred to as Binax-CoV2) is one such assay that detects viral nucleocapsid (N) protein directly from nasal swab samples. The test requires no instrumentation; results are scored visually and returned within 15 minutes. In August 2020, the Food and Drug Administration issued an emergency use authorization for the diagnosis of SARS-CoV-2 infection in symptomatic patients within 7 days of symptom onset [6]. The US Department of Health and Human Services has distributed 150 million test kits. Given the value of a rapid assessment of infectiousness, there is anticipated use in a broad range of subjects, including those who are asymptomatic. Here we present a systematic examination of the performance characteristics of the Binax-CoV2 test in a community screening setting where testing was offered for symptomatic and asymptomatic subjects.
METHODS
Study Population and Specimen Collection
Over 3 days in September 2020, we offered testing in the Mission District, a Latinx-predominant neighborhood, known from prior surveys to have an elevated prevalence of SARS-CoV-2 infection [7, 8]. Walk-up, free testing was conducted at a plaza located at an intersection of the Bay Area-wide subway system (BART) and the San Francisco city bus/streetcar system (MUNI). On the day of testing, participants self-reported symptoms and date of onset, demographics, and contact information, as required by state and federal reporting guidelines. A laboratory technician performed sequential anterior swab (both nares) for the Binax-CoV2 assay followed by a second swab (both nares) for RT-PCR. Participants were notified of RT-PCR test results. For this study, Binax-CoV2 results were not reported back to study subjects.
Laboratory Testing for SARS-CoV-2
RT-PCR detection of SARS-CoV-2 was performed at the Clinical Laboratory Improvement Amendments–certified laboratory operated by the University of California, San Francisco (UCSF), and the Chan Zuckerberg Biohub, as described elsewhere [9, 10].
Field Testing Using Binax-CoV2 Assay
The Binax-CoV2 assay was performed by technicians on site as described by the manufacturer using the supplied swabs. Each assay was read by 2 independent observers, and a site supervisor served as a tiebreaker. Beginning on day 2 of the study, each Binax-CoV2 assay card was scanned onsite using a color document scanner (CanoScan LIDE 400; Canon). Sample bands were retrospectively quantified from image data. Sample and background regions were localized by offset from the control band, and relative mean pixel intensity decreases were calculated from blue and green channels averaged with respect to background.
Titration of in vitro Cultured SARS-CoV-2 on Binax-CoV2 Cards
SARS-CoV-2 from a UCSF clinical specimen was isolated, propagated and plaqued on Huh7.5.1 cells overexpressing angiotensin-converting enzyme 2 and transmembrane serine protease 2 (TMPRSS2) [11]. Viral titers were determined using standard plaque assays [12]. For titration experiments, SARS-CoV-2 was diluted in Dulbecco phosphate-buffered saline, and 40 µL of each dilution was absorbed onto the supplied swab samples. Images of Binax-CoV2 cards were taken with an Apple iPhone 6. All experiments using cultured SARS-CoV-2 were conducted in a biosafety level 3 laboratory.
N Protein Titration Assay
SARS-CoV-2 N protein (1–419) was expressed in BL21(DE3) Escherichia coli and purified by nickel–nitrilotriacetic acid chromatography, incorporating a 1-mol/L sodium chloride, 50-mmol/L imidazole wash to remove bound RNA. Six concentrations of N protein were tested on 10 lots of Binax-CoV2 kits, and 40 µL of N protein was absorbed onto the provided swab sample.
Ethics Statement
The UCSF Committee on Human Research determined that the study met criteria for public health surveillance. All participants provided informed consent for dual testing.
RESULTS
Binax-CoV2 Performance Using a Titration of in vitro Cultured SARS-CoV-2
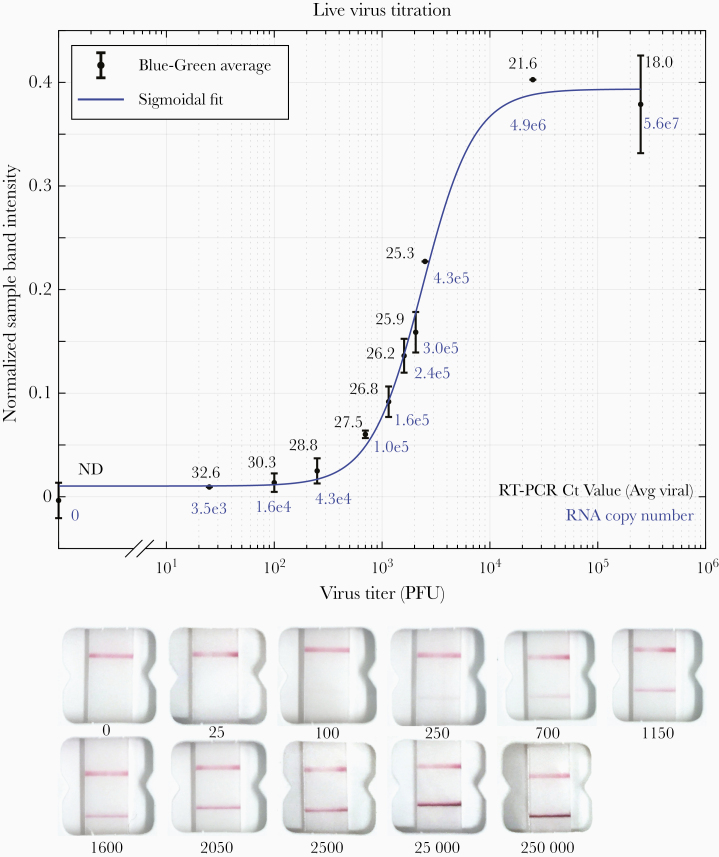
To explore the relationship of RT-PCR cycle threshold (Ct), viral load, and the corresponding visual Binax-CoV2 result, a dilution series of laboratory-cultured SARS-CoV-2 with known titers was assayed with both RT-PCR and Binax-CoV2 (Figure 1). For this stock of virus, the threshold for detectability by human eye on the Binax-CoV2 assay was between 1.6 and 4.3 × 104 viral copies (100–250 plaque-forming units), corresponding to t values (average of N and E genes) of 30.3 and 28.8, respectively, in this assay.
Figure 1.
Titration of in vitro grown severe acute respiratory syndrome coronavirus 2 and detection with Binax-CoV2 assay. Top, Normalized Binax-CoV2 sample band intensity (blue-green average) for cards loaded with a known amount of virus. Error bars represent standard deviation of sample band intensity of technical replicates. Reverse-transcription polymerase chain reaction (RT-PCR) testing was performed at the CLIAHUB consortium [10]. Corresponding RT-PCR cycle threshold (Ct) values (average of N and E gene probes) are shown in black, and the corresponding RNA copy numbers in blue. Note that Ct and genome copy number correlation varies by RT-PCR platform. Bottom, Representative card images from each data point. Abbreviation: PFUs, plaque-forming units.
Community RT-PCR Testing Results
Of the 878 subjects tested, 54% were male, 77% were 18–50 years of age, 81% self-identified as Latinx, and 84% reported no symptoms in the 14 days before testing. Twenty-six persons (3%) were RT-PCR positive; of these, 15 (58%) had a Ct <30, and 6 of the 15 (40%) were asymptomatic. Among asymptomatic individuals with a Ct <30, 4 of 6 developed symptoms within 2 days after testing. Of the 11 persons RT-PCR–positive with a Ct >30, 4 reported symptom onset ≥7 days before testing, 1 reported symptom onset 3 days before testing, and the remainder reported no symptoms.
Comparison of RT-PCR and Binax-CoV2 Testing Results from Community Testing
Because the readout of the Binax-CoV2 assay is by visual inspection, results may be subjective, especially when bands are faint or partial. The manufacturer’s instructions suggest scoring any visible band as positive. On day 1 of testing, these reading instructions were used and 217 samples tested, of which 214 yielded valid Binax-CoV2 results: 7 of 214 (3.3%) were RT-PCR positive; using the manufacturer’s proposed criteria, 5 of these 7 were Binax-CoV2 positive. Of 214, a total of 207 were RT-PCR negative, 9 (4.3%) of which were Binax-CoV2 positive. Thus, using the manufacturer’s criteria, 9 of 14 Binax-CoV2–positive tests (64%) in this population of 217 tests had false-positive results (Binax-CoV2 positive/RT-PCR negative). We thought that these initial criteria used on day 1 of testing were insufficient for classifying faint Binax-CoV2 assay bands, resulting in excessive false-positive calls.
On subsequent testing days, we evaluated additional criteria for classifying a band as positive, in consultation with experts from the manufacturer’s research staff. Optimal performance occurred when the bands were scored as positive, if they extended across the full width of the strip, irrespective of the intensity of the band. Updated scoring criteria were implemented by the third day of testing, when a total of 292 tests were administered. Of this total, 283 were RT-PCR negative, all of which scored Binax-CoV2 negative, demonstrating these updated reading criteria markedly alleviated false-positive readings. Of the 292 total day 3 tests, 9 were RT-PCR positive, of which 5 were Binax-CoV2 positive for antigen with these updated scoring criteria. Of the 9 RT-PCR–positive samples, the 4 that were Binax-CoV2 negative had a Ct >30, consistent with our laboratory-observed limit of detection for Binax-CoV2. We find that scoring a test as positive if bands extend across the full width of the strip, irrespective of band intensity, is the least subjective and easiest method to implement in the field, and we have developed a training tool (https://unitedinhealth.org/binax-training).
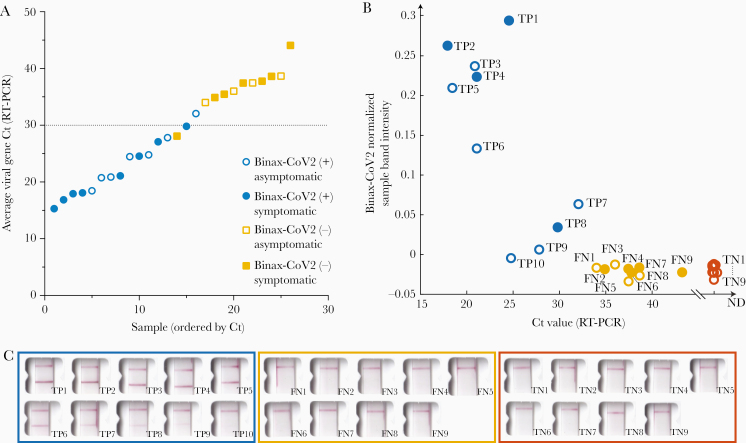
The results of the 26 RT-PCR–positive individuals identified throughout the 3-day study were stratified by RT-PCR test Ct value and categorized according to Binax-CoV2 result (Figure 2). The rapid antigen detection test performed well in samples with higher viral loads: 15 of 16 samples with a Ct<32 were Binax-CoV2 positive (Figure 2A). By contrast, none of the 10 samples with a Ct ≥34 were positive by Binax-CoV2 antigen detection. Retrospective image quantification of Binax-CoV2 sample band intensity is correlated with RT-PCR Ct values for those individuals (Figure 2B). In each case, the corresponding image is shown to demonstrate the correspondence between RT-PCR and the visual result (Figure 2C).
Figure 2.
Comparison of Binax-CoV2 test with quantitative reverse-transcription polymerase chain reaction (RT-PCR) test. A, Average viral cycle threshold (Ct) values from all 26 RT-PCR–positive individuals from the community study, plotted in ascending order. Blue circles indicate Binax-CoV2–positive samples; yellow squares, Binax-CoV2–negative samples. Open symbols represent individuals who were asymptomatic on the day of the test and filled symbols, those who reported symptoms on that day. B, Normalized sample band signal from retrospective image analysis of Binax-CoV2 cards was plotted as a function of Ct value for all available scanner images (19 of 26 RT-PCR–positive samples and a random subset of RT-PCR–negative samples). Binax-CoV2 true-positives are shown in blue and labeled TP; false-negatives, shown in yellow and labeled FN; and true-negatives, shown in red and labeled TN. C, Corresponding Binax-CoV2 card images from the data in B.
Sensitivity and Specificity
RT-PCR is considered a reference standard [3] and, in the RT-PCR assay used in this study, has a limit of detection of 100 viral RNA copies/mL. Direct antigen assays are inherently not as sensitive as RT-PCR. In the context of community based testing, we defined a threshold for high virus levels corresponding to the range of highest probability of transmissibility: a Ct of 30, which corresponds to a viral RNA copy number of approximately 1.9 × 104 in this assay [10, 13]. Using this Ct <30 case definition and 95% confidence intervals (CIs), the sensitivity of the Binax-CoV2 was 93.3% (95% CI, 68.1%–99.8%) (14 of 15), and the specificity was 99.9% (99.4%–99.9%) (855 of 856). Adjusting the threshold to a more conservative Ct value of 33 (2.6 × 103 viral RNA copies), the sensitivity was 93.8% (95% CI, 69.8%–99.8%) (15 of 16), and the specificity was 100% (99.6%–100%) (855 of 855). Without a Ct threshold, the sensitivity of the Binax-CoV2 assay was (57.7%; 95% CI, 36.9%–76.6%) (15 of 26), and the specificity was (100%; 99.6%–100%) (845 of 845). Given that the Binax-CoV2 assay detects infected individuals with high levels of virus (>104), the sensitivity of the assay in the absence of a threshold will largely depend on the viral kinetics within the testing population. Sensitivity and specificity calculations were completed with the final scoring criteria, using retroactive Binax-CoV2 scores from images covering all 3 study days.
Evaluation of Binax-CoV2 Lot-to-Lot Variation
We quantified lot-to-lot variability in 10 different lots of Binax-CoV2 card tests using a dilution series of N protein. (Supplementary Figure 1). At protein concentrations of ≥17.2 ng/mL, a sample band was detected in all lots and thus would not affect the outcome of this binary assay (Supplementary Figure 1A).
DISCUSSION
The data reported here describe the performance characteristics of the Binax-CoV2 antigen detection kit in the context of community testing including asymptomatic subjects. These results indicate a clear relationship between relative viral load and test positivity and provide a practical, real-world criterion to assist calling results in this setting. We found that small training modifications reduced the presence of false-positives, a legitimate concern for the rollout of these tests.
The currently approved emergency use authorization for the Binax-CoV2 assay specifies use only in symptomatic individuals. The results presented here suggest that the Binax-CoV2 test should not be limited to symptomatic testing alone. Many asymptomatic individuals have high viral loads (corresponding to low Ct values) and, therefore, have a high probability of being infectious and transmitting the virus, a feature and likely driver of the pandemic that we and others have observed previously [7, 14]. Limiting use of Binax-CoV2 to symptomatic individuals would have missed nearly half of the SARS-CoV-2 infections in the current study.
Furthermore, the impact of testing on forward transmission is hampered by long wait times. Our group reported previously that in the community setting, by the time a person is tested, counseled, and situated under isolation conditions, the effective isolation period is often nearly over [8]. This is particularly true for many communities of color, where reported delays in accessing tests and results are even longer [4, 15]. Rapid tests could reduce these delays and maximize the time of effective isolation. Limitations of our study include its cross-sectional design and the overall small number of RT-PCR positive cases. Additional field performance of this assay is needed and will help inform optimal use strategies. We recommend evaluating the Binax-CoV2 assay side by side with RT-PCR in each context where it will be used before using Binax-CoV2 without RT-PCR.
During the early stages of infection, viral load may be too low to detect by direct antigen assays such as Binax-CoV2. This inherent lower sensitivity may be offset by faster turn-around and higher frequency of testing, with overall lower cost, relative to RT-PCR methods. That said, for persons who present with a high index of suspicion of coronavirus disease 2019 and a negative Binax-CoV2 result, the test should be complemented with RT-PCR or a repeated Binax-CoV2 test at a later time to make sure cases are not missed.
In summary, under field conditions with supplementary technician training, the Binax-CoV2 assay accurately detected SARS-CoV-2 infection with high viral loads in both asymptomatic and symptomatic individuals. The Binax-CoV2 test could be a valuable asset in an arsenal of testing tools for the mitigation of SARS-CoV-2 spread, as rapid identification of highly infectious individuals is critical.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Bevan Dufty and the BART team, Jeff Tumlin and the San Francisco MUNI, Supervisor Hillary Ronen, Mayor London Breed, Grant Colfax, MD and the Department of Public Health, Salu Ribeiro, MS and Bay Area Phlebotomy and Laboratory services, the PrimaryBio testing platform for coronavirus disease 2019, and our community ambassadors and volunteers. We also thank Don Ganem, MD for writing assistance and critical discussion, Andreas Puschnik, PhD for Huh7.5.1 overexpression cells used for severe acute respiratory syndrome coronavirus disease 2 growth, and Terry Robins, PhD, Stephen Kovacs, and John Hackett Jr, PhD from Abbott Laboratories for their support. Finally, we thank both Abbott Laboratories and the California Department of Public Health for their generous donations of BinaxNOW COVID-19 Ag cards.
Financial support. This study was supported by the University of California, San Francisco, the Chan Zuckerberg Biohub, the Chan Zuckerberg Initiative, the San Francisco Latino Task Force, the National Institute of Allergy and Infectious Diseases (grants T32 AI060530 to L. R. and F31AI150007 to S. S.), and a private donor.
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Johns Hopkins Coronavirus Resource Center. COVID-19 dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html. Accessed 23 October 2020.
- 2. Baker MG, Wilson N, Anglemyer A. Successful elimination of Covid-19 transmission in New Zealand. N Engl J Med 2020; 383:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020; 26:771–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chwe H, Quintana A, Lazer D, et al. . The state of the nation: a 50-state COVID-19 survey. Report #17: COVID-19 test result times. October 2020. http://www.kateto.net/covid19/COVID19%20CONSORTIUM%20REPORT%2017%20TESTING%20OCT%202020.pdf. Accessed 23 October 2020.
- 5. Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity—a strategy for containment. N Engl J Med 2020; 383:e120. [DOI] [PubMed] [Google Scholar]
- 6. Abbott Diagnostics. Abbott BinaxNOW COVID-19 Ag package insert, version 1.6. 2020. https://www.fda.gov/media/141570/download. Accessed 20 October 2020.
- 7. Chamie G, Marquez C, Crawford E, et al. . SARS-CoV-2 community transmission disproportionately affects Latinx population during shelter-in-place in San Francisco [published online ahead of print August 21, 2020]. Clin Infect Dis 2020; doi: 10.1093/cid/ciaa1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kerkhoff AD, Sachdev D, Mizany S, et al. . Evaluation of a novel community-based COVID-19 ‘Test-to-Care’ model for low-income populations. PLoS One 2020; 15:e0239400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crawford ED, Acosta I, Ahyong V, et al. . Rapid deployment of SARS-CoV-2 testing: the CLIAHUB. PLoS Pathog 2020; 16:e1008966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vanaerschot M, Mann SA, Webber JT, et al. . Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by RT-PCR. J Clin Microbiol 2020; 59:e02369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang R, Simoneau CR, Kulsuptrakul J, et al. . Functional genomic screens identify human host factors for SARS-CoV-2 and common cold coronaviruses. bioRxiv [Preprint: not peer reviewed]. 24 September 2020. Available from: https://www.biorxiv.org/content/10.1101/2020.09.24.312298v1. [Google Scholar]
- 12. Honko AN, Storm N, Bean DJ, Vasquez JH, Downs SN, Griffiths A. Rapid quantification and neutralization assays for novel coronavirus SARS-CoV-2 using Avicel R RC-591 semi-solid overlay. 2020. https://www.preprints.org/manuscript/202005.0264/v1. Accessed 23 October 2020.
- 13. Wölfel R, Corman VM, Guggemos W, et al. . Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 14. Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection : a narrative review. Ann Intern Med 2020:362–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim HN, Lan KF, Nkyekyer E, et al. . Assessment of disparities in COVID-19 testing and infection across language groups in Seattle, Washington. JAMA Netw Open 2020; 3:e2021213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.